Abstract
Mesenchymal stem cells (MSCs) have a high self-renewal potential and can differentiate into various types of cells, including adipocytes, osteoblasts, and chondrocytes. Previously, we reported that the enhancer of zeste homolog 2 (EZH2), the catalytic component of the Polycomb-repressive complex 2, and HDAC9c mediate the osteogenesis and adipogenesis of MSCs. In the current study, we identify the role of p38 in osteogenic differentiation from a MAPK antibody array screen and investigate the mechanisms underlying its transcriptional regulation. Our data show that YY1, a ubiquitously expressed transcription factor, and HDAC9c coordinate p38 transcriptional activity to promote its expression to facilitate the osteogenic potential of MSCs. Our results show that p38 mediates osteogenic differentiation, and this has significant implications in bone-related diseases, bone tissue engineering, and regenerative medicine.
Keywords: Osteogenesis, mesenchymal stem cells, p38, HDAC9c, YY1
Introduction
Mesenchymal stem cells (MSCs) are multipotent stromal cells capable of self-renewal and multi-lineage mesenchymal differentiation [1]. MSCs exhibit a highly therapeutic potential in cell-based therapy for graft-versus-host diseases [2,3], multiple sclerosis [4], and orthopedic repair/regeneration [5], among other therapeutic potentials. Using autologous MSCs to engineer specific tissue as well as the attachment of different biomaterials to MSCs have been revealed to repair bone, cartilage, muscle, and tendon lesions in vivo [5-7]. Particularly, bone is one of the most commonly transplanted tissues, with more than 2.2 million bone graft procedures performed annually worldwide [8]. Furthering our understanding of the regulatory mechanism underlying MSCs osteogenesis may identify better approaches to bone tissue engineering.
Several cell signaling cascades regulate MSC pro-osteogenic signaling, including those that control runt-related transcription factor 2 (RUNX2) activity [9], β-catenin-dependent Wnt [10,11], Hedgehog, bone morphogenetic proteins (BMPs) [12,13], and NEL-like protein 1 (NELL-1) [14,15], all of which may trigger and activate the MAPK cascades with a series of phosphorylation on MAPKK kinase (MAP3K), MAPK kinase (MAP2K), and MAPK. Members of the MAPK family, including extracellular signal-related kinases 1/2 (ERK1/2), c-Jun amino (N)-terminal kinases 1/2/3 (JNK1/2/3), and the p38 isoforms (p38α, p38β, p38γ, and p38δ) [16], play important roles in many biological processes, such as propagating extracellular stimuli, e.g., growth factor, cytokines, and environmental stresses, into various cellular actions.
Previous studies have demonstrated MAPKs to be key players in skeletal development and bone homeostasis via regulating osteoblast commitment and differentiation [17]. MEK3-null mice (Map2k3-/-) exhibited severe skeletal defects in their long bones as well as abnormalities in their craniofacial bone structures [18]. The embryonic lethality of Mapk14 (gene encoding p38α) knockout cause to neural and cardiac defects [19], and the lack of Mapk11 (gene encoding p38β) has been implicated in mild bone defects [18]. Both ERK1 and ERK2 are expressed in osteoblasts and have functions associated with bone metabolism. Matsushita et al. demonstrated through the use of a model with Erk1-/-; Erk2prx1:Cre double mutation that both ERK1 and ERK2 are required for osteoblast lineage specification via β-catenin-mediated canonical Wnt signaling [20]. Interestingly, contradictory roles of JNK in osteoblastogenesis have been reported. For instance, interleukin-1β (IL-1β) and tumor necrosis factor α (TNF-1α) activate JNK, which leads to the osteoblast differentiation of human periosteal cells [21]. In contrast, JNK activation has been shown to negatively regulate osteogenesis via its phosphorylation of RUNX2 at Ser104, which inhibits RUNX2 transcriptional activity [22]. Whether phosphorylation of MAPKs plays a direct role in the early stage of osteogenic commitment of MSCs remains unclear.
In the current study, we identify p38 as the major MAPK in regulating osteogenic differentiation through an MAPK antibody array screen, and show that p38 phosphorylation and expression are higher in osteoblasts differentiated from MSCs than their undifferentiated counterparts. In addition, we also show that HDAC9c interacts with YY1, which in turn transcriptionally upregulates p38 expression and is associated with stronger osteogenic differentiation in MSCs. Collectively, these results suggest that HDAC9c and YY1 cooperate to increase p38 transcriptional activity and subsequently enhance the osteogenesis of MSCs.
Materials and methods
Cell culture and differentiation of osteoblasts
hMSCs (3A6) were maintained in low glucose DMEM (Invitrogen) with 10% fetal bovine serum (FBS). Osteoblast differentiation was induced by culturing hMSCs in low glucose DMEM with 10% FBS supplemented with 10-8 M dexamethasone, 50 μg/ml ascorbic acid 2-phosphate, and 10 mM β-glycerophosphate. During differentiation, the medium was replaced every 3 days.
Antibody array
An antibody array (R&D, ARY002) screen was carried out following the manufacturer’s instructions. Un-differentiated and differentiated cells were lysed at 4°C for 30 min. Briefly, 1.5 ml of the diluted sample (300 μg) was placed on each membrane and incubated at 4°C overnight with gentle shaking. The membranes were then washed with 1× washing buffer 3 times. After washing, 1× streptavidin-HRP was added to each membrane and incubated at room temperature for 2 h with gentle shaking. The membranes were washed and then placed in the detection buffer for 2 min, and the signals were detected by autoradiography. Phosphorylation signal intensities of the target proteins were quantified by a densitometer.
Alizarin Red S stain
Osteogenesis was examined by Alizarin Red S (Sigma) staining, and quantitated at A450 as described previously [23]. In brief, the cells were rinsed with PBS, and then fixed with ice-cold 70% ethanol. After a brief wash with water, the cells were stained with 2% Alizarin Red S solution for 30 min at room temperature. The cells were rinsed five times with water followed by a 15-min wash with PBS (with rotation) to reduce nonspecific Alizarin Red S stain.
Real-time RT-PCR
Total RNA was isolated by TRIzol (Invitrogen) based on the manufacturer’s instructions. cDNA was synthesized by SuperScriptTM III First Strand Synthesis kit (Invitrogen). Changes in mRNA expression level were analyzed by real-time PCR (Roche Applied Science, LightCycler 480) using SYBR Green and normalized to β-actin. Primer sequences are listed in Table S1.
Reporter gene assay
The p38 promoter luciferase reporter plasmid was amplified from human genomic DNA containing SacI and XhoI restriction enzyme sites by PCR and then subcloned into pGL3-basic backbone vector. The p38 promoter luciferase reporter plasmid was transfected into 293 cells together with internal control plasmid β-gal or the indicated plasmids using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cell extracts were harvested and luciferase activity measured by luciferase reporter assay system according to the manufacturer’s instructions (Promega).
Western blot analysis and coimmunoprecipitation (CoIP)
Cell lysates were harvested on ice with NETN (150 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, 0.5% Nonidet P-40) buffer supplemented with protease inhibitors. Protein samples (75 μg) were loaded onto SDS-polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membranes for immunoblotting with the following antibodies: HDAC9c, YY1, and p38 (Santa Cruz Biotechnology); pp38 (Cell Signaling). The lysed extracts were subjected to immunoprecipitation with the indicated antibodies. The protein complexes were pulled down by protein G-agarose beads (GE Healthcare) and subjected to immunoblotting using YY1 (Santa Cruz Biotechnology), and HDAC9c (Santa Cruz Biotechnology) antibodies.
Lentiviral infection
Lentiviral shRNA clones were purchased from the National RNAi Core Facility of Academica Sinica (Taiwan). Human MSCs were infected with control (vector alone pLK0.1), p38, HDAC9c, or YY1 shRNA lentivirus in the presence of polybrene (8 μg/ml). After infection, osteogenesis was induced as described above.
Quantitative chromatin immunoprecipitation (qChIP) assay
ChIP assays were performed using the EZ-ChIP kit (Upstate) according to the manufacturer’s instructions. Specific antibodies against YY1 (Santa Cruz Biotechnology) were used for immunoprecipitation. The immunoprecipitated DNA was subjected to RT-PCR using a SYBR Green system according to the manufacturer’s instructions (Roche Applied Science). Data are shown as the fold enrichment of precipitated DNA relative to 2:100 dilution of input chromatin. Primers are listed in Table S1.
Statistical analysis
Student’s t-test was used to compare two groups of independent samples. A P value < 0.05 was considered statistically significant.
Results
p38 stimulates MSC osteogenesis
To investigate the regulation of MAPK in MSC differentiation into osteoblasts, we utilized a human phospho-MAPK array to compare the phosphorylation of MAPKs in both undifferentiated and osteogenic MSC. Phosphorylation of four p38 isoforms (a.k.a. p38α, p38β, p38γ, and p38δ) was substantially increased in differentiated osteoblasts compared with undifferentiated MSCs (Figure 1A). Consistently, Western blot analysis indicated a higher phosphorylation of p38 in differentiated osteoblasts compared with undifferentiated MSCs (Figure 1B). In addition to phosphorylation, the p38 protein expression level was increased in osteogenic MSCs compared with undifferentiated MSC (Figure 1B). To validate the role of p38 in MSC differentiation into osteoblasts, MSCs were treated with or without p38 inhibitor, SB203580, followed by incubation in an osteogenic-inducing medium. The osteogenic potential of MSCs was substantially blocked by p38 inhibitor compared with DMSO treatment (Figure 1C). We also knocked down p38 by short-hairpin RNA (shRNA) in MSCs (Figure 1D), which attenuated their osteogenic potential (Figure 1E). Higher p38 expression and phosphorylation were found in the primary osteoblasts that were isolated than in undifferentiated MSCs (Figure 1F). Collectively, these results provide strong support that p38 expression and phosphorylation are critical for the osteogenic potential of MSCs.
Figure 1.
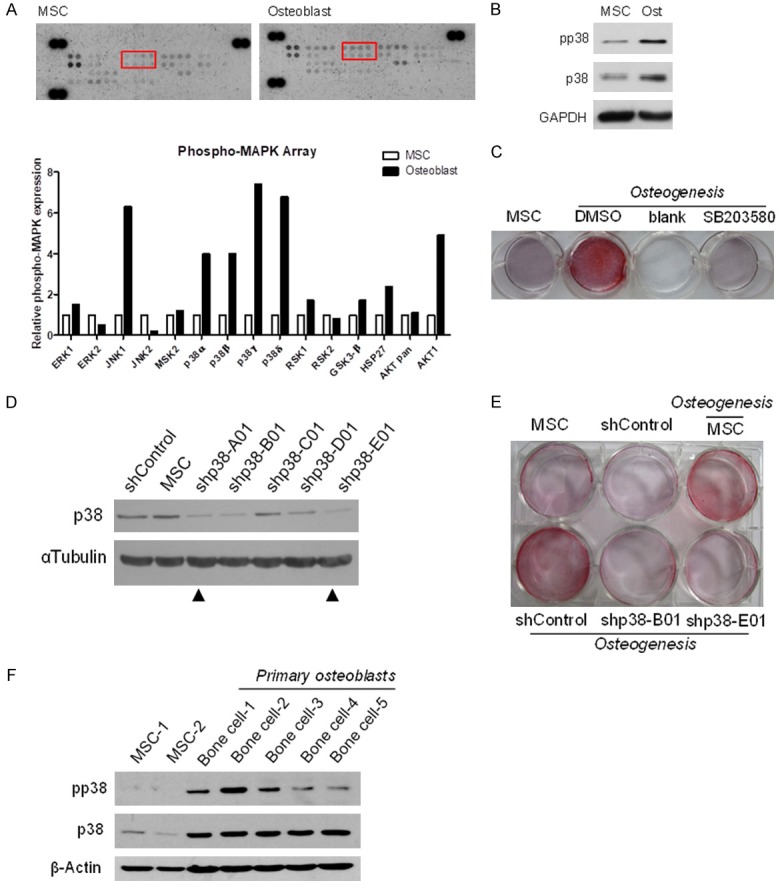
The expression and phosphorylation of p38 in osteogenic MSCs. A. Human MSCs were incubated in osteogenic induction media for 7 days and cell lysate harvested to conduct MAPK antibody array screen. Top, representative images. Bottom, quantification of antibody array. B. Western blot analysis of p38 phosphorylation and expression. GAPDH served as the control. C. MSCs were induced in osteogenic differentiation media containing the p38 inhibitor, SB203580, for 7 days. Following differentiation, osteoblasts were stained with Alizarin Red S. D. Western blot analysis of the efficiency of p38 knockdown by shControl, shp38-A01, shp38-B01, shp38-C01, shp38-D01, and shp38-E01 in human MSCs (hMSCs). E. After infection, hMSCs were cultured in osteogenic differentiation media for 7 days and stained with Alizarin Red S. F. Western blot analysis of p38 expression and phosphorylation in undifferentiated MSCs and bone cells from patients.
p38 and histone deacetylase 9c (HDAC9c) expression correlate positively in differentiated osteoblasts
Members of the HDAC family, HDAC1, HDAC4, and HDAC9c, modulate differentiation of preadipocytes, chondrocytes, and osteoblasts, respectively [23-25]. Previously, we identified HDAC9c, also named MEF-2 Interacting Transcription Repressor (MITR), as an EZH2 target gene by a genome-wide EZH2 ChIP-on-chip study and demonstrated that HDAC9c functions as a co-factor to promote MSC differentiation into osteoblasts by attenuating the transcriptional activity of the peroxisome proliferator-activated receptor gamma 2 (PPARγ-2) [23]. Hence, we explored the role of HDAC9c as it is involved in p38-mediated osteogenesis. We found that the expression of both p38 and HDAC9c were increased in differentiated osteoblasts from MSCs as compared with undifferentiated MSCs (Figure 2A). Moreover, the RNA expression of p38 was significantly reduced when HDAC9c was knocked down by specific shRNA in differentiated osteoblasts (Figure 2B). We further assessed the relationship between p38 and HDAC9c via the lentiviral-based expression of two different shRNAs that specifically target p38 or HDAC9c in differentiated osteoblasts. The expression and phosphorylation of p38 was attenuated in the absence of HDAC9c in differentiated osteoblasts, and no effects were observed when we knocked down p38 (Figure 2C). These results indicate that HDAC9c may serve as an upstream regulator in p38-mediated MSCs osteogenic potential.
Figure 2.
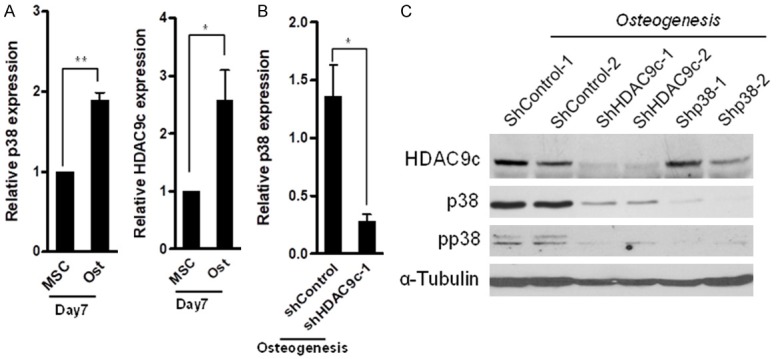
HDAC9c-mediated p38 expression in osteogenic MSCs. A. Relative p38 and HDAC9c mRNA expression in differentiated osteoblasts at the indicated time point. B. Relative p38 mRNA expression in osteogenic MSCs infected with control or HDAC9c shRNA. C. Western blot analysis of HDAC9c and p38 protein expression/phosphorylation in differentiated osteoblasts with the control, HDAC9c, and p38 shRNA. Error bars represent ± SD. *P < 0.05 and **P < 0.01, Student’s t-test.
HDAC9c and YY1 transcriptionally regulate p38-mediated MSC osteogenesis
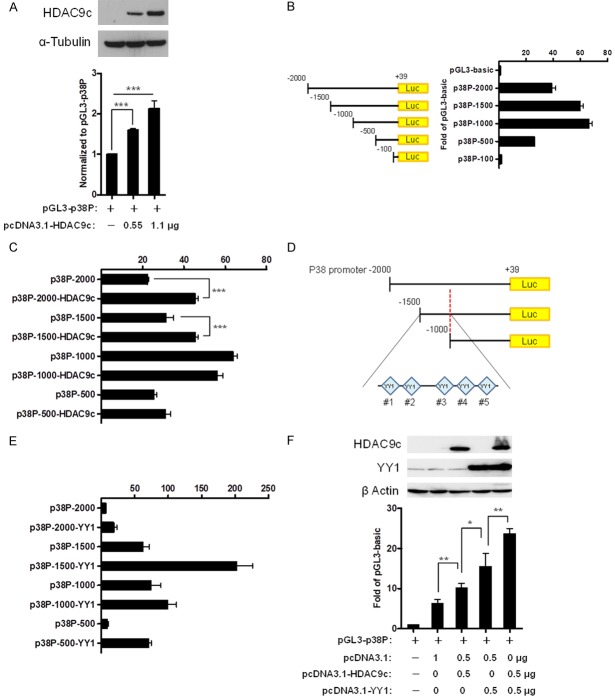
To determine whether HDAC9c transcriptionally regulates p38-mediated MSC osteogenic potential, we examined the p38 promoter activity in 293 cells ectopically expressing HDAC9c via a luciferase reporter assay. The results show that the p38 promoter activity was enhanced when HDAC9c expression was increased (Figure 3A). We then generated serial deletions of the p38 promoter to identify the region with high promoter activities. The results indicate the p38 promoter region at -2000 to -1000 bp of upstream of the transcriptional start site had the highest activity (Figure 3B). To further identify the region within p38 promoter controlled by HDAC9c, serial deletions of the p38 promoter were generated and their activities evaluated in the presence of HDAC9c expression in 293 cells. A comparison of the activities of the different promoter constructs indicated that the activity of the p38 promoter between -2000 to -1500 bp increased with HDAC9c expression. HDAC9c expression did not increase the activity of the p38 promoter region between -1000 and -500 bp (Figure 3C). Therefore, the p38 promoter region between -1500 to -1000 bp is likely critical for HDAC9c activation. Next, we sought to identify the transcription factor binding elements within this region by bioinformatics analysis. We found five Yin Yang 1 (YY1) transcription factor binding sites (referred to as #1, #2, #3, #4, and #5; Figure 3D). In contrast, no HDAC9 binding sites were found. YY1, a ubiquitously expressed transcription factor containing HDAC binding domain, plays a critical role in stem cell differentiation. For instance, YY1 suppresses expression of multiple muscle loci via recruiting histone methyltransferase EZH2 (enhancer of zeste homologue 2) of the polycomb repressive complex 2 (PRC2) [26-28]. A recent study by Zhou et al. showed that Linc-YY1, a lincRNA from the promoter of YY1 gene, may bind to YY1 to facilitate the dissociation of YY1/PRC2 complex at target promoter, and this in turn activates the expression of myogenesis target genes [29]. Moreover, YY1 also contains a specific binding domain for HDAC proteins, e.g., HDAC1 and HDAC2 [30]. Therefore, we hypothesized that YY1 may regulate p38 promoter. To this end, we compared the promoter activities of p38 with or without ectopic expression of YY1 by luciferase reporter assay. The results show that YY1 expression enhanced the p38 promoter activity compared with vector control (Figure 3E). Interestingly, ectopic expression of HDAC9c further increased YY1-mediated p38 promoter activity (Figure 3F). These findings suggest that HDAC9c may serve as a co-factor in YY1-mediated osteoblast differentiation.
Figure 3.
HDAC9c and YY1 coordinate the regulation of p38 promoter activity. (A) Relative promoter activity was measured by reporter assay in 293 cells transfected with HDAC9c expression vector and p38 promoter-driven luciferase plasmid. Top, Western blot analysis of HDAC9c. (B, C) Luciferase reporter assay of the promoter activities of p38 promoter-deletion mutants with (C) or without (B) ectopic expression of HDAC9c in 293 cells. (D) A schematic of YY1 binding site located in p38 promoter region between -1500 bp and -1000 bp. (E) Luciferase reporter assay of the promoter activities of p38 promoter-deletion mutants in 293 cells with ectopic expression of YY1. (F) Luciferase reporter assay of the p38 promoter activity in 293 cells with ectopic expression of both HDAC9c and YY1. Top, Western blot analysis of ectopically expressed HDAC9c and YY1. Error bars represent ± SD. ***P < 0.001, Student’s t-test.
YY1 is required for p38-mediated osteogenic differentiation of MSCs
To determine whether YY1 is essential for MSC differentiation into osteoblasts, we knocked down YY1 expression using five different shRNAs that specifically target YY1 in MSCs. Western blotting analysis indicated that four of the five shRNAs effectively attenuated YY1 expression (Figure 4A). Reducing YY1 expression also inhibited MSC differentiation into osteoblasts as determined by Alizarin Red S staining (Figure 4B). Consistently, YY1 expression was higher in primary osteoblasts than in undifferentiated MSCs (Figure 4C). Knocking down YY1 by shRNA decreased the levels of p38 RNA and protein during MSC osteogenesis (Figure 4D and 4E). These results were similar to those in the HDAC9c knockdown experiments (Figure 2B and 2C). Both YY1 and HDAC9c were highly expressed in differentiated osteoblasts compared with the undifferentiated control (Figure 4F). The results indicate that YY1 and HDAC9c may co-stimulate osteogenic differentiation of MSCs. Next, we asked whether YY1 and HDAC9c physically interact by performing an immunoprecipitation (IP) analysis. The results indicate that YY1 interacted with HDAC9c in differentiated osteoblasts but not in undifferentiated control (Figure 4G), and this interaction can be increased by IP YY1 antibody in a dose-dependent manner in differentiated osteoblasts (Figure 4H).
Figure 4.
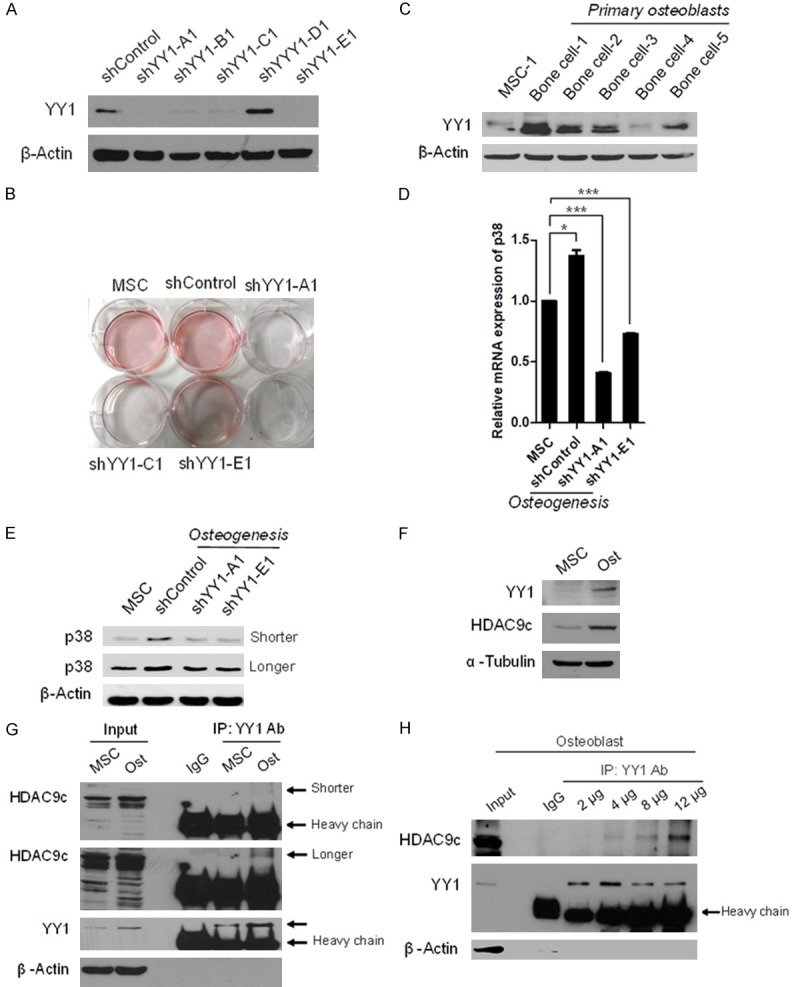
YY1 associates with HDAC9c to govern MSC osteogenic differentiation. (A) Western blot analysis of YY1 knockdown efficiency by shControl, shYY1-A1, shYY1-B1, shYY1-C1, shYY1-D1, or shYY1-E1 in hMSCs. (B) After infection, hMSCs were incubated in osteogenic differentiation media for 7 days and stained with Alizarin Red S. (C) Western blot analysis of YY1 expression in undifferentiated MSCs and primary bone cells. β-Actin served as the control. (D, E) Relative mRNA (D) and protein expression (E) of p38 in osteoblasts infected with two specific YY1 shRNA by qRT-PCR and western blot, respectively. (F) Western blot analysis of YY1 and HDAC9c expression in hMSCs and differentiated osteoblasts. (G) Cell extracts were harvested after induction of osteoblast differentiation of MSCs for 7 days and subjected to immunoprecipitation (IP) with YY1 antibody followed by Western blot analysis with the indicated antibodies. (H) Cells extracts from osteoblasts were immunoprecipitated with different amount of YY1 antibody and subjected to Western blot analysis with the indicated antibodies. Error bars represent ± SD. *P < 0.05 and ***P < 0.001, Student’s t-test.
To identify the key YY1-regulated elements in the p38 promoter, we generated five distinct p38 promoter regions with mutations within the YY1 binding site by site direct mutagenesis followed by luciferase report assay to examine their promoter activities. The results indicate that the activities of the YY1-1 and YY1-5 mutant p38 promoter were substantially reduced (Figure 5A). To validate the above findings, we ectopically expressed YY1 and p38 promoter with or without YY1 binding site mutation and examined the activities of the p38 promoter by luciferase reporter assay. Consistently, the YY1-1 and YY1-5 mutants, but not the wild-type YY1 or YY1-2, YY1-3 or YY1-4 mutant, rendered the p38 promoter inactive in the presence of ectopically expressed YY1 (Figure 5B). The results indicate that the YY1 binding sites, binding site #1 at -1328 to -1324 and binding site #5 at -1038 to -1034 upstream of the p38 promoter are critical for YY1-mediated p38 promoter activation. Finally, we explored the requirement of YY1 activation at the p38 promoter during MSC differentiation into osteoblasts. To this end, we conducted a q-ChIP assay for the p38 promoter by immunoprecipitation with a YY1 antibody. The data showed that YY1 binding to the p38 promoter was strongly reduced when YY1 was knocked down compared with the control in differentiated osteoblasts (Figure 5C). With these results, we propose a model in which HDAC9c and YY1 regulate p38 promoter in osteogenesis (Figure 5D): during MSCs differentiation, HDAC9c acts as a co-activator and cooperates with YY1 to activate the p38 promoter activity through two bindings sites in the promoter to enhance its expression, which stimulates the osteogenesis of MSCs.
Figure 5.
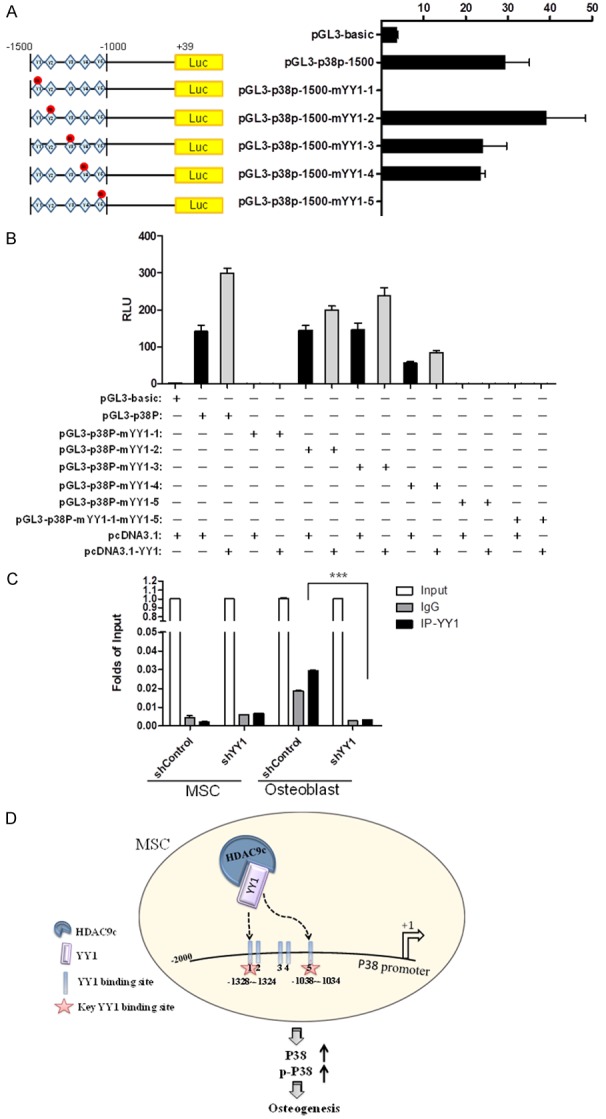
YY1 binds specifically to the p38 promoter in differentiated osteoblasts. A. Five YY1 binding sites located in the p38 promoter were mutated by site direct mutagenesis. Relative p38 promoter activities with or without the YY1 binding site mutations were determined by luciferase reporter assay. B. Reporter assay of p38 promoter activity with single or double YY1-binding site mutations in 293 cells ectopically expressing YY1. C. Undifferentiated MSCs and differentiated osteoblasts were infected with YY1 shRNA or shControl. Cross-linked chromatin was immunoprecipitated by using YY1 or IgG antibodies. The input and immunoprecipitated DNA were subjected to RT- PCR using primers corresponding to the promoter region of p38 (Table S1). D. A proposed model of MSC osteogenesis governed by HDAC9c and YY1. In osteogenic MSCs, HDAC9c and YY1 form a complex, which binds specifically to the indicated regions the p38 promoter to increase its expression and subsequently enhance osteogenesis. Error bars represent ± SD. ***P < 0.001, Student’s t-test.
Discussion
Furthering our understanding of the regulatory mechanisms underlying osteoblast differentiation from MSCs may lead to the development of new cellular strategies for bone tissue engineering and regenerative medicine, which are both promising for the treatment of diseases like osteoporosis, bone cancer, and bone defects. Previously, we reported that the epigenetic modulator EZH2 regulates HDAC9c in osteogenic and adipogenic lineage commitment of MSCs in an age-dependent manner. In the current study, we investigated how YY1 and HDAC9c govern p38 expression in MSC osteogenic differentiation and showed that YY1 plays a critical role in osteoblast differentiation via interaction with HDAC9c.
YY1 is a member of the GLI-Krüppel family of zinc finger transcription factors, also known as delta, NF-E1, UCRBP, or CF1, and it may interact with histone acetyltransferase (HAT) and HDAC cofactors to carry out its transcriptional functions [31]. Additionally, YY1 has dual transcriptional functions that are dependent on the distinct pre-existing YY1-cofactor complexes being recruited to the promoter under certain conditions. The YY1-mediated repression that identified is associated with class I HDACs, such as HDAC1 and HDAC2, which both contain catalytic domains and histone deacetylase activities [32,33]. YY1 associates with other coactivators, such as CBP and p300 protein [34,35], which both contain histone acetyltransferase activity to modify histones and chromatin structure [36]. Intriguingly, we found that YY1 interacted with HDAC9c as a co-activator of the p38 promoter to enhance its expression. Unlike HDAC1 and HDAC2, HDAC9c lacks the catalytic domain and does not possess any deacetylase activities [37], suggesting HDAC9c acts as co-activator but when associated with YY1. Moreover, we previously showed that HDAC9c is highly expressed in differentiated osteoblast and antagonizes PPARγ transcriptional activity, thereby stimulating osteoblasts differentiation potential [23]. Collectively, these findings indicate that HDAC9c enhances MSC osteogenic lineage not only through inhibition of adipogenitic transcription factor activity but also through enhancement of the YY1 transcriptional activity to upregulate p38 expression.
The osteogenic ability of p38 kinase is associated with its function to phosphorylate and induce the activity of several key osteogenic transcription factors. Several studies have shown that multiple p38 osteogenic targets, including RUNX2, DLX5 (Distal-Less Homeobox 5), and OSX (Osterix, osteoblast-specific transcription factor), are osteoblast-specific transcription factors. Phosphorylation of RUNX2 by p38 elevates its transcriptional potential [18,38,39]. Moreover, p38 phosphorylates DLX5, a transactivator of OSX stimulated by BMP-2, at Ser34 and Ser217 to facilitate the recruitment of p300 [40]. Similar p38-activated osteogenic events also occur with the phosphorylation of OSX at Ser77 and Ser33 [41]. Recently, Artigas et al. demonstrated that RUNX2 and OSX physically interact and increase RUNX2-transcriptional ability in a cooperative manner [39]. On the basis of these findings, the induction of p38 expression to phosphorylate and enhance transcriptional activities of key transcription factors is likely important in osteoblast differentiation.
Bone tissue engineering is a potential alternative strategy to overcome drawbacks related to autografts and allografts, including donor-site morbidity, the availability of limited grafting material, and compromised bone quality in patients with osteoporosis [42]. Even though it has been researched and tested for approximately 30 years, few bone tissue-engineering techniques have translated into clinical application, and the standard of care in bone regenerative medicine is still lacking. Our findings that YY1 and HDAC9c positively mediate p38 transcriptional activity to promote the osteogenic potential of MSCs provide a new direction toward the development of MSC cell-based treatment for bone-related degeneration or lesions by inducing YY1 expression and may be applicable in the future of bone tissue engineering.
Acknowledgements
This work was supported in part by the following grants: the Ministry of Science and Technology (MOST 104-2320-B-039-047-MY3 and MOST 106-2320-B-039-047 to Y.-H.C. and MOST 104-2314-B-005-001 and MOST 105-2320-B-005-009 to L.Y. Li.). The authors also appreciate Jennifer L. Hsu and Ian Crews for the critical reading and editing of the paper.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 2.Tolar J, Le Blanc K, Keating A, Blazar BR. Concise review: hitting the right spot with mesenchymal stromal cells. Stem Cells. 2010;28:1446–1455. doi: 10.1002/stem.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Blanc K, Frassoni F, Ball L, Locatelli F, Roelofs H, Lewis I, Lanino E, Sundberg B, Bernardo ME, Remberger M, Dini G, Egeler RM, Bacigalupo A, Fibbe W, Ringden O Developmental Committee of the European Group for Blood and Marrow Transplantation. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. Lancet. 2008;371:1579–1586. doi: 10.1016/S0140-6736(08)60690-X. [DOI] [PubMed] [Google Scholar]
- 4.Freedman MS, Bar-Or A, Atkins HL, Karussis D, Frassoni F, Lazarus H, Scolding N, Slavin S, Le Blanc K, Uccelli A MSCT Study Group. The therapeutic potential of mesenchymal stem cell transplantation as a treatment for multiple sclerosis: consensus report of the International MSCT study group. Mult Scler. 2010;16:503–510. doi: 10.1177/1352458509359727. [DOI] [PubMed] [Google Scholar]
- 5.Caplan AI. New era of cell-based orthopedic therapies. Tissue Eng Part B Rev. 2009;15:195–200. doi: 10.1089/ten.teb.2008.0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan M, Yao W, Liu R, Lam KS, Nolta J, Jia J, Panganiban B, Meng L, Zhou P, Shahnazari M, Ritchie RO, Lane NE. Directing mesenchymal stem cells to bone to augment bone formation and increase bone mass. Nat Med. 2012;18:456–462. doi: 10.1038/nm.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeitouni S, Krause U, Clough BH, Halderman H, Falster A, Blalock DT, Chaput CD, Sampson HW, Gregory CA. Human mesenchymal stem cell-derived matrices for enhanced osteoregeneration. Sci Transl Med. 2012;4:132ra155. doi: 10.1126/scitranslmed.3003396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannoudis PV, Dinopoulos H, Tsiridis E. Bone substitutes: an update. Injury. 2005;36(Suppl 3):S20–27. doi: 10.1016/j.injury.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, Yamana K, Zanma A, Takada K, Ito Y, Komori T. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Alimonte I, Lannutti A, Pipino C, Di Tomo P, Pierdomenico L, Cianci E, Antonucci I, Marchisio M, Romano M, Stuppia L, Caciagli F, Pandolfi A, Ciccarelli R. Wnt signaling behaves as a “master regulator” in the osteogenic and adipogenic commitment of human amniotic fluid mesenchymal stem cells. Stem Cell Rev. 2013;9:642–654. doi: 10.1007/s12015-013-9436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taipaleenmaki H, Abdallah BM, AlDahmash A, Saamanen AM, Kassem M. Wnt signalling mediates the cross-talk between bone marrow derived pre-adipocytic and pre-osteoblastic cell populations. Exp Cell Res. 2011;317:745–756. doi: 10.1016/j.yexcr.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 12.James AW, Leucht P, Levi B, Carre AL, Xu Y, Helms JA, Longaker MT. Sonic Hedgehog influences the balance of osteogenesis and adipogenesis in mouse adipose-derived stromal cells. Tissue Eng Part A. 2010;16:2605–2616. doi: 10.1089/ten.tea.2010.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontaine C, Cousin W, Plaisant M, Dani C, Peraldi P. Hedgehog signaling alters adipocyte maturation of human mesenchymal stem cells. Stem Cells. 2008;26:1037–1046. doi: 10.1634/stemcells.2007-0974. [DOI] [PubMed] [Google Scholar]
- 14.James AW, Pan A, Chiang M, Zara JN, Zhang X, Ting K, Soo C. A new function of Nell-1 protein in repressing adipogenic differentiation. Biochem Biophys Res Commun. 2011;411:126–131. doi: 10.1016/j.bbrc.2011.06.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James AW, Pang S, Askarinam A, Corselli M, Zara JN, Goyal R, Chang L, Pan A, Shen J, Yuan W, Stoker D, Zhang X, Adams JS, Ting K, Soo C. Additive effects of sonic hedgehog and Nell-1 signaling in osteogenic versus adipogenic differentiation of human adipose-derived stromal cells. Stem Cells Dev. 2012;21:2170–2178. doi: 10.1089/scd.2011.0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Carballo E, Gamez B, Ventura F. p38 MAPK signaling in osteoblast differentiation. Front Cell Dev Biol. 2016;4:40. doi: 10.3389/fcell.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenblatt MB, Shim JH, Zou W, Sitara D, Schweitzer M, Hu D, Lotinun S, Sano Y, Baron R, Park JM, Arthur S, Xie M, Schneider MD, Zhai B, Gygi S, Davis R, Glimcher LH. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J Clin Invest. 2010;120:2457–2473. doi: 10.1172/JCI42285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.del Barco Barrantes I, Coya JM, Maina F, Arthur JS, Nebreda AR. Genetic analysis of specific and redundant roles for p38alpha and p38beta MAPKs during mouse development. Proc Natl Acad Sci U S A. 2011;108:12764–12769. doi: 10.1073/pnas.1015013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsushita T, Chan YY, Kawanami A, Balmes G, Landreth GE, Murakami S. Extracellular signal-regulated kinase 1 (ERK1) and ERK2 play essential roles in osteoblast differentiation and in supporting osteoclastogenesis. Mol Cell Biol. 2009;29:5843–5857. doi: 10.1128/MCB.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hah YS, Kang HG, Cho HY, Shin SH, Kim UK, Park BW, Lee SI, Rho GJ, Kim JR, Byun JH. JNK signaling plays an important role in the effects of TNF-alpha and IL-1beta on in vitro osteoblastic differentiation of cultured human periosteal-derived cells. Mol Biol Rep. 2013;40:4869–4881. doi: 10.1007/s11033-013-2586-3. [DOI] [PubMed] [Google Scholar]
- 22.Huang YF, Lin JJ, Lin CH, Su Y, Hung SC. c-Jun N-terminal kinase 1 negatively regulates osteoblastic differentiation induced by BMP2 via phosphorylation of Runx2 at Ser104. J Bone Miner Res. 2012;27:1093–1105. doi: 10.1002/jbmr.1548. [DOI] [PubMed] [Google Scholar]
- 23.Chen YH, Yeh FL, Yeh SP, Ma HT, Hung SC, Hung MC, Li LY. Myocyte enhancer factor-2 interacting transcriptional repressor (MITR) is a switch that promotes osteogenesis and inhibits adipogenesis of mesenchymal stem cells by inactivating peroxisome proliferatoractivated receptor gamma-2. J Biol Chem. 2011;286:10671–10680. doi: 10.1074/jbc.M110.199612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo EJ, Chung JJ, Choe SS, Kim KH, Kim JB. Down-regulation of histone deacetylases stimulates adipocyte differentiation. J Biol Chem. 2006;281:6608–6615. doi: 10.1074/jbc.M508982200. [DOI] [PubMed] [Google Scholar]
- 25.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 26.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Hertlein E, Bakkar N, Sun H, Acharyya S, Wang J, Carathers M, Davuluri R, Guttridge DC. NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol. 2007;27:4374–4387. doi: 10.1128/MCB.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou L, Sun K, Zhao Y, Zhang S, Wang X, Li Y, Lu L, Chen X, Chen F, Bao X, Zhu X, Wang L, Tang LY, Esteban MA, Wang CC, Jauch R, Sun H, Wang H. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun. 2015;6:10026. doi: 10.1038/ncomms10026. [DOI] [PubMed] [Google Scholar]
- 30.Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 32.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 33.Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci U S A. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 35.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 36.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 37.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ge C, Xiao G, Jiang D, Yang Q, Hatch NE, Roca H, Franceschi RT. Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor. J Biol Chem. 2009;284:32533–32543. doi: 10.1074/jbc.M109.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Artigas N, Urena C, Rodriguez-Carballo E, Rosa JL, Ventura F. Mitogen-activated protein kinase (MAPK)-regulated interactions between Osterix and Runx2 are critical for the transcriptional osteogenic program. J Biol Chem. 2014;289:27105–27117. doi: 10.1074/jbc.M114.576793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulsamer A, Ortuno MJ, Ruiz S, Susperregui AR, Osses N, Rosa JL, Ventura F. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J Biol Chem. 2008;283:3816–3826. doi: 10.1074/jbc.M704724200. [DOI] [PubMed] [Google Scholar]
- 41.Ortuno MJ, Ruiz-Gaspa S, Rodriguez-Carballo E, Susperregui AR, Bartrons R, Rosa JL, Ventura F. p38 regulates expression of osteoblast-specific genes by phosphorylation of osterix. J Biol Chem. 2010;285:31985–31994. doi: 10.1074/jbc.M110.123612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neovius E, Engstrand T. Craniofacial reconstruction with bone and biomaterials: review over the last 11 years. J Plast Reconstr Aesthet Surg. 2010;63:1615–1623. doi: 10.1016/j.bjps.2009.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.