Abstract
Integrative and conjugative elements (ICEs) comprise ubiquitous large mobile regions in prokaryotic chromosomes that transmit vertically to daughter cells and transfer horizontally to distantly related lineages. Their evolutionary success originates in maximized combined ICE-host fitness trade-offs, but how the ICE impacts on the host metabolism and physiology is poorly understood. Here we investigate global changes in the host genetic network and physiology of Pseudomonas putida with or without an integrated ICEclc, a model ICE widely distributed in proteobacterial genomes. Genome-wide gene expression differences were analyzed by RNA-seq using exponentially growing or stationary phase-restimulated cultures on 3-chlorobenzoate, an aromatic compound metabolizable thanks to specific ICEclc-located genes. We found that the presence of ICEclc imposes a variety of changes in global pathways such as cell cycle and amino acid metabolism, which were more numerous in stationary-restimulated than exponential phase cells. Unexpectedly, ICEclc stimulates cellular motility and leads to more rapid growth on 3-chlorobenzoate than cells carrying only the integrated clc genes. ICEclc also concomitantly activates the P. putida Pspu28-prophage, but this in itself did not provoke measurable fitness effects. ICEclc thus interferes in a number of cellular pathways, inducing both direct benefits as well as indirect costs in P. putida.
Introduction
Horizontal gene transfer (HGT) is a major driving force for microbial evolution and adaptation, since transmission of genes from a donor bacterium can rapidly confer additional cellular functions and phenotypes to a naive recipient1,2. Typical ecological consequences of HGT are the emergence of bacterial species “suddenly” expressing antibiotic resistance3–5, new virulence factors6,7, or chemical degradation pathways8–10. HGT is frequently, but not exclusively, mediated by mobile DNA vectors, that are permissive for hitchhiking of auxiliary genetic material within their own boundaries. Mobile DNA vectors that promote HGT include, for example, plasmids, integrative and conjugative elements (ICEs) or bacteriophages11.
While acquisition of such auxiliary genes and their relevant phenotypes can be beneficial for hosts to adapt in particular ecological niches, it is generally assumed that the DNA acquisition also imposes various types of fitness costs on the cells. For example, horizontally transmitted genes may come with functional promoters that are active in the host, or intruded transferred regulatory genes may cross-talk to existing host global regulatory networks, resulting in perturbation of the genetic regulatory systems and subsequent fitness loss of the host12. Additional physiological and energetic costs may arise from increased need for replication of newly acquired DNA, transcription and translation of the acquired genes13,14, or cytotoxic effects of misfolded foreign proteins15,16. Costs inflicted by mobile vectors are in principle disadvantageous not only for the host but also for inheritance of the horizontally transmitted genes themselves, if we consider fitness of a mobile vector as maximizing its copy numbers in a given ecological niche. Hence, a variety of systems have evolved that minimize fitness costs of mobile DNAs. Several of these have been described in conjugative plasmids and temperate phages. For example, some conjugative plasmids encode “stealth” proteins to silence derogative functions that may impede plasmid maintenance17–19. Temperate phages express regulatory proteins, such as phage lambda’s cI repressor, that not only stably maintain the lysogenic state but also control host metabolic pathways to ensure host survival and efficient reproduction of phages20–22. In contrast to plasmids and phages, not much is known about the impact of ICEs upon acquisition by a new host.
ICEs are peculiar in that they have two life-styles. In the vertical transmission mode, they reside in the host chromosome23,24. Their segregation to daughter cells is guaranteed by chromosomal replication, like for lysogenic phages, instead of by independent replication, like for plasmids. In the horizontal transmission mode, the ICE excises from the chromosome through site-specific recombination and subsequently circularizes for conjugation, similar as in conjugative plasmid transfer23,24. In the recipient cell, the ICE molecule integrates site-specifically into the chromosome and is again stably maintained. Horizontal transmission of ICEs is proportionally rare, in the order of between 1 per 102–108 donor cells25. ICEs are widespread mobile elements, occurring in several evolutionary distinct families. They can carry ‘cargo’ genes encoding distinct functions, such as antibiotic resistance, heavy-metal resistance, symbiosis, or aromatic compound metabolism, from which the host can benefit under selective conditions24,25.
Here we address the question of measuring the global impact of an ICE in a new bacterial host. The model we use for our study is the ICEclc element, representative of a prevalent ICE-type among proteobacterial genomes26. ICEclc has a size of 103 kb and carries the clc genes, which provide the host with the capacity to degrade 3-chlorobenzoate (3CBA). It can transfer at a relatively high rate (3 per 102 donor cells) from its original host Pseudomonas knackmussii B13 when the host has grown on 3CBA as a sole carbon source27. Transfer is initiated from specialized transfer-competent cells28, which arise during stationary phase as a result of a bistable expression mechanism25 and can conjugate the ICE when presented with new nutrients29. We have previously analyzed the global impact of ICEclc on its non-native host Pseudomonas aeruginosa PAO1. That study indicated less than an estimated 1% fitness loss and only few deviating individual host gene functions compared to PAO1 without ICE30. However, since ICEclc horizontal transmission from PAO1 as donor was very low compared to P. knackmussii (one per 105 donor cells), and since that particular study could not address the impact of the ICE during growth on 3CBA, it may not have captured all relevant ICE-induced effects.
In order to complement previous data and obtain a wider picture of ICEclc-induced effects in a new host, we used here Pseudomonas putida. ICEclc transfer rates from P. putida as host are similar as P. knackmussii B13, and expression of key ICEclc promoters is non-distinguishable between both hosts, confined to a small (3–5%) bistable subpopulation of cells28,31. In order to study the impact of ICEclc during growth on 3CBA, we constructed a P. putida derivative without the ICE but with the clc-genes on the chromosome. We then analyzed genome-wide transcriptome differences in P. putida strains without or with ICEclc by using rRNA-depleted reverse-transcribed RNA sequencing. Cultures were grown on 3CBA as selective carbon source to induce highest ICEclc gene expression. We examined cells both from exponentially–growing conditions (hereafter, EXP phase) and from stationary phase, restimulated for 4.5 hours with fresh 3CBA (regrowth, or REG phase), under which the frequency of ICEclc horizontal transfer is maximal (3–5% of donor cells)29. Identified expression differences were subsequently profiled and substantiated by growth kinetics and physiology experiments. Finally, we verified cross-activation of a prophage in P. putida by ICEclc through single cell gene reporter experiments.
Results
Presence of ICEclc influences P. putida transcriptome differently depending on growth phase
Genome-wide transcription in P. putida with or without ICEclc was assessed from reverse-transcribed ribosomal-RNA depleted RNA samples, which were sequenced at on average 270-fold coverage by Illumina HiSeq and mapped to the P. putida genome (Table S1). Four replicates of the four different experimental treatments (two strains each in EXP or REG phase) clustered more closely together in multidimensional scaling analysis than between treatments, indicating clearly distinguishable global transcriptomes (Fig. S1).
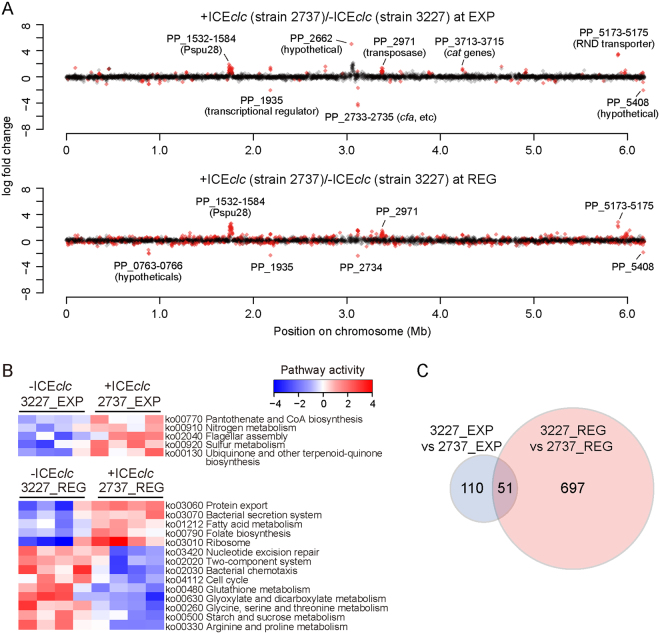
In comparison to P. putida without ICEclc but with the clc genes (strain 3227), a total of 161 genes (outside ICEclc itself) were differentially expressed in P. putida-ICEclc (strain 2737) in EXP phase on 3CBA (FDR with q < 0.05; Fig. 1A, Data S1). Increased transcription of genes encoding RND-family transporter proteins (PP_3425–3427, 5173–5175), several types of transposases (PP_2964, 2971, 2974, 5405-5406), and a series of cryptic prophage Pspu28-related proteins (PP_1532–1584) was noticeable, whereas genes for extracellular-family sigma factors (PP_0352, 0704, 2192, 4208) were markedly lower expressed in strain 2737. Five KEGG–attributable pathways32, including nitrogen (ko00910) and sulfur metabolism (ko00920), flagellar assembly (ko02040), and biosynthesis of co-factors (ko00770, ko00130) were significantly higher expressed in P. putida-ICEclc than in P. putida without ICE in EXP phase (two-tailed Welch t test, p < 0.05; Fig. 1B).
Figure 1.
Gene expression profiles in presence or absence of ICEclc. (A) Log2–fold change of gene expression between P. putida with ICEclc (strain 2737) and that with clc genes only (strain 3227) in exponential (upper) and REG phase (lower). Only genes with an a.value (mean expression values, calculated using the trimmed mean of M values (TMM) normalization method) >4.0 are plotted, according to their positions on the chromosome. Differentially expressed genes with a q-value < 0.05 are coloured in red. (B) Pathway activity in the presence/absence of ICEclc. Activities were inferred from significantly induced or repressed KEGG Pathways identified for each dataset comparison (p < 0.05 in two-tailed Welch t test). Blue/red blocks indicate pathways (rows) that are up/down regulated in quadruplicate samples (columns) of specific strains and growth phases (top). Pathways are clustered based on the similarity of their activities across samples. (C) Venn diagram showing overlap among differentially expressed genes (DEGs) among transcriptome datasets. The intersection shows the DEGs at a q-value < 0.05 that were common to both comparisons and with the same direction of expression change.
In contrast, during REG-phase, P. putida-ICEclc expressed 748 genes statistically significantly differently compared to P. putida without ICEclc (Fig. 1A, Data S1). Only 51 of those overlapped with the previous set of genes observed in EXP phase (Fig. 1C, same direction of expression), indicating that more chromosomal genes respond to the presence of ICEclc in REG than EXP phase. In REG phase P. putida-ICEclc, five KEGG pathways were higher expressed, including ribosome synthesis (ko03010), fatty acid metabolism (ko01212), protein export (ko03060) and secretion system (ko03070), while nine pathways were repressed, such as cell cycle (ko04112) and amino acid metabolism (ko00260, ko00330) (Fig. 1B). None of the 14 KEGG pathways differentially expressed in REG phase overlapped with the five pathways higher expressed in EXP phase ICEclc-carrying cells.
Most of the genes in the ICE-variable region (coordinates 5,000–50,000), which in case of ICEclc contains the clc genes, were indistinguishably expressed between EXP and REG–phase cells, concluded from normalized read counts of the ICEclc region in P. putida 2737 among EXP and REG phase (Fig. S2). On the other hand, the intB13 gene and genes in the ICE core region (coordinates 1–5,000 and 50,000–100,000) involved in horizontal transmission were more highly expressed in REG than EXP phase (Fig. S2), confirming ICE activation in REG-culture conditions.
ICEclc activates acryptic prophage
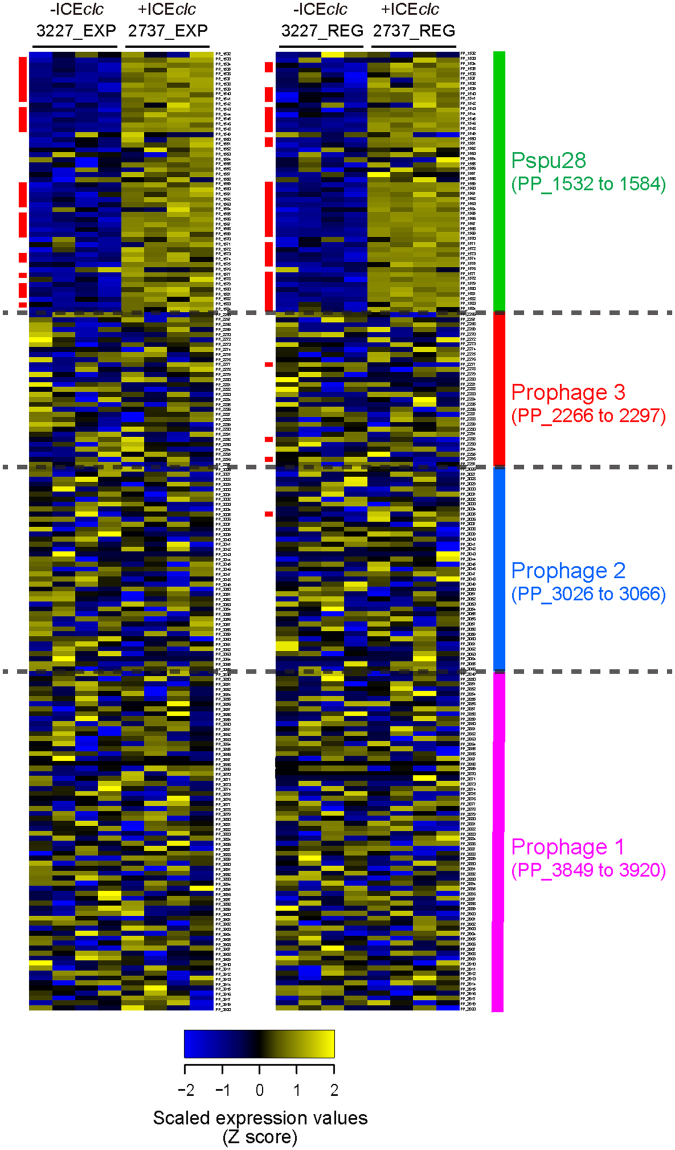
One of the striking unexpected findings in the transcriptome comparisons pointed to a link between ICEclc and expression of one out of four prophages in P. putida (Fig. 2), which has previously been designated Pspu2833,34. Expression of the Pspu28 prophage genes (at position ~1.8 Mb in the genome, Fig. 1) is clearly higher in P. putida-ICEclc (strain 2737) than in P. putida without (strain 3227), and occurs both in EXP and REG phase (Fig. 2). Gene expression in the other prophage regions in the P. putida genome was not different in presence or absence of ICEclc (Fig. 2).
Figure 2.
Differential gene expression profiles in four prophage regions of P. putida in presence or absence of ICEclc. Blue/yellow blocks indicate chromosomal genes (rows) that are up/down regulated in quadruplicate samples (columns) of the indicated P. putida strains and growth phases (top). Genes are clustered according to the four prophage regions as indicated on the right. Vertical red lines on the left side of rows indicate genes with a false-discovery rate q-value < 0.01 on each pairwise comparison.
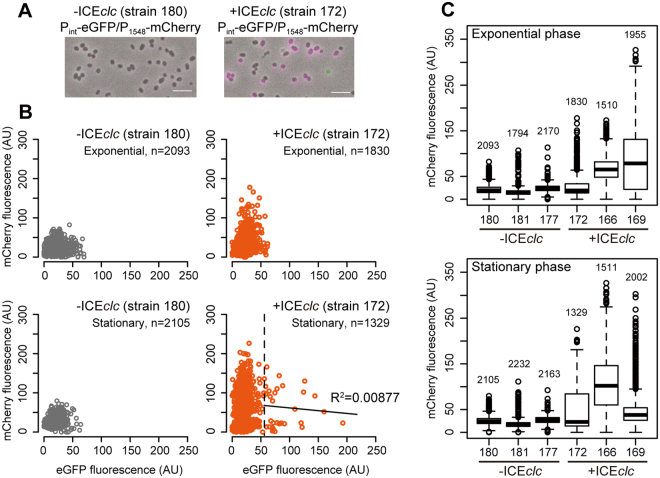
To confirm that phage genes are expressed in P. putida-ICEclc cells, we developed dual fluorescence P. putida reporter strains carrying single copy insertions of an mcherry gene fused with the promoter of PP_1548 (P1548-mcherry), encoding a Pspu28 phage-specific protein. Expression of PP_1548 was easily noticeable in the global transcriptome and therefore deployed as marker here. The reporter strain further contained an egfp gene fused with the ICEclc integrase promoter (Pint-egfp), which becomes active in 3–5% of stationary/REG phase transfer-competent cells where ICEclc horizontal transmission occurs28,35 (Fig. 3A). In agreement with the transcriptome studies, expression levels of P1548-mcherry were higher in P. putida-ICEclc than in P. putida without ICEclc in both EXP and stationary phases on 3CBA (Fig. 3BC). However, the mCherry intensities of single cells among three clones were varying depending on the insertion position of the P1548-mcherry fusion, some of which were not normally distributed (Fig. S3), suggesting that prophage activation occurs in a sub-population. No correlation was observed between P1548-mcherry and Pint-egfp expression at single cell level (R2 < 0.01, Fig. 3B), indicating the phage is indeed activated by the presence of ICEclc, but not specifically induced in the subset of cells that activate Pint-egfp. On the other hand, no obvious differences were observed between P. putida with and without ICEclc growing on LB; regardless of the ICE presence, P1548-mcherry was silent in EXP phase but was expressed in a certain fraction of cells in stationary phase (Fig. S4).
Figure 3.
Differential expression of the Pspu28 phage P1548 promoter at single-cell level in P. putida with or without ICEclc. (A) Representative micrographs of P. putida strains carrying single-copy chromosomal inserts of the P1548-mcherry and the ICEclc bistable Pint–promoter –egfp fusions, via mini-transposon delivery. Images show merged phase–contrast (grey), eGFP (green), and mCherry (magenta). Scale bar indicates 5 μm. (B) Scatter plots showing mCherry (from P1548) and eGFP (from Pint) fluorescence intensities among individual cells (circles) of P. putida without (strain 180) and with ICEclc (strain 172) at exponential and late stationary phases in MM with 3CBA. Threshold (dashed line) between Pint-active and -inactive cells of strain 172 at late stationary phase was calculated according to Reinhard and van der Meer71. Note that mCherry and eGFP fluorescence among the Pint-active cells is not correlated significantly (R2 = 0.00877). (C) Box plots showing mCherry (from P1548) fluorescence intensity among individual cells of P. putida without (strain 180, 181, and 177) and with ICEclc (strain 172, 166, and 169) at exponential and late stationary phases in MM with 3CBA. The number of measured cells is indicated for every sample. Note that the different P. putida strains have randomly inserted fluorescence reporter constructs.
ICEclc influences cell motility depending on growth condition
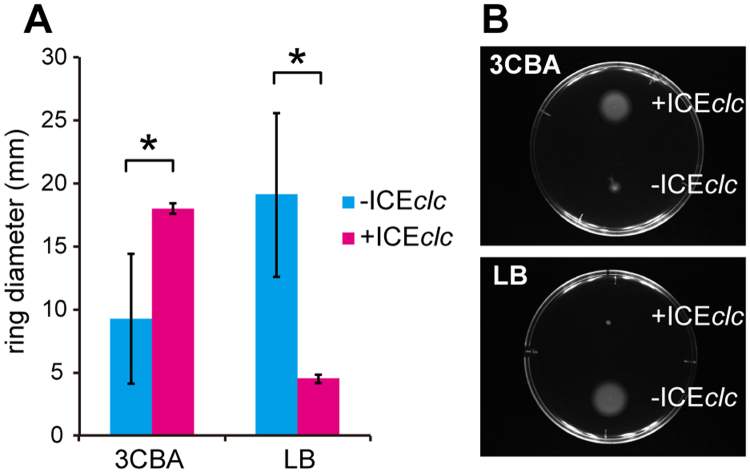
A second surprising finding from transcriptome comparisons was the higher expression of the P. putida flagellar assembly pathway in ICEclc–containing strains in EXP phase (Fig. 1B). P. putida-ICEclc was indeed more motile on agar media than P. putida without ICEclc, but only when pregrown in minimal medium with 3CBA as carbon substrate (Fig. 4). In contrast, motility after growth on complex medium (LB) was reduced in P. putida-ICEclc (Fig. 4). These results indicate that ICEclc increases the host cell’s motility in the presence of substrate that it can metabolize thanks to the clc genes on the ICE.
Figure 4.
Swimming motility of P. putida in the presence/absence of ICE. P. putida strains with (strain 2737) and without ICEclc (strain 3227) were pregrown in MM with 3CBA or LB and tested on swimming plates (PG medium solidified with 0.3% agar). (A) Measurement of the mean colony ring diameter (±1 SD, n = 5) after 19 h incubation. Asterisks above bar diagrams indicate significance of difference (P < 0.01) in one-tailed t tests. (B) Representative images of different colony sizes on swimming plates after 19 h of incubation of P. putida strains precultured in MM with 3CBA (upper panel) or in LB (lower panel).
Host-induced fitness effects by ICEclc are medium dependent
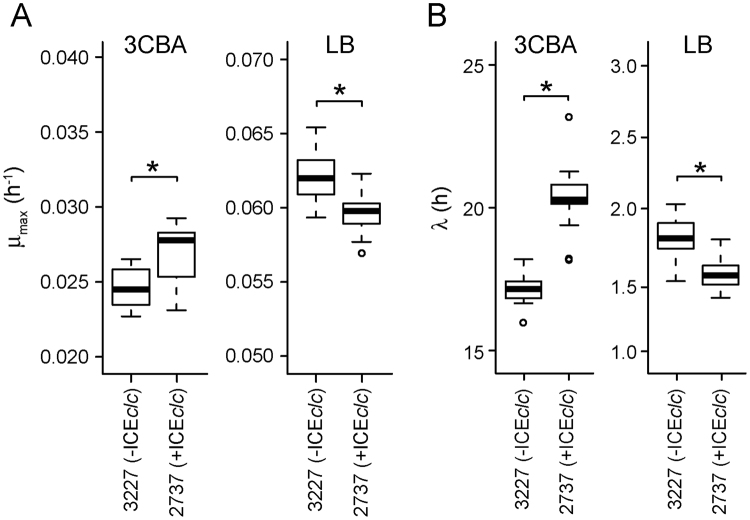
In order to determine whether ICEclc globally impacts fitness of P. putida, we compared growth rates of P. putida with (strain 2737) and without ICEclc (strain 3227) on minimal medium with 3CBA or on complex (LB) medium. The average maximum growth rate (μmax) of P. putida with ICEclc on 3CBA was higher than that without (P < 0.01, Fig. 5A). Given that no difference among the strains was observed in gene expression levels directly involved in 3CBA metabolism, such as the ben and clc genes (Table S2), the beneficial effect of the ICE on P. putida growth could be exerted by other mechanisms. In contrast, the µmax of P. putida-ICEclc was lower than that of P. putida in LB medium (Fig. 5A), indicating that the positive effect of ICEclc on growth rate is carbon source (or medium)-dependent. A longer lag phase on 3CBA was observed in P. putida-ICEclc than P. putida without ICEclc (Figs 5B, S4). This could be the result of growth arrest of 3–5% cells that had activated ICEclc horizontal transmission in the preculture, as previously simulated by modeling31.
Figure 5.
Effect of ICEclc on P. putida growth fitness. (A) Maximum growth rate (μmax, h-1) and (B) length of lag phase ( h), calculated from 20 replicates of each P. putida strain growing in MM with 3CBA (left) or in LB (right). Asterisks above box plots indicate significance of difference on each pair (P < 0.0001) in t test.
Discussion
ICEs are widely present self-transferable elements in bacterial lineages, suggesting adaptive benefits from their presence. However, the global effects of their presence on the host cellular systems and expression networks have been poorly characterized. Our results indicate that, globally, the presence of ICEclc improves P. putida fitness on the carbon substrate 3CBA, which is exclusively metabolizable thanks to the ICE, but impairs fitness on other carbon substrates.
A previous study using micro-array analysis showed very few differences in host gene expression and less than 1% fitness costs of ICEclc on P. aeruginosa PAO1 growing on succinate as a non-selective neutral carbon source30. The absence of immediate detrimental effects to the host PAO1 led us to conclude that ICEclc is a ‘perfect partner leaving very little imprint but with specific selective benefits when needed’30, which might be one of the reasons ICEs of this type are widely distributed among bacterial genomes. However, that study could not test the effect of growing ICEclc–containing hosts on 3CBA, which is the carbon substrate that leads to the highest activation of the ICE in stationary phase (3–5% of cells) and stimulates its excision and horizontal transmission in such cells (REG conditions)29. To complement the impact study of ICEclc in PAO1, therefore, we here used a P. putida host and ensured that both host without and with ICEclc can grow on 3CBA. This was accomplished by introducing the clc genes by single copy Tn7 chromosomal delivery into P. putida (strain 3227). The growth of this strain is very nearly the same as P. putida-ICEclc (strain 2737, see below and Fig. S5).
In cells growing exponentially on 3CBA, expression of 2.8% of chromosomal genes (161 of 5671 total genes) was affected in P. putida with compared to P. putida without the ICE, encompassing five identifiable biological KEGG pathways. In comparison, we previously detected a single expression difference (1 out of 5,900 genes and intergenic regions) between exponentially growing cells on succinate of P. aeruginosa PAO1 with and without ICEclc30. No global pathways of P. putida significantly decreased expression in exponentially growing cells in presence of ICEclc.
In contrast, in REG phase, 13% of P. putida host genes (748 in total) encompassing 14 biological pathways changed expression levels compared to P. putida without ICE. Notably, protein export (ko03060), secretion system (ko03070), and ribosome synthesis (ko03010) were activated, while cell cycle (ko04112) and several metabolic pathways (e.g. ko00500, ko00260, ko00330) were repressed. In P. aeruginosa PAO1, the expression differences among strains with or without ICEclc were also more substantial in stationary than exponential phase on succinate (42 genes significantly changed, out of 5671)30. However, there are only two orthologous genes overlapping between those differentially expressed in P. putida and P. aeruginosa (PP_0356/PA0482, malate synthase G; PP_2351/PA3568, probable acetyl-CoA synthase), suggesting that global changes induced by ICEclc are substantially different between those hosts and conditions. The stronger effect of ICEclc in 3CBA REG conditions in P. putida may be due to the activation of the ICE horizontal transmission program, and might explain the observed reduction in expression of the cell cycle and increased secretion/export systems. Alternatively, there might be specific links between other ICEclc–encoded factors, dependent on the host network that we currently do not understand. The difference in the extent of host global gene expression by ICEclc between P. putida and P. aeruginosa PAO1 as hosts is striking. This may partly be an effect of the host, but may also be an exaggerated effect from the carbon substrate 3CBA as opposed to succinate that results in more active ICEclc expression36.
One of the incidental findings in this study was the observed activation of the cryptic prophage Pspu28 in P. putida by the presence of ICEclc. Although P. putida UWC1 harbors a total of four prophages, only Pspu28 responded to ICEclc, suggesting specific crosstalk between them. To our knowledge, this is the first report to show an ICE–phage global interaction. Pspu28 has a size of 40-kb and encodes a filamentous bacteriophage with a capsid-like hexagonal structure, whose relatives are found among Pseudomonas and Shewanella genomes37. The phage can switch between lysogenic and lytic states, but lacks a lytic enzyme (i.e. endolysin), suggesting that it is incapable of actively killing the host. It releases progeny phage particles when the host eventually dies. A previous study reported that Pspu28 phage lysogeny leads to fitness loss of P. putida in the rhizosphere through an unknown mechanism34. Activation of the Pspu28 phage P1548-promoter was heterogeneous among P. putida cells carrying ICEclc (Fig. S3), but not exclusive to the subpopulation of cells expressing the ICEclc Pint-promoter (Fig. 3). This indicates that the prophage activation is a more general effect of ICEclc presence and not linked to the actual ICE horizontal transmission process from a subset of cells. We currently ignore the exact molecular links between ICE and phage activation. Interestingly, Pspu28 activation has previously also been observed as a side-effect of introduction of the conjugative plasmid pCAR1 into P. putida38. This suggests that the phage is somehow particularly prone for being activated by newly incoming mobile elements, but the ecological reasons or host consequences for this activation are unclear.
Another obvious phenotypic change invoked by the presence of ICEclc was swimming motility. P. putida-ICEclc cells grown on 3CBA more highly expressed genes for flagellar assembly than P. putida without ICE (but with the clc genes for 3CBA metabolism), whereas such cells also displayed stronger motility. In contrast, P. putida-ICEclc cultures pregrown on LB specifically repressed motility. This indicates that ICEclc controls host motility depending on the carbon source. The finding that P. putida-ICEclc cells grown on 3CBA move faster is very suggestive for chemotaxis toward the compound, which could be a selective advantage of carrying the ICE to the host metabolizing 3CBA. Although bacterial chemotaxis toward aromatic compounds has been well characterized in various species39, specific chemotaxis to 3CBA has not been described. 3CBA chemotaxis may be mediated by the orf2848 gene product encoded by ICEclc, which shares the same functional domains throughout the entire protein sequence with PcaK, a bifunctional membrane protein required for both transport of and chemotaxis to 4-hydroxybenzoate40.
In terms of overall fitness, the introduction of ICEclc into P. putida thus caused mixed effects. Despite the differential expression of 2.8% of genes in the P. putida genome in presence of ICEclc in exponentially growing cells, and despite activation of the Pspu28 prophage, the growth rate on 3CBA of P. putida-ICEclc was actually slightly more elevated than that of P. putida 3227 (without ICE, but with the clc genes, Fig. 5A). The lag phase of P. putida-ICEclc on 3CBA, however, is longer than that of P. putida 3227 (Fig. 5B), which is likely due to the limited growth of the 3–5% subpopulation of stationary phase cells that activate ICEclc horizontal transmission and perish upon nutrient restimulation, as previously attested by single-cell microscopy28,29. The poor growth of ICEclc-transmitting cells may thus also be responsible for the observed reduction in REG–phase expression of biological pathways, such as cell cycle (ko04112) and amino acids (ko00260 and ko00330) or carbohydrate (ko00630 and ko00500) metabolism. On average, therefore, ICEclc increases population fitness on 3CBA as carbon substrate (i.e., higher growth rate, increased chemotaxis), while its fitness impacts (i.e., longer lag phase) are minimized by restricting horizontal transmission to a small proportion of cells in the population29. In contrast, growth rates on LB were slightly lower (Fig. 5), without any obvious Pspu28 phage activation (Fig. S4). Although we did not specifically measure global transcriptome changes in LB-grown cells, growth rate differences suggest that ICEclc is imposing a slight fitness costs under such conditions in P. putida. Fitness differences, globally speaking, do not seem to be influenced by the induction or not of Pspu28 phage genes.
Overall, host-mobile element partnerships seem to have evolved multiple times conceptually similar systems to optimize the balance between host-incurred costs and benefits41,42. ICEclc core gene expression remains silent in exponentially-growing cells, which is the result of global repression by the TetR-type MfsR transcription regulator31. In stationary phase, ICEclc horizontal transmission is restricted to a subpopulation of cells as a result of a bistable switch in a hitherto uncharacterized mechanism34. Similarly, but mechanistically different to ICEclc, temperate phages and other ICEs limit horizontal transmission by deploying a strong double negative feedback circuit to repress their lytic cycle21,25,43,44. Conjugative plasmids are known to exploit specific H-NS-like stealth proteins, temporarily silencing their gene expression upon entry into a new recipient cell in order to reduce fitness impairment17–19.
Our study thus indirectly shows the large impact on host cells of ICEclc during its active transmission phase (i.e., REG phase cells). At this stage we can only infer this impact indirectly because of the masking effect of the large proportion of cells in REG phase in which ICEclc remains silent compared to those in which it is active (3–5%). Whereas a number of studies have revealed host genome-wide responses during lytic phage infection45–51, the differences during vertical (lysogenic cycle) and horizontal transmission stages of temperate phages (lytic cycle) have largely remained unexplored20,52,53. Similarly, specific host transcriptome changes during plasmid horizontal transmission have been poorly documented38,54–56. It will be interesting to characterize the host-ICEclc partnership further, in particular in cells actively undergoing ICE horizontal transmission, but this will have to be addressed by separating individual subpopulations.
Methods
Bacterial strains and culture media
Escherichia coli DH5α (Gibco Life Technologies) and DH5αλpir31 were routinely grown at 37 °C on Luria-Bertani (LB) medium57 for plasmid constructions. Derivatives of P. putida UWC1, a spontaneous rifampicin resistant strain of KT2440, used in this study (Table S3) were cultured at 30 °C on LB or type 21 C minimal medium (MM)58 containing 5 mM 3CBA. If necessary, antibiotics were added at the following concentrations; kanamycin 25 μg mL−1, gentamicin 20 μg mL−1, ampicillin 100 μg mL−1, and tetracycline 50 μg mL−1.
RNA-sequencing using bulk populations
P. putida strains were precultured in MM containing 5 mM 3CBA until exponential growth phase (OD = ~0.6) and transferred into fresh medium with 100-fold dilution. The cells of four replicate cultures were collected either at exponential phase (OD = ~0.6) or from late-stationary phase cells grown for 96 h on 3CBA, and then further stimulated for 4.5 h with additional 5 mM 3CBA (REG phase). Collected cells were immediately treated with RNAlater (Ambion). Total RNA was extracted using the hot phenol method59, and the residual co-extracted genomic DNA was digested by incubation with Turbo DNase (Life Technologies) following manufacturer’s instructions. The RNA was further purified using the RNeasy MiniElute cleanup kit (Qiagen), and rRNA was specifically depleted using Ribo-Zero rRNA removal kit Bacteria (Epicentre). The quality and quantity of RNA were measured using a Fragment Analyzer (Advanced Analytical) and a High Sensitivity RNA Analysis Kit (Advanced Analytical). cDNA libraries were generated using a ScriptSeq Complete Kit Bacteria (Epicentre) following strand-specific library preparation protocol. The indexed cDNA libraries were pooled and sequenced on Illumina HiSeq. 2000 platform with 101-nt single-end reads.
Bioinformatics
High quality reads (mean quality score >35) were mapped to the reference sequences of P. putida strain KT2440 chromosome (refseq NC_002947) and ICEclc (Genbank accession AJ617740) using Bowtie allowing 2 bp mismatches for a 101 bp read60. Reads mapped to a unique position were used for subsequent analysis, removing a massive amount of reads from rRNA genes. Mapped data were converted to the BAM format using Samtools61, then read counts for coding sequences (CDSs) were obtained using HTseq62. Coverage was obtained using BEDtools63. We obtained 2.1 to 10 million reads mapped to CDSs per sample.
Differential expression analysis was performed using the TMM normalization-edgeR exact-test iteration protocol of the Bioconductor package TCC64. Multidimensional scaling analysis was performed using the plotMDS function in edgeR with default arguments. A false discovery rate (q.value) of <0.05 was considered statistically significantly differentially expressed. Pathway level activity analysis32 was conducted to detect gene set level expression changes. KEGG orthology ID was assigned to chromosomally encoded genes using blastKOARA of KEGG65, and subsequently a total of 83 pathways, each of which contained more than 10 genes assigned were tested. In this method, pathway activity is essentially the sum of Z-score of gene expression values of gene set members whose expression values were shifted to the major direction. Read counts were normalized across samples using internal functions of TCC package, then used as input data for the Z-score analysis option of Bioconductor package GSVA66.
Fitness assay
P. putida strain 3227 (-ICEclc) and 2737 (+ICEclc) were precultured in MM containing 5 mM 3CBA for 72 h until stationary phase (OD = 1.5), and transferred with 100-fold dilution into fresh LB or MM containing 2 mM 3CBA. The optical densities (OD600) of fresh cultures with 20 replicates were monitored at intervals of 10 min for 48 h, using Tecan Spark 10 M microplate reader. Maximum growth rate (μmax) and length of lag-phase were calculated using the R package grofit67 with smooth.gc parameter 0.5, in which we used logistic model fitting for LB and smoothing spline method with 100 bootstrapping runs for MM containing 3CBA cultures.
Single-cell gene expression assay
A 334-bp promoter region of the PP_1548 gene was amplified with primers 160701 (5′-tttACTAGTCCGAAAGCTCCTACACACTG) and 160702 (5′-tttGGTACCATGGGGTGTTGCTCCGTG) using genomic DNA of P. putida UWC1 as a template, and digested with SpeI and KpnI. An mcherry gene fragment was amplified using primers 160614 (5′-tttGGTACCTTAACTTTTAAGGAGGAAAAACATA) and 160615 (5′-TTATTTGTACAGCTCATCCATG) plus pCK218-Pint-gfp-PinR-mcherry as a template35, and digested with KpnI. The two digested fragments were cloned into SpeI and DraI sites of the mini-Tn5 vector pTnMod-OTc. The resulting plasmid pTnMod-P1548-mcherry and another mini-Tn5 plasmid pCK218-jim1 containing Pint-egfp68 were both introduced into either P. putida strain 3227 or 2737 by electroporation. Three individual clones of each transformant containing the dual reporter genes were cultured in MM plus 5 mM 3CBA or LB and collected at exponential and late stationary phases to use for subsequent microscopic assays. Gene expression analysis at single cell level was performed as previously described69, using a Zeiss AxioObserver.Z1 inverted microscope, equipped with either Zeiss AxioCam MRm or AxioCam 503mono CCD, and 100 x /1.4 oil immersion Plan-Apochromat lens at exposure times of 350 ms for phase contrast and 100 ms for fluorescence images. The light source used for fluorescence imaging was Zeiss Colibri.2. Filters used for eGFP and mCherry were Zeiss 38HE and Semrock mCherry-B-000, respectively.
Motility assay
Bacterial cells were pregrown in LB for 18 h or in MM supplied with 5 mM 3CBA for 72 h until stationary phase, and 1-µl aliquots of precultures adjusted to a culture turbidity of 0.6 at 600 nm were spotted on PG medium (0.5% Difco Proteose Peptone No.3, 0.2% glucose) solidified with 0.3% agar70. After 19 h incubation at 25 °C, diameters of swimming rings were measured (n = 5).
Data availability
RNA-seq datasets generated during this study are deposited and available in the Sequence Read Archives of National Center for Biotechnology Information (NCBI), European Bioinformatics Institute (EBI) and DNA Data Bank of Japan (DDBJ), http://trace.ddbj.nig.ac.jp/DRASearch, under accession number DRA005831.
Electronic supplementary material
Acknowledgements
This work was supported by Japan Society for the Promotion of Science Grant JP26712011 and Japan Science and Technology Agency Grant ERATO JPMJER1502 to RM, and by Swiss National Science Foundation Grants 31003A_144141/1 and 310030B_156926/1 to JRvdM. We thank Nadezda Kryuchkova for her assistance with the edgeR script, Hitomi Matsuo and Kayo Ohkouchi for their technical assistance, and Terumi Horiuchi for the assistance of data deposition. The super-computing resource was provided by Human Genome Center, the Institute of Medical Science, the University of Tokyo.
Author Contributions
R.M. and J.R.v.d.M. designed research; R.M. and V.S. performed research; R.M., H.Y. and J.R.v.d.M. analyzed data; and R.M., H.Y. and J.R.v.d.M. wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Ryo Miyazaki and Hirokazu Yano contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23858-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 2.Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet. 2015;16:472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 3.Bender JK, et al. Population structure and acquisition of the vanB resistance determinant in German clinical isolates of Enterococcus faecium ST192. Sci Rep. 2016;6:21847. doi: 10.1038/srep21847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senn L, et al. The Stealthy Superbug: the Role of Asymptomatic Enteric Carriage in Maintaining a Long-Term Hospital Outbreak of ST228 Methicillin-Resistant Staphylococcus aureus. mBio. 2016;7:e02039–02015. doi: 10.1128/mBio.02039-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wendel AF, Ressina S, Kolbe-Busch S, Pfeffer K, MacKenzie CR. Species Diversity of Environmental GIM-1-Producing Bacteria Collected during a Long-Term Outbreak. Appl Environ Microbiol. 2016;82:3605–3610. doi: 10.1128/AEM.00424-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, et al. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics. 2007;8:121. doi: 10.1186/1471-2164-8-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sangwan N, et al. Reconstructing an ancestral genotype of two hexachlorocyclohexane-degrading Sphingobium species using metagenomic sequence data. ISME J. 2014;8:398–408. doi: 10.1038/ismej.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Top EM, Springael D. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol. 2003;14:262–269. doi: 10.1016/S0958-1669(03)00066-1. [DOI] [PubMed] [Google Scholar]
- 10.Verma, H. et al. Comparative genomic analysis of nine Sphingobium strains: insights into their evolution and hexachlorocyclohexane (HCH) degradation pathways. BMC Genomics15, 1014 (2014). [DOI] [PMC free article] [PubMed]
- 11.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 12.Baltrus DA. Exploring the costs of horizontal gene transfer. Trends Ecol Evol. 2013;28:489–495. doi: 10.1016/j.tree.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Bragg JG, Wagner A. Protein material costs: single atoms can make an evolutionary difference. Trends Genet. 2009;25:5–8. doi: 10.1016/j.tig.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Shachrai I, Zaslaver A, Alon U, Dekel E. Cost of unneeded proteins in E. coli is reduced after several generations in exponential growth. Mol Cell. 2010;38:758–767. doi: 10.1016/j.molcel.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 15.Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narra HP, Cordes MH, Ochman H. Structural features and the persistence of acquired proteins. Proteomics. 2008;8:4772–4781. doi: 10.1002/pmic.200800061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banos RC, et al. Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H-NS. PLoS Genet. 2009;5:e1000513. doi: 10.1371/journal.pgen.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle M, et al. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- 19.Skennerton CT, et al. Phage encoded H-NS: a potential achilles heel in the bacterial defence system. PLoS One. 2011;6:e20095. doi: 10.1371/journal.pone.0020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Golding I, Sawai S, Guo L, Cox EC. Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol. 2005;3:e229. doi: 10.1371/journal.pbio.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feiner R, et al. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol. 2015;13:641–650. doi: 10.1038/nrmicro3527. [DOI] [PubMed] [Google Scholar]
- 22.Paul JH. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008;2:579–589. doi: 10.1038/ismej.2008.35. [DOI] [PubMed] [Google Scholar]
- 23.Johnson CM, Grossman AD. Integrative and Conjugative Elements (ICEs): What They Do and How They Work. Annu Rev Genet. 2015;49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 25.Delavat, F., Miyazaki, R., Carraro, N., Pradervand, N. & van der Meer, J. R. The hidden life of integrative and conjugative elements. FEMS Microbiol Rev (2017). [DOI] [PMC free article] [PubMed]
- 26.Miyazaki R, et al. Comparative genome analysis of Pseudomonas knackmussii B13, the first bacterium known to degrade chloroaromatic compounds. Environ Microbiol. 2015;17:91–104. doi: 10.1111/1462-2920.12498. [DOI] [PubMed] [Google Scholar]
- 27.Miyazaki R, van der Meer JR. A dual functional origin of transfer in the ICEclc genomic island of Pseudomonas knackmussii B13. Mol Microbiol. 2011;79:743–758. doi: 10.1111/j.1365-2958.2010.07484.x. [DOI] [PubMed] [Google Scholar]
- 28.Reinhard F, Miyazaki R, Pradervand N, van der Meer JR. Cell differentiation to “mating bodies” induced by an integrating and conjugative element in free-living bacteria. Curr Biol. 2013;23:255–259. doi: 10.1016/j.cub.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 29.Delavat F, Mitri S, Pelet S, van der Meer JR. Highly variable individual donor cell fates characterize robust horizontal gene transfer of an integrative and conjugative element. Proc Natl Acad Sci USA. 2016;113:E3375–3383. doi: 10.1073/pnas.1604479113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaillard M, Pernet N, Vogne C, Hagenbuchle O, van der Meer JR. Host and invader impact of transfer of the clc genomic island into Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci USA. 2008;105:7058–7063. doi: 10.1073/pnas.0801269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pradervand N, et al. An operon of three transcriptional regulators controls horizontal gene transfer of the integrative and conjugative element ICEclc in Pseudomonas knackmussii B13. PLoS Genet. 2014;10:e1004441. doi: 10.1371/journal.pgen.1004441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee E, Chuang HY, Kim JW, Ideker T, Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput Biol. 2008;4:e1000217. doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieder S, Nikel PI, de Lorenzo V, Takors R. Genome reduction boosts heterologous gene expression in Pseudomonas putida. Microb Cell Fact. 2015;14:23. doi: 10.1186/s12934-015-0207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quesada JM, Soriano MI, Espinosa-Urgel M. Stability of a Pseudomonas putida KT2440 bacteriophage-carried genomic island and its impact on rhizosphere fitness. Appl Environ Microbiol. 2012;78:6963–6974. doi: 10.1128/AEM.00901-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minoia M, et al. Stochasticity and bistability in horizontal transfer control of a genomic island in Pseudomonas. Proc Natl Acad Sci USA. 2008;105:20792–20797. doi: 10.1073/pnas.0806164106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaillard M, et al. Transcriptome analysis of the mobile genome ICEclc in Pseudomonas knackmussii B13. BMC Microbiol. 2010;10:153. doi: 10.1186/1471-2180-10-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. Prophage genomics. Microbiol Mol Biol Rev. 2003;67:238–276. doi: 10.1128/MMBR.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi Y, et al. Modulation of primary cell function of host Pseudomonas bacteria by the conjugative plasmid pCAR1. Environ Microbiol. 2015;17:134–155. doi: 10.1111/1462-2920.12515. [DOI] [PubMed] [Google Scholar]
- 39.Parales RE, Harwood CS. Bacterial chemotaxis to pollutants and plant-derived aromatic molecules. Curr Opin Microbiol. 2002;5:266–273. doi: 10.1016/S1369-5274(02)00320-X. [DOI] [PubMed] [Google Scholar]
- 40.Ditty JL, Harwood CS. Conserved cytoplasmic loops are important for both the transport and chemotaxis functions of PcaK, a protein from Pseudomonas putida with 12 membrane-spanning regions. J Bacteriol. 1999;181:5068–5074. doi: 10.1128/jb.181.16.5068-5074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison E, Brockhurst MA. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012;20:262–267. doi: 10.1016/j.tim.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 42.Turner PE, Cooper VS, Lenski RE. Tradeoff between Horizontal and Vertical Modes of Transmission in Bacterial Plasmids. Evolution. 1998;52:315–329. doi: 10.1111/j.1558-5646.1998.tb01634.x. [DOI] [PubMed] [Google Scholar]
- 43.Trinh JT, Szekely T, Shao Q, Balazsi G, Zeng L. Cell fate decisions emerge as phages cooperate or compete inside their host. Nat Commun. 2017;8:14341. doi: 10.1038/ncomms14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng L, et al. Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell. 2010;141:682–691. doi: 10.1016/j.cell.2010.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doron S, et al. Transcriptome dynamics of a broad host-range cyanophage and its hosts. ISME J. 2016;10:1437–1455. doi: 10.1038/ismej.2015.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavigne R, et al. A multifaceted study of Pseudomonas aeruginosa shutdown by virulent podovirus LUZ19. mBio. 2013;4:e00061–00013. doi: 10.1128/mBio.00061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindell D, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–86. doi: 10.1038/nature06130. [DOI] [PubMed] [Google Scholar]
- 48.Mojardin L, Salas M. Global Transcriptional Analysis of Virus-Host Interactions between Phage varphi29 and Bacillus subtilis. J Virol. 2016;90:9293–9304. doi: 10.1128/JVI.01245-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poranen MM, et al. Global changes in cellular gene expression during bacteriophage PRD1 infection. J Virol. 2006;80:8081–8088. doi: 10.1128/JVI.00065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ravantti JJ, Ruokoranta TM, Alapuranen AM, Bamford DH. Global transcriptional responses of Pseudomonas aeruginosa to phage PRR1 infection. J Virol. 2008;82:2324–2329. doi: 10.1128/JVI.01930-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao X, et al. Global Transcriptomic Analysis of Interactions between Pseudomonas aeruginosa and Bacteriophage PaP3. Sci Rep. 2016;6:19237. doi: 10.1038/srep19237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osterhout RE, Figueroa IA, Keasling JD, Arkin AP. Global analysis of host response to induction of a latent bacteriophage. BMC Microbiol. 2007;7:82. doi: 10.1186/1471-2180-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veses-Garcia M, et al. Transcriptomic analysis of Shiga-toxigenic bacteriophage carriage reveals a profound regulatory effect on acid resistance in Escherichia coli. Appl Environ Microbiol. 2015;81:8118–8125. doi: 10.1128/AEM.02034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lang KS, Johnson TJ. Transcriptome modulations due to A/C2 plasmid acquisition. Plasmid. 2015;80:83–89. doi: 10.1016/j.plasmid.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 55.Paytubi S, et al. A novel role for antibiotic resistance plasmids in facilitating Salmonella adaptation to non-host environments. Environ Microbiol. 2014;16:950–962. doi: 10.1111/1462-2920.12244. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, Xiang L, Shao J, Wegrzyn A, Wegrzyn G. Effects of the presence of ColE1 plasmid DNA in Escherichia coli on the host cell metabolism. Microb Cell Fact. 2006;5:34. doi: 10.1186/1475-2859-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook, J. & Russell, D. W. Molecular Cloning: a laboratory manual. (Cold Spring Harbor Laboratory Press, 2001).
- 58.Gerhardt, P. et al. Manual of Methods for General Bacteriology. (American Society for Microbiology, 1981).
- 59.Baumann B, Snozzi M, Zehnder AJ, Van Der Meer JR. Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J Bacteriol. 1996;178:4367–4374. doi: 10.1128/jb.178.15.4367-4374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun J, Nishiyama T, Shimizu K, Kadota K. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics. 2013;14:219. doi: 10.1186/1471-2105-14-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 66.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kahm, M., Hasenbrink, G., Lichtenberg-Fraté, H., Ludwig, J. & Kschischo, M. grofit: Fitting Biological Growth Curves with R. J Stat Softw33 (2010).
- 68.Sentchilo V, Zehnder AJ, van der Meer JR. Characterization of two alternative promoters for integrase expression in the clc genomic island of Pseudomonas sp. strain B13. Mol Microbiol. 2003;49:93–104. doi: 10.1046/j.1365-2958.2003.03548.x. [DOI] [PubMed] [Google Scholar]
- 69.Miyazaki R, et al. Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet. 2012;8:e1002818. doi: 10.1371/journal.pgen.1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matilla MA, et al. Temperature and pyoverdine-mediated iron acquisition control surface motility of Pseudomonas putida. Environ Microbiol. 2007;9:1842–1850. doi: 10.1111/j.1462-2920.2007.01286.x. [DOI] [PubMed] [Google Scholar]
- 71.Reinhard F, van der Meer JR. Improved statistical analysis of low abundance phenomena in bimodal bacterial populations. PLoS One. 2013;8:e78288. doi: 10.1371/journal.pone.0078288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets generated during this study are deposited and available in the Sequence Read Archives of National Center for Biotechnology Information (NCBI), European Bioinformatics Institute (EBI) and DNA Data Bank of Japan (DDBJ), http://trace.ddbj.nig.ac.jp/DRASearch, under accession number DRA005831.