Abstract
Triple-negative breast cancer (TNBC) has a higher potential for invasion and metastasis than other types of breast cancer. Enhancer of zeste homolog 2 (EZH2) is the catalytic core protein in the polycomb repressive complex 2 (PRC2), which catalyzes the trimethylation of histone H3 at lysine 27 (H3K27me3) and mediates gene silencing of the target genes that are involved in fundamental cellular processes, such as the cell fate decision, cell cycle regulation, senescence, cell differentiation, and cancer formation. A consistent association between TNBC metastasis and EZH2 has not been confirmed. The aim of this study was to investigate the role of EZH2 in the regulation of tissue inhibitor of metalloproteinase (TIMPs) and matrix metalloproteinases (MMPs) to promote metastasis of TNBC cells and to characterize the metastasis-associated genes regulated by EZH2 in TNBC cells. We found that high levels of EZH2 expression induce repression of TIMP2 transcription, leading to increased activity of MMP-2 and MMP-9 and thus to increased invasive activity of TNBC cells.
Keywords: Enhancer of zeste homolog 2, matrix metalloproteinases, tissue inhibitor of metalloproteinase, migration, triple-negative breast cancer cell
Introduction
Triple-negative breast cancer (TNBC) is a heterogeneous disease with distinct molecular subtypes that differentially respond to chemotherapy and targeted agents. Approximately 10-15% of breast carcinomas are known to be of the TNBC subtype, which constitutes ~80% of all ‘basal-like tumors’, which are characterized by the absence of expression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2). Because of the absence of expression of commonly targeted receptors present in other breast tumor subtypes, agents that specifically target TNBC are not yet available. TNBC has a worse prognosis, higher histological grade, more poorly differentiated cell types, and more aggressive characteristics and tends to relapse earlier as compared with other subtypes of breast cancer. Conversely, it displays increased chemosensitivity as compared with other breast tumor subtypes [1-3]. Thus, the need for the accelerated development of effective TNBC therapies is critical. The design of new therapeutic strategies targeting multiple signaling pathways for more effective disease management in TNBC is a primary focus of current research.
The Polycomb group (PcG) protein EZH2 is a transcriptional repressor involved in cell cycle regulation and has been linked to aggressive breast cancer. High expression of EZH2 is strongly associated with the TNBC phenotype as compared with all other non-TNBCs [4]. EZH2 is also a mammalian histone methyltransferase that contributes to the epigenetic silencing of target genes and regulates the survival and metastasis of cancer cells. EZH2 is highly expressed in a wide range of cancer types, including breast, prostate, bladder, colon, lung, and pancreatic cancer; sarcoma; and lymphoma. Overexpression of EZH2 is often correlated with advanced stages of human cancer progression and poor prognosis [5,6].
Invasion and metastasis are the major characteristics of malignancy with poor clinical outcome. These processes require the disruption of several collagen-endowed tissue barriers, among which is the basement membrane that lines vascular endothelial cells and constitutes a continuous physical obstacle to tumor metastasis. The degradative process is mainly mediated by matrix metalloproteinases (MMPs), which are a family of at least 20 zinc-dependent endopeptidases best known for their ability to hydrolyze extracellular matrix (ECM) components [7]. MMP-9 is expressed in large quantities in breast cancer cell line[s] and might play an important role in tumor invasion [8]. The fact that TIMPs are natural inhibitors of MMP activity raised the possibility that an imbalance in local MMP and TIMP concentrations caused by an increase in MMP expression and a concurrent decrease in TIMP production may underlie MMP-dependent ECM remodeling at sites of tumor invasion. During metastasis, tumor cells are involved in numerous interactions with the ECM itself and with those proteins, growth factors, and cytokines associated with the ECM. Generally, cancer invasion, cancer cell migration, and cancer cell-mediated tissue remodeling are the processes that have correlated positively and most highly with MMP levels [9,10].
In this study, we investigated the role of EZH2 in the regulation of TIMPs and MMPs to promote metastasis during TNBC. Using a human cytokine antibody array, we identified the metastasis-associated genes that are regulated by EZH2 in TNBC cells in which EZH2 expression was knocked down. These findings provide insights into the possible mechanisms by which EZH2 regulates TIMPs and MMPs.
Materials and methods
Cell culture
All cell lines used in this study were obtained from American Type Culture Collection. Human TNBC cell lines (MDA-MB-468 and MDA-MB-231) and were maintained in DMEM/F12 (1:1) with 10% FBS. 184A1 and MCF-10A cells were maintained in DMEM/F12 medium supplemented with 1.05 mM CaCl2, 100 ng/ml cholera toxin, 5% horse serum (Gibco), 10 μg/ml insulin, 100 U/ml penicillin, 100 μg/ml streptomycin, 20 ng/ml epidermal growth factor (EGF) (Sigma), and 500 ng/ml hydrocortisone. All cells were maintained in a humidified incubator containing 5% CO2 at 37°C.
Cell extraction and western blotting analysis
For total cell lysate extraction, cells were washed twice with ice-cold PBS and lysed in NETN buffer (20 mM Tris at pH 8.0, 150 mM NaCl, 1 mM EDTA at pH 8.0, 0.5% Nonidet P-40) with protease and phosphatase inhibitors (25 mM NaF, 2 mM Na3VO4, 0.1 mM PMSF, 20 μg/ml aprotinin) by sonication. The soluble extract was collected after centrifugation at 15,000 × g for 15 min at 4°C. The lysates were separated by 6%, 8%, and 15% SDS-polyacrylamide gel electrophoresis, and the proteins were transferred to a polyvinylidene fluoride membrane. Subsequently, the membrane was blocked with 5% skim milk in TBST buffer (TBS containing 0.1% Tween-20) for 1 hour at room temperature and then hybridized with primary antibody with gentle agitation overnight at 4°C. After washing three times with TBST, the membrane was incubated with secondary antibody for 1 hour at room temperature. The band was visualized by the enhanced chemiluminescence detection reagent (GE Healthcare). The following antibodies and chemicals were used: anti-EZH2 (1:1000; BD), anti-TIMP2 (1:1000; GenTex), anti-MMP-2 (1:1000; GenTex), anti-MMP-9 (1:1000; GenTex), anti-H3K27me3 (1:1000; abcam), anti-Histone H3 (1:1000; Santa Cruz), anti-α-tubulin (1:5000; Sigma), and DZNep (CAYMAN CHEMICAL COMPANY). Images were quantified by using Image J software (NIH Imaging).
Gene knockdown by shRNA and lentivirus infection
Knockdown of genes was performed with specific shRNAs delivered by the lentivirus system from the National RNAi Core Facility (Academia Sinica) according to their protocol. 293T cells were co-transfected with 2.5 μg of pLKO.1-Luc, pLKO.1-EZH2, or pLKO.1-TIMP-2 plasmid, with 0.25 μg of pMDG and 2.25 μg of pCMV-ΔR8.91 using Lipofectamine 2000 transfection reagent (Invitrogen). After 6 hours, the medium was changed to DMEM/F12 with 1% bovine serum albumin (BSA) for 24 hours. The supernatants, which contained virus particles, were collected, and the virus particles were harvested by filtration through a 0.22-μm membrane. The virus particle solution was then stored at -80°C until use. For lentivirus infection, cells were transduced with lentivirus in the presence of 8 μg/ml Polybrene. After 24 hours, puromycin was added to the culture medium at a final concentration of 3 μg/ml and the cells were incubated for 3 days to allow selection of infected cells.
Cell invasion and migration assays
A cell invasion assay was conducted using BioCoat Matrigel Invasion Chambers according to the manufacturer’s instructions. Briefly, the Matrigel was added to each chamber to allow hydration of the Matrigel coating for 1 hour immediately before the experiments. Cells (5 × 104) suspended in 100 μl of serum-free medium were then added to the upper chamber of the Matrigel-coated filter inserts. After treatment with surfactin, 700 μl of DMEM/F12 (1:1, v/v) with 10% FBS was added to the bottom as a chemoattractant. The chambers were then incubated for 48 hours at 37°C. The migration assay was conducted as described for the invasion assay but without the Matrigel coating. The chambers were then incubated for 24 hours at 37°C. Migrating cells attached on the lower surface of the filter were fixed and stained with 2% ethanol containing 0.2% crystal violet powder. The cells that invaded or migrated through the membrane were counted under a light microscope (× 40) and by measurement of their absorbance.
Human cytokine antibody arrays
The expression of 10 human cytokines was analyzed using a commercially available antibody array system (RayBio® Human Matrix Metalloproteinase Antibody Array 1 Map, RayBiotech, Inc) that uses membrane-bound cytokine-specific antibodies to capture cytokines in biological fluids. Analysis was performed according to the manufacturer’s instructions. Cells were seeded in 10-cm culture dishes and maintained in serum-free medium for 24 hours. Briefly, the cytokine array membranes were blocked in 2 ml 1 × blocking buffer for 30 minutes and then were incubated with 1 ml of conditioned medium at 4°C overnight. The medium was then decanted from each container, and the membranes were washed three times with 2 ml 1 × wash buffer I, followed by two washes with 2 ml 1 × wash buffer II at room temperature with gentle shaking. Next, the membranes were incubated in biotin-conjugated primary antibodies (diluted 1:200) at 4°C overnight and were washed as described above before incubation in 1:1000-diluted horseradish peroxidase-conjugated streptavidin for 2 hours. The membranes were then washed thoroughly and incubated with a peroxidase substrate at room temperature for 5 min. Finally, the membranes were exposed to X-ray film. The film was scanned, and spots were digitized.
Chromatin immunoprecipitation (ChIP) assay
Chromatin was isolated from MDA-MB-231 cells using the ChIP assay kit (Millipore Inc.) and precipitated with anti-EZH2. Immunoprecipitated DNA was PCR-amplified with the two primer sets that cover specific regions of TIMP2 [11]. To amplify the DNA within a linear range of amplification, the thermal cycling conditions were as follows: 94°C for 5 minutes, 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, for 35 cycles. Alternatively, a serial dilution of input DNA was used. All results shown fell within a linear range. PCR products were analyzed on 2% agarose gels. Q-PCR analysis was performed by a LightCycler 480 II RTPCR system (Roche Applied Sciences, Manheim, Germany) using the Fast Start DNA Master Plus SYBR Green I kit (Roche Applied Sciences).
Gelatin zymography assay
The activity of MMP-2 and MMP-9 in conditioned medium from confluent TNBC cell cultures was determined with the gelatin zymography protease assay. Cells (6 × 105) were seeded in 6-well plates and maintained in serum-free medium for 24 hours and then were concentrated using an Amicon Ultra 10K centrifugation filter (Millipore). The conditioned medium was subsequently collected, cleared by centrifugation and mixed with 2 × SDS sample buffer, followed by electrophoresis on polyacrylamide gels containing 0.1% (w/v) gelatin. After electrophoresis, gels were washed twice for 15 min at room temperature with renaturing buffer (2.5% Triton X-100) with gentle agitation to remove the SDS and were then incubated in developing buffer (1.5 M Tris-HCl at pH 8.8, 5 mM CaCl2, and 0.02% NaN3) at room temperature with gentle agitation for 15 min. After removing the developing buffer, the gels were incubated with fresh developing buffer at 37°C for 16-20 hours. Finally, the gels were stained with 0.1% Coomassie Brilliant Blue for 1 hour and were destained with destaining solution (45% methanol and 10% acetic acid) until clear bands indicative of gelatin digestion appeared.
RNA isolation, RT-PCR and quantitative RT-PCR
Total RNA from cells was extracted with TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. The complementary DNA (cDNA) was synthesized from 2 µg total RNA in a reaction mixture containing oligo (dT) primer, 10 mM dNTP, 10 mM MML-V, RNase inhibitor, 0.1 M DDT, and 5 × first-strand buffer reverse transcriptase. The reaction mixture was incubated at 37°C for 50 minutes, 70°C for 15 minutes, and finally at 4°C until use. The PCR was performed in a reaction mixture containing 2 μl cDNA, 2.5 mM dNTP mixture, 2 μM of each specific primer, 1 U Taq DNA polymerase, and a 10-fold concentration of reaction buffer. Thermal cycling consisted of denaturation at 94°C for 5 minutes, followed by amplification of indicated cycles of 94°C for 30 seconds, 60°C for 30 seconds, and completion at 72°C for 30 seconds. The specific primer sequences for these genes are as follows: EZH2, forward, 5-CAGTAAAAATGTGTCCTGCAAGAA-3 and reverse, 5-TCAAGGGATTTCCATTTCTCTTTCGA-3; TIMP1, forward, 5-ATTCCGACCTCGTCATCAG-3 and reverse, 5-TGCAGTTTTCCAGCAATGAG-3; TIMP-2, forward, 5-TAGTGATCAGGGCCAAAGCG-3 and reverse, 5-TCTTTCCTCCAACGTCCAGC-3; TIMP3, forward, 5-CAAGATGTACACGGGGCTGT-3 and reverse, 5-AGGCGTAGTGTTTGGACTGG-3; TIMP4, forward, 5-CAGACCCTGCTGACACTGAA-3 and reverse, 5-AGGGCTCGATGTAGTTGCAC-3; GAPDH, forward, 5-ACCACAGTCCATGCCATCAC-3 and reverse, 5-TCCACCACCCTGTTGCTGTA-3. Then, 5 μl of each PCR product was run on a 1.6% agarose gel, and bands were visualized under UV light. GAPDH primers were used as an internal control and to confirm equal loading. Q-RT-PCR analysis was performed by a LightCycler 480 II RTPCR system (Roche Applied Sciences, Manheim, Germany) using the Fast Start DNA Master Plus SYBR Green I kit (Roche Applied Sciences). The specific primer sequences for these genes are as follows: EZH2, forward, 5-CAGTAAAAATGTGTCCTGCAAGAA-3 and reverse, 5-TCAAGGGATTTCCATTTCTCTTTCGA-3; TIMP-2, forward, 5-TAGTGATCAGGGCCAAAGCG-3 and reverse, 5-TCTTTCCTCCAACGTCCAGC-3; GAPDH, forward, 5-ACCACAGTCCATGCCATCAC-3 and reverse, 5-TCCACCACCCTGTTGCTGTA-3. Relative quantification of mRNA levels was conducted using the comparative C t method with GADPH as the reference gene and the formula 2-ΔΔCt.
Statistical analysis
Values are expressed as the mean ± SD and were analyzed using an ANOVA with the Bonferroni post hoc test to evaluate differences between multiple groups. All statistical analyses were performed using SPSS for Windows, version 10 (SPSS, Inc.). A value of P < 0.05 was considered to be statistically significant.
Results
EZH2 expression and its correlation with clinicopathologic characteristics
First, we checked the expression of EZH2 by immunohistochemistry (IHC) staining of a tissue array from US Biomax. This triple negative breast carcinoma tissue microarray, containing 42 cases of invasive ductal carcinoma, 2 cases of intraductal carcinoma and medullary carcinoma, 1 case of infiltrating lobular carcinoma and lobular ductal mixed carcinoma, single core per case. We used this array to evaluate the value of EZH2 in a clinical context and to verify whether EZH2 is positively correlated with tumor grade, metastasis, and invasiveness and thus could be a potential tumor biomarker for TNBCs. EZH2 expression was strongly associated with the TNBC phenotype as compared with normal tissues as the negative control (Figure 1). In addition, higher EZH2 expression was more highly associated with grade 3 rather than with grade 1 TNBC tumors.
Figure 1.
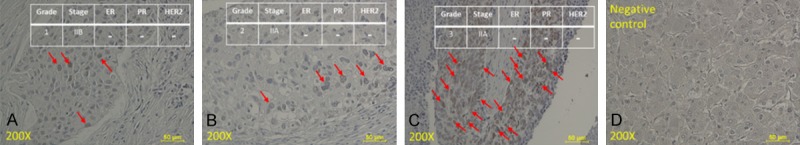
EZH2 expression in tissue samples from TNBC tumors of different grades. (A-D) TNBC tissue sections at different grades and stages are shown. Sections for (A) grade 1, (B) grade 2, (C) grade 3, and (D) normal tissue were immunohistochemically labeled with monoclonal mouse antibody EZH2 (1:2000 dilution). Red arrows indicate positive cells.
EZH2 expression in TNBC cells
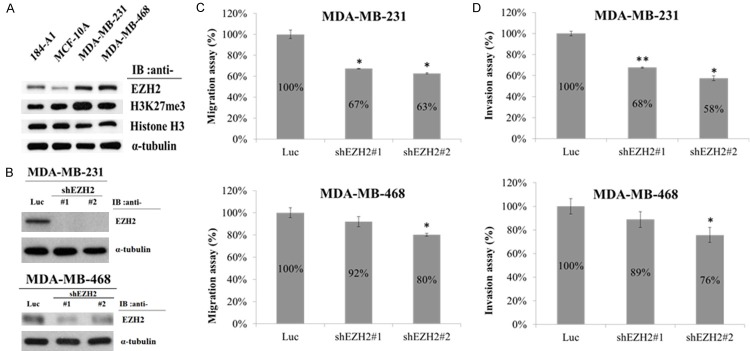
To investigate whether high levels of EZH2 expression are correlated with the invasive phenotype of TNBCs, we first determined the levels of EZH2 in human normal mammary epithelial cell lines (184-A1 and MCF-10A) and in two TNBC cell lines (MDA-MB-231 and MDA-MB-468). Western blot analysis showed that the expression of EZH2 and histone H3K27me3 protein levels were consistently elevated in breast cancer cells as compared with human normal mammary epithelial cells (Figure 2A).
Figure 2.
EZH2 expression and migration and invasion activities of TNBC cells after knockdown of EZH2. (A) Western blot analysis of EZH2 expression in the mammary epithelial cell lines 184-A1 and MCF-10A and the TNBC cell lines MDA-MB-231 and MDA-MB-468. α-Tubulin was used as the internal control. (B) Western blot analysis of EZH2 expression in MDA-MB-231 and MDA-MB-468 after infection with EZH2-specific shRNA (#1 and #2) or Luc-specific shRNA lentivirus. (C) Migration assay of MDA-MB-231 and MDA-MB-468 cells after infection with EZH2-specific shRNA (#1 and #2) or Luc-specific shRNA lentivirus. (D) Invasion assay of MDA-MB-231 and MDA-MB-468 cells after an infection with EZH2-specific shRNA (#1 and #2) or Luc-specific shRNA lentivirus. Values in (C and D) represent the mean ± SD from three independent experiments. *P < 0.05; **P < 0.01.
Suppression of metastasis activity of TNBCs after knockdown of EZH2
Overexpression of EZH2 in immortalized human mammary epithelial cell lines promotes anchorage-independent growth and cell invasion [12]. Here we investigated the functional role of EZH2 in the migration and invasion of breast cancer cell lines (MDA-MB-231 and MDA-MB-468). First we knockdown EZH2 with two different EZH2-specific shRNAs (#1 and #2) (Figure 2B). To examine the metastasis activity of TNBCs expressing EZH2 protein, we used a Transwell Boyden chamber assay. EZH2 expression was knocked down by using two different EZH2-specific shRNAs (#1 and #2), and the knockdown cells were compared with those cells infected with the control lentivirus (shLuc, non-targeting shRNA). EZH2 knockdown significantly decreased the migration and invasion ability of MDA-MB-231 and MDA-MB-468 cells (Figure 2C and 2D). These results suggest that high levels of EZH2 expression promote the invasion and migration of TNBCs.
Identification of the downstream target genes of EZH2 associated with metastasis of TNBCs
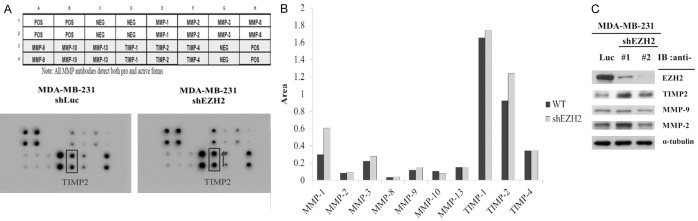
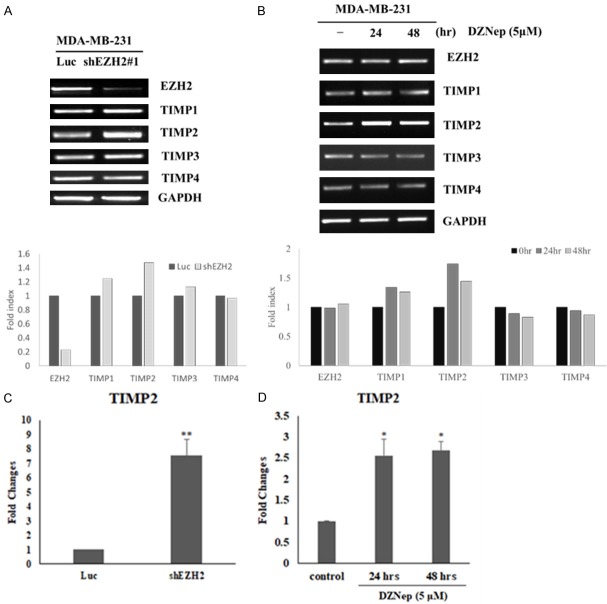
To identify metastasis-associated genes whose expression is regulated by EZH2, we investigated gene expression changes in invasive TNBCs after EZH2 knockdown. The expression of different cytokines in TNBCs was assessed by using a human cytokine antibody array. This array consists of antibodies against 10 proteins (MMP-1, MMP-2, MMP-3, MMP-8, MMP-9, MMP-10, MMP-13, TIMP-1, TIMP-2, and TIMP-4) that are involved in metastasis. Among these proteins, TIMP2 was found to be the protein that is upregulated in MDA-MB-231 cells by knocking down EZH2, thus suggesting that TIMP2 may be a primary target for repression by EZH2 (Figure 3A and 3B). Furthermore, the protein expression of TIMP2 was increased after knockdown of EZH2 (Figure 3C). To examine this hypothesis further, we investigated the transcriptional repression of TIMP2 by EZH2 and analyzed TIMP2 mRNA levels in MDA-MB-231 cells after EZH2 knockdown. The reverse transcription-PCR (RT-PCR) results showed that knockdown of EZH2 induced an increase in TIMP2 mRNA (Figure 4A). And furthermore the quantitative real-time reverse transcription PCR (Q-RT-PCR) results also showed that knockdown of EZH2 induced an increase in TIMP2 mRNA (Figure 4C).
Figure 3.
Identification of the downstream target protein of EZH2. A: Map of the RayBio Human Cytokine Antibody Array (upper) and results from the analysis of MDA-MB-231 cells that express the control (shLuc) or EZH2-specific (sh#2) shRNA (lower). B: Quantification of data from the human cytokine antibody array. TIMP1 and TIMP2 expression was highly affected after EZH2 knockdown in MDA-MB-231 cells. C: Western blot analysis of EZH2, TIMP2, MMP-9 and MMP-2 in MDA-MB-231 after knockdown of EZH2. α-tubulin was used as the internal control.
Figure 4.
TIMP2 expression in TNBC cells increases after EZH2 knockdown or inhibition of EZH2 activity. (A) RT-PCR analysis of mRNA for TIMPs (TIMP-1, TIMP-2, TIMP-3, and TIMP-4) in a TNBC cell line (MDA-MB-231) that expresses the control (shLuc) or EZH2-specific (sh#1) shRNA. (B) RT-PCR analysis of mRNA for TIMPs (TIMP-1, TIMP-2, TIMP-3, and TIMP-4) in MDA-MB-231 cells treated with DZNep (EZH2 inhibitor) for the indicated times. In (A and B) GAPDH was used as an internal control. (C) Q-PCR analysis of RNA levels expression of TIMP2 in the TNBCs (MDA-MB-231) after knockdown of EZH2 or control (Luc). TIMP2 was increased after knockdown EZH2. (D) Q-PCR analysis of RNA levels expression of TIMP2 in the TNBCs (MDA-MB-231) treat with DZNep (EZH2 inhibitor) during different time. TIMP2 was increased after treated with DZNep. Data represent the mean ± standard deviation from three independent experiments. *P < 0.05, **P < 0.01 relative to control.
PRC2-mediated histone methylation plays an important role in aberrant cancer gene silencing and is a potential target for cancer therapy. 3-Deazaneplanocin A (DZNep) induces efficient apoptotic cell death in cancer cells but not in normal cells. DZNep, a unique chromatin-remodeling compound, can deplete the cellular PRC2 proteins and inhibit their associated histone methylation, can selectively inhibit H3K27me3 and trimethylation of lysine 20 on histone H4 (H4K20me3), and can reactivate silenced genes in cancer cells [13]. EZH2 is a component of PRC2 that catalyzes H3K27me3. To explore the underlying mechanism of the epigenetic repression of TIMP2 by EZH2, we investigated the effect of treating breast cancer cells with the histone methylation inhibitor DZNep on TIMP2. DZNep treatment increased TIMP2 mRNA levels (Figure 4B and 4D). These results suggest that transcriptional repression of TIMP2 occurs through the EZH2-mediated H3K27 trimethylation.
Epigenetic silencing of TIMP2 expression by EZH2
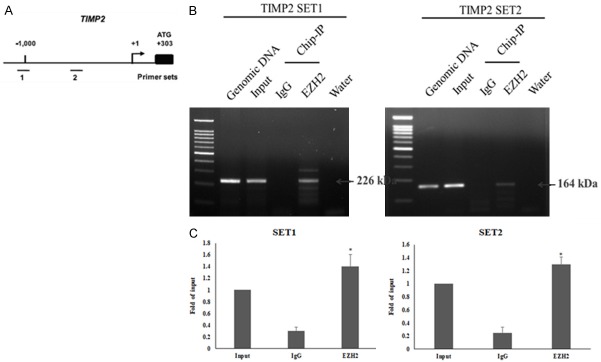
To understand further the molecular mechanism of the transcriptional repression of TIMP2 expression by EZH2, we performed chromatin immunoprecipitation (ChIP) assays in MDA-MB-231 cells using antibody against EZH2.
Because PcG proteins are recruited to DNA via the DNA-binding protein YY1, the immunoprecipitated DNA was analyzed by PCR with primer sets designed to amplify regions containing YY1-binding sites in the TIMP2 promoter [14] (Figure 5A). The results indicate that EZH2 binds to the TIMP2 promoter (Figure 5B and 5C). These observations suggest that in TNBC cells expression of TIMP2 and EZH2 is inversely correlated, probably because EZH2-mediated trimethylation of H3K27 from binding to the TIMP2 promoter and thereby results in transcriptional silencing of the gene.
Figure 5.
EZH2 binds to the promoter of TIMP2. A: Schematic representation of the promoter region of TIMP2. The bent arrow represents the transcription start site (+1). The lines below the TIMP2 locus represent the regions amplified by PCR using primer sets 1 and 2, which were designed to amplify regions containing YY1-binding sites. B: Chromatin immunoprecipitation (ChIP) experiments were performed with chromatin isolated from MDA-MB-231 cells using the two primer sets and antibodies against EZH2. C: Q-PCR analysis chromatin immunoprecipitation (ChIP) experiments were performed with chromatin isolated from MDA-MB-231 cells using the two primer sets and antibodies against EZH2. Data represent the mean ± standard deviation from three independent experiments. *P < 0.05 relative to control.
Transcriptional repression of TIMP2 by EZH2 promotes metastasis of TNBC cells
To further understand the functional link between the downregulation of TIMP2 by EZH2 and the decreased invasive phenotype of TNBC cells after EZH2 knockdown, we knocked down EZH2 and TIMP2 either individually or together in the MDA-MB-231 cell line and analyzed the migration and invasion of those cells using Transwell assays. EZH2 knockdown decreased the invasiveness of MDA-MB-231 cells, but both migration and invasion activities were significantly restored when we knocked down EZH2 and TIM-P2 (Figure 6A and 6B). These findings lend further support that repression of TIMP2 by EZH2 results in increased migration and invasion activities in TNBC cells.
Figure 6.
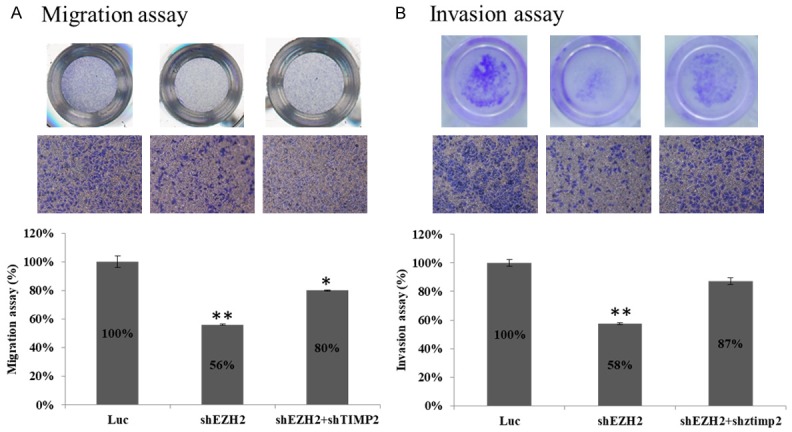
Down-regulation of TIMP2 expression restores metastasis activity after knockdown of EZH2 in TNBC cells. A: MDA-MB-231 cells were infected with either EZH2-specific sh#2 or with sh#2 and a TIMP2-specific shRNA. Luc shRNA served as a control. These cells were then used in Transwell migration and invasion assays. B: Migration and invasion abilities of the shRNA-expressing MDA-MB-231 cells were quantified by counting the number of cells on the outer surface of the polycarbonate membrane under the microscope. Each experiment was carried out in duplicate. Data represent the mean ± standard deviation from three independent experiments. *P < 0.05, **P < 0.01 relative to control.
The balance between MMPs and TIMPs is a critical parameter for the degradation of the ECM and thus is a crucial component of OR, which represents a crucial step in cancer cell invasion and metastasis. We determined mRNA levels for EZH2 and TIMP2 in MDA-MB-231 cells with knockdown of EZH2 and knockdown of EZH2/TIMP2 expression with RT-PCR and Q-PCR (Figure 7A and 7B). Furthermore, we investigated whether enzymatic activity of MMPs is regulated by EZH2 using a gelatin zymography assay. We found that MMP-9 and MMP-2 activity was decreased after knockdown of EZH2. However, activity of MMP-9 and MMP-2 was restore by double knockdown of EZH2 and TIMP2 (Figure 7C). These results support a mechanism whereby EZH2-mediated transcriptional repression of TIMP2 leads to activation of MMP-9 and MMP-2.
Figure 7.
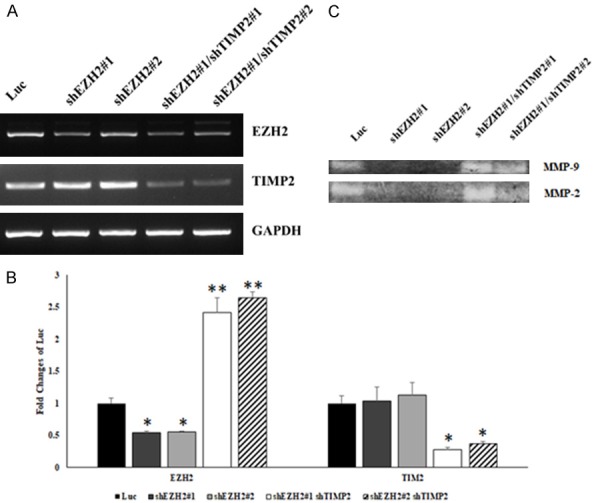
Downregulation of TIMP2 expression restores MMP2 and MMP9 activity after knockdown of EZH2 in TNBC cells. A: PCR analysis of expression of TIMP2 after knockdown of EZH2 and after double knockdown of EZH2 and TIMP2 in MDA-MB-231 cells. GAPDH was used as an internal control. B: Q-PCR analysis of expression of TIMP2 after knockdown of EZH2 and double knockdown of EZH2 and TIMP2 in MDA-MB-231 cells. C: Gelatin zymography assay for MMP-2 and MMP-9 activity in MDA-MB-231 cells after knockdown of EZH2 and double knockdown of EZH2 and TIMP2. Data represent the mean ± standard deviation from three independent experiments. *P < 0.05, **P < 0.01 relative to control.
Discussion
Tumor metastasis is the leading cause of death in cancer patients. Although the genetic basis of tumorigenesis can lead to great variability in metastasis across tumor types, the steps required for metastasis are similar for all tumor cells. Therefore, clinical therapies that block metastasis will be useful in treating cancers of various genetic origins. During the metastatic process, tumor cells need to attach to other cells and/or matrix proteins. Translocation of neoplastic cells across ECM barriers is also part of the metastatic process. The lysis of matrix proteins by specific proteinases is required for invasion [15]. The ratio of MMP and TIMP activities affects the degradation of the ECM and thus can affect cancer cell invasion and metastasis.
PcG proteins are transcriptional repressors that maintain the spatially restricted expression patterns of hox genes in both flies and vertebrates. These PcG proteins which regulate hox gene expression are important for skeletal development and hematopoiesis in mammals. PcG proteins mediate transcription repression via binding to conserved DNA sequence elements known as Polycomb response elements (PREs). PREs contain binding sites for the sequence-specific DNA-binding proteins Pleiohomeotic (PHO) and Pleiohomeotic-like (Phol), which are homologs of the ubiquitous mammalian transcription factor Yin Yang-1 (YY1) [14,16-22]. PcG recruitment functions via the multifunctional transcription factor YY1. This establishes DNA binding by YY1 as a key mechanism for targeting PcG proteins to DNA. The DNA binding by YY1 leads to the recruitment of PcG complexes, which cause deacetylation of histones and methylation of histone H3 at Lys9 and Lys27. Deacetylation could also be mediated by HDACs mediated directly by interaction with YY1 [14]. To identify metastasis-associated genes that are regulated by EZH2, we investigated gene expression changes in an invasive breast cancer cell line after EZH2 knockdown. TIMP2 was upregulated in these cells, suggesting that TIMP2 may be a primary target for repression by EZH2 (Figure 3A and 3B).
Control of gene expression perform at many different levels, one of which is the availability of genes and their control elements for transcriptional mechanisms. The accessibility is largely determined by the chromatin compactness, which is partly affected by the polycomb group proteins. EZH2, SUZ12, and EED form the PRC2, which catalyzes trimethylation of histone H3 lysine 27. PRC2 might recruit other polycomb complexes, DNA methyltransferases, and histone deacetylases, resulting in additional transcriptional inhibition marks and chromatin compactness at key developmental loci. EZH2 is important for transcriptional regulation via chromatin remodeling, nucleosome modification, and interaction with other transcription factors. EZH2 serves as a histone methyltransferase, and disruption of EZH2 expression may lead to the dysregulation of genes. Overexpression of EZH2 is a marker of advanced and metastatic disease in many solid tumors, including breast cancer, and is suggested as a candidate for targeted treatment. Excessive concentrations of EZH2 have been reported as a marker of invasive breast cancer, associated with invasion and cancer progression. Compared with normal or atypical hyperplasia, EZH2 levels in patients with invasive breast cancer increased [4,23,24].
Cancer cell migration inhibition has been commonly studied with the Transwell assay. For example, Luo et al. [25] found that overexpression of MicroRNA-497 inhibits breast cancer cellular migration and invasion. Liu et al. [26] also used the Transwell assay to show that inhibition of the long non-coding RNA HOTAIR by RNA interference decreased the migration and invasion of NSCLC. Here we used the Transwell assay to investigate the migration and invasion activities of MDA-MB-231 cells after knockdown of EZH2 or of EZH2 and TIMP2. We showed that EZH2 knockdown decreased the invasiveness of MDA-MB-231 cells, but the migration and invasion abilities of these cells were significantly restored when we knocked down both EZH2 and TIMP2 (Figure 6A and 6B). These data provide evidence that the repression of TIMP2 by EZH2 results in increased activity of migration and invasion in TNBC cells.
MMP-2 and MMP-9 have been well studied as proteins related to cancer cell migration and invasion. The zymography assay is a common method for detecting their activities. Wyrebska et al. [27] reported a new synthetic α-methylene-δ-lactone that is cytotoxic to and able to inhibit the migration and invasion activity of two breast cancer cell lines. This effect is based on its ability to decrease the secretion of enzymes responsible for the degradation of the extracellular matrix, MMP-9 and urokinase-type plasminogen activator (uPA), which was determined with the zymography assay. Zhu et al. [28] also used a zymography assay to demonstrate that alternol reduces the migration activity of a hepatocellular carcinoma cell line by suppressing MMP-9 activity. Here we used a zymography assay to show that knockdown of EZH2 notably inhibited the activities of MMP-2 and MMP-9 (Figure 7C), suggesting that lower amounts of EZH2 are associated with the inhibition of enzymatically degradative processes of cancer cell migration. We showed that aberrant upregulation of EZH2 expression in TNBC cells shifts the balance between TIMP and MMP activity toward MMP activity and thereby promotes degradation of the ECM. Therefore, the transcriptional repression of TIMP2 by EZH2, which acts as a potential oncogene in various malignancies, plays an active role in the upregulation of MMP activity and promotes ECM degradation and invasion and migration of TNBC cells. To our knowledge, this is the first study to demonstrate that knockdown of EZH2 reduces the biochemical basis of cell migration in TNBC cells.
DNA methylation, associated with histone deacetylation, is a common mechanism used by cancer cells to inhibit the expression of tumor suppressor genes and genes involved in tumor formation. Mucin genes encode large secreted O-glycoproteins that participate in mucus formation and play an important role as a physiological barrier against various intrusions from the underlying epithelia. The genes are located within a 400-kb cluster, on the p15.5 region of chromosome 11 in an area known to be a hot spot for abnormal methylation in cancer. The frequent occurrence of their silencing in cancer cells and the GC-rich structure of their promoters suggest the influence of epigenetics on their expression [29,30]. In addition, a novel molecular mechanism for the manipulation of epidermal growth factor receptor (EGFR)-PI3K-AKT signaling involves the formation of MUC15/EGFR heterodimers. MUC15 may exert its anti-metastatic capabilities by binding EGFR and accelerating EGFR internalization, thereby promoting EGFR degradation and inhibiting EGF-induced PI3K-AKT activation, which not only sheds new light on hepatocellular carcinoma (HCC) progression and metastasis but also provides a potential target for cancer prevention and treatment [31].
Our study thus suggests that overexpression of EZH2 in metastatic TNBC cells may elevate overall MMP activity both by decreasing the level of TIMP2 and by increasing the activity of some MMPs, such as MMP-2 and MMP-9. The ECM degradation by MMPs is of pivotal importance not only for eliminating the barriers to migration and invasion but also for providing breast cancer cells with access to signaling molecules. As demonstrated in this study, EZH2, which acts as a potential oncogene in various malignancies, plays an active role in the upregulation of MMP activity and promotes ECM degradation, providing an insight into the role of EZH2 in breast cancer metastasis (Figure 8). These findings may provide valuable information for the development of future therapies to more effectively prevent TNBC cell migration and invasion.
Figure 8.
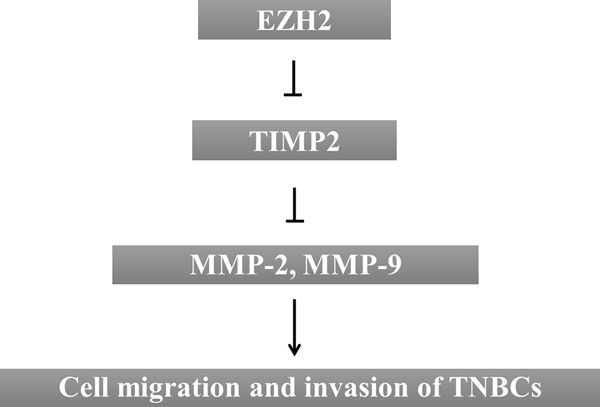
Proposed signaling pathways for EZH2-related modulation of TNBC cell migration and invasion abilities. Schematic model presents the transcriptional repression of TIMP-2 by EZH2, which plays an active role in the upregulation of MMP-2 and MMP-9 activity and promotes invasion and migration of TNBC cells.
Acknowledgements
This study was supported by grants from the Ministry of Science and Technology, Taiwan (MOST106-2320-B-039-051-MY3; MOST106-2320-B-039-048) and the Ministry of Health and Welfare (MOHW106-TDU-B-212-144-003).
Disclosure of conflict of interest
None.
References
- 1.Dawood S. Triple-negative breast cancer: epidemiology and management options. Drugs. 2010;70:2247–2258. doi: 10.2165/11538150-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232:142–150. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, Cristofanilli M, Hortobagyi GN, Pusztai L. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 4.Hussein YR, Sood AK, Bandyopadhyay S, Albashiti B, Semaan A, Nahleh Z, Roh J, Han HD, Lopez-Berestein G, Ali-Fehmi R. Clinical and biological relevance of enhancer of zeste homolog 2 in triple-negative breast cancer. Hum Pathol. 2012;43:1638–1644. doi: 10.1016/j.humpath.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, Brenner JC, Yu J, Kim JH, Han B, Tan P, Kumar-Sinha C, Lonigro RJ, Palanisamy N, Maher CA, Chinnaiyan AM. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Man S, Gao W, Zhang Y, Liu Z, Yan L, Huang L, Liu C. Formosanin C-inhibited pulmonary metastasis through repression of matrix metalloproteinases on mouse lung adenocarcinoma. Cancer Biol Ther. 2011;11:592–598. doi: 10.4161/cbt.11.6.14668. [DOI] [PubMed] [Google Scholar]
- 8.Lirdprapamongkol K, Kramb JP, Suthiphongchai T, Surarit R, Srisomsap C, Dannhardt G, Svasti J. Vanillin suppresses metastatic potential of human cancer cells through PI3K inhibition and decreases angiogenesis in vivo. J Agric Food Chem. 2009;57:3055–3063. doi: 10.1021/jf803366f. [DOI] [PubMed] [Google Scholar]
- 9.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 10.Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol. 2000;10:415–433. doi: 10.1006/scbi.2000.0379. [DOI] [PubMed] [Google Scholar]
- 11.Shin YJ, Kim JH. The role of EZH2 in the regulation of the activity of matrix metalloproteinases in prostate cancer cells. PLoS One. 2012;7:e30393. doi: 10.1371/journal.pone.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivasan L, Atchison ML. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 2004;18:2596–2601. doi: 10.1101/gad.1228204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodhouse EC, Chuaqui RF, Liotta LA. General mechanisms of metastasis. Cancer. 1997;80:1529–1537. doi: 10.1002/(sici)1097-0142(19971015)80:8+<1529::aid-cncr2>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Brown JL, Fritsch C, Mueller J, Kassis JA. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- 17.Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 18.Francis NJ, Kingston RE. Mechanisms of transcriptional memory. Nat Rev Mol Cell Biol. 2001;2:409–421. doi: 10.1038/35073039. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs JJ, van Lohuizen M. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602:151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- 20.Pirrotta V. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 1997;13:314–318. doi: 10.1016/s0168-9525(97)01178-5. [DOI] [PubMed] [Google Scholar]
- 21.Pirrotta V. PcG complexes and chromatin silencing. Curr Opin Genet Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- 22.Pirrotta V. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- 23.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo KH, Hennighausen L. EZH2 methyltransferase and H3K27 methylation in breast cancer. Int J Biol Sci. 2012;8:59–65. doi: 10.7150/ijbs.8.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo Q, Li X, Gao Y, Long Y, Chen L, Huang Y, Fang L. MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. Cancer Cell Int. 2013;13:95. doi: 10.1186/1475-2867-13-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyrebska A, Gach K, Lewandowska U, Szewczyk K, Hrabec E, Modranka J, Jakubowski R, Janecki T, Szymanski J, Janecka A. Anticancer activity of new synthetic alpha-methylene-delta-lactones on two breast cancer cell lines. Basic Clin Pharmacol Toxicol. 2013;113:391–400. doi: 10.1111/bcpt.12120. [DOI] [PubMed] [Google Scholar]
- 28.Zhu XL, Wang YL, Chen JP, Duan LL, Cong PF, Qu YC, Li-Ling J, Zhang MX. Alternol inhibits migration and invasion of human hepatocellular carcinoma cells by targeting epithelial-to-mesenchymal transition. Tumour Biol. 2014;35:1627–1635. doi: 10.1007/s13277-013-1224-y. [DOI] [PubMed] [Google Scholar]
- 29.Pallesen LT, Berglund L, Rasmussen LK, Petersen TE, Rasmussen JT. Isolation and characterization of MUC15, a novel cell membrane-associated mucin. Eur J Biochem. 2002;269:2755–2763. doi: 10.1046/j.1432-1033.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 30.Vincent A, Perrais M, Desseyn JL, Aubert JP, Pigny P, Van Seuningen I. Epigenetic regulation (DNA methylation, histone modifications) of the 11p15 mucin genes (MUC2, MUC5AC, MUC5B, MUC6) in epithelial cancer cells. Oncogene. 2007;26:6566–6576. doi: 10.1038/sj.onc.1210479. [DOI] [PubMed] [Google Scholar]
- 31.Wang RY, Chen L, Chen HY, Hu L, Li L, Sun HY, Jiang F, Zhao J, Liu GM, Tang J, Chen CY, Yang YC, Chang YX, Liu H, Zhang J, Yang Y, Huang G, Shen F, Wu MC, Zhou WP, Wang HY. MUC15 inhibits dimerization of EGFR and PI3K-AKT signaling and is associated with aggressive hepatocellular carcinomas in patients. Gastroenterology. 2013;145:1436–1448. e1–12. doi: 10.1053/j.gastro.2013.08.009. [DOI] [PubMed] [Google Scholar]