Abstract
Breast cancer, the most common malignancy in women worldwide, places a heavy economic burden and mental stress on families and society. Previous research showed that abnormal expression of miRNAs was closely related to the occurrence, metastasis, and angiogenesis of breast cancer. And in this study, the abnormal expression of miR-22 was detected by RT-PCR in the paired breast cancer tissues and adjacent non-tumor tissues. CCK-8 and wound healing assays were performed to evaluate the effects of the proto-oncogene ATP citrate lyase (ACLY) on the growth and metastasis of breast cancer MCF-7 cells. The results showed that miR-22 inhibited the growth and metastasis of MCF-7 cells by down-regulating the expression of ACLY. In conclusion, this study elucidated the roles of miR-22 in regulation of breast cancer differentiation and migration, which provides a target for early diagnose and therapy of breast cancer.
Keywords: miR-22, proto-oncogene, ATP citrate lyase, growth and metastasis, breast cancer
Introduction
Recently, the noncoding RNAs, such as microRNAs, competing endogenous RNA and long noncoding RNA were reported to be closely related to breast cancer proliferation, differentiation, and apoptosis, and attracted the attention of the researchers strongly [1,2]. MicroRNAs (miRNAs) are a class of abundant noncoding small RNAs, which have a complementary function to that of the 3’-untranslated region (3’-UTR) of target mRNA [3-5]. They inhibit protein synthesis and induce mRNA degradation, in addition to inducing negative regulation of the expression of target genes at the post-transcriptional level and regulation of basic physiological processes, such as cell metabolism, proliferation, differentiation, and apoptosis. Previous research showed that miRNA expression levels were altered in tumors, suggesting that miRNAs may play a role as oncogenes or tumor suppressor genes in tumorigenesis [6]. When overexpressed in tumors, miRNAs are known as oncogenes, which promote tumor progression through negative regulation of tumor suppressor genes [7]. They also control cell differentiation and apoptosis genes, such as miRNA-17-92 [7]. MiRNAs with reduced expression in tumors are called tumor suppressor genes, or tumor suppressor miRNAs, and they impede the development of tumors by inhibiting oncogenes or controlling cell differentiation and apoptosis genes, such as Let-7 [8]. MiRNAs are widely involved in the growth and development of the body and other life processes. However, they are also implicated in liver cancer, lung cancer, breast cancer, and other tumors, and miRNA expression can be used in the diagnosis, staging, prognosis, and treatment of some tumors.
Breast cancer is currently the most common malignancy in women, with about 1.2 million women suffering from breast cancer and 500,000 breast cancer-related deaths. Thus, the disease poses a serious threat to women’s health. Breast cancer as a high incidence of swollen, is a lack of a decisive basis for laboratory testing in the early diagnosis [9,10]. The specificities of proto-oncogenes and tumor suppressor genes are not specific, and the early sensitivities of tumor cells are also low. Previous studies showed that abnormal expression of miRNAs was closely related to the occurrence, metastasis, and angiogenesis of breast cancer [11,12]. Improvements in gene chip technology, reverse transcription PCR, and Northern blotting, will enable to combine the miRNA expression levels and the detection of clinical tumor genetic diagnosis [13].
For the mechanism study of breast cancer occurrence, invasion, and metastasis, differential gene analysis of breast cancer tissue and normal tissue revealed abnormal expression of miR-22 in breast cancer tissue. The studies of its effects on the invasion and metastasis of breast cancer cells would provide a theoretical basis for the target treatment and prediction of breast cancer.
Materials and methods
Cell line and animals
The MCF-7 cell line was purchased from the Cell Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in 1640 medium containing 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in an atmosphere of 5% CO2 for 2-3 days. Nude mice were obtained from the Institute of Oncology, Chinese Academy of Medical Sciences. All the experiments were performed in accordance with the guidelines for care and use of experimental animals of the experimental animal research committee of the authors’ institution.
Sequencing analysis
Large samples of breast cancer tissue and surrounding tissue were processed, and RNAs were extracted for sequencing analysis. Sequencing data were obtained using the Illumina HiSeq sequencing platform. The quality of the original sequencing data was evaluated and processed to obtain clean reads. The FPKM method was used for quantitative analysis, and differential gene expression analysis of breast cancer tissues was performed.
Design of mimics and inhibitors for synthesis and transfection
MiR-22 mimics and inhibitors were synthesized by the Gemma gene company (Suzhou, Jiangsu, China). The sequences were as follows: mimics: 5’-agttcttcagtggcaagcttta-3’, mimics-nc: 5’-uucuccgaacgugucacguuu-3’; inhibitors: 5’-uaaagcuugccacugaagaacu-3’, inhibitors-nc: 5’-caguacuuuuguguaguacaa-3’. Centrifuge tubes containing the mimics and inhibitors were added in sterile DEPCs and placed at a concentration of 20 μM. Then, 2 mL of serum-free medium without antibiotics was added to the MCF-7 cells, and a transfection solution containing 250 μL of Opti-MEM and 10 μL of Lipofectamine 2000 was added. The medium was replaced with serum containing antibiotics, the culture was continued for 24 h, and the cells were then collected.
Western blotting analysis
MCF-7 cells were harvested and lysed with ice cold lysis buffer (50 mM Tris-HCl, pH 6.8, 100 mM β-mercaptoethanol, 2 % w/v SDS, 10% glycerol). After centrifugation at 20,000×g for 10 min at 4°C, proteins in the supernatants were quantified and separated by 10% SDS-PAGE and transferred to PVDF membrane (Millipore, MA, USA). After blocking with 10% nonfat milk in PBS, the membranes were immunoblotted with antibodies as indicated, followed by HRP-linked secondary antibodies (Cell Signaling). Antibodies against ALCY (No. #4332, Cell signaling Technology, MA, USA) and GAPDH (No. #5174, Cell signaling Technology, MA, USA). Protein levels were normalized to that of total GAPDH.
Real-time quantitative polymerase chain reaction (Real-time qPCR)
According to the manufacturer’s instructions, total RNA was extracted from cervical tissue using Trizol reagent (Invitrogen, Carlsbad, CA, USA). RNA was quantified according to its absorption at 260 nm. The isolated RNA was then DNase-treated and reverse-transcribed according to manufacturer’s protocol. Briefly, miRNAs were reverse transcribed using a PrimeScript reverse transcription kit, miScript SYBRGreen PCR kit and miScript primer assays according to the manufacturer’s instructions (Qiagen, Valencia, CA, USA). Quantitative real-time PCR was performed using an ABI PRISM 7300 sequence detection system. Cycling parameters were 2 min at 50°C and 10 min at 95°C, followed by a total of 40 cycles of 15 s at 95°C and 1 min at 60°C. All of the reactions were performed in triplicate. The gene expression ΔΔCT values of the miRNA were calculated by normalizing to the internal control β-actin.
Luciferase reporter assay
ACLY double luciferase reporter wild type and mutant type were constructed. The ACLY amplification primers were as follows: h_ACLY_3UTR_F: GGCGGCTCGAGCCATACAGTGATTTACCATT, h_ACLY_3UTR_R: AATGCGGCCGCACAACATTACTTGGGATTTT, h_ACLY _mut_F59: TGTAGGTAATTACAATATTCCCAGAGAATTGTA, h_ACLY_mut_R80: GGGAATATTGTAATTACCTACAGCTGTTTTTA. MCF-7 cells (2×105/well) were seeded in 24-well plates and incubated overnight before transfection. The vector was pmiR-GY-REPORT™ (PsiCHECK-2), the reported fluorescence of the vector was hRluc, the corrected fluorescence was hluc (the internal reference correction). The 3’-UTR region of the gene was cloned into the downstream of the hRluc gene. miRNA is involved in the target gene through the 3’-UTR region. Therefore, miRNA was co-located with the constructed reporter gene vector, and the interaction of miRNA with the target gene was demonstrated by down-regulation of the relative fluorescence value of the reporter gene. Then, 48 h after co-transfection, both firefly and renilla luciferase activities were quantified using a dual luciferase reporter system (Promega, USA), according to the manufacturer’s protocols. Each treatment was performed in triplicate in three independent experiments.
Construction of a lentivirus plasmid
The full-length cDNA sequence of the ACLY gene was cloned into pCDNA3.1. Packaging and infection of the lentivirus were done with the help of the Gemma Gene Company. MCF-7 cells were infected with the overexpressed lentivirus of ACLY and formed stable experimental cell lines.
CCK-8 assay
To detect cell viability, the cells were transfected with miR-22 mimics and inhibitors for 48 h using a CCK-8 kit (Dojindo, Tokyo, Japan) according to the instructions of the manufacturer (Beijing Li Weining Biological Technology Co., Ltd, Beijing, China). The transfected cells were incubated in a cell incubator for 0.5-4 h and tested after 0.5, 1, 2, and 4 h using a microplate reader. Absorbance was measured at 450 nm.
Flow cytometry assay
The apoptoThe cells were digested with trypsin containing no EDTA and centrifuged at 1000 r/min for 5 min. The cells were then harvested and washed twice with precooled PBS. After centrifugation, the cells were stained using an Annexin V-FITC/propidium Iodide Apoptosis Detection Kit (BD Biosciences, MA, USA) according to the manufacturer’s instructions. Briefly, 5 μL of Annexin V and 1 μL of PI were added and incubated at room temperature for about 15 min. Cell apoptosis was measured on a flow cytometer. All of the treatment groups were set up two holes and the experiment was repeated at least three times.
Cell cycle detection
The transfected MCF-7 cells were digested with trypsin according to a predetermined method. After washing with PBS twice, the cells were re-suspended in 300 mL of precooled PBS, followed by the addition of 700 ¼L of precooled ethanol and incubation overnight at -20°C. The cells were centrifuged and washed once with PBS. The supernatant was centrifuged and stained with PI/Rnase staining solution. The cells were re-suspended at room temperature for 15 min. The mixture was measured by flow cytometry. The data were analyzed using ModFit LT software.
Invasion assay
For the migration assay, 5×104 cells were suspended in 1% serum medium, and the upper chamber of the transwell chamber (8 mm; BD, Biosciences, MA, USA) was added to a predeposited matrix gel. Complete medium containing 10% fetal bovine serum was added to the lower chamber. After 48 h of cell culture, the cells migrated to the other side of the membrane and were stained with 0.1% crystal violet. The cells were then stained with and examined by optical microscopy (Olympus, Japan) at 100×. All the experiments were carried out three times.
Wound healing assay
Cultured plates were seeded on the back of the line before the label, cell digestion after access to a 12-well plate, perpendicular to the orifice to create cell scratches. Absorb the cell culture medium, rinse the orifice plate three times with PBS, and wash away the scratches generated cell debris. Serum-free medium was then added, the culture plate into the incubator culture, every 4-6 h to take pictures. Analyze the experimental results based on the collected image data.
Tumor xenograft model in mice
A subcutaneous melanoma tumor model was established by a subcutaneous injection of 1×107 MCF-7 cells. In total, 16 mice were required to establish a melanoma murine model for 4 weeks. The mice were sacrificed on the 26th day after the injection, and the weights of the tumors were measured.
Statistical analysis
The results are presented as the mean ± standard deviation (SD). Data were evaluated using a one-way ANOVA analysis of variance to determine differences between the groups, and P < 0.05 was considered statistically significant.
Results
miR-22 was significantly down-regulated in breast cancer
Analysis of the expression of miR-22 in 40 matched breast tumor tissues and surrounding tissues revealed that miR-22 expression was significantly higher in breast cancer tissues than that in adjacent tissues (Figure 1).
Figure 1.
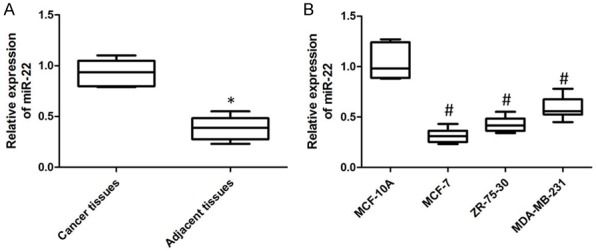
The differentiated expression of miR-22 was detected by using of qPCR. A. The down-regulated expression was detected in the cancer tissues. B. The dysregulated expression was determined in the breast cancer cell lines (P < 0.05).
Inhibition and overexpression of miR-22 influenced ACLY expression
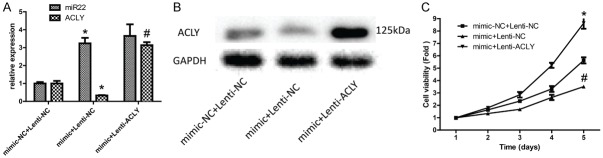
MCF-7 cells were up-regulated and down-regulated miR-22 with transfection with miR-22 mimics and inhibitors, respectively. qPCR was used to detect the transfected efficiencies of them. As shown in Figure 2A, miR-22 expression decreased significantly when MCF-7 cells were transfected with miR-22 inhibitors, and vice versa. Besides, up-regulation of miR-22 reduced the expression of ACLY protein and down-regulation of miR-22 increased ACLY protein level (Figure 2B). As the luciferase reporter assay results shown, up-regulation of miR-22 reduced the fluorescence intensity of wild type ACLY 3’-UTR regions as compared with that of cells co-transfected with miR22 mimics and mutant-type reporter vectors (Figure 2C).
Figure 2.
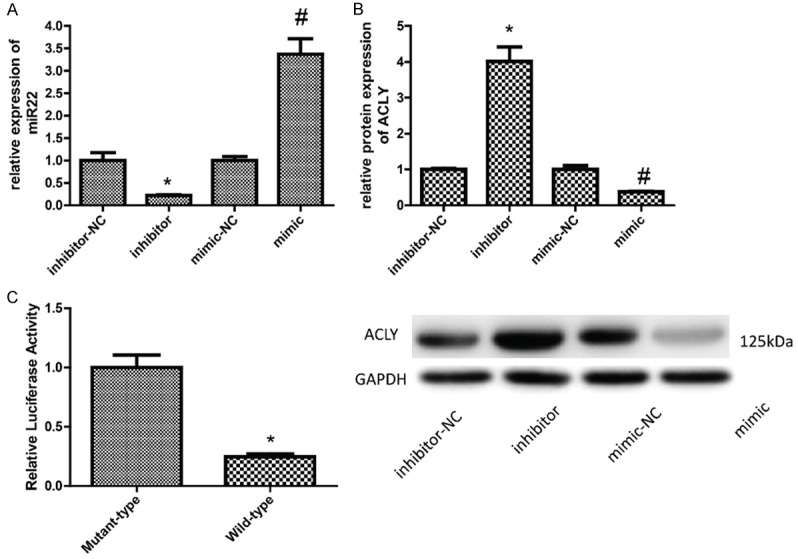
Detection of the effect of miR-22 on the expression and transcriptional activity of ACLY. A. The expression of miR-22 was detected by qPCR 24 h after MCF-7 cells were transfected with miR-22 mimics and inhibitors. B. Western blotting analysis of the protein level of ACLY after MCF-7 cells were transfected with miR-22 mimics and inhibitors for 48 h. C. A dual-luciferase reporter system was used to determine the effect of miR-22 on the transcriptional activity of ACLY (P < 0.05).
Effects of inhibition and overexpression of miR-22 on cell proliferation
The CCK-8 assay was used to detect the proliferation of MCF-7 cells when the cells were transfected with miR-22 mimic and inhibitor. As shown in Figure 3, overexpression of miR-22 inhibited the proliferation of MCF-7 cells, whereas inhibition of miR-182 promoted the proliferation of MCF-7 cells.
Figure 3.
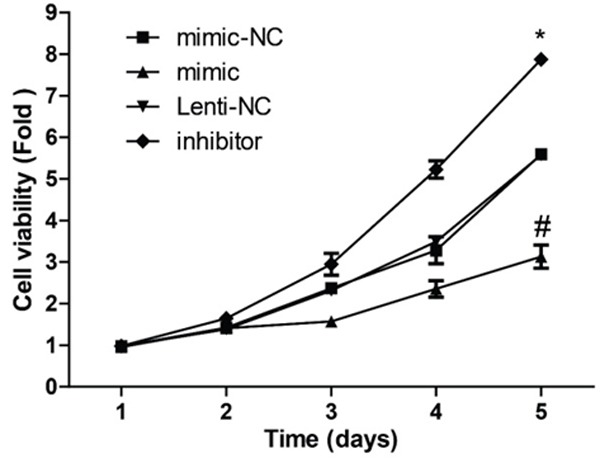
Detection of the effect of miR-22 on cell proliferation. CCK-8 assay was used to detect MCF-7 cell proliferation 24, 48, 72, 96 and 120 h after the cells were transfected with miR-22 mimics and inhibitors (P < 0.05).
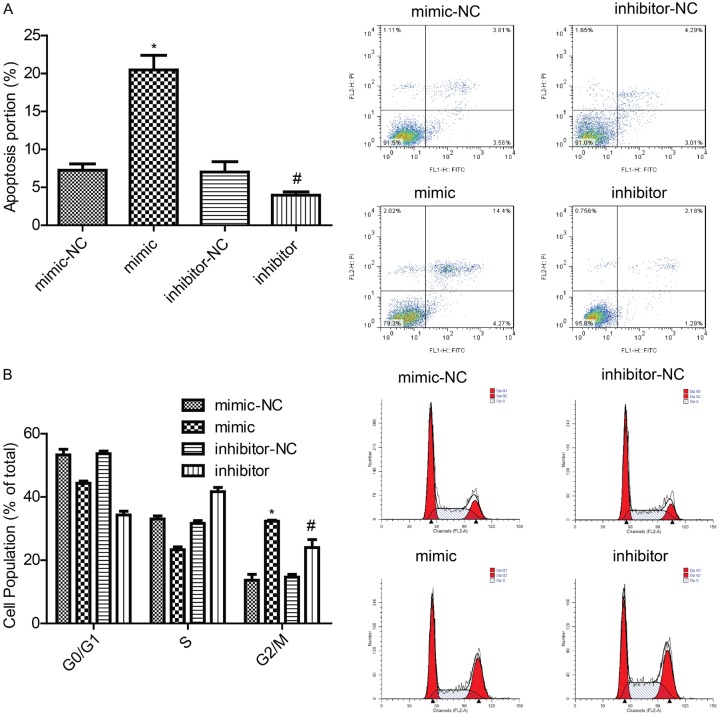
Effects of inhibition and overexpression of miR-22 on cell apoptosis and cell cycle
Cell cycle and apoptosis were detected by flow cytometry. As shown in Figure 4A, overexpression of miR-22 promoted MCF-7 cell apoptosis, whereas down-regulation of miR-22 repressed MCF-7 cell apoptosis. Moreover, overexpression of miR-22 in MCF-7 cells reduced the S phase of the cells, whereas down-regulation of miR-22 had the opposite effect (Figure 4B). These results showed that miR-22 inhibited cell growth and modulated cell cycle in breast cancer.
Figure 4.
Detection of the effects of miR-22 on cell apoptosis and the cell cycle. A, B. A flow cytometry assay was used to detect MCF-7 cell apoptosis and the cell cycle 48 h after the cells were transfected with miR-22 mimics and inhibitors (P < 0.05).
Effects of inhibition and overexpression of miR-22 on cell invasion and migration
Transwell and wound healing assays were performed to detect the invasion (Figure 5) and migration (Figure 6) of MCF-7 cells with inhibition and overexpression of miR-22. And results showed that overexpression of miR-22 weakened the invasion and migration abilities of MCF-7 cells, whereas inhibition of miR-22 enhanced the invasion and migration of MCF-7 cells.
Figure 5.
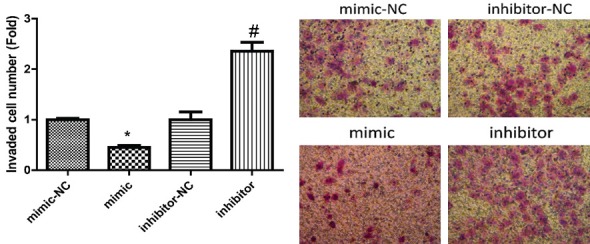
Detection of the effects of miR-22 on cell invasion. Transwell assay was used to detect MCF-7 cell invasion 48 h after the cells were transfected with miR-22 mimics and inhibitors (P < 0.05).
Figure 6.
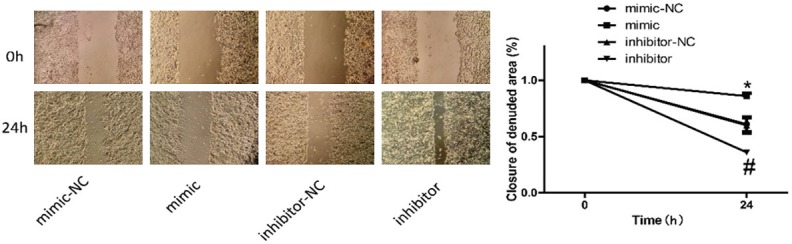
Detection of the effects of miR-22 on cell migration. Wound healing assay was used to detect MCF-7 cell migration 24 h after the cells were transfected with miR-22 mimics and inhibitors (P < 0.05).
Effect of simultaneous overexpression of miR-22 and ACLY on breast cancer progression
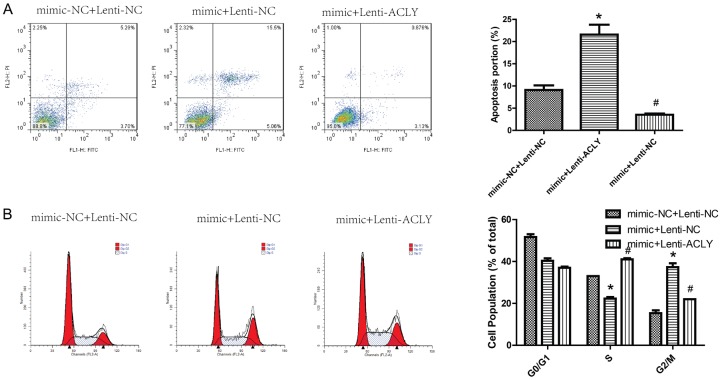
After MCF-7 cells were overexpressed miR-22 and ACLY, qPCR and Western blotting were used to the transfected efficiency. As shown in Figure 7A, 7B, the expression of miR-22 was increased, whereas ACLY expression was decreased after miR-22 was overexpressed in MCF-7 cells. Compared with the control group, overexpression of miR-22 only inhibited cell proliferation, whereas overexpression of both miR-22 and ACLY promoted cell proliferation (Figure 7B), invasion (Figure 8), and migration (Figure 9). Besides, overexpression of miR-22 only promoted cell apoptosis, whereas overexpression of both miR-22 and ACLY impaired this effect (Figure 10A). Moreover, overexpression of miR-22 induced a reduced S phase of MCF-7 cells, and this effect was abolished when overexpression of miR-22 and ACLY together (Figure 10B). Taken together, these results demonstrated that miR-22 inhibited the progression of breast cancer through down-regulating ACLY expression.
Figure 7.
The effect of miR-22/ACLY on MCF-7 cell proliferation, as detected by CCK-8. A. qPCR was used to detect the levels of ACLY and miR-22 in MCF-7 cells after the cells were transfected with mimics-NC + lentiv-NC, mimics + lentiv-NC, and mimics + lentiv-ACLY. B. Western blotting analysis of the protein level of ACLY in MCF-7 cells after the cells were transfected with mimics-NC + lentiv-NC, mimics + lentiv-NC, and mimics + lentiv-ACLY. C. CCK-8 analysis of the proliferation of MCF-7 cells after the cells were transfected with mimics-NC + lentiv-NC, mimics + lentiv-NC, and mimics + lentiv-ACLY (P < 0.05).
Figure 8.
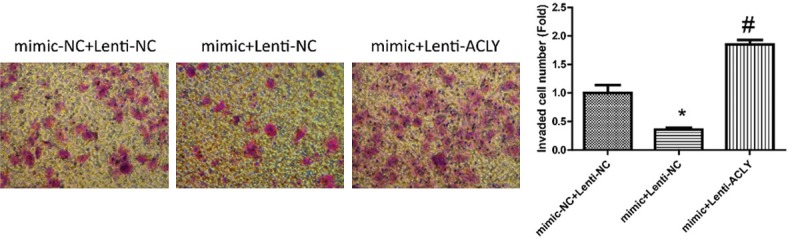
Detection of the effects of miR-22/ACLY on cell invasion. Transwell assay was used to evaluate the invasion of MCF-7 cells after the cells were transfected with mimics-NC + lentiv-NC, mimics + lentiv-NC, and mimics + lentiv-ACLY (P < 0.05).
Figure 9.
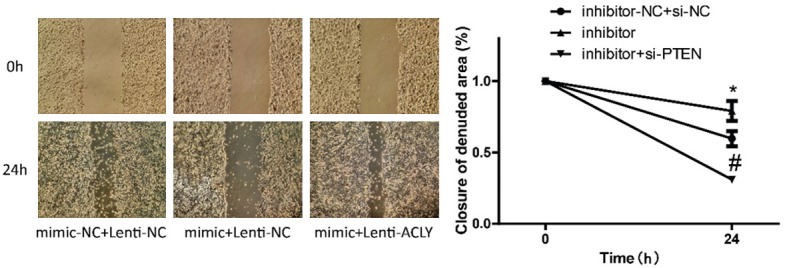
Detection of the effects of miR-22/ACLY on cell migration. Wound healing assay was used to evaluate the migration of MCF-7 cells after the cells were transfected with mimics-NC + lentiv-NC, mimics + lentiv-NC, and mimics + lentiv-ACLY (P < 0.05).
Figure 10.
Detection of the effects of miR-22/ACLY on cell apoptosis and the cell cycle. A, B. A flow cytometry assay was used to evaluate apoptosis and the cell cycle of MCF-7 cells after the cells were transfected with mimics-NC + lentiv-NC, mimics + lentiv-NC, and mimics + lentiv-ACLY (P < 0.05).
Effects of overexpression of miR-22 and ACLY on cell tumorigenesis
MCF-7 cells transfected with miR-22 mimics and lentiv-ACLY were injected into nude mice. After 2 weeks, the mice were sacrificed, and the sizes of the tumors were measured. As shown in Figure 11, overexpression of miR-22 inhibited cell tumor formation ability, whereas this effect was abolished when up-regulation of miR-22 and ACLY together. This result illustrated that miR-22 repressed the carcinogenesis of breast cancer through down-regulating ACLY expression.
Figure 11.
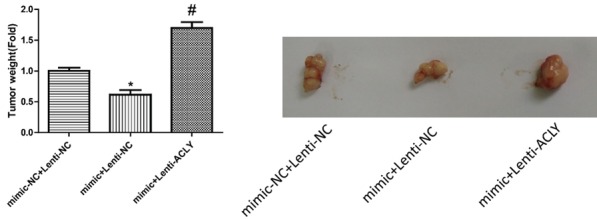
Detection of the effects of miR-22/ACLY on cell tumorigenesis. Tumor xenograft model was established in nude mice to evaluate the tumorigenesis of MCF-7 cells after the cells were transfected with mimics-NC + lentiv-NC, mimics + lentiv-NC, and mimics + lentiv-ACLY (*P < 0.05).
Discussion
In the vast majority of breast cancer patients, effective control of invasion and metastasis is vital to reduce mortality. In recent years, the mechanism underlying the role of miRNAs in invasion and metastasis in breast cancer has received widespread attention. The ontogenesis of breast cancer is a complex process involving multiple genes. Previous research demonstrated that miRNAs had multiple gene regulatory functions and that they were involved in the metastasis of many kinds of tumors, including breast cancer [14,15]. For example, research demonstrated that miR-21, the first miRNA to be recognized in the human genome, was overexpressed in various human tumors [16-18], which also affected cell invasion and metastasis of breast cancer [19,20] and was associated with the prognosis of cancer patients. And the present study demonstrated that miR-22 inhibited the proliferation, migration, invasion and tumorigenesis of breast cancer cells, suggesting that miR-22 served as a tumor suppressor in the progression of breast cancer.
In response to tumor cell proliferation, the demand for energy and macromolecules markedly increases [18]. Thus, changes in metabolic pathways are one of the most important features of tumor cells [21]. Among the series of changes and abnormalities observed in metabolic pathways, the synthesis of new fatty acids is considered crucial [22]. Most normal cells, even under high proliferative conditions, are more likely to use exogenous lipids, such as from the diet and other means. In addition to adipocytes, hepatocytes, and other small cells, in most normal cells, the new fatty acid synthesis pathway is inhibited. However, in tumor cells, inhibition of the fatty acid synthesis pathway is not inhibited. Thus, large amounts of lipids are synthesized, triggering a series of synergistic effects, such as signal transduction abnormalities, changes in gene expression profiles, and susceptibility to drug therapy. Previous research demonstrated that ACLY played a key role in such processes [23]. It also showed that the PI3K/Akt signaling pathway played a key regulatory role in a variety of tumors and that ACLY activated the Akt signaling pathway to promote tumor progression [24].
Previous research demonstrated that ACLY, a cytoplasmic enzyme, catalyzed the conversion of citrate to acetyl-CoA, a key molecule of cell endogenous synthetic fatty acids and cholesterol [23]. It also showed that the ACLY gene played a key role in the control of various metabolic pathways in cells [25]. Furthermore, research showed that acetyl-CoA participated in the modification (acetylation) of proteins, such as histones. A recent study revealed that ACLY was highly expressed in a variety of tumors and inhibition of its activity induced tumor cell proliferation arrest [23], suggesting that ACLY might exert as an oncogene in the progression of cancers. Similarly, the present study showed that the effects of proliferation, migration, invasion and carcinogenesis inhibition induced by miR-22 overexpression were all impaired when up-regulating ACLY, which indicated that miR-22 inhibited the occurrence and development of breast cancer through down-regulating ACLY expression and ACLY served as an oncogene in the progression of breast cancer.
ACLY is dependent on citrate as a substrate. Previous research showed that deletion of ACLY led to citrate accumulation in cytoplasm and mitochondria, followed by glycolysis and mitochondrial function abnormalities, in addition to Akt inactivation, resulting in weakening of the signal related to tumor proliferation [26]. Furthermore, research showed that ACLY affected the methylpenta-dihydroxy acid pathway and that inhibiting ACLY by down-regulating cholesterol and isoprenoid blocked tumor proliferation [26]. In addition, studies demonstrated that ACLY played important roles in the epigenetics of tumors and that it catalyzed the conversion of citrate to acetyl-CoA, which participated in the acetylation of histones [23,27], suggesting that ACLY is an important regulator of the epigenetics of tumors. In addition, research showed that ACLY regulated a series of metabolic-related genes by affecting the acetylation of histones, such as glucose transporter 4, hexokinase 2,6-phosphofructokinase 1, human lactate dehydrogenase A [28]. Therefore, via genetic modification, ACLY might affect the expression of tumor-related genes and promote the progression of tumors. In this study, the expression level of ACLY largely affected the physiological state of tumor cells and played vital roles in the function of breast cancer cells. However, the underlying mechanism of ACLY in the oncogenesis of breast cancer remains largely unclear. Therefore, we will explore the mechanism of ACLY, such as its effects on citrate and metabolic-related genes expression, in breast cancer in our next study.
Previous results based on differential gene analysis showed that the expression level of miR-22 in breast cancer was low, but its underlying mechanism remains unclear. The present study, for the first time, demonstrated that miR-22 inhibited the growth and metastasis of MCF-7 cells by down-regulating the activity of the proto-oncogene ACLY in the 3’-UTR region. This study elucidated the roles of miR-22 in the regulation of breast cancer differentiation and migration and its underlying mechanism. These findings provide an important basis for early prevention and targeted therapy of breast cancer.
Acknowledgements
This work was supported by grants from Suzhou Science and Technology Development Program (SZS201509).
Disclosure of conflict of interest
None.
References
- 1.Tian J, Wang Y, Zhang X, Ren Q, Li R, Huang Y, Lu H, Chen J. Calycosin inhibits the in vitro and in vivo growth of breast cancer cells through WDR7-7-GPR30 Signaling. J Exp Clin Cancer Res. 2017;36:153. doi: 10.1186/s13046-017-0625-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen B, Wang J, Dai D, Zhou Q, Guo X, Tian Z, Huang X, Yang L, Tang H, Xie X. AHNAK suppresses tumour proliferation and invasion by targeting multiple pathways in triple-negative breast cancer. J Exp Clin Cancer Res. 2017;36:65. doi: 10.1186/s13046-017-0522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Y, Li Z, Wang Y, Li L, Wang D, Zhang W, Liu L, Jiang H, Yang J, Cheng J. Overexpression of miR-29b reduces collagen biosynthesis by inhibiting heat shock protein 47 during skin wound healing. Transl Res. 2016;178:38–53. e36. doi: 10.1016/j.trsl.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Oliveto S, Mancino M, Manfrini N, Biffo S. Role of microRNAs in translation regulation and cancer. World J Biol Chem. 2017;8:45–56. doi: 10.4331/wjbc.v8.i1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji W, Sun B, Su C. Targeting microRNAs in cancer gene therapy. Genes (Basel) 2017;8 doi: 10.3390/genes8010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science. 2001;294:797–799. doi: 10.1126/science.1066315. [DOI] [PubMed] [Google Scholar]
- 7.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 8.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 9.Usmani A, Shoro AA, Shirazi B, Memon Z, Hussain M. MiR-16: a novel hereditary marker in breast cancer and their offspring. J Pak Med Assoc. 2017;67:446–450. [PubMed] [Google Scholar]
- 10.Le L, Schairer C, Hablas A, Meza J, Watanabe-Galloway S, Ramadan M, Merajver SD, Seifeldin IA, Soliman AS. Reliability of medical records in diagnosing inflammatory breast cancer in Egypt. BMC Res Notes. 2017;10:126. doi: 10.1186/s13104-017-2433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith B, Agarwal P, Bhowmick NA. MicroRNA applications for prostate, ovarian and breast cancer in the era of precision medicine. Endocr Relat Cancer. 2017;24:R157–R172. doi: 10.1530/ERC-16-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teoh SL, Das S. The role of MicroRNAs in diagnosis, prognosis, metastasis and resistant cases in breast cancer. Curr Pharm Des. 2017;23:1845–1859. doi: 10.2174/1381612822666161027120043. [DOI] [PubMed] [Google Scholar]
- 13.O’Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Negrini M, Calin GA. Breast cancer metastasis: a microRNA story. Breast Cancer Res. 2008;10:203. doi: 10.1186/bcr1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, Li X, Li G, Zeng Z, Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandres E, Agirre X, Ramirez N, Zarate R, Garcia-Foncillas J. MicroRNAs as cancer players: potential clinical and biological effects. DNA Cell Biol. 2007;26:273–282. doi: 10.1089/dna.2006.0544. [DOI] [PubMed] [Google Scholar]
- 17.Subramanian S, Steer CJ. MicroRNAs as gatekeepers of apoptosis. J Cell Physiol. 2010;223:289–298. doi: 10.1002/jcp.22066. [DOI] [PubMed] [Google Scholar]
- 18.Dykxhoorn DM. MicroRNAs and metastasis: little RNAs go a long way. Cancer Res. 2010;70:6401–6406. doi: 10.1158/0008-5472.CAN-10-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song B, Wang C, Liu J, Wang X, Lv L, Wei L, Xie L, Zheng Y, Song X. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res. 2010;29:29. doi: 10.1186/1756-9966-29-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 21.Furuta E, Okuda H, Kobayashi A, Watabe K. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim Biophys Acta. 2010;1805:141–152. doi: 10.1016/j.bbcan.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 23.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Migita T, Narita T, Nomura K, Miyagi E, Inazuka F, Matsuura M, Ushijima M, Mashima T, Seimiya H, Satoh Y, Okumura S, Nakagawa K, Ishikawa Y. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008;68:8547–8554. doi: 10.1158/0008-5472.CAN-08-1235. [DOI] [PubMed] [Google Scholar]
- 25.Zaidi N, Swinnen JV, Smans K. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res. 2012;72:3709–3714. doi: 10.1158/0008-5472.CAN-11-4112. [DOI] [PubMed] [Google Scholar]
- 26.Hanai J, Doro N, Sasaki AT, Kobayashi S, Cantley LC, Seth P, Sukhatme VP. Inhibition of lung cancer growth: ATP citrate lyase knockdown and statin treatment leads to dual blockade of mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3-kinase (PI3K)/AKT pathways. J Cell Physiol. 2012;227:1709–1720. doi: 10.1002/jcp.22895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 28.Deb DK, Chen Y, Sun J, Wang Y, Li YC. ATP-citrate lyase is essential for high glucose-induced histone hyperacetylation and fibrogenic gene upregulation in mesangial cells. Am J Physiol Renal Physiol. 2017;313:F423–F429. doi: 10.1152/ajprenal.00029.2017. [DOI] [PubMed] [Google Scholar]