Abstract
MicroRNAs (miRNAs) serve as major regulators during the tumorigenesis and tumor development of hepatocellular carcinoma (HCC). In addition, miRNAs may serve as new promising biomarkers for the diagnosis and prognosis and as effective therapeutic targets for patients with this malignancy. Therefore, understanding the association between miRNAs and HCC may be beneficial to discover novel therapeutic approaches towards diagnosis and treatments. Results of this study showed that miRNA-337 (miR-337) was markedly downregulated in HCC tissues and cell lines. Decreased miR-337 expression was significantly associated with the TNM stage and lymph node metastasis of HCC. Ectopic expression of miR-337 prohibited the proliferation, colony formation, migration, and invasion of HCC cells. It also promoted the apoptosis in vitro and reduced the tumor growth in vivo of these cells. High-mobility group AT-hook 2 (HMGA2) was identified as a direct target gene of miR-337 in HCC through a series of experiments. HMGA2 was significantly overexpressed in HCC tissues and negatively correlated with miR-337 expression. Moreover, the functions of HMGA2 inhibition were similar to those induced by miR-337 in HCC. Restored HMGA2 expression rescued the tumor-suppressive roles of miR-337 overexpression in HCC. Furthermore, miR-337 overexpression inhibited the activation of PI3K/AKT and Wnt/β-catenin signaling pathways in HCC both in vitro and in vivo. This study demonstrated that miR-337 may play tumor-suppressing roles in HCC, at least partly, via directly targeting HMGA2 and inhibiting the PI3K/AKT and Wnt/β-catenin signaling pathways. Therefore, miR-337 may be a novel and effective target for the therapeutic treatment of patients with HCC.
Keywords: Hepatocellular carcinoma, microRNA-337, tumor suppressor, high-mobility group AT-hook 2
Introduction
Hepatocellular carcinoma (HCC), the most common form of primary liver cancer, ranks as the fifth most common cancer and the third leading cause of cancer-related death worldwide [1]. Over 780,000 novel HCC cases and approximately 745,000 HCC-related deaths were estimated per annum globally [2]. Numerous risk factors, including alcoholic liver disease, non-alcoholic fatty liver disease, liver cirrhosis, and hepatitis B and C infection, influence the occurrence and development of HCC [3-5]. However, the mechanism underlying the pathogenesis of HCC remains largely unknown. Despite great improvements in the diagnosis and treatment of HCC, the therapeutic outcomes for patients with this malignancy remain unsatisfactory with a 5-year survival rate of only 15% [6]. The unfavorable prognosis of HCC patients is largely attributed to the high incidence rates of tumor recurrence and metastasis even after surgery resection, as well as the lack of useful adjuvant therapy [7]. Therefore, understanding the mechanisms underlying HCC tumorigenesis and tumor progression is crucial to develop novel therapeutic methods for patients with this malignancy.
MicroRNAs (miRNAs) are a cluster of endogenous, single-strand, and non-coding short RNA molecules with approximately 19 to 25 nucleotides in length [8]. MiRNAs negatively regulate gene expression through interaction with the 3’-untranslated regions (UTRs) of their target genes in a base-pairing manner to form RNA-induced silencing complexes, resulting in mRNA degradation or translation suppression [9]. Thus far, over 1400 miRNAs have been identified in the human genome and estimated to modulate more than half of all human protein-coding genes [10]. Via regulation of their targets, miRNAs play crucial roles in almost all aspects of physiological and pathological processes, including early embryonic development, immunity, metabolism, cell division, proliferation, apoptosis, differentiation, and metastasis [11,12]. Altered miRNA expression has been identified in numerous types of human neoplasm, including HCC [13,14]. Highly expressed miRNAs may play oncogenic functions by regulation of tumor suppressor genes [15], whereas downregulated miRNAs may perform tumor-suppressive roles via blockade of oncogenes [16]. Hence, further investigation of the biological roles of cancer-related miRNAs may allow for the identification of novel and efficient therapeutic targets for anticancer treatments.
MiR-337 is downregulated in various human cancer types, such as melanoma [17], neuroblastoma [18], colorectal cancer [19], and non-small cell lung cancer [20]. However, the exact expression pattern and biological functions of miR-337 in HCC remain to be fully elucidated. Therefore, this study evaluated the expression level and clinical significance of miR-337 in HCC and investigated the roles and associated regulatory mechanism of miR-337 in HCC.
Materials and methods
Tissue samples and cell lines
Forty-seven paired HCC tissues and adjacent non-cancerous tissues were obtained from HCC patients who underwent surgical resection at Affiliated Zhongshan Hospital of Dalian University between March 2014 and February 2016. Patients who underwent preoperative chemotherapy, radiotherapy, or other treatments were excluded. All tissues were immediately frozen in liquid nitrogen and stored at -80°C until further use. This study was approved by the Ethics Committee of Affiliated Zhongshan Hospital of Dalian University. Written informed consent was also provided by all participants.
Three human HCC cell lines (HepG2, Hep3B, and Huh-7) and a normal human liver cell line LO2 were acquired from the Cell Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All human cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 100 μg/mL streptomycin, and 100 Units/mL penicillin (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and then maintained at 37°C in a humidified incubator with 5% CO2.
Cell transfection
MiR-337 mimics, miRNA mimics negative control (miR-NC), small interfering RNA (siRNA) against the expression of HMGA2 (HMGA2 siRNA), and negative control siRNA (NC siRNA) were chemically synthesized by RiboBio (Guangzhou China). HMGA2 overexpression plasmid pcDNA3.1-HMGA2 and empty pcDNA3.1 plasmid were produced by Integrated Biotech Solutions (Shanghai, China). Cells were plated into 6-well plates at 60%-70% confluence one day prior to transfection. Cell transfection was conducted using LipofectamineTM 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in accordance with the manufacturer’s instructions.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) for miRNAs and mRNAs
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to isolate total RNA from tissues or cells in accordance with the manufacturer’s instructions. For the detection of miR-337 expression, reverse transcription was performed to synthesize complementary DNA (cDNA) form total RNA using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Quantitative PCR (qPCR) was carried out on an ABI7500 Real-Time PCR System (Applied Biosystems, CA, USA) using a TaqMan MicroRNA PCR Kit (Applied Biosystems, Foster City, CA, USA). To quantify HMGA2 mRNA level, cDNA was synthesized with a PrimeScript RT Reagent kit (Takara Bio, Dalian, China) followed by qPCR with a SYBR Premix Ex TaqTM kit (Takara Bio, Dalian, China). U6 snRNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as internal control for miR-337 and HMGA2 mRNA, respectively. Relative expression was calculated according to the 2-ΔΔCt method [21].
Cell Counting Kit-8 (CCK-8) assay
Transfected cells were collected at 24 h post-transfection and seeded into 96-well plates with a density of 3 × 103/well. Cells were then incubated at 37°C for 0, 24, 48 or 96 h. At indicated time points, CCK-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was performed to detect cell proliferation in accordance with the manufacturer’s protocol. In brief, 10 µL of CCK-8 reagent was added into each well. After another incubation at 37°C for 2 h, the absorbance was determined at 450 nm wavelength using an EnSpireTM 2300 Multilabel Reader (PerkinElmer, Inc., Waltham, MA, USA). Each assay was performed in triplicate.
Colony formation assay
Transfected cells were collected at 24 h post-transfection and mechanically suspended into a single cell suspension. Afterward, the transfected cells were counted and plated into 6-well plates with an average 1000 cells per well. After 2 weeks incubation at 37°C in a humidified atmosphere of 5% CO2, the cells were washed with phosphate buffer solution (PBS), fixed with 4% paraformaldehyde, and then stained with Methyal Violet (Beyotime Institute of Biotechnology, Shanghai, China). Finally, the number of colonies (>50 cells/colony) was counted under a light microscope (IX53; Olympus Corporation, Tokyo, Japan).
Transwell cell migration and invasion assays
In brief, 24-well Transwell boyden chambers (8.0 μm pore size; Costar, Cambridge, MA) coated with or without Matrigel (BD Biosciences, San Jose, CA, USA) were applied to evaluate cell migration and invasion, respectively. For both assays, 5 × 104 transfected cells suspended in FBS-free DMEM were plated onto the upper chambers. The lower chambers were filled with 500 µL of DMEM containing 20% FBS to serve as a chemoattractant. Following 24 h incubation at 37°C, the non-migrated or non-invaded cells that remained in the upper chambers were scraped off gently using a cotton swab. The migrated and invaded cells were fixed with 100% methanol, stained with 0.5% crystal violet solution (Beyotime Institute of Biotechnology, Shanghai, China), washed with PBS, and then dried in air. Stained cells were photographed and counted under a light microscope (magnification, 200 ×) with five randomly selected fields per membrane. Each assay contained three replicates and was repeated at least three times.
Flow cytometry analysis
An Annexin V-FITC apoptosis detection kit (Invitrogen Corporation, California, USA) was employed to detect cell apoptosis rate in accordance with the manufacturer’s protocol. Following 48 h incubation, transfected cells were harvested using trypsinization and washed with ice-cold PBS. Subsequently, the cells were suspended in 300 µL of 1 × binding buffer and then incubated for 20 min with 5 µL of FITC-Annexin V and 5 µL of propidium iodide (PI) at room temperature in the dark. Cell apoptosis was detected by flow cytometry (FACScan; BD Biosciences, Franklin Lakes, NJ, USA) and analyzed with CellQuest software version 3.3 (BD Biosciences, Franklin Lakes, NJ, USA). Each assay was performed in triplicate.
Tumor growth assay in nude mice
Ten BALB/c nude mice (4- to 6-weeks of age) were acquired from Changzhou Cavens Laboratory Animal Center (Changzhou, China). Nude mice were randomly divided into two groups (n = 5 for the miR-337 group; n = 5 for the miR-NC group). Cells were collected at 24 h post-transfection and mechanically dissociated into a single cell suspension. A total of 1 × 107 cells in 100 µL of DMEM containing 10% FBS were subcutaneously injected into the nude mice. At 2 weeks after inoculation, the length and width of the tumors were recorded every 2 days. The nude mice were sacrificed at day 30, and the tumors formed by HCC cells were dissected and weighed. Tumor volumes were calculated using the formula: volume = 1/2 × tumor length × tumor width. In vivo studies were approved by the Ethics Review Committee of Affiliated Zhongshan Hospital of Dalian University.
Bioinformatics analysis and luciferase reporter assay
Potential targets of miR-337 were predicted using the publicly available programs TargetScan7.1 (http://www.targetscan.org/) and miRanda (http://www.microrna.org/). HMGA2 was predicted as a major target of miR-337.
The wild-type 3’-UTR of HMGA2 predicted to interact with miR-337, together with mutant binding sequence within the predicted target sites, were synthesized by GenePharma (Shanghai, China), inserted into the pMIR-REPORT luciferase reporter plasmids (Promega, Madison, WI, USA) and then denoted as pMIR-HMGA2-3’-UTR Wt and pMIR-HMGA2-3’-UTR Mut, respectively. Cells were plated into 24-well plates at 60%-70% confluence. After incubation overnight, miR-337 mimics or miR-NC was transfected into cells, together with one of the two reporter plasmids using LipofectamineTM 2000 reagent, in accordance with the manufacturer’s protocol. Following cultivation for 48 h, the transfected cells were collected and evaluated for luciferase activity using a dual luciferase reporter assay kit (Promega, Madison, WI, USA) in accordance with the manufacturer’s manual. The activity of firefly luciferase was normalized to that of Renilla luciferase.
Western blot analysis
Total protein was extracted from tissue samples or cells using radioimmunoprecipitation assay buffer (KeyGen Biotech Co., Ltd. Nanjing, China) containing 0.1 mg/mL phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, and 1 mg/mL aprotinin. A BCA Protein Assay Kit (KeyGen Biotech Co., Ltd. Nanjing, China) was adopted to measure the concentration of total protein. Equal masses of total protein were electrophoresed by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). Following blocking with 5% non-fat milk in Tris-buffered saline containing 0.1% Tween-20 (TBST), the membranes were incubated overnight at 4°C with primary antibodies. After being washed three times with TBST, the membranes were probed with corresponding horseradish peroxidase-conjugated secondary antibodies (1:5000 dilution; sc-2005; Santa Cruz Biotechnology, CA, USA) at room temperature for 2 h. After being washed repeatedly with TBST, the protein bands were visualized using an enhanced chemiluminescence reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Quantity One® software (version 4.62; BioRad Laboratories, Inc., Hercules, CA, USA) was applied to analyze the densities of protein bands. The primary antibodies used in this study include mouse anti-human monoclonal HMGA2 antibody (ab184616; 1:1000 dilution; Abcam, Cambridge, UK), mouse anti-human monoclonal AKT antibody (sc-81434; 1:1000 dilution; Santa Cruz Biotechnology, CA, USA), mouse anti-human monoclonal p-AKT antibody (sc-271966; 1:1000 dilution; Santa Cruz Biotechnology, CA, USA), mouse anti-human monoclonal β-catenin antibody (sc-59737; 1:1000 dilution; Santa Cruz Biotechnology, CA, USA), mouse anti-human monoclonal p-β-catenin antibody (sc-57534; 1:1000 dilution; Santa Cruz Biotechnology, CA, USA) and mouse anti-human monoclonal GAPDH antibody (sc-47724; 1:1000 dilution; Santa Cruz Biotechnology, CA, USA). GAPDH was used as a loading control.
Statistical analysis
Data are expressed as mean ± standard deviation (SD) and analyzed with SPSS software (19.0; SPSS, Inc., Chicago, IL). Data were compared with Student’s t-test or one-way ANOVA plus multiple comparisons combined with a post-hoc analysis (Student-Newman-Keuls test). The chi-square test was used to evaluate the associations between miR-337 expression and clinicopathological factors in HCC. Spearman’s correlation analysis was used to assess the correlation between miR-337 and HAMGA2 mRNA level in HCC tissues. Statistical significance was considered at P<0.05.
Results
MiR-337 is underexpressed in HCC tissues and cell lines
We detected miR-337 expression in 47 paired HCC tissues and adjacent non-cancerous tissues to determine the role of miR-337 in HCC. The data of RT-qPCR analysis revealed that miR-337 was underexpressed in HCC tissues in comparison with that in adjacent non-cancerous tissues (Figure 1A, P<0.05). To clarify the clinical significance of miR-337 in HCC, we divided all HCC patients into low miR-337 expression group (n = 24) or miR-337 high expression group (n = 23) based on the median expression of miR-337. As shown in Table 1, the expression level of miR-337 was significantly correlated with TNM stage (P = 0.006) and lymph node metastasis (P = 0.001). However, no association was found between miR-337 and other clinicopathological features, including age, gender, tumor size and differentiation (all P>0.05).
Figure 1.
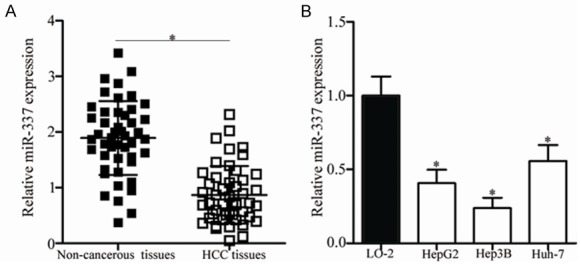
MiR-337 is downregulated in HCC tissues and cell lines. A. The expression level of miR-337 in 47 paired HCC tissues and adjacent non-cancerous tissues was detected using RT-qPCR. *P<0.05 compared with non-cancerous tissues. B. MiR-337 expression was examined in human HCC cell lines HepG2, Hep3B, and Huh-7, and in the normal human liver cell line LO2 (control). *P<0.05 compared with LO2.
Table 1.
The association between miR-337 expression and clinicopathological features of HCC patients
| Clinicopathological features | miR-337 expression | P value | |
|---|---|---|---|
|
| |||
| Low | High | ||
| Age | 0.474 | ||
| <55 years | 8 | 10 | |
| ≥55 years | 16 | 13 | |
| Gender | 0.534 | ||
| Female | 9 | 6 | |
| Male | 15 | 17 | |
| Tumor size | 0.642 | ||
| <5 cm | 11 | 9 | |
| ≥5 cm | 13 | 14 | |
| Differentiation | 0.238 | ||
| Well and Moderate | 12 | 16 | |
| Poor | 12 | 7 | |
| TNM stage | 0.006 | ||
| I-II | 6 | 15 | |
| III-IV | 18 | 8 | |
| Lymph node metastasis | 0.001 | ||
| Negative | 7 | 18 | |
| Positive | 17 | 5 | |
Furthermore, we measured the expression of miR-337 in three HCC cell lines, including HepG2, Hep3B and Huh-7. Compared with that in the normal human liver cell line LO2, miR-337 was downregulated in all examined HCC cell lines at different extents (Figure 1B, P<0.05). These data suggest that miR-337 may play important roles in the regulation of human HCC progression.
MiR-337 overexpression inhibits cell proliferation and promotes apoptosis in HCC
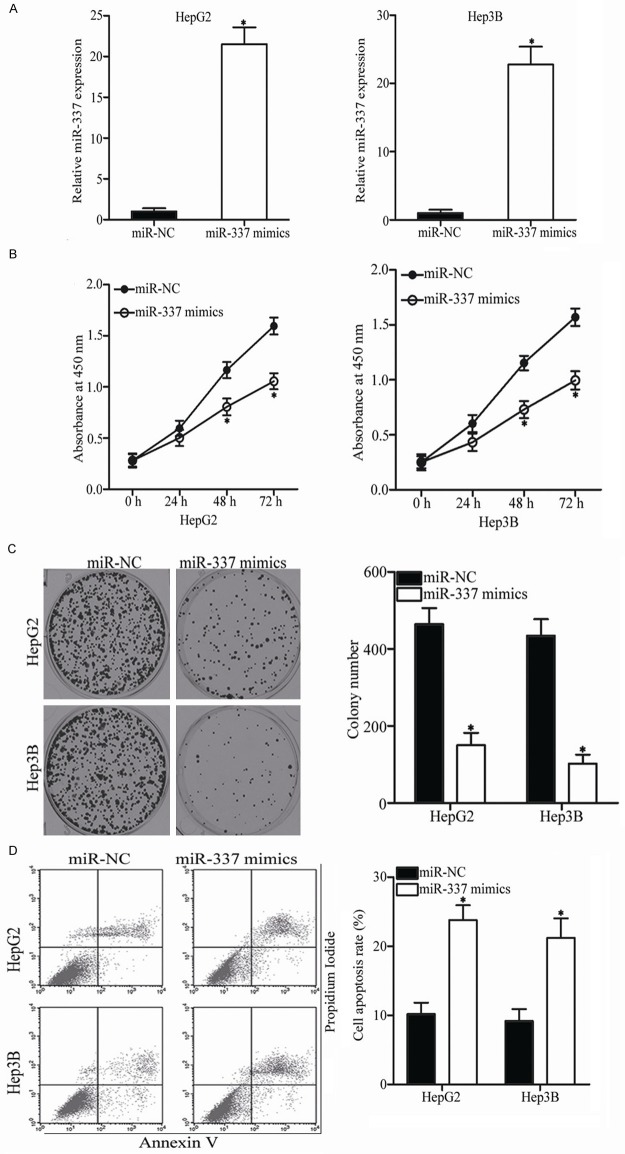
Considering that miR-337 is closely associated with HCC progression, we studied the biological roles of miR-337 in HCC. MiR-337 mimics was transfected into HepG2 and Hep3B cells, which expressed relatively lower miR-337 expression among the three HCC cell lines. RT-qPCR analysis confirmed that miR-337 was effectively improved in the HepG2 and Hep3B cells transfected with miR-337 mimics (Figure 2A, P<0.05). CCK-8 assay was performed to evaluate the effect of miR-337 overexpression on HCC cell proliferation in vitro. As shown in Figure 2B, cell proliferation was obviously suppressed in the HepG2 and Hep3B cells transfected with miR-337 mimics than in the cells transfected with miR-NC (P<0.05). Then, colony formation assay was conducted to confirm the HCC cell proliferation inhibition induced by miR-337. Results revealed that the HepG2 and Hep3B cells transfected with miR-337 mimics exhibited obviously fewer and smaller colonies compared with those transfected with miR-NC (Figure 2C, P<0.05). Furthermore, the effect of miR-337 on cell apoptosis in HCC was determined using flow cytometry analysis. The apoptosis rate of the HepG2 and Hep3B cells transfected with miR-337 mimics was higher than that of the cells transfected with miR-NC (Figure 2D, P<0.05). These results suggest that miR-337 prohibits cell proliferation and promotes apoptosis in HCC.
Figure 2.
Ectopic expression of miR-337 inhibits cell proliferation and induces apoptosis in HCC. A. HepG2 and Hep3B cells transfected with miR-337 mimics or miR-NC were subjected to RT-qPCR for the detection of miR-337 expression. *P<0.05 compared with miR-NC. B. Proliferative activity of HepG2 and Hep3B cells was determined by CCK-8 assay following transfection of miR-337 mimics or miR-NC. *P<0.05 compared with miR-NC. C. The effect of miR-337 overexpression on the colony formation of HepG2 and Hep3B cells was evaluated using colony formation assay. *P<0.05 compared with miR-NC. D. Cell apoptosis rate was detected in HepG2 and Hep3B cells transfected with miR-337 mimics or miR-NC with an Annexin V-FITC apoptosis detection kit. *P<0.05 compared with miR-NC.
Resumption expression of miR-337 attenuates cell migration and invasion in HCC
Considering that the expression of miR-337 is correlated with lymph node metastasis, we hypothesized that this miRNA is involved in HCC metastasis. To confirm this hypothesis, the migration and invasion abilities of HepG2 and Hep3B cells transfected with miR-337 mimics or miR-NC were assessed using Transwell migration and invasion assays, respectively. As expected, upregulation of miR-337 in HepG2 and Hep3B cells significantly decreased their migration capacities compared with the miR-NC group (Figure 3A, P<0.05). Similarly, the numbers of invaded cells were lower in the HepG2 and Hep3B cells transfected with miR-337 mimics than in those transfected with miR-NC (Figure 3B, P<0.05). These results suggest that miR-337 plays an inhibitory role in HCC cell metastasis.
Figure 3.
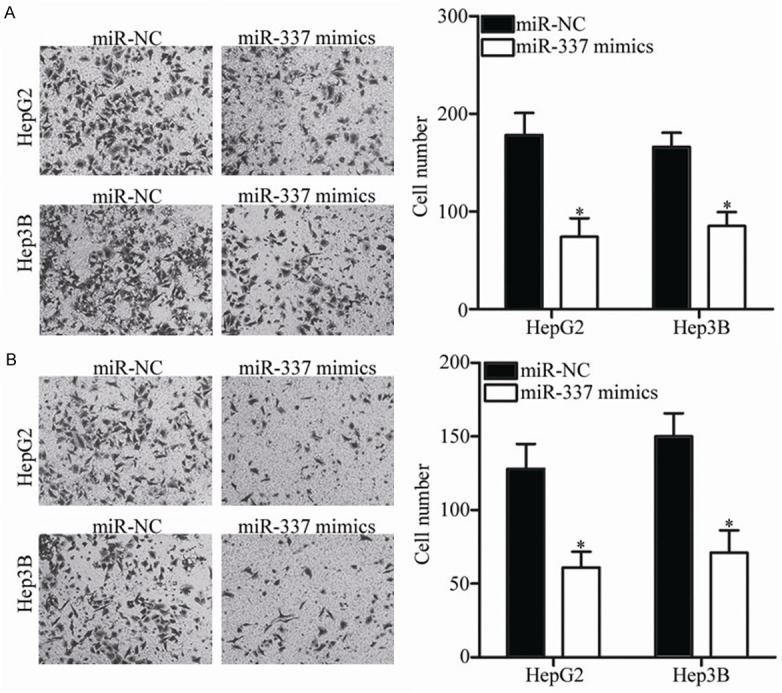
MiR-337 overexpression decreases HCC cell migration and invasion. A, B. Migration and invasion of HepG2 and Hep3B cells were investigated using Transwell migration and invasion assays following transfection of miR-337 mimics or miR-NC. *P<0.05 compared with miR-NC.
HMGA2 is a novel direct target gene of miR-337 in HCC
To elucidate the mechanisms by which miR-337 exerts its tumor-suppressing roles in HCC, bioinformatics analysis was performed to predict the potential target of miR-337. HMGA2, a well-known oncogene in HCC, was predicted to be a hypothetic target gene of miR-337. As presented in Figure 4A, a putative binding site for miR-337 was identified in the 3’-UTR of HMGA2. To validate whether the 3’-UTR of HMGA2 could be directly targeted by miiR-337, luciferase reporter assay was carried out in HepG2 and Hep3B cells cotransfected with miR-337 mimics or miR-NC and pMIR-HMGA2-3’-UTR Wt or pMIR-HMGA2-3’-UTR Mut. Ectopic expression of miR-337 decreased the luciferase activity associated with the pMIR-HMGA2-3’-UTR Wt plasmid (Figure 4B, P<0.05) but did not affect that derived from the pMIR-HMGA2-3’-UTR Mut plasmid. RT-qPCR and Western blot analysis were utilized to explore whether miR-337 affects endogenous HMGA2 expression in HCC. As shown in Figure 4C and 4D, the mRNA and protein expression levels of HMGA2 were significantly inhibited (P<0.05) in the HepG2 and Hep3B cells transfected with miR-337 relative to those transfected with miR-NC. These results demonstrate that HMGA2 is a direct target gene of miR-337 in HCC.
Figure 4.
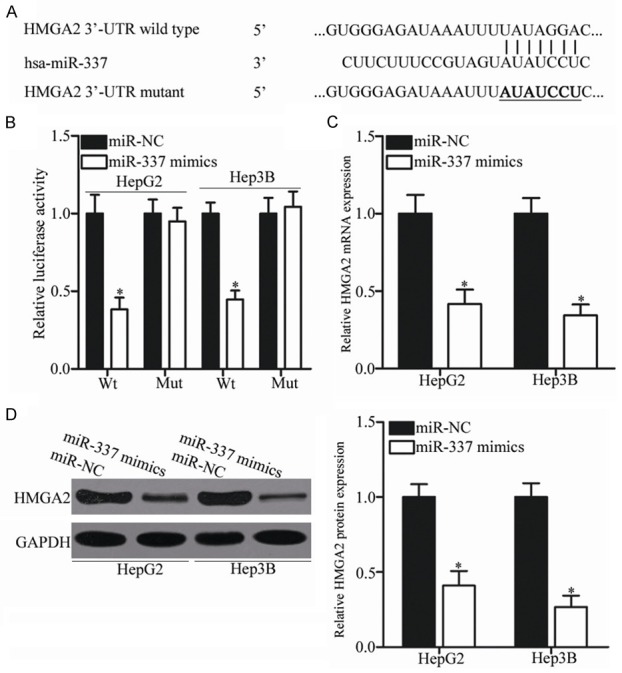
HMGA2 is a direct target of miR-337 in HCC. A. MiR-337 and its putative binding sequences in the 3’-UTR of HMGA2. Mutation was generated on the 3’-UTR of HMGA2 in the complementary sites for the seed region of miR-337. B. HepG2 and Hep3B cells were cotransfected with miR-337 mimics or miR-NC and wild-type or mutant HMGA2 3’-UTR. The relative firefly luciferase activity was determined at 48 h posttransfection. *P<0.05 compared with miR-NC. C, D. HepG2 and Hep3B cells that were transfected with miR-337 mimics or miR-NC were subjected to RT-qPCR and Western blot analysis for detection of HMGA2 mRNA and protein expression, respectively. *P<0.05 compared with miR-NC.
HMGA2 is overexpressed in HCC and inversely correlated with miR-337 expression
To further investigate the association between miR-337 and HMGA2 in HCC, we detected the expression of HMGA2 in HCC tissues and adjacent non-cancerous tissues. RT-qPCR data revealed that the mRNA level of HMGA2 was higher in HCC tissues than in adjacent non-cancerous tissues (Figure 5A, P<0.05). In addition, the protein level of HMGA2 in several paired HCC tissues and adjacent non-cancerous tissues was determined through Western blot analysis. As illustrated in Figure 5B, HMGA2 protein was significantly upregulated in HCC tissues than in adjacent non-cancerous tissues. Furthermore, HMGA2 protein level was also determined in three HCC cell lines, including HepG2, Hep3B and Huh-7. Compared with that in the normal human liver cell line LO2, HMGA2 protein level was overexpressed in all three HCC cell lines (Figure 5C, P<0.05). Besides, an inverse association was validated between miR-337 and HMGA2 mRNA level in HCC tissues (Figure 5D; r = -0.5731, P<0.0001). These results suggest that the upregulation of HMGA2 in HCC is partially caused by the downregulation of miR-337.
Figure 5.
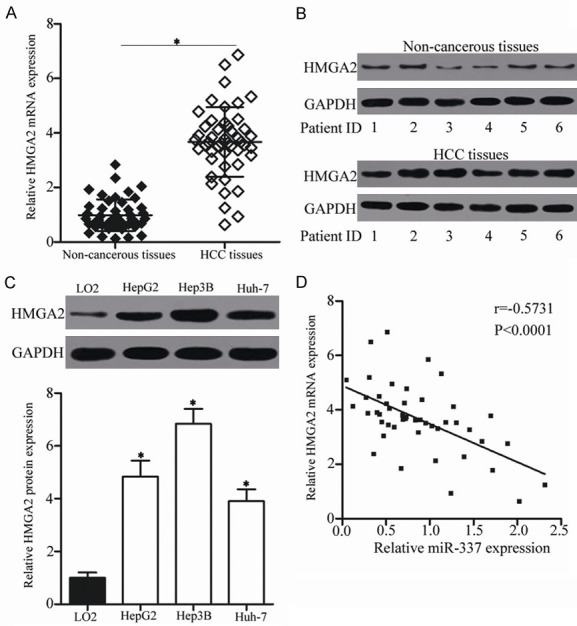
HMGA2 is upregulated in HCC tissues and negatively correlated with miR-337 expression. A, B. RT-qPCR and Western blot analysis were performed to measure HMGA2 mRNA and protein expression in HCC tissues and adjacent non-cancerous tissues. *P<0.05 compared with non-cancerous tissues. C. Western blot analysis was utilized to determine HMGA2 protein expression in three HCC cell lines and the normal human liver cell line LO2 (control). *P<0.05 compared with LO2. D. The association between miR-337 and the mRNA expression of HMGA2 was determined using Spearman’s correlation analysis. r = -0.5731, P<0.0001.
HMGA2 knockdown mimics similar effects to miR-337 overexpression in HCC
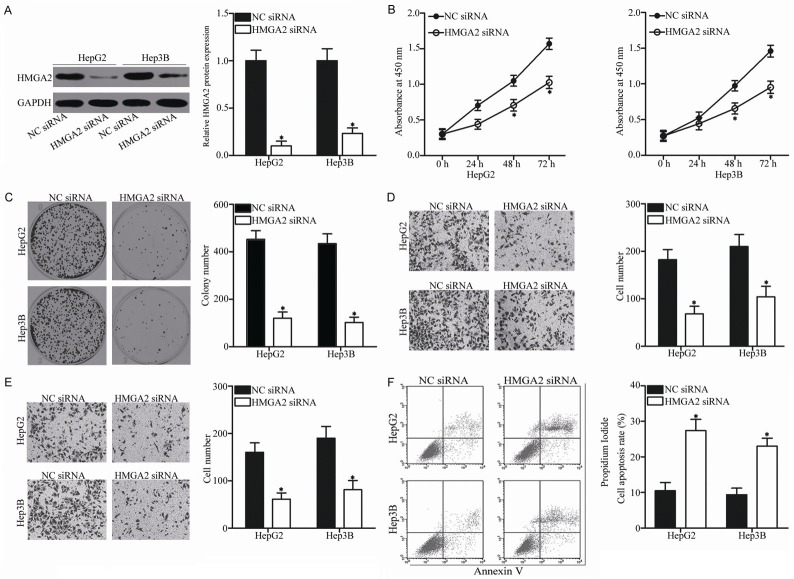
Considering that HMGA2 was identified as a direct target of miR-337 in HCC, we hypothesized that the tumor-suppressive roles of miR-337 in HCC is exhibited by HMGA2 inhibition. To confirm this hypothesis, HMGA2 expression in HepG2 and Hep3B cells was knocked down using HMGA2 siRNA. Western blot analysis showed that HMGA2 protein expression was significantly downregulated in the HepG2 and Hep3B cells transfected with HMGA2 siRNA (Figure 6A, P<0.05). Functional experiments indicated that inhibition of HMGA2 repressed the proliferation (Figure 6B, P<0.05), colony formation (Figure 6C, P<0.05), migration (Figure 6D, P<0.05), and invasion (Figure 6E, P<0.05) while increased the apoptosis (Figure 6F, P<0.05) of HepG2 and Hep3B cells, which was consistent with the effects of miR-337 overexpression. These results further suggest that HMGA2 is a functional downstream target of miR-337 in HCC.
Figure 6.
HMGA2 knockdown decreases the proliferation, migration, and invasion while increases the apoptosis of HCC cells. A. HMGA2 siRNA or NC siRNA was introduced into HepG2 and Hep3B cells. At 72 h after transfection, the transfection efficiency was evaluated using Western blot analysis. *P<0.05 compared with NC siRNA. B-F. CCK-8 assay, colony formation assay, Transwell migration and invasion assays, and cell apoptosis assay were conducted to investigate the effect of HMGA2 knockdown on HepG2 and Hep3B cell proliferation, colony formation, migration, invasion, and apoptosis, respectively. *P<0.05 compared with NC siRNA.
Restored HMGA2 expression reverses the effects of miR-337 overexpression in HCC
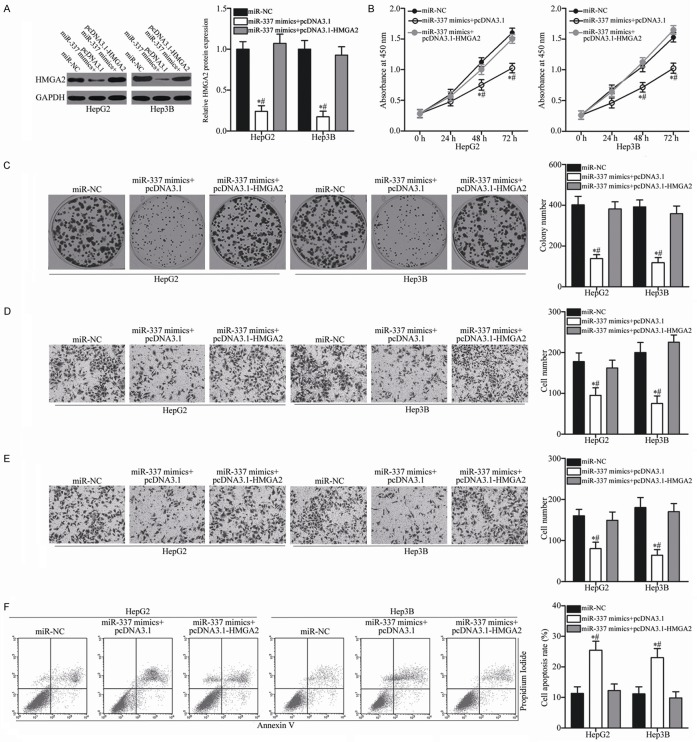
To test whether the tumor-suppressive roles of miR-337 in HCC are mediated by HMGA2, a series of rescue experiments was carried out involving transfection of miR-337 mimics in HepG2 and Hep3B cells together with pcDNA3.1 or pcDNA3.1-HMGA2 lacking the respective 3’-UTR. Following transfection, Western blot analysis confirmed that the decreased HMGA2 protein expression caused by miR-337 overexpression could be recovered by cotransfection with pcDNA3.1-HMGA2 in HepG2 and Hep3B cells (Figure 7A, P<0.05). In addition, restored HMGA2 expression significantly rescued the effects of exogenous miR-337 on the proliferation (Figure 7B, P<0.05), colony formation (Figure 7C, P<0.05), migration (Figure 7D, P<0.05), invasion (Figure 7E, P<0.05), and apoptosis (Figure 7F, P<0.05) of HepG2 and Hep3B cells. These results further confirm that miR-337 exerts tumor-suppressive roles in HCC, at least in part, by inhibiting HMGA2.
Figure 7.
Re-expression of HMGA2 reverses the tumor-suppressing effects of miR-337 on HCC cells. HepG2 and Hep3B cells were transfected with miR-337 mimics along with pcDNA3.1 or pcDNA3.1-HMGA2 lacking the respective 3’-UTR. A. Expression level of HMGA2 protein was analyzed by Western blot analysis in differently treated cells. *P<0.05 compared with miR-NC. #P<0.05 compared with miR-337 mimics + pcDNA3.1-HMGA2. B. CCK-8 assay was carried out to analyze cell proliferation in indicated cells. *P<0.05 compared with miR-NC. #P<0.05 compared with miR-337 mimics + pcDNA3.1-HMGA2. C. Representative images and quantification of colony formation assay in indicated cells. *P<0.05 compared with miR-NC. #P<0.05 compared with miR-337 mimics + pcDNA3.1-HMGA2. D, E. Transwell migration and invasion assays were employed to assess cell migration and invasion in indicated cells, respectively. *P<0.05 compared with miR-NC. #P<0.05 compared with miR-337 mimics + pcDNA3.1-HMGA2. F. Cell apoptosis in differently treated cells was quantified by flow cytometry analysis. *P<0.05 compared with miR-NC. #P<0.05 compared with miR-337 mimics + pcDNA3.1-HMGA2.
MiR-337 affects the PI3K/AKT and Wnt/β-catenin signaling pathways in HCC
HMGA2 is involved in the regulation of the PI3K/AKT and Wnt/β-catenin signaling pathways [22-24]. To determine whether miR-337 affects the PI3K/AKT and Wnt/β-catenin signaling pathways in HCC, we measured the expression levels of AKT, p-AKT, β-catenin and p-β-catenin in HepG2 and Hep3B cells cotransfected with miR-337 mimics and pcDNA3.1-HMGA2 or pcDNA3.1 through Western blot analysis. Results demonstrated that the ectopic expression of miR-337 decreased p-AKT and p-β-catenin expression and that total AKT and β-catenin expression was unaltered in HepG2 and Hep3B cells (Figure 8). Meanwhile, cotransfection of pcDNA3.1-HMGA2 restored the effects of miR-337 overexpression on p-AKT and p-β-catenin expression in HepG2 and Hep3B cells. These results suggest that miR-337 inhibits the activation of the PI3K/AKT and Wnt/β-catenin signaling pathways in HCC by regulation of HMGA2.
Figure 8.
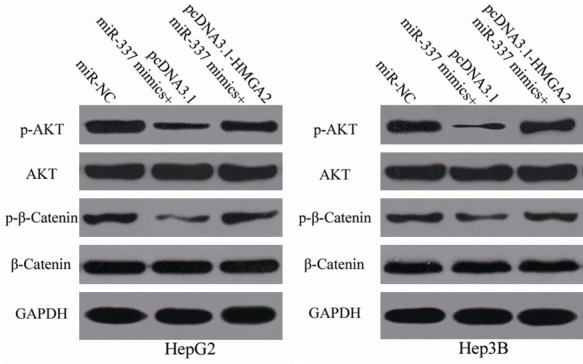
Upregulation of miR-337 suppresses the PI3K/AKT and Wnt/β-catenin signaling pathways in HCC. MiR-337 mimics was transfected into HepG2 and Hep3B cells together with pcDNA3.1 or pcDNA3.1-HMGA2. At 72 h after transfection, Western blot analysis was conducted to measure AKT, p-AKT, β-catenin, and p-β-catenin expression.
MiR-337 suppresses tumor growth of hepatoma xenografts in vivo
To explore the role of miR-337 in tumor growth in vivo, nude mice were treated with HepG2 cells transfected with miR-337 mimics or miR-NC. The tumor volume was measured every 2 days. At 30 days after inoculation, all nude mice were sacrificed and the xenografts were weighed. We firstly detected miR-337 expression in tumor xenografts. The data of RT-qPCR analysis showed that miR-337 was significantly upregulated in miR-337 mimics-transfected tumor xenografts compared with that in miRNC-transfected tumor xenografts (Figure 9A, P<0.05). The tumor volume of the miR-337 mimics groups was smaller than that of the miR-NC groups from day 20 to day 30 (Figure 9B and 9C, P<0.05). In addition, the tumor weight of the miR-337 groups significantly decreased compared with that of the miR-NC groups (Figure 9D, P<0.05). Furthermore, Western blot analysis indicated that the expression levels of HMGA2, p-AKT, and p-β-catenin in the xenograft tumor tissues transfected with miR-337 mimics were reduced compared with those in the xenograft tumor tissues transfected with miR-NC (Figure 9E). These results suggest that miR-337 represses HCC cell growth in vivo by directly targeting HMGA2 and regulating the PI3K/AKT and Wnt/β-catenin signaling pathways.
Figure 9.
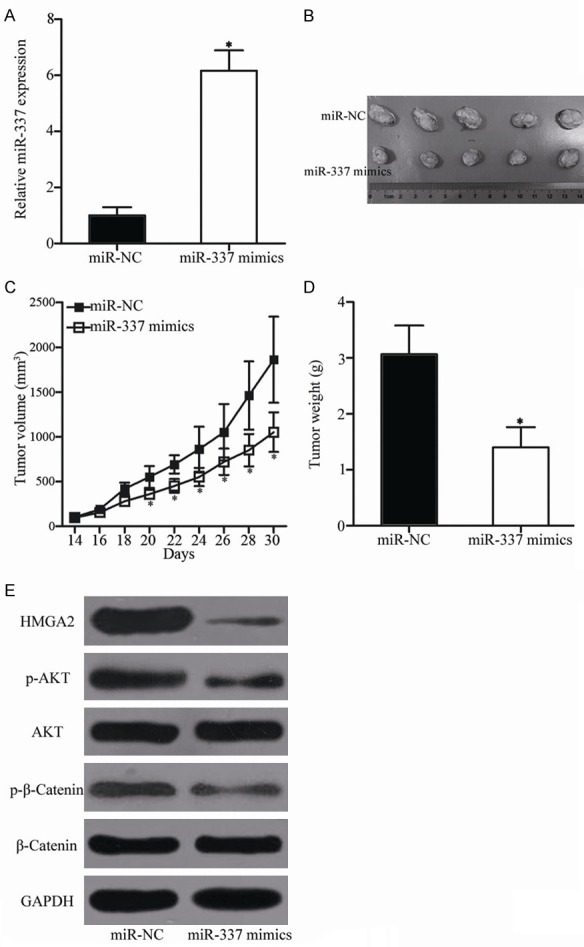
MiR-337 reduces HCC tumor growth in vivo. A. MiR-337 expression was determined in tumor xenografts using RT-qPCR. *P<0.05 compared with miR-NC. B. Mice were sacrificed, and representative images of the miR-337 mimics and miR-NC xenograft tumors were obtained. C. Tumor volume was calculated every 2 days after inoculation for 2 weeks. *P<0.05 compared with miR-NC. D. Tumor weight in the different groups. *P<0.05 compared with miR-NC. E. Western blot analysis was performed to detect HMGA2, AKT, p-AKT, β-catenin, and p-β-catenin expression in xenograft tumor tissues of the miR-337 and miR-NC groups.
Discussion
MiRNAs serve as major regulators during the tumorigenesis and tumor development of HCC [25-27]. In addition, miRNAs may serve as new promising biomarkers for the diagnosis and prognosis and as effective therapeutic targets for patients with this malignancy [28,29]. Therefore, understanding the association between miRNAs and HCC may be beneficial to discover novel therapeutic approaches towards diagnosis and treatments. In the present study, the expression level of miR-337 was downregulated in HCC tissues and cell lines. Decreased miR-337 expression was significantly correlated with TNM stage and lymph node metastasis. In addition, resumption expression of miR-337 inhibited cell proliferation, colony formation, migration, and invasion; promoted apoptosis in vitro; and reduced tumor growth in vivo. Furthermore, HMGA2 was identified as a direct target of miR-337 in HCC. HMGA2 was overexpressed in HCC and inversely correlated with miR-337 expression. Moreover, HMGA2 knockdown exhibited similar effects to miR-337 overexpression in HCC cells. Restored HMGA2 expression rescued the tumor-suppressive roles of miR-337 overexpression in HCC. Furthermore, upregulation of miR-337 inhibited the activation of the PI3K/AKT and Wnt/β-catenin signaling pathways in HCC in vitro and in vivo. Taken together, miR-337 may serve as a tumor suppressor in HCC by directly targeting HMGA2 and regulating the PI3K/AKT and Wnt/β-catenin signaling pathways.
MiR-337 is commonly aberrantly expressed in multiple types of human malignancy. For example, miR-337 expression is underexpressed in gastric cancer tissues and cell lines. Its aberrant expression is correlated with lymph node metastasis [30,31]. Gastric cancer patients with low miR-337 level have worse prognosis than those patients with high miR-337 level [31]. In pancreatic ductal adenocarcinoma, the expression level of miR-337 is downregulated in tumor tissues and positively correlated with TNM stage and lymph node status [32]. Low miR-337 expression could be an independent prognostic factor for the favorable outcome of patients with pancreatic ductal adenocarcinoma [32]. In cervical cancer, miR-337 is decreased in tumor tissues and cell lines. Downregulation of miR-337 is significantly associated with the tumor size, International Fe|deration of Gynecology and Obstetrics stage, and lymph node metastasis of cervical cancer [33]. Decreased miR-337 expression is also observed in melanoma [17], neuroblastoma [18], colorectal cancer [19], and non-small cell lung cancer [20]. These findings suggest that deregulation of miR-337 is a promising prognostic biomarker in these types of human cancer.
MiR-337 is involved in malignant progression of several cancer types. For instance, Zheng et al. reported that restoration expression of miR-337 prohibits the growth, metastasis, invasion, and angiogenesis of gastric cancer cells in vitro and in vivo [31]. Zhang et al. revealed that miR-337 overexpression attenuates the cell proliferation, colony formation, and invasion of pancreatic ductal adenocarcinoma [34]. Dong et al. found that ectopic expression of miR-337 decreases the proliferation and invasion of cervical cancer cells [33]. Xiang et al. demonstrated that miR-337 re-expression suppresses the proliferation, migration, invasion, and angiogenesis of neuroblastoma cells in vitro and in vivo [18]. Kim et al. indicated that enforced expression of miR-337 increases cellular senescence in vitro [19]. Du et al. showed that resumption expression of miR-337 increases the taxane chemosensitivity of non-small cell lung cancer [20]. These findings suggest that miR-337 is a novel and efficient therapeutic target for the treatment of these malignancies.
Previous studies have identified several targets of miR-337, including MMP14 in gastric cancer [31] and neuroblastoma [18], HOXB7 in pancreatic ductal adenocarcinoma [34], SP1 in cervical cancer [33], STAT3 in melanoma [17], CKIIα in colorectal cancer [19], and STAT3 and RAP1A in non-small cell lung cancer [20]. The current study confirmed HMGA2 as a direct target of miR-337 in HCC. HMGA2, a membrane of high mobility group A proteins, is a non-histone chromatin-binding protein [36]. It is overexpressed in various types of human cancer and regulates the carcinogenesis and progression of different cancer types, such as breast [38], thyroid [35], lung [36], colorectal [39], and prostate [40] cancers. In addition, dysregulation of HMGA2 correlates with clinicopathologic factors and prognosis in multiple human cancer types. For example, HMGA2 expression is increased in bladder cancer and correlated with tumor grade and stage. Bladder cancer patients with high HMGA2 level have shorter recurrence-free survival and progression-free survival in contrast to those with low HMGA2. Multivariate analysis also demonstrates HMGA2 as an independent predictor of tumor recurrence and progression in bladder cancer [37]. Therefore, HMGA2 may serve as a potential target for the monitoring and treatment of human cancers.
HMGA2 is highly expressed in HCC tissues and cell lines [38,39]. Upregulation of HMGA2 is strongly associated with tumor size, capsule invasion, and vascular invasion. The overall survival rate of HCC patients with high HMGA2 levels is shorter than that of HCC patients with low HMGA2 level. In addition, univariate and multivariate analyses demonstrates that MGA2 is an independent prognostic and postoperative survival predictor factor for HCC patients [38]. Further studies illustrates that HMGA2 may serve important roles in the occurrence and development of HCC through regulating cell proliferation, apoptosis, migration, invasion, metastasis, and epithelial-to-mesenchymal transition [39-41]. In consideration of the important roles of HMGA2 in HCC, the miR-337/HMGA2 axis may provide novel and efficient therapeutic opportunities to treat patients with this disease.
In conclusion, miR-337 is underexpressed in HCC tissues and cell lines, and its reduced expression is correlated with TNM stage and lymph node metastasis. MiR-337 plays tumor-suppressive roles in HCC by directly targeting HMGA2 and regulating the PI3K/AKT and Wnt/β-catenin signaling pathways. The results of this study may provide novel evidence for the potential utility of a miR-337/HMGA2-based targeted therapy for patients with HCC.
Acknowledgements
The present study was supported by Funds for doctors of Dalian University (grant no. 20172QL018).
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Vertemati M, Moscheni C, Petrella D, Lamperti L, Cossa M, Gambacorta M, Goffredi M, Vizzotto L. Morphometric analysis of hepatocellular nodular lesions in HCV cirrhosis. Pathol Res Pract. 2012;208:240–244. doi: 10.1016/j.prp.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Liang T, Chen EQ, Tang H. Hepatitis B virus gene mutations and hepatocarcinogenesis. Asian Pac J Cancer Prev. 2013;14:4509–4513. doi: 10.7314/apjcp.2013.14.8.4509. [DOI] [PubMed] [Google Scholar]
- 5.Gao J, Xie L, Yang WS, Zhang W, Gao S, Wang J, Xiang YB. Risk factors of hepatocellular carcinoma--current status and perspectives. Asian Pac J Cancer Prev. 2012;13:743–752. doi: 10.7314/apjcp.2012.13.3.743. [DOI] [PubMed] [Google Scholar]
- 6.Yin JM, Sun LB, Zheng JS, Wang XX, Chen DX, Li N. Copper chelation by trientine dihydrochloride inhibits liver RFA-induced inflammatory responses in vivo. Inflamm Res. 2016;65:1009–1020. doi: 10.1007/s00011-016-0986-2. [DOI] [PubMed] [Google Scholar]
- 7.Llovet JM, Schwartz M, Mazzaferro V. Resection and liver transplantation for hepatocellular carcinoma. Semin Liver Dis. 2005;25:181–200. doi: 10.1055/s-2005-871198. [DOI] [PubMed] [Google Scholar]
- 8.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 10.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 12.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2017;9:852. doi: 10.15252/emmm.201707779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Duan X, Hu J, Wang Y, Gao J, Peng D, Xia L. MicroRNA-145: a promising biomarker for hepatocellular carcinoma (HCC) Gene. 2014;541:67–68. doi: 10.1016/j.gene.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Yang T, Thakur A, Chen T, Yang L, Lei G, Liang Y, Zhang S, Ren H, Chen M. MicroRNA-15a induces cell apoptosis and inhibits metastasis by targeting BCL2L2 in non-small cell lung cancer. Tumour Biol. 2015;36:4357–4365. doi: 10.1007/s13277-015-3075-1. [DOI] [PubMed] [Google Scholar]
- 16.Wu G, Liu J, Wu Z, Wu X, Yao X. MicroRNA-184 inhibits cell proliferation and metastasis in human colorectal cancer by directly targeting IGF-1R. Oncol Lett. 2017;14:3215–3222. doi: 10.3892/ol.2017.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao W, Yao E, Zheng W, Tian F, Tian L. miR-337 can be a key negative regulator in melanoma. Cancer Biol Ther. 2017;18:392–399. doi: 10.1080/15384047.2017.1323581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang X, Mei H, Zhao X, Pu J, Li D, Qu H, Jiao W, Zhao J, Huang K, Zheng L, Tong Q. miRNA-337-3p suppresses neuroblastoma progression by repressing the transcription of matrix metalloproteinase 14. Oncotarget. 2015;6:22452–22466. doi: 10.18632/oncotarget.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SY, Lee YH, Bae YS. MiR-186, miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular senescence by targeting alpha subunit of protein kinase CKII in human colorectal cancer cells. Biochem Biophys Res Commun. 2012;429:173–179. doi: 10.1016/j.bbrc.2012.10.117. [DOI] [PubMed] [Google Scholar]
- 20.Du L, Subauste MC, DeSevo C, Zhao Z, Baker M, Borkowski R, Schageman JJ, Greer R, Yang CR, Suraokar M, Wistuba II, Gazdar AF, Minna JD, Pertsemlidis A. miR-337-3p and its targets STAT3 and RAP1A modulate taxane sensitivity in non-small cell lung cancers. PLoS One. 2012;7:e39167. doi: 10.1371/journal.pone.0039167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Ou W, Lv J, Zou X, Yao Y, Wu J, Yang J, Wang Z, Ma Y. Propofol inhibits hepatocellular carcinoma growth and invasion through the HMGA2-mediated Wnt/beta-catenin pathway. Exp Ther Med. 2017;13:2501–2506. doi: 10.3892/etm.2017.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Liang Y, Shen L, Shen L. MicroRNA-204 modulates colorectal cancer cell sensitivity in response to 5-fluorouracil-based treatment by targeting high mobility group protein A2. Biol Open. 2016;5:563–570. doi: 10.1242/bio.015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan L, Wei X, Zheng L, Zeng J, Liu H, Yang S, Tan H. Amplified HMGA2 promotes cell growth by regulating Akt pathway in AML. J Cancer Res Clin Oncol. 2016;142:389–399. doi: 10.1007/s00432-015-2036-9. [DOI] [PubMed] [Google Scholar]
- 25.Drakaki A, Hatziapostolou M, Iliopoulos D. Therapeutically targeting microRNAs in liver cancer. Curr Pharm Des. 2013;19:1180–1191. doi: 10.2174/138161213804805658. [DOI] [PubMed] [Google Scholar]
- 26.Huang S, He X. The role of microRNAs in liver cancer progression. Br J Cancer. 2011;104:235–240. doi: 10.1038/sj.bjc.6606010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, Tang ZY, Wang XW. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 28.Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 29.Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Wang J, Yang Y, Hao B, Wang R, Li Y, Wu Q. Loss of has-miR-337-3p expression is associated with lymph node metastasis of human gastric cancer. J Exp Clin Cancer Res. 2013;32:76. doi: 10.1186/1756-9966-32-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng L, Jiao W, Mei H, Song H, Li D, Xiang X, Chen Y, Yang F, Li H, Huang K, Tong Q. miRNA-337-3p inhibits gastric cancer progression through repressing myeloid zinc finger 1-facilitated expression of matrix metalloproteinase 14. Oncotarget. 2016;7:40314–40328. doi: 10.18632/oncotarget.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang R, Zheng S, Du Y, Wang Y, Zang W, Zhao G. Levels of HOXB7 and miR-337 in pancreatic ductal adenocarcinoma patients. Diagn Pathol. 2014;9:61. doi: 10.1186/1746-1596-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong W, Li B, Wang J, Song Y, Zhang Z, Fu C. MicroRNA-337 inhibits cell proliferation and invasion of cervical cancer through directly targeting specificity protein 1. Tumour Biol. 2017;39:1010428317711323. doi: 10.1177/1010428317711323. [DOI] [PubMed] [Google Scholar]
- 34.Zhang R, Leng H, Huang J, Du Y, Wang Y, Zang W, Chen X, Zhao G. miR-337 regulates the proliferation and invasion in pancreatic ductal adenocarcinoma by targeting HOXB7. Diagn Pathol. 2014;9:171. doi: 10.1186/s13000-014-0171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belge G, Meyer A, Klemke M, Burchardt K, Stern C, Wosniok W, Loeschke S, Bullerdiek J. Upregulation of HMGA2 in thyroid carcinomas: a novel molecular marker to distinguish between benign and malignant follicular neoplasias. Genes Chromosomes Cancer. 2008;47:56–63. doi: 10.1002/gcc.20505. [DOI] [PubMed] [Google Scholar]
- 36.Gao X, Dai M, Li Q, Wang Z, Lu Y, Song Z. HMGA2 regulates lung cancer proliferation and metastasis. Thorac Cancer. 2017;8:501–510. doi: 10.1111/1759-7714.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang GL, Zhang LH, Bo JJ, Hou KL, Cai X, Chen YY, Li H, Liu DM, Huang YR. Overexpression of HMGA2 in bladder cancer and its association with clinicopathologic features and prognosis HMGA2 as a prognostic marker of bladder cancer. Eur J Surg Oncol. 2011;37:265–271. doi: 10.1016/j.ejso.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Wu L, Wang Z, Lu R, Jiang W. Expression of high mobility group A2 is associated with poor survival in hepatocellular carcinoma. Pathol Oncol Res. 2012;18:983–987. doi: 10.1007/s12253-012-9514-z. [DOI] [PubMed] [Google Scholar]
- 39.Luo Y, Li W, Liao H. HMGA2 induces epithelial-to-mesenchymal transition in human hepatocellular carcinoma cells. Oncol Lett. 2013;5:1353–1356. doi: 10.3892/ol.2013.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang W, Li J, Guo X, Zhao Y, Yuan X. miR-663a inhibits hepatocellular carcinoma cell proliferation and invasion by targeting HMGA2. Biomed Pharmacother. 2016;81:431–438. doi: 10.1016/j.biopha.2016.04.034. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Liang H, Jiang X. MiR-1297 promotes apoptosis and inhibits the proliferation and invasion of hepatocellular carcinoma cells by targeting HMGA2. Int J Mol Med. 2015;36:1345–1352. doi: 10.3892/ijmm.2015.2341. [DOI] [PubMed] [Google Scholar]