Abstract
Non-small-cell carcinoma (NSCLC) is one of the most lethal malignancies of lung cancers and its prognosis remains dismal due to the paucity of effective therapeutic targets. Recent reports show that Golgi membrane protein 1 (GOLM1) is highly expressed in a variety of tumor cells, functions as a negative regulator of T cells and then promotes tumor progression. However, its expression and role in NSCLC remain unclear. Herein, we showed that GOLM1 was markedly up-regulated in NSCLC cell lines and clinical tissues. Clinically, NSCLC patients with high expression of GOLM1 had shorter overall survival (OS) and high GOLM1 expression in tumor samples was significantly related to malignant phenotype, such as lymph node metastasis and high tumor stage. Ectopic expression of GOLM1 in NSCLC cells induced epithelial-to-mesenchymal transition (EMT) and promoted proliferation, migration, and invasion of NSCLC cells in vitro. Furthermore, GOLM1 overexpressing significantly promoted the tumorigenicity of NSCLC cells in vivo whereas silencing endogenous GOLM1 caused an opposite outcome. Moreover, we demonstrated that GOLM1 enhanced NSCLC aggressiveness by activating matrix metalloproteinase-13 (MMP13) signaling. Together, our results provided new evidence that GOLM1 overexpression promoted the progression of NSCLC and might represent a novel therapeutic target for its treatment.
Keywords: GOLM1, NSCLC, metastasis, MMP13
Introduction
Non-small-cell carcinoma (NSCLC) is among the most threatening malignancies with a high incidence and metastasis rate [1]. The investigation of molecular mechanism of NSCLC facilitates the development of the rational design of corresponding targeted therapies. Nowadays, the survival of patients with malignant NSCLC has improved owing to the development of advanced treatment options [2]. Unfortunately, tumor metastasis remains one of the main causes of death among malignant tumor patients. Metastasis is a major hallmark of cancer and yet remains the most poorly understood component of cancer pathogenesis [3]. It is a complex multistep process involving alterations in the dissemination, invasion, survival, and growth of new cancer cell colonies, which are regulated by a complex network of intra- and inter-cellular signal transduction cascades [4]. Previous studies regarding tumor metastasis have primarily focused on the adhesion and migration ability of cancer cells themselves. Recently, many therapeutic strategies have been designed to target tumor cells.
GOLM1 (Golm 1, NM_016548) is a resident cis-Golgi membrane protein of unknown function. GOLM1 has a single N-terminal transmembrane domain and an extensive C-terminal, coiled-coil domain that faces the luminal surface of the Golgi apparatus [5]. N-terminal cleavage by a furin proprotein convertase resulted in the release of the C-terminal ectomain and its appearance in serum [6]. Golgi has been shown to play an active role in cell migration through posttranslational modification and prominent changes in the Golgi apparatus, as evidenced by the disruption of biochemical composition, structure and functional levels observed in human carcinogenesis and metastasis [7]. Consistently, another Golgi-associated protein, GOLPH3 has recently been shown to act as an oncogene by linking cancer to Golgi and DNA damage signaling. The cleaved form of GOLM1 was detectable in the serum of patients with hepatocellular cancer, a finding that may have diagnostic value [8]. In addition, previous studies demonstrate that GOLM1 mRNA levels can serve as significant predictors of prostate cancer [9]. In prostate cancer, GOLM1 acts as a critical oncogene by promoting prostate cancer cell proliferation, migration and invasion, and inhibiting apoptosis, mainly through activating PI3K-AKT-mTOR signaling pathway [10].
Matrix metalloproteinases (MMPs) are members of zinc-dependent endopeptidases implicated in a variety of physiological and pathological processes [11]. Over the decades, MMPs have been studied for their role in cancer progression, migration, and metastasis. As a result, accumulated evidence of MMPs incriminating role has made them an attractive therapeutic target [12]. Overexpression of MMP13 was observed in esophageal squamous cell carcinoma (ESCC) clinical tissues, and the expression of MMP13 promoted cancer cell aggressiveness [13]. In addition, MMP13 was overexpressed in nasopharyngeal cancer (NPC) cells and exosomes purified from conditioned medium (CM) as well as NPC patients’ plasma and MMP13-containing exosomes in NPC progression which might offer unique insights for potential therapeutic strategies for NPC progressions [14]. High level of MMP13 protein expression is proved to be significant correlation with lymph node metastasis and tumor staging of oral squamous cell carcinoma (OSCC). Multivariate Cox regression model analysis revealed that high level of mRNA and protein expressions of MMP13 were significantly associated with poor prognosis of OSCC [15]. Taken together, these observations indicate that the MMP13 overexpression could be considered as a prognostic marker and potential target of cancer.
Although GOLM1 is believed to positively correlate to several tumor types, including hepatocellular carcinoma metastasis, the regulation mechanism and function of GOLM1 in human NSCLC is poorly understood. In this study, we demonstrated that GOLM1 is over-expression in NSCLC tissues and cell lines. Meanwhile, we also confirmed that GOLM1 promotes NSCLC metastasis directly by enhancing migration and invasion. Collectively, our results suggest that GOLM1 is a vital factor for NSCLC progression, and provide GOLM1 as a potential target for NSCLC treatment.
Materials and methods
Bioinformatics analysis
Gene expression datasets used for statistical analysis were acquired from the National Center for Biotechnology Information GEO database with the accession codes GSE69732 and GSE89039. The screening was performed in GEO datasets which contained both the lung tumor samples and the matched adjacent normal lung samples. The value of logFC (Tumor/Normal) was calculated for each selected probe and listed in the rank order. In protein-protein interaction (PPI) network construction, STRING 10.0 software (http://string-db.org/) is a web-based database for providing comprehensive interactions information for the already known or predicted proteins.
Tissue specimens and cell culture
A total of 37 primary NSCLC cases were obtained between 2010 and 2016 from the First Affiliated Hospital of Chongqing Medical University (Chongqing, China) and Inner Mongolia People’s Hospital (Inner Mongolia, China). All patients were treatment naïve before surgery. The morphology was confirmed by two pathologists. This study was approved by Inner Mongolia People’s Hospital Research Ethics Committee and written informed consent was obtained from each patient. The human NSCLC cell lines H1975, A549, HCC827 and H1650 were purchased from the Shanghai Cancer Institute. H1975, A549 and HCC827 cells were cultured in RPMI-1640 medium and H1650 cells were grown in DMEM (Gibco BRL, Grand Island, NY, USA). All media were supplemented with 10% fetal bovine serum (FBS). Cells were maintained at 37°C with 5% CO2.
Establishment of GOLM1-knock down cell lines
GOLM1 expression construct was generated by sub-cloning PCR-amplied full-length human GOLM1 cDNA into the pMSCV retrovirus plasmid, and human GOLM1 shRNA lentiviral transduction particles were cloned into pSuperretro-puro to generate pSuper-retro-TRIM14-shRNA(s). The shRNA sequences were: shRNA #1: CCGGGTGAATAACATCACCACAGGTCTCGAGACCTGTGGTGATGTTATTCACTTTTTTG, shRNA #2: CCGGGCAGGGAATGACAGAAACATACTCGAGTATGTTTCTGTCATTCCCTGCTTTTTTG, shRNA #3: CCGGGAACAGTGTGAGGAGCGAATACTCGAGTATTCGCTCCTCACACTGTTCTTTTTTG (synthesized by Sigma). siRNA of MMP13 were synthesized by GenePharma Co. Ltd. (GenePharma, Shanghai, China) and the targeted sequences was: sense, 5’-GGAGAUAUGAUGAUACUAAdTdT-3’; antisense, 5’-UUAGUAUCAUCAUAUCUCCdTdT-3’. Scrambled siRNA (sc-37007; Santa Cruz Biotechnology, Inc.) was used as a negative control.
Cell proliferation assay
NSCLC cells were seeded on 96-well microplates (1 × 105 cells per well) and cultured for 24 h, 48 h, 72 h, or 96 h respectively. 100 μl of MTT reagent (Biotium, Fremont, CA, USA) were added in each well and incubated at 37°C. After 4 h, cell viability was revealed through the conversion of the water-soluble MTT to insoluble formazan. Formazan was solubilized adding 200 μl DMSO and its concentration was measured by optical density at 450 nm.
Colony formation assay
NSCLC cells were placed into 6-well plates (1000 cells/well), incubated at 37°C for four weeks, fixed and stained with crystal violet. The mean ± SD number of colonies was counted under a microscope from three independent replicates [16].
Wound-healing assay
NSCLC cells were transfected with GOLM1-specific shRNA or a scrambled shRNA. Cells were seeded in 6-well cell culture plates and after 24 h, the cell monolayer was scratched through the central axis of the plate. Migration of the cells into the scratch was digitally documented 0 h and 24 h after being made, and percentage of migratory was calculated [17].
Transwell invasion assay
3 × 104 cells in RPMI-1640 medium with 2% FBS were seeded into the upper chamber of Matrigel-coated Transwells, which containing 8 μm pores (Corning Costar). Media with 10% FBS were added to the lower chamber. After 24 h, cells were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet. The number of invaded cells was quantified in five randomly fields [18].
Immunofluorescence
Cells were seeded on glass coverslips, fixed with 4% paraformaldehyde and incubated with primary antibodies against E-cadherin, N-cadherin and Vimentin (CST) overnight at 4°C, followed by Alexa Fluor-594 or -488- conjugated secondary antibodies (Proteintech, Chicago, IL, USA).
Quantitative real-time PCR (qRT-PCR)
Trizol reagents (Invitrogen) were applied for total RNA extraction. The mRNA levels of target genes were determined by ReverTra Ace qPCR RT kit and SYBR Green PCR kit (TakaraBio, Tokyo, Japan). qRT-PCR was performed using IQTM SYBR Green supermix and the iQ5 real-time detection system (Bio-Rad Laboratories, Hercules, CA). The comparative cycle threshold (Ct) method was applied to quantify the expression levels through calculating the 2(-ΔΔCt) method. The primers used for PCR were as follows (sense and antisense, respectively): GAPDH: TGGATTTGGACGCATTGGTC and t TTTGCACTG-GTACGTGTTGAT; GOLM1: TGGCCTGCATCATCGTCTTG and CCCTGGAACTCGTTCTTCTTCA. cDNAs amplification and relative expression values were obtained from three independent experiments. GAPDH was used as an endogenous control for mRNA.
Western blot analysis
Whole-cell lysates were prepared with RIPA buffer containing protease and phosphatase inhibitors. 30 μg cell lysates were loaded on 8% SDS-PAGE and transferred onto PVDF membranes. After membranes were blocked, they were incubated with monoclonal antibody against GOLM1 (1:1000, Signalway Antibody), GPADH (1:1000, Bioworld Technology) followed by incubation with horseradish peroxidase-conjugated IgGs (1:10000, Bioworld Biotechnology). Target proteins were detected by the ECL system (Millipore, Braunschweig, Germany) and visualized with the ChemiDoc XRS system (Bio-Rad, Hercules, CA, USA).
Tumor xenografts
Male athymic nude mice were purchased from Weitonglihua Biotechnology (Beijing, China) and maintained in a specific pathogen-free environment. The experimental protocol was approved by the First Affiliated Hospital of Chongqing Medical University Animal Care Committee and all procedures were performed in compliance with the institutional guidelines. 1 × 106 cells (n = 6/group) in 100 μl PBS were inoculated subcutaneously into the nude mice. Tumor size was measured by Vernier calipers and the tumor volume was calculated with the formula: length × (width)2 × 1/2. The mice were observed over 6 weeks for tumor formation [19]. For experimental metastasis analysis, the mice were injected at the lateral tail vein with (5 × 105) indicated NSCLC cells. Mice were sacrificed 4 weeks after inoculation and all organs were examined for the presence of macroscopic metastases. Lung metastatic nodules were determined under a dissecting microscope. Furthermore, the lung metastases were confirmed by hematoxylin and eosin staining. Animal handling and experimental procedures were approved by the First Affiliated Hospital of Chongqing Medical University Animal Care Committee.
Statistical analysis
All the analyses were completed by Graphpad prism 5 software, with P < 0.05 being considered statistically significant. The two tailed unpaired t-test was used to evaluate statistical significance between the mean values of the two groups.
Results
GOLM1 was highly expressed in NSCLC
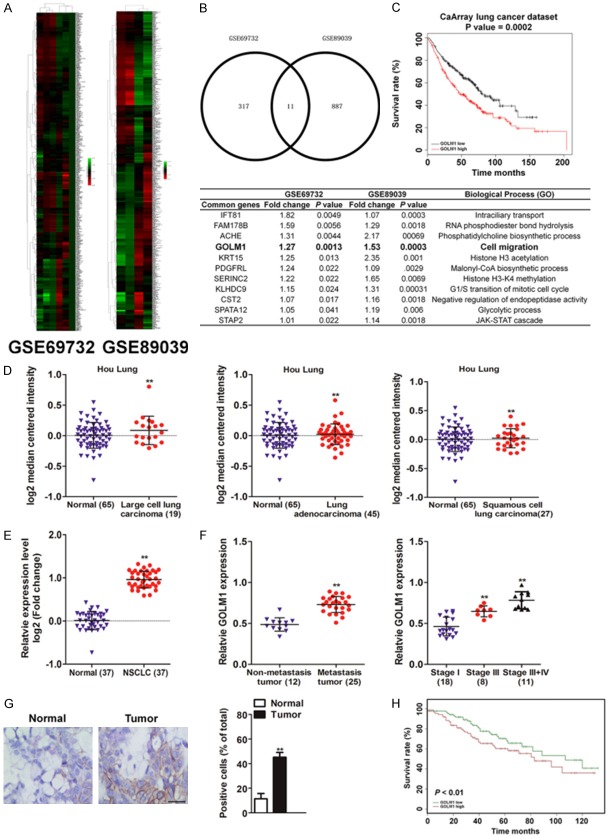
Gene expression datasets used for statistical analysis were acquired from the GEO database with the accession codes GSE69732 and GSE89039. The screening was performed in GEO datasets which contained both the lung tumor samples and the matched adjacent normal lung samples (Figure 1A). The 11 common potential over-expression genes in lung cancer were screened out based on the logFC were summarized in Figure 1B (Fold change > 1 and P < 0.05). Among these candidates, we focus on GOLM1, which was previous proved closely associated with several cancer metastases. To explore the correlation of GOLM1 with lung cancer prognosis, the relation was analyzed by the Kaplan-Meier plotter (http://kmplot.com/analysis) and the data suggested that GOLM1 was associated with poor prognosis of lung cancer patients (Figure 1C). Further Oncomine analysis (Hou J Lung dataset) [20] showed that the expression of GOLM1 was higher in tumor tissue (including lung adenocarcinoma, squamous cell Lung carcinoma, large cell lung carcinoma) than in normal tissue (Figure 1D and Supplementary Table 1) (P < 0.01). In addition, we detected the GOLM1 expression in tumor tissues and their corresponding adjacent non-tumor tissues from 37 NSCLC patients by quantitative real-time PCR (qRT-PCR). As shown in Figure 1E, the GOLM1 expression in tumor tissues was much higher than that in adjacent non-tumor tissues (P < 0.01). Furthermore, GOLM1 expression was significantly positively associated with metastasis and stages of the patients (Supplementary Table 2 and Figure 1F, P < 0.01). We further used immunohistochemistry (IHC) to investigate the expression and location of GOLM1 in tumor samples from 37 NSCLC patients. The results showed that positive staining for GOLM1 protein was evidently stronger in lung cancer tissues that in corresponding adjacent non-tumor tissues (Figure 1G). Finally, Kaplan-Meier analysis indicated that patients with higher GOLM1 expression showed poor overall survival in patient with NSCLC (Figure 1H). Altogether, these data suggest that the expression of GOLM1 was increased in NSCLC and its down-regulation is associated with poor prognosis.
Figure 1.
High expression of GOLM1 in tumor tissues of NSCLC patients. A. Expression profiling of dys-regulated mRNAs in NSCLC cancer tissues compared to normal tissues (NCBI/GEO/GSE69732 and NCBI/GEO/GSE89039). B. Summary of the common genes in GSE69732 and GSE89039 (lower panel). C. Kaplan-Meier analysis of overall survival in lung cancer patients was from Kaplan-Meier plotter (http://kmplot.com/analysis), N = 468, P < 0.01, HR = 1.6 (1.24-2.05). D. The expression of GOLM1 mRNA in primary NSCLC tissues vs. normal tissues in Oncomine database (Hou J Lung dataset). E. qRT-PCR showed that the expression of GOLM1 mRNA in tumor tissues was higher than that in correspondingly peritumoral tissues. F. High-level expression of GOLM1 was associated with lymph node metastasis of NSCLC (left panel) and TNM classification of NSCLC (right panel). G. Representative image showed the expression of GOLM1 protein in tumor samples and peritumoral tissues from NSCLC patients by IHC staining. Scale bar represents 100 μm. H. Kaplan-Meier survival test was used to test the relationship between GOLM1 expression and overall survival of NSCLC patients. **P < 0.01 as compared with normal.
High expression of GOLM1 promoted metastasis and invasion of NSCLC cells in vitro
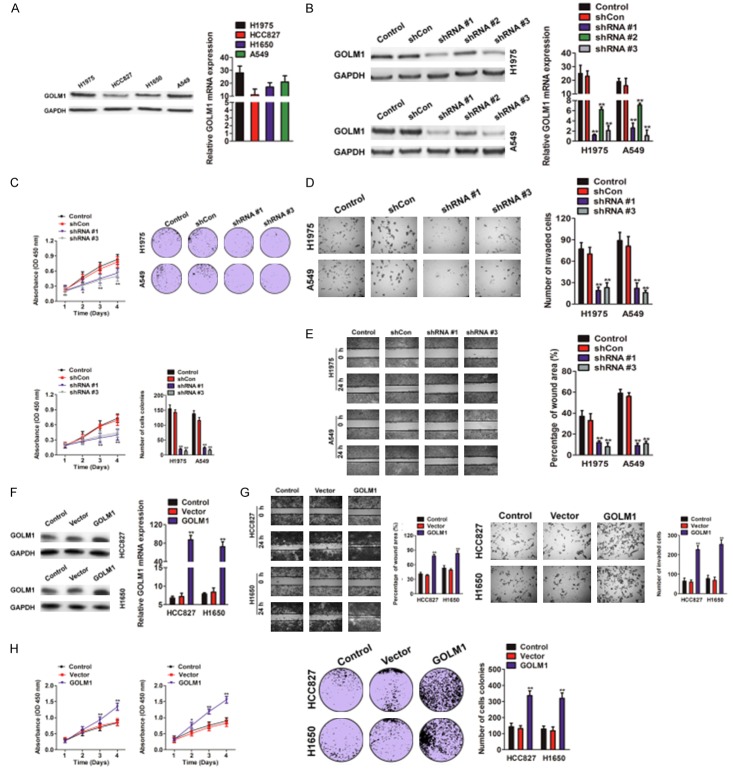
In order to investigate the role of GOLM1 in NSCLC, we first examined the expression of GOLM1 in four NSCLC cell lines by western blotting and qRT-PCR assays, and found that NSCLC cell line H1975 and A549 had higher expression of GOLM1 compared with HCC827 and H1650 cells (Figure 2A). Then, we chose H1975 and A549 cells with high expression of GOLM1, to construct stably knockdown expression of GOLM1 cells by short hairpin RNA (shRNA). shRNA # 1 and shRNA # 3 target sequence with highly interfered efficiency were chosen for function study, which verified by western blot analysis (Figure 2B). GOLM1 knocked-down slightly reduced cell proliferation and generated a decrease in the number of colony formation (Figure 2C). Transwell assay showed the impaired invasion of H1975 and A549 cells after GOLM1 interference (Figure 2D). Wound-healing assay also showed that knockdown of GOLM1 in H1975 and A549 cells dramatically inhibited the ability of migration (Figure 2E). After successful transfection with GOLM1-cDNA in NSCLC cell line HCC827 and H1650 with low level of GOLM1 (Figure 2F), the elevated GOLM1 expression significantly enhanced the ability of motility and invasion of both HCC827 and H1650 cells (Figure 2G). We also investigated the role of GOLM1 in cell proliferation and found that over-expression of GOLM1 in NSCLC cells resulted in the increased proliferation rate and colony formation (Figure 2H). These data revealed that high expression of GOLM1 promoted proliferation, invasion, and migration of NSCLC cells in vitro.
Figure 2.
Role of GOLM1 expression in the invasion and metastasis of NSCLC cells in vitro. A. The expression of GOLM1 in four NSCLC cell lines was determined by western blotting (left panel) and qRT-PCR assay (right panel). B. Western blotting and qRT-PCR were performed to assay the interference efficiency of three sequence of GOLM1 shRNA in H1975 and A549 cells. The sequence 1 and 3 GOLM1 shRNA was validated as high interference efficiency. C. H1975 and A549 cells transfected with shGOLM1 and was subjected to MTT (left panel) and colony formation (right panel) assays. D. Migration capacity of GOLM1 in H1975 and A549 cells was examined by wound healing assay. E. H1975 and A549 cells transfected with shGOLM1 and invasion ability of cells was determined by Transwell invasion assay. F. Western blotting and qRT-PCR showed up-regulated expression of GOLM1 in HCC827 and H1650 cells transfected with GOLM1 cDNA. G. Wound healing (left panel) and Transwell invasion assay (right panel) showed the enhanced mobility and invasion of HCC827 and H1650 cells transfected with GOLM1 cDNA. H. GOLM1 over-expressing HCC827 and H1650 cells were subjected to proliferation assay and colony formation analysis.
High expression of GOLM1 promoted tumor growth and progression of NSCLC cells in vivo
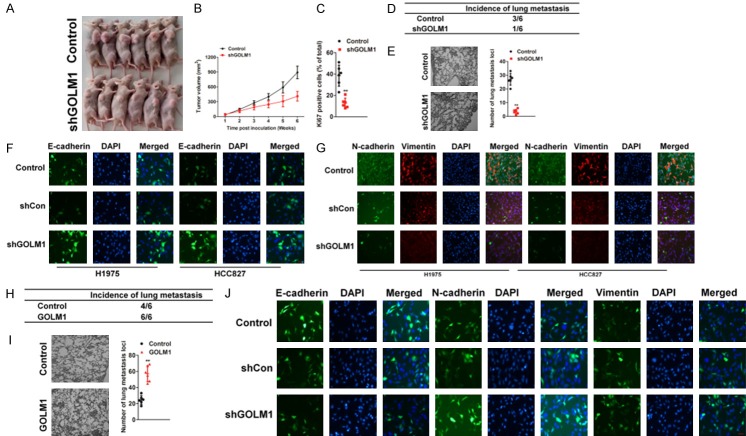
Next we constructed a subcutaneous xenograft model using H1975-GOLM1 shRNA cells and their controls. Four weeks after inoculation, all mice successfully formed palpable tumors. Tumor growth curve showed that tumors derived from H1975-control cells grew faster than those in H1975 shGOLM1 cells (Figure 3A). After inoculation for 6 weeks, the mice were killed. Tumor volume of H1975 shGOLM1 cells was 411±99 mm3, which was significantly less than that derived from and control-derived xenografts (892.15±127 mm3, P < 0.01, Figure 3B). Moreover, the Ki67 percentage score of tumor cells in shGOLM1 group was relatively decreased compared with that in control group (Figure 3C). Then, we investigated the lung metastasis rate using serial section and found that the pulmonary metastasis rate was 50% (3/6) in the H1975-control group and 16% (1/6) in H1975 shGOLM1 group (P < 0.05) (Figure 3D). Furthermore, the number of lung metastasis nodules declined significantly in tumor xenografts with GOLM1 knocked-down (Figure 3E). It is well documented that epithelial-mesenchymal transition (EMT) enables cancer cells with invasive and metastatic properties and plays critical roles in tumor progression. Here, we showed that GOLM1 knocked-down increased epithelial marker expression (E-cadherin) (Figure 3F), but suppressed mesenchymal markers levels (N-cadherin and Vimentin) in H1975 and HCC827 cells (Figure 3G). Conversely, GOLM1 overexpression displayed the opposite effect in NSCLC H1975 cells (Figure 3H-J). Collectively, these results indicate that GOLM1 could significantly promote tumor growth and tumor progression of NSCLC cells in vivo.
Figure 3.
High expression of GOLM1 promoted tumor progression of NSCLC cells in vivo. A. Representative images of tumor-bearing mice. B. Growth curve showed that tumor derived from parental cells grew faster than that of H1975-shGOLM1 group (n = 6). C. The expression of Ki67 was evaluated by IHC staining utilizing Ki67 percentage score. Data were expressed as Mean ± SD. The results were representative of three independent experiments. D. Summary of the total numbers of intravenous tumor xenografts with distant lung metastasis after injection of parental or H1975-shGOLM1 cells. E. Quantification of effects of GOLM1 down-expression on experimental metastasis in vivo. The tumor lesions confirmed by H&E staining from five sections were counted as the number of metastatic foci inside the lung tissues. F. Immunofluorescence staining images of E-cadherin were shown in H1975 and HCC827 cells transfected with shGOLM1. The cells were fixed and stained with indicated antibodies, and then detected by anti-rabbit-Alexa-488 (green). Cell nuclei were visualized by DAPI staining. G. Immunofluorescence staining images of N-cadherin and Vimentin were shown in H1975 and HCC827 cells transfected with shGOLM1. Indicated cells were fixed and stained with indicated antibodies, and then detected by anti-rabbit-Alexa-594 (red) or-488 (green). Cell nuclei were visualized by DAPI staining. H. Summary of the total numbers of intravenous tumor xenografts with distant lung metastasis at 28 days after injection of parental or GOLM1 over-expressing H1975 cells. I. Quantification of effects of GOLM1 over-expression on H1975 cells experimental metastasis in vivo. J. Immunofluorescence staining images of E-cadherin, N-cadherin and Vimentin were shown in H1975 cells transfected with GOLM1. Cells were fixed and stained with indicated antibodies, and then detected by anti-rabbit-Alexa-488 (green). Cell nuclei were visualized by DAPI staining.
Identification of MMP13 as a target gene of GOLM1 in NSCLC
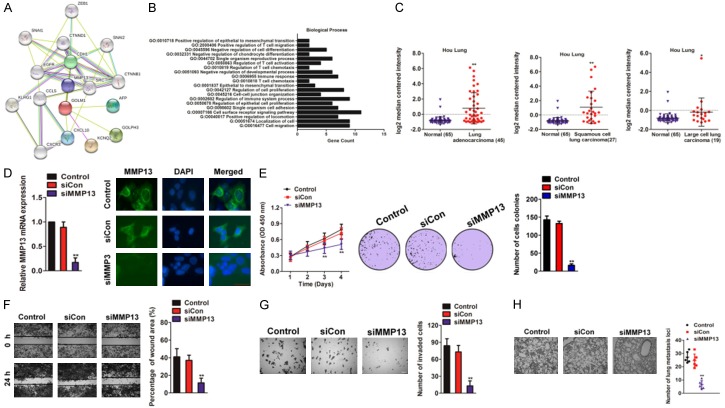
To better understand the molecular mechanism of GOLM1 in cancer metastasis, we mapped GOLM1 onto STRING database to build a PPI network. By using the ‘+ more proteins’ option, additional 17 predicted functional partners were allowed into the network. As shown in Figure 4A, MMP13 acted as a bridge to connect GOLM1. Functional enrichments of the PPI network by STRING software, using and biological process GO analysis, revealed that GOLM1 was involved in cell migration and regulation of epithelial cell proliferation (Figure 4B). Oncomine analysis of neoplastic vs. normal tissue showed that MMP13 was significantly over-expressed in different types of lung cancer in different datasets (Figure 4C and Supplementary Table 3). To investigate the biological functions of MMP13 in NSCLC cells, endogenous MMP13 was knocked-down in H1975 cells with a specific siRNA against MMP13 (siMMP13) (Figure 4D). MMP13 Knocked-down in H1975 cells inhibited cell growth and colony formation in vitro (Figure 4E). Consistently, the mobility and invasion was significantly suppressed by siMMP13 and (Figure 4F, 4G). To investigate the role of MMP13 in metastasis of NSCLC cells, the experimental metastasis assay was conducted. MMP13 knocked-down or control cells were injected into nude mice via the lateral tail vein. Four weeks post inoculation, injection of parental cells resulted in the formation of numerous metastasis loci whereas silencing of MMP13 markedly inhibited pulmonary metastasis (Figure 4H). These results implied that MMP13 silencing indeed perturbed the growth and metastasis of NSCLC cells and has a similar effect with down-expressed GOLM1.
Figure 4.
Loss of expression of MMP13 inhibits NSCLC cells growth and metastasis. A. Protein interaction network of MMP13, CDH1, CXCL10, CCL5, CHEK2, CDH1, and TP53. The colored lines between the proteins indicate the various types of evidence demonstrating the interaction. B. The significantly enriched annotation of the biological process analysis of the genes relevant to GOLM1. C. The expression of MMP13 mRNA in primary NSCLC tissues vs. normal tissues in Oncomine database (Hou Lung dataset). D. H1975 cells were transfected with either the negative control siRNA (siCon) or siMMP13. The expression levels of MMP13 were detected by qRT-PCR and immunofluorescence assay. E. After transfected with siCon or siMMP13, the cell proliferation rates were determined by the MTT assay (left panel). Colony formation assays of H1975 cells. Representative images were shown in right panel. F. H1975 cells were transfected with siMMP13 and the ability of migration was determined by wound scratch assay. Scale bar: 200 μm. G. H1975 cells with silent expression of MMP13 exhibit less invasive abilities in Transwell invasion assay. The invaded cells were stained with crystal violet and counted. Scale bar: 200 μm. H. siMMP13 cells or control cells were injected into nude mice via lateral vein. Representative pictures of lungs from mice were taken after four weeks. Numbers of lung metastasis were quantified.
Knock-down of MMP13 reverses the effects of GOLM1 in NSCCL cells
We next explored whether MMP13 knocked-down could reverse the effects of GOLM1 over-expression on H1975 and HCC827 cell proliferation, migration and invasion. H1975 and HCC827 cells were co-transfected with GOLM1 and/or MMP13 siRNA (siMMP13). The levels of MMP13 in two cell lines were verified on mRNA level (Figure 5A). Then, MTT and colony formation assays were conducted to determine whether siMMP13 reversed the acceleration effects of GOLM1 on H1975 and HCC827 cell growth in vivo. As shown in Figure 5B and 5C, knocked-down of MMP13 reversed all the effects of GOLM1 over-expression on cell proliferation and colonies formation. Consistently, cells transfected with siMMP13 inhibited the mobility and invasion in the presence GOLM1 (Figure 5D and 5E). These results suggested that GOLM1 influenced the growth and aggressiveness of H1975 and HCC827 cells by targeting MMP13.
Figure 5.
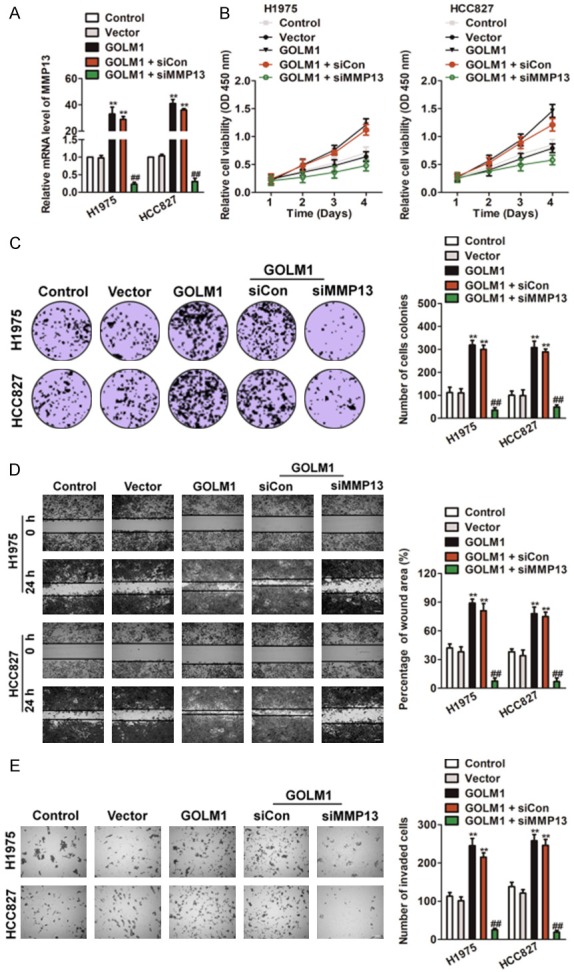
MMP13 knocked-down reversed the effects of GOLM1 in NSCLC. A. H1975 and HCC827 cells were transfected with GOLM1 or co-transfected with siMMP13 and GOLM1. The expression of MMP13 was determined by qRT-PCR analysis. B. Both H1975 and HCC8271 cells were transfected with GOLM1, or co-transfected with siMMP13 and GOLM1, and then were seeded into 96 well plates. After 24 h, 48 h, 72 h and 96 h, the MTT assay was performed to analysis cell proliferation. C. Colony formation assays of H1975 and HCC827 cells. Representative images for each treatment were shown. D. H1975 and HCC827 cells co-transfected with siMMP13 and GOLM1 were subjected to wound closure assay. The percentage of wound closure was quantified (right panel). Scale bar: 200 μm. E. Transwell invasion assay was performed after transfection of H1975 and HCC827 cells with siMMP13 and GOLM1. The invaded cells were stained with crystal violet and counted. Scale bar: 200 μm. *P < 0.01, **P < 0.01 as compared to control and ##P < 0.01 as compared to GOLM1.
Overexpression of MMP13 rescues the effects of GOLM1 down-regulation in NSCLC
To investigate the functional relevance of MMP13 targeting by GOLM1, we assessed whether MMP13 over-expression could rescue the inhibitory effects of shGOLM1 on H1975 and HCC827 cell proliferation, migration and invasion. H1975 and HCC827 cells were co-transfected with shGOLM1 and MMP13 overexpression plasmids. qRT-PCR analysis was used to validate the MMP13 mRNA in the rescue experiment (Figure 6A). The MTT analysis suggested that the exogenous expression of MMP13 rescued the inhibitory effect of shGOLM1 on cell proliferation in vitro (Figure 6B). To elucidate the functions of GOLM1/MMP13, we conducted colony formation assay. As shown in Figure 6C, shGOLM1 significantly inhibited colony formation in H1975 and HCC827 cells whereas MMP13 rescued the colony formation in NSCLC cells. After co-transfected with shGOLM1 and MMP13 overexpression plasmid, both H1975 and HCC827 cells were subjected to wound healing and Transwell invasion assay. As shown in Figure 6D, 6E, the mobility and invasion capacity inhibited by shGOLM1 was remarkably rescued by the overexpression of MMP13.
Figure 6.
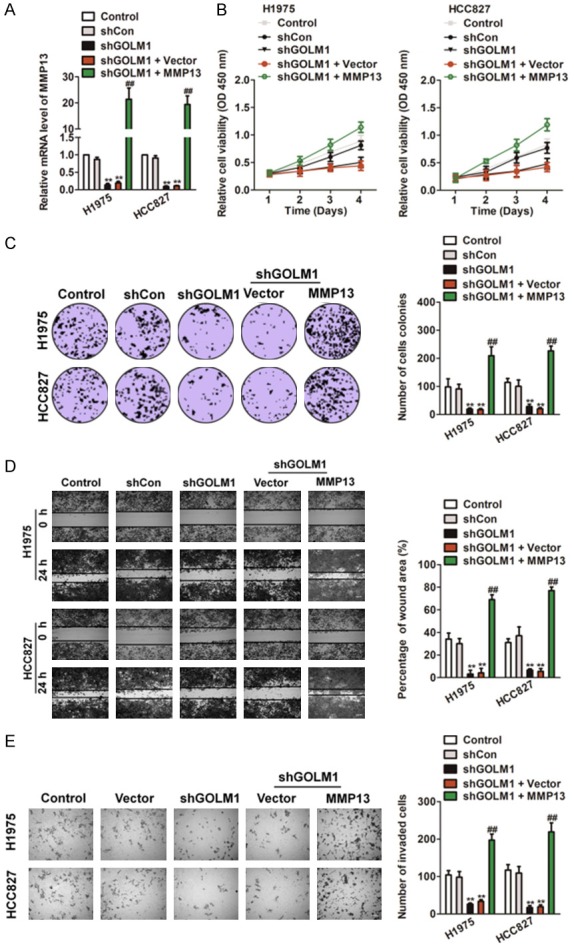
Up-regulation of MMP13 rescues the effects of GOLM1 down-expression in NSCLC. A. H1975 and HCC827 cells were transfected with shGOLM1, or co-transfected with MMP13 over-expression plasmid and shGOLM1. MMP13 expression on mRNA levels by qRT-PCR assay. B. Cells were transfected with shGOLM1 alone, or co-transfected with MMP13 and shGOLM1, and then were seeded into 96 well plates. After 24 h, 48 h, 72 h and 96 h, the MTT assay was performed to analysis cell proliferation. C. Colony formation assays of H1975 and HCC827 cells. Representative images for each treatment were shown. D. H1975 and HCC827 cells co-transfected with MMP13 over-expression plasmid and shGOLM1 were subjected to wound healing assay and images were taken at 0 and 24 h (left panel). The percentage of wound closure was quantified (right panel). E. Transwell invasion assay was performed after transfection of H1975 and HCC827 cells with MMP13 plasmid and shGOLM1. The invaded cells were stained with crystal violet and counted. *P < 0.05, **P < 0.01 as compared to control, #P < 0.05, ##P < 0.01 as compared to shGOLM1 group.
Discussion
Non-small-cell carcinoma (NSCLC) metastasis is the main cause of NSCLC-related mortality. However, its mechanism remains poorly understood. In an attempt to identify genes that are specific for NSCLC metastasis, we recently conducted a genome-wide profiling analysis by comparing primary tumors to normal. We have characterized one of the common genes in the NSCLC metastasis signature, i.e., GOLM1, for further functional studies. In the present study, we show that GOLM1 encodes a Golgi associated protein whose function is critical for NSCLC metastasis. Mechanistically, GOLM1 may promote NSCLC metastasis through its regulation of MMP13. Our study indicates that GOLM1 may serve as an effective molecular target to block NSCLC metastasis. Increasing evidences have suggested that GOLM1 is overexpressed or amplified in hepatocellular carcinoma, prostate cancer, glioma and esophageal adenocarcinoma [21,22].
In the present study, our data showed that GOLM1 expression was overexpressed in NSCLC tissues. Abundant evidences based on wound-healing assay and Transwell assay have shown that Inhibition of GOLM1 expression impaired invasion and proliferation of NSCLC cells. Importantly, inhibition of GOLM1 expression in NSCLC cells markedly impaired tumor growth and lung metastasis in vivo. It is well known that malignant tumor cells usually undergo the EMT process to gain the invasive and metastatic properties [23]. Our data showed that NSCLC cells with high expression of GOLM1 presented with high ability of metastasis and invasion. Moreover, in vitro experiment showed that elevated expression of GOLM1 in NSCLC cells could up-regulate the expression of mesenchymal marker N-cadherin, and down-regulated the expression of epithelial marker E-cadherin. Thus, our results definitely suggest that the high level of GOLM1 in NSCLC cells promote tumor progression through EMT of tumor cells. Clinically, high expression of GOLM1 in tumor tissues positively correlated to malignant phenotype and the poor prognosis of NSCLC patients.
However, the mechanisms of GOLM1 in direct metastasis capability needs to be further explored, in consideration of the migration capability is concerned with MMPs such as MMP13. Moreover, because of the significant role of MMPs in metastasis, some clinical drugs, such as Batimastat [24] and Marimastat [25], may have novel therapeutic effects for malignant tumor patients. The MMPs family proteins includes more than 20 members, which have been reported to be fundamentally involved in many biological processes, such as cell proliferation, apoptosis, angiogenesis, and invasion. For example, MMP-9 has been reported to be up-regulated in multiple tumor tissues and its overexpression can promote cancer development and progression. Additionally, MMP-2 was also significantly up-regulated in human prostate cancers and found to play an important role in prostate cancer progression. Herein, we found that MMP13 was up-regulated in NSCLC and that MMP13 overexpression promoted NSCLC aggressiveness both in vitro and in vivo, which is in agreement with oncogenic-effect of MMPs family members. Interestingly, we found that a high level of MMP13 was positively corrected with the level of GOLM1in NSCLC.
Recently, several studies reported that GOLM1 had an oncogenic role in multiple tumor types and high level of GOLM1 could serve as a biomarker for predicting poor prognosis of cancer patients [26]. In the present study, our results also showed that NSCLC patients with high expression of GOLM1 in tumor tissues had poor prognosis. Moreover, high expression of GOLM1 was significantly related to malignant phenotype, such as metastasis, high TNM stage, and poor prognosis. Overexpression of GOLM1 augmented NSCLC aggressiveness in vitro and in vivo and activated the MMP13. Therefore, understanding the biological function of GOLM1 in NSCLC progression both advance our knowledge of the mechanisms that underlie NSCLC aggressiveness, and establish GOLM1 as a potential therapeutic target for the treatment of NSCLC.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Rossi D. What can we save for the first-line treatment of NSCLC in 2016? World J Oncol. 2017;8:31–33. doi: 10.14740/wjon1017w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Postmus PE. Changing health care costs for NSCLC, what does it mean? Lung Cancer. 2018;117:62–63. doi: 10.1016/j.lungcan.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Lee G, Gardner BK, Elashoff DA, Purcell CM, Sandha HS, Mao JT, Krysan K, Lee JM, Dubinett SM. Elevated levels of CXC chemokine connective tissue activating peptide (CTAP)-III in lung cancer patients. Am J Transl Res. 2011;3:226–233. [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Q, Hu H, Jiang F, Guo CY, Yang XW, Liu X, Kuang YK. Meta-analysis of incidence and risk of severe adverse events and fatal adverse events with crizotinib monotherapy in patients with ALK-positive NSCLC. Oncotarget. 2017;8:75372–75380. doi: 10.18632/oncotarget.18536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donizy P, Kaczorowski M, Biecek P, Halon A, Szkudlarek T, Matkowski R. Golgi-related proteins GOLPH2 (GP73/GOLM1) and GOLPH3 (GOPP1/MIDAS) in cutaneous melanoma: Patterns of expression and prognostic significance. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17101619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong Y, Long Q, Xie H, Zhang T, Peng T. Cloning and characterization of human Golgi phosphoprotein 2 gene (GOLPH2/GP73/GOLM1) promoter. Biochem Biophys Res Commun. 2012;421:713–720. doi: 10.1016/j.bbrc.2012.04.067. [DOI] [PubMed] [Google Scholar]
- 7.Hu L, Li L, Xie H, Gu Y, Peng T. The Golgi localization of GOLPH2 (GP73/GOLM1) is determined by the transmembrane and cytoplamic sequences. PLoS One. 2011;6:e28207. doi: 10.1371/journal.pone.0028207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen MH, Jan YH, Chang PM, Chuang YJ, Yeh YC, Lei HJ, Hsiao M, Huang SF, Huang CY, Chau GY. Expression of GOLM1 correlates with prognosis in human hepatocellular carcinoma. Ann Surg Oncol. 2013;20(Suppl 3):S616–624. doi: 10.1245/s10434-013-3101-8. [DOI] [PubMed] [Google Scholar]
- 9.Varambally S, Laxman B, Mehra R, Cao Q, Dhanasekaran SM, Tomlins SA, Granger J, Vellaichamy A, Sreekumar A, Yu J, Gu W, Shen R, Ghosh D, Wright LM, Kladney RD, Kuefer R, Rubin MA, Fimmel CJ, Chinnaiyan AM. Golgi protein GOLM1 is a tissue and urine biomarker of prostate cancer. Neoplasia. 2008;10:1285–1294. doi: 10.1593/neo.08922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan G, Ru Y, Wu K, Yan F, Wang Q, Wang J, Pan T, Zhang M, Han H, Li X, Zou L. GOLM1 promotes prostate cancer progression through activating PI3K-AKT-mTOR signaling. Prostate. 2018;78:166–177. doi: 10.1002/pros.23461. [DOI] [PubMed] [Google Scholar]
- 11.Cao C, Xu N, Zheng X, Zhang W, Lai T, Deng Z, Huang X. Elevated expression of MMP-2 and TIMP-2 cooperatively correlates with risk of lung cancer. Oncotarget. 2017;8:80560–80567. doi: 10.18632/oncotarget.20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao XY, Lang XP. Correlation between MMP-7 and bFGF expressions in non-small cell lung cancer tissue and clinicopathologic features. Cell Biochem Biophys. 2015;73:427–432. doi: 10.1007/s12013-015-0656-y. [DOI] [PubMed] [Google Scholar]
- 13.Zhang DH, Zhang LY, Liu DJ, Yang F, Zhao JZ. Expression and significance of MMP-9 and MDM2 in the oncogenesis of lung cancer in rats. Asian Pac J Trop Med. 2014;7:585–588. doi: 10.1016/S1995-7645(14)60099-7. [DOI] [PubMed] [Google Scholar]
- 14.You Y, Shan Y, Chen J, Yue H, You B, Shi S, Li X, Cao X. Matrix metalloproteinase 13-containing exosomes promote nasopharyngeal carcinoma metastasis. Cancer Sci. 2015;106:1669–1677. doi: 10.1111/cas.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent-Chong VK, Salahshourifar I, Karen-Ng LP, Siow MY, Kallarakkal TG, Ramanathan A, Yang YH, Khor GH, Rahman ZA, Ismail SM, Prepageran N, Mustafa WM, Abraham MT, Tay KK, Cheong SC, Zain RB. Overexpression of MMP13 is associated with clinical outcomes and poor prognosis in oral squamous cell carcinoma. ScientificWorldJournal. 2014;2014:897523. doi: 10.1155/2014/897523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu YX, Wu HJ, Tan BX, Qiu C, Liu HZ. CSF-1R regulates non-small cell lung cancer cells dissemination through Wnt3a signaling. Am J Cancer Res. 2017;7:2144–2156. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Zhu P, Zhao N, Sheng D, Hou J, Hao C, Yang X, Zhu B, Zhang S, Han Z, Wei L, Zhang L. Inhibition of growth and metastasis of colon cancer by delivering 5-Fluorouracil-loaded pluronic P85 copolymer micelles. Sci Rep. 2016;6:20896. doi: 10.1038/srep20896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geng R, Tan X, Wu J, Pan Z, Yi M, Shi W, Liu R, Yao C, Wang G, Lin J, Qiu L, Huang W, Chen S. RNF183 promotes proliferation and metastasis of colorectal cancer cells via activation of NF-kappaB-IL-8 axis. Cell Death Dis. 2017;8:e2994. doi: 10.1038/cddis.2017.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiwari R, Pandey SK, Goel S, Bhatia V, Shukla S, Jing X, Dhanasekaran SM, Ateeq B. SPINK1 promotes colorectal cancer progression by downregulating Metallothioneins expression. Oncogenesis. 2015;4:e162. doi: 10.1038/oncsis.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye QH, Zhu WW, Zhang JB, Qin Y, Lu M, Lin GL, Guo L, Zhang B, Lin ZH, Roessler S, Forgues M, Jia HL, Lu L, Zhang XF, Lian BF, Xie L, Dong QZ, Tang ZY, Wang XW, Qin LX. GOLM1 modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell. 2016;30:444–458. doi: 10.1016/j.ccell.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu W, Qin L. GOLM1-regulated EGFR/RTK recycling is a novel target for combating HCC metastasis. Sci China Life Sci. 2017;60:98–101. doi: 10.1007/s11427-016-0311-x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Cui P, Ding B, Guo Y, Han K, Li J, Chen H, Zhang W. Netrin-1 elicits metastatic potential of non-small cell lung carcinoma cell by enhancing cell invasion, migration and vasculogenic mimicry via EMT induction. Cancer Gene Ther. 2017 doi: 10.1038/s41417-017-0008-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 24.Chirivi RG, Garofalo A, Crimmin MJ, Bawden LJ, Stoppacciaro A, Brown PD, Giavazzi R. Inhibition of the metastatic spread and growth of B16-BL6 murine melanoma by a synthetic matrix metalloproteinase inhibitor. Int J Cancer. 1994;58:460–464. doi: 10.1002/ijc.2910580326. [DOI] [PubMed] [Google Scholar]
- 25.Ulasov I, Thaci B, Sarvaiya P, Yi R, Guo D, Auffinger B, Pytel P, Zhang L, Kim CK, Borovjagin A, Dey M, Han Y, Baryshnikov AY, Lesniak MS. Inhibition of MMP14 potentiates the therapeutic effect of temozolomide and radiation in gliomas. Cancer Med. 2013;2:457–467. doi: 10.1002/cam4.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Lin W, Kannan K, Luo L, Li J, Chao PW, Wang Y, Chen YP, Gu J, Yen L. Aberrant chimeric RNA GOLM1-MAK10 encoding a secreted fusion protein as a molecular signature for human esophageal squamous cell carcinoma. Oncotarget. 2013;4:2135–2143. doi: 10.18632/oncotarget.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.