Abstract
Host specialization after host shifting is traditionally viewed as the pathway to speciation in parasitic plants. However, geographical and environmental changes can also influence parasite speciation, through hybridization processes. Here we investigated the impact of past climatic fluctuations, environment, and host shifts on the genetic structure and patterns of hybridization and gene flow between Psittacanthus calyculatus and P. schiedeanus, a Mesoamerican species complex. Using microsatellites (408 individuals), we document moderate genetic diversity but high genetic differentiation between widespread parental clusters, calyculatus in dry pine-oak forests and schiedeanus in cloud forests. Bayesian analyses identified a third cluster, with admixture between parental clusters in areas of xeric and tropical dry forests and high levels of migration rates following secondary contact. Coincidently host associations in these areas differ from those in areas of parental species, suggesting that past hybridization played a role in environmental and host shifts. Overall, the observed genetic and geographic patterns suggest that these Psittacanthus populations could have entered a distinct evolutionary pathway. The results provide evidence for highlights on the importance of the Pleistocene climate changes, habitat differences, and potential host shifts in the evolutionary history of Neotropical mistletoes.
Introduction
The speciation process is viewed as a continuum (the ‘speciation continuum’) with many stages that vary in space and time1–3, particularly for closely related taxa with varying levels of divergence4. The duration of different stages along the speciation continuum depends on the magnitude and timing of gene flow and on the balance among other evolutionary forces including genetic drift, recombination and selection3,5, which in turn are likely affected by historical and environmental factors6. Understanding population divergence and species boundaries is a complex task because it needs the integration of historical, demographic and ecological data5,7,8. For instance, knowledge of historical changes of distributions of populations is essential to understand current geographic patterns of genetic structure and species diversification9.
Based on a three-gene mitochondrial DNA phylogeny of seed plants, researchers inferred at least 11 independent origins of parasitism in Angiosperms (1%), eight of which consist entirely of holoparasitic species that lack photosynthetic ability10. Modern-day parasites have disproportionately evolved in certain lineages and the endoparasitic habit has arisen by convergence in four clades10. In addition, single gene analyses revealed multiple horizontal transfers from host to parasite lineage, making parasitic plants a very interesting model for studying the effects of the parasitic lifestyle on plant diversification10. Mistletoe parasitism has been postulated as a major driver of speciation because the parasite’s success depends on multiple, sometimes specialized biotic interactions (hosts, pollinators and seed dispersers) that could indirectly promote the strength and evolution of traits reinforcing the long-term relationship between species enough to isolate parasite populations into distinct races11–19.
The parasitic Loranthaceae (Santalales) are mostly aerial mistletoes very common in temperate and tropical plant communities, and typically use a broad range of host tree species15,20–22. The life cycle of most loranth mistletoes starts with frugivorous birds dispersing the seeds from tree to tree. Once the seeds are regurgitated or defecated by the bird, these adhere to the host tree with the natural ‘glue’ viscin, and then penetrate the woody host tissue with a structure named haustorium13,15. Upon reaching sexual maturity, the pollen is typically moved from flower to flower by a wide range of pollinators, including several species of insects, birds and bats16,23–27. Given that most avian seed dispersers and pollinators of loranth mistletoes are not sufficiently specialized21, it is unlikely that gene dispersal vectors reproductively isolate mistletoes into populations growing on different hosts within a community (for alternative scenarios see16,28,29). Instead, the genetic structuring of mistletoe populations is more likely to be influenced by host-parasite interactions12,30–32.
Mistletoes are expected to establish and survive on higher-quality hosts (‘host quality’ hypothesis33) and, therefore, variation in host quality would account for non-random occurrence patterns of parasitic plants. Beyond these mechanisms determining the distribution at local and large geographical scales, the diversification of mistletoe species has been explained through different mechanisms linked to host-parasite interactions12. Accordingly, the most accepted explanation of mistletoe diversification is that of ‘host-race formation’12,34–36, where genetic differentiation, and eventually host-race formation, is acquired through isolation-by-distance or by ecological adaptation following the ‘invasion’ of a different host species (‘host-switching’ hypothesis12). In addition, the geographic structuring of genetic variation in some mistletoe species has been explained as the result of past climate changes31,32,37,38, landscape fragmentation39, emergence of biogeographic barriers30–32,38, and by the parasites’ own climatic niche preferences40,41.
Although hybridization and introgression have been acknowledged as potentially important mechanisms for adaptive evolution42–44, their effects on the genetic structuring of mistletoe populations remain unexplored. Under changing climatic conditions, hybridization and introgression could have played an important role in diversification by enhancing species’ responses to environmental changes45. More specifically, the strong impact of Pleistocene climate cycles on distributions of populations of North American plant communities46–50, provided the opportunity for populations of recently diverged species to come into secondary contact, leading to hybridization, introgression, and the presence of heterogeneous admixed populations11,51–54. In parasitic species, the presence of hybrid intermediates can affect patterns of host specificity by facilitating host shifts, enhancing virulence or increasing the transmission rates of among different hosts (‘hybrid bridge’ hypothesis55,56). Thus, the evaluation of the temporal and geographical patterns of hybridization in mistletoe species may shed light onto complex evolutionary scenarios of species divergence and their association with host preferences and geographic isolation of populations.
Psittacanthus (Loranthaceae, c. 119 species) are characteristic stem parasites (hemiparasites) throughout the Neotropics21. Most species are host generalists and ecologically very important because they provide food resources (e.g., fruits and nectar) to many animals13 and indirectly influence community structure in low productivity systems33,57. The recently diverged species complex of Psittacanthus calyculatus and P. schiedeanus (c. 2.5–1.8 Ma32,38) can be found infecting different host tree species under distinct environmental conditions along their wide geographical distributions29,41,58–60. Kuijt21 reported that these species are distributed, more or less sympatrically, from Mexico to Panama, but recent molecular data suggest that these are allopatric and restricted to Mexico41. In addition, P. calyculatus mainly parasitizes Quercus (Fagaceae) trees in the central highlands of Mexico, whereas P. schiedeanus parasitizes cloud forest trees, mainly Liquidambar styraciflua (Altingiaceae) in eastern Mexico32,38,41. Patterns of genetic structure in chloroplast DNA (cpDNA) sequences and nuclear microsatellites showed two main lineages within the species complex32,38,41, with further genetic structuring of populations and evidence of genetic admixture in particular regions41. However, the presence of highly admixed individuals within particular populations has not been explored or discussed further41. Despite clear geographic and ecological differences between the ‘calyculatus’ and ‘schiedeanus’ genetic clusters, molecular evidence suggests population admixture within the Tehuacán-Cuicatlán Valley and the Central Valleys of Oaxaca, and the Central Depression of Chiapas32,38,41. Overall, these regions have a drier climate and more xeric vegetation types than those occurring elsewhere at higher elevations. Interestingly, populations within these two regions tend to infect a different set of host species16,21,41. Furthermore, past and present distribution models predict geographic overlap between the two species in the Central Valleys of Oaxaca during the last glacial cycle (ca. 100–200 kya32,38). Therefore, we would hypothesize that past migrations of the two species into the more xeric lowlands led to a secondary contact zone, possibly associated with shifts in host-preferences.
In this study we re-analysed previously reported microsatellite data41 to estimate levels of hybridization among groups of populations and rates of migration and directionality over contemporary and historical timescales. We then inferred the most likely scenario and timing of secondary contact and introgression using historical demography and Approximate Bayesian Computation (ABC) methods. Specifically, we addressed the following questions: (1) Are levels of hybridization and introgression geographically concordant with differences in the climatic and host preferences of populations? (2) Are the predicted temporal changes in migration and range shifts associated with the presence of varying levels of hybridization and introgression? We discuss the main causes that could account for the complex phylogeographic patterns observed and the ecological context that could provide valuable clues for the understanding of the evolutionary course of introgressive hybridization in a parasitic plant species’ complex.
Results
Genetic differentiation and population structure
Descriptive statistics for the two groups defined by previous species assignments (CALY and SCHI) were remarkably similar to groups defined by habitat, admixture level or geography (Table S2), suggesting shared patterns of demographic history between the CALY and SCHI groups. Allelic richness was generally high for all groups, yet the observed heterozygosity (HO) was lower than expected and inbreeding coefficient values ranged from 0.038 to 0.64 (Table S2). The existence of two major clusters was supported by AMOVA, in which 5.8% of the total genetic variation was explained by significant differentiation between the CALY and SCHI genetic groups (Table S3). When AMOVA was used to explore for geographic structure between groups separated by habitat (three groups), admixture (three groups) or geography (four groups), group differences contributed significantly to 5.9%, 6.6% and 6.38% of the total variance, respectively (Table S3).
Genetic differentiation (FST-NA) and allele frequencies (Jost’s D) showed the same pattern of differentiation among groups (Table 1). The highest values of genetic differentiation were observed between populations from the TMVB and SMOr, corresponding to group-comparisons by species and admixture level (CALY vs. SCHI) and habitat (XTF vs. CF) (Table 1). Interestingly, the CALY and SCHI groups are clearly differentiated from each other and the HYBR group was more similar to CALY (Table 1).
Table 1.
Genetic differentiation (FST) corrected by the presence of null alleles and absolute allele frequencies differences (Jost’s D) between groups with 95% CI.
| FST-ENA | Jost’s D | |
|---|---|---|
| Species | ||
| CALY vs. SCHI | 0.107 (0.055–0.183) | 0.61 (0.57–0.65) |
| Habitat type | ||
| XTF vs. CF | 0.133 (0.074–0.214) | 0.64 (0.60–0.68) |
| XTF vs. TDF | 0.076 (0.044–0.129) | 0.56 (0.51–0.62) |
| CF vs. TDF | 0.058 (0.030–0.111) | 0.27 (0.20–0.35) |
| Admixture level | ||
| CALY vs. HYBR | 0.082 (0.044–0.153) | 0.53 (0.47–0.58) |
| CALY vs. SCHI | 0.152 (0.068–0.264) | 0.60 (0.56–0.65) |
| SCHI vs. HYBR | 0.101 (0.066–0.148) | 0.65 (0.57–0.70) |
| Geography | ||
| TMVB vs. OAX | 0.084 (0.045–0.155) | 0.48 (0.41–0.53) |
| TMVB vs. SMOr | 0.157 (0.077–0.260) | 0.61 (0.56–0.65) |
| TMVB vs. CHIS | 0.113 (0.067–0.174) | 0.67 (0.58–0.76) |
| OAX vs. SMOr | 0.117 (0.082–0.165) | 0.68 (0.60–0.75) |
| OAX vs. CHIS | 0.074 (0.046–0.110) | 0.54 (0.45–0.64) |
| SMOr vs. CHIS | 0.099 (0.049–0.168) | 0.50 (0.39–0.61) |
TMVB = calyculatus group, SMOr = schiedeanus group, OAX = Oaxaca region, CHIS = Chiapas region. CF = cloud forest, XTF = temperate forest and xeric groups, TDF = Tropical dry forest. HYBR = admixed populations from Oaxaca and Chiapas regions. Estimates were done over 9 loci.
Admixture analysis and hybrid identification
As expected, two genetic groups were recovered by STRUCTURE (best-supported K = 2 determined by the Evanno’s method61; Figs 1 and 2A), partially corresponding to previously recognized cpDNA lineages, P. calyculatus and P. schiedeanus32,38,41. Further sub-structuring was observed within each of the two clusters, in the P. calyculatus sub-structure corresponded to populations from the western-central portion of the Trans-Mexican Volcanic Belt (TMVB) and populations from central Mexico (Tlaxcala, Puebla) and Oaxaca (Fig. 2B), whereas in the P. schiedeanus sub-structure corresponded to populations from the Sierra Madre Oriental (SMOr, central Veracruz) and populations from the northernmost distribution (Puebla and San Luis Potosí), Oaxaca, and Chiapas (Fig. 2C). At K = 2, several individuals showed signs of admixture and were identified as individuals with an apparent hybrid ancestry between the two main clusters (assignment uncertainty).
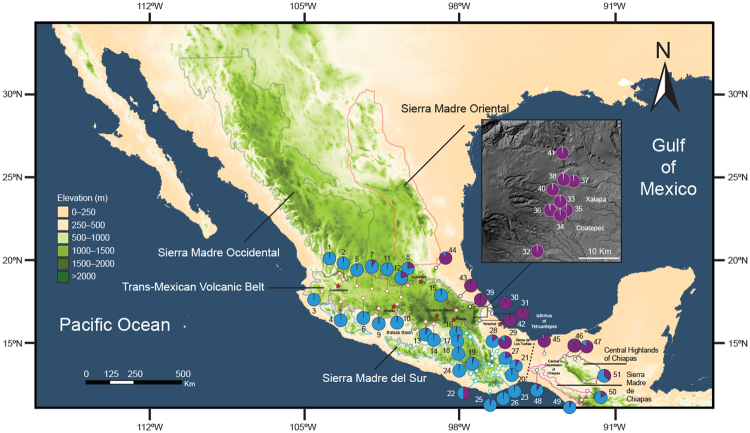
Figure 1.
Relief map showing collection sites and assignment probabilities of individuals to populations of the Psittacanthus calyculatus/P. schiedeanus complex in Mexico. In the inset are the collection sites located in central Veracruz. Numbers refer to collection sites according to Table S1. Average assigning probabilities of individuals to putative populations at K = 2 according to STRUCTURE analysis (see Results). Pie colour coding is the same as in Fig. 2. The studied populations are located in the map with specific colours corresponding to the genetic groups: CALY = P. calyculatus (blue), SCHI = P. schiedeanus (purple). Stars represent main cities along the Trans-Mexican Volcanic Belt. The Mexican mountain systems are highlighted by contour lines corresponding to Sierra Madre Occidental (dark green), Sierra Madre Oriental (orange), Trans-Mexican Volcanic Belt (violet), Sierra Madre del Sur (light blue), Sierra Madre de Chiapas (pink), and Central Highlands of Chiapas (black). This map was generated using the ‘raster’ package in R (https://CRAN.R-project.org/package=raster) and the Global 30 arc-second elevation (GTOPO30) model at a 30-arc seconds spatial resolution (c. 1 km) developed with data through a collaborative effort led by the U.S. Geological Survey’s (USGS) Center for Earth Resources Observation and Science (EROS) Center (https://lta.cr.usgs.gov/GTOPO30). The map in the inset is based on digital elevation model (DEM) available from the Instituto Nacional de Estadística y Geografía (INEGI; http://www.inegi.org.mx/). The figure was drawn using Adobe Illustrator CS6 v16.0.0 (Adobe Systems, Inc.).
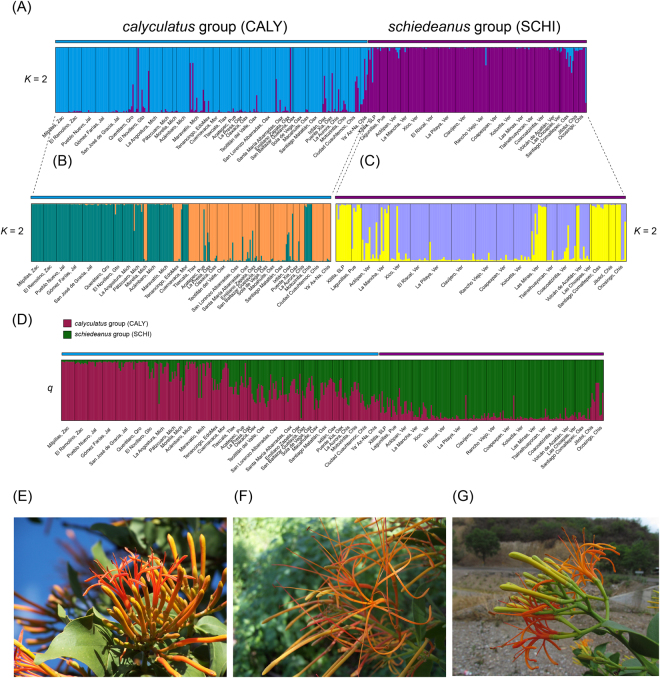
Figure 2.
Assignment probabilities (q) of 408 P. calyculatus/schiedeanus individuals at K = 2 using STRUCTURE (A), and assignment probabilities of individuals to putative sub-structure at K = 2 within each first-level clustering, with individuals of the P. schiedeanus (B) and P. calyculatus (C) excluded. (D) Assignment probabilities of individuals to pure species (P. calyculatus, P. schiedeanus) using the POPFLAG prior information and correlated allele frequencies. Each individual is represented by a vertical line that is partitioned into coloured sections, with the length of each section proportional to the estimated membership coefficient. Photos show flowers of (E) Psittacanthus calyculatus (by Eduardo Ruiz Sanchez) from Jalisco, (F) Psittacanthus schiedeanus (by Juan Francisco Ornelas) from Veracruz, and (G) Psittacanthus putative hybrid (by Eduardo Ruiz Sanchez) from the contact zone in Oaxaca. Note their morphological similarities, with P. schiedeanus larger leaves and fruits, and flowers of typically much longer and more slender than flowers of P. calyculatus21. Flower size is variable in P. schiedeanus and sometimes appears to be intermediate to P. calyculatus.
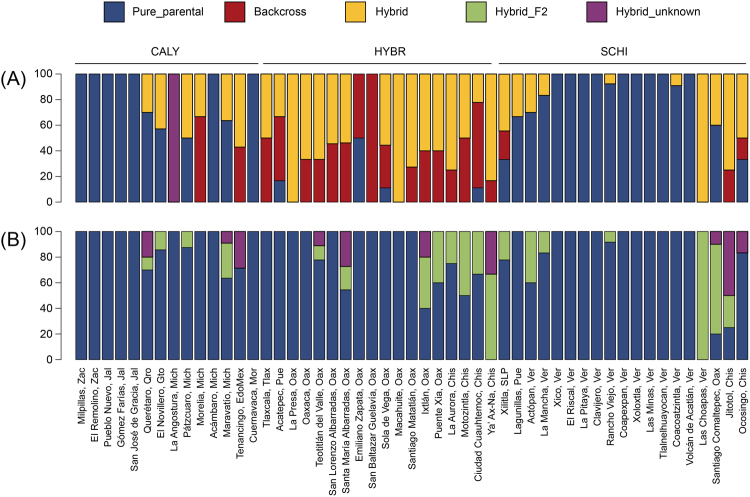
Admixture analysis using STRUCTURE revealed high levels of interspecific admixture in the Central Valleys of Oaxaca and the Central Depression of Chiapas (Fig. 2D). In general, we found a higher proportion of individuals with some degree of hybridization in these two regions, compared to populations from the TMVB (CALY) and the Sierra Madre Oriental (SCHI) (Fig. S1 in Supporting information). When comparing group assignments to the simulated data, we found that several populations varied in the percentage of individuals belonging to each hybrid category (Parental, Hybrid and Backcrosses; Fig. 3). NEWHYBRIDS identified fewer hybrid individuals and these were classified as F2 hybrids or unknown. No F1 or first generation backcrosses were found and some individuals were classified as hybrids of unknown hybrid origin (latter generation hybrids or generation backcrosses; Fig. 3).
Figure 3.
Percentage of hybrids in each population estimated with (A) STRUCTURE and (B) NEWHYBRIDS. CALY = calyculatus group, SCHI = schiedeanus group, OAX = Oaxaca region, CHIS = Chiapas region.
Contemporary and historical migration rates
BAYESASS runs yielded low levels of contemporary gene flow between the two main genetic groups (<10%). The highest migration rates were inferred from the CALY population to the SCHI and HYBR populations and from the SCHI population to the CALY and HYBR populations (Table 2). Migration rates from some populations were not significantly different from zero and migration between groups was very low (<2%).
Table 2.
Contemporary and historical migration rates estimated with BAYESASS and MIGRATE between the two species (CAL = P. calyculatus, SCHI = P. schiedeanus) and the hybrid (HYBR) region (OAX = Oaxaca region and CHIS = Chiapas region). CI 95% intervals are given for contemporary migration (m). Mean and 25% and 75% posterior distribution percentiles of are given for historical migration (M).
| Recipient population | Source population | ||
|---|---|---|---|
| CALY | HYBR | SCHI | |
| BAYESASS | |||
| Recent migration rates (m) | |||
| CALY | 0.9910 (0.0051) | 0.0034 (0.0033) | 0.0026 (0.0025) |
| HYBR | 0.0048 (0.0038) | 0.9866 (0.0072) | 0.0037 (0.0038) |
| SCHI | 0.0037 (0.0034) | 0.0101 (0.0065) | 0.9937 (0.0045) |
| MIGRATE | |||
| Historical migration rates (M) | |||
| CALY | — | 0.12 (0–0.63) | 0.36 (0.15–0.87) |
| HYBR | 0.23 (0.03–0.78) | — | 0.36 (0.12–0.90) |
| SCHI | 0.11 (0–0.6) | 0.20 (0–0.36) | — |
| Historical migration rates (m) | |||
| CALY | — | 0.00006 (0–0.0003) | 0.0002 (0.00007–0.0004) |
| HYBR | 0.0001 (0.00001–0.0003) | — | 0.00018 (0.00006-0.0004) |
| SCHI | 0.000055 (0-0.0003) | 0.0001 (0–0.00018) | — |
| Migrants per generation (θ × M) | |||
| CALY | — | 1.40 (0–8.6) | 2.06 (0.36–3.05) |
| HYBR | 1.79 (0.3–6.1) | — | 2.03 (0.29–3.15) |
| SCHI | 0.83 (0–4.7) | 2.37 (0–4.9) | — |
Theta values: CALY (θ1) = 7.56; HYBRID (θ2) = 11.49; SCHI (θ3) = 5.56.
Estimates of historical migration rates (M) calculated using MIGRATE revealed generally low migration rates among populations, except for the slightly higher migration rates from the SCHI population to the CALY and HYBR populations and from the CALY population to the HYBR population. Estimates of scaled migration rates (m) ranged from 0.00018 to 0.00005 and showed the same pattern among populations. The number of migrants per generation (Nem) ranged from 0.83 to 2.37 (Table 2). Despite the low migration rates, the Mantel test showed that the historical and contemporary migration matrices were not significantly correlated (r = 0.0196, P = 0.5), implying that the rate and intensity of migration have changed over time between groups, with increased gene flow from past to present.
Inference of divergence and secondary contact scenarios
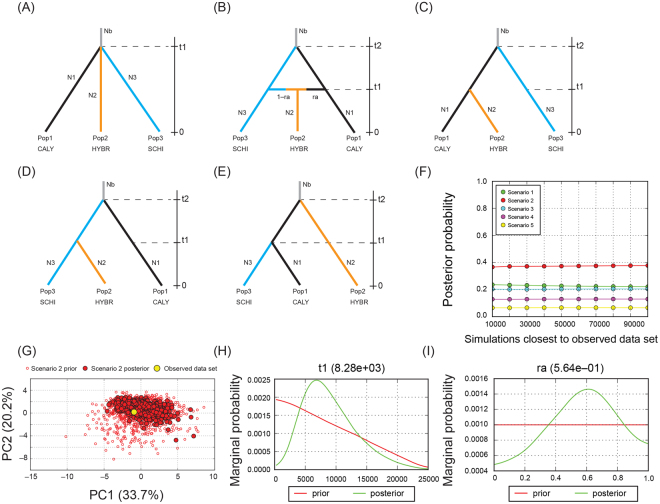
DIYABC analyses indicated that isolation with admixture is the best-supported scenario (scenario 2; Fig. 4), with a higher posterior probability value and 95% confidence intervals that did not overlap with those obtained for the other scenarios (Table 3). Under this scenario, divergence between CALY and SCHI occurred 192.5 kya (t2) followed by secondary contact, and admixture between CALY and SCHI giving rise to the HYBR populations in the Central Valleys of Oaxaca and Central. Depression of Chiapas. Posterior mean parameter estimates suggest that secondary contact occurred 9,010 generations ago (t1). Considering an 11-yr generation time for Psittacanthus32, this translates into 99.1 kya (Table S4 in Supporting information), consistent with a period in which the ranges of CALY and SCHI are predicted to overlap. Finally, the most probable admixture mean rate (ra = 0.547) was higher between the CALY and HYBR populations than admixture between the SCHI and HYBR populations (1-ra = 0.453; Table S4 in Supporting information).
Figure 4.
Competing demographic scenarios of Psittacanthus calyculatus divergence. Five evolutionary scenarios were built and tested using DIYABC: (A) simple split model (scenario 1), in which CALY (Pop1), HYBR (Pop2) and SCHI (Pop3) diverged simultaneously at t1; (B) isolation with admixture model (scenario 2), in which Pop2 (HYBR) was generated by admixture between Pops 1 (CALY) and 3 (SCHI) at t1, then CALY merged with SCHI at t2; (C) hierarchical split model 1 (scenario 3), in which HYBR merged with CALY at t1, then both populations merged with SCHI at t2; (D) hierarchical split model 2 (scenario 4), in which HYBR merged with SCHI at t1, then both populations merged with CALY at t2; (E) hierarchical split model 3 (scenario 5), in which CALY merged with SCHI at t1, then both populations merged with HYBR at t2. The posterior probability of scenarios was assessed using a weighted logistic regression on the 1% of simulated datasets closest to the observed data and, for the best-supported scenario (scenario 2); (F) Results of a logistical model comparing the posterior probability of each scenario with the number of simulations used to calculate it; (G) A model checking procedure was applied using a PCA on test statistic vectors to visualize the fit between the simulated and observed datasets. Note the large cloud of data from the prior and observed datasets centred on a small cluster from the posterior predictive distribution, suggesting that the best-supported scenario explained the observed data well. Prior and posterior probabilities of parameters t1 (H) and ra (=admixture rate) are provided (I).
Table 3.
Posterior probability and 95% confidence intervals (CI) using a logistic regression in the Approximate Bayesian Computation (ABC) analysis for five scenarios of Psittacanthus calyculatus/schiedeanus divergence and hybridization. Type I and Type II error rates were estimated for scenario 2.
| Scenario | Posterior probability | 95% CI | Type I error rate | Type II error rate |
|---|---|---|---|---|
| Simple split model (scenario 1) | 0.2212 | 0.2170–0.2255 | ||
| Isolation with admixture model (scenario 2) | 0.3778 | 0.3729–0.3828 | 0.28 | 0.058 |
| Hierarchical split model (scenario 3) | 0.2000 | 0.2008–0.2029 | ||
| Hierarchical split model (scenario 4) | 0.1300 | 0.1266–0.1335 | ||
| Hierarchical split model (scenario 5) | 0.0659 | 0.0631–0.0686 |
Discussion
Patterns of genetic structure in the Psittacanthus species complex
We investigated patterns of genetic structure and levels of hybridization among groups of populations and estimated rates and directionality of migration over contemporary and historical timescales between P. calyculatus and P. schiedeanus using nuclear microsatellites. Our study revealed that the high levels of admixture are best explained by past introgression as a result of gene flow during secondary contact.
The distribution of individuals with signs of admixture was not restricted to the area of sympatry in the Central Valleys of Oaxaca, but also occurred in the Central Depression of Chiapas, which is separated by warmer and drier conditions along the Isthmus of Tehuantepec from most other populations of the species41. These two regions presented a higher proportion of intermixed individuals, likely F2 or later generation hybrids. Although genetic differentiation between P. calyculatus and P. schiedeanus is moderate (FST-NA = 0.107), the genetic differentiation between groups increases (FST-NA = 0.152) when data for populations of the HYBR group are excluded, indicating that this group represents a genetic bridge between the two parental species. Overall, the admixture results confirm our prediction that the more arid Central Valleys of Oaxaca and Central Depression of Chiapas represent hybrid zones. However, the genetic distinction between parental species can be maintained by host association (environmental context) and by the geographic characteristics of their ranges, even in the presence of gene flow.
Interestingly, the very low genetic differentiation and high admixture levels between populations in the Central Valleys of Oaxaca and the Central Depression of Chiapas suggest that the Isthmus of Tehuantepec may not be strong barrier to gene flow for these mistletoes, as observed for other bird-dispersed plant species62,63, but not in other species with different dispersal mechanisms48,64–66. In addition, the Chiapas region (and adjacent Guatemala) is also home to other species of Psittacanthus closely related with P. calyculatus and P. schiedeanus41: P. breedlovei and P. angustifolius21. Thus, it is possible that hybridization with one of these other species may be the reason for the apparent high admixture and the low genetic differentiation between mistletoes in these two regions. However, these two species can be easily recognizable based on leaf morphology and host preferences21. In addition we observed, albeit using few samples, that P. angustifolius and P. breedlovei appeared to be genetically indistinguishable from their widespread sister species41. Therefore, further exploration of the genetic patterns including more samples from the other Psittacanthus species distributed in Chiapas is needed to understand their evolutionary history within this region.
Species boundaries and differential admixture
The observed genetic structure and migration patterns seem to be linked to the demographic history of the species complex. For instance, migration rates clearly increased from past to present between geographical regions. Migration rates showed that historical and contemporary gene flow was directional from the SCHI and CALY populations to the HYBR population, whereas contemporary gene flow occurred also from the CALY population to the SCHI population (Table 2). The low differentiation between the HYBR populations in Oaxaca and the CALY group might be due to asymmetrical gene flow, resulting in higher levels of introgression from P. schiedeanus into P. calyculatus (Fig. 2d). In theory, minimal differences in the timing of flowering or fruiting phenologies may promote pre-mating isolation and prevent hybridization between P. schiedeanus and P. calyculatus. However, the flowering and fruiting periods of the two species in the arid Central Valleys of Oaxaca greatly overlap23,24,58,60. Thus, further mechanisms for such considerable degree of gene flow have to be considered.
Díaz Infante and collaborators16 investigated the reproductive biology and phylogenetic relationships between a population of P. calyculatus from the Trans-Mexican Volcanic Belt and two sympatric populations of P. calyculatus and P. auriculatus from the xeric Central Valleys of Oaxaca. Flowers of the two species in sympatry were reciprocally pollinated to assess post-mating components of reproductive isolation (RI), with fewer heterospecific matings observed than expected by chance in P. calyculatus compared to P. auriculatus. When considering other factors of ecological isolation that affect co-occurrence, the RI values indicated that isolation by hummingbird pollinators was less effective than isolation by host tree species and seed dispersers, suggesting that host usage is the most important ecological isolation factor between the two species and that the host tree species’ barrier is currently contributing the most to maintaining these species in sympatry16.
For the origin of the hybrid forms located in the Central Valleys of Oaxaca and the Central Depression of Chiapas regions, a late Pleistocene scenario and assumed hybridization processes seem to be a plausible alternative for some reasons. The most likely DIYABC scenario supported a hybrid origin of the populations occurring c. 99 k years ago, corresponding approximately to the early last glacial period (Wisconsin glaciation; 120–110 ka). This is consistent with the estimated initial diversification of the species complex occurring 200 kya32, followed by dramatic changes in the distribution of species during the last glacial period32,38. Under this scenario, populations of the Psittacanthus calyculatus and P. schiedeanus may have expanded into the currently xeric lowlands of Oaxaca during glacial periods, with secondary contact and formation of hybrid zones among otherwise genetically differentiated populations (sympatric stage). During the Holocene, populations of the two parental species contracted back into temperate and cloud forests at higher elevation where little genetic structure is observed (allopatric stage). Alternatively, ancestral hybrid populations of P. calyculatus and P. schiedeanus remained in situ (in the arid Central lowlands of Oaxaca and Chiapas) and differentiation of these populations occurred under more xeric conditions and specializing on different host species (host shifting hypothesis).
Genetic status of the Oaxaca populations
Individuals with signatures of admixture were most frequent in populations located in the central arid regions of Oaxaca and Chiapas (HYBR group). The observed historical and contemporary patterns of gene flow shed some light on how historical gene flow between these closely related species occurred and what geographical barriers have maintained their genetic integrity. The non-significant correlation between historical and contemporary migration rates implies that migration rates between groups have changed over the evolutionary history of the species complex. Although rates of historical and contemporary migration were low among populations, we found evidence of past migration from the CALY and SCHI populations into the HYBR populations, supporting the demographic model for the hybrid origin of these populations. Results of the DIYABC analysis supported one of the allopatric speciation scenarios and indicated that divergence between groups of populations occurred at least 100 kya, consistent with previous evidence using nuclear and chloroplast DNA sequences32,38. Although we tested several scenarios of speciation with gene flow, future work incorporating genomic data might give more accurate estimates of the dynamics of gene flow and test alternative and more complex scenarios of allopatric speciation followed by hybridization during secondary contact67.
Previous phylogeographic studies using cpDNA sequences, accompanied by ecological niche modelling, provided evidence that individuals of P. schiedeanus and P. calyculatus from the Central Valleys of Oaxaca formed a genetic cluster different from those corresponding to the distribution of each of the two species, and recognized this region as a potential area of secondary contact32,38. Our study revealed that the arid Central regions of Oaxaca, as well as the Central Depression of Chiapas, represents a secondary contact hybrid zone between P. calyculatus and P. schiedeanus. An interesting feature of the overlapping distribution area in Oaxaca for these mistletoes is that the more xeric climatic conditions contrast with the adjacent pine-oak forests (P. calyculatus) and cloud forests (P. schiedeanus) that parental species inhabit32,38,41. In addition, the main host species used by HYBR individuals in the secondary contact zone are also different, with a higher prevalence (57%) on Celtis caudata (Ulmaceae) at one locality16 or strongly associated with Anacardiaceae hosts based on herbarium records21,41. Therefore, we propose that expansion of P. calyculatus and P. schiedeanus towards lower elevations during the last glaciation32,38, promoted different events of hybridization in secondary contact zones, creating a hybrid swarm, probably with low hybrid frequencies. Once the glaciation period was over, admixed individuals prevailed within the same area because these had the capacity to confront new environmental conditions with different host species, with drier climatic conditions and more xeric vegetation, whereas non-admixed individuals contracted into the highlands to their present distribution along the TMVB and SMOr associated with different sets of host species. This hypothesis does not exclude the possibility of current gene flow (though restricted) to areas where cloud forests are very close to the tropical dry forests and xeric zones, like in the Central Valleys of Oaxaca. Secondary contact and hybridization could have weakened the reproductive barriers and maintained low genetic differentiation between populations from the temperate (TMVB) and cloud forests (SMOr). In light of the weak reproductive barriers between P. calyculatus and P. auriculatus, the sister group of the P. calyculatus and P. schiedeanus species complex16,32, we believe that hybridization very likely occurs among populations within the complex due to close genetic relationships between species. In addition, over the 20 bird species recognized as seed dispersers and more than 20 hummingbird species that pollinate the flowers of these mistletoe species23,24,58,60,68, several species are shared16 thus increasing the probabilities of current gene flow between species. It is possible that the present hybrid population has resulted from contemporary gene flow between parental species facilitated by bird seed-dispersal, rather than the result of historical gene flow by secondary contact after the differentiation of the two parental Psittacanthus species in late Pleistocene. Our analyses showed that contemporary gene flow is present between the parental and the hybrid genetic clusters. However, we believe that if hybrid populations result only from recent gene flow, we would also have observed a significant proportion of pure parental individuals within the hybrid zone. That is, birds would transport (in independent events) pure parentals into one region and only after their establishment would these begin to hybridize. Nonetheless, first generation hybrids were almost absent in the analyses and the timing of admixture by the DIYABC analysis was congruent with the idea of secondary contact after the differentiation of the two parental Psittacanthus species in late Pleistocene.
In parasitic plants the colonization of a new environment and further differentiation could be associated to adaptations to biotic factors such as new host associations, pollinators and dispersers or through adaptations to abiotic factors (like the parasite’s own niche) or both41,69. In mistletoes, controversial evidence exists about the role of host race formation as the main diversifying force. Here gene flow can be interrupted or diminished if mating, dispersal and establishment occur only among mistletoes adapted to specific hosts36. Particularly, cross-dispersal experiments have provided some evidence for host-race formation in P. schiedeanus and P. calyculatus growing on distantly related host species in sympatry, in which seedling development was greatest when seeds were placed on their source host species60,70. The hypothesis of host-race formation is supported by the observed low heterozygosity values that might be produced by subpopulation structure, i.e. mistletoes that aggregate in one or adjacent hosts by geographic or behavioural barriers of gene dispersal vectors to gene flow followed by genetic drift in the subpopulations (Wahlund effect14). The significant population differentiation and genetic structure in the P. calyculatus and P. schiedeanus species complex has been attributed to climatic variables, rather than to host association41. Evidence in plant, fungi and animal species has shown the potential of hybridization to the adaptation to different environmental conditions44,71–73. In our study, we found that the area with higher values of admixture corresponds to individuals from locations with drier climatic conditions, more xeric vegetation types and different associated host species, suggesting that colonization of a ‘new’ habitat is linked to hybridization and further genetic differentiation.
With the evidence in hand, many questions arise about the role of hybridization, reinforcement and introgression for the evolution of this system, including the colonization of new environments, host shifts, host race formation, and a possible case of hybrid speciation. If the reproductive barriers are weak and there is no reinforcement (selection against hybrids), differentiation between species will be eroded and the effect of host race formation would be diluted74. On the other hand, strong selection to different environments could maintain genetic differences between populations, despite the presence of gene flow1,3. Further studies combining genomic scans and experimental data of host-specific relationships through cross-inoculation experiments, or reciprocal transplants to different environments comparing parental and hybrid individuals would shed light on the degree of reproductive isolation and the role of introgressive hybridization in the evolution of host specialization and environmental shifts in Psittacanthus mistletoes in particular, and other aerial parasites in general. The processes of reinforcement, introgression and transgressive segregation along with evolutionary forces like genetic drift and selection could influence the chances that the hybrid population adapts to new host species. For example, the gain of alleles in the hybrids through introgression and the presence of transgressive traits (those exceeding the values from parental species) could benefit hybrid individuals to adapt to the new environment by creating a phenotype able to colonize a new host or tolerate different temperatures. Also, if hybrid mistletoes tend to aggregate in different hosts, the role of genetic drift will could be important to reproductively isolate hybrids3. We do not yet know the specific forces leading the hybrid populations into a new host as the initial stage towards speciation. It maybe directly related to the hybrid origin, by gaining some genetic advantage permitting the invasion of a new host. Or perhaps the adaptation into a new host is related to the in situ survival of the hybrids in the region, which was subjected to floristic turnover during the last-glacial/Holocene transition. In this case, the increasing abundance of arid-adapted hosts (such as Anacardiaceae) would lead to the specialization of these mistletoes and the host shifting leading to host race formation12. Results of this study suggest that hybridization could have been important for the colonization to a new environment under past climatic changes and could help to understand the range of outcomes of climate alterations and recently human-altered environmental changes shaping current patters of diversity.
Methods
Study sites and molecular data
Our analyses are based on the molecular data provided by Ramírez-Barahona and collaborators41. These data included information of 10 microsatellite loci for 415 individuals in 54 populations throughout the species’ distribution ranges in Mexico41, an area spreading about 14° in longitude and 10° in latitude, that includes three biogeographical regions. However, due to the few individuals genotyped for some populations we pooled populations in close proximity, ending up with 51 populations for this study (Table S1). The sampling included individuals from allopatric populations from the TMBV and eastern slopes of the SMOr, individuals from sympatric populations in the Central Valleys of Oaxaca and the Central Depression of Chiapas, and individuals of the range-restricted P. breedlovei and P. angustifolius in Chiapas (Fig. 1).
All the individuals included in the dataset were successfully genotyped for at least five of the 10 microsatellites, with 70% of the individuals successfully genotyped for seven or more loci. Microsatellite data are available in the Supporting Information of Ramírez-Barahona and collaborators (nph14471-sup-0003-NotesS2.csv)41. To confirm the presence of genetic clusters in the sample, we carried out a two-step Bayesian Markov chain Monte Carlo (MCMC) clustering analysis of microsatellite data using STRUCTURE 2.3.475, as previously implemented41. The most likely number of populations was determined estimating the DeltaK (∆K) and the log likelihood of K, ln P(K) = L(K) between successive K values61. According to Evanno’s method61, we confirmed the presence of two clusters, CALY and SCHI, and additional genetic sub-structuring was observed within each of these clusters in the second-step analysis (see also41 for similar findings). Interestingly, several individuals scattered throughout the STRUCTURE plot show signs of admixture at K = 2 (Fig. 2A,C).
Admixture analysis and hybrid identification
Given the observed patterns of genetic structure at K = 2 and the distribution of admixed individuals scattered throughout the STRUCTURE plot, we decided to reanalyse the full dataset particularly because a high number of admixed individuals (mean membership coefficient <0.75) were not included in previous analyses41. To explore the occurrence of hybridization (see76,77 for more details), we assigned individuals from the allopatric CALY and SCHI clusters as ‘pure parental’ when the membership coefficient (q) was q > 0.9 for either of the two clusters. Individuals from the sympatric regions (Central Valleys of Oaxaca and the Central Depression of Chiapas) were not considered for the ‘pure parental’ assignment because the suspected high admixture in these regions could bias further assignment. The possibility that P. auriculatus, geographically restricted to the Oaxaca dry valleys, can be the donor for the hybrid population given its sympatric distribution with the P. calyculatus/schiedeanus complex is unlikely. The specific microsatellites used here41 were designed using populations of the species complex of P. schiedeanus and P. calyculatus and previous work by our lab group has shown low success in transferring these microsatellite loci into other species due to poor amplification.
We then performed a STRUCTURE analysis using the ‘pure parental’ information, implemented through the POPFLAG prior, to define individuals belonging to the parent populations. In order to uncover admixture between the two species, all individuals were assigned to either CALY or SCHI populations by setting K = 2. We set the allele frequencies to be correlated and performed ten replicates with 100 000 MCMC after 50 000 burning period. The outputs of each replicate were combined in CLUMPP for visualization78.
We then used the Bayesian model-based program NEWHYBRIDS79 to calculate the posterior probability of individuals belonging to one of six categories: (1) pure CALY, (2) pure SCHI, (3) first generation hybrids, (4) second generation hybrids, (5) CALY backcrosses, and (6) SCHI backcrosses. The analysis focuses on first generation hybrids, thus helping to detect on-going hybridization between species. Assignments were done using Jeffrey’s-like prior for allele frequencies and mixing proportions as suggested in the program’s manual. We ran three replicates with 500 000 sweeps and a burn-in period of 20 000, and the prior information of pure parental individuals obtained from previous STRUCTURE included in the analysis.
To define assignment thresholds to each hybrid category used for our results, we performed hybridization simulations in HYBRIDLAB80. For this, we used the individuals classified as parental in the STRUCTURE analysis as the starting populations and simulated 50 hybrids for several generations: first, second and third generation hybrids; and first, second and third generation backcrosses to each parental species. Data obtained for each simulation were run in STRUCTURE with the same parameters as above. The results for K = 2 were combined with CLUMPP78 and the proportion of membership was visualized using R 3.1.3 (R Development Core Team, 2015). From the simulations, we identified three categories of individuals: (1) pure species, individuals with an assignment probability of q ≥ 0.80 to one cluster; (2) backcrosses, individuals with an assignment probability between q = 0.6 and q = 0.8 to one cluster; and (3) hybrids (including the indistinguishable first, second and third generations), individuals with an assignment probability between 0.4 and 0.6 (Fig. S1 in Supporting information).
Finally, the simulated data were also run in NEWHYBRIDS and the program correctly assigned the pure species individuals with a probability >0.9. For the first- and second-generation hybrid individuals, we considered correctly assigned individuals those with probabilities higher than 0.5. Subsequent hybrid generations and backcrosses were not identified in NEWHYBRIDS, thus we considered hybrid individuals of unknown generation those with assignment probabilities between 0.1 and 0.9. Finally, we estimated the proportion of individuals belonging to each category for each population to assess the level of admixture across populations.
Genetic differentiation and population structure
To analyse genetic differentiation and population structure, we used the classification provided by the initial STRUCTURE runs and grouped populations into two groups, CALY and SCHI. Because of the high levels of admixture observed in the Central Valleys of Oaxaca and the Central Depression of Chiapas, we also grouped samples into three groups as follows: (1) CALY, samples from temperate forests along the TMVB; (2) SCHI, samples from cloud forests in eastern Mexico and three populations in Chiapas; and (3) HYBR, samples from the xeric Central Valleys of Oaxaca and Central Depression of Chiapas. Given that most genetic structure has been explained by environmental factors41, we also grouped samples according to habitat type: (1) cloud forests; (2) xeric temperate forests; and (3) tropical dry forests. Finally, populations were also grouped according to geography into four groups: (1) the Chiapas region; (2) the Oaxaca region; (3) Sierra Madre Oriental; and (4) Trans-Mexican Volcanic Belt.
For each of the four groupings (species, geography, admixture, and habitat), we estimated gene diversity and absolute allele frequency differentiation (Jost’s D) using the R package “DiveRsity”81 and rarefied allelic richness using the R package “hierfstat”82. Hardy-Weinberg equilibrium departures and linkage disequilibrium among loci were estimated in GENEPOP 1.283. We used FreeNA84 to estimate the frequencies of null alleles with the EM algorithm85 and the pairwise genetic differentiation (Weir’s FST86) among groups of populations using the ENA correction. Note that genetic structure below the level of our sampling (population subdivision) can produce significant departures from Hardy-Weinberg even in the absence of null alleles (i.e. Wahlund effect: reduction of heterozygosity in a population caused by subpopulation structure). For these analyses, locus four was eliminated because it was only present in the SCHI populations.
To explore whether the distribution of the genetic variance in our populations is related to species differences, geography, or to environmental differentiation between groups of populations, we performed an analysis of molecular variance (AMOVA87) as implemented in ARLEQUIN 3.0188. Four AMOVAs were performed with different groups of populations: (a) two clusters corresponding to each species (CALY and SCHI); three clusters corresponding to (b) habitat type (cloud forest, xeric temperate forest, tropical dry forest) or (c) level of admixture (CALY, SCHI, HYBR); and (d) four clusters corresponding to geography (CHIS, OAX, SMOr, TMVB; Table S1). Significance of each AMOVA was evaluated with 10 000 permutations.
Contemporary and historical migration rates
We compared migration rates over contemporary and historical timescales89 using unlinked microsatellite data with BAYESASS90 and MIGRATE91 for the three groups identified with the admixture analysis: pure CALY, pure SCHI, and HYBR from the Central Valleys of Oaxaca and the Central Depression of Chiapas. Using Bayesian inference, BAYESASS estimates recent migration rates between populations within the last few generations (m), whereas MIGRATE uses the coalescent approach to jointly estimate the relative effective population size θNe (4Neµ) and asymmetrical gene flow M (m/µ) between pairs of populations over much longer periods of time, approximately thousands of years (ca. 4Ne generations in the past91).
BAYESASS was initially run with the default delta values (Δ) for allelic frequency (A), migration rate (M), and inbreeding (F). Subsequent runs incorporated different Δ to ensure that the acceptance rate for proposed changes in parameters were between 20–40% for each parameter. Adjusted final delta values used were ΔA = 0.2 (41% acceptance rate), ΔM = 0.8 (22%) and ΔF = 0.2 (48%), respectively. To ensure convergence, we performed five independent runs (50 million iterations, 5 million burn-in, and sampling frequency of 2000) each with a different seed number, comparing the posterior mean parameter estimates for convergence. We also analysed the trace file of each run with TRACER 1.592 to ensure an appropriate mixing of parameters and burn-in number. We give estimates of m from one randomly chosen run out of the three final runs as their parameter estimates were similar. We ran MIGRATE incorporating Bayesian inference analyses to estimate historical migration rates (M) among groups of populations. We used a Brownian-motion model with a constant mutation rate and FST to estimate θ. Several short runs were performed to search for the appropriate priors. After finding suitable priors, MIGRATE was run three times to confirm convergence. These final runs consisted of one long chain, 100 000 sampled trees, 1000 recorded, with a burn-in of 10000 with three replicates and each run with a different seed number. We set the minimum and maximum boundaries for theta (θ) and migration (M) as 0.0 and 30.0, with a delta value of 3. A four-chain heating at temperatures of 1, 1.5, 3 and 10000 was implemented to increase the efficiency of the MCMC89. Lastly, we performed a Mantel test with 5000 permutations to test for similarity between contemporary and historical values of m. For this analysis, we used the values of m directly generated by BAYESASS and estimated m from values of M (m/µ) generated by MIGRATE by multiplying all M values by an estimated mutation rate of 5 × 10−4 for microsatellites93. The number of migrants per generation was estimated by multiplying the θ value from the source populations to M91.
Inference of divergence and secondary contact scenarios
Approximate Bayesian Computation (ABC) analysis was performed using samples in the three clusters defined based on genetic differentiation and genetic sub-structuring: CALY, SCHI, and HYBR. Five possible divergence scenarios leading to these three clusters were included in the ABC analysis to test if the admixed populations have a hybrid origin (see Results). In each scenario, t refers to timescale ranging from 300 to 25000 generations and with a conditional prior t2 > t1, the maximum t included the predicted split between P. schiedeanus and P. calyculatus around 2.18 × 105 years ago32. The effective size (N) of the corresponding populations (Pops 1, 2, 3 or the ancestral population) during each time period (e.g., 0–t1, t1–t2) was set to a maximum of 30000.
We used DIYABC 2.194 to simulate ten million datasets (2 million per scenario) with the same number of populations, loci and individuals. The most likely scenario was evaluated by comparing posterior probabilities using the logistic regression approach95,96. Temporal and demographic parameters were estimated with a logistic regression for the best-supported model with the 1% simulated data closest to the observed data95,96. Finally, to assess confidence in the model selection we simulated 500 pseudo-observed datasets (PODs) to estimate type I and type II error rates95,96.
Data availability
The dataset (microsatellites) generated and analysed during the current study are available in the Supporting Information of Ramírez-Barahona et al. (nph14471-sup-0003-NotesS2.csv)41.
Electronic supplementary material
Acknowledgements
We thank Cristina Bárcenas, Sergio Díaz Infante, Etelvina Gándara, Luis Manuel García-Feria, Clementina González, Antonio González-Rodríguez, Carlos Lara, Yuyini Licona-Vera, María Teresa Mejía-Saules, G. Merino-Díaz, Andrés E. Ortiz-Rodríguez, María José Pérez-Crespo, Flor Rodríguez-Gómez, Eduardo Ruiz-Sanchez, Carlos Soberanes, and Antonio A. Vásquez-Aguilar for field and laboratory assistance. Permission to conduct our fieldwork was granted by the Mexican government (Instituto Nacional de Ecología, Secretaría del Medio Ambiente y Recursos Naturales, SGPA/DGGFS/712/1299/12). This project was funded by a competitive grant (155686) from the Consejo Nacional de Ciencia y Tecnología (CONACyT; http://www.conacyt.mx) and research funds (20030/10563) from the Departamento de Biología Evolutiva, Instituto de Ecología, AC awarded to JFO. FB and SRB were supported by a postdoctoral fellowship (225799, 155686, respectively) from CONACyT. The publication costs were financed by the Dirección General of the INECOL (20029/60813).
Author Contributions
F.B.D. and J.F.O. conceived the study. F.B.D., S.R.B. and J.F.O. collected the samples. S.R.B. designed and conducted the experiments. F.B.D. and S.R.B. analysed the data. F.B.D. and J.F.O. constructed the images. All authors contributed intellectually to the interpretation of the results and writing of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23707-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mallet J. Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Phil Trans R Soc Lond B Biol Sci. 2008;363:2971–2986. doi: 10.1098/rstb.2008.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nosil, P. Ecological Speciation. Oxford University Press, New York (2012).
- 3.Abbott R, et al. Hybridization and speciation. J Evol Biol. 2013;26:229–46. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 4.Seehausen O, et al. Genomics and the origin of species. Nat Rev Genet. 2014;15:176–92. doi: 10.1038/nrg3644. [DOI] [PubMed] [Google Scholar]
- 5.Baack E, Melo MC, Rieseberg LH, Ortiz-Barrientos D. The origins of reproductive isolation in plants. New Phytol. 2015;207:968–984. doi: 10.1111/nph.13424. [DOI] [PubMed] [Google Scholar]
- 6.Sexton JP, Hangartner SB, Hoffmann AA. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution. 2014;68:1–15. doi: 10.1111/evo.12258. [DOI] [PubMed] [Google Scholar]
- 7.Wu CI. The genic view of the process of speciation. J Evol Biol. 2001;14:851–865. doi: 10.1046/j.1420-9101.2001.00335.x. [DOI] [Google Scholar]
- 8.Sobel JM, Chen GF, Watt LR, Schemske DW. The biology of speciation. Evolution. 2010;64:295–315. doi: 10.1111/j.1558-5646.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- 9.Pettengill, J. B. & Moeller, D. A. Phylogeography of speciation: allopatric divergence and secondary contact between outcrossing and selfing Clarkia. Mol Ecol21, 4578–92, 10.1111/j.1365-294X.2012.05715.x (2012). [DOI] [PubMed]
- 10.Barkman TJ, et al. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol. 2007;7:248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton, D. A: R., Hipp, A. L., González-Rodríguez, A. & Cavender-Bares, J. Historical introgression among the American live oaks and the comparative nature for introgression. Evolution69, 2587–2601, 10.1111/evo.12758 (2015). [DOI] [PubMed]
- 12.Norton DA, Carpenter MA. Mistletoes as parasites: host specificity and speciation. Trends Ecol Evol. 1998;13:101–105. doi: 10.1016/S0169-5347(97)01243-3. [DOI] [PubMed] [Google Scholar]
- 13.Watson DM. Mistletoe–A keystone resource in forests and woodlands worldwide. Ann Rev Ecol Syst. 2001;32:219–249. doi: 10.1146/annurev.ecolsys.32.081501.114024. [DOI] [Google Scholar]
- 14.Huyse T, Poulin R, Théron A. Speciation in parasites: a population genetics approach. Trends Parasitol. 2005;21:469–475. doi: 10.1016/j.pt.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Mathiasen RL, Nickrent DL, Shaw DC, Watson DM. Mistletoes: pathology, systematics, ecology, and management. Plant Dis. 2008;92:988–1006. doi: 10.1094/PDIS-92-7-0988. [DOI] [PubMed] [Google Scholar]
- 16.Díaz Infante S, Lara C, Arizmendi MC, Eguiarte LE, Ornelas JF. Reproductive ecology and isolation of Psittacanthus calyculatus and P. auriculatus mistletoes (Loranthaceae) PeerJ. 2016;4:e2491. doi: 10.7717/peerj.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brooker RW, et al. Don’t dis integration: a comment on Ricklefs’ disintegrating communities. Am Nat. 2009;174:919–927. doi: 10.1086/648058. [DOI] [PubMed] [Google Scholar]
- 18.Kursar TA, et al. The evolution of antiherbivore defenses and their contribution to species coexistence in the tropical tree genus Inga. Proc Natl Acad Sci USA. 2009;106:18073–18078. doi: 10.1073/pnas.0904786106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorpe AS, Aschehoug ET, Atwater DA, Callaway RM. Interactions among plants and evolution. J Ecol. 2011;99:729–740. doi: 10.1111/j.1365-2745.2011.01802.x. [DOI] [Google Scholar]
- 20.Kuijt, J. The biology of parasitic flowering plants. University of California Press, Berkeley, California (1969).
- 21.Kuijt J. Monograph of Psittacanthus (Loranthaceae). Syst. Bot Monogr. 2009;86:1–362. [Google Scholar]
- 22.Musselman, L. J. & Press, M. C. Introduction to parasitic plants. In: Parasitic plants (Press M. C., Graves, J.D., eds). Chapman & Hall, London, pp. 1–13 (1995).
- 23.Azpeitia F, Lara C. 2006. Reproductive biology and pollination of the parasitic plant Psittacanthus calyculatus (Loranthaceae) in central Mexico. J Torrey Bot Soc. 2006;133:429–438. doi: 10.3159/1095-5674(2006)133[429:RBAPOT]2.0.CO;2. [DOI] [Google Scholar]
- 24.Ramírez MM, Ornelas JF. Pollination and nectar production of Psittacanthus schiedeanus (Loranthaceae) in central Veracruz, Mexico. Bol Soc Bot Méx. 2010;87:61–67. [Google Scholar]
- 25.Guerra, T. J., Galetto, L. & Silva, W. R. Nectar secretion dynamic links pollinator behavior to consequences for plant reproductive success in the ornithophilous mistletoe Psittacanthus robustus. Pl Biol16, 956–966, https://doi.org/10.1111/plb.12146 (2014). [DOI] [PubMed]
- 26.Pérez-Crespo, M. J., Ornelas, J. F., Martén-Rodríguez, S., González-Rodríguez, A. & Lara, C. Reproductive biology and nectar production of the Mexican endemic Psittacanthus auriculatus (Loranthaceae), a hummingbird-pollinated mistletoe. Pl Biol18, 73–83, 10.1111/plb.12365 (2016). [DOI] [PubMed]
- 27.Fadini, R. F. et al. Bat and bee pollination in Psittacanthus mistletoes, a genus regarded as exclusively hummingbird-pollinated. Ecology00, 00–00 10.1002/ecy.2140 (2018). [DOI] [PubMed]
- 28.Ollerton J, et al. Pollination niche overlap between a parasitic plant and its host. Oecologia. 2007;151:473–485. doi: 10.1007/s00442-006-0605-y. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Crespo MJ, Lara C, Ornelas JF. Uncorrelated mistletoe infection patterns and mating success with local host specialization in Psittacanthus calyculatus (Loranthaceae) Evol Ecol. 2016;30:1061–1080. doi: 10.1007/s10682-016-9866-z. [DOI] [Google Scholar]
- 30.Amico GC, Vidal-Russell R, Garcia MA, Nickrent DL. Evolutionary history of the South American mistletoe Tripodanthus (Loranthaceae) using nuclear and plastid markers. Syst Bot. 2012;37:218–225. doi: 10.1600/036364412X616783. [DOI] [Google Scholar]
- 31.Lira-Noriega A, Toro-Nuñez O, Oaks JR, Mort ME. The roles of history and ecology in chloroplast phylogeographic patterns of the bird-dispersed plant parasite Phoradendron californicum (Viscaceae) in the Sonoran Desert. Amer J Bot. 2015;102:149–164. doi: 10.3732/ajb.1400277. [DOI] [PubMed] [Google Scholar]
- 32.Ornelas JF, et al. A mistletoe tale: postglacial invasion of Psittacanthus schiedeanus (Loranthaceae) to Mesoamerican cloud forests revealed by molecular data and species distribution modeling. BMC Evol Biol. 2016;16:78. doi: 10.1186/s12862-016-0648-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watson DM. Determinants of parasitic plant distribution: the role of host quality. Botany. 2009;87:16–21. doi: 10.1139/B08-105. [DOI] [Google Scholar]
- 34.Jerome CA, Ford BA. The discovery of three genetic races of the dwarf mistletoe Arceuthobium americanum (Viscaceae) provides insight into the evolution of parasitic angiosperms. Mol Ecol. 2002;11:387–405. doi: 10.1046/j.0962-1083.2002.01463.x. [DOI] [PubMed] [Google Scholar]
- 35.Zuber D, Widmer A. Phylogeography and host race differentiation in the European mistletoe (Viscum album L.) Mol Ecol. 2009;18:1946–1962. doi: 10.1111/j.1365-294X.2009.04168.x. [DOI] [PubMed] [Google Scholar]
- 36.Yule KM, Koop JAH, Alexandre NM, Johnston LR, Whiteman NK. 2016. Population structure of a vector-borne plant parasite. Mol Ecol. 2016;25:3332–3343. doi: 10.1111/mec.13693. [DOI] [PubMed] [Google Scholar]
- 37.Amico GC, Nickrent DL. Population structure and phylogeography of the mistletoes Tristerix corymbosus and T. aphyllus (Loranthaceae) using chloroplast DNA sequence variation. Amer J Bot. 2009;96:1571–1580. doi: 10.3732/ajb.0800302. [DOI] [PubMed] [Google Scholar]
- 38.Pérez-Crespo MJ, et al. Phylogeography and population differentiation in the Psittacanthus calyculatus (Loranthaceae) mistletoe: a complex scenario of the climate-volcanism interaction along the Trans-Mexican Volcanic Belt. J Biogeogr. 2017;44:2501–2514. doi: 10.1111/jbi.13070. [DOI] [Google Scholar]
- 39.Stanton S, Honnay O, Jacquemyn H, Roldán-Ruiz I. A comparison of the population genetic structure of parasitic Viscum album from two landscapes differing in degree of fragmentation. Pl Syst Evol. 2009;281:161–169. doi: 10.1007/s00606-009-0198-0. [DOI] [Google Scholar]
- 40.Lira-Noriega A, Peterson AT. Range-wide ecological niche comparisons of parasite, hosts and dispersers in a vector-borne plant parasite system. J. Biogeogr. 2014;41:1664–1673. doi: 10.1111/jbi.12302. [DOI] [Google Scholar]
- 41.Ramírez-Barahona S, González C, González-Rodríguez A, Ornelas JF. The influence of climatic niche preferences on the population genetic structure of a mistletoe species complex. New Phytol. 2017;214:1751–1761. doi: 10.1111/nph.14471. [DOI] [PubMed] [Google Scholar]
- 42.Anderson E, Stebbins GL. Hybridization as an evolutionary stimulus. Evolution. 1954;8:378–388. doi: 10.1111/j.1558-5646.1954.tb01504.x. [DOI] [Google Scholar]
- 43.Stebbins GL. The role of hybridization in evolution. Proc Am Phil Soc. 1959;103:231–251. [Google Scholar]
- 44.Arnold M. Transfer and origin of adaptations through natural hybridization: were Anderson and Stebbins right? The Plant Cell. 2004;16:562–570. doi: 10.1105/tpc.160370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamilton J, Miller J. Adaptive introgression as a resource for management and genetic conservation in changing climate. Conserv Biol. 2015;30:33–41. doi: 10.1111/cobi.12574. [DOI] [PubMed] [Google Scholar]
- 46.Jaramillo-Correa JP, Beaulieu J, Khasa DP, Bousquet J. Inferring the past from the present phylogeographic structure of North American forest trees: seeing the forest for the genes. Can J Pl Res. 2009;39:286–307. doi: 10.1139/X08-181. [DOI] [Google Scholar]
- 47.Gugger, P. F., Sugita, S. & Cavender-Bares, J. Phylogeography of Douglas-fir based on mitochondrial and chloroplast DNA sequences: testing hypotheses from the fossil record. Mol Ecol19, 1877–1897, 10.1111/j.1365-294X.2010.04622.x (2010). [DOI] [PubMed]
- 48.González C, Ornelas JF, Gutiérrez-Rodríguez C. Selection and geographic isolation influence hummingbird speciation: genetic, acoustic and morphological divergence in the wedge-tailed sabrewing (Campylopterus curvipennis) BMC Evol Biol. 2011;11:38. doi: 10.1186/1471-2148-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramírez-Barahona S, Eguiarte LE. Changes in the distribution of cloud forests during the last glacial predict the patterns of genetic diversity and demographic history of the tree fern Alsophila firma (Cyatheaceae) J Biogeogr. 2014;41:2396–2407. doi: 10.1111/jbi.12396. [DOI] [Google Scholar]
- 50.Ruiz-Sanchez E, Ornelas JF. Phylogeography of Liquidambar styraciflua (Altingiaceae) in Mesoamerica: survivors of a Neogene widespread temperate forest (or cloud forest) in North America? Ecol Evol. 2014;4:311–328. doi: 10.1002/ece3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rieseberg, L. & Wendel, J. F. Introgression and its consequences in plants. In: Hybrid zones and the evolutionary process (Harrison, R., ed.). Oxford University Press, New York, pp. 70–109 (1993).
- 52.Vallejo-Marin M, Hiscock SJ. Hybridization and hybrid speciation under global change. New Phytol. 2016;211:170–1187. doi: 10.1111/nph.14004. [DOI] [PubMed] [Google Scholar]
- 53.Avise, J. C. Phylogeography: the history and formation of species. Harvard University Press Cambridge, MA (2000).
- 54.Petit RJ, Excoffier L. Gene flow and species delimitation. Trends Ecol Evol. 2009;24:386–393. doi: 10.1016/j.tree.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 55.Floate, K. & Whitman, T. The “hybrid bridge” hypothesis: host shifting via plant hybrid swarms. Am Nat141, 651–662 10.1086/285497 (1993). [DOI] [PubMed]
- 56.Detwiler J, Criscione C. An infectious topic in reticulate evolution: Introgression and hybridization in animal parasites. Genes. 2010;1:102–123. doi: 10.3390/genes1010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watson DM, Herring M. On pluralism in ecology: seeing the forest and the trees. Proc R Soc B Biol Sci. 2014;281:20132696. doi: 10.1098/rspb.2013.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.López de Buen L, Ornelas JF. Frugivorous birds, host selection and the mistletoe Psittacanthus schiedeanus, in central Veracruz, Mexico. J Trop Ecol. 1999;15:329–340. doi: 10.1017/S0266467499000851. [DOI] [Google Scholar]
- 59.López de Buen L, Ornelas JF. Host compatibility of the cloud forest mistletoe Psittacanthus schiedeanus (Loranthaceae) in central Veracruz, Mexico. Amer J Bot. 2002;89:95–102. doi: 10.3732/ajb.89.1.95. [DOI] [PubMed] [Google Scholar]
- 60.Lara C, Pérez G, Ornelas JF. Provenance, guts, and fate: field and experimental evidence in a host-mistletoe-bird system. Ecoscience. 2009;16:399–407. doi: 10.2980/16-3-3235. [DOI] [Google Scholar]
- 61.Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology14, 2611–2620, 10.1111/j.1365-294X.2005.02553.x (2005). [DOI] [PubMed]
- 62.Ornelas JF, Ruiz-Sanchez E, Sosa V. Phylogeography of Podocarpus matudae (Podocarpaceae): pre-Quaternary relicts in northern Mesoamerican cloud forests. J Biogeogr. 2010;37:2384–2396. doi: 10.1111/j.1365-2699.2010.02372.x. [DOI] [Google Scholar]
- 63.Ornelas JF, Rodríguez-Gómez F. Influence of Pleistocene glacial/interglacial cycles on the genetic structure of the mistletoe cactus Rhipsalis baccifera (Cactaceae) in Mesoamerica. J Hered. 2015;106:196–210. doi: 10.1093/jhered/esu113. [DOI] [PubMed] [Google Scholar]
- 64.Gutiérrez-Rodríguez C, Ornelas JF, Rodríguez-Gómez F. Chloroplast DNA phylogeography of a distylous shrub (Palicourea padifolia, Rubiaceae) reveals past fragmentation and demographic expansion in Mexican cloud forests. Mol Phylogenet Evol. 2011;61:603–615. doi: 10.1016/j.ympev.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 65.Ornelas JF, et al. Comparative phylogeographic analyses illustrate the complex evolutionary history of threatened cloud forests of northern Mesoamerica. PLoS One. 2013;8:e56283. doi: 10.1371/journal.pone.0056283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodríguez-Gómez F, Gutiérrez-Rodríguez C, Ornelas JF. 2013. Genetic, phenotypic and ecological divergence with gene flow at the Isthmus of Tehuantepec: the case of the azure-crowned hummingbird (Amazilia cyanocephala) J Biogeogr. 2013;40:1360–1373. doi: 10.1111/jbi.12093. [DOI] [Google Scholar]
- 67.Payseur BA, Rieseberg LH. A genomic perspective on hybridization and speciation. Mol Ecol. 2016;25:2337–2360. doi: 10.1111/mec.13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.López de Buen L, Ornelas JF. Seed dispersal of the mistletoe Psittacanthus schiedeanus by birds in central Veracruz, Mexico. Biotropica. 2001;33:487–494. doi: 10.1111/j.1744-7429.2001.tb00202.x. [DOI] [Google Scholar]
- 69.Arce-Acosta I, Suzán-Azpiri H, García-Rubio O. Biotic factors associated with the spatial distribution of the mistletoe Psittacanthus calyculatus in a tropical deciduous forest of central Mexico. Bot Sci. 2016;94:89–96. doi: 10.17129/botsci.263. [DOI] [Google Scholar]
- 70.Ramírez MM, Ornelas JF. Cross-Infection Experiments of Psittacanthus schiedeanus: effects of host provenance, gut passage, and host fate on mistletoe seedling survival. Plant Dis. 2012;96:780–787. doi: 10.1094/PDIS-06-11-0509. [DOI] [PubMed] [Google Scholar]
- 71.Brasier CM, Cooke DEL, Duncan JM. Origin of a new Phytophthora pathogen through interspecific hybridization. Proc Natl Acad Sci USA. 1999;96:5878–5883. doi: 10.1073/pnas.96.10.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin N, Bouck A, Arnold M. Detecting adaptive trait introgression between Iris fulva and I. brevicaulis in highly selective field conditions. Genetics. 2006;4:2481–2489. doi: 10.1534/genetics.105.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arnold M, Martin N. Adaptation by introgression. J Biol. 2009;8:82. doi: 10.1186/jbiol176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hopkins R. Reinforcement in plants. New Phytol. 2013;197:1095–1103. doi: 10.1111/nph.12119. [DOI] [PubMed] [Google Scholar]
- 75.Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics155, 945–959 (2000). [DOI] [PMC free article] [PubMed]
- 76.Garroway CJ, et al. 2010. Climate change induced hybridization in flying squirrels. Global Change Biol. 2010;16:113–121. doi: 10.1111/j.1365-2486.2009.01948.x. [DOI] [Google Scholar]
- 77.Tsy LP. J. M., et al. Nuclear microsatellite variation in Malagasy baobabs (Adansonia, Bombacoideae, Malvaceae) reveals past hybridization and introgression. Ann Bot. 2013;112:1759–1773. doi: 10.1093/aob/mct230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 79.Anderson EC, Thompson EA. A model-based method for identifying species hybrids using multilocus data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nielsen EE, Bach LA, Kotlicki P. HYBRIDLAB (version 1.0): a program for generating simulated hybrids from population samples. Mol Ecol Notes. 2006;6:971–973. doi: 10.1111/j.1471-8286.2006.01433.x. [DOI] [Google Scholar]
- 81.Keenan K, Mcginnity P, Cross TF, Crozier WW, Prodöhl PA. DiveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol. 2013;4:782–788. doi: 10.1111/2041-210X.12067. [DOI] [Google Scholar]
- 82.Goudet, J. & Jombart, T. hierfstat: Estimation and tests of hierarchical F-statistics. R package version 0.04-22. https://CRAN.R-project.org/package=hierfstat (2015).
- 83.Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. doi: 10.1093/oxfordjournals.jhered.a111573. [DOI] [Google Scholar]
- 84.Chapuis MP, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol. 2007;24:621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- 85.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc B. 1977;39:1–38. [Google Scholar]
- 86.Weir, B. S. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sinauer Associates, Sunderland, MA (1996).
- 87.Excoffier L, Smouse P, Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 89.Chiucchi JE, Gibbs HL. Similarity of contemporary and historical gene flow among highly fragmented populations of an endangered rattlesnake. Mol Ecol. 2010;19:5345–5358. doi: 10.1111/j.1365-294X.2010.04860.x. [DOI] [PubMed] [Google Scholar]
- 90.Wilson GA, Rannala B. 2003. Bayesian inference of recent migration rates using multilocus genotypes. Genetics. 2003;163:1177–91. doi: 10.1093/genetics/163.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beerli P. Estimation of the population scaled mutation rate from microsatellite data. Genetics. 2007;177:1967–1968. doi: 10.1534/genetics.107.078931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garza JC, Williamson EG. Detection of reduction in population size using data from microsatellite loci. Mol Ecol. 2001;10:305–318. doi: 10.1046/j.1365-294x.2001.01190.x. [DOI] [PubMed] [Google Scholar]
- 94.Cornuet, J. M. P. et al. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics30, 1187–1189, 10.1093/bioinformatics/btt763 (2014). [DOI] [PubMed]
- 95.Cornuet JM, et al. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics. 2008;24:2713–2719. doi: 10.1093/bioinformatics/btn514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beaumont MA, Zhang W, Balding DJ. 2002. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset (microsatellites) generated and analysed during the current study are available in the Supporting Information of Ramírez-Barahona et al. (nph14471-sup-0003-NotesS2.csv)41.