Abstract
Cigarette smoking is a leading cause of preventable mortality worldwide. Nicotine dependence, which reduces the likelihood of quitting smoking, is a heritable trait with firmly established associations with sequence variants in nicotine acetylcholine receptor genes and at other loci. To search for additional loci, we conducted a genome-wide association study (GWAS) meta-analysis of nicotine dependence, totaling 38,602 smokers (28,677 Europeans/European Americans and 9,925 African Americans) across 15 studies. In this largest ever GWAS meta-analysis for nicotine dependence and the largest-ever cross-ancestry GWAS meta-analysis for any smoking phenotype, we reconfirmed the well-known CHRNA5-CHRNA3-CHRNB4 genes and further yielded a novel association in the DNA methyltransferase gene DNMT3B. The intronic DNMT3B rs910083-C (frequency=44%–77%) associates with increased risk of nicotine dependence at P=3.7×10−8 (odds ratio [OR]=1.06 and 95% confidence interval [CI]=1.04–1.07 for severe vs mild dependence). The association was independently confirmed in the UK Biobank (N=48,931) using heavy vs never smoking as a proxy phenotype (P=3.6×10−4, OR=1.05, and 95% CI=1.02–1.08). Rs910083-C is also associated with increased risk of squamous cell lung carcinoma in the International Lung Cancer Consortium (N=60,586, meta-analysis P=0.0095, OR=1.05, and 95% CI=1.01–1.09). Moreover, rs910083-C was implicated as a cis-methylation quantitative trait locus (QTL) variant associated with higher DNMT3B methylation in fetal brain (N=166, P=2.3×10−26) and a cis-expression QTL variant associated with higher DNMT3B expression in adult cerebellum from the Genotype-Tissue Expression project (N=103, P=3.0×10−6) and the independent Brain eQTL Almanac (N=134, P=0.028). This novel DNMT3B cis-acting QTL variant highlights the importance of genetically influenced regulation in brain on the risks of nicotine dependence, heavy smoking, and consequent lung cancer.
Introduction
Cigarette smoking is a leading cause of preventable death, resulting annually in nearly 6 million premature deaths worldwide.1 Smoking-related deaths are most often attributed to increased rates of cancer, cardiovascular disease, and chronic obstructive pulmonary disease (COPD).2 Despite the well-known adverse health effects, an estimated 45.3 million U.S. adults smoke cigarettes, of whom over 68% report wanting to quit.3, 4
Cigarette smoking is a complex multi-step behavior involving initiation, regular smoking, nicotine dependence, cessation, and relapse. Some regular smokers maintain low-level smoking without developing symptoms of dependence,5 while others become heavily dependent smokers and experience the most difficulty with cessation and the highest risk of relapse.6, 7 Nicotine dependence has high heritability (estimates up to 75%),8, 9 and besides reducing the likelihood of quitting smoking, it is predictive of withdrawal severity,10 response to treatment,11 and smoking-related health outcomes.12, 13
GWAS analyses of nicotine dependence phenotypes14–24 have firmly established associations with several loci, including nicotinic acetylcholine receptor genes on chromosomes 15q25 (CHRNA5-CHRNA3-CHRNB4), 8p11 (CHRNB3-CHRNA6), and 20q13 (CHRNA4). The largest GWAS meta-analyses relied on widely ascertained phenotypes such as cigarettes per day (CPD),16–18 which represents only one of several components of nicotine dependence.25 Focusing GWAS on nicotine dependence rather than CPD may improve statistical power for identifying variants that influence the broader construct of dependence.19 This idea is supported by our prior nicotine dependence GWAS meta-analysis (total N=17,074 ever-smokers of European/European American ancestry [EUR]) that discovered associations with CHRNA4 SNPs that were driven by time to first cigarette in the morning (TTFC) and had not been detected in GWAS meta-analyses of CPD with much larger sample sizes.23 To improve statistical power further and to search for additional loci, we more than doubled our sample size to perform the largest GWAS meta-analysis of nicotine dependence to date, including 38,602 ever-smokers (28,677 of EUR and 9,925 of African American [AA] ancestries) across 15 studies. We extended our study to include correlations with DNA methylation (DNAm) and RNA expression (RNAexp) of nearby genes across human brain tissues and evaluated associations with a critical smoking-related outcome: lung cancer (total N=81,821 cases and controls).
Materials and Methods
Study protocols received institutional review board approval at their respective sites. All study participants provided written informed consent.
We included the five studies from our prior GWAS meta-analysis23 and 10 additional studies. Details of their study design, genotyping, quality control, 1000 Genomes (1000G) imputation, and analysis are provided in Supplementary Methods and Supplementary Table 1.
Nicotine dependence phenotype
We included studies with SNP genotypes and Fagerström Test for Nicotine Dependence (FTND) data26 collected among smokers. FTND scores range from 0 (no dependence) to 10 (highest dependence level). As before,23 we used FTND to categorize nicotine dependence as mild (scores 0–3), moderate (scores 4–6), or severe (scores 7–10). Two of the 15 studies additionally included low-intensity smokers who reported CPD as ≤10 but had no data available on the other FTND items and were defined as mildly dependent. Concordance rates between these FTND and CPD categories showed minimal phenotype misclassification (Supplementary Methods).
Nicotine dependence GWAS meta-analysis and independent follow-up
We used linear regression to test SNP/indel associations with categorical nicotine dependence (mild=0/moderate=1/severe=2) in each separate study and ancestry group. Covariates included age, sex, principal components, and study-specific covariates (as needed); additional adjustment for family structure was made in studies with relatives included (Supplementary Methods).
We combined GWAS results, using METAL27 with fixed-effects inverse variance-weighting meta-analysis, across all studies with FTND data to maximize statistical power. Genomic control was applied to the deCODE results to adjust for inflation due to relatedness among participants; all other studies had low inflation values (λ≤1.02). We excluded SNPs/indels with minor allele frequency (MAF) <1% in 1000G EUR or African (AFR) panels, depending on the ancestry group analyzed. The standard threshold (P<5×10−8), originally based on 1 million independent tests genome-wide as computed using HapMap-based imputation for EUR studies,28 has been validated for 1000G-imputed GWAS of common variants.29 Rather than imposing a more stringent, yet to be consistently determined, threshold when analyzing common variants across EUR and AA studies,29–32 we carried forward novel variation implicated at P<5×10−8 and relied on confirmation in an independent study to declare genome-wide significance. For this confirmation step, we utilized UK Biobank (N=48,931 EUR participants) results with heavy, defined as pack-years ([CPD/20 cigarettes per pack] × years smoked) ≥10, vs never smoking as a proxy phenotype.24 This prior GWAS was designed as a nested case-control analysis that sampled the extremes of smoking behavior, and thus did not encompass light smoking.
Regional association plots were created using LocusZoom33 with linkage disequilibrium (LD) estimates of r2 and D’ based on 1000G EUR and AFR panels. Allele frequencies were weighted by sample size. Odds ratio (OR) estimates were computed using the β estimate from the SNP term in the linear regression model (e[2×β] for severe vs mild dependence) and then compared across studies using the Forest Plot Viewer.34 Heterogeneity across studies was assessed using the I2 index.35
SNP associations with DNAm and RNAexp
The top novel nicotine dependence-associated variant was assessed for cis-acting regulatory effects, using previously published methylation quantitative trait locus (meQTL) and expression QTL (eQTL) studies of postmortem brain tissues (Supplementary Methods). We used meQTL results from 166 fetal brain samples in the Human Developmental Biology Resource and UK Medical Research Council Brain Banks network; these samples were not dissected further into different tissues.36 SNP genotypes were imputed using 1000G, and DNAm was measured using the Illumina HumanMethylation450 BeadChip. meQTL results were obtained at http://epigenetics.essex.ac.uk/mQTL/.
We assessed SNP associations with RNAexp of nearby genes (+/− 1Mb around the transcription start sites) across adult brain tissues from the Genotype-Tissue Expression (GTEx) project37 with 1000G-imputed genotypes and RNA sequencing measures from up to 103 deceased persons. We followed up cis-eQTL SNP results in the Brain eQTL Almanac dataset (N=134 EUR deceased persons, http://www.braineac.org/), using RNAexp probes measured on Affymetrix Human Exon 1.0 ST arrays across brain tissues, many of which overlap with tissues available in GTEx.38
SNP associations with lung cancer
The top novel nicotine dependence-associated variant was tested for association with lung cancer using 27,349 cases and 54,472 controls from >30 EUR studies in the Transdisciplinary Research for Cancer in Lung of the International Lung Cancer Consortium (TRICL-ILCCO). Their genotyping, quality control, 1000G imputation, and statistical analyses were previously described.39 Briefly, the SNP associations were derived from a meta-analysis of logistic regression model results that compared lung cancer cases and controls, adjusting for age, sex, and the first two principal components. Histological subtypes of adenocarcinoma and squamous cell lung carcinoma cases were compared, separately, to controls. We removed the EAGLE study from the original set of lung cancer studies to avoid overlap in the nicotine dependence and lung cancer meta-analyses. All lung cancer analyses included ever- and never-smokers; our follow-up association testing made additional adjustments for smoking history (ever vs never) and pack-years.
Results
We performed GWAS analyses across 15 studies, totaling 38,602 (28,677 EUR and 9,925 AA) ever-smokers (Supplementary Table 2), with nicotine dependence defined as mild (N=17,796; 46.1%), moderate (N=13,527; 35.0%), or severe (N=7,279; 18.9%). More than 99% of the participants were ≥18 years old. Males constituted 53.2% of the total sample size.
Our GWAS meta-analysis tested nearly 18 million genotyped and 1000G-imputed SNPs/indels for association with mild/moderate/severe dependence (λ=1.027, Supplementary Figure 1A). We observed SNP/indel associations at meta-analysis P<5×10−8 at two loci (Figure 1): the well-known chromosome 15q25 locus spanning CHRNA5-CHRNA3-CHRNB4 (smallest meta-analysis P=1.3×10−25) and a novel chromosome 20q11 locus spanning the DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B) gene (smallest meta-analysis P=3.7×10−8). No additional loci were identified at P<5×10−8 in the ancestry-specific GWAS meta-analyses (Supplementary Figures 1B–C and 2A–B).
Figure 1. Manhattan plot of SNP and indel associations with nicotine dependence from GWAS meta-analysis across 15 studies (total N=38,602 European/European Americans and African Americans).
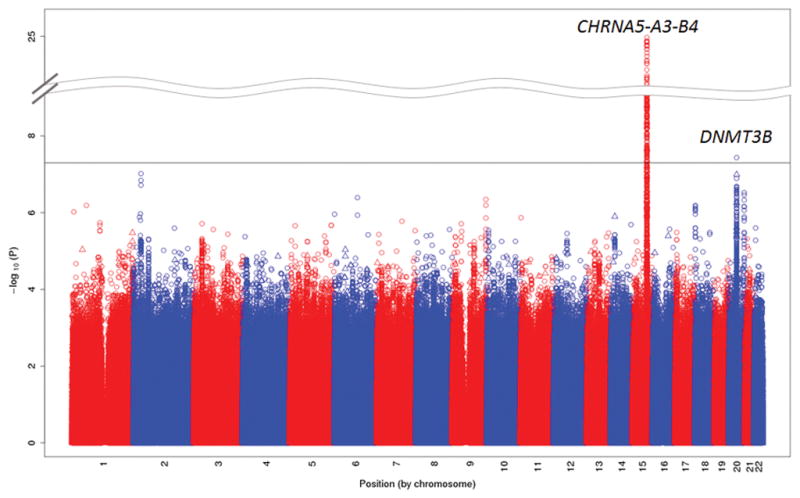
The –log10 meta-analysis p-values are plotted by chromosomal position of SNPs (depicted as circles) and indels (depicted as triangles). The genome-wide statistical significance threshold (P<5×10−8) is shown as a solid black line.
DNMT3B SNP associations with nicotine dependence
Rs910083, an intronic DNMT3B SNP, was identified across both ancestries: meta-analysis P=3.7×10−8 and β (standard error)=0.032 (0.0057) for the C allele (Table 1), corresponding to OR (95% confidence interval [CI]) of 1.06 (1.04–1.07) for severe vs mild dependence (Supplementary Figure 3). Rs910083 was imputed well (quality scores=0.98–1), and it showed no evidence of heterogeneity across studies (P=0.71).
Table 1.
SNP associations in the novel DNMT3B gene and previously reported genes that were identified at P<5×10−7 in the nicotine dependence GWAS meta-analysis. The SNP with the smallest nicotine dependence GWAS meta-analysis P is shown, along with nearby SNPs previously reported in other GWAS of smoking phenotypes. Results from the UK Biobank heavy vs never smoking GWAS are also presented.
| Chr | Gene/nearest gene | Prior GWAS reported smoking phenotype | SNP (allele) | Nicotine dependence meta-analyses among ever regular smokers | UK Biobank heavy vs never smokers (N=48,931)24 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EUR studies (N=28,677) | AA studies (N=9,925) | EUR and AA studies (N=38,602) | |||||||||
| β (SE) | P | β (SE) | P | β (SE) | P | β (SE) | Pa | ||||
| 20q11 | DNMT3B | None | rs910083 (C) | 0.027 (0.0065) | 4.2×10−5 | 0.047 (0.012) | 7.3×10−5 | 0.032 (0.0057) | 3.7×10−8 | 0.048 (0.014) | 3.6×10−4 |
| NOL4L | Heavy vs never smoking24 | rs57342388 (CACGG) | 0.021 (0.0075) | 0.0059 | 0.032 (0.020) | 0.10 | 0.022 (0.0070) | 0.0017 | 0.094 (0.016) | 4.7×10−9 | |
| 20q13 | CHRNA4 | None | rs6062901b (G) | 0.043 (0.0088) | 8.3×10−7 | 0.022 (0.010) | 0.035 | 0.034 (0.0067) | 3.0×10−7 | 0.048 (0.018) | 0.0075 |
| Nicotine dependence23 | rs2273500b (C) | 0.046 (0.0098) | 2.0×10−6 | 0.0085 (0.016) | 0.59 | 0.036 (0.0083) | 1.6×10−5 | 0.091 (0.019) | 1.4×10−6 | ||
| Heavy vs never smoking24 | rs11697662b (C) | 0.033 (0.0085) | 9.4×10−5 | 0.013 (0.011) | 0.20 | 0.025 (0.0066) | 1.2×10−4 | 0.091 (0.017) | 1.0×10−7 | ||
| 9q 34 | DBH | None | rs56116178c (G) | 0.053 (0.010) | 4.5×10−7 | NA | NA | 0.053 (0.010) | 4.5×10−7 | 0.097 (0.022) | 9.3×10−6 |
| Heavy vs never smoking24 | rs111280114c (G) | 0.052 (0.010) | 6.4×10−7 | NA | NA | 0.052 (0.0098) | 2.5×10−5 | 0.099 (0.022) | 6.0×10−6 | ||
| Smoking cessation17 | rs3025343c (A) | 0.043 (0.0099) | 1.7×10−5 | NA | NA | 0.042 (0.0098) | 2.5×10−5 | 0.091 (0.021) | 1.1×10−5 | ||
NA, not available due to minor allele frequency <1%.
P-values correspond to genomic control being applied.
Rs2773500 and rs11697662 were originally implicated in EUR studies, where they exist in moderate LD (r2=0.64 and D′=0.95 in 1000G EUR). Rs6062901 had r2=0.47–0.57 and D′=0.73–0.84 with rs2273500 and rs11697662 in 1000G EUR and r2=0.11–0.51 and D′=0.72–0.92 in 1000G AFR.
rs56116178 and rs111280114 had r2=0.98 and D′=1 with one another and r2=0.72–0.73 and D′=0.96 with rs3025343 in 1000G EUR.
Rs910083-C, the minor allele for EUR (frequency=44%) but the major allele for AA (frequency=77%), is associated with increased nicotine dependence risk (Table 1): EUR-specific meta-analysis P=4.1×10−5 and OR (95% CI)=1.06 (1.03–1.08); and AA-specific meta-analysis P=7.3×10−5 and OR (95% CI)=1.10 (1.05–1.15). Many SNPs/indels were in moderate to high LD (r2>0.4) with rs910083 in 1000G EUR, spanning 220 kb (chr20:31,268,924–31,488,466) and including DNMT3B and its neighboring genes, microtubule-associated protein, RP/EB family, member 1 (MAPRE1) and COMM domain containing 7 (COMMD7). However, in 1000G AFR, SNPs in LD (r2>0.4) with rs910083 were localized to a 47 kb region (chr20:31,356,560–31,403,394) including only DNMT3B (Figure 2A–B, NCBI build 37 positions).
Figure 2. Novel DNMT3B SNP associations with nicotine dependence from GWAS meta-analysis of EUR and AA studies.
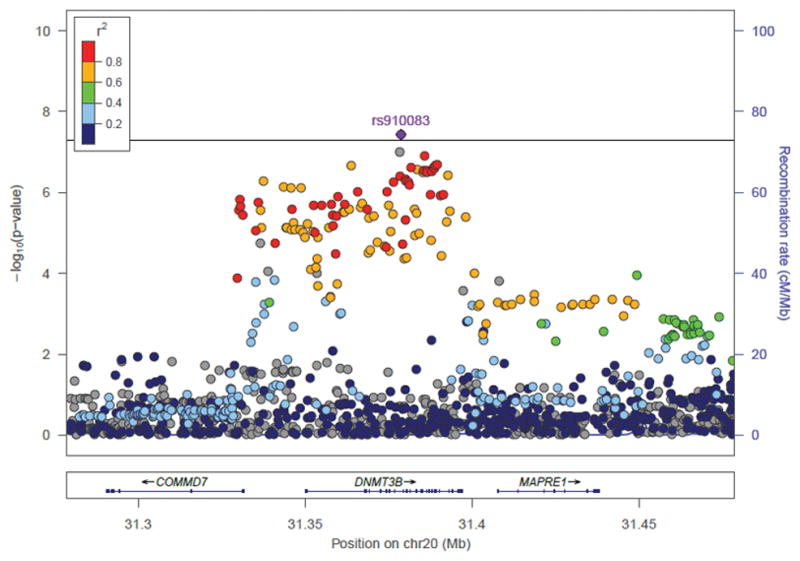
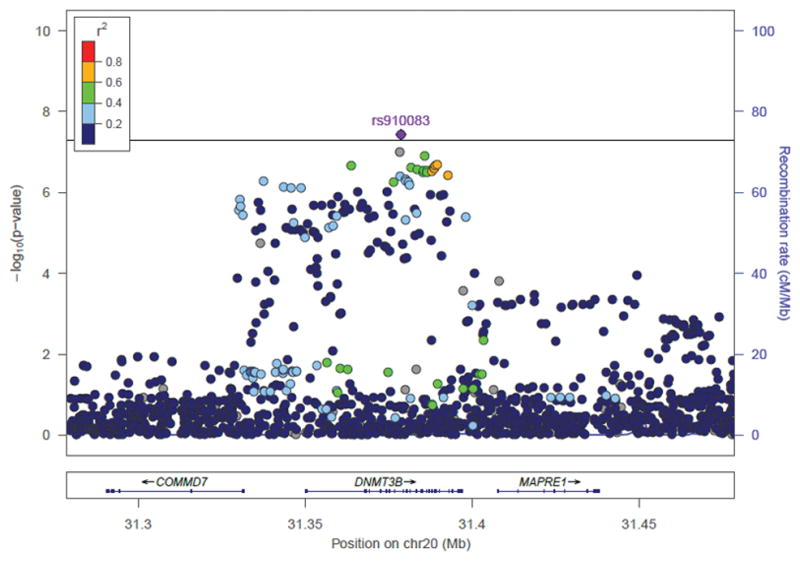
SNP and indel associations are shown across DNMT3B and its 100kb flanking region (NCBI build 37 positions presented). r2 values between the top SNP rs910083 and all other SNPs are shown in reference to 1000 Genomes panels: (A) European (EUR) and (B) African (AFR). Indels with missing r2 values are indicated in grey. The p-value threshold of 5×10−8 is marked by the solid black line.
No DNMT3B variants have been implicated previously for any substance use disorder (SUD) phenotype. Upstream of DNMT3B, chromosome 20q11 also harbors the nucleolar protein 4-like (NOL4L) gene, which was reported at genome-wide significance for heavy vs never smoking in the UK Biobank for the indel rs57342388.24 This indel was associated at meta-analysis P=0.0017 in our study (Table 1): OR (95% CI)=1.04 (1.02–1.07) for severe vs mild dependence for the insertion allele, consistent with the prior result. Rs57342388 is located 216 kb upstream of our top DNMT3B SNP rs910083. The two variants are weakly correlated (r2=0.11 in 1000G EUR where MAF=2% for rs57342388 vs 18% for rs910083, r2=0.0022 in AFR where MAF=19% vs 42%) but are in moderate to high LD (D′=0.57 in EUR, D′=1 in AFR). In follow-up testing with both SNPs included in the same model, both were associated with nicotine dependence (meta-analysis P=1.7×10−6 for rs910083 and 8.3×10−3 for rs57342388), showing that our observed DNMT3B association signal is not explained by the previously reported NOL4L signal.
We tested rs910083-C for association with each of the specific FTND items, as presented in Supplementary Table 3. The rs910083-C association was driven most strongly by TTFC (meta-analysis P=1.2×10−4). Its next most significantly associated FTND item was CPD (meta-analysis P=0.0011). TTFC is an indicator of withdrawal severity upon awakening40 and behavioral automaticity (habitual smoking without awareness or cognitive control7). Although TTFC has its distinct features (e.g., strongest predictor of cessation success among the FTND items), it is correlated with heaviness of smoking as captured by CPD.7
DNMT3B SNP association with heavy smoking in an independent study
Using heavy vs never smoking GWAS results from the UK Biobank (N=24,457 heavy and 24,474 never smokers),24 we found that rs910083-C is associated at P=3.6×10−4 and OR (95% CI)=1.05 (1.02–1.08) for heavier smoking. Although effect sizes were small, its associations with risks of nicotine dependence and heavy smoking were consistent (Table 1).
DNMT3B SNP associations with DNAm and RNAexp
Using a genome-wide meQTL study of 166 fetal brain samples,36 we found that rs910083-C associates with higher DNAm levels of the probe cg13636640, located 252 base pairs upstream of the DNMT3B gene (β=0.082, P=2.3×10−26). This probe was the only one observed in the region with significant mQTL variants (Supplementary Figure 4). There were a total of 140 SNPs associated with DNAm of this probe (smallest P=1.8×10−45), and we observed associations of these SNPs with nicotine dependence with meta-analysis p-values ranging from 3.66×10−8 for rs910083 to 0.0051 (Supplementary Table 4). To our knowledge, there are no similar data to assess cis-meQTL effects in brain tissues from adults with no psychiatric disease. However, we assessed the cis-acting effect of rs910083 on RNAexp across several adult brain tissues using two independent datasets, GTEx37 followed by Brain eQTL Almanac.38
Across the 13 brain tissues in GTEx, we observed the highest DNMT3B gene expression levels in cerebellum (median log10 RPKM=0.304) and cerebellar hemisphere (median log10 RPKM=0.298), compared to median log10 RPKM≤−0.325 for all other brain tissues (Supplementary Figure 5). Moreover, across the brain tissues, rs910083 was most significantly associated with DNMT3B gene expression levels in cerebellum (P=3.0×10−6) and cerebellar hemisphere (P=7.0×10−7), with the C allele associated with higher DNMT3B expression (Figure 3 and Supplementary Table 5). We replicated this pattern in the Brain eQTL Almanac, where DNMT3B mRNA transcript expression levels were highest in cerebellar cortex (Supplementary Figure 6)—the outer layer of the cerebellum that comprises most of its volume. Consistent with GTEx, rs910083-C was associated with increased DNMT3B mRNA transcript expression levels specifically in cerebellar cortex (P=0.028).
Figure 3. Normalized DNMT3B gene expression levels as a function of rs910083 genotype in cerebellum from the Genotype-Tissue Expression (GTEx) project.
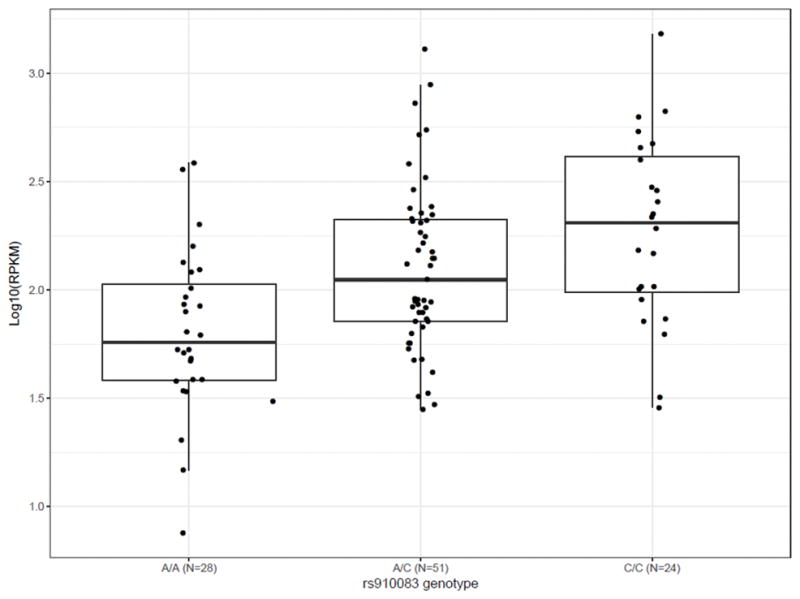
The box lines mark the first quartile, median, and third quartile; and the whiskers are marked by the highest and lowest data points within the 1.5 × inter-quartile range (third – first quartile) to show outliers that fall outside of these boundaries.
Beyond the brain tissues in GTEx, rs910083 is associated with RNAexp of other genes within 1 MB (Supplementary Table 6): MAPRE1 (smallest P=7.3×10−17 in sun exposed skin), COMMD7 (smallest P=1.0×10−6 in colon), and BPI fold containing family B, member 2 (BPIFB4, smallest P=5.4×10−5 in artery).
DNMT3B SNP associations with lung cancer
We assessed rs910083 for association with lung cancer using a GWAS meta-analysis of EUR studies from TRICL-ILCCO39 (N=27,349 cases and 54,472 controls, Supplementary Table 7). Rs910083-C is significantly associated with increased risk of squamous cell carcinoma (N=6,937 cases and 53,649 controls, meta-analysis P=0.0095 and OR [95% CI]=1.05 [1.01–1.09]), consistent with the increased nicotine dependence risk. Rs910083 is not associated with adenocarcinoma.
We evaluated the effect of adjusting for smoking on the rs910083 association with squamous cell carcinoma in the studies with smoking data readily available. We found that the level of statistical significance and magnitude of association were both weakened with adjustment for ever/never smoking and pack-years (Supplementary Table 8), suggesting that the rs910083 association with squamous cell carcinoma is mediated by smoking.
Discussion
This largest-ever GWAS meta-analysis for nicotine dependence, and the largest-ever cross-ancestry GWAS meta-analysis for any smoking phenotype, identified rs910083 as a novel SNP that regulates DNMT3B in human brain and contributes to risks of nicotine dependence and heavy smoking. Rs910083 was discovered via meta-analysis of two ancestry groups; the association signal includes SNPs in high LD with rs910083 across the COMMD7, DNMT3B, and MAPRE1 genes in EUR ancestry, but LD is localized to the DNMT3B gene in AA ancestry. Moreover, rs910083 was implicated as a cis-acting QTL SNP that influences DNMT3B DNAm in fetal brain and DNMT3B RNAexp in adult cerebellum, with the C allele being associated with higher DNAm and RNAexp levels. While this pattern might contrast the traditional view of higher DNAm being correlated with lower RNAexp, the observed effects reflect temporal and spatial differences, thus limiting our ability to draw direct correlations. Nonetheless, genome-wide QTL comparisons in human brain have shown that almost half of SNPs that act as both an eQTL and meQTL show the same direction of association for DNAm and RNAexp,41 as we observed here for DNMT3B and before for CHRNA5.42 The previously established nicotinic acetylcholine receptor genes also harbor nicotine dependence-associated SNPs with important consequences for gene regulation, including noncoding SNPs that correlate with DNAm,42 splicing,23 and/or RNAexp43, 44 in brain tissues that are frequently studied for nicotine and other SUDs because of their role in primary reward pathways and executive function, such as prefrontal cortex.45 This newly identified DNMT3B SNP association highlights changes in DNAm in fetal brain and with RNAexp specifically in cerebellum, a part of the brain that has often been overlooked despite some indications for its involvement in the neurobiology of addiction.46–48
DNMT3B encodes a DNA methyltransferase prominently involved in de novo DNAm that establishes patterns early in development; it may also contribute to maintenance DNAm.49 Although initially reported for maintaining DNAm at imprinted loci,50 later evidence showed that other members of the DNMT family (DNMT3A and DNMT3L) are required for imprinting, while DNMT3B may not play an essential role.51 Mouse models have shown that complete loss of DNMT3B function is embryonically lethal.50 However, recessive inheritance of rare mutations that render DNMT3B partially functional are known to cause Immunodeficiency, Centromeric instability and Facial dysmorphism (ICF) syndrome, which manifests with growth and neurodevelopmental abnormalities.49 Because of the critical role that DNMT3B plays in establishing methylation, altered expression has been associated with ICF syndrome at >700 genes involved in brain development and other processes.52 These and other genes that are regulated by DNMT3B methylation represent candidate genes that may directly contribute to nicotine dependence susceptibility.
DNMT3B has not previously been connected with the biology underlying the risk of nicotine dependence or any other SUD. However, there is evidence that in vitro cigarette smoke exposure leads to increased DNMT3B expression in human respiratory epithelial cells, and DNMT3B overexpression results in downstream hypermethylation that has been widely implicated in lung cancer.53 Given this indication, DNMT inhibition has been an active area of research for cancer treatment; two inhibitor agents are currently approved by the US Food and Drug Administration (decitabine, which shows high affinity for DNMT3A/3B over DNMT1,54 and azacytidine), and at least one other promising agent (zebularine) awaits clinical trial testing.55 With the DNMT3B variant discovery for nicotine dependence, DNMT3B inhibition may merit future study for smoking cessation treatment.
56Rs910083 resides in an active promoter marked by H3K9ac in several brain regions examined in the Roadmap Epigenomics Project,56 and it associates with DNMT3B RNAexp specifically in the cerebellum. This finding does not negate the importance of other brain regions known to be involved in SUDs but brings up the possibility of altered gene regulation in the cerebellum contributing to the complex neurobiological pathway leading to dependence. A main function of the cerebellum is motor coordination, but it is also involved in non-motor functions relevant to SUD, including reward.46–48 The cerebellum responds to acute and long-term exposures to nicotine57–62 and other substances,47 and it makes functional connections with the prefrontal cortex and other brain tissues that are widely recognized for their involvement in SUDs.47
Our discovery of the rs910083 association with nicotine dependence was made possible by assembling the largest possible sample size of FTND studies comprised of ever-smokers to maximize statistical power. No large FTND studies were left for replication, but because CPD is a central component of both the FTND and the heavy vs never smoking definition used in the UK Biobank, we tested for independent confirmation of the novel association with heavy vs never smoking and found that rs910083-C also conferred risk in the UK Biobank.
Larger GWAS meta-analyses have been reported for other smoking phenotypes such as ever vs never smoking, but these studies were comprised only of EUR participants and based on HapMap imputation.16–18 Rs910083, a 1000G-imputed SNP, was not captured in these studies, but 9 of the 18 DNMT3B SNPs associated with nicotine dependence at meta-analysis P<5×10−7 in the present study were HapMap-imputed (Supplementary Table 9); these SNPs were in strong LD with rs910083 among EURs (r2=0.78–0.99 in 1000G EUR) but weaker LD among AAs (r2=0.29–0.76 in 1000G AFR). Using results from the largest GWAS meta-analysis of CPD (Tobacco and Genetics [TAG] consortium, N=38,181 EUR ever smokers independent of the ones included here), we found that the 9 HapMap-imputed DNMT3B SNPs were associated with CPD at p-values ranging from 0.027 to 0.059 and a consistent direction of association with nicotine dependence; in comparison, p-values for ever vs never smoking (N=74,035 in the TAG consortium) ranged from 0.049 to 0.34 (Supplemental Table 9). We caution that the best DNTM3B signal in the TAG consortium was observed for CPD at only nominal significance (smallest P=0.027), despite having a nearly equivalent sample size as our study. However, our study yielding more statistically significant DNMT3B SNP associations with nicotine dependence (smallest P=3.7×10−8) is likely due to a combination of factors, including (1) reliance on FTND, a multi-dimensional phenotype that encompasses CPD and other important features of smoking behavior including TTFC that drove the rs910083 association, and (2) 1000G imputation which has been shown to strengthen association signals for some loci due to the finer mapping available.63, 64 We have similarly observed more statistically significant associations with nicotine dependence and stronger effect sizes, compared to CPD, in prior studies of CHRNB319 and CHRNA4.23
Until now, the only common DNMT3B variant implicated by GWAS was identified in a study of inflammatory bowel disease (IBD; intronic rs4911259). This SNP was associated with nicotine dependence in our meta-analysis (P=3.5×10−6) and is in LD with our top SNP rs910083 (r2=0.76 and D′=1 in 1000G EUR, r2=0.16 and D′=1 in 1000G AFR).65 Cigarette smoking is the environmental factor most consistently associated with IBD,66 and these shared SNP association signals suggest that DNMT3B SNPs may exert pleiotropic effects. Alternatively, it is possible that smoking mediates the DNMT3B SNP association with IBD, but the IBD GWAS did not include adjustment for smoking.65
Beyond finding DNMT3B and (as expected) CHRNA5-CHRNA3-CHRNB4, our GWAS meta-analysis resulted in P<5×10−7 for two other loci previously implicated in smoking, CHRNA4 on chromosome 20q1323, 24 and dopamine β-hydroxylase (DBH) on chromosome 9q34.17, 24 Our prior nicotine dependence GWAS meta-analysis of EUR studies identified the CHRNA4 splice site SNP rs2773500.23 Rare CHRNA4 variants have also been found to associate with nicotine dependence.67, 68 Our study supported common CHRNA4 SNP associations among EUR samples, but no association was detected for these SNPs among AA studies (Table 1).
DBH is a strong functional candidate for influencing nicotine dependence. The dopaminergic system lies at the core of the brain’s reward pathway, and the DBH enzyme converts dopamine into norepinephrine. An upstream DBH SNP (rs3025343) was identified in a GWAS of smoking cessation (current vs former smokers)16–18 and later independently replicated.69, 70 Consistent with rs3025343-A being associated with reduced success of quitting smoking, its phenotypic profile has been expanded to include associations with: (1) heavier smoking (N=48,931 in the UK Biobank, P=1.2×10−5);24 (2) higher FTND scores (N=1,430 EUR participants, P=0.023);71 and (3) higher nicotine dependence risk in our EUR studies (N=28,677, meta-analysis P=1.7×10−5). Smaller p-values were found for other 1000G-imputed upstream DBH SNPs in the UK Biobank and our study (rs111280114 and rs56116178, respectively; Table 1); the minor alleles of these SNPs were similarly associated with increased risks among EUR studies (Supplementary Figure 7 for rs56116178). Because these DBH SNPs all occur at <1% frequency among AAs, studying DBH variation on nicotine dependence risk in this ancestry group will require larger sample sizes or an alternative study design.
Nicotine dependence-associated variants in CHRNA5-CHRNA3-CHRNB444, 72 and CHRNA423, 67 have been previously shown to associate with lung cancer and other smoking-related diseases. Our study shows that the nicotine dependence-associated SNPs in DNMT3B and DBH are also associated with lung cancer (Supplementary Table 7). These findings may reflect the SNPs acting indirectly on lung through their influence on smoking (Supplementary Table 8). Alternatively, because DBH is expressed in the lung73 and DNMT3B overexpression has been shown in lung cancer, we cannot exclude the possibility that either of these SNPs act directly to promote lung cancer through an unknown mechanism.53 The DNMT3B and DBH SNPs were both associated with squamous cell lung carcinoma. This histological subtype has a strong association with smoking and occurs infrequently in never-smokers. In contrast, neither SNP is associated with adenocarcinoma, a subtype that has a weaker association with smoking74 and an increasing prevalence over time among never-smokers.75 Histology-specific associations are not uncommon for lung cancer genetic loci.39
Our findings expand the known genetic architecture of nicotine dependence, by showing that the DNMT3B SNP rs910083 increases the likelihood of developing nicotine dependence as observed across two different ancestries, smoking heavily, and consequently incurring a heightened risk of lung cancer.53, 55 The convergence of prior and current findings indicate that the complex neurobiology underlying nicotine dependence involves several sequence variants with functional and regulatory effects across distinct brain tissues.
Supplementary Material
Acknowledgments
We thank the many study participants. We also thank Michael E. Hall for reviewing the manuscript. This work was supported by the National Institute on Drug Abuse grant numbers R01 DA035825, R01 DA036583, and R01 DA042090. Acknowledgments for the nicotine dependence studies are included in the Supplementary Information. Funding for lung cancer studies was provided by the National Cancer Institute grant number U19 CA148127.
Footnotes
Conflicts of Interest
Dr. Bierut and the spouse of Dr. Saccone are listed as inventors on U.S. Patent 8,080,371,”Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. Authors listed with the affiliation deCODE Genetics/AMGEN are employees of deCODE genetics/AMGEN. Although unrelated to this research, Dr. Kranzler has been a consultant or advisory board member for Lundbeck and Indivior and is a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last three years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor, and Amygdala Neurosciences. Dr. Kaprio has consulted for Pfizer in 2012–2014 on nicotine dependence.
References
- 1.World Health Organization. WHO report on the global tobacco epidemic, 2011: Warning about the dangers of tobacco. World Health Organization; Geneva, Switzerland: 2011. [Google Scholar]
- 2.U.S. Department of Health and Human Services. The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General. Atlanta (GA): 2014. [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) Vital signs: current cigarette smoking among adults aged ≥18 years--United States, 2005–2010. MMWR Morb Mortal Wkly Rep. 2011;60(35):1207–1212. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC) Quitting smoking among adults--United States, 2001–2010. MMWR Morb Mortal Wkly Rep. 2011;60(44):1513–1519. [PubMed] [Google Scholar]
- 5.Shiffman S, Tindle H, Li X, Scholl S, Dunbar M, Mitchell-Miland C. Characteristics and smoking patterns of intermittent smokers. Exp Clin Psychopharmacol. 2012;20(4):264–277. doi: 10.1037/a0027546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breslau N, Johnson EO. Predicting smoking cessation and major depression in nicotine-dependent smokers. Am J Public Health. 2000;90(7):1122–1127. doi: 10.2105/ajph.90.7.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker TB, Piper ME, McCarthy DE, Bolt DM, Smith SS, Kim SY, et al. Time to first cigarette in the morning as an index of ability to quit smoking: implications for nicotine dependence. Nicotine Tob Res. 2007;9(Suppl 4):S555–570. doi: 10.1080/14622200701673480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan PF, Kendler KS. The genetic epidemiology of smoking. Nicotine Tob Res. 1999;1(Suppl 2):S51–57. doi: 10.1080/14622299050011811. discussion S69–70. [DOI] [PubMed] [Google Scholar]
- 9.Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35(4):397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- 10.Baker TB, Piper ME, Schlam TR, Cook JW, Smith SS, Loh WY, et al. Are tobacco dependence and withdrawal related amongst heavy smokers? Relevance to conceptualizations of dependence. J Abnorm Psychol. 2012;121(4):909–921. doi: 10.1037/a0027889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zelman DC, Brandon TH, Jorenby DE, Baker TB. Measures of affect and nicotine dependence predict differential response to smoking cessation treatments. J Consult Clin Psychol. 1992;60(6):943–952. doi: 10.1037//0022-006x.60.6.943. [DOI] [PubMed] [Google Scholar]
- 12.Gu F, Wacholder S, Kovalchik S, Panagiotou OA, Reyes-Guzman C, Freedman ND, et al. Time to smoke first morning cigarette and lung cancer in a case-control study. J Natl Cancer Inst. 2014;106(6):dju118. doi: 10.1093/jnci/dju118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guertin KA, Gu F, Wacholder S, Freedman ND, Panagiotou OA, Reyes-Guzman C, et al. Time to First Morning Cigarette and Risk of Chronic Obstructive Pulmonary Disease: Smokers in the PLCO Cancer Screening Trial. PLoS One. 2015;10(5):e0125973. doi: 10.1371/journal.pone.0125973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice JP, Hartz SM, Agrawal A, Almasy L, Bennett S, Breslau N, et al. CHRNB3 is more strongly associated with Fagerstrom Test for Cigarette Dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results. Addiction. 2012;107(11):2019–2028. doi: 10.1111/j.1360-0443.2012.03922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David SP, Hamidovic A, Chen GK, Bergen AW, Wessel J, Kasberger JL, et al. Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry. 2012;2:e119. doi: 10.1038/tp.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loukola A, Wedenoja J, Keskitalo-Vuokko K, Broms U, Korhonen T, Ripatti S, et al. Genome-wide association study on detailed profiles of smoking behavior and nicotine dependence in a twin sample. Mol Psychiatry. 2014;19(5):615–624. doi: 10.1038/mp.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R, et al. Genome-wide association study of nicotine dependence in American populations: identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry. 2015;77(5):493–503. doi: 10.1016/j.biopsych.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hancock DB, Reginsson GW, Gaddis NC, Chen X, Saccone NL, Lutz SM, et al. Genome-wide meta-analysis reveals common splice site acceptor variant in CHRNA4 associated with nicotine dependence. Transl Psychiatry. 2015;5:e651. doi: 10.1038/tp.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wain LV, Shrine N, Miller S, Jackson VE, Ntalla I, Artigas MS, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3(10):769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker TB, Breslau N, Covey L, Shiffman S. DSM criteria for tobacco use disorder and tobacco withdrawal: a critique and proposed revisions for DSM-5. Addiction. 2012;107(2):263–275. doi: 10.1111/j.1360-0443.2011.03657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86(9):1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 27.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 29.Fadista J, Manning AK, Florez JC, Groop L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur J Hum Genet. 2016;24(8):1202–1205. doi: 10.1038/ejhg.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanai M, Tanaka T, Okada Y. Empirical estimation of genome-wide significance thresholds based on the 1000 Genomes Project data set. J Hum Genet. 2016;61(10):861–866. doi: 10.1038/jhg.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Vries PS, Sabater-Lleal M, Chasman DI, Trompet S, Ahluwalia TS, Teumer A, et al. Comparison of HapMap and 1000 Genomes Reference Panels in a Large-Scale Genome-Wide Association Study. PLoS One. 2017;12(1):e0167742. doi: 10.1371/journal.pone.0167742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sveinbjornsson G, Albrechtsen A, Zink F, Gudjonsson SA, Oddson A, Masson G, et al. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat Genet. 2016;48(3):314–317. doi: 10.1038/ng.3507. [DOI] [PubMed] [Google Scholar]
- 33.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyles AL, Harris SF, Rooney AA, Thayer KA. Forest Plot Viewer: a new graphing tool. Epidemiology. 2011;22(5):746–747. doi: 10.1097/EDE.0b013e318225ba48. [DOI] [PubMed] [Google Scholar]
- 35.Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11(2):193–206. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- 36.Hannon E, Spiers H, Viana J, Pidsley R, Burrage J, Murphy TM, et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci. 2016;19(1):48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large scale genetic analysis identifies novel loci and histological variability in susceptibility to lung cancer. Nat Genet. doi: 10.1038/ng.3892. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fagerstrom KO. Measuring degree of physical dependence to tobacco smoking with reference to individualization of treatment. Addict Behav. 1978;3(3–4):235–241. doi: 10.1016/0306-4603(78)90024-2. [DOI] [PubMed] [Google Scholar]
- 41.Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6(5):e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hancock DB, Wang JC, Gaddis NC, Levy JL, Saccone NL, Stitzel JA, et al. A multiancestry study identifies novel genetic associations with CHRNA5 methylation in human brain and risk of nicotine dependence. Hum Mol Genet. 2015;24(20):5940–5944. doi: 10.1093/hmg/ddv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang JC, Spiegel N, Bertelsen S, Le N, McKenna N, Budde JP, et al. Cis-Regulatory Variants Affect CHRNA5 mRNA Expression in Populations of African and European Ancestry. PLoS One. 2013;8(11):e80204. doi: 10.1371/journal.pone.0080204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang JC, Cruchaga C, Saccone NL, Bertelsen S, Liu P, Budde JP, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009;18(16):3125–3135. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldstein RZ, Volkow ND. Dysfunction of the prefrontal cortex in addiction: neuroimaging findings and clinical implications. Nat Rev Neurosci. 2011;12(11):652–669. doi: 10.1038/nrn3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miquel M, Vazquez-Sanroman D, Carbo-Gas M, Gil-Miravet I, Sanchis-Segura C, Carulli D, et al. Have we been ignoring the elephant in the room? Seven arguments for considering the cerebellum as part of addiction circuitry. Neurosci Biobehav Rev. 2016;60:1–11. doi: 10.1016/j.neubiorev.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Moulton EA, Elman I, Becerra LR, Goldstein RZ, Borsook D. The cerebellum and addiction: insights gained from neuroimaging research. Addict Biol. 2014;19(3):317–331. doi: 10.1111/adb.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strick PL, Dum RP, Fiez JA. Cerebellum and nonmotor function. Annu Rev Neurosci. 2009;32:413–434. doi: 10.1146/annurev.neuro.31.060407.125606. [DOI] [PubMed] [Google Scholar]
- 49.Weissman J, Naidu S, Bjornsson HT. Abnormalities of the DNA methylation mark and its machinery: an emerging cause of neurologic dysfunction. Semin Neurol. 2014;34(3):249–257. doi: 10.1055/s-0034-1386763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 51.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 52.Jin B, Tao Q, Peng J, Soo HM, Wu W, Ying J, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet. 2008;17(5):690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- 53.Teneng I, Tellez CS, Picchi MA, Klinge DM, Yingling CM, Snider AM, et al. Global identification of genes targeted by DNMT3b for epigenetic silencing in lung cancer. Oncogene. 2015;34(5):621–630. doi: 10.1038/onc.2013.580. [DOI] [PubMed] [Google Scholar]
- 54.Oka M, Meacham AM, Hamazaki T, Rodic N, Chang LJ, Terada N. De novo DNA methyltransferases Dnmt3a and Dnmt3b primarily mediate the cytotoxic effect of 5-aza-2′-deoxycytidine. Oncogene. 2005;24(19):3091–3099. doi: 10.1038/sj.onc.1208540. [DOI] [PubMed] [Google Scholar]
- 55.Gnyszka A, Jastrzebski Z, Flis S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res. 2013;33(8):2989–2996. [PubMed] [Google Scholar]
- 56.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Domino EF, Minoshima S, Guthrie S, Ohl L, Ni L, Koeppe RA, et al. Nicotine effects on regional cerebral blood flow in awake, resting tobacco smokers. Synapse. 2000;38(3):313–321. doi: 10.1002/1098-2396(20001201)38:3<313::AID-SYN10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 58.Zubieta JK, Heitzeg MM, Xu Y, Koeppe RA, Ni L, Guthrie S, et al. Regional cerebral blood flow responses to smoking in tobacco smokers after overnight abstinence. Am J Psychiatry. 2005;162(3):567–577. doi: 10.1176/appi.ajp.162.3.567. [DOI] [PubMed] [Google Scholar]
- 59.Brody AL, Mandelkern MA, Jarvik ME, Lee GS, Smith EC, Huang JC, et al. Differences between smokers and nonsmokers in regional gray matter volumes and densities. Biol Psychiatry. 2004;55(1):77–84. doi: 10.1016/s0006-3223(03)00610-3. [DOI] [PubMed] [Google Scholar]
- 60.Gallinat J, Meisenzahl E, Jacobsen LK, Kalus P, Bierbrauer J, Kienast T, et al. Smoking and structural brain deficits: a volumetric MR investigation. Eur J Neurosci. 2006;24(6):1744–1750. doi: 10.1111/j.1460-9568.2006.05050.x. [DOI] [PubMed] [Google Scholar]
- 61.Kuhn S, Romanowski A, Schilling C, Mobascher A, Warbrick T, Winterer G, et al. Brain grey matter deficits in smokers: focus on the cerebellum. Brain Struct Funct. 2012;217(2):517–522. doi: 10.1007/s00429-011-0346-5. [DOI] [PubMed] [Google Scholar]
- 62.Sutherland MT, Riedel MC, Flannery JS, Yanes JA, Fox PT, Stein EA, et al. Chronic cigarette smoking is linked with structural alterations in brain regions showing acute nicotinic drug-induced functional modulations. Behav Brain Funct. 2016;12(1):16. doi: 10.1186/s12993-016-0100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wood AR, Perry JR, Tanaka T, Hernandez DG, Zheng HF, Melzer D, et al. Imputation of variants from the 1000 Genomes Project modestly improves known associations and can identify low-frequency variant-phenotype associations undetected by HapMap based imputation. PLoS One. 2013;8(5):e64343. doi: 10.1371/journal.pone.0064343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang J, Ellinghaus D, Franke A, Howie B, Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur J Hum Genet. 2012;20(7):801–805. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ananthakrishnan AN. Environmental risk factors for inflammatory bowel disease. Gastroenterol Hepatol (N Y) 2013;9(6):367–374. [PMC free article] [PubMed] [Google Scholar]
- 67.Thorgeirsson TE, Steinberg S, Reginsson GW, Bjornsdottir G, Rafnar T, Jonsdottir I, et al. A rare missense mutation in CHRNA4 associates with smoking behavior and its consequences. Mol Psychiatry. 2016;21(5):594–600. doi: 10.1038/mp.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie P, Kranzler HR, Krauthammer M, Cosgrove KP, Oslin D, Anton RF, et al. Rare nonsynonymous variants in alpha-4 nicotinic acetylcholine receptor gene protect against nicotine dependence. Biol Psychiatry. 2011;70(6):528–536. doi: 10.1016/j.biopsych.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Siedlinski M, Cho MH, Bakke P, Gulsvik A, Lomas DA, Anderson W, et al. Genome-wide association study of smoking behaviours in patients with COPD. Thorax. 2011;66(10):894–902. doi: 10.1136/thoraxjnl-2011-200154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hirvonen K, Korhonen T, Salomaa V, Männistö S, Kaprio J. Effect of the DBH polymorphism rs3025343 on smoking cessation in a large population-based sample. Nic Tob Research. doi: 10.1093/ntr/ntx066. In press. [DOI] [PubMed] [Google Scholar]
- 71.Yang J, Wang S, Yang Z, Hodgkinson CA, Iarikova P, Ma JZ, et al. The contribution of rare and common variants in 30 genes to risk nicotine dependence. Mol Psychiatry. 2015;20(11):1467–1478. doi: 10.1038/mp.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25. 1. Nat Genetics. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barrie ES, Weinshenker D, Verma A, Pendergrass SA, Lange LA, Ritchie MD, et al. Regulatory polymorphisms in human DBH affect peripheral gene expression and sympathetic activity. Circ Res. 2014;115(12):1017–1025. doi: 10.1161/CIRCRESAHA.116.304398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee PN, Forey BA, Coombs KJ. Systematic review with meta-analysis of the epidemiological evidence in the 1900s relating smoking to lung cancer. BMC Cancer. 2012;12:385. doi: 10.1186/1471-2407-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee PN, Forey BA. Indirectly estimated absolute lung cancer mortality rates by smoking status and histological type based on a systematic review. BMC Cancer. 2013;13:189. doi: 10.1186/1471-2407-13-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


