Abstract
The dirigent (DIR and DIR-like) proteins involved in lignification, play a pivotal role against biotic and abiotic stresses in plants. However, no information is available about DIR gene family in pepper (Capsicum annuum L.). In this study, 24 putative dirigent genes (CaDIRs) were identified, their gene structure, genome location, gene duplication and phylogenetic relationship were elucidated. Tissue-specific expression analysis displayed the highest transcription levels in flower, stem and leaf. Some CaDIRs were up-regulated by virulent (CaDIR2, 3, 6, 7, 11, 14, 16, 22 and 23) and avirulent (CaDIR3, 5, 7, 16, 20, 22, 23 and 24) Phytophthora capsici strains, as well as by Methyl jasmonate, salicylic acid, NaCl and mannitol stresses. Acid-soluble lignin content increased (103.21%) after P. capsici inoculation (48-hour). Silencing of CaDIR7 weakened plant defense by reducing (~50%) root activity and made plants more susceptible (35.7%) to P. capsici and NaCl (300 mM). Leaf discs of the CaDIR7:silenced plants exposed to NaCl and mannitol (300 mM each), exhibited a significant decrease (56.25% and 48% respectively) in the chlorophyll content. These results suggested that CaDIR7 is involved in pepper defense response against pathogen and abiotic stresses and the study will provide basic insights for future research regarding CaDIRs.
Introduction
Pepper (Capsicum annuum L.) is an important solanaceous vegetable worldwide. Many vegetables are affected by the oomycete pathogen P. capsici, which has been reported to infest pepper, eggplant, tomato, all cucurbits, and more recently snap and limabeans1,2. In general, with a sessile and autotrophic lifestyle, plants grown in the open environment are always vulnerable to various biotic and abiotic stresses, such as pathogen infections, pest attacks, extreme temperatures, drought, salinity, and heavy metals3,4. These stresses cause a significant reduction in yield and quality. In response, plants have evolved some sophisticated defense mechanisms, including oxidative burst, the deposition of lignin and callose into the cell wall and regulation of signaling networks, to combat these stresses5–8. To control pathogen growth and invasion in the host tissues, inducible biochemical reactions create a protective physiological condition9. The inducible defense responses of plant include the synthesis of signaling molecules such as salicylic acid, ethylene and jasmonates, which in turn regulate gene expression and produce defense molecules such as reactive oxygen species (ROS), phenylpropanoids, phytoalexins and pathogenesis-related (PR)10. In response to biotic and abiotic challenges, plants activate a variety of genes, including the DIR gene9,11. DIR is a disease resistance responsive gene (DRRG), which enhances the stress resistance in different crop plants11.
The word dirigent comes from the Latin word dirigere (to guide or align), and dirigent proteins were found for the first time in Forsythia intermedia12. DIR proteins from Forsythia suspensa13, Podophyllum peltatum14, and Thuja plicata (western red cedar)15 are biochemically involved in directing the stereo-specific coupling of E-coniferyl alcohol to produce the lignan ( + )-pinoresinol. Based on this activity and its associated cellular localization in F. intermedia, DIR proteins could also function in the formation of lignin16. If DIR proteins are absent, then at the 8–8′, 8–5′, or 8-O-4′ positions a non-specific radical-radical coupling occurs, resulting in racemic lignan products13,17. DIR proteins share a dirigent-conserved domain with disease resistance response (DRR) family proteins18, and they are believed to mediate the free radical coupling of monolignol plant phenols in plants to yield lignans and lignins13,16; thus, DIRs have been culpably involved in disease resistance responses19,20. Lignan, either constitutive or inducible, has antifungal properties, which seems to be primarily involved in plant defense21. In defense response against pathogen infection, lignin deposition is considered to function as a physical barrier22. Moura et al.23 suggested that an increase in lignification occurs in response to pathogen attacks. Fang et al.24 noted that in mature leaves the thickened structure contains lignin, which acts as an effective physical barrier to pathogen attacks. As a non-degradable mechanical barrier for most microorganisms, lignin makes the host less susceptible. Lignin is an imperative compound that is primarily deposited in terminally differentiated cells of supportive and water-conducting tissues, and it is mainly associated with mechanical support, water transport in the xylem vessels and defense against pests and microorganisms25.
Earlier studies have shown that DIR proteins exist in almost all vascular plants, including lichens, ferns, gymnosperms and angiosperms11,26–29. In canola, when the DIR gene was expressed constitutively, increased resistance was observed against a broad range of fungal pathogens including Rhizoctonia solani and Leptosphaeria maculans19. The BhDIR1 transcripts in Boea hygrometrica were found to be expressed in response to various abiotic stresses including dehydration, CaCl2, ABA, H2O2, EGTA, and temperature stresses28. Ralph and colleagues suggested that DIR proteins can be further divided into five subgroups, i.e., DIR-a, DIR-b, DIR-c, DIR-d and DIR-e11. A further increase in the number of DIR proteins and the appearance of two other subfamilies i.e. DIR-f and DIR-g, the DIR-b and DIR-d subfamilies were clustered together27.
Understanding the functions of the dirigent gene family in biological and physiological process would be a possible practicable approach to analyzing and improving the defense response of crops against biotic and abiotic stresses. However, no study has been conducted on the DIR gene family in pepper until now. Hence, the present study was designed to explore the DIR and DIR-like protein (CaDIRs) family in pepper through a genome-wide identification and gene expression analysis. A total of 24 CaDIRs from the pepper genome were identified through a bioinformatics analysis and PCR testing. Subsequently, we also performed a detailed analysis of the gene structure, conserved domains, tertiary structures, chromosomal distribution, gene duplication, cis-acting elements in the promoter regions and phylogenetic relationships to explore the evolutionary history of DIR and DIR-like protein expansion in pepper. In addition to ascertaining the probable role of CaDIRs in pepper, the expression patterns of CaDIRs in different plant tissues were evaluated, as well as their responses to P. capsici and various abiotic and hormonal stresses. Furthermore, the acid-soluble lignin content after P. capsici inoculation was also measured to confirm the role of CaDIRs in lignin biosynthesis. The function of the CaDIR7 gene in pepper plants against P. capsici and salt stress was characterized through VIGS. The current study will provide a basis for the role of CaDIRs in pepper defense response against P. capsici and abiotic stresses and future insights for research on the DIR gene family.
Materials and Methods
Identification and Annotation of CaDIRs in Pepper
For identification of the CaDIR domain with accession no. PF03018, the methodology of our previous study was followed30. The alignment of the candidate CaDIRs from CM334 and Zunla-1 databases were conducted by DNAMAN software (Version 5.0) to select genes with differences in sequences in the two databases31. Gene-specific primer pairs (Supplementary Table S1) were designed by Primer Premier 5.0 (Premier Biosoft International, CA, U.S.A.) to amplify the different target regions, which were then aligned with sequences of the same gene from the CM334 and Zunla-1 databases to confirm the reliable sequences. Nomenclature of the putative CaDIRs was assigned based on their chromosome orders.
Bioinformatics analysis, exon/intron structure determination and phylogenetic tree construction
Multiple sequence alignments of amino acid sequences were performed using DNAMAN software (Version 5.0). To compute the molecular formula, the total number of items, instability index, protein molecular weight (MW) and theoretical isoelectric point (pI), the amino acid sequences were blast in Expasy ProtoParam (http://web.expasy.org/protparam/). WoLF PSORT II (http://www.genscript.com/wolf-psort.html)32 and TargetP (http://www.cbs.dtu.dk/services/TargetP/)33 were used to predict the subcellular locations. N-glycosylation sites (Asn) of the CaDIRs were searched online using NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/)34. For phylogenetic tree construction, the amino acid sequences of DIR and DIR-like genes from different plant species were aligned using CLUSTALW as described by Guo et al.35 while MEGA 6.0 was used for tree construction36. The exon/intron structures of the CaDIRs were searched and presented as described by Kang et al.37. Prediction of the tertiary structures and homologs of CaDIRs was conducted through the online server Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id = index)38 as described by Li et al.29.
Chromosomal Location and Duplication Analysis of CaDIRs
Information about the chromosomal location of CaDIRs was obtained from the Pepper Genome Platform (PGP) (http://peppergenome.snu.ac.kr/) as described by Zhang et al.30, and the genes were mapped on chromosomes using MapDraw39. A duplication analysis within the pepper genome was performed with the criteria depicted by Gu et al.40: (1) the FASTA-alignable region between the two proteins should be longer than 80% of the longer protein, and (2) the identity between the two proteins (I) should be I ≥ 30% if the alignable region is longer than 150 aa and I ≥ 0.01n + 4.8 L−0.32(1+exp(−L/1000) 41 if otherwise, where n = 6 and L is the alignable length between the two proteins40,41.
Analysis of Conserved Motifs of CaDIRs
The CaDIR-conserved domains were confirmed by Pfam (http://pfam.xfam.org/) and SMART (http://smart.embl-heidelberg.de/). The identification of the conserved motifs was determined as described by Guo et al.35 with the maximum number of motifs = 10.
Search for cis-acting Elements in the Promoters of CaDIRs
As described by Guo et al.35, 1500 bp upstream regions from the start codon (ATG) of the CaDIRs were derived from PGD and used as a query to search the Cis-acting regulatory elements. Cis-element analysis of CaDIRs was conducted using the online program PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)42.
Plant Materials and Seedlings Treatment
The plant materials, P. capsici strains and inoculation method used in the current research work are the same as those described by Zhang et al.30. Root samples were collected at 0, 6, 12, 24 and 48 hours post inoculation (hpi) for RNA extraction. For the plant-signaling molecule treatments, 5 mM of salicylic acid (SA) or 50 μM of methyl jasmonate (MeJA) solutions and sterile ddH2O were sprayed on the pepper plants. The leaf samples for RNA extraction were collected at different time points (0, 3, 6, 9, 12, 24 and 48 hours post treatment (hpt)). For drought and salt stress, the roots of seedlings were watered with 300 mM each of mannitol and NaCl solutions, respectively, while sterile water was used in the control plants, and the root samples from the treated and control plants were collected at various times (0, 3, 6, 9, 12, 24 and 48 hpt) for RNA extraction. For tissue-specific expression, samples were collected from leaves, stems, flowers, roots, green fruits and red fruits. All of the collected samples were immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction. All treatments were performed and analyzed thrice in separate experiments.
P. capsici Preparation and Inoculation
P. capsici was prepared as described by Zhang et al.43. The detached leaf inoculation assay was conducted as described in our previous study44 and stored in a growth chamber at 28 °C with 60% relative humidity and a 16 h light/8 h dark photoperiod.
RNA Extraction and qRT-PCR analysis
Total RNA was extracted using the method of Guo et al.45, and the synthesis of cDNA was conducted according to the instructions of the manufacturer’s Prime Script Kit (Takara, Dalian, China). To check the quality and concentration of the cDNA, the Nano-Drop (Thermo Scientific Nano-Drop 2000C, USA) instrument was used. For the real-time quantitative PCR (qRT-PCR) analysis, Primer Premier 5.0 software was used to design the primer pairs (Supplementary Table S2) for CaDIRs, and their specificities was evaluated online using NCBI Primer BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The internal control used in the study was a pepper ubiquitin-conjugating protein gene (CaUbi3)46. The qRT-PCR was performed as described by Guo et al.31 with a slight modification of the annealing temperature (59 °C for 30 s). The relative expression levels of all of the CaDIRs were calculated using the 2−ΔΔCT method47.
Measurement of the acid-soluble lignin content after P. capsici inoculation
The samples (whole plants) were collected at different times (0, 6, 12, 24, 48, and 72 h) with P. capsici post-inoculation (pi) and oven dried at 80 °C for 24 h. The acid-soluble lignin content was measured by the Klason method as described by Wu et al.28.
VIGS Assay of CaDIR7 in Pepper
The VIGS system was used for the knock-down of the CaDIR7 gene in the pepper plant cultivar AA3, and the VIGS assay was performed as described by Liu et al.48. Briefly, the 3′-untranslated region (UTR) of the CaDIR7 gene was cloned into a pTRV2 vector to construct the recombinant plasmid pTRV2:CaDIR7, which was further used in the subsequent research to ensure the specific silencing of CaDIR7 (primer pairs used for vector construction are shown in Supplementary Table S3). Afterwards, the freeze-thaw method was used to transform pTRV1, pTRV2 (negative control), and pTRV2:CaPDS (positive control) along with the combined vector pTRV2:CaDIR7 into an Agrobacterium tumefaciens strain (GV3101). A. tumefaciens strain (GV3101) harboring pTRV1 was mixed at a 1:1 ratio with pTRV2, pTRV2-CaPDS and pTRV2-CaDIR7. The agrobacterium inocula suspensions harboring pTRV1, pTRV2:00, pTRV2:CaPDS or pTRV2:CaDIR7 (OD600 = 1.0) were infiltrated into the cotyledons of pepper plants using a 1.0 mL sterilized needleless syringe. Then, these infiltrated pepper plants were grown in a growth chamber under the same growing conditions as described by Wang et al.49. Forty-five days post-infiltration, leaf samples from the control and CaDIR7-silenced plants were collected to measure the silencing efficiency. The experiment was conducted with three biological replicates.
Determination of Root Activity
The triphenyltetrazolium chloride (TTC) method was used to measure the root activity50. Before the TTC test, root tips (approximately 0.2 g) from the control (TRV:00) and CaDIR7-silenced (pTRV2:CaDIR7) plants were collected at various times after the inoculation with the P. capsici avirulent strain (PC), washed with ddH2O and then gently dried with moisture-absorbent paper. Afterwards, a modified TTC method was used to measure the root activity as described by Jin et al.44. The treatments were conducted in three biological replications, and measurements were repeated thrice.
Measurement of relative electrolyte leakage (REL) and chlorophyll content
The relative electrolyte leakage (REL) was measured according to the method described by Yin et al.51. REL was computed as follow: REL (%) = C1/C2 × 100. To measure the chlorophyll content, leaf disks (0.5 cm) from the control and CaDIR7-silenced plants were floated in different concentrations (0, 100, 300 mM) of NaCl and Mannitol solutions at 25 °C for 72 hours. The total chlorophyll contents of the samples (0.2 g) were spectrophotometrically measured after extracting into 80% (v/v) acetone as described by Guo et al.52.
Statistical analysis
The data were subjected to an analysis of variance (ANOVA) using SPSS software (SPSS version 16.0, SPSS Inc., U.S.A.). The analyzed data were expressed as the means ± standard deviation (SD) of three replicates in all measured parameters. A least significant difference (P < 0.05) test was used to identify significant differences among the treatments.
Results
Identification and Annotation of Dirigent Genes in Pepper
The Hidden Markov Model (HMM) profile of the dirigent domain (Accession no. PF03018) was blast-searched in the pepper genome to identify the dirigent gene family members in pepper. As a result, 26 and 25 dirigent/dirigent-like genes were found in the CM334 and Zunla-1 databases, respectively. Next, these gene sequences were aligned to avoid repetition and alternative splicing. In the case of Zunla-1, out of 25 genes, the single gene (Capana01g000149) sequence exhibited poor identification with CM334 (CA01g01690). When this was used as a query to blast in NCBI Conserved Domain Database (CDD), its domain (Uncharacterized protein family UPF0016) was found to be other than dirigent. Subsequently, primer pairs (Supplementary Table S1) were designed to amplify and confirm the doubtful gene sequences through cloning and sequencing. Finally, all of the predicted and sequenced genes protein sequences were confirmed through a blast search in NCBI CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). In conclusion, 24 CaDIRs were confirmed in pepper and assigned names on the basis of their chromosomal position and order (Table 1).
Table 1.
List of 24 CaDIR and CaDIR-like genes identified in pepper and their sequence characteristics.
| No. | Name | Gene ID | Chr | ORF | AA | MW | pI | Instability index | Introns | N-Glyc (Asn) position | Localization predicted |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CaDIR1 | CA01g01700 | 1 | 465 | 154 | 17.50 | 5.62 | 27.94 | 0 | 17, 30, 105 | cyto |
| 2 | CaDIR2 | CA01g01710 | 1 | 570 | 189 | 21.23 | 7.68 | 27.65 | 0 | 55, 68, 143 | chlo |
| 3 | CaDIR3 | CA01g01720 | 1 | 558 | 185 | 20.88 | 6.55 | 23.96 | 0 | 48, 56, 61, 121 | chlo |
| 4 | CaDIR4 | CA01g15980 | 1 | 522 | 173 | 19.07 | 8.78 | 12.81 | 0 | 5 | chlo |
| 5 | CaDIR5 | CA01g16630 | 1 | 573 | 190 | 21.12 | 9.30 | 29.24 | 0 | 130 | extr |
| 6 | CaDIR6 | CA02g19860 | 2 | 579 | 192 | 20.95 | 9.25 | 24.22 | 0 | 53 | vacu |
| 7 | CaDIR7 | CA03g02390 | 3 | 750 | 249 | 26.09 | 5.46 | 33.6 | 1 | 46 | nucl |
| 8 | CaDIR8 | Capana04g001233★ | 4 | 573 | 190 | 21.32 | 8.76 | 24.49 | 0 | 59, 69, 130, 174 | chlo |
| 9 | CaDIR9 | CA05g07510 | 5 | 561 | 186 | 20.50 | 8.71 | 37.16 | 0 | 31, 79, 179 | chlo |
| 10 | CaDIR10 | CA05g18980 | 5 | 687 | 228 | 25.14 | 6.83 | 43.93 | 0 | 24, 35, 62, 92 | chlo |
| 11 | CaDIR11 | CA05g18990 | 5 | 675 | 224 | 23.90 | 5.22 | 30.92 | 0 | — | nucl |
| 12 | CaDIR12 | Capana06g000465★ | 6 | 993 | 330 | 33.04 | 4.66 | 24.92 | 1 | 120 | chlo |
| 13 | CaDIR13 | CA06g10690 | 6 | 576 | 191 | 20.99 | 6.03 | 27.77 | 0 | 94, 131 | cyto, extr |
| 14 | CaDIR14 | CA07g11710 | 7 | 531 | 176 | 19.39 | 5.52 | 28.73 | 0 | 31, 44, 116, 170 | extr |
| 15 | CaDIR15 | Capana08g002690★ | 8 | 552 | 183 | 18.67 | 8.00 | 12.29 | 0 | 51, 65, 85, 132, 176 | cyto, extr |
| 16 | CaDIR16 | CA08g18950 | 8 | 582 | 193 | 20.71 | 6.72 | 17.47 | 0 | 62, 72, 186 | vacu |
| 17 | CaDIR17 | CA08g18960 | 8 | 321 | 106 | 11.58 | 5.16 | 24.3 | 0 | 48, 91 | extr |
| 18 | CaDIR18 | CA09g04150 | 9 | 567 | 188 | 20.83 | 6.36 | 36.71 | 0 | 57, 67, 128, 172 | extr |
| 19 | CaDIR19 | CA09g06650 | 9 | 489 | 162 | 17.78 | 5.94 | 33.41 | 1 | — | cyto |
| 20 | CaDIR20 | CA10g05650 | 10 | 579 | 192 | 21.20 | 9.28 | 18.19 | 0 | 59, 95, 130 | extr |
| 21 | CaDIR21 | CA11g13500 | 11 | 690 | 228 | 22.73 | 4.76 | 23.76 | 1 | — | cyto |
| 22 | CaDIR22 | CA11g18860 | 11 | 450 | 149 | 16.40 | 8.63 | 28.21 | 0 | 23,31,89 | cyto |
| 23 | CaDIR23 | Capana12g000353★ | 12 | 759 | 252 | 26.46 | 6.17 | 27.17 | 1 | 20, 53 | chlo, extr |
| 24 | CaDIR24 | CA12g07560 | 12 | 585 | 194 | 20.85 | 9.07 | 27.49 | 0 | — | chlo |
Chr: chromosome; ORF: open reading frame; AA: amino acid; MW: molecular weight (kDa); pI: isoelectric point. Sequenced IDs marked with Pentagram (★) are from Zunla-1 genome and others are from CM334 genome.
Bioinformatics analysis, exon/ intron structure determination and phylogenetic tree construction
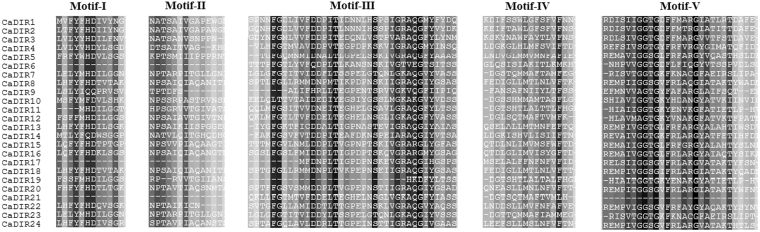
Five well-conserved motifs were found in the amino acid sequences of 24 CaDIRs (Fig. 1). The CDS of CaDIRs ranged from 321 bp (CaDIR17) to 993 bp (CaDIR12), whereas the deduced proteins had 106 to 330 amino acids. The predicted pI values ranged from 4.66 to 9.30, MW ranged from 11.58 to 33.04 kDa and the instability index varied from 12.29 to 43.93 (Table 1). Most of the CaDIRs (except CaDIR11, 19, 21 and 24) contained Asn sites, indicating that CaDIRs are probably secreted proteins. We anticipated all 24 CaDIRs through the special pathway for extracellular release or their probable final localization in the chloroplast, extra cellular region, nucleus, and cytoplasmic and vacuolar locations (Table 1). The molecular formula and total number of items of the deduced protein are shown in (Supplementary Table S4). The SMART results show that transmembrane regions existed in CaDIR2, 3, 4, 9, 10, 12, 13, 14, 16, 23 and 24. The molecular formula showed that CaDIR3 contained the highest (11) sulfur elements while CaDIR11 and 19 had only two sulfur elements each. To elucidate the topological structures and evolutionary relationships of the CaDIRs, multiple sequence alignments of amino acid sequences were used to build a Neighbor-Joining (NJ) tree (Fig. 2A). The exon/intron structure showed that out of 24 CaDIRs, 19 genes (79.17%) had no introns while five (20.83%) genes contained only one intron (Fig. 2B). Interestingly, all of the intron containing CaDIRs belonged to the DIR-e group.
Figure 1.
Five conserved characteristic motifs (I-V) of dirigent proteins in pepper CaDIR protein sequences.
Figure 2.
Phylogenetic tree and exon-intron analysis of pepper CaDIRs (A) Phylogenetic tree (B) exon-intron analysis.
To better understand the similarities and differences in CaDIRs between pepper and other plants, an unrooted phylogenetic tree was generated using 220 DIR and DIR-like gene protein sequences from various plant species. These sequences were from Arabidopsis thaliana, Agrostis stolonifera, Arachis hypogaea, Forsythia intermedia, Picea glauca, Hordeum vulgare, Nicotiana benthamiana, Gossypium barbadense, Brassica rapa, Isatis indigotica, Oryza sativa, Pisum sativum, Podophyllum peltatum, Saccharum officinarum, Sesamum indicum, Sorghum bicolor, Tamarix androssowii, Thuja plicata, Triticum aestivum, Tsuga heterophylla and Zea mays (Supplementary Table S5). The analysis showed that 24 CaDIRs were clearly separated into three distinct groups (Fig. 3). Five CaDIRs (CaDIR1, 2, 3, 6 and 9) were clustered into subfamily DIR-a with five A. thaliana and five I. indigotica genes. Previously, IiDIR12 was assigned to DIR-e, but in the current study it was close to BrDIR4 which lies in the DIR-a group; thus, IiDIR12 was designated a DIR-a. Another seven CaDIRs (CaDIR7, 10, 11, 12, 19, 21 and 23) were clustered into the subfamily DIR-e with six A. thaliana and seven I. indigotica genes. The other 12 CaDIRs (CaDIR4, 5, 8, 13, 14, 15, 16, 17, 18, 20, 22 and 24) were clustered into another subfamily DIR-b/d with fourteen A. thaliana and seven I. indigotica genes. The subfamily branches were colored following the previous studies27,29. The 24 CaDIRs were classified into three clusters based on sequence relatedness, which suggested that the two methods were in good agreement.
Figure 3.
Phylogenetic tree of DIR and DIR-like genes from pepper and other plant species. Protein sequences of 220 dirigent or dirigent-like (DIR) proteins were analyzed by the Maximum Likelihood (ML) test using MEGA 6.0. The subfamilies DIR-a, DIR-b/d, DIR-c, DIR-e, DIR-f and DIR-g are indicated by light pink, yellow, green, purple, sky cyanide and dark pink branch lines, respectively. The nomenclature of the DIRs used in the tree is as follows: Ah, Arachis hypogaea; As, Agrostis stolonifera; At, Arabidopsis thaliana; Br, Brassica rapa; Ca, Capsicum annuum; Fi, Forsythia intermedia; Gb, Gossypium barbadense; Hv, Hordeum vulgare; Ii, Isatis indigotica; Nb, Nicotiana benthamiana; Os, Oryza sativa; P, Picea glauca, Picea sitchensis or P. glauca x engelmannii; Pp, Podophyllum peltatum; Ps, Pisum sativum; Sb, Sorghum bicolor; Si, Sesamum indicum; So, Saccharum officinarum; Ta, Triticum aestivum; Tan, Tamarix androssowii; Th, Tsuga heterophylla; Tp, Thuja plicata and Zm, Zea mays. Different species are labeled with different color display markers while CaDIRs are labeled with red circle display markers.
Tertiary structures and homologs of CaDIRs
The predicted three-dimensional structures of 24 CaDIR proteins are shown in Fig. 4. Four identical top-scoring proteins for all 24 inquired sequences were found. The hypothetical protein c4revB belonged to disease resistance response protein 206 (drr206) while the other three belonged to the allene oxide cyclase-like protein (AOC) family. Disease resistance response protein 206 (c4revB) shared 20–68% sequence identity with CaDIRs, which was anticipated as a homolog of DIR with 100% probability except CaDIR21 (99.9%). On the other hand, AOC barrel-like protein d2brja1 shared only 15–27% sequence identity with CaDIRs, and its probability as a DIR homolog was approximately 98.3%, while the probabilities for the other two proteins d1zvca1 and c4h69A were 98.2% and 98.1%, respectively (Table 2).
Figure 4.
Predicted tertiary structures of pepper CaDIR proteins.
Table 2.
The probability and identity of homologous relationship of CaDIRs.
| c4revB (drr206) | d2brja1 (AOC) | c4h69A (AOC) | d1zvca1 (AOC) | |||||
|---|---|---|---|---|---|---|---|---|
| % confidence | % identity | % confidence | % identity | % confidence | % identity | % confidence | % identity | |
| CaDIR1 | 100.0 | 33 | 97.9 | 21 | 97.9 | 22 | 97.8 | 21 |
| CaDIR2 | 100.0 | 33 | 97.9 | 24 | 97.9 | 18 | 97.8 | 26 |
| CaDIR3 | 100.0 | 31 | 97.7 | 18 | 97.7 | 22 | 97.7 | 20 |
| CaDIR4 | 100.0 | 33 | 97.8 | 19 | 97.7 | 24 | 97.6 | 21 |
| CaDIR5 | 100.0 | 30 | 98.3 | 22 | 98.0 | 20 | 98.2 | 27 |
| CaDIR6 | 100.0 | 29 | 97.8 | 23 | 97.7 | 20 | 97.7 | 20 |
| CaDIR7 | 100.0 | 34 | 97.9 | 21 | 97.8 | 18 | 97.8 | 18 |
| CaDIR8 | 100.0 | 32 | 97.7 | 24 | 97.7 | 22 | 97.7 | 25 |
| CaDIR9 | 100.0 | 68 | 97.7 | 15 | 97.7 | 25 | 97.6 | 19 |
| CaDIR10 | 100.0 | 60 | 97.7 | 22 | 97.7 | 23 | 97.7 | 22 |
| CaDIR11 | 100.0 | 53 | 97.8 | 22 | 97.7 | 20 | 97.7 | 20 |
| CaDIR12 | 100.0 | 31 | 98.1 | 22 | 97.9 | 22 | 98.1 | 24 |
| CaDIR13 | 100.0 | 31 | 98.0 | 19 | 97.9 | 23 | 97.9 | 18 |
| CaDIR14 | 100.0 | 36 | 98.0 | 26 | 98.0 | 17 | 98.0 | 20 |
| CaDIR15 | 100.0 | 35 | 98.0 | 26 | 97.8 | 21 | 97.9 | 22 |
| CaDIR16 | 100.0 | 33 | 98.0 | 24 | 97.9 | 22 | 98.0 | 24 |
| CaDIR17 | 100.0 | 30 | 97.7 | 27 | 97.5 | 28 | 97.3 | 27 |
| CaDIR18 | 100.0 | 40 | 98.0 | 20 | 97.8 | 21 | 98.0 | 20 |
| CaDIR19 | 100.0 | 23 | 98.3 | 19 | 98.1 | 20 | 98.2 | 22 |
| CaDIR20 | 100.0 | 29 | 97.9 | 21 | 97.9 | 20 | 97.9 | 21 |
| CaDIR21 | 99.9 | 28 | ||||||
| CaDIR22 | 100.0 | 24 | 97.6 | 24 | 97.3 | 16 | 97.3 | 21 |
| CaDIR23 | 100.0 | 23 | 97.6 | 18 | 97.4 | 19 | 97.4 | 18 |
| CaDIR24 | 100.0 | 20 | 97.8 | 23 | 97.4 | 23 | 97.4 | 23 |
The c4revB are disease resistance response proteins 206 (drr206), and d2brja1, c4h69A and d1zvca1 are allene oxide cyclase-like proteins (AOC barrel-like).
Chromosomal Location and Gene Duplication of CaDIRs
According to the chromosomal location of dirigent genes in pepper, the 24 CaDIRs were distributed across all 12 chromosomes of pepper (Fig. 5). The results showed that chromosome 1 had the highest number of genes (20.83%) compared with the other chromosomes. There were three genes (12.5%) on chromosome 5, while two genes each were found on chromosome 8, 9 and 11. The remaining chromosomes (2, 3, 4, 6, 7, 10 and 12) each contained one gene. Three genes (CaDIR12, 15 and 23) located on chromosome 6, 8 and 12, respectively, were from the Zunla-1 database, so they were not mentioned in the figure.
Figure 5.
Distribution of CaDIRs on twelve chromosomes of pepper. Three genes (CaDIR12, 15 and 23) were from the Zunla-1 database and located on chromosome 6, 8 and 12, respectively, so they were not mentioned in the figure. The scale represents mega bases (Mb), and the chromosome numbers are indicated at the top of each bar.
The duplication analysis in Table 3 shows that CaDIR5 had more segmental duplication events than other genes, and they occurred on chromosome 6, 8, 9 and 10. CaDIR4, 14 and 16 each had three segmental duplications. Another three genes (CaDIR7, 13 and 18) had undergone two segmental duplications, respectively. Meanwhile, CaDIR11 and 20 had one segmental duplication event each. There were 2 clusters of tandemly duplicated CaDIRs, which occurred on chromosome 1. CaDIR15 also had one segmental duplication event (not shown in the figure). Taken together, our findings suggest that in the expansion of pepper CaDIRs, tandem and segmental duplication have an important contribution.
Table 3.
Paralogous groups of CaDIRs in pepper.
| Paralogous Group | Gene name | |
|---|---|---|
| Segmentally duplicated | 1 | CaDIR4, CaDIR13, CaDIR14, CaDIR18 |
| 2 | CaDIR5, CaDIR13, CaDIR16, CaDIR18, CaDIR20 | |
| 3 | CaDIR7, CaDIR11 | |
| 4 | CaDIR8, CaDIR15 | |
| 5 | CaDIR11, CaDIR21 | |
| 6 | CaDIR13, CaDIR2, CaDIR18 | |
| 7 | CaDIR14, CaDIR13, CaDIR16, CaDIR3 | |
| 8 | CaDIR16, CaDIR18, CaDIR20, CaDIR24 | |
| 9 | CaDIR18, CaDIR20, CaDIR24 | |
| 10 | CaDIR20, CaDIR24 | |
| Tandemly duplicated | 1 | CaDIR1, CaDIR2, CaDIR3 |
| 2 | CaDIR4, CaDIR2 |
Paralogous groups of segmentally and tandemly duplicated CaDIRs in pepper.
Identification of conserved motifs of CaDIRs in pepper
The conserved motifs of CaDIR proteins were found through the online MEME server (http://meme-suite.org/tools/meme). Ten distinct motifs were identified (Fig. 6, Table 4). The details of the putative motifs are shown in Supplementary Table S6. Motif1 (except CaDIR17 and 19) and motif2 (except CaDIR21) were found in nearly all of the CaDIRs. Motif3 was also present in most of the CaDIRs except CaDIR1, 6, 9, 11, 17, 19 and 21. Motif4 and 6 were present in 11 CaDIRs. Motif5 and motif10 were each found in 5 CaDIRs. Motif8 and motif9 existed in 7 and 6 CaDIRs, respectively, while motif7 only existed in 3 CaDIRs. CaDIR6 and 9 contained the lowest number of motifs (2), and they were motif1 and motif2.
Figure 6.
Distribution of conserved motifs of pepper CaDIR members. Each of the ten putative motifs is represented by a number in the colored box. The names of all CaDIRs along with their combined E-values are shown at the left side of the figure, and the motif size scale is at the bottom of the figure. For more details on the motifs refer to Table 5 and Supplementary Table S5.
Table 4.
Motif sequences identified by the MEME Suite.
| Motif | Length (aa) | Sequence |
|---|---|---|
| 1 | 41 | FGTLTMCDDPLTEGPEPNSKIIGRAQGMYVYSSQDDLSQWM |
| 2 | 41 | GSSLSFFGCNPIMHKYREMPIIGGTGKFRMARGYATAKTYW |
| 3 | 27 | KQKMTKLHFYWHDWLSGKNPSAIPICQ |
| 4 | 15 | TGDAIVEYNVVVLHY |
| 5 | 20 | DQHTTDGVETILHITVYLTY |
| 6 | 11 | MNFVFTEGKYN |
| 7 | 14 | YFRLCVDIKLYECW |
| 8 | 21 | PTTHGWEQEPKGVEKWFKRLP |
| 9 | 21 | NGNNPIVNNNNYPFLTGLSGT |
| 10 | 21 | YSGQVPFAKPNGQQPPKNGGV |
Motif numbers corresponded to the motifs in Fig. 6.
Cis-acting elements in the promoter regions of CaDIRs
To investigate the possible cis-elements involved in the activation of defense-related genes, upstream regions of all CaDIRs were analyzed online with Plant CARE. The silico analysis revealed that CaDIRs contained cis-elements conferring responsiveness to plant hormones and biotic and abiotic stresses. As shown in Tables 5 and 6, the defense and stress responsiveness elements (TC-rich repeats) were found in the promoters of all CaDIRs except CaDIR4, 14, 15 and 17, in which the TC-rich repeats in the promoter region of CaDIR3 was highest (6) followed by CaDIR5 and 9 (each having 5). MeJA-responsiveness elements (TGACG-motif) were found in the promoter region of 19 CaDIRs, where CaDIR2 and 9 had the highest number (4) of elements followed by CaDIR7, 11, 12, 15, 16, 18 and 21 (each having 3). Cis-acting elements involved in salicylic acid responsiveness (TCA-element) and heat stress responsiveness elements (HSE-element) were found in the promoter regions of 16 CaDIRs. The MYB binding site involved in drought-inducibility (MBS), GA-responsive element (GARE-motif) and fungal elicitor-responsive element (W box) were found in the promotor regions of 15, 13 and 12 CaDIRs, respectively. Abscisic acid responsiveness elements (ABRE), Ethylene-responsive element (ERE) and GA-responsive element (P box) were found in the promoter regions of 11 CaDIRs. The cis-acting element involved in low temperature responsiveness (LTR) was found in the promoter region of 10 CaDIRs. In addition, wound-responsive element (WUN-motif) and auxin-responsive elements (TGA-element and AuxRR-core) were found in some of the CaDIRs promoter regions. All of the predicted cis-elements were involved in the response to signaling hormones and stresses.
Table 5.
Cis-acting elements in the promoter regions of CaDIR genes.
| Gene name | TGACG-motif | GARE-motif | TC-rich repeats | TCA-element | MBS | W box | ERE | HSE | ABRE | TGA-element | AuxRR-core | LTR | WUN-motif | P-box |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CaDIR1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CaDIR2 | 4 | 1 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CaDIR3 | 0 | 1 | 6 | 0 | 2 | 2 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 1 |
| CaDIR4 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 0 |
| CaDIR5 | 1 | 0 | 5 | 3 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| CaDIR6 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| CaDIR7 | 3 | 1 | 2 | 1 | 0 | 3 | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 1 |
| CaDIR8 | 1 | 0 | 1 | 2 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| CaDIR9 | 4 | 0 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| CaDIR10 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 3 | 1 | 1 | 0 | 1 | 0 | 0 |
| CaDIR11 | 3 | 1 | 1 | 3 | 5 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 |
| CaDIR12 | 3 | 1 | 2 | 0 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 3 |
| CaDIR13 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 5 | 1 | 0 | 0 | 1 | 0 | 1 |
| CaDIR14 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| CaDIR15 | 3 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| CaDIR16 | 3 | 1 | 1 | 1 | 2 | 0 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| CaDIR17 | 0 | 1 | 0 | 0 | 3 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| CaDIR18 | 3 | 0 | 2 | 1 | 0 | 3 | 4 | 2 | 0 | 1 | 0 | 1 | 0 | 0 |
| CaDIR19 | 0 | 2 | 2 | 2 | 1 | 0 | 0 | 3 | 1 | 0 | 0 | 1 | 1 | 1 |
| CaDIR20 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 2 |
| CaDIR21 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| CaDIR22 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| CaDIR23 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 1 |
| CaDIR24 | 1 | 0 | 2 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 3 |
Table 6.
Function of the cis-elements found in the promoters of CaDIRs.
| Cis-acting element | Motif | Function | Reference |
|---|---|---|---|
| Defense-related and stress stimulative elements | TC-rich repeats | Defense and stress responsiveness | 83 |
| W box | Fungal elicitor-responsive element | 84 | |
| WUN-motif | Wound-responsive element | 85 | |
| HSE | Heat stress responsiveness | 85 | |
| LTR | Low temperature responsiveness | 86 | |
| MBS | MYB binding site involved in drought-inducibility | 87 | |
| Plant hormones responsive elements | TGACG-motif | MeJA responsiveness | 75 |
| GARE-motif | GA-responsive element | 88 | |
| TCA-element | Salicylic acid responsiveness | 76 | |
| TGA-element | Auxin-responsive element | 85 | |
| ERE | Ethylene-responsive element | 89 | |
| ABRE | ABA responsiveness | 90 | |
| AuxRR-core | Auxin responsiveness | 91 | |
| P-box | GA-responsive element | 92 |
Expression Analysis of CaDIRs under P. capsici, abiotic and signaling molecules challenges
To explore the effect of P. capsici infestation on CaDIRs, the pepper cultivar “AA3” was inoculated with virulent and avirulent strains via root drenching, and the expression levels were analyzed by qRT-PCR (Fig. 7). The results revealed that different expression patterns were observed for CaDIRs; therefore, they were categorized into eight groups based on their expression levels. Group-I contained CaDIR13, 15 and 18, which showed complete down-regulation with both strains pi. Another three CaDIRs (CaDIR1, 10 and 21) belonged to Group-II, showed up-regulation in the case of avirulent strain pi at 6 h and then down-regulated. Meanwhile, with the virulent strain pi, they exhibited total down-regulation except CaDIR1, which was slightly up-regulated at 12 h and then down-regulated. Group-III (CaDIR2, 3, 22 and 24) showed up-regulation after avirulent strain pi, in which CaDIR2, 22 and 24 were up-regulated at 6 h, followed by down-regulation and then up-regulation to the maximum at 48 h (12.06, 21.69 and 8.92 respectively), while CaDIR3 was peaked (25.46) at 48 h. In the case of virulent strain pi, CaDIR22 and 24 were up-regulated, reached a maximum at 12 h (40.89 and 2.71 respectively) and then down-regulated, whereas CaDIR2 and 3 were up-regulated at 6 h, then showed a downward trend at 12 h, and again up-regulated. Group-IV members comprised CaDIR8, 11, 16 and 23. The transcription levels of all members of this group were elevated with virulent strain inoculation and reached a maximum at 48 hpi (9.87, 31.02 and 51.71 respectively) except CaDIR8, which was initially down-regulated, then up-regulated and finally reached a peak at 48 h (2.60). Meanwhile, in the case of avirulent strain inoculation, CaDIR8 (1.43) and 11 (2.71) were up-regulated to a maximum at 24 hpi, whereas CaDIR16 and 23 were up-regulated to a maximum at 12 hpi (38.83) and 6 (35.91) hpi, respectively. Group-V contained six CaDIRs (CaDIR5, 9, 12, 17, 19 and 20), which showed up-regulation at 6 hpi with the avirulent strain and then down-regulated at other time points; meanwhile, with the virulent strain pi, down-regulation was observed for CaDIR9, 12 and 19, whereas CaDIR17, 5 and 20 showed slight up-regulation at 12 h and 24 h. Group-VI (CaDIR4 and 6) initially exhibited down-regulation after inoculation with the virulent strain, up-regulated to the maximum (4.01 and 12.27 respectively) at 24 h and then down-regulated again, whereas with avirulent pi, both CaDIRs showed down-regulation. Group-VII contained CaDIR14, which showed up-regulation to a maximum at 6 h for both strains and then down-regulated. Group-VIII contained CaDIR7, which exhibited progressive up-regulation for both virulent and avirulent strains and reached a peak (51.43 and 36.28 respectively) at 48 hpi.
Figure 7.
Expression profiles of CaDIRs in response to inoculation with virulent and avirulent Phytophthora capsici strains. Mean values and SDs for three replicates are shown. Small letters represent significant differences (p < 0.05).
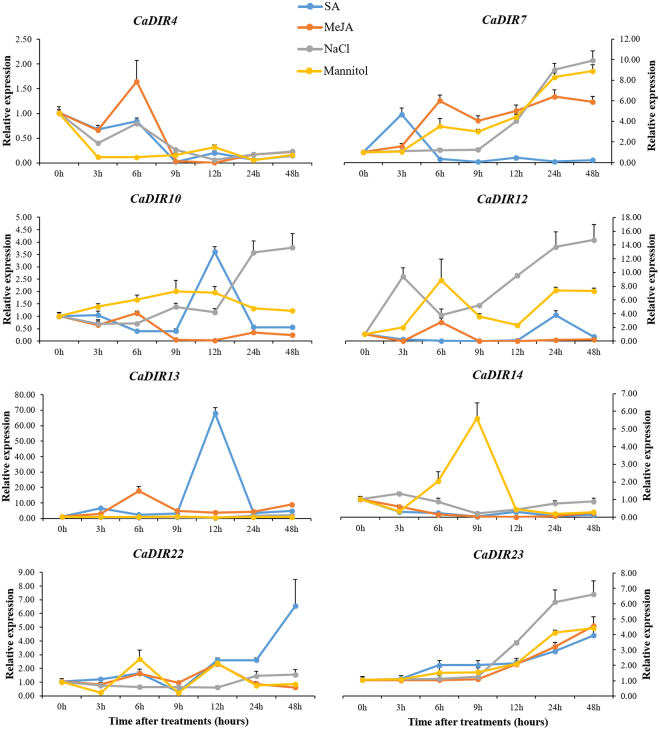
To investigate the effects of abiotic stresses on the expression patterns of CaDIRs, eight representative genes (CaDIR4, CaDIR7, CaDIR10, CaDIR12, CaDIR13, CaDIR14, CaDIR22 and CaDIR23) from each of the eight groups above were subjected to NaCl and mannitol stresses (Fig. 8). The results showed that two candidate CaDIRs (CaDIR4 and CaDIR13) showed no significant response to both stresses, and CaDIR4 was completely down-regulated. Similarly, CaDIR14 also showed down-regulation in response to NaCl, whereas it up-regulated at 6 h and 9 h and then down-regulated in response to mannitol. CaDIR7 and CaDIR23 were gradually up-regulated by abiotic stresses and reached to peak at 48 hours post treatment (hpt). CaDIR10 and CaDIR12 were up-regulated in response to mannitol and were highest at 9 (2.01) hpt and 6 (8.87) hpt, respectively, whereas both peaked (3.77 and 14.73 respectively) at 48 h in response to NaCl. CaDIR22 was initially down-regulated in response to NaCl but then up-regulated at 24 h and 48 h, whereas in response to mannitol concomitant up- and down-regulation occurred.
Figure 8.
Expression profiles of CaDIRs in response to treatment with SA, MeJA, NaCl and Mannitol. Mean values and SDs for three replicates are shown.
The selected CaDIRs from the eight groups above were also exposed to hormonal stress (MeJA and SA). The expression results revealed that in response to signaling molecules, CaDIR4, CaDIR10 and CaDIR14 showed no significant response, but CaDIR4 and CaDIR10 showed slight up-regulation at 6 h and 12 h post MeJA and SA treatments, respectively (Fig. 8). CaDIR7 and CaDIR22 showed a significant response to MeJA and SA respectively, reaching a maximum at 24 and 48 hpt, respectively. CaDIR12 and CaDIR13 responded to SA and reached a peak at 24 and 12 hpt, respectively, whereas in response to MeJA both peaked at 6 hpt and then showed down-regulation. CaDIR23 exhibited a progressive increment in the expression levels and reached a maximum (4.56 and 3.94) at 48 hpt.
Expression analysis of CaDIRs in different tissues of pepper
To investigate the involvement of CaDIRs in the growth and development of pepper, the expression levels in various vegetative and reproductive organs of cultivar AA3 grown under normal conditions were analyzed through qRT-PCR using gene-specific primer pairs (Supplementary Table S2). As shown in Fig. 9, the expression pattern of each CaDIR was different in various organs and stages. The analysis showed that out of 24 CaDIRs, two genes (CaDIR2 and 5) were not expressed in all of the tested organs. Five CaDIRs were expressed in one organ only: CaDIR6 and 13 were expressed in the flower while CaDIR12, 19 and 22 were expressed in the stem, root and red fruit, respectively. CaDIR1, 3, 8, 9, 10, 14, 17, 18 and 23 were expressed in all of the tested tissues except the root, and CaDIR4 and 20 were expressed in all tissues except red fruit. CaDIR7 was expressed in the leaf, stem, flower and red fruit but showed no expression in the root and green fruit. Similarly, CaDIR11 was also expressed in the leaf, stem, flower and green fruit. The other CaDIRs were expressed in two or three organs. Among all CaDIRs, CaDIR23 was expressed at the highest level in the stem (71.55) and green fruit (66.48). Collectively, CaDIRs showed the highest expression in flowers, followed by the stem, leaf, green fruit and red fruit, whereas they were expressed at the lowest levels in roots.
Figure 9.
Tissue-specific expression analysis of pepper CaDIRs. The analyzed tissues included root, stem, leaf, flower, green fruit and red fruit.
Changes in acid-soluble lignin content after P. capsici inoculation
The acid-soluble lignin content was measured at various times (0, 6, 12, 24, 48 and 72 h) after inoculation with the P. capsici strains. The results showed that in the case of the avirulent strain, the acid-soluble lignin content was increased by 14.46%, 36.09%, 25.67%, 90.40% and 43.93%, whereas with the virulent strain, the increase was 21.68%, 50.57%, 33.07%, 116.02% and 68.10% compared with control plants at 6, 12, 24, 48 and 72 hpi, respectively (Table 7).
Table 7.
Changes in the acid soluble (Klason) lignin content of the pepper plants after inoculation with PC and HX-9 strains of Phytophthora capsici at different time points.
| 0 h (Untreated) | 6 h | 12 h | 24 h | 48 h | 72 h | |
|---|---|---|---|---|---|---|
| Mock-inoculated | 6.29 ± 0.44 | 7.02 ± 0.49 | 4.77 ± 0.33 | 5.32 ± 0.37 | 3.52 ± 0.25 | 5.02 ± 0.35 |
| PC strain inoculated | 6.29 ± 0.44 | 8.04 ± 0.24 | 6.50 ± 0.13 | 6.68 ± 0.27 | 6.70 ± 0.07 | 7.23 ± 0.36 |
| Increased Percentages | 0 | 14.46% | 36.09% | 25.67% | 90.40% | 43.93% |
| HX-9 strain inoculated | 6.29 ± 0.44 | 8.54 ± 0.34 | 7.19 ± 0.14 | 7.08 ± 0.28 | 7.60 ± 0.38 | 8.44 ± 0.25 |
| Increase Percentages | 0 | 21.68% | 50.57% | 33.07% | 116.02% | 68.10% |
Data are the mean value of three replications with standard deviation (mean ± SD for triplicates).
Reduced tolerance of CaDIR7-silenced pepper plants to P. capsici
To determine the loss of function of the CaDIR7 gene in pepper, the VIGS approach was used for gene knock-down in the pepper cultivar AA3. To confirm the visual success of VIGS, a positive control vector (pTRV2:CaPDS) was used for the silencing of the CaPDS gene, which produced a typical white color as a result of the photo-bleaching phenotype in the leaves, while the negative control used in the study was pTRV2:00. After 45 days of inoculation, the photobleaching phenotypes observed in the leaves of CaPDS-silenced plants (Fig. 10A), demonstrating that VIGS was successful. At that point, the silencing efficiency of CaDIR7 was investigated through a qRT-PCR analysis, and the results showed that the expression levels of CaDIR7 gene in the silenced plants (pTRV2-CaDIR7) were much lower than in the control plants (pTRV-00) (Fig. 10B). The expression of CaDIR7 was induced significantly with the interaction of the avirulent strain of P. capsici. Thus, to ensure the role of CaDIR7 in defense response, the fourth to fifth leaves from the top of the silenced (pTRV2:CaDIR7) and control (pTRV2:00) pepper plants were removed and infiltrated with 20 µL zoospore suspension (104 zoospores mL−1) of the avirulent strain of P. capsici. Three days post inoculation (dpi), disease lesions were detected on the leaves of both CaDIR7-silenced (a) and control (b) plants, but numerous and larger lesions were found on CaDIR7-silenced plants leaves (Fig. 10C). The quantitative analysis of the lesion area showed that detached leaves of the silenced plants exhibited significantly larger lesion area than the control (Fig. 10D). Additionally, the expression level of the CaDIR7 gene during P. capsici infection was checked, and the time was expanded to 10 dpi. A significant increase in the expression levels of CaDIR7 was observed at 2, 4, 7 and 10 dpi in the empty vector control plants versus the CaDIR7-silenced plants (Fig. 11A). Moreover, for P. capsici pi, the root activity of the CaDIR7-silenced and empty vector control plants was also measured by the TTC method. The root activity of the silenced plants was lower than the control, and a significant difference was observed at 4, 7 and 10 dpi between the silenced and control plants (Fig. 11B).
Figure 10.
The phenotypes and loss of function analysis of CaDIR7 in leaves of pepper. (A) The phenotypes of TRV2:00, TRV2:CaPDS and TRV2:CaDIR7, (B) The relative expression of CaDIR7 in leaves of CaDIR7-silenced and control (TRV2:00) plants, (C) disease symptoms developed on the detached leaves of CaDIR7-silenced (a) and control (TRV2:00) (b) plants at 3 days post inoculation (dpi), (D) percentage of the lesion area of leaves exposed to P. capsici infection from TRV2:00 and TRV2:CaDIR7. Error bars represent the mean ± SD of three independent biological replicates. Small letters represent significant differences (p < 0.05).
Figure 11.
The CaDIR7-silenced pepper plants exhibit reduced resistance to P. capsici. (A) The expression level of CaDIR7 in control (TRV2:00) and CaDIR7-silenced plants, (B) root activity of the control and CaDIR7-silenced plants after inoculation with the avirulent strain of P. capsici. Values are the means ± SD from three independent experiments. Small letters represent significant differences (p < 0.05).
Reduced tolerance of CaDIR7-silenced pepper plants to salt
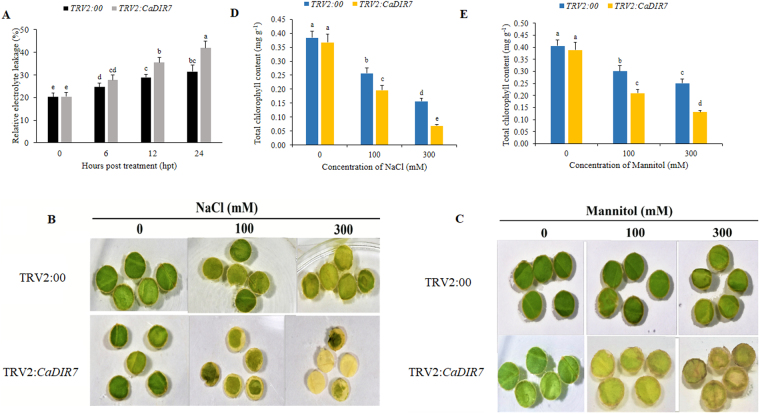
To assess the function of CaDIR7 under salt stress, the empty vector control and CaDIR7-silenced plants were exposed to NaCl stress (300 mM). As shown in Fig. 12A, the loss of function of CaDIR7 significantly compromised salt stress tolerance. A higher expression of CaDIR7 was recorded in the empty vector control plants than in the CaDIR7-silenced plants. In addition, the expression levels of other defense-related genes were also examined to see whether the silencing of CaDIR7 alters their expression. It was found that with the passage of time after salt stress, the transcript levels of CaDEF1 (a JA dependent gene)53 and CaPO1 (a peroxidase gene)54 were elevated, but their elevation in the empty vector control plants was higher than in the CaDIR7-silenced plants (Fig. 12B,C). Furthermore, root activity was also measured, and the results showed a significant reduction in the root activity after salt stress. At 24 h post NaCl stress, the root activity of the CaDIR7-silenced plants (2.46) was less than half of that of the empty vector control (1.22) (Fig. 12D).
Figure 12.
The CaDIR7-silenced pepper plants compromised tolerance to NaCl stress. (A) The expression level of CaDIR7, (B) expression level of CaDEF1, and (C) expression level of CaPO1 genes in control (TRV2:00) and CaDIR7-silenced plants, (D) root activity of the control and CaDIR7-silenced plants. Values are the means ± SD from three independent experiments. Small letters represent significant differences (p < 0.05).
Effect of silencing of CaDIR7 on REL and chlorophyll content
The extent of membrane damage caused by NaCl stress was assessed by an indirect measurement of electrolyte leakage. After salt stress, leaf discs were collected from TRV2:00 and TRV2:CaDIR7 plants at different times for REL measurement. The results showed excessive electrolyte leakage in TRV2:CaDIR7 versus TRV2:00 plants. At 24 h post salt stress, the REL was significantly higher in the silenced plants (41.9) than in the control (31.5) (Fig. 13A). It is well known that photosynthetic efficiency has a direct effect on the chlorophyll content55. To assess whether the knock-down of CaDIR7 reduces the tolerance of pepper plants to salt and dehydration stress, leaf discs from TRV2:00 and CaDIR7-silenced plants were exposed to different concentrations (0, 100 and 300 mM) of NaCl and Mannitol. The results revealed that after 72 hours, the 0 mM-treated leaf discs had no significant differences in the chlorophyll content of both TRV2:00 and TRV2:CaDIR7, whereas in response to 100 and 300 mM NaCl and mannitol stresses, there was a significant decrease in the total chlorophyll content of TRV2:CaDIR7 versus TRV2:00. Phenotypically, the leaf discs of TRV2:00 remained green and that of silenced plants exhibited a loss of chlorophyll (Fig. 13B and C). Intriguingly, as shown in Fig. 13D,E, the chlorophyll content of the CaDIR7-silenced plants leaf discs decreased drastically compared with TRV2:00 at the 300 mM concentration.
Figure 13.
Reduced tolerance of CaDIR7-silenced pepper plants to salt and mannitol stresses. (A) REL post salt stress of the CaDIR7-silenced and control (TRV2:00) plants, (B,C) leaf discs phenotypes (0.5 cm in diameter), (D,E) chlorophyll contents of the CaDIR7-silenced and control plants in response to 100 mM and 300 mM NaCl and mannitol stresses after 48 h, respectively. Values are the means ± SD from three independent experiments. Small letters represent significant differences (p < 0.05).
Discussion
Dirigent proteins are an important disease resistance responsive multigene family in plants29,56,57. They play an important role in increasing resistance against stresses in different crops11. DIRs and their homologous have been described in all vascular plants26, and they are speculated to be involved with the production of lignin and lignan26,58. The number of DIR protein genes varies in different plant species. Previously, 25 DIRs in Arabidopsis, 19 DIRs in I. indigotica, 54 DIRs in rice, 35 DIRs in Picea glauca and 29 DIRs in B. rapa have been reported9,11,27,29. In the past, no study has been conducted on a genome-wide identification and characterization of DIR and DIR-like proteins in response to biotic and abiotic stresses in pepper. In the current study, we identified 24 CaDIRs for the first time in ‘CM334′ and ‘Zunla-1′ pepper genomes. Consistent with previous studies9,11,27,29, five well-conserved motifs were found in the amino acid sequence alignments of all 24 CaDIRs including CaDIR7 (Fig. 1). The structural analysis showed that only 20.83% of CaDIRs contained one intron which contrasts with a previous study on the dirigent family in rice56. The ORFs analysis revealed that the amino acid (aa) sequences ranged from 106 aa (CaDIR17) to 330 aa (CaDIR12). The members of the DIR-e subfamily had an aa length longer than the CaDIRs from other subfamilies, ranging from 162 aa (CaDIR19) to 330 aa (CaDIR12). These results are supported by Li et al.29, who also found that the aa length of IiDIRs in the DIR-e subfamily was longer than in other subfamilies.
Previous studies have suggested that Asn sites are the features of secreted proteins observed in FiDIR1, the first and best characterized DIR protein12,16. In the present study, we found that 20 of the 24 CaDIRs had Asn sites, and 15 CaDIRs had more than one Asn site, which indicates that most of the CaDIRs seem to be secreted (Table 1). The X-ray or NMR structures of the dirigent proteins remain unavailable because DIRs have not yet been crystallized, and the tertiary structure of the DIRs are unknown59,60. Thus, to model DIRs, their homologous proteins with known structures can be used as templates29,59. Consequently, Phyre2 was used for the prediction of the tertiary structures as well as for the search for homologous proteins to CaDIRs. It was found that all of the CaDIRs are barrel-like proteins (Table 2). Meanwhile, Burlat et al.16 identified DIR genes and described their cellular localization and exclusive domain of lignification in Forsythia intermedia tissues. In the current study the subcellular location of all CaDIRs were predicted, and we found that pepper DIR proteins are targeted for extracellular release or possible final localization in the chloroplast, extracellular region, nucleus, cytoplasmic and vacuolar locations (Table 1).
The chromosomal locations revealed that CaDIRs were located on all 12 chromosomes of pepper, with chromosome 1 containing the highest number of CaDIRs (5). Similarly, the most BrDIRs (6) were found on chromosome A01 in B. rapa9. In the expansion of a gene family, tandem duplication, segmental duplication and transposition are the main evolutionary mechanisms61. Compared with tandem duplication and transposition, segmental duplication occurs frequently because of polyploidy in most plants, which conserve several duplicated chromosomal blocks in their genomes61. In the duplication analysis of CaDIRs, we obtained ten clusters of segmental duplication and two pairs of tandem duplication events (Table 3). In support of our study, tandem and segmental duplication events were also found in rice OsDIRs56. The results suggest that segmental and tandem duplications contribute to the expansion of the dirigent gene family in pepper.
Ralph et al.27 proposed that the DIR gene family can be grouped into six distinct subfamilies: DIR-a, DIR-b/d, DIR-c, DIR-e, DIR-f and DIR-g. Consistent with previous results from I. indigotica29, the 24 CaDIRs were separated into 3 subfamilies, DIR-a had 5 CaDIRs, DIR-b/d had 12 CaDIRs and DIR-e had 7 CaDIRs (Fig. 2A). The members of the DIR-a subfamily are considered to be dirigent genes, involved in lignification and primarily associated with defense and resistance against pathogens and insects13,27. On the other hand, the biochemical functions of members of the DIR-b/d, DIR-c, DIR-f and DIR-g subfamilies are not known; thus, these genes are referred to as dirigent-like genes11,27. The present findings regarding CaDIRs belonging to the DIR-a, DIR-b/d and DIR-e subfamilies suggest that they share a common DIR ancestor and some similar biological functions11,27,28.
To further elucidate the potential roles of CaDIRs in the growth and development of pepper, the expression profiles of CaDIRs were examined in different tissues. The RT-qPCR analysis of the organ-specific expression showed that most of the CaDIRs exhibited the highest expression in flowers, followed by the stem, leaf, green fruit and red fruit, whereas the roots expressed the genes at the lowest levels of all organs. This is in contrast with previous studies on B. rapa and I. indigotica, where comparatively higher expression levels were observed in roots9,29; however, it is consistent, up to some extent, with the findings of Jin-long et al.57 and Damaj et al.62. In F. intermedia, the expression of DIR genes was also detected in different tissues16. In B. hygrometrica, it was anticipated that DIR proteins promote lignin biosynthesis, and the stem, root and flower buds were found to contribute to the biosynthesis of lignin28. Moreover, Rogers and Campbell63 also reported that lignification occurred throughout normal tissue development, and at specific sites lignin biosynthesis was induced by several biotic and abiotic stresses. The pepper CaDIRs were expressed in an organ-specific way, signifying the probable function of CaDIRs in lignin formation via specific organs and contribution to developmental processes.
In the development of vascular plants, lignin is crucial, and it mainly deposits in the vascular tissues; during plant development, it provides additional strength and imperviousness to the cell wall64. Moreover, lignin deposition in plants is considered a physical barrier against the penetration of pathogens22. Xu et al.65 also concluded that lignin had a critical role in the disease resistance of cotton. Among members of the A. thaliana DIR-a subfamily, AtDIR5 and AtDIR6 were different from other DIRs observed earlier59,66, and wherever AtDIR6 was present, the final product of E-coniferyl alcohol was enantiomer (−)-pinoresinol. Later, Kim et al.67 confirmed that AtDIR5 and AtDIR6 engendered the preferential formation of (−)-pinoresinol. A recent study suggested that the dirigent protein family in plants contain several proteins involved in lignification and the response to pathogen infection as well as abiotic stresses57. In several plants, the changes in lignin content were observed to be caused by various biotic as well as abiotic stresses23. Lignin synthesis is a complex process because several gene families with many members are involved in the process8. Previous studies have reported changes in acid-soluble lignin content after biotic and abiotic stresses8,9,28. In the current study, the CaDIRs exhibited higher similarities with A. thaliana, indicating their involvement in the biosynthesis of lignin. In particular, members of the DIR-a subfamily showed identity based on topological tree (Fig. 3) and sequence alignments (Figure S1–S3) with AtDIR5 and AtDIR6, which might have the ability to produce (−)-lignans. In light of this, we measured the acid-soluble lignin content of pepper plants inoculated with P. capsici. The results showed an increase after inoculation with the P. capsici strains; at 48 hpi, a greater-than-90% increase was noticed in the lignin content. Our results are supported by the findings of Zhang et al.8. After inoculation with P. capsici, most CaDIRs were also expressed at higher levels at 48 h, which further corroborate the acid-soluble lignin content results of the present study. Perhaps the CaDIRs enhanced the coupling of monolignols, thus increasing the lignin content of pepper plants. Taken together, these findings suggest that pepper CaDIRs might play a key role in the biosynthesis of lignin and consequently participate in the defense response of pepper.
Mostly, CaDIR expression was elevated with the inoculation of virulent and avirulent strains of P. capsici. Specifically, CaDIR2, CaDIR3, CaDIR6, CaDIR7, CaDIR8, CaDIR11, CaDIR16, CaDIR22, CaDIR23 and CaDIR24 showed significantly higher expression levels (Fig. 7). Moreover, the transcript levels of CaDIR3, CaDIR16 and CaDIR24 were higher in plants inoculated with the avirulent strain versus the virulent strain, whereas CaDIR2, CaDIR6, CaDIR7, CaDIR8, CaDIR11, CaDIR22 and CaDIR23 had abundant transcript levels after inoculation with the virulent strain versus the avirulent strain. An earlier study also showed that two G. barbadense dirigent-like Gbd1 and Gbd2 genes were induced by a fungal disease V. dabliae20. The expression of the CABPR1 gene in pepper was higher in the virulent strain interaction versus the avirulent interaction68. However, in our previous study30, it was found that the expression of most CaSBPs was comparatively higher in the avirulent interaction than in the compatible interaction. Meanwhile, other studies have revealed that the expression of oxysterol-binding protein gene (CanOBP) and a novel peroxidase gene (CanPOD) were higher in the incompatible interaction49,69. Some other defense-related genes such as b-1,3-glucanase gene (CABGLU), disease-associated protein gene (CABPR1), and peroxidase gene (CAPO1) were expressed in a similar pattern in the roots of pepper after inoculation with virulent and avirulent strains of P. capsici70. A phylogenetic analysis revealed that CaDIR4 and CaDIR14 showed a closer relationship with two G. barbadense genes (Gbd1 and Gbd2), which were induced by V. dahliae infection20. Similarly, overexpression of the GhDIR1 gene in cotton delayed the spread of the fungal pathogen V. dahliae in transgenic plants64. Moreover, CaDIR6 and CaDIR9 were adjacent to the B. rapa DIR-like gene (BrDIR2), which showed differential expression in F. oxysporum infection9. The differences in the expression patterns of CaDIRs and other defense-related genes might be due to the differences in the inoculation of P. capsici strains or the variation in their compatibility systems.
SA and MeJA intercede the responses of plants to biotic and abiotic stresses via a signal transduction pathway71–73. The basic defense to biotrophic pathogens is mediated by SA72, whereas it is controlled by MeJA in response to necrotrphic pathogens74. The cis-acting elements responsible for MeJA (TGACG-motif)75 and SA (TCA-element)76 were found in most of the CaDIRs promoters. In light of this, the pepper plants were exposed to SA and MeJA stresses in the current study to investigate their effects on the expression levels of representative CaDIRs (CaDIR4, CaDIR7, CaDIR10, CaDIR12, CaDIR13, CaDIR14, CaDIR22 and CaDIR23) from each group. The qRT-PCR analysis confirmed that the expression of four CaDIRs was induced by MeJA and another four showed no significant response, whereas five CaDIRs were up-regulated by SA and three were down-regulated (Fig. 8). Our results corroborate the former studies on the inducibility of several DIR homologs. In B. hygrometrica, it was found that the expression of the BhDIR1 gene was induced by SA28. Meanwhile, a study on I. indigotica reported that some liDIRs were up- and down-regulated by MeJA stress29. Similarly, Damaj et al.62 found that SHDIR11 and SHDIR16 were up-regulated by SA and MeJA. Moreover, the expression of four DIRs in sugarcane roots other than SHDIR1677, five DIRs in spruce stem11, and nineteen DIRs in the bark of the conifer P. glauca27 were also reported to be induced by MeJA treatment. The CaDIRs above were slightly expressed in the leaves under normal conditions but responded significantly to the hormonal treatments, especially CaDIR7 and 23, signifying their role in the defense response of pepper.
Earlier studies have shown that dirigent genes were expressed in different patterns in response to various abiotic stresses9,28,56,78. Hence, we extended our study to investigate the expression analysis of eight representative CaDIRs after salt (NaCl) and drought (mannitol) stresses (Fig. 8). The results revealed that five of these genes showed a significant response to one or both of these stresses. Wu et al.28 also reported a similar finding in one of their studies. Similarly, differential expression of 13 OsDIRs was noticed in rice after salt stress compared with mock-treated control seedlings56. Moreover, in sugarcane an increased ScDIR gene expression was reported in response to NaCl, PEG and oxidative stress treatments57. These findings show the potential roles of CaDIRs in the regulation of abiotic stress.
In phylogenetic analysis, the CaDIR7 was clustered into DIR-e subfamily with six A. thaliana and seven I. indigotica genes (Fig. 3). CaDIR7 existing on chromosome 3, having a 750 bp ORF, with MW of 26.09 kDa and pI 5.46, containing one intron, is predicted to localized in the nucleus (Table 1). CaDIR7 also containing the Asn site, a feature of secreted proteins, indicating that it is likely to be a secreted protein (Table 1). The probability and identity analysis of CaDIR7 homologous relationship revealed that it has 34% sequence identity with hypothetical protein c4revB belongs to disease resistance response protein 206 (drr206), while 21, 18 and 18% identity with AOC barrel-like proteins i.e. d2brja1, c4h69A and d1zvca1 respectively (Table 2). It has one segmental duplication (CaDIR11), while no tandem duplication was observed (Table 3). In conserved motif analysis of the CaDIR proteins through MEME server, it was found that CaDIR7 containing the highest number of motifs (6) (Fig. 6). In its promoter region, a number of cis-acting elements including MeJA and SA responsiveness, defense and stress responsiveness, fungal elicitor-responsive element, heat stress responsiveness, ethylene-, auxin- and GA-responsive elements were found (Table 5). Besides CaDIR7 showed the highest expression levels in response to P. capsici, MeJA, NaCl and mannitol stresses (Figs 7 and 8), while in tissue specific expression, it showed relatively higher transcription levels in stem and red fruit (Fig. 9). This further attracted our attention towards the importance of this gene. Thus, to affirm the role of CaDIR7 against P. capsici, it was successfully knocked down in pepper plants through VIGS. An assay of the detached leaves of the CaDIR7-silenced and control plants with P. capsici inoculation was conducted. Larger lesions were observed in the leaves of silenced plants than in the control, indicating that silencing CaDIR7 made the pepper leaves more prone to infection by P. capsici, following the same pattern as reported in our previous study44. The current finding was also supported by Ma and Liu79 on transgenic wheat lines. Our results are also consistent with findings that over-expression of the GhDIR1 gene in cotton significantly delayed the invasion of the fungal pathogen V. dahlia64.
In addition, significant differences in CaDIR7 expression levels were observed at 2, 4, 7 and 10 dpi in the control and silenced pepper plants. Wang et al.80 found that the root activity of pepper plants was higher with infection by the avirulent strain than by the virulent strain of P. capsici. In the current study on P. capsici pi, the root activity of the silenced plants decreased significantly compared with the control plants. This significant reduction in the root activity of the silenced pepper plants signifies severe injury to the roots caused by P. capsici. This concurs with our previous research work on pepper with a different gene CaPTI144. Taken together, the larger leaf lesion areas and lessened root activity of the CaDIR7-silenced plants suggest the significant role of CaDIR7 in the defense response of pepper against P. capsici infection.
In the present study, the leaf discs of CaDIR7-silenced plants showed evidently bleached phenotypes when exposed to salt and osmotic stresses compared with the control, whereas a significant decrease in chlorophyll content was observed in the silenced plants compared with the control. The REL assay in the current study revealed that after salt stress (300 mM), a significant increase in electrolyte leakage of the silenced plant leaf discs was recorded compared with the control. Consistent with our study, Chen et al.81 observed the same results for chlorophyll content after 3 days of salt and osmotic treatments, while electrical conductivity was significantly increased in CaDHN1-silenced plants after cold stress. Furthermore, the silenced and control plants exposed to NaCl stress (300 mM) showed a significant difference in the expression of CaDIR7. Furthermore, salt stress also altered the expression of the defense-related CaDEF1 (a JA dependent) and CaPO1 (a peroxidase) genes in the roots of silenced and control plants, indicating their role in salt stress tolerance. In response to high salinity, strong accumulation of the CADEF1 transcript was observed in pepper53. A previous study also reported a marked increase in CAPO1 mRNA levels after 4 days of treatment with copper in pepper stems, suggesting that this gene participates in the formation of defensive barriers54. More recently, CaPO2 transgenic A. thaliana plants exhibited more tolerance to high salt, drought, and oxidative stress82. Additionally, we measured the root activity after salt stress, and a significant reduction in the root activity of the silenced plants was observed compared with the control. This reduction in root activity is due to the stress injury caused by salt stress. Therefore, we speculate that CaDIR7 is involved in the defense response of pepper against salt and drought stress.
Electronic supplementary material
Acknowledgements
This work was supported through funding from National Key R&D Program of China (No. 2016YFD0101900), the National Natural Science Foundation of China (No. 31272163, No. U1603102) and Applied Basic Research Program of Shanxi Province (No. 201601D021126).
Author Contributions
A.K. and Z.-H.G. designed the research. A.K., M.A., F.-M., R.-J.L. and J.-T.S. conducted the research. A.K., H.-X.Z. and J.-H.J. performed the statistical analyses. A.K. wrote the manuscript. A.K., S.H. and Z.-H.G. revised the manuscript. J.-E.W. and Z.-H.G. provided the reagents/materials/analysis tools. All authors discussed the data and made comments on the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23761-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lamour KH, Stam R, Jupe J, Huitema E. The oomycete broad-host-range pathogen Phytophthora capsici. Mol. Plant Pathol. 2012;13:329–337. doi: 10.1111/j.1364-3703.2011.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hausbeck MK, Lamour KHR. Progress and Management Challenges Phytophthora capsici on Vegetable Crops: Plant Dis. 2004;88:1292–1303. doi: 10.1094/PDIS.2004.88.12.1292. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- 4.Mittler R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006;11:15–19. doi: 10.1016/j.tplants.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Jones JDG, Dangl L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 6.Singh A, Jha SK, Bagri J, Pandey GK. ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS One. 2015;10:1–24. doi: 10.1371/journal.pone.0125168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002;5:325–331. doi: 10.1016/S1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 8.Zhang SH, Yang Q, Ma RC. Erwinia carotovora ssp. carotovora infection induced ‘defense lignin’ accumulation and lignin biosynthetic gene expression in Chinese cabbage (Brassica rapa L. ssp. pekinensis) J. Integr. Plant Biol. 2007;49:993–1002. doi: 10.1111/j.1672-9072.2007.00478.x. [DOI] [Google Scholar]
- 9.Thamil Arasan SK, et al. Characterization and expression analysis of dirigent family genes related to stresses in Brassica. Plant Physiol. Biochem. 2013;67:144–153. doi: 10.1016/j.plaphy.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 10.Bowles DJ. Defense-Related Proteins in Higher Plants. Annu. Rev. Biochem. 1990;59:873–907. doi: 10.1146/annurev.bi.59.070190.004301. [DOI] [PubMed] [Google Scholar]
- 11.Ralph S, Park JY, Bohlmann J, Mansfield SD. Dirigent proteins in conifer defense: Gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.) Plant Mol. Biol. 2006;60:21–40. doi: 10.1007/s11103-005-2226-y. [DOI] [PubMed] [Google Scholar]
- 12.Gang DR, et al. Regiochemical control of monolignol radical coupling: A new paradigm for lignin and lignan biosynthesis. Chem. Biol. 1999;6:143–151. doi: 10.1016/S1074-5521(99)89006-1. [DOI] [PubMed] [Google Scholar]
- 13.Davin LB, et al. Stereoselective Bimolecular Phenoxy Radical Coupling by an Auxiliary (Dirigent) Protein Without an Active Center. Science (80-.). 1997;275:362–367. doi: 10.1126/science.275.5298.362. [DOI] [PubMed] [Google Scholar]
- 14.Xia ZQ, Costa MA, Proctor J, Davin LB, Lewis NG. Dirigent-mediated podophyllotoxin biosynthesis in Linum flavum and Podophyllum peltatum. Phytochemistry. 2000;55:537–549. doi: 10.1016/S0031-9422(00)00242-9. [DOI] [PubMed] [Google Scholar]
- 15.Kim MK, Jeon JH, Fujita M, Davin LB, Lewis NG. The western red cedar (Thuja plicata) 8-8′ DIRIGENT family displays diverse expression patterns and conserved monolignol coupling specificity. Plant Mol. Biol. 2002;49:199–214. doi: 10.1023/A:1014940930703. [DOI] [PubMed] [Google Scholar]
- 16.Burlat V, Kwon M, Davin LB, Lewis NG. Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry. 2001;57:883–897. doi: 10.1016/S0031-9422(01)00117-0. [DOI] [PubMed] [Google Scholar]
- 17.Halls SC, Lewis NG. Secondary and quaternary structures of the (+)-pinoresinol-forming dirigent protein. Biochemistry. 2002;41:9455–9461. doi: 10.1021/bi0259709. [DOI] [PubMed] [Google Scholar]
- 18.Culley DE, Horovitz D, Hadwiger L. a. Molecular characterization of disease-resistance response gene DRR206-d from Pisum sativum (L.) Plant Physiol. 1995;107:301–302. doi: 10.1104/pp.107.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Fristensky B. Transgenic canola lines expressing pea defense gene DRR206 have resistance to aggressive blackleg isolates and to Rhizoctonia solani. Mol. Breed. 2001;8:263–271. doi: 10.1023/A:1013706400168. [DOI] [Google Scholar]
- 20.Zhu L, et al. Isolation and characterization of two novel dirigent-like genes highly induced in cotton (Gossypium barbadense and G. hirsutum) after infection by Verticillium dahliae. J. Plant Pathol. 2007;89:41–45. [Google Scholar]
- 21.Lewis NG, Davin LB. Evolution of Lignan and Neolignan BiochemicalPathways. Isopentenoids Other Nat. Prod. 1994;562:10–202. [Google Scholar]
- 22.Moerschbacher BM, Noll U, Gorrichon L, Reisener HJ. Specific inhibition of lignification breaks hypersensitive resistance of wheat to stem rust. Plant Physiol. 1990;93:465–470. doi: 10.1104/pp.93.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moura JCMS, Bonine CAV, de Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010;52:360–376. doi: 10.1111/j.1744-7909.2010.00892.x. [DOI] [PubMed] [Google Scholar]
- 24.Fang Y, et al. De novo Transcriptome analysis reveals distinct defense mechanisms by young and mature leaves of Hevea brasiliensis (Para Rubber Tree) Sci. Rep. 2016;6:33151. doi: 10.1038/srep33151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Lee C, Zhong R, Ye Z-H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21:248–66. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davin LB, Lewis NG. Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 2000;123:453–461. doi: 10.1104/pp.123.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ralph SG, Jancsik S, Bohlmann J. Dirigent proteins in conifer defense II: Extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.) Phytochemistry. 2007;68:1975–1991. doi: 10.1016/j.phytochem.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 28.Wu R, et al. Cloning and expression analysis of a dirigent protein gene from the resurrection plant Boea hygrometrica. Prog. Nat. Sci. 2009;19:347–352. doi: 10.1016/j.pnsc.2008.07.010. [DOI] [Google Scholar]
- 29.Li Q, et al. The dirigent multigene family in Isatis indigotica: gene discovery and differential transcript abundance. BMC Genomics. 2014;15:388. doi: 10.1186/1471-2164-15-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H-X, et al. Genome-wide identification and analysis of the sbp-box family genes under phytophthora capsici stress in pepper (Capsicum annuum L.) Front. Plant Sci. 2016;7:1–14. doi: 10.3389/fpls.2016.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo M, et al. Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.) BMC Plant Biol. 2015;15:151. doi: 10.1186/s12870-015-0512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horton P, et al. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007;35:1–3. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J.Mol.Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 34.Gupta, R., Jung, E. & Brunak, S. Prediction of N-glycosylation sites in human proteins. (2004).
- 35.Guo M, et al. Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 2016;252:246–256. doi: 10.1016/j.plantsci.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang W-H, Kim S, Lee H-A, Choi D, Yeom S-I. Genome-wide analysis of Dof transcription factors reveals functional characteristics during development and response to biotic stresses in pepper. Sci. Rep. 2016;6:33332. doi: 10.1038/srep33332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 39.Liu R, Meng J. MapDraw: A microsoft Excel Macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas. 2003;25:317–321. [PubMed] [Google Scholar]
- 40.Gu Z, Cavalcanti A, Chen F-C, Bouman P, Li W-H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002;19:256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- 41.Rost B. Twilight zone of protein sequence alignments. Protein Eng. Des. Sel. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 42.Lescot M, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang YL, et al. Characteristic of the pepper CaRGA2 gene in defense responses against Phytophthora capsici leonian. Int. J. Mol. Sci. 2013;14:8985–9004. doi: 10.3390/ijms14058985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin J-H, et al. A New Ethylene-Responsive Factor CaPTI1 gene of pepper (Capsicum annuum L.) involved in the regulation of defense response to Phytophthora capsici. Front. Plant Sci. 2016;6:1–12. doi: 10.3389/fpls.2015.01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo WL, Chen RG, Gong ZH, Yin YX, Ahmed SS. Exogenous abscisic acid increases antioxidant enzymes and related gene expression in pepper (Capsicum annuum L.) leaves subjected to chilling stress. Genet. Mol. Res. 2012;11:4063–4080. doi: 10.4238/2012.September.10.5. [DOI] [PubMed] [Google Scholar]
- 46.Wan H, et al. Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.) Biochem. Biophys. Res. Commun. 2011;416:24–30. doi: 10.1016/j.bbrc.2011.10.105. [DOI] [PubMed] [Google Scholar]
- 47.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 48.Liu Z, et al. SGT1 is required in PcINF1/SRC2-1 induced pepper defense response by interacting with SRC2-1. Sci. Rep. 2016;6:21651. doi: 10.1038/srep21651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang JE, et al. A novel peroxidase CanPOD gene of pepper is involved in defense responses to Phytophtora capsici infection as well as abiotic stress tolerance. Int. J. Mol. Sci. 2013;14:3158–3177. doi: 10.3390/ijms14023158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ou LJ, Dai XZ, Zhang ZQ, Zou XX. Responses of pepper to waterlogging stress. Photosynthetica. 2011;49:339–345. doi: 10.1007/s11099-011-0043-x. [DOI] [Google Scholar]
- 51.Yin YX, et al. Cloning and characterisation of a pepper aquaporin, CaAQP, which reduces chilling stress in transgenic tobacco plants. Plant Cell. Tissue Organ Cult. 2014;118:431–444. doi: 10.1007/s11240-014-0495-3. [DOI] [Google Scholar]
- 52.Guo W-L, et al. Characterization and expression profile of CaNAC2 pepper gene. Front. Plant Sci. 2015;6:1–9. doi: 10.3389/fpls.2015.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Do HM, Lee SC, Jung HW, Sohn KH, Hwang BK. Differential expression and in situ localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Plant Sci. 2004;166:1297–1305. doi: 10.1016/j.plantsci.2004.01.008. [DOI] [Google Scholar]
- 54.Chmielowska J, Veloso J, Gutiérrez J, Silvar C, Díaz J. Cross-protection of pepper plants stressed by copper against a vascular pathogen is accompanied by the induction of a defence response. Plant Sci. 2010;178:176–182. doi: 10.1016/j.plantsci.2009.11.007. [DOI] [Google Scholar]
- 55.Dai Y, et al. Effects of shade treatments on the photosynthetic capacity, chlorophyll fluorescence, and chlorophyll content of Tetrastigma hemsleyanum Diels et Gilg. Environ. Exp. Bot. 2009;65:177–182. doi: 10.1016/j.envexpbot.2008.12.008. [DOI] [Google Scholar]
- 56.Liao Y, et al. Genome-wide analysis and environmental response profiling of dirigent family genes in rice (Oryza sativa). Genes. Genomics. 2016;39:47–62. [Google Scholar]
- 57.Jin-long G, et al. A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses. Plant Cell Rep. 2012;31:1801–1812. doi: 10.1007/s00299-012-1293-1. [DOI] [PubMed] [Google Scholar]
- 58.Kim MK, Jeon JH, Davin LB, Lewis NG. Monolignol radical-radical coupling networks in western red cedar and Arabidopsis and their evolutionary implications. Phytochemistry. 2002;61:311–322. doi: 10.1016/S0031-9422(02)00261-3. [DOI] [PubMed] [Google Scholar]
- 59.Pickel B, et al. A model of dirigent proteins derived from structural and functional similarities with allene oxide cyclase and lipocalins. FEBS J. 2012;279:1980–1993. doi: 10.1111/j.1742-4658.2012.08580.x. [DOI] [PubMed] [Google Scholar]
- 60.Pickel B, Schaller A. Dirigent proteins: Molecular characteristics and potential biotechnological applications. Appl. Microbiol. Biotechnol. 2013;97:8427–8438. doi: 10.1007/s00253-013-5167-4. [DOI] [PubMed] [Google Scholar]
- 61.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Damaj MB, et al. Sugarcane DIRIGENT and O-METHYLTRANSFERASE promoters confer stem-regulated gene expression in diverse monocots. Planta. 2010;231:1439–1458. doi: 10.1007/s00425-010-1138-5. [DOI] [PubMed] [Google Scholar]
- 63.Rogers LA, Campbell MM. The genetic control of lignin deposition during plant growth and development. New Phytol. 2004;164:17–30. doi: 10.1111/j.1469-8137.2004.01143.x. [DOI] [PubMed] [Google Scholar]
- 64.Shi, H. et al. Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. 44, 555–564 (2012). [DOI] [PubMed]
- 65.Xu L, et al. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 2011;62:5607–5621. doi: 10.1093/jxb/err245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pickel B, et al. Angew. Chemie - Int. Ed. 2010. An enantiocomplementary dirigent protein for the enantioselective lacease-catalyzed oxidative coupling of phenols; pp. 202–204. [DOI] [PubMed] [Google Scholar]
- 67.Kim KW, et al. Opposite stereoselectivities of dirigent proteins in Arabidopsis and Schizandra species. J. Biol. Chem. 2012;287:33957–33972. doi: 10.1074/jbc.M112.387423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim YJ, Hwang BK. Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol. Plant. 2000;108:51–60. doi: 10.1034/j.1399-3054.2000.108001051.x. [DOI] [Google Scholar]
- 69.Liu, K. K. Studies on the resistance and its mechanisms of pepper to Phytophthora capsici. Ph.D. thesis Northwest A&F University, Yangling District (2009).
- 70.Wang, J. E. Expression analysis and functional identification of CaRGA1 and CaPOD genes induced by Phytophthora capsici in Pepper. Ph.D. thesis Northwest A&F University, Yangling District. (2013).
- 71.Glazebrook J, et al. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 2003;34:217–228. doi: 10.1046/j.1365-313X.2003.01717.x. [DOI] [PubMed] [Google Scholar]
- 72.Thomma BPHJ, Penninckx IAMA, Broekaert WF, Cammue BPA. The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 2001;13:63–68. doi: 10.1016/S0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 73.An, S. H. et al. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 61–78, 10.1007/s00425-008-0719-z (2008). [DOI] [PMC free article] [PubMed]
- 74.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 75.Rouster J, Leah R, Mundy J, Cameron-Mills V. Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain. Plant J. 1997;11:513–523. doi: 10.1046/j.1365-313X.1997.11030513.x. [DOI] [PubMed] [Google Scholar]
- 76.Merkouropoulos G, Barnett DC, Shirsat AH. The Arabidopsis extensin gene is developmentally regulated, is induced by wounding, methyl jasmonate, abscisic and salicylic acid, and codes for a protein with unusual motifs. Planta. 1999;208:212–219. doi: 10.1007/s004250050552. [DOI] [PubMed] [Google Scholar]
- 77.Bower NI, et al. Transcriptional response of sugarcane roots to methyl jasmonate. Plant Sci. 2005;168:761–772. doi: 10.1016/j.plantsci.2004.10.006. [DOI] [Google Scholar]
- 78.Behr M, Legay S, Hausman JF, Guerriero G. Analysis of cell wall-related genes in organs of Medicago sativa L. under different abiotic stresses. Int. J. Mol. Sci. 2015;16:16104–16124. doi: 10.3390/ijms160716104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma, Q. H. & Liu, Y. C. TaDIR13, a Dirigent protein from wheat, promotes lignan biosynthesis and enhances pathogen resistance. Plant Mol. Biol. Report. 143–152, 10.1007/s11105-014-0737-x (2015).
- 80.Wang JE, et al. Defence responses of pepper (Capsicum annuum L.) infected with incompatible and compatible strains of Phytophthora capsici. Eur. J. Plant Pathol. 2013;136:625–638. doi: 10.1007/s10658-013-0193-8. [DOI] [Google Scholar]
- 81.Chen R. gang et al. Silencing of dehydrin CaDHN1 diminishes tolerance to multiple abiotic stresses in Capsicum annuum L. Plant Cell Rep. 2015;34:2189–2200. doi: 10.1007/s00299-015-1862-1. [DOI] [PubMed] [Google Scholar]
- 82.Choi HW, Hwang BK. The pepper extracellular peroxidase CaPO2 is required for salt, drought and oxidative stress tolerance as well as resistance to fungal pathogens. Planta. 2012;235:1369–1382. doi: 10.1007/s00425-011-1580-z. [DOI] [PubMed] [Google Scholar]
- 83.Diaz-De-Leon F, Klotz KL, Lagrimini LM. Nucleotide sequence of the tobacco (Nicotiana tabacum) anionic peroxidase gene. Plant Physiol. 1993;101:1117–1118. doi: 10.1104/pp.101.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rushton PJ, et al. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996;15:5690–5700. [PMC free article] [PubMed] [Google Scholar]
- 85.Pastuglia M, Roby D, Dumas C, Cockagi JM. Rapíd induction by wounding and bacterial infection of an s gene family receptor-like kinase gene in Brassica oleracea. Plant Cell Am. Soc. Plant Physiol. 1997;9:49–60. doi: 10.1105/tpc.9.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.White AJ, Dunn MA, Brown K, Hughes MA. Comparative analysis of genomic sequence and expression of a lipid transfer protein gene family in winter barley. 1994;45:1885–1892. [Google Scholar]
- 87.Nash J, Luehrsen KR, Walbot V. Bronze-2 gene of maize: reconstruction of a wild-type allele and analysis of transcription and splicing. Plant Cell. 1990;2:1039–1049. doi: 10.1105/tpc.2.11.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Skriver K, Olsen FL, Rogers JC, Mundy J. cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc. Natl. Acad. Sci. USA. 1991;88:7266–70. doi: 10.1073/pnas.88.16.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Itzhaki H, Woodson WR. Characterization of an ethylene-responsive glutathione S-transferase gene cluster in carnation. Plant Mol. Biol. 1993;22:43–58. doi: 10.1007/BF00038994. [DOI] [PubMed] [Google Scholar]
- 90.Baker SS, Wilhelm KS, Thomashow MF. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- 91.Sakai T, Takahashi Y, Nagata T. Analysis of the promoter of the auxin-inducible gene, parC, of tobacco. Plant Cell Physiol. 1996;37:906–13. doi: 10.1093/oxfordjournals.pcp.a029038. [DOI] [PubMed] [Google Scholar]
- 92.Kim JK, Cao J, Wu R. Regulation and interaction of multiple protein factors with the proximal promoter regions of a rice high pI α-amylase gene. Mol Gen Genet. 1992;232:383–393. doi: 10.1007/BF00266241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.