Abstract
Cold stress is a major environmental factor that impairs plant growth and development, geographic distribution, and crop productivity. The C-repeat binding factor (CBF) regulatory pathway has an essential role in response to cold stress. Here, we characterized a bHLH transcription factor from Nicotiana tabacum, NtbHLH123, in response to cold stress (4°C). Overexpression of NtbHLH123 enhanced cold tolerance in transgenic tobacco plants. Based on yeast one-hybrid, chromatin immunoprecipitation PCR, and transient expression analysis assays, NtbHLH123 binds directly to the G-box/E-box motifs in the promoter of the NtCBF genes and positively regulates their expression. Furthermore, NtbHLH123-overexpressing plants showed lower electrolyte leakage, reduced malondialdehyde contents, H2O2 and reactive oxygen species (ROS) accumulation under cold stress, which contributed to alleviating oxidative damage to the cell membrane after cold stress treatment. And NtbHLH123 increased stress tolerance by improving the expression of a number of abiotic stress-responsive genes to mediate the ROS scavenging ability and other stress tolerance pathways. Taken together, we present a model suggesting that NtbHLH123 is a transcriptional activator that functions as a positive regulator of cold tolerance by activating NtCBF, ROS scavenging-related, and stress-responsive genes.
Keywords: NtbHLH123, NtCBF pathway, reactive oxygen species (ROS), transcriptional regulation, cold stress, Nicotiana tabacum
Introduction
Many environmental factors (e.g., high or low temperature, salt and drought) limit the growth and development, geographic distribution, yield, and quality of crop plants (Xiong et al., 2002). Low temperature (cold stress) is a key environmental stress (Chinnusamy et al., 2007). Cold stress affects physiological metabolic reactions, oxidative damage, poor germination, accelerated senescence, membrane damage, and tissue breakdown among others (Chinnusamy et al., 2007; Sanghera et al., 2011). Plants have developed sophisticated mechanisms to adapt to and tolerate cold stress by altering biochemical and physiological processes (Thomashow, 1999; Chinnusamy et al., 2007; Zhu, 2016). Many cold-responsive genes have been identified that are involved in the cold tolerance of plants, including phosphatases, protein kinases, transcription factors (TFs), and so on (Pearce, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). Among them, cold stress can rapidly induce the expression of many TFs, such as C-repeat-binding factor/dehydration-responsive binding protein (CBF/DREB), ABA-responsive element-binding protein/ABA-binding factor, APETALA2 (AP2)/ethylene responsive factor, basic region/leucine zipper motif (bZIP), MYB, basic helix-loop-helix (bHLH), and NAM, ATAF1, 2, and CUC2 (NAC) families, which in turn bind the promoter of numerous stress-responsive genes and regulate their expression (Chinnusamy et al., 2003; Vogel et al., 2005; Agarwal et al., 2006; Yamaguchi-Shinozaki and Shinozaki, 2006; Puranik et al., 2012; Shi et al., 2012).
The bHLH TF superfamily is a large group of functionally diverse proteins found in plants and animals (Garrell and Campuzano, 1991; Ledent and Vervoort, 2001). These proteins have a 60-amino-acid conserved domain, which contains two functionally distinct regions; a basic region containing 13–17 amino acids at the N-terminus that functions as a DNA-binding domain, and the HLH region at the C-terminus, which contributes to the formation of homodimers or heterodimers (Heim et al., 2003; Toledo-Ortiz et al., 2003). Research has demonstrated that plant bHLHs have important roles in regulating gene transcriptional networks involving light signaling (Pedmale et al., 2016), hormone signaling (Zhang et al., 2009), flavonoid biosynthesis (Xu et al., 2015), flowering time (Song et al., 2015), stomata, trichome and root hair formation (Feller et al., 2011), and abiotic stress responses including cold, heat (Chinnusamy et al., 2007; Shi et al., 2015; Zhu, 2016).
To date, 167 and 177 bHLH genes have been identified in Arabidopsis and rice, respectively (Mao et al., 2017). However, characterization of the functions of most bHLHs is relatively limited. Only 64 of 167 and 19 of 177 bHLHs have been characterized or partially characterized functionally in Arabidopsis and rice (Ji et al., 2016). For instance, AtbHLH104, CmbHLH1, and MdbHLH104 enhance iron deficiency tolerance in Arabidopsis, chrysanthemum, and apple (Zhao et al., 2014, 2016b; Zhang et al., 2015). MUTE, FAMA, SPEECHLESS, OsMUTE, OsFAMA, OsSPCH2, and ZmMUTE regulate stomata differentiation in Arabidopsis, rice, and maize (Torii, 2015). PIF1/PIL5, PIF3, PIF4, PIF5/PIL6, and OsbHLH102 are involved in light and gibberellin signaling in Arabidopsis and rice (Leivar and Monte, 2014). Recent studies have indicated that a number of bHLH genes are involved in abiotic stress responses. For example, MYC2 functions as a TF in regulating wound, oxidative, drought, and salt stress responses (Fujita et al., 2006). OsbHLH1 is involved in the cold signal transduction pathway (Wang et al., 2003). PtrbHLH has been shown to confer cold tolerance in Poncirus trifoliata (Huang et al., 2013). OrbHLH2 overexpression improved tolerance to salt and osmotic stress in rice (Zhou et al., 2009). Overexpression of OrbHLH001 confers freezing and salt tolerance (Li et al., 2010). OsbHLH148 overexpression in rice increases drought tolerance (Seo et al., 2011). AtbHLH112 is involved in abiotic stress tolerance (Liu et al., 2015). However, the functional roles of bHLH genes in response to cold stress have not been demonstrated in tobacco (Nicotiana tabacum).
Although several cold-related plant bHLH TFs have been characterized in Arabidopsis, rice, maize, Pyrus ussuriensis, Populus suaveolens, Tamarix hispida, and apple (Chinnusamy et al., 2007; Feng et al., 2012; Ji et al., 2016; Jin et al., 2016; Zhu, 2016). However, the functions of bHLH genes in cold tolerance remain poorly characterized in tobacco. As a model plant, tobacco has a key role in plant molecular research on cold tolerance and is sensitive to cold stress (Jin et al., 2017). In this study, we report the molecular cloning and functional characterization of NtbHLH123 isolated from tobacco to identify and characterize the molecular mechanism of the tobacco response to cold stress. NtbHLH123 was induced by cold stress (4°C), and overexpression in tobacco plants under the control of the cauliflower mosaic virus (CaMV) 35S promoter enhanced cold resistance. These data suggest that NtbHLH123 has a positive role in cold resistance in tobacco, and may be an important candidate gene for the molecular breeding of cold-tolerant plants.
Materials and Methods
Plant Materials
Nicotiana tabacum L. ‘NC89’ was used as the wild type (WT). The seeds of the WT and transgenic lines were sterilized with 3.0% NaClO, and then germinated on plates containing Murashige and Skoog (MS) medium containing 3% sucrose and 0.6% agar at 25°C (Murashige and Skoog, 1962). The seedlings were transferred to soil and cultured in greenhouse (16-h light/8-h dark cycle at 25°C) and used for gene cloning, expressional analysis.
Vector Construction and Genetic Transformation in Tobacco
To construct a reporter vector to analyze the expression of NtbHLH123 in response to cold treatment, the full length of the promoter was amplified by PCR and cloned into pCXGUS-P to drive expression of the β-glucuronidase (GUS) reporter gene. For overexpression of NtbHLH123 in tobacco, the NtbHLH123 open reading frame (ORF) was cloned into the pRI 101-GFP vector containing a CaMV 35S promoter. These constructs were transferred into Agrobacterium strain LBA4404.
For tobacco transformation, the leaves of young seedlings from shoots grown in vitro were excised and cut into small strips. The leaf strips were immersed into Agrobacterium suspension culture for 15 min, and then dried with sterile filter paper. Then, the leaf strips were transferred onto MS medium with 0.1 mg L-1 1-naphthaleneacetic acid (NAA) + 1.0 mg L-1 6-benzylaminopurine (6-BA) for co-cultivation at 25 ± 1°C in the dark. A total of 3 days later, the leaf strips were subsequently transferred to selection medium containing 0.1 mg L-1 NAA + 1.0 mg L-1 6-BA + 100 mg L-1 kanamycin + 250 mg L-1 carbenicillin. After 3 weeks, adventitious shoots were regenerated and transferred to rooting medium (1/2 MS + 150 mg L-1 kanamycin + 250 mg L-1 carbenicillin). Rooted plants were transplanted into soil. Homozygous seedlings were used for further investigation.
Expression Analysis
For expression analysis, the uniformly sized seedlings were treated with control (25°C) or cold stress (4°C). Then the samples were collected from the various treated plants at specific time points and were immediately frozen in liquid nitrogen and stored at -80°C. Total RNA from the samples was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions. First-strand cDNA was synthesized using the PrimeScript First Strand cDNA Synthesis Kit transcriptase (TaKaRa, Dalian, China) according to manufacturer’s instructions.
Quantitative reverse transcription (qRT)-PCR was performed to determine transcript levels of NtbHLH123 and stress-related genes in the transgenic and control plants. Reactions were carried out in a total volume of 20 μL containing 10 μL of SYBR Premix Ex Taq II, 0.8 μL of both forward and reverse primers, 0.4 μL of ROX Reference Dye II, and 100 ng of cDNA template. qRT-PCR reactions (95°C, 30 s; 95°C, 5 s; 60°C, 34 s; 40 cycles) were performed using the SYBR Green method on an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). Gene expression levels were calculated based on the full-quantification method, and NtACTIN (GenBank accession number: U60495) was used as the internal control for normalizing gene expression. All of the primers used are shown in Supplementary Table S1.
Protein Extraction and Western Blot
Total proteins of WT and three transgenic plants were extracted in 1 × SDS buffer. For Western blot, the proteins were separated with SDS–PAGE gel and transferred onto PVDF membrane (Roche, United States). The gel blot was probed with anti-GFP (Beyotime, China) and the signals were visualized by chemiluminescence using the ECL plus kit (Millipore, Bedford, MA, United States) according to the manufacturer’s instructions. Anti-Actin served as a loading control.
Cold Tolerance Analysis
For germination rate analysis, the aseptic seeds of control and three transgenic lines were planted on MS medium, and were incubated for 8 days under 25 ± 1 or 4 ± 1°C. Changes of germination (testa rupture) were measured.
For the root length measurements, 10-days-old seedlings were maintained at 25 ± 1 or 4 ± 1°C. The treatment plates were placed vertically. The relative root length was calculated as follows: elongation of root length/original of root length.
For adult plant analysis, the seedlings were first transplanted into plastic containers filled with a mixture of soil and sand (1:1) and grown under normal conditions. The 40-days-old tobacco plants of WT and transgenic plants were directly exposed to cold stress (4°C) for 2 days without cold acclimation, and then moved to recover under normal conditions for 14 days. All the results are based on the average of three independent biological replicates.
Yeast One-Hybrid Assay
The yeast one-hybrid (Y1H) assay was performed using the Matchmaker One-Hybrid Library and Screening Kit (Clontech, Mountain View, CA, United States). The full length of the NtbHLH123 gene was amplified from tobacco cDNA using PCR and recombined into the pGADT7 vector (Clontech, Mountain View, CA, United States). NtCBF promoter fragments and mutated fragments were amplified from tobacco genomic DNA and cloned into the pAbAi vector (Clontech, Mountain View, CA, United States). Both the pAbAi bait vector and the pGADT7 prey vector were introduced into Y1H Gold Yeast (Clontech, Mountain View, CA, United States) and cultured on SD/–Leu medium with or without 150 ng mL-1 aureobasidin A (AbA) for 3–5 days at 30°C. The primer sequences used are listed in Supplementary Table S2.
Chromatin Immunoprecipitation (ChIP)-PCR Analysis
ChIP analysis was carried out using the Chromatin Immunoprecipitation Assay Kit (Millipore, MA, United States) following the manufacturer’s instructions. Protein-DNA complexes were cross-linked and incubated with GFP antibody (Beyotime, China). IP protein-DNA complexes were precipitated with Protein A-Sepharose beads overnight at 4°C. The DNA fragments in the IP complex were purified as described by Zhao et al. (2016a). DNA fragment enrichment was analyzed using qRT-PCR. The primers used are listed in Supplementary Table S2.
Transient Expression Assay in Tobacco Leaves
The NtbHLH123 coding region was inserted into pGreenII 62-SK to generate effector plasmids. The promoter fragments and mutant promoter fragments were cloned into pGreenII 0800-LUC to create reporter plasmids. The vectors of the effector and reporter were separately transformed into Agrobacterium tumefaciens LBA4404 cells, and the transformed LBA4404 cells were used to infiltrate the leaves of Nicotiana benthamiana for transient expression, which was sampled after 2 days. Firefly (LUC) and Renilla (REN) luciferase were detected with dual luciferase assay reagents (Promega, Madison, WI, United States) using an Infinite M200 plate reader (Tecan, Männedorf, Switzerland). The Promoter activities were calculated as the ratio of LUC/REN.
GUS Staining and Activity Analysis
The GUS staining was carried out according to the method of Zhao et al. (2016b). Transgenic tobacco plants were immersed into GUS staining buffer at 37°C for 4 h in the dark. After staining, the transgenic tobacco plants were de-stained and photographed.
For quantitative analysis of GUS activity, fluorescence was measured using a VersaFluor spectrofluorometer (excitation wavelength of 365 nm and emission wavelength of 450 nm).
Histochemical Staining Analysis
The H2O2 accumulation in leaves was stained by 3,3′-diaminobenzidine (DAB). Details of assay have been described by Zhao et al. (2016a).
The reactive oxygen species (ROS) accumulation was detected using the fluorescent probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFH2-DA, Sigma-Aldrich, United States) according to Zhao et al. (2016a). For DCFH2-DA staining, leaves were treated with 10 μM DCFH2-DA for 20 min and then washed with distilled H2O. The images were obtained using the confocal microscope (Ex, 488 nm; Em, 522 nm) (Leica Microsystems, Wetzlar, Germany).
Physiological Measurements
The leaves, which were exposed to control or cold stress conditions, were sampled from the WT and three transgenic plants (L1, L2, and L3). For electrolyte leakage (EL) measurement, the leaves were placed in 30 ml distilled water and shaken at room temperature for 3 h, the initial conductivity (E1) was measured with electric conductivity meter. Then the leaves were boiled for 20 min and were cooled to room temperature, the electrolyte conductivity (E2) was measured, and the electrolyte leakage (%) was calculated as follows: Electrolyte leakage (%) = 100 × E1/E2. The malondialdehyde (MDA) accumulation was measured using the thiobarbituric acid–based method with an MDA assay kit (A003-3, Jiancheng, Nanjing, China). The Chlorophyll content was measured as described by Feng et al. (2012).
For the enzyme assays, the SOD activities were determined by measuring the inhibiting rate of the enzyme to O2-⋅ produced by the xanthine morpholine with xanthine oxidase using the SOD Detection Kit (A001, Jiancheng, Nanjing, China). The CAT activity was measured as the degradation of H2O2 at 405 nm according to the CAT Detection Kit (A007, Jiancheng, Nanjing, China). The reaction mixture, which contained 0.1 mM H2O2 and 60 mM potassium phosphate buffer (pH 7.0), was incubated at 37°C for 1 min. Then, the reaction was stopped by adding 32.5 mM ammonium molybdate, and the yellow complex of H2O2 and ammonium molybdate was detected at 405 nm. The POD activity was measured based on the change of absorbance at 420 nm by catalyzing H2O2 according to POD Detection Kit for plant (A084-3, Jiancheng, Nanjing, China).
Statistical Analysis
Cold stress treatment of the WT and transgenic lines was repeated at least three independent biological replicates with consistent results. All experimental data are averages of at least three independent biological replicates. The data were analyzed by Duncan’s multiple range tests in the ANOVA program of SPSS (IBM SPSS 22), taking ∗P < 0.01, ∗∗P < 0.001 as significantly different.
Results
NtbHLH123 Transcription Is Induced by Cold Stress
Transcriptome analysis, which was conducted using cDNA samples extracted from tobacco seedlings treated with or without cold stress, showed that NtbHLH123 was upregulated by cold treatment (Jin et al., 2017), suggesting that NtbHLH123 might be a cold-responsive bHLH TF gene. Bioinformatics analysis showed that NtbHLH123, which has seven exons and six introns and is localized on the chromosome 1 of the tobacco genome, contains a 1389-bp ORF and encodes 463 amino acids with a predicted molecular weight of 50.72 kDa and a pI of 6.40 (Supplementary Figure S1A). Motif analysis showed that NtbHLH123 has a highly conserved bHLH domain containing 61 amino acids in its C-terminal regions (Supplementary Figure S1B).
To examine NtbHLH123 transcript levels in response to cold stress, we performed qRT-PCR analysis. NtbHLH123 expression changed slightly during the studied period under normal condition; however, under cold stress (4°C), NtbHLH123 expression was highly induced and reached a peak (eightfold) at 12 h, and decreased at 24 h (Figure 1A). In addition, GUS staining and GUS activity showed that NtbHLH123 expression increased in both the leaves and roots of plants expressing ProNtbHLH123::GUS in response to cold stress (Figures 1B,C). These results suggest that cold stress activates NtbHLH123 transcription in tobacco.
FIGURE 1.
NtbHLH123 expression profiles in response to cold stress. (A) Analysis of NtbHLH123 expression using real-time PCR. Tobacco plants were subjected to 4°C for the indicated times. The relative expression levels were normalized to the expression of the NtACTIN gene. Data are the means ± SD of three independent biological replicates. (B,C) Analysis of NtbHLH123 expression in response to cold treatment using (B) β-glucuronidase (GUS) histochemical staining and (C) GUS activity measurement. The ProNtbHLH123::GUS transgenic seedlings were grown in Murashige and Skoog medium with or without treatment at 4°C for 4 h, and GUS histochemical staining and GUS activity measurement were performed. Data are expressed as the mean ± SD as determined from three independent biological replicates. Asterisks indicate that the value is significantly different from that of the control at the same time point (∗∗P < 0.001).
NtbHLH123 Overexpression Increases Cold Tolerance
To investigate the function of NtbHLH123, 35S::NtbHLH123 transgenic tobacco plants were obtained via Agrobacterium-mediated transformation. In total, eight T0 independent transgenic lines were confirmed by PCR with CaMV 35S and NPTII primers (Supplementary Figure S2). Among them, three independent lines (L1, L2, and L3) were used for functional characterization, while the WT was used as a control. qRT-PCR and Western blot assays indicated that all three transgenic tobacco lines had increased transcript levels and produced more NtbHLH123-GFP fusion protein (Figures 2A,B), indicating that NtbHLH123 was overexpressed in these transgenic tobacco lines.
FIGURE 2.
Seed germination and seedling root lengths of NtbHLH123 transgenic tobacco were insensitive to cold stress. (A) NtbHLH123 transcript levels in transgenic tobacco. WT, wild-type; L1, L2, and L3, transgenic tobacco lines. (B) NtbHLH123-GFP fusion protein levels in 35S::NtbHLH123-GFP transgenic plants as determined by immunoblot analysis using an anti-GFP antibody. Anti-actin antibody was used as a loading control. (C,D) Seed germination rates were determined in the overexpression lines and WT. All tests were repeated at least three independent biological replicates, and approximately 50 seeds were counted for each experiment. Data are expressed as the means ± SD. (E,F) Comparison of transgenic and WT plant growth on plates. Tobacco seedlings were grown vertically for 8 days and the root lengths were measured under normal or cold-stress conditions. Data are expressed as the mean ± SD as determined from three independent biological replicates of 30 seedlings. Asterisks indicate that the value is significantly different from that of the WT at the same time point (∗P < 0.01; ∗∗P < 0.001).
To examine whether NtbHLH123 has a role in the response to cold stress, the seeds of transgenic tobacco plants and the WT control were sown on MS medium and exposed to 25 or 4°C, and the germination rates were examined. At 25°C condition, the transgenic and WT seeds have the same germination rates and all seeds germinated at 8 days. At 4°C condition, seed germination of both the WT and the transgenic lines was inhibited. However, 47.7–58.5% of the three transgenic lines seeds germinated, while only 12.3% of the WT seeds germinated, indicating that overexpression of NtbHLH123 in tobacco leads to reduced sensitivity of seed germination to cold stress (Figures 2C,D). Moreover, we measured the relative root lengths after cold treatment. At 25°C condition, there is not much difference in the relative root lengths of three transgenic seedlings (2.21, 2.32, and 2.36) and WT control (2.18) (Figures 2E,F). In contrast, the relative root lengths of three transgenic seedlings were 1.01, 1.16, and 1.53, respectively, while that of WT control was 0.58 at 4°C condition (Figures 2E,F). Therefore, NtbHLH123 overexpression enhanced cold tolerance in transgenic tobacco seedlings.
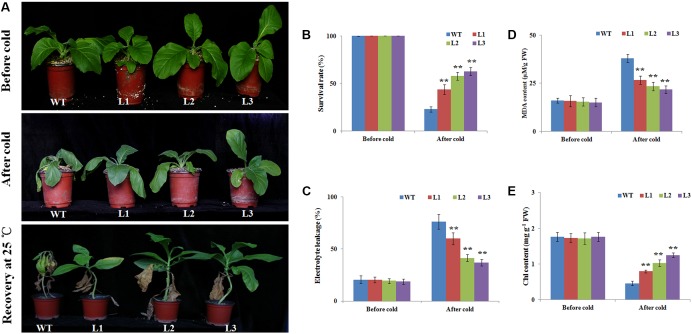
Adult plants (40-days-old seedlings) were also used to assess cold stress tolerance. Before cold treatment, there were no morphological differences between the transgenic and WT plants (Figure 3A). When seedlings were exposed to cold stress (4°C) for 2 days, cold injuries were observed in the WT plants more serious than the transgenic plants. After recovery for 14 days under 25°C, the survival rates of the WT plants was 10%, while the transgenic plants were 39.3, 50.1, and 59.6%, respectively (Figures 3A,B). EL and MDA levels, important indicators of cell damage, can reflect the extent of membrane injure (Huang et al., 2013). The EL in the WT plants (63.1%) was significantly higher than in three transgenic plants (49.9, 38.8, and 33.4%) (Figure 3C). While the MDA content in the WT were 1.47–1.81 times higher than in the transgenic plants (Figure 3D), suggesting that membrane damage was alleviated in the transgenic plants under cold stress condition. We also quantitatively measured the chlorophyll levels. After the cold treatment, the chlorophyll contents of L1 (0.79 mg g-1 FW), L2 (1.02 mg g-1 FW), and L3 (1.24 mg g-1 FW) transgenic plants were higher than the WT plants (0.46 mg g-1 FW) (Figure 3E). Overall, the results indicated that NtbHLH123 overexpression in tobacco resulted in increased protection of membrane integrity and enhanced cold tolerance compared with the WT control.
FIGURE 3.
Overexpression of NtbHLH123 enhances cold tolerance in transgenic tobacco. (A) Phenotypes of 40-days-old seedlings of wild type (WT) and transgenic plants subjected to cold treatment (4°C) 2 days followed by recovery at 25°C for 14 days. At least three independent biological replicates were performed for each individual experiment. (B) Survival ratios of the WT and transgenic plants after recovery from cold stress treatment. Data represent the means ± SD of at least three independent biological replicates. Asterisks indicate that the value is significantly different from that of the WT at the same time point (∗P < 0.01; ∗∗P < 0.001). (C,D) Malondialdehyde (MDA) content and electrolyte leakage, respectively. FW, fresh weight. Data represent the means ± SD of at least three independent biological replicates. Asterisks indicate that the value is significantly different from that of the WT at the same time point (∗P < 0.01; ∗∗P < 0.001). (E) Effects of cold stress on the chlorophyll (chl) content in WT and transgenic tobacco leaves. Data are the means ± SD of three independent biological replicates. Asterisks indicate that the value is significantly different from that of the WT at the same time point (∗∗P < 0.001).
Next, the transcript levels in both transgenic plants and WT were analyzed under control and cold stress (Supplementary Figure S3). The relative mRNA levels of NtbHLH123 in the transgenic lines were more than twofold, significantly higher than that of WT under cold stress condition. These results indicate that the transgenic plants improve the cold stress tolerant, mainly through the overexpression of NtbHLH123 in tobacco.
NtbHLH123 Directly Regulates NtCBF Gene Expression
Using the Tobacco Genomic Database1, we identified 21 NtCBF genes (Supplementary Figure S4A). A phylogenetic tree indicated that the 21 NtCBF genes were closely related to four Arabidopsis AtCBF genes (AP2 family) (Supplementary Figure S4B). To determine whether the NtbHLH123-regulated plant response to cold stress is dependent on the CBF pathway, we examined the expression levels of NtCBF genes in NtbHLH123 transgenic tobacco plants after cold treatment using qRT-PCR. NtbHLH123 overexpression significantly induced the expression of NtCBF1–NtCBF8 genes (Figure 4A). These results demonstrated that NtbHLH123 positively regulates plant cold tolerance by upregulating NtCBF expression.
FIGURE 4.
NtbHLH123 binds to the promoters of NtCBF genes. (A) Expression analysis of NtCBF genes in the wild type (WT) and three transgenic tobacco lines after cold treatment. Data are the means ± SD of three independent biological replicates. The value for the WT control was set to 1. Asterisks indicate that the value is significantly different from that of the WT at the same time point (∗∗P < 0.001). (B) Schematic diagrams of the NtCBF1, NtCBF7, and NtCBF8 promoter regions showing the presence of E-box DNA motifs. Transverse lines show the positions of primers used in the chromatin immunoprecipitation (ChIP)-PCR experiment. (C) ChIP-qPCR analyses of the DNA binding ratio of NtbHLH123 to the promoters of NtCBF1, NtCBF7, and NtCBF8. Data represent the means ± SD of three independent biological replicates. Asterisks indicate that the value is significantly different from that of the 35S::GFP at the same time point (∗∗P < 0.001). (D) Analysis of the binding of NtbHLH123 to the promoters of NtCBF1, NtCBF7, and NtCBF8 based on a yeast one-hybrid study. The normal or mutant promoters co-transformed with AD-NtbHLH123 and the positive controls were grown on selection medium with or without 150 ng mL-1 aureobasidin A (AbA).
In our previous works, NtbHLH123 proteins specifically bind to the E-box and G-box sequence ‘CANNTG’ (unpublished data). Using the plantCARE and PLACE programs, we found many G-box or E-box motifs in the promoters of the NtCBF1–NtCBF8 genes (Figure 4B). To detect the binding ability of NtbHLH123 to the NtCBF promoters, we performed ChIP-qPCR using 35S::GFP and 35S::NtbHLH123-GFP transgenic plants. After immunoprecipitation with anti-GFP, the DNA fragments containing G-box or E-box motifs were amplified with qRT-PCR. A clear band was detected around 1 (NtCBF1), 6 (NtCBF7), and 9 (NtCBF8) when NtbHLH123-GFP was precipitated (Figure 4C), demonstrating that NtCBF1, NtCBF7, and NtCBF8 are target genes of NtbHLH123. In addition, E-box elements were present in other NtCBF genes (NtCBF2–NtCBF6). However, the ChIP-PCR results showed that none of these recruited MdbHLH104-GFP proteins (Supplementary Figure S5). The Y1H assay was used to confirm the results of the ChIP-qPCR assay. The NtbHLH123 ORF and promoter fragments containing the E-box motifs were cloned into plasmids pGADT7 and pAbAi, respectively. pGADT7-NtbHLH123 and pAbAi-1/6/9 were co-transformed into Y1H Gold, which were selected on SD/–Leu medium with or without 150 ng mL-1 AbA. When 150 ng mL-1 AbA was added, only the cells co-transformed with pGADT7-NtbHLH123 and pAbAi-1/6/9 grew (Figure 4D). These results indicate that the NtbHLH123 protein can bind to the E-box elements in the NtCBF promoters.
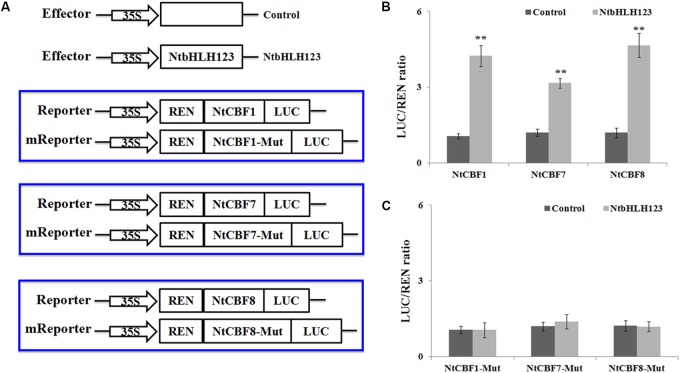
We performed transient expression assays to examine how NtbHLH123 regulates the expression of NtCBFs (NtCBF1, NtCBF7, and NtCBF8). The NtbHLH123 ORF was cloned into pGreenII 62-SK, and the of the NtCBF1, NtCBF7, and NtCBF8 promoter fragments were cloned into pGreenII 0800-LUC (Figure 5A). A significant increase in luminescence intensity was observed when NtbHLH123 was co-expressed with the NtCBF1, NtCBF7, or NtCBF8 promoters, whereas 35Spro:NtbHLH123 failed to activate NtCBF1, NtCBF7, or NtCBF8 expression when the E-box motifs were mutated (Figure 5B). These findings reveal that NtbHLH123 can activate NtCBF1, NtCBF7, and NtCBF8 expression.
FIGURE 5.
NtbHLH123 directly activates the expression of NtCBF genes. (A) Illustration of the effector and reporter constructs used for transient expression assay. The mReporter vector was constructed using the mutated E-box motifs. (B,C) Transient expression assay of the promoter activity in tobacco leaves co-transformed with the effector and reporter constructed using the normal or mutated E-box motif. Data represent the means ± SD of three independent biological replicates. Asterisks indicate that the value is significantly different from that of the control at the same time point (∗∗P < 0.001).
Analysis of ROS Levels, and Antioxidant Enzyme Activities
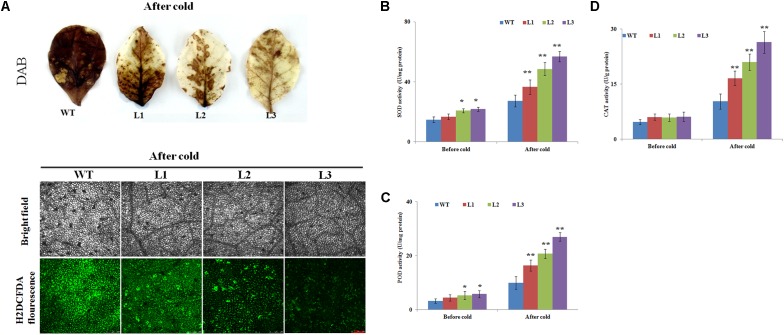
It is well documented that cold stresses usually results in the excessive accumulation of ROS (e.g., O2- or H2O2), which can be used as indicators of cell damage (Mittler, 2002). To test whether overexpression of NtbHLH123 genes also affects ROS levels, leaves from WT plants and transgenic plants were stained with DAB or H2DCFDA after treatment with cold stress. Histochemical staining indicated that WT plants showed higher level of ROS than transgenic plants, after the plants were subjected to cold stress (Figure 6A).
FIGURE 6.
Histochemical staining analysis and activities of antioxidant enzymes in wild type (WT) and transgenic plants before and after cold treatment. (A) H2O2 and ROS accumulation in transgenic and WT plants based on histochemical staining before and after cold treatment. (B–D) Activities of (B) superoxide dismutase (SOD), (C) peroxidase (POD), and (D) catalase (CAT) in the WT and transgenic plants under normal or cold stress conditions. Data represent the means ± SD of at least three independent biological replicates. Asterisks indicate that the value is significantly different from that of the WT at the same time point (∗P < 0.01; ∗∗P < 0.001).
The above results showed that the WT plants had higher ROS levels than the transgenic plants; therefore, three key antioxidant enzymes (e.g., CAT, POD, and SOD), which play critical roles in ROS scavenging and affect cellular ROS levels (Mittler, 2002), were measured before and after cold stress treatment. The transgenic plants showed slightly higher activities of these enzymes than the WT plants before cold treatment (Figures 6B–D). After cold treatment, SOD activities were markedly enhanced in the transgenic lines, whereas the WT plants showed only a slight increase (Figure 6B). Exposure to cold condition resulted in notably rise of POD activity in transgenic lines, which was 1.65-, 2.09-, and 3.11-fold of that in WT, respectively (Figure 6C). The CAT activities of the transgenic plants (between 16.5 U g-1 protein and 26.3 U g-1 protein) were significantly higher than WT (10.2 U g-1 protein) (Figure 6D).
Expression Analysis of ROS Scavenging-Related and Stress-Responsive Genes in Transgenic and WT Plants Before and After Cold Treatment
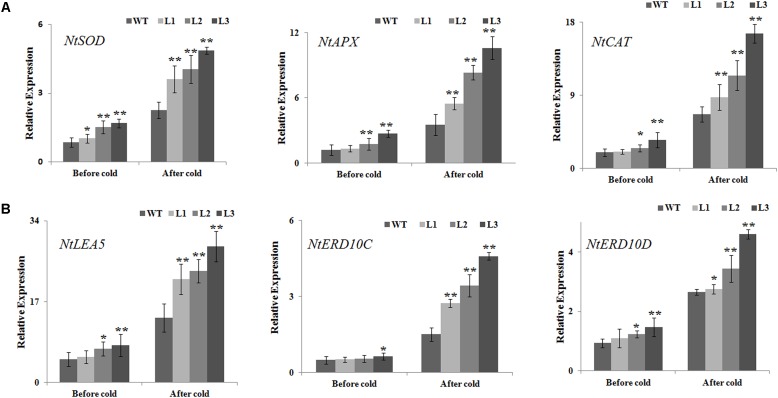
The positive effect of NtbHLH123 overexpression on antioxidant enzyme activities suggested that NtbHLH123 might be involved in the regulation of ROS homeostasis under cold stress treatment. Therefore, the expression levels of ROS-related genes encoding enzymes for direct ROS detoxification were detected in the WT and transgenic plants before and after cold treatment. The transcript levels of NtAPX, NtSOD, and NtCAT were slightly enhanced in the transgenic lines compared with those of WT in the absence of cold stress. However, all three genes were dramatically upregulated by 1.5- to 5-fold in the NtbHLH123-overexpressing plants after cold treatment (Figure 7A), suggesting that NtbHLH123 may be a key regulator upstream of some ROS-related genes, and overexpression of NtbHLH123 could trigger the induction of a series of ROS-related genes to cope positively with adverse environmental conditions. In addition, we analyzed the transcript levels of several cold stress defensive proteins (NtLEA5, NtERD10C, and NtERD10D). Under normal conditions, the mRNA levels of all three genes were slightly higher in the transgenic plants than in the WT plants. After the cold treatment, the expression levels of three genes increased rapidly in the WT and transgenic plants, but the NtLEA5 and NtERD10C caused a 2- to 5-fold increase in transgenic plants and NtERD10D resulted only in a 0.8- to 1.7-fold upregulation transgenic plants compared with the WT (Figure 7B). These results suggest that NtbHLH123 overexpression enhances the mRNA levels of ROS scavenging-related and stress-responsive genes with cold stress treatment.
FIGURE 7.
Analysis of relative gene expression of reactive oxidation species scavenging-related (A) and stress-responsive genes (B) based on quantitative reverse transcription PCR in WT and transgenic plants before and after cold treatment. Data represent the means ± SD of at least three independent biological replicates. Asterisks indicate that the value is significantly different from that of the WT at the same time point (∗P < 0.01; ∗∗P < 0.001).
Discussion
Cold stress is one of the major environmental factors which restricts the growth, development, productivity, and distribution of crops worldwide. Cold tolerance is the major mechanism by which plants adapt to stress through the regulation of a wide range of cold-responsive genes (Thomashow, 1999). Numerous studies have indicated that TFs have significant roles in regulating various biological processes to protect plant cells against cold-induced damage (Yamaguchi-Shinozaki and Shinozaki, 2006; Chinnusamy et al., 2007; Shi et al., 2015; Zhu, 2016). Among them, bHLH TFs contain a conserved bHLH domain, which involved in DNA binding through sequence-specific interactions in the promoter regions of target genes (Carretero-Paulet et al., 2010). However, only a few bHLH proteins have been characterized involving in abiotic stress in plants, especially in tobacco. Here, NtbHLH123 was separated from tobacco based on its differential expression in response to stresses treatment (unpublished data). NtbHLH123 was found to have a highly conserved bHLH domain of bHLH proteins, suggesting that it is a putative tobacco bHLH protein (Supplementary Figure S1). In addition, NtbHLH123 expression was induced by cold stress (Figure 1), suggesting that NtbHLH123 may have a regulatory role during cold stress.
Studies have shown that the overexpression of ICE1, which results in insensitivity to chilling and freezing stresses, positively regulates CBF gene expression under cold stress (Chinnusamy et al., 2003; Budhagatapalli et al., 2016). In our work, the stress cold tolerance assay showed that the transgenic plants exhibited enhanced tolerance to cold stress compared with the WT, by measuring survival rate, EL (%), MDA, and chlorophyll contents, in accordance with phenotypic observation, suggesting that overexpression of NtbHLH123 conferred cold stress. However, it is worth mentioning that the transcript level of NtbHLH123 is induced by cold stress in both WT and transgenic plants. The possible explanation for this is that the NtbHLH123 transgenic lines exhibited high levels than in the WT under normal condition, so the overexpression plants could ensure a rapid response by plants to resist the cold stress under non-cold-acclimated conditions. While the resistance of WT is gradually enhanced, it was cold damage in the early stages of responses to cold stress. In addition, the transcript levels of NtbHLH123 were induced in WT, while was lower than the transgenic plants upon exposure to cold stress (Supplementary Figure S3), it may be because of CaMV 35S promoter can be induced by some stresses (Czechowski et al., 2005; Wang et al., 2010; Jin et al., 2016). In addition, CBF genes are induced by cold, and by overexpression enhance cold tolerance in transgenic Arabidopsis, rice, tobacco, rapeseed, tomato, and apple (Gilmour et al., 2000; Hsieh et al., 2002; Ito et al., 2006; Yang et al., 2011). The ICE1-CBF-COR transcriptional cascade is the best-understood cold acclimation signaling pathway (Chinnusamy et al., 2007). In this pathway, CBFs/DREBs are activated by cold and directly regulate the expression of cold-responsive genes by binding to their promoter regions (Thomashow, 1999, 2010; Chinnusamy et al., 2007). Recent evidence has indicated that CBF-dependent pathways are mediated by many protein kinases or TFs at transcriptional, posttranslational, and posttranscriptional levels (Shi et al., 2015). In Arabidopsis, AtICE1 and AtICE2, respectively, bind the E-box (or G-box) motifs in the AtCBF3 and AtCBF1 promoters to trigger expression (Chinnusamy et al., 2003; Lee et al., 2005; Fursova et al., 2009). Here, NtbHLH123 overexpression resulted in the elevated expression of NtCBF genes (Figure 4A), suggesting that NtbHLH123 may act as a signal transduction component in a potential CBF-dependent pathway and is associated with cold tolerance in tobacco. ChIP-PCR and Y1H assays indicated that NtbHLH123 protein could bind to the E-box motifs of NtCBFs promoters, further determining that it acts as transcriptional activator (Figures 4, 5). However, in tobacco, NtbHLH123 only bound to the promoters of the NtCBF1, NtCBF7, and NtCBF8 genes (Figure 4 and Supplementary Figure S5). In conformity to the specific binding, the increases in expression were observed for NtCBF1, NtCBF7, and NtCBF8, although NtCBF2, NtCBF3, NtCBF4, NtCBF5, and NtCBF6 expression was also enhanced (Figure 4A). This may be explained either by indirect regulation of the expression of NtCBF genes by NtbHLH123 or by regulation of the expression of other NtCBF genes by NtCBF1, NtCBF7, and NtCBF8, as is the case for AtCBFs in Arabidopsis (Novillo et al., 2004).
Several reports have shown that ROS (e.g., O2- or H2O2) production is triggered following exposure to abiotic stresses (Suzuki and Mittler, 2006; Suzuki et al., 2012). When plants are exposed to cold stress, high ROS levels result in oxidative stress, leading to lipid peroxidation and membrane damage in plants (Mittler, 2002; Choudhury et al., 2017). Therefore, plants have evolved complex mechanisms to scavenge the overproduction of ROS and adapt to low-temperature-induced damage. MDA and EL are related to the membrane system (Huang et al., 2013). In this study, higher MDA and EL levels in the WT plants implied that they might have been subjected to more oxidative stress than the 35S::NtbHLH123 transgenic plants (Figures 3C,D). Furthermore, the WT plants exhibited more intense histochemical staining compared with the transgenic plants after cold stress (Figure 6A), suggestive of less ROS accumulation in the transgenic plants than the WT plants. However, ROS accumulation relies greatly on ROS-scavenging systems under different abiotic stress conditions (Mittler et al., 2004; Choudhury et al., 2017). To reduce the influence of oxidative stress, antioxidant enzymes (e.g., SOD, CAT, and POD) have essential roles in maintaining ROS homeostasis in plants (Mittler, 2002, 2017; Suzuki and Mittler, 2006). The activities of SOD, POD, and CAT in the transgenic lines were slightly higher in the transgenic plants than in the WT under normal conditions (Figures 6B–D), which this may be due to the fact that remaining a basal level of ROS in cells for life and oxidative stress was not serious in WT and transgenic plants (Mittler, 2017). However, SOD, CAT, and POD activities were significantly higher in the transgenic lines than in the WT after cold stress (Figures 6B–D). In line with the antioxidant enzyme activities, ROS-scavenging-related genes (e.g., NtSOD, NtCAT, and NtAPX) were expressed at higher levels in the NtbHLH123-overexpressing plants than in the WT plants before and after cold stress (Figure 7A). The expression levels of these genes were consistent with the higher activities of antioxidant enzymes described above. These results suggest that overexpressing NtbHLH123 could increase cold tolerance, partially through a better ROS-scavenging system.
Furthermore, the mRNA levels of stress-responsive genes (i.e., NtLEA5, NtERD10C, and NtERD10D) were analyzed using qRT-PCR before and after cold treatment. In previous studies, most of these genes or their homologs have been shown to respond to abiotic stresses (Hundertmark and Hincha, 2008; Jin et al., 2016). Stronger induction of these genes was detected in the transgenic plants than in the WT plants, suggestive of less damage to transgenic plants, which was supported by the lower amount of membrane damage under cold stress (Figure 7B). In the future, additional research is needed to determine whether NtbHLH123 directly regulates stress-responsive genes to enhance cold tolerance.
Conclusion
We characterized a TF, NtbHLH123, which functions as a positive regulator to activate stress-responsive gene expression, conferring cold tolerance in a CBF-dependent manner and resulting in increased cold tolerance in tobacco (Figure 8). These results provide new information that helps to clarify the complex CBF-dependent pathway of transcriptional control upon cold stress in plants. Moreover, NtbHLH123 may be a valuable gene candidate for cold-tolerance trait improvement.
FIGURE 8.
Model of the regulatory network of NtbHLH123 involved in the cold stress response. Cold stress activates the expression of NtbHLH123. Cold-activated NtbHLH123 subsequently binds to E-box (or G-box) motifs to regulate the expression of its target genes (e.g., NtCBFs) or regulates reactive oxidative species (ROS) scavenging-related and stress-responsive genes, leading to improved cold stress tolerance.
Author Contributions
QZ and YW conceived and designed the research. QZ performed most of the experiments. QZ, XX, DL, AY, and YW performed the research. QZ and YW analyzed the data and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer SS and handling Editor declared their shared affiliation.
Funding. This work was supported by Natural Science Foundation of Shandong Province (ZR2016CB07), China Postdoctoral Science Foundation (2016M602214), Special Support for Post-doc Creative Funding in Shandong Province (201702056), the Agricultural Science and Technology Innovation Program (ASTIP-TRIC01), and Science Foundation for Young Scholars of Tobacco Research Institute of CAAS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00381/full#supplementary-material
References
- Agarwal M., Hao Y., Kapoor A., Dong C. H., Fujii H., Zheng X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281 37636–37645. 10.1074/jbc.M605895200 [DOI] [PubMed] [Google Scholar]
- Budhagatapalli N., Narasimhan R., Rajaraman J., Viswanathan C., Nataraja K. N. (2016). Ectopic expression of AtICE1. J. Plant Biochem. Biotehnol. 25 285–293. 10.1007/s13562-015-0340-8 [DOI] [Google Scholar]
- Carretero-Paulet L., Galstyan A., Roig-Villanova I., Martínez-García J. F., Bilbao-Castro J. R., Robertson D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153 1398–1412. 10.1104/pp.110.153593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B. H., Hong X., Agarwal M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17 1043–1054. 10.1101/gad.1077503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12 444–451. 10.1016/j.tplants.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Choudhury F. K., Rivero R. M., Blumwald E., Mittler R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90 856–867. 10.1111/tpj.13299 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E. L., Grotewold E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66 94–116. 10.1111/j.1365-313X.2010.04459.x [DOI] [PubMed] [Google Scholar]
- Feng X. M., Zhao Q., Zhao L. L., Qiao Y., Xie X. B., Li H. F., et al. (2012). The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol. 12:22. 10.1186/1471-2229-12-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Noutoshi Y., Takahashi F., Narusaka Y., Yamaguchi-Shinozaki K., et al. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9 436–442. 10.1016/j.pbi.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Fursova O. V., Pogorelko G. V., Tarasov V. A. (2009). Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 429 98–103. 10.1016/j.gene.2008.10.016 [DOI] [PubMed] [Google Scholar]
- Garrell J., Campuzano S. (1991). The helix-loop-helix domain: a common motif for bristles, muscles and sex. Bioessays 13 493–498. 10.1002/bies.950131002 [DOI] [PubMed] [Google Scholar]
- Gilmour S. J., Sebolt A. M., Salazar M. P., Everard J. D., Thomashow M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124 1854–1865. 10.1104/pp.124.4.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim M. A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P. C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20 735–747. 10.1093/molbev/msg088 [DOI] [PubMed] [Google Scholar]
- Hsieh T., Lee J., Charng Y., Chan M. (2002). Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol. 130 618–626. 10.1104/pp.006783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. S., Wang W., Zhang Q., Liu J. H. (2013). A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol. 162 1178–1194. 10.1104/pp.112.210740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M., Hincha D. K. (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118. 10.1186/1471-2164-9-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Katsura K., Maruyama K., Taji T., Kobayashi M., Seki M., et al. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47 141–153. 10.1093/pcp/pci230 [DOI] [PubMed] [Google Scholar]
- Ji X., Nie X., Liu Y., Zheng L., Zhao H., Zhang B., et al. (2016). A bHLH gene from Tamarix hispida improves abiotic stress tolerance by enhancing osmotic potential and decreasing reactive oxygen species accumulation. Tree Physiol. 36 193–207. 10.1093/treephys/tpv139 [DOI] [PubMed] [Google Scholar]
- Jin C., Huang X. S., Li K. Q., Yin H., Li L. T., Yao Z. H., et al. (2016). Overexpression of a bHLH1 transcription factor of Pyrus ussuriensis confers enhanced Cold tolerance and increases expression of stress-responsive genes. Front. Plant Sci. 7:441. 10.3389/fpls.2016.00441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Zhang H., Zhang J., Liu P., Chen X., Li Z., et al. (2017). Integrated transcriptomics and metabolomics analysis to characterize cold stress responses in Nicotiana tabacum. BMC Genomics 18:496. 10.1186/s12864-017-3871-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent V., Vervoort M. (2001). The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 11 754–770. 10.1101/gr.177001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H., Henderson D. A., Zhu J. K. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17 3155–3175. 10.1105/tpc.105.035568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E. (2014). PIFs: systems integrators in plant development. Plant Cell 26 56–78. 10.1105/tpc.113.120857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Guo S., Zhao Y., Chen D., Chong K., Xu Y. (2010). Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from Dongxiang Wild Rice confers freezing and salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 29 977–986. 10.1007/s00299-010-0883-z [DOI] [PubMed] [Google Scholar]
- Liu Y., Ji X., Nie X., Qu M., Zheng L., Tan Z., et al. (2015). Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 207 692–709. 10.1111/nph.13387 [DOI] [PubMed] [Google Scholar]
- Mao K., Dong Q., Li C., Liu C., Ma F. (2017). Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant Sci. 8:480. 10.3389/fpls.2017.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7 405–410. 10.1016/S1360-1385(02)02312-9 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2017). ROS are good. Trends Plant Sci. 22 11–19. 10.1016/j.tplants.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9 490–498. 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Novillo F., Alonso J. M., Ecker J. R., Salinas J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1and CBF3/DREB1expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101 3985–3990. 10.1073/pnas.0303029101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce R. S. (1999). Molecular analysis of acclimation to cold. Plant Growth Regul. 29 47–76. 10.1023/A:1006291330661 [DOI] [Google Scholar]
- Pedmale U. V., Huang S. S. C., Zander M., Cole B. J., Hetzel J., Ljung K., et al. (2016). Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164 233–245. 10.1016/j.cell.2015.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S., Sahu P. P., Srivastava P. S., Prasad M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17 369–381. 10.1016/j.tplants.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Sanghera G. S., Wani S. H., Hussain W., Singh N. B. (2011). Engineering cold stress tolerance in crop plants. Curr. Genomics 12 30–43. 10.2174/138920211794520178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J. S., Joo J., Kim M. J., Kim Y. K., Nahm B. H., Song S. I., et al. (2011). OsbHLH48, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65 907–921. 10.1111/j.1365-313X.2010.04477.x [DOI] [PubMed] [Google Scholar]
- Shi Y., Ding Y., Yang S. (2015). Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 56 7–15. 10.1093/pcp/pcu115 [DOI] [PubMed] [Google Scholar]
- Shi Y., Tian S., Hou L., Huang X., Zhang X., Guo H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24 2578–2595. 10.1105/tpc.112.098640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant. Biol. 3 217–223. 10.1016/S1369-5266(00)00067-4 [DOI] [PubMed] [Google Scholar]
- Song Y. H., Shim J. S., Kinmonth-Schultz H. A., Imaizumi T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66 441–464. 10.1146/annurev-arplant-043014-115555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Koussevitzky S., Mittler R. O. N., Miller G. A. D. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35 259–270. 10.1111/j.1365-3040.2011.02336.x [DOI] [PubMed] [Google Scholar]
- Suzuki N., Mittler R. (2006). Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol. Plant. 126 45–51. 10.1111/j.0031-9317.2005.00582.x [DOI] [Google Scholar]
- Thomashow M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 50 571–599. 10.1146/annurev.arplant.50.1.571 [DOI] [PubMed] [Google Scholar]
- Thomashow M. F. (2010). Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154 571–577. 10.1104/pp.110.161794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P. H. (2003). The Arabidopsis basic/helixloop-helix transcription factor family. Plant Cell 15 1749–1770. 10.1105/tpc.013839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K. U. (2015). Stomatal differentiation: the beginning and the end. Curr. Opin. Plant. Biol. 28 16–22. 10.1016/j.pbi.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Vogel J. T., Zarka D. G., Van Buskirk H. A., Fowler S. G., Thomashow M. F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41 195–211. 10.1111/j.1365-313X.2004.02288.x [DOI] [PubMed] [Google Scholar]
- Wang L., Xie W. B., Chen Y., Tang W. J., Yang J. Y., Ye R. J., et al. (2010). A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 61 752–766. 10.1111/j.1365-313X.2009.04100.x [DOI] [PubMed] [Google Scholar]
- Wang Y. J., Zhang Z. G., He X. J., Zhou H. L., Wen Y. X., Dai J. X., et al. (2003). A rice transcription factor OsbHLH1 is involved in cold stress response. Theor. Appl. Genet. 107 1402–1409. 10.1007/s00122-003-1378-x [DOI] [PubMed] [Google Scholar]
- Xiong L., Schumaker K. S., Zhu J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 S165–S183. 10.1105/tpc.000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Dubos C., Lepiniec L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20 176–185. 10.1016/j.tplants.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803. 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- Yang W., Liu X. D., Chi X. J., Wu C. A., Li Y. Z., Song L. L., et al. (2011). Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 233 219–229. 10.1007/s00425-010-1279-6 [DOI] [PubMed] [Google Scholar]
- Zhang J., Liu B., Li M., Feng D., Jin H., Wang P., et al. (2015). The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27 787–805. 10.1105/tpc.114.132704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Y., Bai M. Y., Wu J., Zhu J. Y., Wang H., Zhang Z. G., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21 3767–3780. 10.1105/tpc.109.070441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Song A., Li P., Chen S., Jiang J., Chen F. (2014). A bHLH transcription factor regulates iron intake under Fe deficiency in chrysanthemum. Sci. Rep. 4:6694 10.1038/srep06694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Ren Y. R., Wang Q. J., Wang X. F., You C. X., Hao Y. J. (2016a). Ubiquitination-related MdBT scaffold Proteins Target a bHLH transcription factor for Iron Homeostasis. Plant Physiol. 172 1973–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Ren Y. R., Wang Q. J., Yao Y. X., You C. X., Hao Y. J. (2016b). Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple. Plant Biotechnol. J. 14 1633–1645. 10.1111/pbi.12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li F., Wang J. L., Ma Y., Chong K., Xu Y. Y. (2009). Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt-and osmotic stress in Arabidopsis. J. Plant Physiol. 166 1296–1306. 10.1016/j.jplph.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Zhu J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167 313–324. 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.