Abstract
Histomorphometry and Micro-CT are commonly used to assess bone remodeling and bone microarchitecture. These approaches typically require separate cohorts of animals to analyze 3D morphological changes and involve time-consuming immunohistochemistry preparation. Intravital Microscopy (IVM) in combination with mouse genetics may represent an attractive option to obtain bone architectural measurements while performing longitudinal monitoring of dynamic cellular processes in vivo. In this study we utilized two-photon, multicolor fluorescence IVM together with a lineage tracing reporter mouse model to image skeletal stem cells (SSCs) in their calvarial suture niche and analyze their differentiation fate after stimulation with an agonist of the canonical Wnt pathway (recombinant Wnt3a). Our in vivo histomorphometry analyses of bone formation, suture volume, and cellular dynamics showed that recombinant Wnt3a induces new bone formation, differentiation and incorporation of SSCs progeny into newly forming bone. IVM technology can therefore provide additional dynamic 3D information to the traditional static 2D histomorphometry.
Introduction
Bone histomorphometry is commonly used to quantitatively assess changes in bone microarchitecture, bone morphogenesis, remodeling, and metabolism1. It offers the opportunity to perform a broad range of measures, including bone formation rate and quantification of cellular compositions; therefore, it has been an important tool in evaluating bone turnover2 under genetic manipulations3, presence of cancer4, or pharmacological intervention in bone metabolic diseases5. Histomorphometry relies on ex vivo post-mortem processing and typically involves time-consuming sample preservation and histological preparation, which potentially requires optimization trials to minimize distortion of microstructures1. In addition, as thin-sliced bones of 5 to 10 μm are usually used, the conventional approach inherently does not provide 3-dimensional parameters. Volumetric analysis of bone microarchitecture can be accomplished using micro-computed tomography (μCT) aided by finite element analysis6,7 and advanced reconstruction algorithms. Recent advances such as high-resolution in vivo μCT8–10 and high-resolution peripheral quantitative computed tomography (HR-pQCT)11,12 further allow time-lapse studies, with spatial resolution of tens of micrometers. However, radiation dose minimization remains an issue because of the potential impact of radiation on bone metabolism13. In addition, Micro-CT only provides morphological information, thus immunohistochemistry is still required to identify various cell populations.
Intravital microscopy (IVM) can overcome the limitations mentioned above14,15. Laser scanning confocal (LSCM) or multi-photon fluorescence microscopy (MPM) is capable of achieving optical sectioning with diffraction-limited spatial resolution, which renders 3D imaging of sub-cellular resolution after reconstruction16. This feature makes IVM a suitable tool for evaluating histology in vivo. For example, LSCM-based IVM has been used in clinical settings such as in dermatology17 and gastroenterology18. IVM, by acquiring real-time videos or time-lapse movies, can help visualizing and tracking biological processes at various time scales from milliseconds to days and weeks14,19. With advancement in transgenic mouse models15,20 and fluorescent probes to visualize cellular and molecular targets, IVM has been used as a powerful tool in examining dynamic cellular events, such as cell division21, migration22,23, and cell fate24 in the fields of immunology, stem cell biology, and bone research. For instance, IVM enables characterization of stem cell niche in the bone marrow cavities14,25, where expression of fluorescent proteins or exogenous fluorescent probe labeling at separable spectra allows for identification of various cells and cell lineages. Bone is readily identified by second harmonic generation (SHG) of type I collagen due to the organization of the collagen fibrils and the noncentrosymmetric molecular structures26. IVM has been used with fluorescent pH sensors27, or transgenic pH reporter mice28, to quantify local acidity and study bone resorption as well as osteoblast-osteoclast interactions29. More recently, IVM has also been employed to investigate the dynamic interactions between angiogenesis and osteoblast cells expressing 2.3Col1-GFP over a 9-week time-course study of calvarial defect healing30.
The cranial suture has recently been identified as a stem cell niche for calvarial development31–33. Our recent finding using IVM revealed that calvarial skeletal stem cells expressing Prx1 (Prx1 + SSCs), a transcription factor essential in early limb bud development34,35, are located exclusively in the cranial sutures, and respond by up-regulating osteogenesis markers upon calvarial subperiosteal injection of mouse recombinant Wnt3a (rmWnt3a), an agonist of the canonical Wnt/β-catenin pathway33.
Canonical Wnt/β-catenin signaling has been shown to tightly regulate skeletal development and is well known to facilitate self-renewal of multi-potent mesenchymal stem cells (MSCs), direct the MSC cell fate towards osteoblastic lineage, enhance proliferation of osteoblast precursors, and inhibit adipogenesis, chondrogenesis, and osteoclastic bone resorption in a context dependent manner36–38. In calvarial development, genetic39 or pharmaceutical activation40 of Wnt signaling increases differentiation of adult osteoblasts and osteogenesis.
Here we describe an in vivo histomorphometric approach to evaluate the effects of the activation of Wnt signaling on calvarial bone formation and cell composition. To do so we examine the effect of canonical Wnt signaling on Prx1 + SSCs and their progeny of the calvarial bone using two-photon IVM to simultaneously (i) visualize two different cell populations, (ii) track their cellular fate, and (iii) quantify cell number and dynamics of bone growth. We used a lineage tracing reporter mouse model based on the Prx1-creER-EGFP;Rosa-loxP-STOP-loxP-tdTomato mouse line33. In this mouse line, upon treatment with tamoxifen, the CreER recombinase excises a STOP cassette inducing overexpression of tdTomato fluorescent proteins in Prx1 + cells and their progeny. We evaluated Prx1 + cells and their progeny in the coronal suture for their contribution in bone formation when exogenous rmWnt3a is administered locally, by calvarial subperiosteal injections. Dynamic changes in terms of cell incorporation rate into the new bone, bone growth, population expansion, and suture volume are measured in live animals before and after administration of rmWnt3a.
Results
In vivo, multi-color two-photon imaging enables longitudinal tracking of cell differentiation and bone morphometry
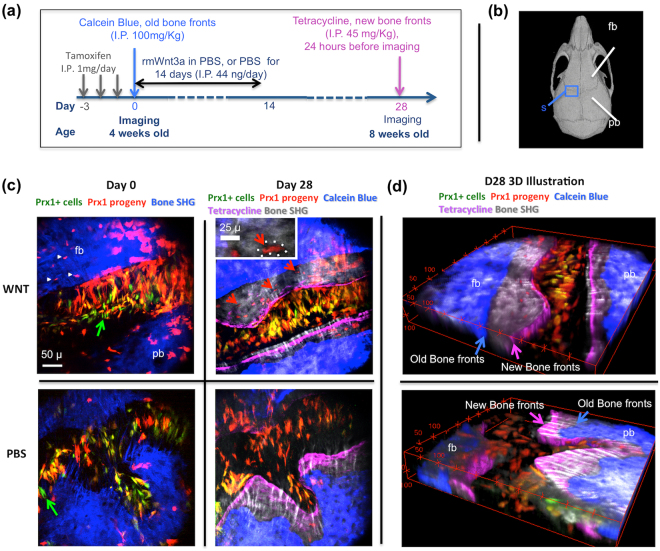
Lineage tracing reporter mice (Prx1-creER-EGFP mice crossed into Rosa26LoxP-STOP-loxP-tdtomato) were treated with Tamoxifen for 3 days to allow visualization of Prx1 + cells and their progeny with tdTomato fluorescence (Fig. 1a). Multi-color two-photon images of the coronal suture space (Fig. 1b, blue rectangle) were acquired at Day 0 and Day 28 using IVM before and after treating the animals with rmWnt3a or PBS for 14 days. Tetracycline and calcein, two calcium-binding fluorochromes that stain the bone mineralization fronts41, are administered intraperitoneally on Day 0 and Day 28, respectively, to define the old and new bone fronts. As shown in Fig. 1c, IVM revealed dynamic changes of bone structures visualized by SHG (Blue at Day0, Gray at Day28), old bone fronts stained by calcein blue (Blue, which corresponds to the SHG structure at Day0), new bone fronts labeled by tetracycline (Purple), the skeletal stem cells expressing Prx1-creER-EGFP (Green), their progeny expressing tdTomato fluorescence (Red or Yellow if co-expressing GFP), and the contribution of Prx1 progeny to bone formation shown as tdTomato expressing (tdTomato+) osteocytes that reside in the lacunae42,43 in the newly formed bones (red arrows). As observed in Fig. 1c, a higher number of osteocytes were descendants of Prx1 + cells in animals treated with rmWnt3a. IVM stack-images up to a depth of 50-μm enable quantification of histomorphometric parameters including cell number, bone volume, and suture volume (Fig. 1d).
Figure 1.
In vivo, multi-color two-photon imaging. (a) Injection regimens and imaging time points for in vivo histomorphometry. (b) The coronal suture space (S) between frontal (fb) and parietal (pb) bones. (c) Animals were treated with either rmWnt3a (in PBS, 44 ng/day) or just PBS for 14 days. Maximum intensity projection of a 25-μm layer was used for demonstration. Images on Day0 show Prx1 + cells (Green), their progeny (Red or yellow, co-expression of tdTomato and EGFP). In addition, on Day 28, the initial bone structure was identified by calcein blue (Blue) and the new bone fronts were demarcated by tetracycline (purple), as indicated by blue and purple arrows in Fig. 1(d). Spontaneous recombination (white arrowheads) and incomplete recombination (green arrows) may be present. It is noted that a higher number of osteocytes originated from Prx1 + cells were embedded in lacunae (the red arrows and the inset) from rmWnt3a treatment. (d) Quantifications of bone morphometry and cellular dynamics were performed over a 50-μm layer, as shown in 3D reconstructed images.
Of note, we observed various extent of incomplete (Prx1 + cells without tdTomato fluorescence) and spontaneous recombination (tdTomato + osteocytes present in the old bone) between regions and animals before treatment with rmWnt3a or PBS, as seen on Day 0 images in Fig. 1c. IVM therefore presents a major advantage that enables normalization of the measured parameters on D28 to the baseline values on Day 0, thus minimizing inter-regional and inter-subject variability during result interpretation.
Image processing and quantitative analysis
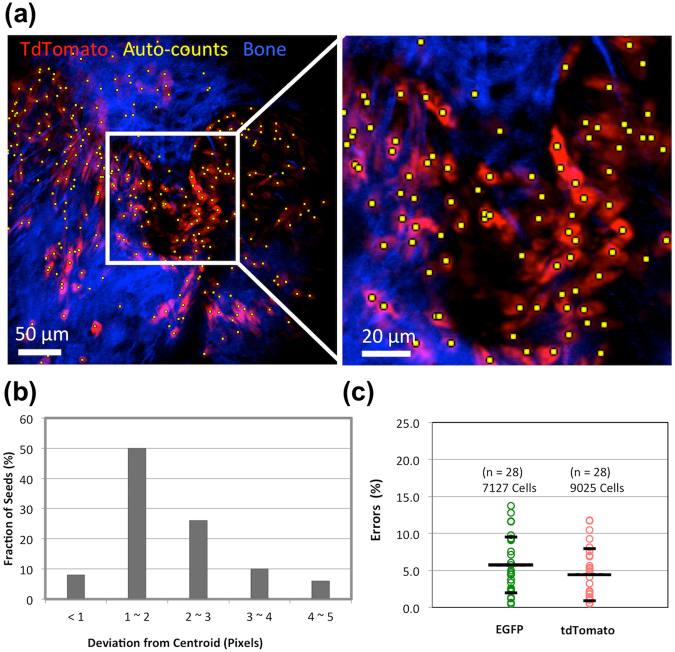
To quantify cell dynamics, image processing and automatic cell counting were performed to retrieve dynamic changes of each cell population. Figure 2a displays representative images showing “seeds” identified by automatic cell counting (yellow dots) overlaid on the tdTomato + cells (red). The identified seed reflects the 3D location of the strongest fluorescence emission found in each cell (Supplementary video 1). For 2D demonstration, the tdTomato + cells within the region of interest are displayed by maximum intensity projection (from 0 to 50 μm) and then superimposed with all the identified seeds. A zoomed region shows that the seeds identify the center of each cell in 3D, with a mean deviation of only 2 pixels in xyz coordinates when validated against the centroid of individual cells manually segmented then measured using Image J (3D Object Counter\Centroid\Statistics) (Fig. 2b).
Figure 2.
Image processing and quantitative analysis. Automatic cell counting adapted from Image J Plugins (a) Overlay of the identified seeds (yellow) with tdTomato + cells. Images were presented using maximum intensity projection. (b) Validation of automatic seed identification against 50 manually identified cells. (c) The counting accuracy of 5.7% ± 3.8% in the EGFP group, and 4.4% ± 3.5% in the tdTomato group was validated by 28 randomly picked regions (a total of 7127 and 9025 cells, respectively). Error bars represent standard deviation.
To investigate the accuracy of automatically counted values, the results were compared with manual annotations of Prx1 + and tdTomato + cells from 28 randomly picked regions, corresponding to a total of 7127 (Prx1) and 9025 (tdTomato) cells. In Fig. 2c, we show the counting accuracy between automatic counting and the ground truth was 5.7% ± 3.8% in the GFP group, and 4.4% ± 3.5% in the tdTomato group. We used standardized threshold adjustment based on Tukey’s anomaly (Methods and Supplementary Fig. 1), which performed better than global thresholding, enabling better identification of the seeds of dim cells (Supplementary Fig. 2). An algorithm that considers size variation of cells was also utilized to significantly improve the counting accuracy (Supplementary Fig. 3).
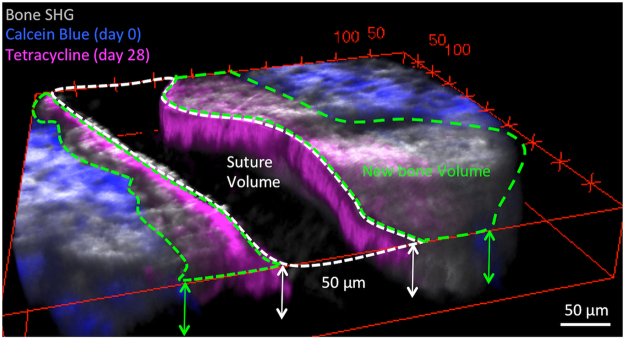
To assess bone formation and bone volume, the bone fronts at Day 0 and Day 28 were manually segmented based on calcein blue and tetracycline margins, then the bound area between the bone fronts were calculated and summed over all the 3D slices to yield the volumes of the total newly formed bones, growth of frontal or parietal bones, and the coronal suture (Fig. 3). Automatic segmentation was not performed for volumetric analysis due to discrete staining pattern typically present by tetracycline labeling (Supplementary Fig. 4., indicated by purple arrows). In regions of missing tetracycline signal, the simultaneously acquired bone SHG signal has served to delineate the new bone fronts. Automated segmentation of the SHG images was hampered however by the bright collagen fibers in the suture space (Supplementary Fig. 4., indicated by white arrows), typically at perpendicular orientation to the bone fronts.
Figure 3.
3D quantifications of bone morphometry. Volume measurement is demonstrated by a 3D reconstructed image. The bone fronts were manually segmented in each z-slice. The white dashed lines represent the suture area, and the green dashed lines represent the new bone area in the first slice. All the measured areas were summed over 50 μm in z dimension and converted to mm3.
Dynamic changes of bone histomorphometry in response to rmWnt3a
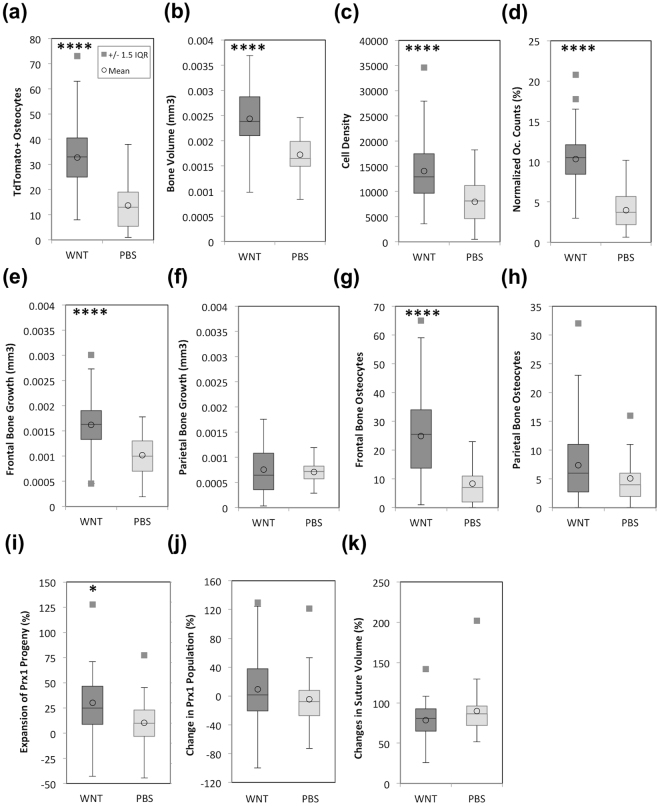
It is observed in Fig. 1 that local delivery of rmWnt3a promotes incorporation of Prx1 progeny during bone formation, as a higher number of tdTomato + osteocytes were found in the newly formed bone (red arrows, Fig. 1c). In Fig. 4, we processed the acquired intravital images to yield quantitative analysis. The box plots demonstrate the mean, median, the interquartile range, the whiskers showing the total range of the measurements, and the individual data points greater or less than by 1.5 times the interquartile range.
Figure 4.
Dynamic changes of bone histomorphometry in response to rmWnt3a. Quantitative measurement of dynamic changes through a period of 28 days in response to rmWnt3a treatment compared to the control group. The box plots demonstrate the mean, median, the interquartile range, the whiskers showing the total range of the measurements, and the individual data points greater or less than by 1.5 times the interquartile range. (a) tdTomato + osteocytes derived from Prx1 progeny (b) Bone growth (c) Cell density, tdTomato + osteocytes normalized to bone growth (d) tdTomato + osteocytes normalized to initial cell count at Day 0 (e) Frontal bone growth (f) Parietal bone growth (g,h) Osteocytes incorporated to frontal bone and parietal bone, respectively (i) Expansion of Prx1 progeny (j) Changes of Prx1 + population (k) Changes in suture volume. (N = 6 and 5 animals in rmWnt3a and PBS treated groups, respectively. 6 to 8 regions in the coronal suture of each animal were acquired).
As shown in Fig. 4(a–c), compared to PBS treated animals, rmWnt3a treated animals showed an increased number of Prx1 progeny as osteocytes (Fig. 4a) (33 ± 14 vs. 14 ± 9, p < 0.00005). In addition, 3-dimensional bone volume measurements through a 50-μm z-stack revealed that treatment with rmWnt3a increased bone apposition by 41% (p < 0.00005)(Fig. 4b). The increase in the number of Prx1 progeny as osteocytes within the newly formed bone was not simply due to bone growth, as the cell density, calculated by normalizing the contribution of Prx1 progeny to bone growth, also increased by 77% (P < 0.0005) compared to the control group (Fig. 4c). Note that the quantification is not affected by pre-existing tdTomato + osteocytes in the old bone, (Fig. 1c), since only the ones within the new bone volume (Fig. 3) were included in the analysis. By imaging the same animal at different time points, IVM further avoids bias that might arise from variations of baseline cell numbers before rmWnt3a treatment. Figure 4d presents the normalized cell counts of tdTomato + osteocytes to the initial tdTomato + cell number of the region, and shows a significantly higher fraction of tdTomato + cells that have committed cell fate to become osteocytes (10% ± 4% vs. 4% ± 2%, p < 0.00005).
Interestingly, and confirming the differential effects of agonist Wnt signals on the neuro-ectodermal derived frontal bone versus the mesoderm derived calvarial bone44, rmWnt3a had a differential effect on frontal and parietal bones: the former showed an increase of 59% (p < 0.00005) compared to the control group whereas the latter showed no significant increase (Fig. 4e–f). This discrepancy in bone growth correlated with the number of Prx1 progeny as osteocytes (Fig. 4g,h, p < 0.00005). Incorporation rate of tdTomato + cells into osteocytes in frontal bones (tdTomato + Osteocytes/new frontal bone volume) in rmWnt3a treated animals was found to be 56% higher than the rate in parietal bones. In contrast, only 15% increase was found in the control group treated with PBS (data not shown).
The total number of tdTomato + cells, representing Prx1 + skeletal stem cells and their progeny, was found to go through substantial expansion (30%) after rmWnt3a treatment in comparison with the PBS treated animals (10%). In contrast, no significant difference was observed when just comparing the Prx1 + population (9% in the group given rmWnt3a as opposed to −5% in the control group) (Fig. 4i,j). In analyzing the expansion of Prx1 progeny, all tdTomato + cells in the 3D volume were counted without excluding the pre-existing cells in the old bone, which were present in small numbers and did not significantly affect the outcome of the analysis. However, changes in population were normalized to the baseline cell number in order to take into account regional variations in cell numbers at Day 0. Lastly, treatment with rmWnt3a yielded little or no changes in suture volume at this age (Fig. 4k).
Discussions
This work used two-photon, multicolor IVM as a novel histomorphometric tool to quantify changes in bone anatomical structures and track cell fate in 3D. We measured histomorphometric parameters through a 50 μm-thick bone layer using live animals, and obtained dynamic 3D volumetric analysis via image acquisition and processing at longitudinal time points. Bone morphology in 3D is visualized by SHG of collagen, with the old and new bone fronts demarcated by double staining with calcein blue (depicted in blue in the figures) and tetracycline (depicted in purples in the figures). In vivo tracking of cell function is further enabled by the lineage tracing reporter mouse model. Using IVM and image processing, we showed that local delivery of rmWnt3a promotes expansion of Prx1 progeny and their incorporation in bone formation at the adolescent age, and that this effect is more prominent in the frontal bone compared to the parietal bone.
Canonical Wnt signaling has been extensively studied and known for facilitating osteogenesis or inhibiting endochondral bone formation in calvarial sutures. Its acts on specific skeletal cell population in a context dependent manner3,37,40,45–47. In the shown experiments, the quantitative results from IVM are consistent with the results of our previous study, where post-natal Prx1 + cells were shown to go through osteo-differentiation when treated with rmWnt3a in vitro and in vivo33. Interestingly, adding exogenous rmWnt3a led to significantly more bone growth of the frontal bone, whereas such a change was not observed in the parietal bone compared to PBS treated group. Our results suggest that elevated canonical Wnt signaling may control predominantly morphogenesis of the anterior skull.
A number of factors may affect the quantifications. Mounting and stabilization of mouse calvaria is critical to obtain consistent field of view for cell counting between measurements. Additional attention should be paid to spectral separation between fluorochromes. For example, Prx1-EGFP fluorescence is much weaker than tdTomato fluorescence in this study; hence the detection gate of EGFP fluorescence was moved towards shorter wavelengths (480 nm–520 nm) instead of the typical green channel (500 nm–550 nm) to avoid leakage of tdTomato fluorescence into the EGFP channel. Careful selections of fluorochrome combinations for labeling bone fronts is also important to avoid spectral overlap with other cells of interest.
In Table 1, we summarize and compare pros and cons of our newly developed IVM 3D histomophometry with traditional ex vivo histomorphometry. Briefly, traditional histomorphometry is performed using separate cohorts of animals for analysis at longitudinal time points and lengthy sample preparation for histomorphometric analysis is also required. More importantly, by means of IVM, dynamic changes of bone and cellular events are acquired with minimal perturbation of natural physiological conditions. Although numerous advantages are present in the IVM approach, it should be noted that conventional histomorphometry possesses inherent flexibility in exploring multiple cellular/molecular targets of interest. For instance, fluorescence labeling of multiple cell populations can be achieved by immunohistochemistry, with fewer limitations in terms of the “types” of targets, as long as the associated biological markers can be identified and labeled. Ex vivo methodology with the help of a variety of fluorescent or chromogenic contrast agents can provide contrast between cartilages, mineralized bones, and osteoids. Similarly, IVM with additions of various modalities will allow studies of these structures. For example, SHG has been used in characterizing bone ultrastructure48, aging effects of cortical bones49, and the extracellular matrix50. IVM equipped with Raman scattering detection has allowed direct observation of mineralization processes51,52. In addition, many common chromogenic reagents are fluorescent41,53, such as Toluidine Blue for cartilage staining, and Alizarin Red utilized in discriminating mineralized bone and osteoid seam, and therefore they could become feasible for IVM 3D histomorphometry. Furthermore, IVM is also capable of imaging other non-cellular structures such as lipids (commonly stained with Oil Red in ex vivo histomorphometry) via the use of third harmonic generation (THG), Coherent Anti-stokes Raman Scattering (CARS) and development of optical biosensors54–57. In many occasions, in vivo immunohistochemistry can be achieved by local33 or systemic delivery of fluorescent antibodies58,59, given that specific surface receptors are present in vivo. In terms of tissue accessibility (i.e. tissue depth of the analyses), in conventional ex vivo histomophometry, a series of sections obtained throughout the specimens is commonly used. IVM has been shown to efficiently identify cell activity throughout various tissues and bone, such as in tibial bone27,29, in tibial bone marrow60, as well as in calvarial bones61. Signal attenuation of SHG in bone is more apparent due to its high scattering coefficient. The thickness of 50-μm used in this study ensured clear visualization of bone structures, and also standardized the measured suture areas as the coronal suture at this depth fits in one single field of view. Overall, the achievable imaging depth from two-photon excitation is typically from tens to hundreds of microns based on tissue optical properties62,63. Breakthrough in imaging depth will be an upmost advantage to allow thorough 3D analysis. For instance, the image depth can be further improved with 3-photon microscopy64. Last but not least, continuing advances in image processing of microscopic data, including the development of novel quantifying 3D algorithms65, would make IVM 3D histomophometry a promising tool to study bone metabolism and bone regeneration.
Table 1.
Comparison of ex vivo and in vivo histomorphometry.
| Pros | Cons | |
|---|---|---|
| IVM | Allowing longitudinal tracking of dynamic cell behavior. Simultaneous acquisition of histology and morphometric parameters. Well established in imaging calvaria. Also allows short-term tracking (within a week) of long bones. Retrieving 3D information is relatively more straightforward. | Relying on genetic mouse models and/or multi- modalities to probe targets of interest. Access to deep tissue is limited by penetration depth of light. |
| Ex vivo | Generating good contrast of mineralized bone and osteoids by chromogenic staining. Well established in imaging both long bones and calvaria. Flexibility in exploring a wide range of cell targets by using immunohistochemistry. | Requiring separate cohorts of animals for longitudinal histological studies although some morphometric parameters can be analyzed by micro-CT. Relatively more time-consuming in sample preparation. |
Conclusion
When tracking the cell fate and bone growth for a period of 28 days before and after rmWnt3a administration, IVM revealed an increased bone growth and an increased incorporation rate of osteolineage cells. Using customized cell counting algorithms, we showed rmWnt3a led to expansion of Prx1 progeny, but not the Prx1 + cells. We conclude that in vivo IVM histomorphometry analysis allows for simultaneous visualization of histology and morphometric information and may be utilized to study the effects of anabolic or catabolic signaling during calvarial bone formation.
Materials and Methods
Animal models
All animal experiments were conducted in compliance with the institutional guidelines and approved by the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School (IACUC approval IS00000535) and Massachusetts General Hospital (IACUC approval 2015N000098). Lineage tracing was performed using Prx1-creER-EGFP mice crossed into Rosa26LoxP-STOP-loxP-tdtomato (tdTomato) mice33. Both heterozygous and homozygous Prx1-creER-EGFP mice were used for this study. As homozygous Prx1-creER-EGFP mice may present more GFP cells, homozygous animals were divided evenly to Wnt and PBS groups. Also, since Prx1 is expressed in ovary and the creER is sensitive to estrogen, to minimize spontaneous recombination only male mice were utilized throughout the studies.
Reagents
10 mg/mL tamoxifen (T5648, Sigma, St. Louis, MO) was prepared in sterile oil. Calcein blue power (M1255, Sigma) was dissolved in 2% sodium bicarbonate solution to make a 3% working concentration. Mouse recombinant Wnt3a (1324-WN-002, R&D systems, Minneapolis, MN) was prepared at a concentration of 0.44 μg/mL in DPBS. 7.5 mg/mL tetracycline (T7660, Sigma) was prepared in sterilized water and NaCl was added to obtain isotonic solution immediately before injection.
In vivo histomorphometry
As illustrated in Fig. 1, 4-week-old Prx1-creER-EGFP;tdTomato male mice were injected with tamoxifen (40 mg/kg in sterile oil, I.P.) for 3 consecutive days. Starting at Day 0, animals were treated with calvarial subperiosteal injections of either rmWnt3a (in PBS, 44ng/day) or the vehicle control (PBS) for 14 days. Calcein blue (100 mg/Kg, I.P.) was used to label the existing bone fronts on Day 0, and tetracycline (45 mg/Kg, I.P.) was administered 24 hours before the follow-up imaging (Day 28) to stain the new bone fronts.
Intravital microscopy
IVM was performed as previously described at the time points Day 0 and Day 2814. In the IVM setup, the mouse head was stabilized in a mouse restrainer and maintained in anesthesia using 1.25% vaporized isofluorane (Fig. 2(a)). A skin flap was created without damaging the periosteum for en face acquisition of the coronal suture, the patent space between frontal (fp) and parietal bones (pb), as marked by the blue box in Fig. 2(b). Figure 2(c) shows 3D visualization of a sample region, containing the coronal suture and the Prx1 + cells in a live animal. Specifically, the excitation beam was focused into the sample plane using a 60x objective lens (NA = 1), yielding a field of view of 400 × 400 μm. Fluorescence emission was then collected by photomultiplier tubes with proper dichroic and filter settings corresponding to fluorophores of interest. A Ti:Sapphire laser pulsing at 900 nm was used for acquiring SHG of collagen, fluorescence of EGFP and tdTomato, which was detected using 415–455 nm, 480–520 nm, and 605–650 nm band pass filters, respectively. The settings of the emission filters were intended to avoid signal crosstalks between EGFP and tdTomato fluorescence, which could be a confounding factor in cell counting. Using the same set of blue and green emission filters, 775 nm laser pulses were used to excite calcein blue (blue channel) and tetracycline (green channel). All image stacks were acquired at 1 μm interval starting from the surface of the skull. Image analysis was performed within the 50-μm layer, where the coronal suture was placed at the center of the field of view with no obvious tilt horizontally.
Quantitative analysis
Multi-channel z-stack images were rendered and analyzed using Fiji66. Calcein blue emission leaked into the green detector was first subtracted before overlaying all emission channels. Osteocytes and mature osteoblasts expressing tdTomato were identified in the newly formed bone demarcated by calcein blue and tetracycline staining. When evaluating the incorporation of Prx1 progeny in bone formation, tdTomato expressing (tdTomato+) mature osteoblast cells localizing right at the border lines marked by calcein blue and tetracycline were included, whereas cells appeared in the vessel walls, bone marrow cavities, and in the old bones were excluded. The incorporation rate was presented as “cell density”, calculated by the number of tdTomato+ osteocytes and osteoblasts divided by the new bone volume (shown in Fig. 4). The fractional changes of all the measured parameters (ΔM), including expansion of different cell populations and the suture volume, were calculated by:
Image processing for automatic cell counting
Automatic cell counting of Prx1 + and tdTomato + cells in each stack was obtained using Image J Macros containing multiple Fiji plugins, including Moment segmentation, 3D image J suite, and 3D object counter. Specifically, cell areas were first segmented using the built-in segmentation algorithm “Moment” in Image J, followed by standardized contrast enhancement to allow 1.2% of saturated pixels in each stack. This step minimizes oversaturation, improves visibility of dim cells while not over-enhancing background intensity from autofluorescence or crosstalks between emission channels. Then, detection of the “seed” of each cell was achieved from a two-step process: (i) Using the 3D image J suite plugins67, image stacks were processed with a combination of filters, including 3D Mean (kernel size in all dimensions was set to 1 pixel), followed by 3D Maximum Local to retrieve local maxima of individual cells. The kernel size in each dimension (e.g. x = 6; y = 8; z = 10 pixels) of the 3D Maximum Local filter was determined based on estimated cell size. The x dimension is a particularly useful to separate cells located very close to each other (Supplementary Fig. 3). (ii) From the identified local maxima, 3D object counter68 was applied as a second step to identify the centroid of each local maxima cluster and return the “seed”, which then represented the location of a single cell. It should be noted that a threshold was required in 3D object counter to disregard “non-cell” local maxima clusters. To standardize threshold determination, we measured the outlier of “cell surrounding” (Supplementary Fig. 1a, shown in red) in each image, and the outlier was then considered as a true cell. Specifically, the seeds to be included (“the outliers” of cell surrounding) was determined based on Tukey’s anomaly69, as calculated in equations (1–3), where Q1, Q3, and x are the first quartile, third quartile, and the mean value obtained from the histogram of cell surroundings (Supplementary Fig. 1b). Eventually, T, the estimated outlier calculated in equation 3, was then used as a threshold for 3D object counter. It should be noted that the image that had gone through more rigorous contrast enhancement (e.g. the max. intensity was only between 20 to 25 in a raw 8-bit image, Supplementary Fig. 1c) inherently yielded higher background intensity. To standardize segmentation of cell surrounding without taking excessive background noise, “step background gating” was applied. Specifically, the cut-off value at the lower bound of the histogram shown in Supplementary Fig. 1b was determined using the mode intensity (the background intensity) measured in each raw image, with step-adjustment according to the signal level (Supplementary Fig. 1c).
| 1 |
| 2 |
| 3 |
Statistics
5 control (PBS) and 6 experimental (rmWnt3a) animals were used for the study. Six to eight regions in the coronal suture were acquired from each animal. Statistical analysis for mean and standard deviation was performed by pooling regions from all animals without excluding outliers. The plotted outlier was defined as a value less than or greater than 1.5 times the interquartile range. Statistical significant difference was obtained using two-tailed distribution equal variance Student’s t-test and results are shown as means ± S.D in figures unless specified otherwise. Statistical significant difference is indicated as *(p < 0.05), **(p < 0.01), ***(p < 0.005) or ****(p < 0.0005).
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This project was supported by grant no. R00DE021069 (NIH/NIDCR) to Dr. Giuseppe Intini, and by the HSDM Dean’s Scholar Award to S.-C.A.Y. We acknowledge Dr. Shunichi Murakami (Case Western Reserve University) for kindly providing the Prx1-creER-EGFP mice. We also appreciate Dr. Roland Baron, Dr. Francesca Gori, Dr. Nagano Kenichi, and Dr. Dorothy Wu for advice in bone histomorphometry. We acknowledge Dr. Zahra Aldawoods for her help in animal maintenance.
Author Contributions
S.-C.A.Y. performed the vast majority of the experiments described in this manuscript. K.W. performed most Wnt administration and mouse colony maintenance. C.P.L. and G.I. supervised the studies and directed the interpretations of the results.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23785-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Charles P. Lin, Email: lin@helix.mgh.harvard.edu
Giuseppe Intini, Email: Giuseppe_Intini@hsdm.harvard.edu.
References
- 1.Vidal B, et al. Bone histomorphometry revisited. Acta Reumatol. Port. 2012;37:294–300. [PubMed] [Google Scholar]
- 2.Ott SM. Histomorphometric measurements of bone turnover, mineralization, and volume. Clin. J. Am. Soc. Nephrol. 2008;3(Suppl 3):151–156. doi: 10.2215/CJN.04301206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morvan F, et al. Deletion of a single allele of the Dkk1 gene leads to an increase in bone formation and bone mass. J. Bone Miner. Res. 2006;21:934–945. doi: 10.1359/jbmr.060311. [DOI] [PubMed] [Google Scholar]
- 4.Nemeth JA, et al. Matrix metalloproteinase activity, bone matrix turnover, and tumor cell proliferation in prostate cancer bone metastasis. J. Natl. Cancer Inst. 2002;94:17–25. doi: 10.1093/jnci/94.1.17. [DOI] [PubMed] [Google Scholar]
- 5.Kulak CAM, Dempster DW. Bone histomorphometry: a concise review for endocrinologists and clinicians. Arq. Bras. Endocrinol. Metabol. 2010;54:87–98. doi: 10.1590/S0004-27302010000200002. [DOI] [PubMed] [Google Scholar]
- 6.Rhee Y, et al. Assessment of bone quality using finite element analysis based upon micro-CT images. Clin. Orthop. Surg. 2009;1:40. doi: 10.4055/cios.2009.1.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stauber M, Müller R. Volumetric spatial decomposition of trabecular bone into rods and plates - A new method for local bone morphometry. Bone. 2006;38:475–484. doi: 10.1016/j.bone.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Burghardt AJ, Link TM, Majumdar S. High-resolution computed tomography for clinical imaging of bone microarchitecture. Clin. Orthop. Relat. Res. 2011;469:2179–2193. doi: 10.1007/s11999-010-1766-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambers FM, Schulte FA, Kuhn G, Webster DJ, Müller R. Mouse tail vertebrae adapt to cyclic mechanical loading by increasing bone formation rate and decreasing bone resorption rate as shown by time-lapsed in vivo imaging of dynamic bone morphometry. Bone. 2011;49:1340–1350. doi: 10.1016/j.bone.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 10.Muller R, et al. Morphometric analysis of human bone biopsies: A quantitative structural comparison of histological sections and micro-computed tomography. Bone. 1998;23:59–66. doi: 10.1016/S8756-3282(98)00068-4. [DOI] [PubMed] [Google Scholar]
- 11.Nyman JS, et al. Predicting mouse vertebra strength with micro-computed tomography-derived finite element analysis. Bonekey Rep. 2015;4:664. doi: 10.1038/bonekey.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klintstrom E, et al. Predicting trabecular bone stiffness from clinical cone-beam CT and HR-pQCT Data; an In vitro study using finite element analysis. PLoS One. 2016;11:1–19. doi: 10.1371/journal.pone.0161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waarsing JH, Day JS, Weinans H. Longitudinal micro-CT scans to evaluate bone architecture. J. Musculoskelet. Neuronal Interact. 2005;5:310–312. [PubMed] [Google Scholar]
- 14.Lo Celso, C. et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature457, 92–96 (2009). [DOI] [PMC free article] [PubMed]
- 15.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147:983–991. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003;21:1369–77. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 17.Non invasive diagnostic techniques in clinical dermatology. 10.1007/978-3-642-32109-2 (Springer Berlin Heidelberg, 2014).
- 18.Kiesslich R, et al. In vivo histology of Barrett’s esophagus and associated neoplasia by confocal laser endomicroscopy. Clin. Gastroenterol. Hepatol. 2006;4:979–87. doi: 10.1016/j.cgh.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Veilleux I, Spencer JA, Biss DP, Cote D, Lin CP. In vivo cell tracking with video rate multimodality laser scanning microscopy. IEEE J. Sel. Top. Quantum Electron. 2008;14:10–18. doi: 10.1109/JSTQE.2007.912751. [DOI] [Google Scholar]
- 20.Elefteriou F, Xiangli Yang. Genetic mouse models for bone studies—Strengths and limitations. Bone. 2011;49:1242–1254. doi: 10.1016/j.bone.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orth JD, et al. Analysis of mitosis and antimitotic drug responses in tumors by In Vivo microscopy and single-cell pharmacodynamics. Cancer Res. 2011;71:4608–4616. doi: 10.1158/0008-5472.CAN-11-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sumen C, Mempel TR, Mazo IB, Von Andrian UH. Intravital microscopy: Visualizing immunity in context. Immunity. 2004;21:315–329. doi: 10.1016/j.immuni.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Ishii M, Kikuta J, Shimazu Y, Meier-Schellersheim M, Germain RN. Chemorepulsion by blood S1P regulates osteoclast precursor mobilization and bone remodeling in vivo. J. Exp. Med. 2010;207:2793–2798. doi: 10.1084/jem.20101474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park D, et al. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10:259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spencer JA, et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 2014;508:269–73. doi: 10.1038/nature13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Nadiarynkh O, Plotnikov S, Campagnola PJ. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat. Protoc. 2012;7:654–669. doi: 10.1038/nprot.2012.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sano H, et al. Intravital bone imaging by two-photon excitation microscopy to identify osteocytic osteolysis in vivo. Bone. 2015;74:134–139. doi: 10.1016/j.bone.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Kowada T, et al. In vivo fluorescence imaging of bone-resorbing osteoclasts. J. Am. Chem. Soc. 2011;133:17772–17776. doi: 10.1021/ja2064582. [DOI] [PubMed] [Google Scholar]
- 29.Ishii M, Fujimori S, Kaneko T, Kikuta J. Dynamic live imaging of bone: Opening a new era with ‘bone histodynametry’. J. Bone Miner. Metab. 2013;31:507–511. doi: 10.1007/s00774-013-0437-x. [DOI] [PubMed] [Google Scholar]
- 30.Huang C, et al. Spatiotemporal analyses of osteogenesis and angiogenesis via intravital imaging in cranial bone defect repair. J. Bone Miner. Res. 2015;30:1217–30. doi: 10.1002/jbmr.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao H, et al. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat. Cell Biol. 2015;17:386–396. doi: 10.1038/ncb3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maruyama T, Jeong J, Sheu T-J, Hsu W. Stem cells of the suture mesenchyme in craniofacial bone development, repair and regeneration. Nat. Commun. 2016;7:10526. doi: 10.1038/ncomms10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilk K, et al. Postnatal calvarial skeletal stem cells expressing PRX1 reside exclusively in the calvarial sutures and are required for bone regeneration. Stem Cell Reports. 2017;8:933–946. doi: 10.1016/j.stemcr.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ten Berge D, Brouwer A, Korving J, Martin JF, Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development. 1998;125:3831–3842. doi: 10.1242/dev.125.19.3831. [DOI] [PubMed] [Google Scholar]
- 35.Ouyang Z, et al. Prx1 and 3.2kb Col1a1 promoters target distinct bone cell populations in transgenic mice. Bone. 2014;58:136–145. doi: 10.1016/j.bone.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat. Med. 2013;19:179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 37.Ling L, Nurcombe V, Cool SM. Wnt signaling controls the fate of mesenchymal stem cells. Gene. 2009;433:1–7. doi: 10.1016/j.gene.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 39.Behr, B., Longaker, M. T. & Quarto, N. Absence of endochondral ossification and craniosynostosis in posterior frontal cranial sutures of Axin2−/−mice. PLoS One8, (2013). [DOI] [PMC free article] [PubMed]
- 40.Behr B, Longaker MT, Quarto N. Differential activation of canonical Wnt signaling determines cranial sutures fate: A novel mechanism for sagittal suture craniosynostosis. Dev. Biol. 2010;344:922–940. doi: 10.1016/j.ydbio.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 41.Pautke C, et al. Polychrome labeling of bone with seven different fluorochromes: Enhancing fluorochrome discrimination by spectral image analysis. Bone. 2005;37:441–445. doi: 10.1016/j.bone.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Tokarz D, et al. Intravital imaging of osteocytes in mouse calvaria using third harmonic generation microscopy. PLoS One. 2017;12:1–15. doi: 10.1371/journal.pone.0186846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tokarz, D. et al. Hormonal regulation of osteocyte perilacunar and canalicular remodeling in the hyp mouse model of X-linked hypophosphatemia. J. Bone Miner. Res. 33, 499–509 (2018). [DOI] [PMC free article] [PubMed]
- 44.Quarto N, et al. Origin matters: Differences in embryonic tissue origin and Wnt signaling determine the osteogenic potential and healing capacity of frontal and parietal calvarial bones. J. Bone Miner. Res. 2010;25:1680–94. doi: 10.1359/jbmr.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin H, et al. Anti-DKK1 antibody promotes bone fracture healing through activation of beta-catenin signaling. Bone. 2015;71:63–75. doi: 10.1016/j.bone.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song L, et al. Loss of wnt/beta-catenin signaling causes cell fate shift of preosteoblasts from osteoblasts to adipocytes. J. Bone Miner. Res. 2012;27:2344–2358. doi: 10.1002/jbmr.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhamdhere GR, et al. Drugging a stem cell compartment using Wnt3a protein as a therapeutic. PLoS One. 2014;9:e83650. doi: 10.1371/journal.pone.0083650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Georgiadis, M., Mu, R. & Schneider, P. Techniques to assess bone ultrastructure organization: orientation and arrangement of mineralized collagen fibrils. J. R. Soc. Interface13, (2016). [DOI] [PMC free article] [PubMed]
- 49.Ambekar R, Chittenden M, Jasiuk I, Toussaint KC. Quantitative second-harmonic generation microscopy for imaging porcine cortical bone: Comparison to SEM and its potential to investigate age-related changes. Bone. 2012;50:643–650. doi: 10.1016/j.bone.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 50.Zoumi A, Yeh A, Tromberg BJ. Imaging cells and extracellular matrix in vivo by using second-harmonic generation and two-photon excited fluorescence. Proc. Natl. Acad. Sci. USA. 2002;99:11014–11019. doi: 10.1073/pnas.172368799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellis R, Green E, Winlove CP. Structural analysis of glycosaminoglycans and proteoglycans by means of Raman microspectrometry. Connect. Tissue Res. 2009;50:29–36. doi: 10.1080/03008200802398422. [DOI] [PubMed] [Google Scholar]
- 52.Hashimoto A, et al. Time-lapse Raman imaging of osteoblast differentiation. Sci. Rep. 2015;5:12529. doi: 10.1038/srep12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jebaramya J, Ilanchelian M, Prabahar S. Spectral studies of toluidine blue o in the presence of sodium dodecyl sulfate. Dig. J. Nanomater. Biostructures. 2009;4:789–797. [Google Scholar]
- 54.Débarre D, et al. Imaging lipid bodies in cells and tissues using third-harmonic generation microscopy. Nat. Methods. 2006;3:47–53. doi: 10.1038/nmeth813. [DOI] [PubMed] [Google Scholar]
- 55.Hirata E, Kiyokawa E. Future perspective of single-molecule FRET biosensors and intravital FRET microscopy. Biophys. J. 2016;111:1103–1111. doi: 10.1016/j.bpj.2016.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Manen H-J, Kraan YM, Roos D, Otto C. Single-cell Raman and fluorescence microscopy reveal the association of lipid bodies with phagosomes in leukocytes. Proc. Natl. Acad. Sci. USA. 2005;102:10159–10164. doi: 10.1073/pnas.0502746102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le TT, Huff TB, Cheng J-X. Coherent anti-Stokes Raman scattering imaging of lipids in cancer metastasis. BMC Cancer. 2009;9:42. doi: 10.1186/1471-2407-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Runnels JM, et al. Imaging molecular expression on vascular endothelial cells by in vivo immunofluorescence microscopy. Mol. Imaging. 2006;5:31–40. doi: 10.2310/7290.2006.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cummings RJ, Mitra S, Lord EM, Foster TH. Antibody-labeled fluorescence imaging of dendritic cell populations in vivo. J Biomed Opt. 2008;13:44041. doi: 10.1117/1.2966122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim, S., Lin, L., Brown, G. A. J., Hosaka, K. & Scott, E. W. Extended time-lapse in vivo imaging of tibia bone marrow to visualize dynamic hematopoietic stem cell engraftment. Leukemia 31, 1582–1592 (2017). [DOI] [PMC free article] [PubMed]
- 61.Le V-H, et al. In vivo longitudinal visualization of bone marrow engraftment process in mouse calvaria using two-photon microscopy. Sci. Rep. 2017;7:44097. doi: 10.1038/srep44097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ntziachristos V. Going deeper than microscopy: the optical imaging frontier in biology. Nat. Methods. 2010;7:603–614. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 63.Dunn AK, Wallace VP, Coleno M, Berns MW, Tromberg BJ. Influence of optical properties on two-photon fluorescence imaging in turbid samples. Appl. Opt. 2000;39:1194–1201. doi: 10.1364/AO.39.001194. [DOI] [PubMed] [Google Scholar]
- 64.Horton NG, et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat. Photonics. 2013;7:205–209. doi: 10.1038/nphoton.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klauschen F, et al. Quantifying cellular interaction dynamics in 3D fluorescence microscopy data. Nat. Protoc. 2009;4:1305–1311. doi: 10.1038/nprot.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ollion J, Cochennec J, Loll F, Escudé C, Boudier T. TANGO: A generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics. 2013;29:1840–1841. doi: 10.1093/bioinformatics/btt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bolte S, Cordelieres FP. A guided tour into subcellular colocalisation analysis in light microscopy. J. Microsc. 2006;224:13–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 69.Tukey, J. Exploratory Data Analysis. (Addison Wesley, 1977).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.