Abstract
Tissue specific biosynthesis of secondary metabolites is a distinguished feature of medicinal plants. Withania somnifera, source of pharmaceutically important withanolides biosynthesizes withaferin-A in leaves and withanolide-A in roots. To increase the in planta withanolides production, a sustainable approach needs to be explored. Here, we isolated endophytes from different parts of W. somnifera plants and their promising role in in planta withanolide biosynthesis was established in both in-vivo grown as well in in-vitro raised composite W. somnifera plants. Overall, the fungal endophytes improved photosynthesis, plant growth and biomass, and the root-associated bacterial endophytes enhanced the withanolide content in both in-vivo and in-vitro grown plants by modulating the expression of withanolide biosynthesis genes in leaves and roots. Surprisingly, a few indole-3-acetic acid (IAA)-producing and nitrogen-fixing root-associated endophytes could induce the biosynthesis of withaferin-A in roots by inducing in planta IAA-production and upregulating the expression of withanolide biosynthesis genes especially MEP-pathway genes (DXS and DXR) in roots as well. Results indicate the role of endophytes in modulating the synthesis and site of withanolides production and the selected endophytes can be used for enhancing the in planta withanolide production and enriching roots with pharmaceutically important withaferin-A which is generally absent in roots.
Introduction
Withania somnifera well-known as Indian ginseng or Ashwagandha, is an important medicinal plant widely distributed around the globe and used in traditional medicine systems like Ayurveda, Siddha, Unani and Chinese. All parts of this plant like roots, stem, bark, leaves, flowers and seeds have medicinal importance1,2. Medicinal properties of W. somnifera include anti-hyperglycemic, neuropharmacological, immunomodulatory, cardioprotective, musculotropic, hepatoprotective, radiosensitizing, chemoprotective, anti-aging, macrophage-activating, diuretic, hypocholesterolemic, aphrodisiac, rejuvenating and hemopoietic3–7. Withania leaves are used in the treatment of fever, tumours and ulcers8. Medicinal importance of different parts of W. somnifera is due to the presence of pharmaceutically active steroidal lactones withanolides including withanolide-A and withaferin-A as major bioactive molecules. Withaferin-A has anti-leukemic, anti-invasive, anti-metastatic, apoptotic, anti-inflammatory, radiosensitizing and antidiabetic activity and also act as a potential leptin-sensitizer9–12. Withanolide-A is a potential neurological, immunological and anti-stress agent13–15. Roots of W. somnifera plants are rich in withanolide-A however withaferin-A is present in leaves in large amount and totally absent or present in traces in roots of W. somnifera plant16–20. Withanolides are terpenoids and synthesised in plants using the precursor isoprenoids which are synthesized via mevalonate (MVA) and 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway. Genes encoding key enzymes of withanolide biosynthesis have been characterized21,22 and consistent efforts are being made to increase the production of withanolides in roots as well as leaves. Biotechnological tools like genetic engineering are being focused for enhancing the withanolide production. Overexpression of squalene synthase (SQS), a key regulatory gene of withanolide biosynthesis in W. somnifera could increase the content of withaferin A and withanolide A in the leaves up to 4–4.5 fold23. Overexpression of cycloartenol synthase (CAS) in W. somnifera increased the withanolide content to the extent of 1.06 to 1.66 fold24. Use of cell suspension25, adventitious26,27, and hairy root culture28 for improvement of withanolide production have also been tried. Overexpression of SQS in the Withania suspension cultures enhanced the withanolide A content up to 2.5 fold compared to non-transformed culture29. Enhanced production of withanolides could be achieved by the treatment of salicylic acid and methyl jasmonate in Withania hairy roots28. It has been observed that treatment with the extract of sea weeds on Withania hairy roots increased the production of withanolides by increasing the expression of key genes of withanolide biosynthesis such as squalene epoxidase (SQE), SQS, 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) and farnesyl pyrophosphate synthase (FPPS)30. Generation of transgenics by genetic manipulation of withanolide biosynthetic pathway and the cell culture approaches have limitations related to practical and economic feasibility and social acceptability, therefore, requires exploration of new sustainable approach for enhancement of withanolide production.
Endophytes are known to promote plant growth, protect plants to environmental stresses, and as the source of therapeutic compounds31–38. Endophytes are found to be associated with different parts of a plant and are involved in modulation of primary and secondary metabolism of the host plant39,40. Occasionally, endophytes are also found to be involved in improving secondary metabolite biosynthesis of the host plant39–43. Therefore, endophyte-mediated improvement of crop yield and secondary metabolite biosynthesis of medicinal plants could be a sustainable approach for producing the higher yield of therapeutically important chemicals. In the present study, efforts were made to identify the most potent endophytes from W. somnifera plants to decipher their roles in enhancing the plant growth and in parallel, the overall yield of therapeutically important withanolides, through their judicious screening on in-vivo grown plants as well as in in-vitro raised composite plants (i.e., plants with wild-type shoots and wild type Agrobacterium rhizogenes induced transgenic roots, as illustrated previously44). Since A. rhizogenes is reputed to stimulate robust root formation upon infection and roots of W. somnifera plants are considered as the major sites of its desired metabolite synthesis, the envisaged rationale of its use in raising the composite plants during the present course of study imparts the overall intention of expanding the host-endophyte association prospect for better comprehension of the plant-endophyte interaction. To the best of our knowledge, the use of composite plant for serving this specific goal remained uncharted so far. Instead, composite plants had earlier been successfully utilized as a suitable alternative for the functional characterization of any targeted gene(s) in the host plants to overcome their prevailing tedious and lengthy transformation and regeneration processes45. Then again, several other studies, such as nutrient/hormone uptake, interactions with root nodulating bacteria and mycorrhizal symbiotic association, have also documented the competence of this composite plant based technology46.
Results
Isolation and molecular characterization of endophytes
Endophytes were isolated from leaves, roots and seeds of W. somnifera plant. We performed 16S rRNA and ITS sequencing for identification of bacterial and fungal endophytes, respectively. A total of 40 isolates including 29 bacteria and 11 fungi were isolated and identified (Table 1); of these 25 endophytes (18 bacteria and 7 fungi) from roots, 12 endophytes (8 bacteria and 4 fungi) from leaves and 3 bacterial endophytes from seeds were isolated.
Table 1.
List of endophytes isolated from Withania somnifera cv. Poshita.
| Plant part | Strain | Culture name | Accession no. |
|---|---|---|---|
| Bacterial endophytes | |||
| Leaves | WPL1 | Bacillus amyloliquefaciens | KY000380 |
| WPL2 | Bacillus horneckiae | KY000381 | |
| WPL3 | Staphylococcus haemolyticus | KY000382 | |
| WPL4 | Bacillus sp. | KY000383 | |
| WPL5 | Micrococcus luteus | KY000384 | |
| WPL6 | Pseudomonas putida | KY000385 | |
| WPL7 | Bacillus firmus | KY000386 | |
| WPL8 | Bacillus licheniformis | KY000387 | |
| Root | WPR12 | Bacillus muralis | KY000391 |
| WPR13 | Brevibacterium frigoritolerans | KY000392 | |
| WPR14 | Bacillus pumilis | KY000393 | |
| WPR15 | Bacillus aryabhattai | KY000394 | |
| WPR16 | Bacillus megaterium | KY000395 | |
| WPR17 | Pseudomonas sp. | KY000396 | |
| WPR18 | Bacillus pseudomycoides | KY000397 | |
| WPR19 | Bacillus aquimaris | KY000398 | |
| WPR20 | Bacillus thuringiensis | KY000399 | |
| WPR21 | Bacillus indicus | KY000400 | |
| WPS23 | Streptomyces sp. | KY454621 | |
| WPR26 | Paenibacillus sp. | KY000401 | |
| WPR27 | Bacillus cereus | KY000402 | |
| WPR28 | Bacillus thuringiensis | KY000403 | |
| WPR29 | Pseudomonas sp. | KY000404 | |
| WPR30 | Rhizobium sullae | KY000405 | |
| WPR31 | Sinorhizobium fredii | KY000406 | |
| WPR32 | Pantoea sp. | KY000407 | |
| Seed | WPS9 | Bacillus subtilis | KY000388 |
| WPS10 | Bacillus cereus | KY000389 | |
| WPS11 | Bacillus tequilensis | KY000390 | |
| Fungal endophytes | |||
| Leaves | WPLF1 | Penicillium sp. | KX928761 |
| WPLF2 | Aspergillus terreus | KX928762 | |
| WPLF3 | Trametes versicolor | KX928763 | |
| WPLF4 | Sarocladium implicatum | KX928764 | |
| Root | WPRF5 | Penicillium oxalicum | KX928765 |
| WPRF6 | Colletotrichum capsici | KX928766 | |
| WPRF7 | Ceratobasidium sp. | KX928767 | |
| WPRF8 | Penicillium sp. | KX928768 | |
| WPRF9 | Aspergillus brasiliensis | KX928769 | |
| WPRF10 | Colletotrichum truncatum | KX928770 | |
| WPRF11 | Hypocrea lixii | KX928771 | |
Photosynthetic pigments, photosynthesis, stomatal conductance and transpiration rate
To study the individual effect of a particular endophyte on photosynthetic efficiency of W. somnifera plants, each endophyte was inoculated to the endophyte-free plants and their photosynthetic pigment content, net CO2 assimilation, transpiration rate and stomatal conductance were measured, and compared with the non-inoculated endophytes free control plants. Interestingly, inoculation with fungal endophytes, in general, improved the photosynthetic efficiency of W. somnifera plants. WPLF4, WPRF5, WPRF6, WPRF7, WPRF8, WPRF9, WPRF10, WPRF11 inoculated plants had higher chlorophyll (17–29%), carotenoid (29–46%), net CO2 assimilation (17–43%), transpiration rate (18–31%) and stomatal conductance (10–42%) than that of non-inoculated endophyte-free control plants (Table 2). WPLF2 and WPLF3 inoculated plants had 16.4% and 22.4% higher transpiration rate respectively than that of non-inoculated endophyte free control plants. WPLF3 inoculation could also increase net CO2 assimilation by 12.2% compared to endophyte free control plants. Inoculation with bacterial endophytes could not improve the measured photosynthesis parameters significantly (Supplementary Table S1).
Table 2.
Effect of inoculation with fungal endophytes on physiological parameters of Withania somnifera plants.
| Treatment | Chlorophyll (mg gFW−1) | Carotenoids (mg gFW−1) | A (μ mol m−2 s−1) | E (m mol m−2 s−1) | gS (m mol m−2 s−1) |
|---|---|---|---|---|---|
| Control | 0.550 ± 0.009d | 0.087 ± 0.006b | 23.50 ± 0.35e | 10.98 ± 0.51c | 498.00 ± 9.81 f |
| WPLF1 | 0.608 ± 0.039bc | 0.094 ± 0.004b | 25.50 ± 1.09de | 11.22 ± 0.60c | 510.00 ± 21.55ef |
| WPLF2 | 0.595 ± 0.017 cd | 0.084 ± 0.005b | 25.27 ± 0.78de | 12.77 ± 0.51 b | 514.00 ± 9.29ef |
| WPLF3 | 0.603 ± 0.010 bc | 0.091 ± 0.005b | 26.37 ± 0.33 d | 13.43 ± 0.44 ab | 489.67 ± 3.84 f |
| WPLF4 | 0.646 ± 0.011 ab | 0.111 ± 0.007 a | 27.93 ± 0.88 bc | 13.04 ± 0.45 ab | 629.00 ± 13.45 bc |
| WPRF5 | 0.663 ± 0.019 ab | 0.117 ± 0.007 a | 27.53 ± 0.44 cd | 14.02 ± 0.42 ab | 645.00 ± 9.87 b |
| WPRF6 | 0.701 ± 0.048 a | 0.116 ± 0.008 a | 30.83 ± 0.87 ab | 13.20 ± 0.17 ab | 663.67 ± 17.91 ab |
| WPRF7 | 0.712 ± 0.027 a | 0.122 ± 0.004 a | 30.20 ± 1.49 bc | 13.31 ± 0.32 ab | 592.00 ± 24.50 cd |
| WPRF8 | 0.692 ± 0.023 a | 0.123 ± 0.002 a | 33.60 ± 0.55 a | 14.35 ± 0.60 a | 707.00 ± 13.53 a |
| WPRF9 | 0.695 ± 0.011 a | 0.127 ± 0.002 a | 31.03 ± 1.86 ab | 13.02 ± 0.33 ab | 664.67 ± 9.70ab |
| WPRF10 | 0.678 ± 0.022 ab | 0.126 ± 0.005 a | 30.30 ± 1.16 bc | 12.97 ± 0.52 ab | 574.33 ± 2.85 d |
| WPRF11 | 0.703 ± 0.013 a | 0.125 ± 0.005 a | 28.10 ± 0.90 bc | 13.23 ± 0.43 ab | 548.00 ± 27.02 de |
A-Net CO2 assimilation, E-Transpiration rate, gS-Stomatal conductance. Values are the means of six biological replicates ± S.E. Values with different letters are significantly different at P ≤ 0.05 (Duncan’s multiple range test). Values significantly higher than that of control plants are highlighted in bold.
Plant biomass
Some of the isolated fungal endophytes were able to enhance the shoot and root biomass of W. somnifera (Fig. 1). It was observed that inoculation with isolated bacterial endophytes could not improve the shoot and root biomass of W. somnifera plants significantly (Supplementary Table S2). WPLF4, WPRF5, WPRF6, WPRF7, WPRF8, WPRF9, WPRF10 and WPRF11 inoculated plants had 32–189% higher shoot biomass than that of non-inoculated endophyte free control plants (Fig. 1). Inoculation with WPRF7, WPRF8, and WPRF9 could also enhance the root biomass by 62–130% (Fig. 1). The endophytes, WPRF7, WPRF8 and WPRF9 were found to be effective in enhancing both shoot and root biomass.
Figure 1.
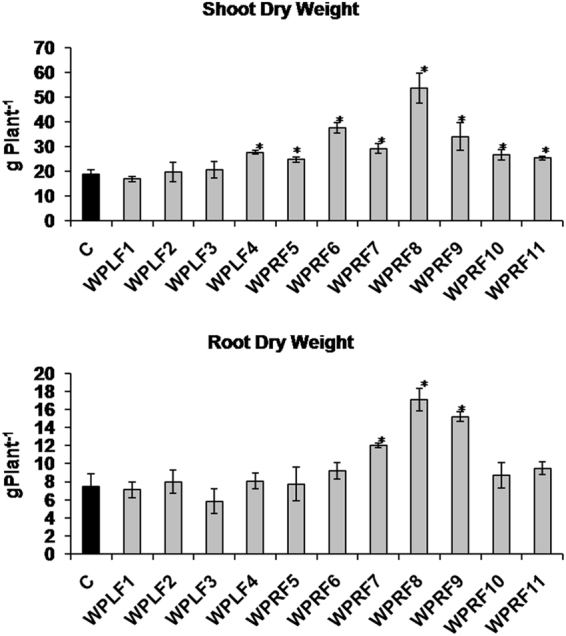
Effect of endophyte-inoculation on shoot and root biomass of Withania somnifera plant. One month old endophyte-free W. somnifera plants that originated from seeds treated with bactericides and fungicides were inoculated with endophytes and grown in pots filled with autoclaved soil and vermicompost mixture. Non-inoculated endophyte-free plants were used as a control. At 90 days stage shoot and root biomass was measured. Each data point is an average of six replicates and the error bars represent standard errors. Asterisks indicate significant differences between the control and endophyte inoculation (Duncan’s multiple range test P ≤ 0.05).
Plant growth promotion activity of selected endophytes
Plant growth promotion traits such as phosphate solubilization and indole acetic acid (IAA) production was tested in the selected bacterial and fungal endophytes able to promote the growth of the W. somnifera plant and enhance the content of withanolides. Fungal endophytes WPLF1, WPLF2, WPLF3, WPRF5, WPRF7, WPRF8, WPRF9 and WPRF10 were found positive for phosphate solubilization and WPLF3, WPRF6, WPRF7, WPRF9, WPRF10 and WPRF11were positive for IAA production (Supplementary Table S3). Among bacterial endophytes, WPR12, WPR16, WPR17, WPS23 and WPR32 were found positive for IAA production and nitrate reduction activity, and negative for phosphate solubilization (Supplementary Table S4).
Withanolides
Effect of endophyte inoculation on withanolide production was studied by measuring the content of withaferin A (WFA), 12-deoxy withstramonolide (DWL) and withanolide A (WLA) in leaves and roots of Withania plant. Endophytes (WPS11, WPR13, WPR16, WPR17, WPR19, WPR20, WPS23, WPR26, WPR27, WPR28, WPR30, WPR31, WPR32, WPLF1, WPLF2, WPLF4, WPRF5, WPRF6, WPRF7, WPRF9 and WPRF11) inoculated Withania plants had 20–232% higher WFA content in leaves compared to non-inoculated endophyte free control plants (Table 3). WPL3, WPS10, WPR12, WPR21 inoculated plants had 108–383% higher leaf DWL content. Endophyte inoculations, however, could not improve the WLA content in leaves of Withania plant significantly, interestingly, however, some endophyte-inoculations (WPL5, WPL6, WPL7, WPR12, WPR16, WPR17, WPR18, WPS23, WPR28, WPR32 and WPLF1) induced the biosynthesis of WFA in roots which could not be detected in non-inoculated endophyte free control plants (Table 3). On the other hand, DWL which could not be detected in roots of endophyte-free control Withania plants was detected in roots of WPL2, WPL3, WPL4, WPL5, WPS10, WPR12, WPR17 inoculated plants (Table 3). In roots, WLA content enhanced considerably by 75–298% with the inoculation of few endophyte (WPL1, WPL4, WPS10, WPR15, WPR18, WPR21, WPS23, WPR26, WPR29, WPR30, WPR31, WPR32, WPLF1, WPLF2 and WPRF10). None of the endophytes produced secondary metabolites of the host plant in culture medium i.e. independent of the host (data not shown).
Table 3.
Withanolide content in leaves and roots of endophyte-inoculated Withania plants.
| Treatment | Leaf | Root | ||||
|---|---|---|---|---|---|---|
| %WFA | %DWL | %WLA | %WFA | %DWL | %WLA | |
| Control | 0.56 ± 0.04hi | 0.12 ± 0.009ef | 0.35 ± 0.029ab | nd | nd | 0.067 ± 0.012gh |
| WPL1 | 0.39 ± 0.05 lm | 0.09 ± 0.009ef | 0.21 ± 0.035hi | nd | nd | 0.143 ± 0.015 cd |
| WPL2 | 0.32 ± 0.09mn | 0.03 ± 0.009ij | 0.04 ± 0.006p | nd | 0.012 ± 0.002 d | 0.043 ± 0.009hi |
| WPL3 | 0.50 ± 0.07ij | 0.43 ± 0.055 b | 0.09 ± 0.006op | nd | 0.013 ± 0.003 d | 0.070 ± 0.010gh |
| WPL4 | 0.52 ± 0.12ij | 0.09 ± 0.009ef | 0.35 ± 0.026ab | nd | 0.013 ± 0.003 d | 0.177 ± 0.009 bc |
| WPL5 | 0.58 ± 0.14gh | 0.13 ± 0.019e | 0.27 ± 0.067 cd | 0.013 ± 0.002 d | 0.012 ± 0.002 d | 0.067 ± 0.012gh |
| WPL6 | 0.56 ± 0.13hi | 0.01 ± 0.003j | 0.08 ± 0.007op | 0.012 ± 0.002 d | nd | 0.063 ± 0.009hi |
| WPL7 | 0.53 ± 0.05ij | 0.13 ± 0.009ef | 0.25 ± 0.043de | 0.015 ± 0.003 cd | nd | 0.073 ± 0.007gh |
| WPL8 | 0.58 ± 0.15gh | 0.07 ± 0.010ef | 0.40 ± 0.086a | nd | nd | 0.057 ± 0.012hi |
| WPS9 | 0.48 ± 0.15ij | 0.13 ± 0.015e | 0.20 ± 0.041hi | nd | nd | 0.053 ± 0.012hi |
| WPS10 | 0.59 ± 0.10gh | 0.58 ± 0.067 a | 0.44 ± 0.086a | nd | 0.022 ± 0.004 b | 0.170 ± 0.015 bc |
| WPS11 | 0.85 ± 0.03 cd | 0.13 ± 0.018e | 0.22 ± 0.018gh | nd | nd | 0.060 ± 0.006hi |
| WPR12 | 0.15 ± 0.04n | 0.25 ± 0.018 d | nd | 0.012 ± 0.002 d | 0.018 ± 0.002 c | 0.077 ± 0.015gh |
| WPR13 | 1.31 ± 0.17 b | 0.11 ± 0.015ef | Nd | nd | nd | 0.060 ± 0.010hi |
| WPR14 | 0.58 ± 0.12gh | 0.07 ± 0.015ef | 0.17 ± 0.009hi | nd | nd | 0.053 ± 0.009hi |
| WPR15 | 0.45 ± 0.09ij | 0.05 ± 0.012hi | 0.23 ± 0.012fg | nd | nd | 0.130 ± 0.015 de |
| WPR16 | 0.98 ± 0.10 bc | 0.08 ± 0.003ef | 0.18 ± 0.019hi | 0.043 ± 0.003 a | nd | 0.073 ± 0.009gh |
| WPR17 | 1.86 ± 0.15 a | 0.07 ± 0.015ef | 0.42 ± 0.075a | 0.012 ± 0.002 d | 0.038 ± 0.002a | 0.070 ± 0.012gh |
| WPR18 | 0.44 ± 0.17jl | 0.11 ± 0.015ef | 0.26 ± 0.045 cd | 0.015 ± 0.003 cd | nd | 0.183 ± 0.026 bc |
| WPR19 | 1.00 ± 0.08 bc | 0.09 ± 0.012ef | 0.24 ± 0.007ef | nd | nd | 0.060 ± 0.012hi |
| WPR20 | 1.05 ± 0.06 bc | 0.06 ± 0.018ef | 0.26 ± 0.009 cd | nd | nd | 0.067 ± 0.009gh |
| WPR21 | 0.56 ± 0.10hi | 0.32 ± 0.078 c | 0.16 ± 0.020ij | nd | nd | 0.117 ± 0.013 ef |
| WPS23 | 0.79 ± 0.06 de | 0.06 ± 0.010fg | 0.14 ± 0.012 lm | 0.018 ± 0.002 bc | nd | 0.267 ± 0.018 a |
| WPR26 | 0.67 ± 0.14 fg | 0.08 ± 0.006ef | 0.33 ± 0.026ab | nd | nd | 0.157 ± 0.024 cd |
| WPR27 | 0.75 ± 0.06 ef | 0.09 ± 0.006ef | 0.11 ± 0.012mn | nd | nd | 0.043 ± 0.012hi |
| WPR28 | 1.11 ± 0.09 bc | 0.09 ± 0.012ef | 0.23 ± 0.021fg | 0.022 ± 0.004 b | nd | 0.060 ± 0.015hi |
| WPR29 | 0.41 ± 0.13 lm | 0.08 ± 0.010ef | 0.39 ± 0.032ab | nd | nd | 0.217 ± 0.027 b |
| WPR30 | 1.05 ± 0.08bc | 0.10 ± 0.006ef | 0.15 ± 0.015jl | nd | nd | 0.160 ± 0.015 cd |
| WPR31 | 0.94 ± 0.06bc | 0.09 ± 0.006ef | 0.19 ± 0.010hi | nd | nd | 0.160 ± 0.015 cd |
| WPR32 | 1.17 ± 0.13bc | 0.07 ± 0.009ef | 0.28 ± 0.015bc | 0.010 ± 0.001d | nd | 0.117 ± 0.022ef |
| WPLF1 | 0.73 ± 0.11 ef | 0.06 ± 0.015gh | 0.27 ± 0.030 cd | 0.012 ± 0.002d | nd | 0.130 ± 0.015de |
| WPLF2 | 0.71 ± 0.11fg | 0.06 ± 0.009fg | 0.28 ± 0.021bc | nd | nd | 0.127 ± 0.023de |
| WPLF3 | 0.62 ± 0.13gh | 0.08 ± 0.006ef | 0.37 ± 0.029ab | nd | nd | 0.073 ± 0.019gh |
| WPLF4 | 1.20 ± 0.10bc | 0.12 ± 0.012ef | 0.22 ± 0.023gh | nd | nd | 0.027 ± 0.009ij |
| WPRF5 | 0.85 ± 0.18 cd | 0.12 ± 0.017ef | 0.18 ± 0.015hi | nd | nd | 0.060 ± 0.006hi |
| WPRF6 | 1.03 ± 0.13bc | 0.10 ± 0.009ef | 0.34 ± 0.035ab | nd | nd | 0.057 ± 0.018hi |
| WPRF7 | 0.68 ± 0.10fg | 0.09 ± 0.003ef | 0.27 ± 0.061 cd | nd | nd | 0.030 ± 0.012ij |
| WPRF8 | 0.43 ± 0.12 lm | 0.03 ± 0.009ij | 0.37 ± 0.045ab | nd | nd | 0.050 ± 0.006hi |
| WPRF9 | 0.68 ± 0.11fg | 0.09 ± 0.009ef | 0.11 ± 0.026no | nd | nd | 0.043 ± 0.018hi |
| WPRF10 | 0.46 ± 0.16ij | 0.06 ± 0.017fg | 0.22 ± 0.028gh | nd | nd | 0.177 ± 0.037bc |
| WPRF11 | 0.69 ± 0.12fg | 0.11 ± 0.010ef | 0.25 ± 0.022ef | nd | nd | 0.047 ± 0.018hi |
WFA- withaferin A, DWL- 12-deoxy withstramonolide and WLA- withanolide A. Values are the means of six biological replicates ± S.E. Values with different letters are significantly different at P ≤ 0.05 (Duncan’s multiple range test).Values significantly higher than that of control plants are highlighted in bold. nd- not detected.
Quantitative Real-time PCR (qRT-PCR) analysis of withanolide biosynthetic genes
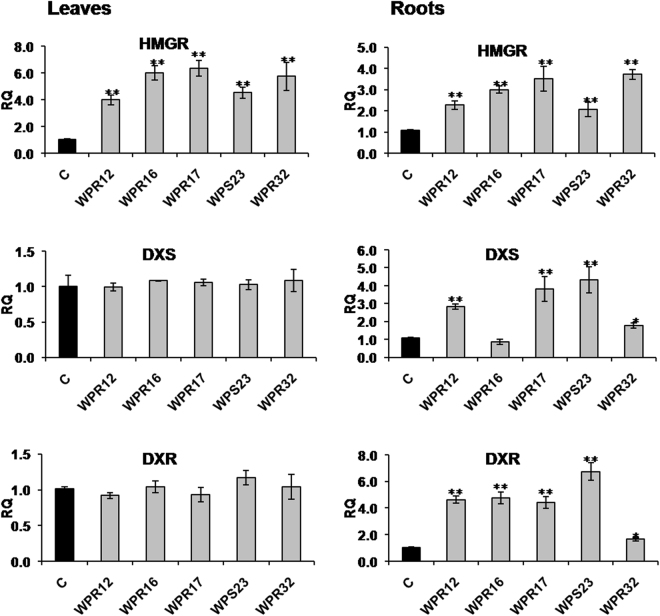
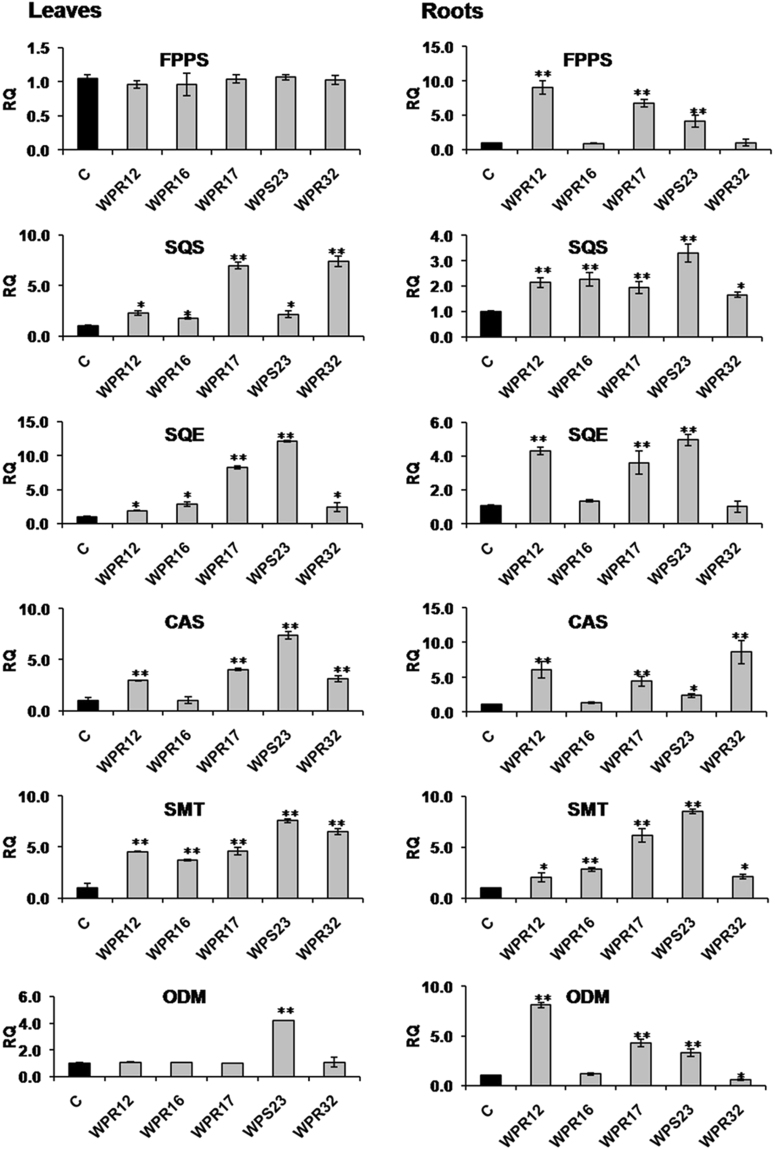
To understand the mechanism of the increased withanolide production in leaves and roots of Withania plants because of endophyte inoculation, expression of genes involved in withanolide biosynthesis was studied using qRT-PCR. We selected WPR12, WPR16, WPR17, WPS23, and WPR32 inoculated plants to understand the endophyte-mediated modulation of withanolide biosynthesis because these endophytes could substantially enhance biosynthesis of more than one withanolide simultaneously. A total of 12 genes transcripts were quantified in leaves and root tissues. Expression of MVA-pathway gene HMGR was found to be upregulated in leaves and roots of the selected endophyte-inoculated plants (Fig. 2). Expression of MEP-pathway gene DXS and DXR were not affected in leaves of endophyte-inoculated plants (Fig. 2). However, WPR12, WPR17, WPS23 and WPR32 inoculation upregulated the expression of DXS in roots. The expression of DXR in roots of all the selected endophyte-inoculated plants was upregulated (Fig. 2). In leaves, the expression of FPPS remained unaffected in endophyte-inoculated plants (Fig. 3). However, WPR12, WPR17, and WPS23 inoculated plants showed upregulated FPPS expression. WPR12, WPR16, WPR17, WPS23 and WPR32 inoculated plants had increased expression of SQS in both leaves and root (Fig. 3). All the selected endophyte upregulated the expression of SQE in leaves of Withania plant (Fig. 3). However, its expression in roots could be upregulated in plants inoculated with WPR12, WPR17 and WPS23. CAS expression was upregulated in leaves and roots of WPR12, WPR17, WPS23 and WPR32 inoculated plants whereas SMT expression was upregulated in leaves and roots of all the selected endophyte-inoculated plants (Fig. 3). Expression of ODM was upregulated in leaves of WPS23 inoculated plants, however, its expression in roots was observed to be higher in WPR12, WPR17 and WPS23 inoculated plants compared to non-inoculated endophyte free control plants (Fig. 3).
Figure 2.
Effect of endophyte inoculation on the expression of genes involved in Mevalonate (MVA) and 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway. Expression of MVA pathway gene (3-hydroxy-3-methylglutaryl-coenzyme A reductase; HMGR), and MEP pathway genes (1-Deoxy-D-xylulose-5-phosphate synthase; DXS and 1-deoxy-D-xylulose-5-phosphate reductase; DXR) was analyzed. Results were normalized to actin (reference transcript) and are shown relative to the level in non-inoculated endophyte-free control plants (calibrator). Data are means ± SD (n = 3 biological replicates) and Y-axis represents relative quantity (RQ). Asterisks indicate significant differences between the control and endophyte inoculation (Duncan’s multiple range test *P ≤ 0.05, **P ≤ 0.01).
Figure 3.
Effect of endophyte inoculation on the expression of genes involved in withanolide biosynthesis. Expression of FPPS, SQS, SQE, CAS, SMT and ODM was analyzed. Results were normalized to actin (reference transcript) and are shown relative to the level in non-inoculated endophyte-free control plants (calibrator). Data are means ± SD (n = 3 biological replicates) and Y-axis represents relative quantity (RQ). Asterisks indicate significant differences between the control and endophyte inoculation (Duncan’s multiple range test *P ≤ 0.05, **P ≤ 0.01).
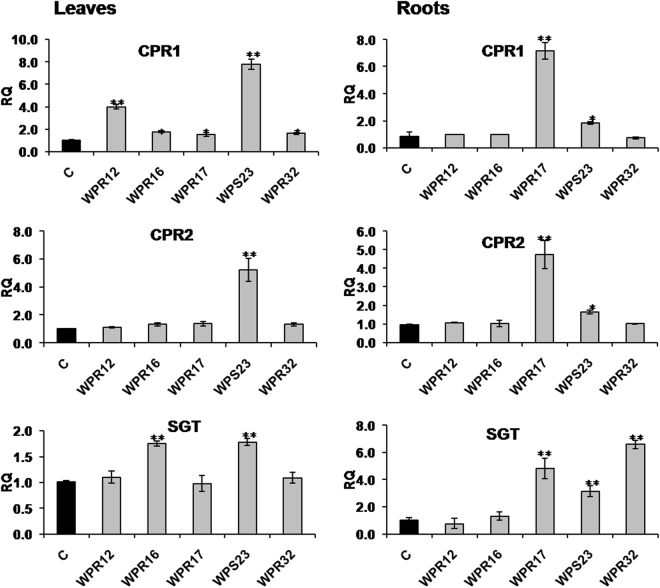
Expression of CPR1 was found to be upregulated in leaves of all the selected endophyte-inoculated plants (Fig. 4). Leaves of WPS23-inoculated plants showed increased expression of CPR2 than that of endophyte free control plants (Fig. 4). Expression of CPR1 and CPR2 was significantly increased in roots of WPR17, and WPS23 inoculated plants. Expression of SGT was upregulated in WPR16, and WPS23 inoculated plant leaves (Fig. 4). WPR17, WPS23 and WPR32 inoculation significantly increased the expression of SGT in Withania roots. Differential modulation of expression of different genes of withanolide biosynthesis by inoculated endophytes is represented in Fig. 5.
Figure 4.
Effect of endophyte inoculation on the expression of genes encoding cytochrome P450 reductases and sterol glucosyltransferases. Expression of cytochrome P450 reductase (CPR1 and CPR2) and sterol glucosyltransferases (SGT) was analyzed. Results were normalized to actin (reference transcript) and are shown relative to the level in non-inoculated endophyte-free control plants (calibrator). Data are means ± SD (n = 3 biological replicates) and Y-axis represents relative quantity (RQ). Asterisks indicate significant differences between the control and endophyte inoculation (Duncan’s multiple range test *P ≤ 0.05, **P ≤ 0.01).
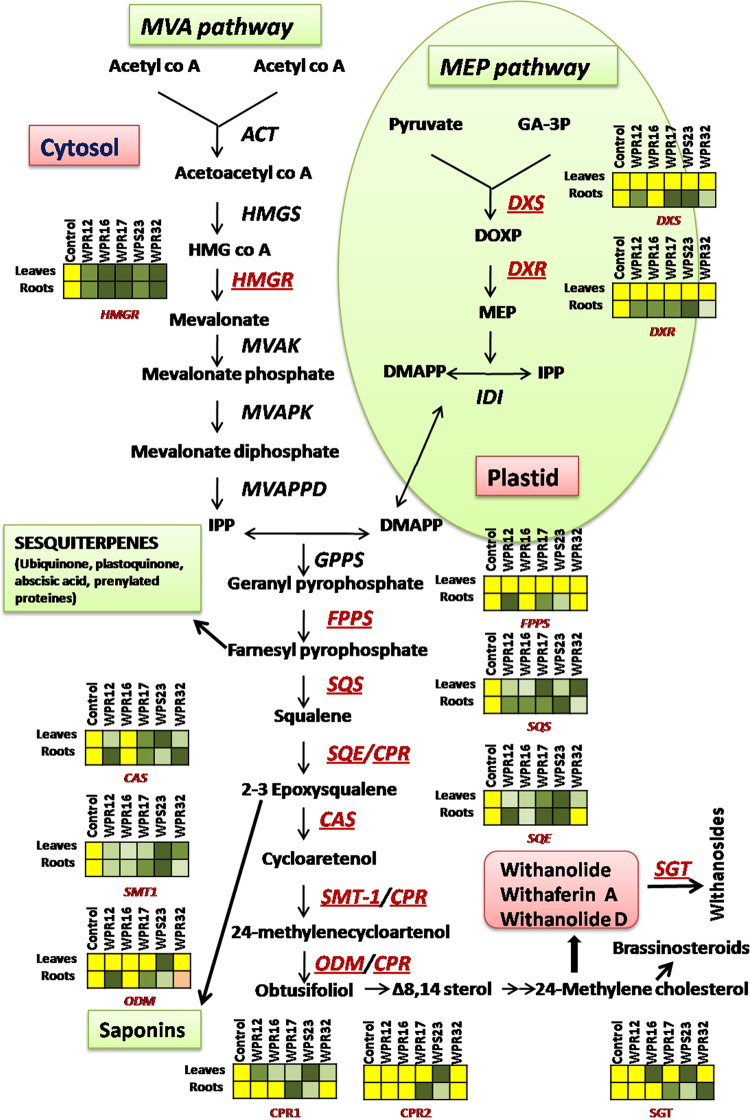
Figure 5.
Differential modulation of withanolide biosynthesis by different endophytes. Endophytes isolated from Withania somnifera plants modulated the expression of most of the genes of withanolide biosynthetic pathway. Expression of HMGR in both leaves and roots was increased by all the selected endophytes. Most of the selected endophyte-inoculation upregulated the expression of SQS, SQE, CAS and SMT that could enhance the production of withanolide in leaves and root of W. somnifera plants. WPR17 and WPS23 could upregulate all the key genes of withanolide biosynthetic pathway in roots of W. somnifera plant. Expression of different genes was presented in square boxes. Green colour of boxes indicate upregulated expression (intensity of green colour shows the level of expression i.e. more green more expression and vice versa), yellow colour shows the level of expression in non-inoculated endophyte free control plants and orange colour shows downregulated expression. Enzyme abbreviations: HMGR, 3-hydroxy-3-methylglutaryl-coenzyme A reductase; DXS, 1-Deoxy-D-xylulose-5-phosphate synthase; DXR, 1-deoxy-D-xylulose-5-phosphate reductase; FPPS, farnesyl diphosphate synthase; SQS, squalene synthase; SQE, squalene epoxidase; CAS, cycloartenol synthase; CPRs (1,2), cytochrome P450 reductase; SMT, sterol methyl transferase; ODM, obtusifoliol-14 –demethylase.
Effect of endophyte-inoculation on in planta IAA content
Selected endophytes (WPR12, WPR16, WPR17, WPS23 and WPR32) having the potential to enhance in planta withanolide production were found to be positive for IAA production (Supplementary Table S4). Therefore, to study the effect of these endophyte-inoculations on IAA level of plants, in planta IAA content in roots as well as leaves was quantified. Endophyte inoculation affected the IAA content significantly in both leaves and root tissue (Fig. 6). IAA content was found to be higher in roots than in leaves, and endophyte inoculated plants always had higher IAA content in both leaves and roots than that of non-inoculated endophyte free control plants (Fig. 6).
Figure 6.
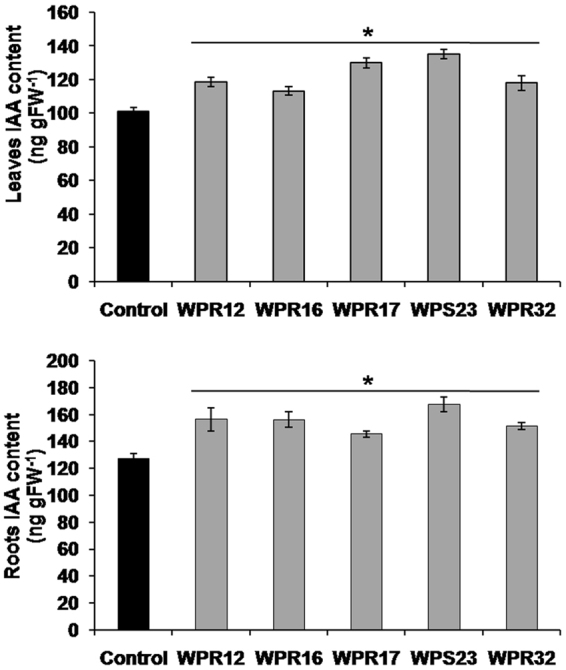
Effect of endophyte-inoculation on indole-3-acetic acid content of leaves and roots of Withania plants. Indole-3-acetic acid (IAA) content was estimated in leaves (third leaf from top) and roots of 90 days old endophyte inoculated and non-inoculated endophyte free control Withania plants. Each data point is an average of six replicates and the error bars represent standard errors. Asterisks indicate significant differences between the control and endophyte inoculation (Duncan’s multiple range test P ≤ 0.05).
Effect of endophyte-inoculation on in-vitro grown W. somnifera plant
In-vitro plants were developed from nodal explants of W. somnifera. Multiple shoot regeneration through axillary bud proliferation of the nodal explants was observed on MS medium containing 2 mg L−1 BAP and 0.1 mg L−1 NAA, within 3–4 weeks of culture. Rooting was induced on transfer of this plantlets to half strength MS medium without any plant growth regulator (Fig. 7).
Figure 7.
Generation of in-vitro grown Withania somnifera plant. Establishment of normal plant (a) shoot formation in MS medium supplimented with 2 mg L −1 BAP & 0.1 mg L−1 NAA; (b) rooting initiation on ½ MS medium; (c) Complete in-vitro establishment of normal plant (d) shoot and root after harvesting; (e) hardening of normal plant under glass house condition) Establishment of composite plant [(f) shoot initiation in MS medium without PGR infected with A. rhizogenes; (g) transformed root initiation after 15–20 days of infection and root elongation after 35–40 days of inoculation; (h) complete in-vitro raised composite plant; (i) transformed root morphology after harvesting; (j) established composite plant under glass house condition].
Composite plants were raised by inoculating the wild type A4 strain of A. rhizogenes to the cut ends of the in-vitro raised shoots, where profuse root emergence could be observed after 15–20 days of bacterial inoculation (Fig. 7). These composite plantlets with A. rhizogenes induced roots were subsequently transferred to the pre-optimized half strength MS medium for their further establishment and were maintained under the confined environment of glass house for further experimentations with the endophytes (data not shown). PCR analysis showed the presence of the Rol A, B and C genes in the transformed roots of these composite W. somnifera plants (Supplementary Fig. S1). The areal parts of these composite plants did not reveal any observable morphological difference with that of the normal plants. However, a striking difference was noticeable in their rooting behavior as the composite plants demonstrated enormous proliferation and elongation of root growth (Fig. 7). The fresh weight of the harvested roots of the composite plants was found to be 4.29 ± 0.29 g plant−1 which was 2.66 fold higher than that of the normal plant roots (1.61 ± 0.21 g plant−1) (Supplementary Fig. S2).
Subsequent experimentation through inoculation with the three selected endophytes (WPR16, WPR17 and WPS23) to the in-vitro raised normal and composite W. somnifera plants (which were maintained under the confined environment of glass house) revealed diversified influence on the biosynthetic potentials of both their leaves and roots pertaining to all the three targeted withanolides (i.e., WFA, DWL and WLA). At the outset, the leaves of the composite plants exhibited higher contents of WFA (71.2%), DWL (50%) and WLA (196.7%), compared to that of the normal plants (Table 4). Noticeably, the roots of normal plants reveal the sole presence of WLA with no traces of WFA and DWL at the start while the presence of all the three withanolides could be noted in significant amount in the roots of composite plants (Table 4). Interestingly, the roots of composite plants showed 593% higher WLA content compared to that of the normal plants’ roots without any endophyte treatment.
Table 4.
Withanolide content in leaves and roots of endophyte-inoculated in-vitro grown Withania somnifera plants.
| Leaves | Roots | |||||
|---|---|---|---|---|---|---|
| %WFA | %DWL | %WLA | %WFA | %DWL | %WLA | |
| Normal | ||||||
| Control | 0.59 ± 0.02c | 0.08 ± 0.01c | 0.30 ± 0.01c | nd | nd | 0.057 ± 0.002d |
| WPR16 | 0.88 ± 0.09b | 0.08 ± 0.02c | 0.69 ± 0.03b | nd | nd | 0.139 ± 0.013b |
| WPR17 | 1.11 ± 0.06a | 0.36 ± 0.02a | 1.09 ± 0.06a | nd | nd | 0.223 ± 0.015a |
| WPS23 | 1.15 ± 0.07a | 0.27 ± 0.01b | 0.71 ± 0.01b | nd | nd | 0.096 ± 0.001c |
| Composite | ||||||
| Control | 1.01 ± 0.13c | 0.12 ± 0.007b | 0.89 ± 0.01b | 0.082 ± 0.012d | 0.01 ± 0.002d | 0.395 ± 0.006c |
| WPR16 | 0.67 ± 0.12d | 0.12 ± 0.001b | 0.48 ± 0.09c | 0.321 ± 0.018a | 0.018 ± 0.003c | 0.638 ± 0.009a |
| WPR17 | 1.94 ± 0.09b | 0.16 ± 0.006a | 1.88 ± 0.05a | 0.131 ± 0.009c | 0.027 ± 0.002b | 0.414 ± 0.004c |
| WPS23 | 2.3 ± 0.13a | 0.15 ± 0.007a | 1.86 ± 0.04a | 0.153 ± 0.008b | 0.053 ± 0.002a | 0.566 ± 0.008b |
WFA- withaferin A, DWL- 12-deoxy withstramonolide and WLA- withanolide A. Values are the means of six biological replicates ± S.E. Values with different letters are significantly different at P ≤ 0.05 (Duncan’s multiple range test). nd- not detected.
Then again, inoculation of the in-vitro raised normal plants with the three selected endophytes (WPR16, WPR17 and WPS23) led to the enhancements in the contents of the WFA and WLA in their leaves at 49–95% and 130–263% higher levels respectively as compared to that in the endophyte non-inoculated control plants (Table 4). At the same time, except for WPR16, the other two endophytes (i.e., WPR17 and WPS23) inoculation caused 237–350% stimulation in the DWL content in leaves of normal plants (Table 4). Alternatively, all the three tested endophytes enhanced the WLA content in roots of in-vitro raised normal plants by a range of 68–291% as compared to that in the endophyte non-inoculated control plants (Table 4).
On the other hand, inoculation with these two endophytes (WPR17 and WPS23) to the in-vitro raised composite W. somnifera plants enhanced the foliar contents of all the three withanolides, i.e., WFA, DWL and WLA, by 92–128%, 25–33% and 109–111% respectively, whereas the WPR16 proved ineffective for improvement in DWL content (Table 4). However, every single one of the three tested endophytes stimulated the production of all WFA, DWL and WLA at variable levels in the roots of the composite plants which ranged at 60–291%, 80–430% and 43–61% higher yields respectively, compared to that in the non-inoculated endophyte free control plants’ roots (Table 4).
The overall analysis of the results in terms of the influence of the three tested endophytes on the biosynthetic potentials of the normal and composite plants (in their leaves and roots) pertaining to the three desired withanolides revealed very interesting outcomes which highlights the future applicability of this study. The WPS23 endophyte proved best for enhancing the WFA and WLA contents in the leaves of both the normal and composite plants, but the latter proved most responsive towards this endophyte, resulting in 100% and 162% stimulation in their WFA and WLA productivity respectively compared to that in the former (Table 4). The WPR17 endophyte demonstrated the second optimum efficiency which also boosted the foliar production trend of these two above-stated withanolides (WFA and WLA) in both the control and composite plants, where the latter superseded the former by 74.77% and 72.5% better productivities respectively (Table 4). Noticeably, this WPR17 endophyte had additionally enhanced the foliar content of DWL in the normal plant, which was 125% higher compared to that in the leaves of the treated composite plants (Table 4). On the other hand, the potential of the roots of the composite plants towards the synthesis of all the three desired withanolides in the background of their total absence in the roots of the normal plants, needs special mention, which showed an up-ward augmentation by the inoculation with all the three endophytes with observable variability (Table 4).
The expression of different genes involved in withanolide biosynthesis was studied by qRT-PCR in in-vitro grown W. somnifera plants inoculated individually with selected endophytes (WPL16, WPL17, WPS23). Expression of SQS, SE, CAS, FPPS, SMT, ODM, HMGR, SGT, DXS, DXR, CPR1 and CPR2 genes were quantified in leaves as well as in roots of in-vitro grown normal and composite plants. Expression of CAS, SMT, HMGR, SGT, DXS, DXR, CPR1 and CPR2 genes was increased in leaves of endophyte inoculated in-vitro normal plants compared to non-inoculated endophyte free control plants, while expression of SQS, SE, FPPS, and ODM was upregulated by WPL17-, WPS23-inoculation (Fig. 8). Expression of all studied genes in roots of in-vitro normal plants was increased by selected endophyte inoculation (Fig. 8). Similarly, leaves and roots of endophytes inoculated in-vitro composite plants also had increased expression of studied pathway genes compared to non-inoculated endophyte-free control plants (Fig. 8).
Figure 8.
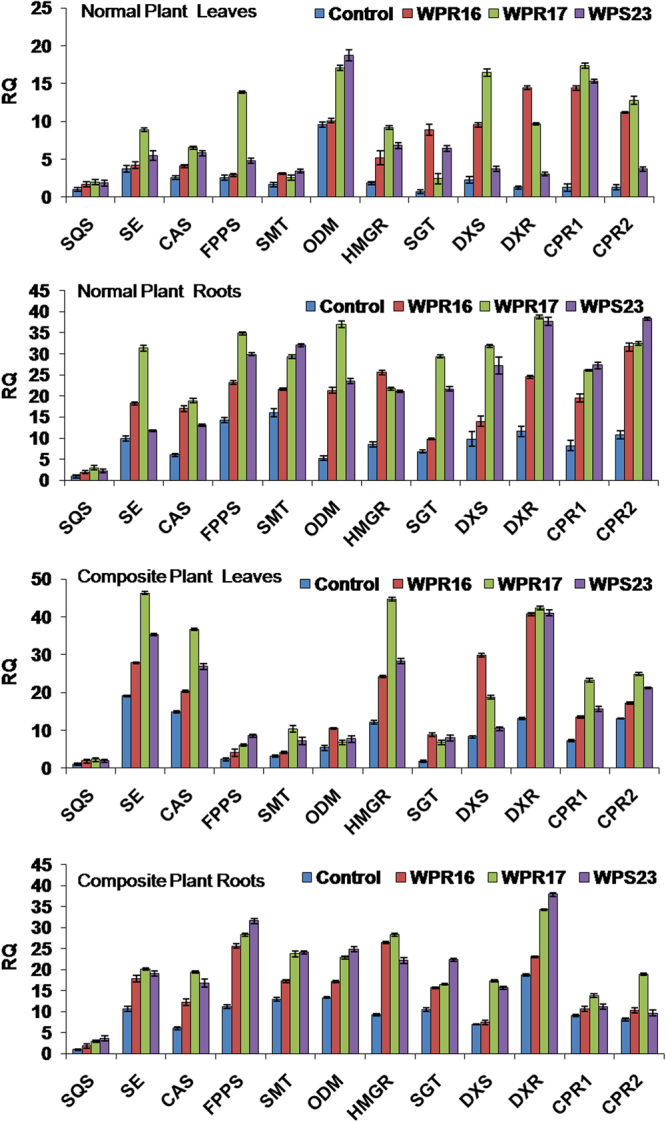
Effect of endophyte inoculation on the expression of withanolide biosynthesis genes in in-vitro grown normal and composite Withania somnifera plant. Expression of different withanolide biosynthesis genes was analyzed. Results were normalized to actin (reference transcript) and are shown relative to the level in non-inoculated endophyte-free control plants (calibrator). Data are means ± SD (n = 3 biological replicates) and Y-axis represents relative quantity (RQ).
Discussion
Endophytes have promising potential for sustainable agriculture as they can promote plant growth and confer tolerance to plant from environmental stresses31–35. They are also the source of therapeutically important novel chemicals36–38. Earlier work from our laboratory has indicated that some endophytes may enhance the secondary metabolite production of the medicinal plants39,40,42,43. We have shown that medicinal plant opium poppy harbour numerous endophytes residing in different parts of the plants and play a role in a tissue-specific manner39. Endophytes associated with opium leaves were involved in improving photosynthetic efficiency of the plant while the endophytes associated with capsule (major site for secondary metabolites production) were involved in improving secondary metabolites specifically benzylisoquinoline alkaloid production39. No such relation was observed in Withania plants as far as shoot growth is concerned as the endophytes isolated both from leaf and root could enhance the shoot growth. However, endophytes isolated from roots could significantly enhance the root growth, supporting our earlier study suggesting their tissue specific roles39. Two fungal endophytes Curvularia sp. and Choanephora infundibulifera isolated from Catharanthus roseus enhanced in planta production of vindoline by regulating the expression of key structural and regulatory genes of terpenoid indole alkaloid biosynthesis40. In the present study, we demonstrated an important role of endophytes in improving plant yield and secondary metabolite production in medicinally important W. somnifera plant. We could isolate and identify a total of 29 bacterial endophytes and 11 fungal endophytes. The role of these isolated endophytes was studied by inoculating them in endophyte-free plants and compared with the non-inoculated endophyte-free control plant. It was observed that few isolated fungal endophytes could improve the photosynthetic efficiency of W. somnifera plant by improving the content of photosynthetic pigments (Chlorophyll and carotenoids), net CO2 assimilation rate, transpiration rate and stomatal conductance. Improved photosynthetic efficiency due to endophyte inoculation resulted in increased biomass of Withania plant that was evident from the higher shoot and root biomass. As both leaves and roots are the economic parts, used for the extraction of withanolides, therefore, these fungal endophytes can be used for the substantial improvement of biomass. Improved growth in endophyte-inoculated Withania plants may be due to indole acetic acid production and phosphate solubilization activity of endophytes.
Withanolides are the major secondary metabolites responsible for pharmacological properties of different parts of W. somnifera plant. Inoculation with endophytes modulated the content of withanolides in leaves and roots of W. somnifera plant. Surprisingly, some endophytes induced the synthesis of withaferin A in roots (0.010–0.043%; Table 3) which is abundantly synthesized in leaves17–19 and totally absent16 or present in traces (0.0005–0.0007%) in roots of some varieties20 of W. somnifera. This clearly shows a strong involvement of endophytes in modulating the biosynthetic pathway. Expression study of different genes involved in withanolide biosynthesis revealed the possible mechanism associated with endophyte mediated changes in withanolide biosynthesis. We observed that endophyte inoculation modulated the expression of genes of withanolide biosynthesis. Withanolides biosynthesis involves both MVA and MEP pathways which regulate the flux of the isoprene units (isopentenylnpyrophosphate; IPP and dimethylallyl pyrophosphate; DMAPP) for the synthesis of intermediates of withanolide biosynthesis47. Endophytes inoculation upregulated the expression of HMGR (both in leaves and roots), involved in the biosynthesis of isopentenylnpyrophosphate (IPP) via MVA pathway which is the predominant pathway for biosynthesis of withanolides48, and this might be a key reason for the higher production of withanolides in endophyte-inoculated plants. The expression of DXR and DXS, involved in Methylerythritol 4-phosphate (MEP) pathway, was not affected in leaves of the endophyte-inoculated plant which might be due to the existence of distinct/specific transcription regulatory mechanism for genes encoding for enzymes involved in MEP pathway49. However, endophytes could upregulate the expression of DXS and DXR in roots. It might be due to the constant association of these endophytes with roots (their site of isolation) that could specifically modulate their expression in roots. The expression of DXS, DXR and FPPS remained unaffected in the leaves of endophyte inoculated plants, however, their expression was upregulated in roots and this might be a possible reason for induction of withaferin A synthesis in roots. This also indicates the importance of MEP-pathway genes DXS and DXR in withaferin A biosynthesis. Plant growth promoting activity such as IAA production and nitrogen fixation ability of these root associated endophytes (WPR12, WPR16, WPR17, WPS23 and WPR32) might also facilitate production of withaferin A in roots. Selected endophyte inoculation could increase the in planta content of IAA in leaves and roots that could enhance withanolide biosynthesis. Improvement of withaferin A production by application of nitrogen significantly upregulating structural and regulatory genes of withanolide biosynthesis also supports our present observation50. The previous report demonstrated that IAA favours the biosynthesis of withaferin A in in-vitro root cultures of W. somnifera51 consolidates the role of IAA producing endophytes in enhancing the content of withaferin A in the present study. Expression of SQS, encoding squalene synthase catalysing the condensation of farnesyl pyrophosphate (FPP) to produce squalene (a key intermediate for biosynthesis of various triterpenoids) was found to be upregulated in leaves and roots of all the selected endophyte-inoculated Withania plant. Overexpression of SQS in W. somnifera resulted in increased production of withanolides23 and virus-induced gene silencing (VIGS) of SQS in W. somnifera led to the reduction in withanolide synthesis confirmed the crucial role of SQS in withanolide biosynthesis52. Overexpression of SQS in cell suspension culture of W. somnifera substantially enhanced (2.5 fold as compared to non-transformed cell cultures) withanolide production and also started withaferin A biosynthesis which was absent in non-transformed cell cultures29. Therefore, upregulated expression of SQS (up to 3.3 folds) in endophyte inoculated plants might induce the synthesis of withanolides especially withaferin A in roots which was clearly visible in WPR 16 and WPS 23 inoculated plants. Endophyte-inoculated plants had upregulated expression of SQE which encodes squalene epoxidase which is a rate limiting enzyme in the withanolide biosynthetic pathway53,54. Cascading influence of SQE on the upregulation of downstream genes and its genetic manipulation in W. somnifera plant suggest a promising way for the production of therapeutically important triterpenoid molecules55,56. Therefore, higher expression of SQE (up to 4.9 folds in roots and 12.1 folds in leaves) in the endophyte-inoculated plants might enhance the expression of downstream genes that could have resulted in higher withanolide production both in roots and leaves.
Endophyte-inoculation could upregulate the expression of CAS which forms cycloartenol (the precursor for withanolide biosynthesis) and acts at metabolic branching point for synthesis of withanolides and various triterpenes such as saponins (Fig. 5). Previous reports show that CAS overexpression and silencing resulted in an increase and decrease of withanolide content respectively24. In endophyte-inoculated plants, increased expression of CAS, SMT and ODM were correlated with enhanced withanolides production. Few endophytes modulated the expression of CPRs which encode cytochrome P450 reductase. P450s are involved in the primary and secondary metabolism of a plant by catalysing various reaction such as hydroxylations, epoxidations, sulfoxidations, dealkylations, peroxidations, reductive dehalogenations and isomerisation reactions57,58. Endophytes also modulated the expression of SGT (sterol glucosyltransferases) responsible for glyco-transformations of steroids.
Figure 5 shows differential regulation of withanolide biosynthetic pathway by inoculated endophytes. All the selected endophyte-inoculations upregulated the expression of HMGR in both leaf and root tissues, indicating that it may be a promising candidate gene that can be used for increasing in planta withanolide production. Previously, HMGR as rate limiting enzyme of isoprenoids biosynthesis of eukaryotes has also been suggested59. Piriformospora indica elicitation increased withaferin A biosynthesis by upregulating the regulatory genes of MEP, MVA and withanolide biosynthetic pathway and showed the highest expression of HMGR in cell suspension cultures of Withania somnifera60. Most of the endophyte-inoculation increased the expression of SQS, SQE, CAS and SMT that could enhance the production of withanolides in both leaves and root tissues. The expression of DXS, DXR and FPPS which was not affected in leaves, indicates the presence of different regulatory components/factors that were not targeted by endophytes. WPR17 and WPS23-inoculation upregulated the expression of all the key genes of withanolide biosynthesis in roots. Therefore, these are the most promising candidates that can be used for the improvement of in planta withanolide production.
As roots of the W. somnifera plants are the major source of withanolides and used for herbal therapeutics, therefore, in the present study we have evaluated the effect of the selected endophytes on in-vitro grown composite plants having higher root biomass than that of the normal plants. The utilization of composite plants had earlier been reported as a successful alternative approach for the functional characterization of any targeted gene(s) in the host plants, where the transformation and regeneration are tedious and lengthy45. This technology has also been found competent for several other studies such as nutrient and hormone uptake, interactions with root nodulating bacteria and mycorrhizal symbiotic association46. The present study has first time explored the potentials of the composite plants with respect to evaluating the influence of external applications of targeted endophytes with the overall intention of amplifying the host-endophyte interaction interphase (to best of our knowledge). Here, we developed the composite plant of W. somnifera to evaluate the potentials of such transformed roots towards the biosynthesis of secondary metabolites with and without treatment of selected endophytes isolated from W. somnifera and found to enhance the secondary metabolite production.
Normal plants were regenerated in-vitro from nodal explants, and composite plants were generated by inoculating wild-type A. rhizogenes A4 strain to the cut ends of in-vitro raised shoots resulting in the production of profuse rooting. Successful integration of rol gene in the genome of normal plants could result in the development of composite plants having more roots than the normal plants. Shoot and root regeneration in nodal explants was found to be similar as reported previously61–66. Generation of composite plants also enhanced the biosynthesis of all the measured withanolide (WFA, DWL and WLA) in both the leaf and root tissues. Furthermore, in general the inoculation with selected endophytes (WPL16, WPL17 and WPS23) enhanced the content of all the measured withanolide (WFA, DWL and WLA) in the leaves and roots of the composite plants, which is due to the upregulation of the expression of key genes of withanolide biosynthesis as observed in the case of in-vivo normal plants. qRT-PCR study showed the upregulation of HMGR in both the leaves and root tissues of normal in-vivo as well as in-vitro grown normal and composite plants. This observation strongly indicates that HMGR could be the key gene that can be considered as the target for enhancing the withanolide content in Withania for genetic manipulation/transgenic considerations. Reduction in the content of WFA in leaves of WPR16 inoculated composite plants, DWL in WPR17 and WPS23 inoculated plants and WLA in WPR16 inoculated plants indicates endophyte mediated change of flux of withanolide biosynthesis from leaves to roots as these alkaloids accumulated in increased amounts in their root tissue. Similarly presence of WFA and DWL in roots of composite plant (which was not detected in normal plants) in both endophyte inoculated and non-inoculated plants also indicates the change of flux of withanolide biosynthesis from leaves to roots, which might have resulted due to the Ri T-DNA mediated changed nature of the composite plant.
To summarize, results of the present study suggest that the medicinal plant W. somnifera harbours various bacterial and fungal endophytes associated with leaves, roots and seeds. These endophytes may play an important role in the biosynthesis of important secondary metabolites and also have the potential to improve plant shoot and root biomass by improving the photosynthetic pigments, photosynthesis rate, transpiration rate and stomatal conductance through growth promotion activities such as IAA production and phosphate solubilization. We also demonstrated that W. somnifera have endophytes which have the ability to modulate the withanolide biosynthesis in the leaves and roots of W. somnifera by modulating the expression of key genes of withanolide biosynthetic pathway. The presence of few IAA-producing and nitrogen-fixing root-associated endophytes that can induce the production of leaf alkaloids (withaferin A) in roots by upregulating the expression of withanolide biosynthesis genes especially MEP pathway genes (DXS and DXR) as promising candidates for enhancing the withaferin A biosynthesis in Withania root is also suggested. Application of these endophytes in Withania agriculture will benefit growers to harvest higher amounts of both withanolide A and withaferin A from roots. Although the mechanism associated with endophyte-mediated modulation of host plant metabolism is really difficult to unravel, their possible role in promoting plant growth and yields, and pharmaceutically important withanolide biosynthesis cannot be ignored. Formulation and testing of microbial consortia consisted of endophytes enhancing plant biomass and endophytes enhancing withanolide content may empower host plant to produce higher biomass coupled with more withanolide content.
Methods
Plant material and growth conditions
Seeds of Withania somnifera genotype Poshita were obtained from the National Gene Bank for Medicinal and Aromatic Plants, CSIR-Central Institute of Medicinal and Aromatic Plants (CSIR-CIMAP), Lucknow, India and grown in a greenhouse in natural photoperiod of 16 h day/8 h night cycle under natural light intensity at 25 °C ± 2 °C. Withania plants were grown in earthen pots (22 cm top diameter × 12 cm bottom diameter × 17 cm height and 3.7 L volume) filled with potting mixture (3.5 kg) consisted of autoclaved soil and vermicompost (2:1, v/v) and watered with sterile water.
Isolation and identification of endophytes
Endophytes were isolated from healthy green leaves, root and seeds of field-grown Withania somnifera genotype Poshita plant following the procedure described previously39,40. Isolation of endophytes was done in triplicate. Each replicate of leaves (1 g) and roots (1 g) was collected from three individual plants. Approximately 100 mg seeds was used in triplicate for endophyte isolation. Initially, surface sterilization of plant tissues was done by placing tissues in 1% sodium hypochlorite solution for 10 min and then washed (five times) in 0.02 M sterile potassium phosphate buffer (pH 7.0). Sterility check of tissues was performed by adding 100 µl of an aliquot of the final buffer wash into 5 ml nutrient broth (g l−1; 5 g peptone, 2 g beef extract) and incubated in an incubator shaker (200 rpm at 28 °C) for 72 h. Samples were discarded if the growth was detected in the sterility check samples. Surface sterilized tissues were then homogenised in a sterile pestle and mortar with sterile distilled water (10 ml). Homogenate was serially diluted (10−1, 10−2 and 10−3) in sterile distilled water and plated on nutrient agar (NA) (g L−1; 5 g peptone, 2 g beef extract, 20 g agar, pH 5.0) and potato dextrose agar (PDA) (g L−1; 200 g potato infusion, 20 g dextrose, 15 g agar, pH 5.6) plates in triplicates. Bacterial endophytes were selected on NA plates, incubated at 28 °C for 24–72 h and fungal endophytes were selected on PDA plates incubated at 28 °C for 10 days. A representative of each endophyte (as distinct from their colony morphology) was transferred to a fresh plate and pure culture was established. Identification of bacterial and fungal endophytes was performed by 16 S rRNA and internal transcribed spacer (ITS) sequencing respectively. Amplification and sequencing of 16 S rRNA and ITS fragments were performed following the procedure described earlier39,40. Similarities in nucleotide sequences were determined by using NCBI BLAST, version 2.0. Partial sequence data of 16 S rRNA and ITS fragment have been submitted to the NCBI GenBank, and accession numbers were obtained (Table 1).
Inoculum production for endophytes
Bacteria were grown in nutrient broth to the mid-log phase and then pelleted by centrifugation at 4000 rpm for 10 min at 4 °C temperature. The pellet was washed and resuspended (1 × 108 CFU mL−1) in phosphate buffer saline (PBS) (g L−1; 0.24 g potassium dihydrogen phosphate, 1.44 g disodium hydrogen phosphate, 8 g sodium chloride, 0.2 g potassium chloride, pH 7.4). For fungi inoculum preparation, the individual fungus was grown in potato dextrose broth for one week at 28 °C. Numbers of conidia/spore were counted on microscope and dilution (1 × 108 spores/conidia mL−1) was made in PBS.
Treatment of Withania plants with endophytes
Endophyte-free Withania plants were used to study the effects of treatment with isolated endophytes. Endophytes-free Withania plants were generated following previously established procedure39,40. In brief, Withania seeds were extensively washed in water and then incubated in Bavistin (a fungicide containing carbendazim 50% W.P., BASF India Limited) and K-Cycline (a bactericide containing streptomycin sulphate 90% w/w and tetracycline hydrochloride 10% w/w, Karnataka Antibiotics & Pharmaceuticals Ltd. Bangalore, India) solution at 28 °C temperature, 120 rpm shaking for 72 h and then washed extensively in sterile water and homogenized in a sterile pestle and mortar with sterile PBS. Homogenate was plated on NA and PDA and incubated at 28 °C for 10 days. No microbes were obtained on incubated plates. Therefore, these W. somnifera seeds were used as endophyte free seeds and used for growing endophyte-free nursery. The nursery was grown in earthen pots (30 cm top diameter × 20 cm bottom diameter × 7 cm height) filled with of autoclaved potting mixture (3.0 kg) consisted of soil and vermicompost (2:1 v/v) and watered with sterile water. These pots were grown under greenhouse condition (natural photoperiod and light intensity at 25 °C ± 2 °C) for one month, and grown seedlings were re-confirmed for their endophyte-free status. These seedlings were uprooted delicately to minimize the damage to their roots and inoculated with isolated endophytes. For endophyte inoculation, roots of seedlings were dipped in individual endophyte suspension (1 × 108 CFU mL−1 for bacteria and 1 × 108 spore/conidia mL−1 for fungus) prepared in PBS for 3 h. These endophyte-inoculated seedlings were re-planted in pots (22 cm top diameter × 12 cm bottom diameter × 17 cm height) filled with autoclaved soil and vermicompost mixture (2:1 v/v) and grown under greenhouse conditions. The roots of non-inoculated endophyte-free control were dipped in PBS for the same duration. Plants were watered with sterile water as and when required. In all experimental analyses, endophyte-inoculated plants were compared with the non-inoculated endophyte-free control plants that developed from endophyte-free Withania seeds. Inoculation (10 mL pot−1 containing 1 × 108 CFU mL−1 bacterial cell or 1 × 108 fungal spore/conidia mL−1) with individual endophyte was again repeated after 15-days of the first inoculation to maintain the presence of an adequate number of inoculated endophytes in the soil. Presence of inoculated endophytes in Withania plants was confirmed by re-isolating the endophytes from tissues of endophyte-inoculated plants before carrying out further experimental analyses.
Sampling for all analyses was carried out at the same stage (90 d which is an intermediate stage neither too young nor too old) and position of leaf (third leaf from top) for minimizing the variations dependent on developmental stage of plant and other variables.
Photosynthetic pigments and photosynthesis parameters analysis
Photosynthetic pigments (Chlorophyll and carotenoids) and photosynthetic efficiency of third leaves of 90-days old Withania plants (n = 3) were determined. Pigments were extracted in chilled 100% methanol, and the content of chlorophyll and carotenoids was measured following to previously established procedure67. Photosynthesis parameters (net CO2 assimilation, transpiration rate and stomatal conductance) were measured in the attached leaves using a portable photosynthesis system (CIRAS-3, PP Systems, USA). For photosynthesis measurement, leaf was pre-exposed for 15 min at 400 µmol photons m−2 s−2 light, 400 ppm CO2 and 25 °C temperature.
Establishment of in-vitro grown normal plants of W. somnifera
W. somnifera cv. Poshita, cultivated in the field of CSIR-CIMAP, Lucknow were used as explants source for establishment of in-vitro culture. In-vitro culture of W. somnifera was established using nodal explants according to the previously published protocol68. In short, the nodal explants were initially washed with 10% (v/v) Tween-20 for 5 min followed by washing with running tap water. Surface sterilization of explants was done with 0.1% HgCl2 and then inoculated on MS medium69 supplemented with 0.1 mg L−1 NAA and 2 mg L−1 BAP for multiple shooting and further transferred in half strength of MS medium without any supplementation of plant growth regulator for the establishment of the complete plant. After the successful establishment of in-vitro culture (with roots and shoots) they were transferred to sterilized soil, hardened and acclimatized under confined glasshouse condition (natural photoperiod and light intensity at 25 °C ± 2 °C and 80% relative humidity)
Establishment of in-vitro grown composite plants of W. somnifera
The transformation experiments for the development of composite plants were carried out following previously published protocol62 with slight modification utilizing the wild type of A4 strain of A. rhizogenes (a kind gift from Prof. D. Tepfer, INRA, Versailles Cedex, France). In this study, the needle-pricking method was applied at the cut end of nodal explant by inoculation of over-night grown bacterial suspension (O.D 600 = 1.0). After 2–3 days of co-cultivation of explants on half strength MS medium with the Agrobacterium, the explants were transferred to ½ MS containing 100 mg L−1 of Cefotaxime (Alkem, India). Similarly, the complete composite plants (with normal shoots & transformed roots) were transferred to sterilized soil, hardened and acclimatized under glasshouse condition.
Inoculation of the selected endophytes (WPL16, WPL17, WPS23) was done after the transfer of invitro grown plants (normal and composite) to sterilized soil. The roots of invitro grown plants were dipped in individual endophyte suspension (1 × 108 CFU mL−1) prepared in PBS for 3 h and then planted in the pot filled with autoclaved soil. For control plants, only PBS (without endophyte) was used. Plants were grown under confined glasshouse condition.
Confirmation for Rol genes in transformed roots of composite plants
DNA was extracted from the in-vitro raised transformed roots of composite plants (CPR) and normal plant root (NPR) according to the protocol described previously70. Rol A, B and C genes were PCR amplified using the rol specific primer for Rol A (Forward: 5′-GGAATTAGCCGGACTAAACG-3′ and Reverse 5′-CCGGCGTGGAAATGAATCG-3′), Rol B (Forward: 5′-ATGGATCCCAAATTGCTATTCC-3′ and Reverse: 5′-GTTTACTGCAGCAGCAGCAGGCTTCATG-3′) Rol C (Forward: 5′-ATGGCTGA AGACGACCTGTGT-3′ and Reverse: 5′-CCGATTGCAAACTTGCACTC-3′) were used. The PCR conditions were followed as described previously70.
Measurement of plant biomass
Shoot and root biomass of 90-days old W. somnifera plant (n = 6) were measured. Entire shoots (aboveground plant parts) and entire roots (belowground plant parts), were harvested and completely dried at 70 °C for 5 d, and their dry weight was measured.
Analysis of withanolide
Withanolide A (WLA), withaferin A (WFA) and dideoxywithanoliside (DWL) content was estimated in leaves and roots of Withania plant by HPLC. Leaves (third from top) and roots of 90-days old Withania plant were taken for alkaloid extraction. In the case of in-vitro grown normal and composite Withania plant leaves and roots samples were taken at 60 d stage after glass house acclimatization. Tissues were dried and ground to fine powder. 100 mg of powdered samples were extracted with 2 mL warm (50 °C) methanol for 4 h. The extraction process was repeated three times. Methanol extract was filtered through Whatman No. 1 filter paper and concentrated by drying and re-dissolved in methanol for HPLC analysis. The withanolide content was estimated by HPLC equipment (Shimadzu, Japan) containing Spherisorbs® C18 (250 mm × 4.6 mm i.d.) 10 μm particle size ODS2 column (Waters, Milford, MA), pumps (LC-10AT), an auto-injector (SIL-10AD) and PDA (SPD-M10A) detector71,72. Mobile phase composition was 40:60 (v/v) mixture of acetonitrile: trifluoroacetic acid [0.1% (v/v) in water] with the flow rate of 1.0 mL min−1 and detector wavelength of 220 nm was used throughout the analysis. All standard withanolides were procured from ChromaDex (Irvine, CA) and all solvents used for analysis were of HPLC grade. The specificity of the measured compounds (WLA, WFA and DWL) was established through peak purity analysis using a PDA detector, matching UV-Vis spectra and spiking with reference compound of known concentration (Supplementary Fig. S3).
Quantitative real time-PCR (qRT-PCR) analysis of withanolide biosynthesis genes
The qRT-PCR analysis was performed in third leaves (from top), and roots of 90-days old non-inoculated endophyte free control and endophytes inoculated Withania plant. In the case of in-vitro grown normal and composite Withania plants leaves and roots samples were taken at the 60-d stage after glass house acclimatization. Total RNA was isolated using TRI-reagent (Sigma-Aldrich). Concentration of RNA was determined using NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). RNase-free enzyme DNase I (Thermo Scientific) was used to eliminate genomic-DNA contamination. First-strand cDNA was synthesized using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) following manufacturer’s protocol. Transcripts of a total of 12 genes involved in withanolide biosynthesis were quantified. qRT-PCR was performed using SYBR-Green I chemistry on triplicate technical replicates of triplicate biological samples. Primers used for qRT-PCR analysis are described in Supplementary Table S5. PCR mixtures consisted of 300 nM of both forward and reverse primers, 5 μL SYBR Premix Ex Taq (TAKARA BIO INC.), 0.2 μL ROX Reference Dye and 1 µL of 10 times diluted cDNA synthesis reaction in a reaction volume of 10 μL. qRT-PCR condition was an initial denaturation at 95 °C for 10 min; 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min. Fluorescent signal intensities were recorded and analysed on an Applied Biosystems StepOnePlusTM Real-Time PCR System. Melt-curve analysis using the dissociation method (Applied Biosystems) was included to verify the specificity of RT-qPCR. The actin of W. somnifera was used as reference transcript. Non-inoculated endophyte free control plants were used as calibrator, and the relative quantification 2−∆∆Ct method was used73,74.
Screening for growth promotion activity
The phosphate utilization, IAA production and nitrate reduction test was performed following previously established method75–78.
Indole-3-acetic acid measurement
IAA content was estimated using the Phytodetek-IAA Immunoassay kit (Agdia, Elkhart, IN) as described previously79. Leaves and root samples were frozen in liquid nitrogen and ground to fine powder. 0.5 g of the powdered tissue was suspended in 5 ml of extraction solution containing 80% methanol, 0.5 g L−1 citric acid monohydrate and 100 mg L−1 butylated hydroxy toluene, and stirred overnight at 4 °C in the dark. The solution was centrifuged at 1000 g for 20 min at 4 °C and then the supernatant was dried under vacuum. The dried residue was dissolved with 100% methanol (100 µl) and 900 µl of tris-buffered saline (pH 7.8). The IAA concentration in the filtrate was determined using the Phytodetek-IAA Immunoassay kit (Agdia, Elkhart, IN) as per the manufacturer’s instructions.
Statistical analysis
Statistical analysis of data was done by applying ANOVA, suitable to completely randomised design (CRD), using the software ASSISTAT Version 7.7 beta. Significant differences among different treatments were carried out using Duncan’s multiple range tests (DMRTs) at a significance level of P ≤ 0.05. For measurement of chlorophyll, carotenoids, net CO2 assimilation, stomatal conductance, transpiration rate, plant shoot and root biomass, withanolide and IAA content six replicates were used, and for expression analysis, three biological replicates for each treatment were used. Three independent experiments were performed for three consecutive years under greenhouse condition and similar results were obtained.
Electronic supplementary material
Acknowledgements
This work was supported by Council of Scientific and Industrial Research (CSIR), India, through grant NWP BSC0117 (XII Five Year Plan Network Project). The authors are thankful to Director, CSIR-Central Institute of Medicinal and Aromatic Plants, Lucknow, India for his support and encouragement in providing necessary facilities during investigation. SSP and HP thank Council of Scientific and Industrial Research (CSIR, New Delhi, India) for financial assistance in the form of Senior Research Associateship (SRA) and Senior Research Fellowship respectively. SS gratefully acknowledge Indian Council of Medical Research (ICMR, New Delhi, India) for financial support in the form of fellowships. Authors also acknowledge the help of Dr. Rakesh K Shukla of CSIR-CIMAP for critically going through the manuscript. SS and HP are thankful to Academy of Scientific and Innovative Research (AcSIR-CIMAP).
Author Contributions
A.K. and S.S.P. conceived and designed the experiments. S.S.P., S.S., H.P., M.S., S. Soni, T.R., A.P. performed the experiments. A.K., M.M.G., S.B., K.S., C.S.V., S.S.P. analyzed the data. A.K., S.B. and S.S.P. wrote the paper. All authors read and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23716-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kumar A, Kaul MK, Bhan MK, Khanna PK, Suri KA. Morphological and chemical variation in 25 collections of the Indian medicinal plant, Withania somnifera (L.) Dunal (Solanaceae) Genet. Resour. Crop Evol. 2007;54:655–660. doi: 10.1007/s10722-006-9129-x. [DOI] [Google Scholar]
- 2.Sharma V, Sharma S, Pracheta., Paliwal R. Withania somnifera: a rejuvenating Ayurvedic medicinal herb for the treatment of various human ailments. Int. J. Pharmtech Res. 2011;3:187–192. [Google Scholar]
- 3.Singh G, Sharma PK, Dudhe R, Singh S. Biological activities of Withania somnifera. Annals Biol. Res. 2010;1:56–63. [Google Scholar]
- 4.Ahlawat P, Khajuria A, Bhagwat DP, Kalia B. Therapeutic benefits of Withania somnifera: An exhaustive review. Int. J. Pharma. Chem. Sci. 2012;1:491–496. [Google Scholar]
- 5.Jain R, Kachhwaha S, Kothari SL. Phytochemistry, pharmacology, and biotechnology of Withania somnifera and Withania coagulans: A review. J. Med. Plants Res. 2012;6:5388–5399. doi: 10.5897/JMPR12.704. [DOI] [Google Scholar]
- 6.Sehgal N, et al. Withania somnifera reverses Alzheimer disease pathology by enhancing low-density lipoprotein receptor related protein in liver. Proc. Natl. Acad. Sci. USA. 2012;109:3510–3515. doi: 10.1073/pnas.1112209109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uddin Q, Samiulla L, Singh VK, Jamil SS. Phytochemical and pharmacological profile of Withania somnifera Dunal: A Review. J. Appl. Pharma. Sci. 2012;2:170–175. [Google Scholar]
- 8.Gauttam VK, Kalia AN. Development of polyherbal antidiabetic formulation encapsulated in the phospholipids vesicle system. J. Adv. Pharm. Technol. Res. 2013;4:108–117. doi: 10.4103/2231-4040.111527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devi PU, Sharada AC, Solomon FE. In vivo growth inhibitory and radiosensitizing effects of withaferin A on mouse Ehrlich ascites carcinoma. Cancer Letters. 1995;95:189–93. doi: 10.1016/0304-3835(95)03892-Z. [DOI] [PubMed] [Google Scholar]
- 10.Sen N, et al. Apoptosis is induced in leishmanial cells by a novel protein kinase inhibitor withaferin A and is facilitated by apoptotic topoisomerase I-DNA complex. Cell Death Differ. 2007;14:358–367. doi: 10.1038/sj.cdd.4402002. [DOI] [PubMed] [Google Scholar]
- 11.Shi LH, Wu XJ, Liu JS, Gao YB. Withaferin A activates stress signalling proteins in high risk acute lymphoblastic leukemia. Int. J. Clin. Exp. Pathol. 2015;8:15652–15660. [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, et al. Withaferin A is a leptin sensitizer with strong antidiabetic properties in mice. Nat. Med. 2016;22:1023–1032. doi: 10.1038/nm.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuboyama T, Tohda C, Komatsu K. Neuritic regeneration and synaptic reconstruction induced by withanolide A. Br. J. Pharmacol. 2005;144:961–971. doi: 10.1038/sj.bjp.0706122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kour K, et al. Restoration of stress-induced altered T cell function and corresponding cytokines patterns by withanolide A. Int Immunopharmacol. 2009;9:1137–1144. doi: 10.1016/j.intimp.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Malik T, Pandey DK, Dogra N. Ameliorative potential of aqueous root extract of Withania somnifera against paracetamol induced liver damage in mice. Pharmacologia. 2013;4:89–94. doi: 10.5567/pharmacologia.2013.89.94. [DOI] [Google Scholar]
- 16.Gupta AP, Verma RK, Misra HO, Gupta MM. Quantitative determination of withaferin A in different plant parts of Withania somnifera by TLC densitometry. Journal of Medicinal and Aromatic Plant Sciences. 1996;18:788–790. [Google Scholar]
- 17.Chaurasiya ND, Sangwan RS, Misra LN, Tuli R, Sangwan NS. Metabolic clustering of a core collection of Indian ginseng Withania somnifera Dunal through DNA, isoenzyme, polypeptide and withanolide profile diversity. Fitoterapia. 2009;80:496–505. doi: 10.1016/j.fitote.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Bharti SK, Bhatia A, Tewari SK, Sidhu OP, Roy R. Application of HRMAS NMR spectroscopy for studying chemotype variations of Withania somnifera (L.) Dunal. Magn. Reson. Chem. 2011;49:659–667. doi: 10.1002/mrc.2817. [DOI] [PubMed] [Google Scholar]
- 19.Gupta P, Akhtar N, Tewari SK, Sangwan RS, Trivedi PK. Differential expression of farnesyl diphosphate synthase gene from Withania somnifera in different chemotypes and in response to elicitors. Plant Growth Regul. 2011;65:93–100. doi: 10.1007/s10725-011-9578-x. [DOI] [Google Scholar]
- 20.Siriwardane AS, Dharmadasa RM, Samarasinghe K. Distribution of withaferin A, an anticancer potential agent, in different parts of two varieties of Withania somnifera (L.) Dunal. grown in Sri Lanka. Pak. J. Biol. Sci. 2013;16:141–144. doi: 10.3923/pjbs.2013.141.144. [DOI] [PubMed] [Google Scholar]
- 21.Dhar N, et al. A decade of molecular understanding of withanolide biosynthesis and in-vitro studies in Withania somnifera (L.) Dunal: Prospects and perspectives for pathway engineering. Front. Plant Sci. 2015;6:1031. doi: 10.3389/fpls.2015.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senthil K, et al. Transcriptome analysis reveals in vitro cultured Withania somnifera leaf and root tissues as a promising source for targeted withanolide biosynthesis. BMC Genomics. 2015;16:14. doi: 10.1186/s12864-015-1214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel N, Patel P, Kendurkar SV, Khan BM. Overexpression of squalene synthase in Withania somnifera leads to enhanced withanolide biosynthesis. Plant Cell Tiss. Organ Cult. 2015;122:409–420. doi: 10.1007/s11240-015-0778-3. [DOI] [Google Scholar]
- 24.Mishra S, et al. RNAi and homologous over-expression based functional approaches reveal triterpenoid synthase gene cycloartenol synthase is involved in downstream withanolide biosynthesis in Withania somnifera. PLoS One. 2016;11:e0149691. doi: 10.1371/journal.pone.0149691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciddi V. Withaferin A from cell cultures of Withania somnifera. Indian J. Pharm. Sci. 2006;68:490–492. doi: 10.4103/0250-474X.27824. [DOI] [Google Scholar]
- 26.Sivanandhan G, et al. Chitosan enhances withanolides production in adventitious root cultures of Withania somnifera (L.) Dunal. Ind. Crop Prod. 2012;37:124–129. doi: 10.1016/j.indcrop.2011.11.022. [DOI] [Google Scholar]
- 27.Sivanandhan G, et al. Optimization of elicitation conditions with methyl jasmonate and salicylic acid to improve the productivity of withanolides in the adventitious root culture of Withania somnifera (L.) Dunal. Appl. Biochem. Biotechnol. 2012;168:681–696. doi: 10.1007/s12010-012-9809-2. [DOI] [PubMed] [Google Scholar]
- 28.Sivanandhan G, et al. Increased production of withanolide A, withanone, and withaferin A in hairy root cultures of Withania somnifera (L.) Dunal elicited with methyl jasmonate and salicylic acid. Plant Cell Tiss. Organ Cult. 2013;114:121–129. doi: 10.1007/s11240-013-0297-z. [DOI] [Google Scholar]
- 29.Grover A, Samuel G, Bisaria VS, Sundar G. Enhanced withanolide production by overexpression of squalene synthase in Withania somnifera. J. Biosci. Bioeng. 2013;115:680–685. doi: 10.1016/j.jbiosc.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Sivanandhan G, et al. Expression of important pathway genes involved in withanolides biosynthesis in hairy root culture of Withania somnifera upon treatment with Gracilaria edulis and Sargassum wightii. Plant Physiol. Biochem. 2015;91:61–64. doi: 10.1016/j.plaphy.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Redman RS, Sheehan KB, Stout RG, Rodriguez RJ, Henson JM. Thermotolerance generated by plant/fungal symbiosis. Science. 2002;298:1581. doi: 10.1126/science.1072191. [DOI] [PubMed] [Google Scholar]
- 32.Waller F, et al. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. USA. 2005;102:13386–13391. doi: 10.1073/pnas.0504423102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez RJ, et al. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008;2:404–416. doi: 10.1038/ismej.2007.106. [DOI] [PubMed] [Google Scholar]
- 34.Quecine MC, et al. Sugarcane growth promotion by the endophytic bacterium Pantoea agglomerans 33.1. Appl. Environ. Microbiol. 2012;78:7511–7518. doi: 10.1128/AEM.00836-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuklinsky-Sobral J, et al. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ. Microbiol. 2004;6:1244–1251. doi: 10.1111/j.1462-2920.2004.00658.x. [DOI] [PubMed] [Google Scholar]
- 36.Strobel G, Daisy B, Castillo U, Harper J. Natural products from endophytic microorganisms. J. Nat. Prod. 2004;67:257–268. doi: 10.1021/np030397v. [DOI] [PubMed] [Google Scholar]
- 37.Staniek A, Woerdenbag HJ, Kayser O. Endophytes: exploiting biodiversity for the improvement of natural product-based drug discovery. J. Plant Interact. 2008;3:75–93. doi: 10.1080/17429140801886293. [DOI] [Google Scholar]
- 38.Aly AH, Debbab A, Kjer J, Proksch P. Fungal endophytes from higher plants: a prolific source of phytochemicals and other bioactive natural products. Fungal Diversity. 2010;41:1–16. doi: 10.1007/s13225-010-0034-4. [DOI] [Google Scholar]
- 39.Pandey SS, et al. Endophytes of opium poppy differentially modulate host plant productivity and genes for the biosynthetic pathway of benzylisoquinoline alkaloids. Planta. 2016;243:1097–1114. doi: 10.1007/s00425-016-2467-9. [DOI] [PubMed] [Google Scholar]
- 40.Pandey SS, et al. Fungal endophytes of Catharanthus roseus enhance vindoline content by modulating structural and regulatory genes related to terpenoid indole alkaloid biosynthesis. Sci Rep. 2016;6:26583. doi: 10.1038/srep26583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.del Giudice L, et al. The microbial community of Vetiver root and its involvement into essential oil biogenesis. Environ. Microbiol. 2008;10:2824–2841. doi: 10.1111/j.1462-2920.2008.01703.x. [DOI] [PubMed] [Google Scholar]
- 42.Tiwari R, et al. Endophytic bacteria from Ocimum sanctum and their yield enhancing capabilities. Curr. Microbiol. 2010;60:167–71. doi: 10.1007/s00284-009-9520-x. [DOI] [PubMed] [Google Scholar]
- 43.Tiwari R, et al. Bacterial endophyte-mediated enhancement of in planta content of key terpenoid indole alkaloids and growth parameters of Catharanthus roseus. Industrial Crops and Products. 2013;43:306–310. doi: 10.1016/j.indcrop.2012.07.045. [DOI] [Google Scholar]
- 44.Collier R, Fuchs B, Walter N, Kevin-Lutke W, Taylor CG. Ex vitro composite plants: an inexpensive, rapid method for root biology. Plant J. 2005;43:449–457. doi: 10.1111/j.1365-313X.2005.02454.x. [DOI] [PubMed] [Google Scholar]
- 45.Mellor KE, Hoffman AM, Timko MP. Use of ex vitro composite plants to study the interaction of cowpea (Vigna unguiculata L.) with the root parasitic angiosperm Striga gesnerioides. Plant Methods. 2012;8:1–22. doi: 10.1186/1746-4811-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor CG, Fuchs B, Collier R, Lutke WK. Generation of composite plants using. Agrobacterium rhizogenes. Methods Mol. Biol. 2006;343:155–168. doi: 10.1385/1-59745-130-4:155. [DOI] [PubMed] [Google Scholar]
- 47.Bhat WW, et al. Molecular cloning, bacterial expression and promoter analysis of squalene synthase from Withania somnifera (L.) Dunal. Gene. 2012;499:25–36. doi: 10.1016/j.gene.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Singh S, et al. Sterol partitioning by HMGR and DXR for routing intermediates toward withanolide biosynthesis. Physiol. Plant. 2014;152:617–633. doi: 10.1111/ppl.12213. [DOI] [PubMed] [Google Scholar]
- 49.Cordoba E, Salmi M, León P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 2009;60:2933–2943. doi: 10.1093/jxb/erp190. [DOI] [PubMed] [Google Scholar]
- 50.Pal S, et al. Nitrogen treatment enhances sterols and withaferin A through transcriptional activation of jasmonate pathway, WRKY transcription factors, and biosynthesis genes in Withania somnifera (L.) Dunal. Protoplasma. 2017;254:389–399. doi: 10.1007/s00709-016-0959-x. [DOI] [PubMed] [Google Scholar]
- 51.Pankajavalli T, Kalaiselvi R, Pradeepa D, Kalaiselvi S. Effect of exogenous indole-3-butyric acid and indole-3-acetic acid on biomass and legendary withanolides from in vitro root cultures of Withania somnifera-jawahar 20 cultivar. Int. J. Pharm. Bio. Sci. 2014;5:971–979. [Google Scholar]
- 52.Singh AK, et al. Virus induced gene silencing of Withania somnifera squalene synthase negatively regulates sterol and defence related genes resulting in reduced withanolides and biotic stress tolerance. Plant Biotechnol. J. 2015;13:1287–1299. doi: 10.1111/pbi.12347. [DOI] [PubMed] [Google Scholar]
- 53.Ryder N. Terbinafine: mode of action and properties of the squalene epoxidase inhibition. Br. J. Dermatol. 1992;126:2–7. doi: 10.1111/j.1365-2133.1992.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 54.He F, Zhu Y, He M, Zhang Y. Molecular cloning and characterization of the gene encoding squalene epoxidase in Panax notoginseng. DNA Seq. 2008;19:270–273. doi: 10.1080/10425170701575026. [DOI] [PubMed] [Google Scholar]
- 55.Han JY, In JG, Kwon YS, Choi YE. Regulation of ginsenoside and phytosterol biosynthesis by RNA interferences of squalene epoxidase gene in Panaxginseng. Phytochemistry. 2010;71:36–46. doi: 10.1016/j.phytochem.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 56.Takemura T, Chow YL, Todokoro T, Okamoto T, Sato F. Over-expression of rate-limiting enzymes to improve alkaloid productivity. Methods Mol. Biol. 2010;643:95–109. doi: 10.1007/978-1-60761-723-5_7. [DOI] [PubMed] [Google Scholar]
- 57.Guengerich F. Uncommon P450-catalyzed reactions. Curr. Drug Metab. 2001;2:93–115. doi: 10.2174/1389200013338694. [DOI] [PubMed] [Google Scholar]
- 58.Hrycay EG, Bandiera SM. The monooxygenase, peroxidase, and peroxygenase properties of cytochrome P450. Arch. Biochem. Biophys. 2012;522:71–89. doi: 10.1016/j.abb.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 59.Friesen JA, Rodwell VW. The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome Biol. 2004;5:248. doi: 10.1186/gb-2004-5-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahlawat S, Saxena P, Ali A, Abdin MZ. Piriformospora indica elicitation of withaferin A biosynthesis and biomass accumulation in cell suspension cultures of Withania somnifera. Symbiosis. 2016;69:37–46. doi: 10.1007/s13199-015-0364-9. [DOI] [Google Scholar]
- 61.Alpizar E, et al. Efficient production of Agrobacterium rhizogenes-transformed root and composite plants for studying gene expression in coffee roots. Plant Cell Rep. 2006;25:959–967. doi: 10.1007/s00299-006-0159-9. [DOI] [PubMed] [Google Scholar]
- 62.Sinharoy S, Saha S, Chaudhury SR, DasGupta M. Transformed hairy roots of Arachis hypogea: A tool for studying root nodule symbiosis in a non-infection thread legume of the Aeschynomeneae tribe. Mol. Plant-Microbe Int. 2009;22:132–142. doi: 10.1094/MPMI-22-2-0132. [DOI] [PubMed] [Google Scholar]
- 63.Geng L, et al. Efficient production of Agrobacterium rhizogenes-transformed roots and composite plants in peanut (Arachis hypogaea L.) Plant Cell Tiss. Organ Cult. 2012;109:491–500. doi: 10.1007/s11240-012-0113-1. [DOI] [Google Scholar]
- 64.Kulkarni AA, Thengane SR, Krishnamurthy KV. Direct shoot regeneration from node, internode, hypocotyl and embryo explants of Withania somnifera. Plant Cell Tiss. Organ Cult. 2000;62:203–209. doi: 10.1023/A:1006413523677. [DOI] [Google Scholar]
- 65.Saritha KV, Naidu CV. In vitro flowering of Withania somnifera Dunal.-An important antitumor medicinal plant. Plant Science. 2007;172:847–851. doi: 10.1016/j.plantsci.2006.12.016. [DOI] [Google Scholar]
- 66.Rao S, Teesta VK, Bhattrai A, Khushi K, Bhat S. In vitro propagation of Withania somnifera and estimation of withanolides for neurological disorders. J. Pharmacog. 2012;3:85–87. [Google Scholar]
- 67.Lichtenthaler, H. K. & Buschmann, C. in Current Protocols in Food Analytical Chemistry (eds Wrolstad, R. E et al.) F4.3.1–F4.3.8 (New York: John Wiley & Sons, 2001).
- 68.Pandey H, Pandey P, Pandey SS, Singh S, Banerjee S. Meeting the challenge of stevioside production in the hairy roots of Stevia rebaudiana by probing the underlying process. Plant Cell Tiss. Organ Cult. 2016;126:511–521. doi: 10.1007/s11240-016-1020-7. [DOI] [Google Scholar]
- 69.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 70.Pandey P, et al. Long-term stability in biomass and production of terpene indole alkaloids by hairy root culture of Rauvolfia serpentina and cost approximation to endorse commercial realism. Biotechnol. Lett. 2014;36:1523–1528. doi: 10.1007/s10529-014-1495-4. [DOI] [PubMed] [Google Scholar]
- 71.Scartezzini. Genetic and phytochemical difference between some Indian and Italian plants of Withania somnifera (L.) Dunal. Nat. Prod. Res. 21, 923–932 (2007). [DOI] [PubMed]
- 72.Srivastava P, et al. Simultaneous quantification of withanolides in Withania somnifera by a validated high-performance thin-layer chromatographic method. J. AOAC Int. 2008;91:1154–1161. [PubMed] [Google Scholar]
- 73.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2∆∆Ct method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 74.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 75.Pikovskaya RI. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Microbiologiya. 1948;17:362–370. [Google Scholar]
- 76.Tang YW, Bonner J. The enzymatic inactivation of indoleacetic acid; some characteristics of the enzyme contained in pea seedlings. Arch. Biochem. 1947;13:11–25. [PubMed] [Google Scholar]
- 77.Conn HJ, Breed RS. The use of the nitrate-reduction test in characterizing bacteria. J Bacteriol. 1919;4:267–290. doi: 10.1128/jb.4.3.267-290.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gusberti FA, Syed SA. Development of a miniaturized nitrate reduction test for the identification of oral bacteria. J. Microb. Methods. 1984;2:333–338. doi: 10.1016/0167-7012(84)90052-6. [DOI] [Google Scholar]
- 79.Nhan LV, et al. Phytotoxic mechanism of nanoparticles: Destruction of chloroplasts and vascular bundles and alteration of nutrient absorption. Sci. Rep. 2015;5:11618. doi: 10.1038/srep11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.