Abstract
Substantial evidence has shown that numerous microRNAs (miRNAs) are deregulated in colorectal cancer (CRC) and that their dysregulation is involved in CRC formation and progression. miRNA-based targeted therapy that inhibits or restores expression may be a promising therapeutic approach for anti-cancer therapy. Therefore, a comprehensive investigation of the mechanisms underlying CRC occurrence and development may help identify effective therapeutic targets for the therapy of CRC, thus improving the prognosis of patients with this disease. This study showed that miRNA-532 (miR-532) was significantly down-regulated in CRC tissues and cell lines. Low miR-532 expression strongly correlated with aggressive clinicopathological characteristics, including tumor size, lymphatic metastasis and TNM stage. Exogenous expression of miR-532 restricted cell proliferation, colony formation, migration and invasion; promoted cell apoptosis in vitro; and reduced tumor growth in vivo. Mechanistically, insulin-like growth factor 1 receptor (IGF-1R) was determined to be a novel direct target gene of miR-532 in CRC. In clinical CRC tissues, the expression of miR-532 was inversely correlated with that of IGF-1R, which was clearly overexpressed in CRC tissues. Furthermore, IGF-1R silencing simulated the tumor-suppressing roles of miR-532 in CRC. Moreover, recovered IGF-1R expression antagonized the inhibitory effects of miR-532 overexpression on CRC cells. Notably, miR-532 overexpression inhibited activation of the PI3K/Akt signaling pathway in CRC, both in vitro and in vivo. These results indicate that miR-532 plays an important role in CRC development, partly by directly targeting IGF-1R and regulating the PI3K/Akt signaling pathway. Thus, the miR-532/IGF-1R axis has clinical significance in the therapy of patients with CRC.
Keywords: Colorectal cancer, microRNA-532, insulin-like growth factor 1 receptor, PI3K/Akt pathway
Introduction
Colorectal cancer (CRC) is the third most frequently diagnosed cancer and the fourth most common cause of cancer-related deaths worldwide [1]. Approximately 1.36 million novel CRC cases and 694,000 mortalities are attributed to CRC annually worldwide [2]. Numerous risk factors for CRC were previously identified, including age, hereditary components, chronic intestinal inflammation, obesity, excessive intake of alcohol and red meat, smoking and lack of physical exercise [3-5]. Currently, surgery followed by radiotherapy and chemotherapy remains the primary therapeutic strategy for patients with CRC; however, 25%-30% of patients are diagnosed at advanced stages and are unsuitable for surgical resection [6]. Despite the advancement in comprehensive therapy, the long-term survival of CRC patients remains unsatisfactory [7]. The poor therapeutic outcome of patients with CRC is mainly due to local recurrence and distant metastases, particularly liver metastasis [8]. Therefore, a comprehensive understanding of the mechanisms underlying CRC onset and development is particularly urgent for the identification of novel therapeutic strategies for patients with this malignant tumor.
microRNAs (miRNAs) are a cluster of endogenously expressed, noncoding, single-stranded and short RNA molecules 19-24 nucleotides long [9]. miRNAs are implicated in gene regulation through imperfect or perfect binding to the 3’-untranslated regions (UTRs) of their target genes, resulting in translation inhibition and/or corresponding transcript degradation [10]. To date, more than 1400 mature miRNAs have been identified in the human genome, and bioinformatics and cloning studies predicted that these miRNAs interact with approximately 60% of all human protein-coding genes [11]. More than half of miRNAs are located at cancer-associated fragile sites and genomic regions, and this phenomenon indicates that miRNAs are involved in the pathogenesis of human cancers [12]. Recent studies reported that a variety of miRNAs, such as miR-30d [13], miR-145 [14], miR-455 [15] and miR-600 [16], are differentially expressed during CRC progression. Dysregulated miRNAs in human cancer may have tumor-suppressing or oncogenic roles, which mainly depend on the biological functions of their target genes [17]. These findings indicate that miRNAs are a new research focus for the development of effective diagnosis and therapy of patients with CRC.
miRNA-532 (miR-532) is differentially expressed in several human cancers, such as ovarian cancer [18,19], lung adenocarcinoma [20] and hepatocellular carcinoma [21]. However, the expression pattern and detailed roles of miR-532 in CRC remain unknown. Here, we report that miR-532 inactivates the PI3K/Akt signaling pathway by inhibiting IGF-1R expression and consequently plays tumor-suppressing roles in CRC progression.
Material and methods
Tissue specimens
This study was approved by the Ethics Committee of The Second Hospital of Jilin University. Written informed consent was also provided by all patients enrolled in this study. In total, 58 pairs of CRC tissues and corresponding adjacent normal tissues were collected from CRC patients who were treated with surgical resection at The Second Hospital of Jilin University between May 2014 and January 2017. None of the patients had received chemotherapy, radiotherapy, or other therapeutic strategies prior to surgery. All tissues were quickly frozen in liquid nitrogen after resection and stored at -80°C for further use.
Cell lines and culture conditions
The normal human colon epithelium cell line FHC and four human CRC cell lines (SW480, SW620, HCT116 and HT29) were purchased from American Type Culture Collection (Manassas, VA, USA). All cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% v/v fetal bovine serum (FBS) and 1% v/v penicillin/streptomycin mixture (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and were maintained at 37°C in a humidified incubator containing 5% CO2.
Oligonucleotides, siRNA, plasmids and transfection
miR-532 mimics, miRNA mimics negative control (miR-NC), small interfering RNAs (siRNAs) targeting the expression of IGF-1R (IGF-1R siRNA) and the negative control siRNA (NC siRNA) were chemically produced by GenePharma Co., Ltd. (Shanghai, China). The IGF-1R overexpressing pcDNA3.1-IGF-1R plasmid and empty pcDNA3.1 plasmid were acquired from GeneCopoeia (Guangzhou, China). For the transfection assays, cells were inoculated into six-well plates at 60%-70% confluence. The transfection was performed using Lipofectamine® 2000 (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. After transfection for 6 h, the culture medium was discarded and replaced with fresh DMEM containing 10% FBS.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA from clinical tissues or cells was extracted with TRIzol® reagent (Invitrogen Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA, USA) following the manufacturer’s protocol. For the detection of miR-532 expression, total RNA was reverse transcribed using the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Subsequently, quantitative polymerase chain reaction (qPCR) was performed with the TaqMan MicroRNA Assay Kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) on the Applied Biosystems 7500 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). To analyze the IGF-1R mRNA level, the PrimeScript RT Reagent Kit (Takara Bio, Dalian, China) was utilized to synthesize complementary DNA. Then, qPCR was performed with the SYBR Premix Ex Taq™ Kit (Takara Bio, Dalian, China). The relative expression levels of miR-532 and IGF-1R mRNA were calculated using the 2-ΔΔCt method [22] and were normalized to U6 snRNA and GAPDH, respectively.
Cell counting kit-8 (CCK-8) and colony formation assays
The CCK-8 assay was utilized to determine cell proliferation. In brief, the transfected cells were collected at 24 h post-transfection and prepared as a single-cell solution. A total of 3 × 103 cells were added to 96-well plates and cultured at 37°C in a humidified incubator for 0, 24, 48 and 72 h. At the end of culturing, 10 µL of CCK-8 solution (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added per well, and the cells were incubated at 37°C for an additional 2 h. Then, the absorbance value was detected at a wavelength of 450 nm using a SpectraMax Microplate® Spectrophotometer (Molecular Devices LLC, Sunnyvale, CA, USA).
The colony formation assay was used to determine colony formation. After transfection for 24 h, cells were harvested, resuspended and plated into six-well plates at a density of 1000 cells per well. The cells were incubated at 37°C in a humidified incubator for 14 days. On day 15, colony formation assays were performed, and surviving colonies (> 50 cells per colony) were counted following fixation with 4% paraformaldehyde and staining with methyl violet (Beyotime Institute of Biotechnology, Inc., Shanghai, China). All experiments were repeated at least three times.
Flow cytometric analysis
The cell apoptosis rate was evaluated using flow cytometric analysis. After transfection for 48 h, cells were harvested and washed with cold PBS. Cell apoptosis was assessed using the Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (Biolegend, San Diego, CA, USA) in accordance with the manufacturer’s protocol. In brief, transfected cells were resuspended in 100 μL of binding buffer, stained with 5 µL of Annexin V-FITC and 5 µL of propidium iodide at room temperature in the dark for 20 min and then analyzed using flow cytometry (FACScan; BD Biosciences, Bedford, MA, USA).
Transwell assay
The effects of miR-532 on CRC cell migration and invasion were assessed using 24-well 8-µm Transwell plates coated with or without Matrigel (both from BD Biosciences, Bedford, MA, USA), respectively. The lower chambers of the Transwell plates were covered with 500 µL of DMEM containing 20% FBS to serve as a chemoattractant. A total of 1 × 105 transfected cells resuspended in 200 µL of FBS-free DMEM were seeded into the upper chambers. After incubation at 37°C with 5% CO2 for 24 h (migration assay) or 36 h (invasion assay), the non-migrated or non-invaded cells were removed carefully from the upper surface of the membrane with a cotton swab. The migrated and invaded cells were fixed in 4% paraformaldehyde and stained with 0.5% crystal violet (Beyotime Institute of Biotechnology, Inc., Shanghai, China). Subsequent to washing with PBS and drying in air, the number of migrated or invaded cells was counted under an IX71 inverted microscope (Olympus Corporation, Tokyo, Japan) in five randomly selected visual fields from each chamber.
Xenograft experiment
For the in vivo tumor growth assay, eight four-week-old BALB/c nude mice were ordered from the Shanghai Laboratory Animal Center (Chinese Academy of Sciences, Shanghai, China). All experiments involving animals were approved by the Ethics Review Committee of The Second Hospital of Jilin University. All nude mice were maintained under special pathogen-free conditions and randomly divided into two groups (n = 4 mice per group). A total of 5 × 106 SW480 cells transfected with miR-532 mimics or miR-NC were suspended in 100 µL of culture medium and subcutaneously injected into the dorsal flank of each nude mouse. At 2 weeks after inoculation, the xenograft tumor size was measured using vernier calipers every 2 days, and the tumor volume was calculated according to the following formula: 1/2 × tumor length × tumor width. Mice were sacrificed 30 days after inoculation. The tumor xenografts were excised, weighed, frozen in liquid nitrogen and then stored at -80°C for further analysis.
miR-532 target prediction
Bioinformatics algorithms, TargetScan7.1 (http://www.targetscan.org/) and microRNA.org (http://www.microrna.org/microrna/home.do), were utilized to predict the potential targets of miR-532.
Luciferase reporter assay
The 3’-UTR of IGF-1R containing the wild-type (Wt) miR-532 putative binding sites and mutant (Mut) binding sites was designed and chemically synthesized by GenePharma Co., Ltd. and inserted into the pGL3 luciferase vector (Promega Corporation, Madison, WI, USA) to construct pGL3-IGF-1R-3’-UTR Wt and pGL3-IGF-1R-3’-UTR Mut, respectively. Cells were inoculated in triplicate in 24-well plates and allowed to settle for 24 h. Next, the cells were co-transfected with miR-532 mimics or miR-NC and with pGL3-IGF-1R-3’-UTR Wt or pGL3-IGF-1R-3’-UTR Mut using Lipofectamine® 2000 in accordance with the manufacturer’s protocol. Then, the cells were harvested at 48 h post-transfection and the relative luciferase activities were measured using the Dual-Luciferase Reporter Assay Kit (Promega Corporation, Madison, WI, USA) and normalized to those of Renilla luciferase activities.
Western blot analysis
The primary antibodies used in this study included mouse anti-human monoclonal IGF1R antibody (sc-81464; 1:1000 dilution; Santa Cruz Biotechnology, CA, USA), rabbit anti-human monoclonal p-pi3k antibody (ab182651; 1:1000 dilution; Abcam, Cambridge, UK), mouse anti-human monoclonal pi3k antibody (ab86714; 1:1000 dilution; Abcam, Cambridge, UK), mouse anti-human monoclonal p-Akt antibody (sc-81433; 1:1000 dilution; Santa Cruz Biotechnology, CA, USA), mouse anti-human monoclonal Akt antibody (sc-56878; 1:1000 dilution; Santa Cruz Biotechnology, CA, USA), and mouse anti-human monoclonal GAPDH antibody (sc-66163; 1:1000 dilution; Santa Cruz Biotechnology, CA, USA). Transfected cells or homogenized tissues were lysed in radioimmunoprecipitation buffer, and the total protein concentration was evaluated using the Bicinchoninic Acid Assay Kit (Beyotime Institute of Biotechnology, Inc., Shanghai, China) according to the manufacturer’s instructions. Equal amounts of protein were loaded, separated by 10% SDS-PAGE electrophoresis and then transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA), followed by blocking at room temperature in 5% non-fat dry milk dissolved in Tris-buffered saline containing 0.1% Tween 20 (TBST) for 1 h. Then, the membranes were incubated overnight at 4°C with the primary antibodies. On the second day, the membranes were washed three times with TBST and further probed with the corresponding horseradish-peroxidase-conjugated secondary antibodies (Abcam, Cambridge, UK) at room temperature for 2 h. Finally, the protein bands were visualized by an enhanced chemiluminescence plus reagent (GE Healthcare, Chicago, IL, USA) in accordance with the manufacturer’s recommendation. GAPDH served as a control for normalization.
Statistical analysis
All data were presented as the mean ± standard deviation and analyzed with the Statistical Package for Social Sciences (version 19.0; IBM SPSS, Inc., Armonk, NY, USA). The chi-square test was used to assess the relationship between expression levels of miR-532 and various clinicopathological characteristics of CRC. The association between the miR-532 and IGF-1R mRNA level was determined using Spearman’s correlation analysis. Student’s t test or one-way ANOVA plus multiple comparisons combined with Tukey’s post hoc test was utilized to evaluate the difference between groups. A value of P < 0.05 was considered statistically significant.
Results
miR-532 expression is down-regulated in CRC tissues and cell lines
To determine the role of miR-532 in CRC carcinogenesis, miR-532 expression in 58 pairs of CRC tissues and the corresponding adjacent normal tissues was analyzed using RT-qPCR. The results showed that miR-532 expression was significantly decreased in CRC tissues relative to the corresponding adjacent normal tissues (P < 0.05; Figure 1A). The expression levels of miR-532 in four CRC cell lines and the normal human colon epithelium cell line FHC were examined to confirm this observation. The RT-qPCR analysis showed that miR-532 expression was lower in the CRC cell lines than in FHC (P < 0.05; Figure 1B). Altogether, these results indicate that miR-532 expression is decreased in CRC and that its down-regulation may be associated with CRC development.
Figure 1.
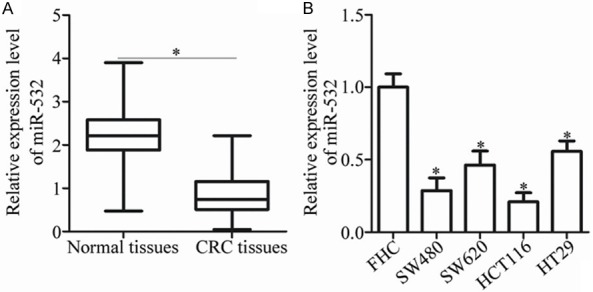
miR-532 expression is down-regulated in CRC tissues and cell lines. A. Total RNA was extracted from 58 pairs of CRC tissues and the corresponding adjacent normal tissues and subjected to RT-qPCR for the detection of miR-532 expression. *P < 0.05 vs. adjacent normal tissues. B. RT-qPCR analysis was utilized to detect miR-532 expression in four CRC cell lines and the normal human colon epithelium cell line FHC. *P < 0.05 vs. FHC.
Down-regulation of miR-532 is correlated with the adverse clinicopathological characteristics of CRC
We further investigated the relationship between miR-532 expression and the clinicopathological features of CRC patients to explore the clinical significance of decreased miR-532 expression in CRC. All patients were subdivided into either miR-532 low (n = 29) or high (n = 29) expression groups according to the median expression of miR-532. Low miR-532 expression was strongly correlated with the tumor size (P = 0.017), lymphatic metastasis (P = 0.034) and TNM stage (P = 0.007) of CRC. However, it showed no significant association with gender, age, or tumor differentiation (all P > 0.05; Table 1). Therefore, miR-532 expression may serve as a prognostic biomarker for patients with CRC.
Table 1.
The relationship between miR-532 and clinicopathological factors of colorectal cancer patients
| Clinicopathologic factors | miR-532 low group (n = 29) | miR-532 high group (n = 29) | P |
|---|---|---|---|
| Gender | 0.565 | ||
| Male | 19 | 22 | |
| Female | 10 | 7 | |
| Age (years) | 0.141 | ||
| < 55 | 5 | 11 | |
| ≥ 55 | 24 | 18 | |
| Tumor differentiation | 0.592 | ||
| Well and Moderate | 13 | 10 | |
| Poor | 16 | 19 | |
| Tumor size (cm) | 0.017 | ||
| < 5 | 8 | 18 | |
| ≥ 5 | 21 | 11 | |
| Lymphatic metastasis | 0.034 | ||
| Absence | 11 | 20 | |
| Presence | 18 | 9 | |
| TNM stage | 0.007 | ||
| I-II | 6 | 17 | |
| III-IV | 23 | 12 |
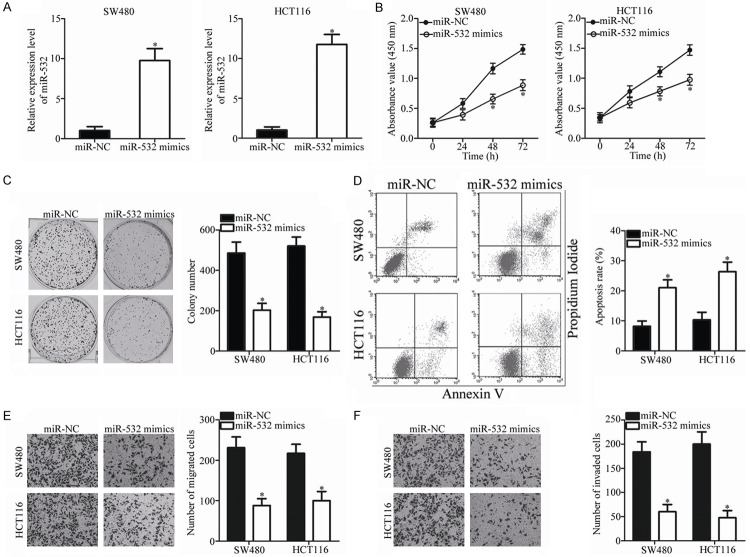
miR-532 attenuates the proliferation, migration and invasion and promotes the apoptosis of CRC cells
To observe the role of miR-532 in CRC, SW480 and HCT116 cells, which exhibited relatively lower endogenous miR-532 expression among the four CRC cell lines, were selected for the subsequent experiments and transfected with miR-532 mimics or miR-NC. After transfection for 48 h, significant overexpression of miR-532 was confirmed by RT-qPCR analysis (P < 0.05; Figure 2A). After confirming the efficacy of transfection, the CCK-8 assay was used to examine the effect of miR-532 overexpression on CRC cell proliferation. As shown in Figure 2B, the proliferation of SW480 and HCT116 cells transduced with miR-532 mimics was significantly suppressed compared with that in cells transfected with miR-NC (P < 0.05). Similarly, transfection with miR-532 mimics significantly decreased the number and size of surviving colonies (P < 0.05; Figure 2C). The effect of miR-532 overexpression on CRC cell apoptosis was determined using flow cytometric analysis. The results indicated that miR-532 up-regulation significantly increased the apoptosis rate of SW480 and HCT116 cells (P < 0.05; Figure 2D). Furthermore, Transwell assays showed that exogenous miR-532 expression significantly reduced the migration (P < 0.05; Figure 2E) and invasion (P < 0.05; Figure 2F) of SW480 and HCT116 cells. Together, these results indicate that miR-532 plays a tumor-suppressing role in CRC progression.
Figure 2.
miR-532 inhibits proliferation, colony formation, migration and invasion and promotes the apoptosis of SW480 and HCT116 cells. A. The expression level of miR-532 was determined in SW480 and HCT116 cells after transfection with miR-532 mimics or miR-NC for 48 h. *P < 0.05 vs. miR-NC. B, C. Ectopic expression of miR-532 reduced the proliferation and colony formation capabilities of SW480 and HCT116 cells. *P < 0.05 vs. miR-NC. D. The apoptosis rate was increased in miR-532-mimics-transfected SW480 and HCT116 cells in comparison with the miR-NC-transfected cells. *P < 0.05 vs. miR-NC. E, F. The number of migrated and invaded cells was significantly decreased after miR-532 restoration in SW480 and HCT116 cells. *P < 0.05 vs. miR-NC.
IGF-1R is a direct target of miR-532 in CRC cells
We searched for potential target genes of miR-532 using bioinformatics analysis to identify the mechanisms responsible for the tumor-suppressing effects of miR-532 on CRC. IGF-1R, a well-known oncogene, is predicted as a major target of miR-532 and attracted our attention because of its important roles in CRC initiation and progression [23-27]. The binding regions between the miR-532 and the 3’-UTR of IGF-1R are shown in Figure 3A. Luciferase reporter assays were performed on SW480 and HCT116 cells, which were co-transfected with miR-532 mimics or miR-NC and pGL3-IGF-1R-3’-UTR Wt or pGL3-IGF-1R-3’-UTR Mut to confirm this hypothesis. Restoring the expression of miR-532 significantly decreased the luciferase activities of the luciferase plasmid harboring the Wt binding sites (P < 0.05), whereas mutation of the miR-532 binding site blocked this suppressive effect (Figure 3B).
Figure 3.
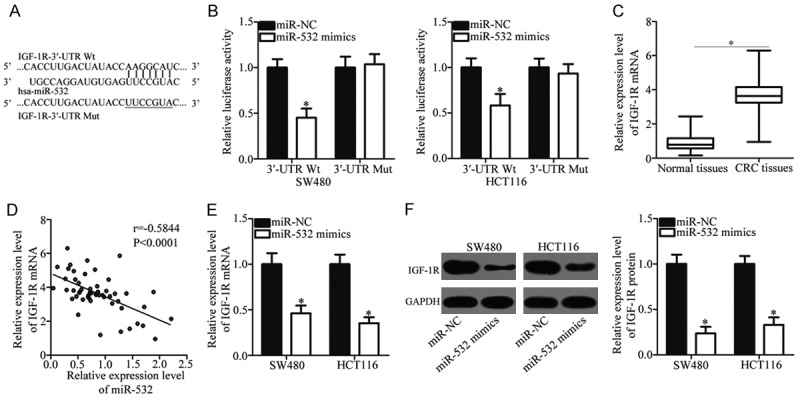
IGF-1R is a direct target of miR-532 in CRC. A. Putative wild-type (Wt) and mutant (Mut) miR-532 binding sites in the 3’-UTR of IGF-1R. B. Relative luciferase activities were analyzed in SW480 and HCT116 cells cotransfected with Wt or Mut reporter plasmids and miR-532 mimics or miR-NC. *P < 0.05 vs. miR-NC. C. RT-qPCR analysis of IGF-1R mRNA expression in 58 pairs of CRC tissues and corresponding adjacent normal tissues. *P < 0.05 vs. adjacent normal tissues. D. Spearman’s correlation analysis was performed to evaluate the relationship between miR-532 and IGF-1R mRNA expression in CRC tissues. r = -0.5844, P < 0.0001. E, F. IGF-1R expression at the mRNA and protein levels in SW480 and HCT116 cells after transfection with miR-532 mimics or miR-NC was determined by RT-qPCR and western blot analysis. *P < 0.05 vs. miR-NC.
We measured the expression of IGF-1R in 58 pairs of CRC tissues and the corresponding adjacent normal tissues using RT-qPCR to further investigate the association between miR-532 and IGF-1R in CRC. As shown in Figure 3C, the mRNA level of IGF-1R was increased in CRC tissues compared with the adjacent normal tissues (P < 0.05). Furthermore, a strong inverse relationship between miR-532 and IGF-1R mRNA levels in CRC tissues was observed using Spearman’s correlation analysis (r = -0.5844, P < 0.0001; Figure 3D). Moreover, we detected IGF-1R expression at the mRNA and protein levels in SW480 and HCT116 cells after transfection with miR-532 mimics or miR-NC. The results showed that IGF-1R mRNA (P < 0.05; Figure 3E) and protein (P < 0.05; Figure 3F) expression levels were down-regulated in the SW480 and HCT116 cells transfected with miR-532 mimics compared with cells transfected with miR-NC. In summary, these results indicate that IGF-1R is a direct target of miR-532 in CRC cells.
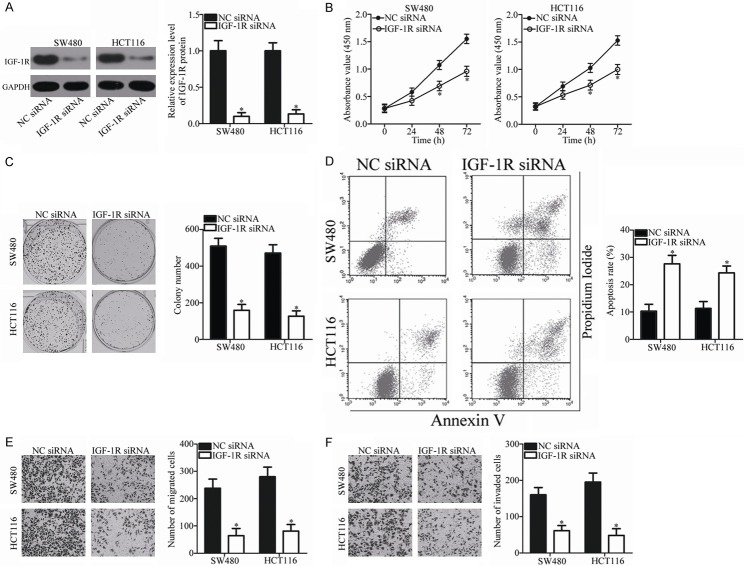
Silencing of IGF-1R simulates the tumor-suppressing roles of miR-532 in CRC
Because IGF-1R is a direct target of miR-532 in CRC, we hypothesized that the tumor-suppressing roles of miR-532 in CRC cells could be imitated by IGF-1R knockdown. To confirm this hypothesis, we silenced IGF-1R expression in SW480 and HCT116 cells by transfection with IGF-1R siRNA. Western blot analysis was used to evaluate the siRNA-mediated knockout efficiency in SW480 and HCT116 cells. As shown in Figure 4A, the protein level of IGF-1R was silenced effectively in SW480 and HCT116 cells after transfection with IGF-1R siRNA (P < 0.05). Similar to miR-532 restoration, IGF-1R knockdown decreased proliferation (P < 0.05; Figure 4B) and colony formation (P < 0.05; Figure 4C) and promoted apoptosis (P < 0.05; Figure 4D) of SW480 and HCT116 cells. Additionally, inhibition of IGF-1R decreased the migration (P < 0.05; Figure 4E) and invasion (P < 0.05; Figure 4F) of SW480 and HCT116 cells. These results further indicate that IGF-1R is a direct functional target of miR-532 in CRC.
Figure 4.
Down-regulation of IGF-1R suppresses the proliferation, colony formation, migration and invasion and increases the apoptosis of SW480 and HCT116 cells. A. Transfection with IGF-1R siRNA efficiently knocked down IGF-1R expression in SW480 and HCT116 cells. *P < 0.05 vs. NC siRNA. B, C. CCK-8 and colony formation assays were conducted to examine the effects of IGF-1R knockdown on the proliferation and colony formation of SW480 and HCT116 cells. *P < 0.05 vs. NC siRNA. D. Flow cytometric analysis was conducted to investigate the effect of IGF-1R knockdown on SW480 and HCT116 cell apoptosis. *P < 0.05 vs. NC siRNA. E, F. Effect of IGF-1R knockdown on the migration and invasion of SW480 and HCT116 cells was assessed using Transwell assay. *P < 0.05 vs. NC siRNA.
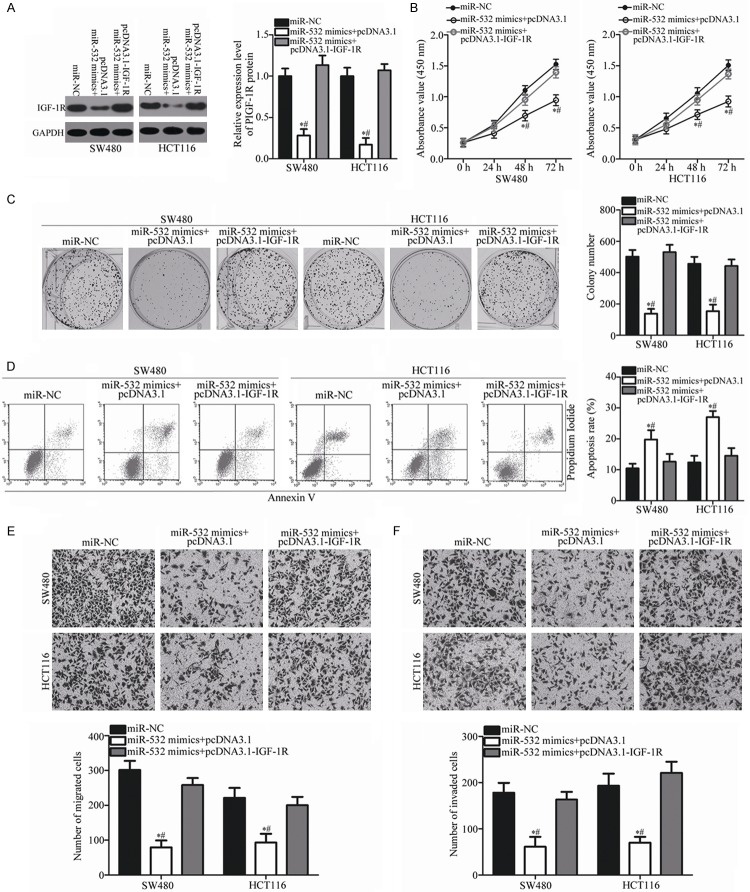
Restored IGF-1R expression rescues the effects of miR-532 overexpression on CRC cells
A series of rescue experiments were performed to further clarify whether IGF-1R mediates the function of miR-532 in CRC cells. pcDNA3.1-IGF-1R or pcDNA3.1 was transfected into SW480 and HCT116 cells in the presence of miR-532 mimics. Western blot analysis showed that the decreased level of IGF-1R caused by miR-532 overexpression was restored in SW480 and HCT116 cells after co-transfection with pcDNDA3.1-IGF-1R (P < 0.05; Figure 5A). Functional experiments indicated that the effects of miR-532 overexpression on the proliferation (P < 0.05; Figure 5B), colony formation (P < 0.05; Figure 5C), apoptosis (P < 0.05; Figure 5D), migration (P < 0.05; Figure 5E), and invasion (P < 0.05; Figure 5F) of SW480 and HCT116 cells were significantly rescued by re-expressing exogenous IGF-1R. These results indicate that miR-532 exerts tumor-suppressing roles in CRC by directly targeting and inhibiting IGF-1R.
Figure 5.
Tumor-suppressing roles of miR-532 on CRC cells are rescued by IGF-1R overexpression. SW480 and HCT116 cells were cotransfected with miR-532 mimics and pcDNA3.1-IGF-1R or pcDNA3.1. After incubation for different times, transfected cells were used in the subsequent assays. A. At 72 h post-transfection, the protein level of IGF-1R in the indicated cells was measured using western blot analysis. *P < 0.05 vs. miR-NC. #P < 0.05 vs. miR-532 mimics + pcDNA3.1-IGF-1R. B-F. The CCK-8 assay, colony formation assay, flow cytometric analysis and Transwell assay were applied to determine the proliferation, colony formation, apoptosis, migration, and invasion of the previously described cells. *P < 0.05 vs. miR-NC. #P < 0.05 vs. miR-532 mimics + pcDNA3.1-IGF-1R.
miR-532 inhibits the PI3K/Akt signaling pathway in CRC by targeting IGF-1R
IGF-1R was previously implicated in the PI3K/Akt signaling pathway [28-30]. To explore whether miR-532 contributes to the regulation of the PI3K/Akt pathway, western blot analysis was used to detect the expression levels of p-PI3K, PI3K, p-Akt and Akt in SW480 and HCT116 cells after co-transfection with miR-532 mimics and pcDNA3.1-IGF-1R or pcDNA3.1. The cellular protein levels of p-PI3K and p-Akt were significantly reduced in SW480 and HCT116 cells expressing miR-532 mimics. However, the changes in PI3K and Akt were not statistically significant. Co-transfection with pcDNA3.1-IGF-1R also restored the miR-532-inhibited cellular levels of p-PI3K and p-Akt (Figure 6). These data indicate that miR-532 inactivates the PI3K/Akt signaling pathway in CRC by targeting IGF-1R.
Figure 6.
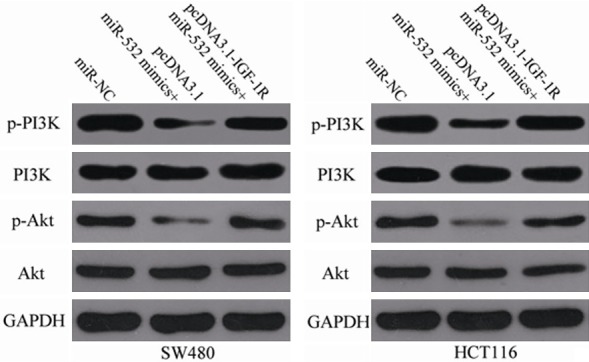
miR-532 inhibits the PI3K/Akt signaling pathway in CRC by targeting IGF-1R. SW480 and HCT116 cells were transfected with miR-NC, miR-532 mimics + pcDNA3.1 or miR-532 mimics + pcDNA3.1-IGF-1R. After transfection for 72 h, the expression levels of p-PI3K, PI3K, p-Akt and Akt were measured with western blot analysis.
miR-532 inhibits tumor growth of CRC in vivo by targeting IGF-1R
SW480 cells transfected with miR-532 mimics or miR-NC were subcutaneously inoculated into nude mice. The xenograft tumor volumes were detected every 2 days at 2 weeks after inoculation. Nude mice were sacrificed on day 30 following inoculation, and xenografts were excised and weighed. The tumor weight of the miR-532 mimics group was significantly reduced compared with the miR-NC group (P < 0.05; Figure 7A). The tumor volume of the miR-532 mimics group was also significantly smaller than the miR-NC group (P < 0.05; Figure 7B and 7C). Furthermore, the total RNA and protein were isolated from the xenograft tumor tissues. Subsequent RT-qPCR analysis indicated that the expression level of miR-532 was significantly higher in the miR-532 mimics group than the miR-NC group (P < 0.05; Figure 7D). The western blot analysis indicated that the up-regulation of miR-532 decreased the expression levels of IGF-1R, p-PI3K and p-Akt in vivo (Figure 7E). These results indicate that miR-532 inhibits CRC tumor growth in vivo by targeting IGF-1R and regulating the PI3K/Akt signaling pathway.
Figure 7.
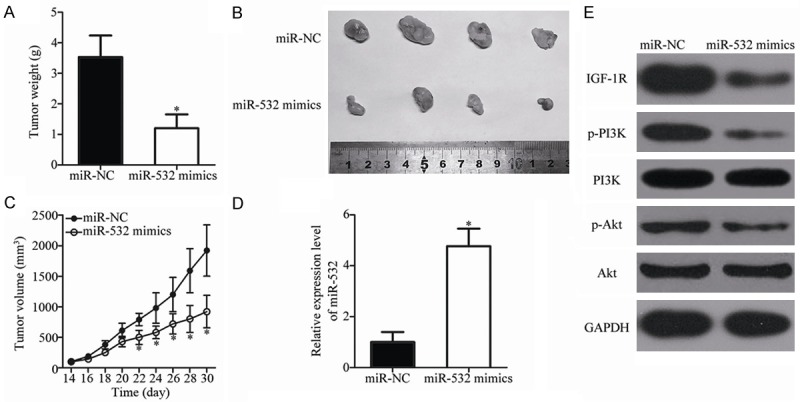
miR-532 inhibits CRC tumor growth in vivo by repressing IGF-1R. A. Mice were sacrificed, and the weights of the xenograft tumors from the miR-532 mimics and miR-NC groups were weighed. *P < 0.05 vs. miR-NC. B. Representative images of the miR-532 mimics and miR-NC xenograft tumors. C. The volume of the xenograft tumor from the miR-532 mimics and miR-NC groups was calculated every 2 days after inoculation for 2 weeks. *P < 0.05 vs. miR-NC. D. Expression level of miR-532 was detected in xenograft tumor tissues from the miR-532 mimics and miR-NC groups. *P < 0.05 vs. miR-NC. E. Relative protein levels of IGF-1R, p-PI3K, PI3K, p-Akt and Akt were determined in xenograft tumor tissues from the miR-532 mimics and miR-NC groups.
Discussion
Substantial evidence has shown that numerous miRNAs are deregulated in CRC and that their dysregulation is involved in CRC formation and progression [31-33]. miRNA-based targeted therapy that inhibits or restores expression may also provide a promising therapeutic approach for anti-cancer therapy [34]. Therefore, a comprehensive investigation of the mechanisms underlying CRC occurrence and development may help to identify effective therapeutic targets for CRC therapy, thereby improving the prognosis of patients with this disease. Here, we identified that miR-532 expression was significantly down-regulated in CRC tissues and cell lines. Reduced miR-532 expression was strongly correlated with aggressive clinicopathological characteristics, including tumor size, lymphatic metastasis and TNM stage. Functional experiments also indicated that exogenous miR-532 expression inhibited CRC cell proliferation, migration and invasion; induced cell apoptosis in vitro; and decreased tumor growth in vivo. Furthermore, IGF-1R was validated as a novel target gene of miR-532 in CRC. Notably, the up-regulation of miR-532 inactivated the PI3K/Akt signaling pathway in CRC in vitro and in vivo. Thus, miR-532 may serve as a tumor suppressor in CRC by directly targeting IGF-1R and regulating the PI3K/Akt pathway, indicating that miR-532 may be a novel promising therapeutic target for CRC patients.
miR-532 is differentially expressed in several types of human cancers. For example, miR-532 is down-regulated in ovarian cancer. Ovarian cancer patients with low miR-532 levels exhibit a worse therapeutic outcome than patients with high miR-532 levels [18,19]. miR-532 is also weakly expressed in the tissues and plasma of lung adenocarcinoma [20]. However, miR-532 is overexpressed in hepatocellular carcinoma tissues and cell lines [21]. Aberrantly highly expressed miR-532 is also observed in triple-negative breast cancer [35] and gastric cancer [36,37]. These conflicting findings indicate that the expression pattern of miR-532 in human cancers exhibits tissue specificity and that miR-532 may be a new and effective non-invasive approach for the diagnosis of patients with these specific tumor types.
Dysregulation of miR-532 is closely associated with tumorigenesis and tumor development in diverse human malignancies. For example, ectopic miR-532 expression suppresses cell proliferation, invasion and the epithelial-mesenchymal transition in ovarian cancer [18]. Griesing et al. determined that miR-532 overexpression increases cell apoptosis in lung adenocarcinoma cells [38]. Nevertheless, Song et al. reported that miR-532 plays oncogenic roles in hepatocellular carcinoma by promoting cell growth and metastasis [21]. Hu and Xu et al. reported that restoration of miR-532 expression promotes the colony formation and motility of gastric cancer cells in vitro, inhibits cell apoptosis and cell cycle arrest in vitro and enhances the lung metastasis of gastric cancer in vivo [36,37]. These observations in dicate that the biological functions of miR-532 in carcinogenesis and cancer progression are tissue specific and that miR-532 has potential as an effective target for cancer therapy.
Multiple direct targets of miR-532 have been identified, including hTERT [18] in ovarian cancer, CXCL2 [21] in hepatocellular carcinoma, KRAS [38] and MKL2 [38] in lung adenocarcinoma and NKD1 [36] and RUNX3 [37] in gastric cancer. In the present study, IGF-1R was identified as a novel target of miR-532 in CRC. IGF-1R, a transmembrane tyrosine kinase receptor of the insulin receptor family, is up-regulated in multiple types of human cancer, including bladder cancer [39], nasopharyngeal carcinoma [40], osteosarcoma [41], lung cancer [42] and endometrial cancer [43]. IGF-1R activates multiple downstream signaling cascades, including the PI3K/Akt and MAPK/ERK signaling pathways, and participates in the regulation of cell growth, cycle, apoptosis, angiogenesis, invasion and metastasis [42,44-46]. Moreover, IGF-1R is overexpressed in CRC tissues and cell lines, and its overexpression is significantly correlated with tumor size, depth of invasion, venous invasion and liver metastasis [23,24]. Abnormal activation of IGF-1R contributes to CRC cell growth, apoptosis, migration, metastasis and radiotherapy sensitivity [25-27]. Given the importance of IGF-1R in CRC, the miR-532/IGF-1R pathway may provide novel therapeutic opportunities for treating this aggressive cancer.
In conclusion, this study showed that miR-532 is significantly down-regulated in CRC tissues and cell lines. This miRNA can potently inhibit CRC cell proliferation and metastasis, increase cell apoptosis in vitro and reduce tumor growth in vivo by directly targeting IGF-1R and inactivating the PI3K/Akt signaling pathway. These results suggest that miR-532 is a new therapeutic target for CRC patients.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Andrews L. Dietary flavonoids for the prevention of colorectal cancer. Clin J Oncol Nurs. 2013;17:671–672. doi: 10.1188/13.CJON.671-672. [DOI] [PubMed] [Google Scholar]
- 4.Altobelli E, Lattanzi A, Paduano R, Varassi G, di Orio F. Colorectal cancer prevention in Europe: burden of disease and status of screening programs. Prev Med. 2014;62:132–141. doi: 10.1016/j.ypmed.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Sugarbaker PH. Colorectal cancer: prevention and management of metastatic disease. Biomed Res Int. 2014;2014:782890. doi: 10.1155/2014/782890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheer A, Auer RA. Surveillance after curative resection of colorectal cancer. Clin Colon Rectal Surg. 2009;22:242–250. doi: 10.1055/s-0029-1242464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22:191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies RJ, Miller R, Coleman N. Colorectal cancer screening: prospects for molecular stool analysis. Nat Rev Cancer. 2005;5:199–209. doi: 10.1038/nrc1569. [DOI] [PubMed] [Google Scholar]
- 9.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R, Xu J, Zhao J, Bai J. Mir-30d suppresses cell proliferation of colon cancer cells by inhibiting cell autophagy and promoting cell apoptosis. Tumour Biol. 2017;39:1010428317703984. doi: 10.1177/1010428317703984. [DOI] [PubMed] [Google Scholar]
- 14.Sheng N, Tan G, You W, Chen H, Gong J, Chen D, Zhang H, Wang Z. MiR-145 inhibits human colorectal cancer cell migration and invasion via PAK4-dependent pathway. Cancer Med. 2017;6:1331–1340. doi: 10.1002/cam4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao QD, Zhang W, Zhao K, Cao B, Yuan H, Wei LZ, Song MQ, Liu XS. MicroRNA-455 suppresses the oncogenic function of HDAC2 in human colorectal cancer. Braz J Med Biol Res. 2017;50:e6103. doi: 10.1590/1414-431X20176103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P, Zuo Z, Wu A, Shang W, Bi R, Jin Q, Wu J, Jiang L. miR-600 inhibits cell proliferation, migration and invasion by targeting p53 in mutant p53-expressing human colorectal cancer cell lines. Oncol Lett. 2017;13:1789–1796. doi: 10.3892/ol.2017.5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 18.Bai L, Wang H, Wang AH, Zhang LY, Bai J. MicroRNA-532 and microRNA-3064 inhibit cell proliferation and invasion by acting as direct regulators of human telomerase reverse transcriptase in ovarian cancer. PLoS One. 2017;12:e0173912. doi: 10.1371/journal.pone.0173912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, Chang JT, Kao CJ, Huang RS. High expression of miR-532-5p, a tumor suppressor, leads to better prognosis in ovarian cancer both in vivo and in vitro. Mol Cancer Ther. 2016;15:1123–1131. doi: 10.1158/1535-7163.MCT-15-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Zhao H, Gao X, Wei F, Zhang X, Su Y, Wang C, Li H, Ren X. Identification of a three-miRNA signature as a blood-borne diagnostic marker for early diagnosis of lung adenocarcinoma. Oncotarget. 2016;7:26070–26086. doi: 10.18632/oncotarget.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song X, Wang Z, Jin Y, Wang Y, Duan W. Loss of miR-532-5p in vitro promotes cell proliferation and metastasis by influencing CXCL2 expression in HCC. Am J Transl Res. 2015;7:2254–2261. [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Shiratsuchi I, Akagi Y, Kawahara A, Kinugasa T, Romeo K, Yoshida T, Ryu Y, Gotanda Y, Kage M, Shirouzu K. Expression of IGF-1 and IGF-1R and their relation to clinicopathological factors in colorectal cancer. Anticancer Res. 2011;31:2541–2545. [PubMed] [Google Scholar]
- 24.Oshima T, Akaike M, Yoshihara K, Shiozawa M, Yamamoto N, Sato T, Yamada R, Fujii S, Rino Y, Kunisaki C, Tanaka K, Masuda M, Imada T. Clinicopathological significance of the gene expression of matrix metalloproteinase-7, insulin-like growth factor-1, insulin-like growth factor-2 and insulin-like growth factor-1 receptor in patients with colorectal cancer: insulin-like growth factor-1 receptor gene expression is a useful predictor of liver metastasis from colorectal cancer. Oncol Rep. 2008;20:359–364. [PubMed] [Google Scholar]
- 25.Huang F, Xu LA, Khambata-Ford S. Correlation between gene expression of IGF-1R pathway markers and cetuximab benefit in metastatic colorectal cancer. Clin Cancer Res. 2012;18:1156–1166. doi: 10.1158/1078-0432.CCR-11-1135. [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Zhu Z, Gao W, Jiang Q, Yu J, Fu C. Systemic analysis of different colorectal cancer cell lines and TCGA datasets identified IGF-1R/EGFR-PPAR-CASPASE axis as important indicator for radiotherapy sensitivity. Gene. 2017;627:484–490. doi: 10.1016/j.gene.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, Li X, Ren Q, Tian J, Chen J. Calycosin induces apoptosis in colorectal cancer cells, through modulating the ERbeta/MiR-95 and IGF-1R, PI3K/Akt signaling pathways. Gene. 2016;591:123–128. doi: 10.1016/j.gene.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Xue M, Cao X, Zhong Y, Kuang D, Liu X, Zhao Z, Li H. Insulin-like growth factor-1 receptor (IGF-1R) kinase inhibitors in cancer therapy: advances and perspectives. Curr Pharm Des. 2012;18:2901–2913. doi: 10.2174/138161212800672723. [DOI] [PubMed] [Google Scholar]
- 29.Wojtalla A, Salm F, Christiansen DG, Cremona T, Cwiek P, Shalaby T, Gross N, Grotzer MA, Arcaro A. Novel agents targeting the IGF-1R/PI3K pathway impair cell proliferation and survival in subsets of medulloblastoma and neuroblastoma. PLoS One. 2012;7:e47109. doi: 10.1371/journal.pone.0047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Moerkens M, Ramaiahgari S, de Bont H, Price L, Meerman J, van de Water B. Elevated insulin-like growth factor 1 receptor signaling induces antiestrogen resistance through the MAPK/ERK and PI3K/Akt signaling routes. Breast Cancer Res. 2011;13:R52. doi: 10.1186/bcr2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirafkan N, Mansoori B, Mohammadi A, Shomali N, Ghasbi M, Baradaran B. MicroRNAs as novel biomarkers for colorectal cancer: new outlooks. Biomed Pharmacother. 2018;97:1319–1330. doi: 10.1016/j.biopha.2017.11.046. [DOI] [PubMed] [Google Scholar]
- 32.Stiegelbauer V, Perakis S, Deutsch A, Ling H, Gerger A, Pichler M. MicroRNAs as novel predictive biomarkers and therapeutic targets in colorectal cancer. World J Gastroenterol. 2014;20:11727–11735. doi: 10.3748/wjg.v20.i33.11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aravalli RN, Steer CJ. Circulating microRNAs: novel biomarkers for early detection of colorectal cancer. Transl Res. 2015;166:219–224. doi: 10.1016/j.trsl.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Christopher AF, Kaur RP, Kaur G, Kaur A, Gupta V, Bansal P. MicroRNA therapeutics: discovering novel targets and developing specific therapy. Perspect Clin Res. 2016;7:68–74. doi: 10.4103/2229-3485.179431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai HP, Huang SF, Li CF, Chien HT, Chen SC. Differential microRNA expression in breast cancer with different onset age. PLoS One. 2018;13:e0191195. doi: 10.1371/journal.pone.0191195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu S, Zheng Q, Wu H, Wang C, Liu T, Zhou W. miR-532 promoted gastric cancer migration and invasion by targeting NKD1. Life Sci. 2017;177:15–19. doi: 10.1016/j.lfs.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 37.Xu X, Zhang Y, Liu Z, Zhang X, Jia J. miRNA-532-5p functions as an oncogenic microRNA in human gastric cancer by directly targeting RUNX3. J Cell Mol Med. 2016;20:95–103. doi: 10.1111/jcmm.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griesing S, Kajino T, Tai MC, Liu Z, Nakatochi M, Shimada Y, Suzuki M, Takahashi T. Thyroid transcription factor-1-regulated microRNA-532-5p targets KRAS and MKL2 oncogenes and induces apoptosis in lung adenocarcinoma cells. Cancer Sci. 2017;108:1394–1404. doi: 10.1111/cas.13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie QX, Lin XC, Zhang MF, Han CX, Guo YH. [Expression of IGF-I and IGF-IR in bladder cancer] . Ai Zheng. 2004;23:707–709. [PubMed] [Google Scholar]
- 40.Yuan Y, Zhou X, Song J, Qiu X, Li J, Ye L, Meng X, Xia D. Expression and clinical significance of epidermal growth factor receptor and type 1 insulin-like growth factor receptor in nasopharyngeal carcinoma. Ann Otol Rhinol Laryngol. 2008;117:192–200. doi: 10.1177/000348940811700306. [DOI] [PubMed] [Google Scholar]
- 41.Wang YH, Han XD, Qiu Y, Xiong J, Yu Y, Wang B, Zhu ZZ, Qian BP, Chen YX, Wang SF, Shi HF, Sun X. Increased expression of insulin-like growth factor-1 receptor is correlated with tumor metastasis and prognosis in patients with osteosarcoma. J Surg Oncol. 2012;105:235–243. doi: 10.1002/jso.22077. [DOI] [PubMed] [Google Scholar]
- 42.Zhou F, Nie L, Feng D, Guo S, Luo R. MicroRNA-379 acts as a tumor suppressor in non-small cell lung cancer by targeting the IGF1R-mediated AKT and ERK pathways. Oncol Rep. 2017;38:1857–1866. doi: 10.3892/or.2017.5835. [DOI] [PubMed] [Google Scholar]
- 43.Pengchong H, Tao H. Expression of IGF-1R, VEGF-C and D2-40 and their correlation with lymph node metastasis in endometrial adenocarcinoma. Eur J Gynaecol Oncol. 2011;32:660–664. [PubMed] [Google Scholar]
- 44.Xiao Y, Tian Q, He J, Huang M, Yang C, Gong L. MiR-503 inhibits hepatocellular carcinoma cell growth via inhibition of insulin-like growth factor 1 receptor. Onco Targets Ther. 2016;9:3535–3544. doi: 10.2147/OTT.S106351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramani R, Lopez-Valdez R, Arumugam A, Nandy S, Boopalan T, Lakshmanaswamy R. Targeting insulin-like growth factor 1 receptor inhibits pancreatic cancer growth and metastasis. PLoS One. 2014;9:e97016. doi: 10.1371/journal.pone.0097016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heidegger I, Kern J, Ofer P, Klocker H, Massoner P. Oncogenic functions of IGF1R and INSR in prostate cancer include enhanced tumor growth, cell migration and angiogenesis. Oncotarget. 2014;5:2723–2735. doi: 10.18632/oncotarget.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]