Abstract
Objective: The expression level and clinical significances of long non-coding RNAs (LncRNAs) are presently unknown in the early-stage cervical cancer (CC). This study was aimed to explore the expression signatures of lncRNAs between normal and cervix carcinoma tissues and the prognostic value of LncRNAs in early-stage CC patients. Materials and Methods: The patients diagnosed with FIGO stage I-IIb CC of the First Affiliated Hospital of Sun Yat-sen University between January 1st 2006 and December 31st 2009 were retrospectively reviewed. Molecular microarray was conducted to identify differentially expression profiles of LncRNAs. In situ hybridization was applied for detection of candidate lncRNAs in cervical tissues. Results: A total of 2574 upregulated lncRNAs and 3270 downregulated lncRNAs with significantly differential expression (≥2.0-fold) were identified. Among the differentially expressed lncRNAs, RP11-396F22.1 expression was one of the most significantly overexpressed in the CC tissues compared to nomal cervical tissues (P<0.001). In situ hybridization confirmed RP11-396F22.1 expression was highly expressed in cancerous tissues. The results of Scratch and Transwell test showed that the migration ability decreased remarkably in transfected group (P<0.001). Moreover, the coding gene cpne8 was significantly upregulated by RP11-396F22.1 knockdown (P=0.035). Conclusions: These findings demonstrate that LncRNA RP11-396F22.1 might be a potent biomarker for CC progression.
Keywords: Long noncoding RNA RP11-396F22.1, cervical cancer, prognosis, apoptosis, cpne8
Introduction
Cervical cancer (CC) is known as the fourth common cause of cancer death worldwide, and the second pandemic malignant tumor in developing countries [1,2]. According to data of 2012, an estimated 527,600 new CCs and 265,700 CC deaths occurred each year in the world, and 25 percent of those (about 131,500 cases) were diagnosed in China. It’s reported that the tumor markers such as squamous cell carcinoma (SCC), carbohydrate antigen 125 (CA125) and ccarcinoembryonic antigen (CEA) raise in patients with terminal CC, which are associated with disease activity, inflammation, and other physiological and pathological processes [3-5]. Cervical cytology and high risk-HPV detection lead to an earlier discovery and treatment of CC and precancerous lesions, but five years survival rate maintained at 69~70% has not decreased significantly over the past 40 years [6]. One of the most prominent parts is accurate diagnosis of preoperative staging and prognosis for patients with stage IIB CC [7]. Therefore, it is vital to determine the invasion degree and lymph node metastasis and establish an accurate operation or radiochemotherapy for the improvement of cervical cancer patients. The previous evidences indicated that long non-encoding RNAs (LncRNAs) had functions of regulation similar to proto oncogene or tumor suppressor gene, which were associated with biological behavior of malignant tumors [8-10]. Though many researches prove that a regulation of LncRNAs will affect the growth and invasion ability of tumors, the relation between incidence of CC and LncRNAs is still lacking. This study intends to obtain LncRNAs expression profiling in CC tissues using molecular microarray screening for discovery of differential expression of LncRNAs. Then the target LncRNAs linked to incidence and prognosis of CC are traced with retrospective analysis of clinical cases for exploring the potential of LncRNAs that act as the sensitive markers for poor prognosis and metastasis of cervical cancer.
Materials and methods
Clinical patient data
The 141 patients diagnosed with FIGO stag I~IIb CC were obtained from the department of gynaecology of the First Affiliated Hospital of Sun Yat-sen University between January 2006 and December 2009. Pathologic types of CC in this study included squamous cell carcinomas, adenocarcinoma and adenosquamous carcinomas. Normal cervical tissues collected from benign gynecological disease resection were regarded as controls. All tissues had no visible and microscopic changes with pathological confirmation. The average age of participators was 48.8 years. In addition, the general exclusion criteria for the patients are as follows: 1st: rare pathological types (e.g. sarcoma of the cervix and cervical small cell carcinoma); 2nd: CC patients complicated with other malignant tumor of organs; 3rd: CC patients with end-stage medical disease (e.g. renal failure and cardiac dysfunction); 4th: patients died from accidental death. 141 cases were enrolled in this study followed up for 6 years. The shortest follow-up patient died 12 month after surgery, and the longest follow-up time was 108 months. This study was approved by the Medical Ethics Committee of The First Affiliated Hospital, Sun Yat-sen University with written informed consent obtained from all patients.
Cell culture
Four human cervical carcinoma cell lines (SiHa, HeLa, C33a and Ms751) were cultured in DMEM/F-12 (Gibco, USA) with 10% FBS (Gibco, USA) at 37°C in a humidified 5% CO2. miRNA-32 mimics and inhibitors and Cpne8 siRNA were from Dharmacon.
qRT-PCR
Total RNAs were isolated from tissue samples of 141 patients diagnosed with FIGO stag I~IIb CC (141 para-carcinoma control tissues) and cells (SiHa, HeLa, C33a and Ms751), respectively. Reverse transcription was carried out according to the protocol of TaKaRa PrimeScriptTM RT Master Mix Kit. qRT-PCR assay was performed using FastStart Universal SYBR Green Master (ROX) Kit (Roche, Switzerland). The primers were synthesized by Shanghai Generay Biotech Co., Ltd. RP11-396F22.1 sense primer was 5’-GCAAGAACTGAGACCTGACG-3’ and the anti-sense primer was 5’-TAAGCACACCACTCCACTGC-3’; cpne8 sense primer was 5’-CAATGGTCGAATTGGATGGAGA-3’ and the anti-sense primer was 5’-GCCATGCTCAGTATGTGGTTTC-3’; miRNA-32 sense primer was: 5’-CGGTATTGCACATTACTAAGTTGCA-3’ and the anti-sense primer was: 5’-CTCGCTTCGGCAGCACA-3’. The Actin was the internal reference, and its sense primer was 5’-GCAAAGACCTGTACGCCAA-3’ and the anti-sense primer was 5’-GGAGGAGCAATGATCTTGATCTTC-3’ (Shanghai GenePharma Co., Ltd., Shanghai, China). Relative expressions of RP11-396F22.1 in the cancer tissues and cell lines were calculated using the 2-ΔΔct method.
siRNA transfection
Small interfering RNA that targeted RP11-396F22.1 (si-RP11-396F22.1) and a scrambled negative control (si-NC) were designed and synthesized by Genepharma (Shanghai, China). The sequences of si-RP11-396F22.1 were si-RP11-396F22.1-1: 5’-CGUUUCCAAAGGCAUGAAATT-3’, 5’-UUUCAUGCCUUUGGAAACGTT-3’; si-RP11-396F22.1-2: 5’-CCAAAGGCAAAGAAUAUCATT-3’ and 5’-UGAUAUUCUUUGCCUUUGGTT-3’; si-NC: 5’-UUCUCCGAACGUGUCACGUTT-3’, 5’-ACGUGACACGUUCGGAGAATT-3’. Human cervical cancer cells were transfected with either 50 nmol si-RP11-396F22.1 or si-NC using HiPerFect Transfection (cat NO. 301705) according to the manufacturer’s instruction (QIAGEN, Germany).
Biochip hybridization
Molecular microarray was used in this procedure according to the protocol of Agilent Gene Expression Hybridization Kit. 10× Blocking Agent was made up to a volume of 500 μl with nuclease-free water, then votex the agent gently. Fragmentation mixcontaining cRNA was mixed with 2×GE Hybridization buffer HI-RPM. The samples were spun in a microcentrifuge at 13,000 rpm for 1 min at room temperature to drive any residual sample from the walls and lid of the tubes and to help with bubble reduction. Tissue samples of 141 CC patients and para-carcinoma control tissues were pipetted into the gasket slide well of the Agilent SureHyb chamber and the ‘active side’ of the array placed directly on top of the gasket slide to form a sandwich pair. The SureHyb chamber cover was placed on the sandwich slides and the clamp assembly tightened onto the chamber. The assembled slide chamber was then placed in a rotisserie hybridization oven at 10 rpm and the samples allowed to hybridize at 65°C for 17 h. The slides were then washed with Wash Buffer 2 which was pre-warmed to 37°C. In addition, 100% acetonitrile was added to staining dish with medium speed stir. After elution for 5 min, the slides were dried and stained. Then the slides were scanned using GenePix 4000B microarray Scanner. Each determination of fluorescence intensity in a single sample was performed three times. The data were analyzed with Gene Spring GX v11.5.1 (Agilent Technologies). The differences of data among parallel experiments were normalized using quantile normalization. The quality of the genetic data obtained after filtering and the distribution of data sets were assessed and visualized by creating box plots, which showed that there were no significant differences in the distributions of log2 ratios among samples.
Bioinformatics analysis
In order to identify differentially expressed lncRNAs with statistical significance, a Volcano Plot filtering was performed between CC group and normal control group. The threshold we used to screen Up or Down regulated lncRNAs was Fold Change ≥2.0 and p-value ≤0.05. The result of cluster shows distinguishable lncRNA expression profiling among 12 samples.
In situ hybridization
Slices were processed using EmiRCURY LNA™ microRNA ISH Optimization Kit (FFPE) (Denmark) according to the manufacturer’s protocol. The In situ hybridization probes designed by Exiqon technology (Denmark) were as follows: LncRNA RP11-396F22.1 Detection probe (DNA oligo/5DigN/AGCTGCGTTATATGGTTGTGA/3Dig_N/), LncRNA Detection control probe, beta actin hasmmurno (/5DigN/CTCATTGTAGAAGGTGTGGTGCCA/), LncRNA Detection scramble probe, (DNA oligo/5DigN/GTGTAACACGTCTATACGCCCA/3Dig_N/). Briefly, tissue samples were treated with Proteinase K (15 μg/mL, 300 μl per section) for 10 min at 37°C. After washed and dehydrated, the slices were dried for 10 min at room temperature. The samples were incubated for 60 min at 50°C with 25 μl hybridization mix containing target RNA and primers. After washed with SSC buffer, the sections were incubated with blocking solution for 15 min at room temperature. Anti-digoxigenin-AP (anti-DIG-AP, 1:200) reagent was added to tissue slices at 4°C overnight. After washed three times for 3 min each with PBS-T, the samples were then stained, incubated with KTBT, and washed. Finally, dehydration, neutral balata and camera observation were performed. A scoring criterion for in situ hybridization was used in which the blue staining proportion indicated the number of tumor cells (positive) and the degree of color represented ISH signal intensity. The scoring was graded as 0 (negative), 1 (<10% positive, light blue), 2 (10%-50% positive, blue), or 3 (>50% positive, dark blue). The final ISH scores (proportion score multiplied by intensity score) were regarded as low expression (0, 1, 2, 3, 4) and high expression [6,9].
Scratch test
Cell culture and transfection were performed as described above. At 24 h after transfection, cells were digested with 0.25% trypsin, centrifuged at 1000 rpm for 10 min, and resuspended in 1640 medium supplemented with 10% fetal bovine serum. Cells were seeded as single-cell suspensions of 1×105/ml, 1 ml into 6-well plates or as whole-cell monolayers cultured in serum-containing medium (1-2 ml). Adherent cells in the medium were aspirated with a micropipette-like tip to create a 1-mm cell-free zone that was washed with serum-free medium or PBS to remove any remaining cells. Images were acquired at different time points during the incubation.
Transwell assay
Cell culture and transfection were performed as described above. At 24 h after transfection, cells were digested with 0.25% trypsin, centrifuged at 1000 rpm for 10 min, and resuspended in 1640 medium supplemented with 10% fetal bovine serum. Cells were seeded as single-cell suspensions of 1×105 cells/well/200 µl in Transwell invasion chambers with 700 µl of 1640 medium supplemented with 10% fetal bovine solution. Cells from the two groups (empty vector control group and experimental group) were incubated for 48 h at 37°C with 5% CO2. Cells were fixed in a neutral formalin solution and examined on an inverted microscope at low magnification; three fields were chosen at random, and the number of cells was counted under high magnification to determine the mean number of cells per field.
Apoptosis assay
Cell apoptosis was detected using APC-conjugated Annexin V (Annexin V-APC) Kit (cat. no: 550474, BD Biosciences, San Jose, CA, USA) and 7-aminoactinomycin D (7-AAD; cat. no: AP104-60-AAD, Multisciences) according to the protocol of previous study [11].
Western blotting
Cells were lysed in lysis buffer containing protease inhibitors. After incubation on ice for 20 min, the lysate was centrifuged at 12,000 rpm for 20 min at 4°C, and the supernatant was collected. Protein samples were separated on 10% SDS-PAGE gel and transferred onto polyvinylidene difluoride membranes. After blocking with 5% milk in TBST (0.1% Tween-20), the membrane was incubated with rimary anti-Cpne8 antibody (Thermofisher) or anti-GAPDH antibody (Cell signaling technology) followed by horseradish peroxidase (HRP)-conjugated secondary antibodies. After washed with PBS, protein bands were detected using the ECL Western Blotting Kit (Pierce).
Statistical analysis
All statistical analyses were performed using the SPSS 20.0 software (Chicago, IL, USA). Quantitative data were presented as Mean ± SD. A Chisquared test was used to analyze the relationship between RP11-396F22.1 expression levels and the clinicopathological characteristics. A one-way ANOVA was used to compare the RP11-396F22.1 expression levels between the CC of different clinical stages. Survival curves were plotted using the Kaplan-Meier method and compared using the log-rank test. The survival data were evaluated using univariate and multivariate Cox regression analyses. A p-value of less than 0.05 was considered statistically significant.
Results
LncRNA expression profiles
The scatter-plot is a visualization that showed the distribution and reproducibility between chips were nearly the same (Figure 1). To identify CC-relevant lncRNAs, we compared the expression profiles of lncRNAs between cases and controls. A total of 2574 upregulated lncRNAs and 3270 downregulated lncRNAs with significantly differential expression (≥2.0-fold) were identified. Among the differentially expressed lncRNAs, RP11-396F22.1 expression was one of the most significantly overexpressed in the CC tissues compared to adjacent non-tumor tissues. To confirm the role of RP11-396F22.1 in CC progression, we detected the RP11-396F22.1 expression levels in CC cell lines (SiHa, Hela, C33a, Ms751) by qRT-PCR, and normalizing to actin. Results showed that the transcript levels of RP11-396F22.1 in cervical carcinoma cells were significantly higher than those in adjacent normal tissues (Figure 2, P<0.05), which was very similar to that of the microarray analysis.
Figure 1.
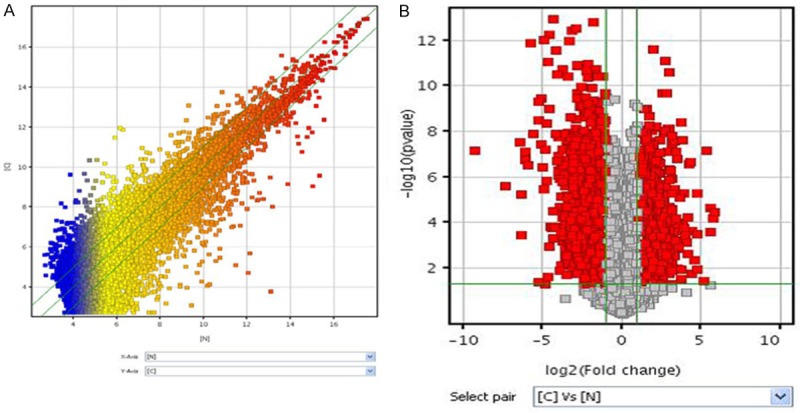
LncRNA expression profiles. A: The scatter-plot of lncRNAs chip; B: The Volcano Plot of differential expression profiles of lncRNAs.
Figure 2.
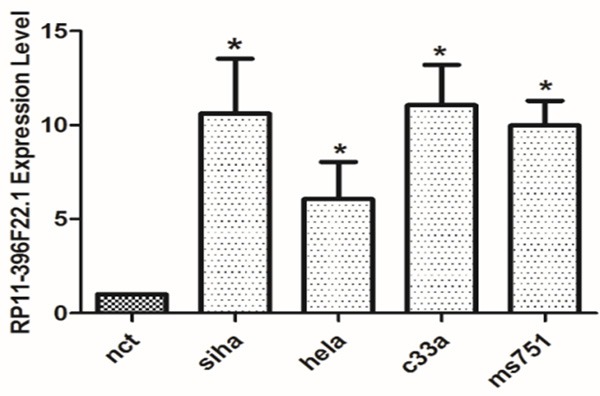
The expression of RP11-396F22.1 in CC cells *P<0.05 vs Negative control group (nct). The expression level of RP11-396F22.1 was significantly higher in Siha, hela, c33a and ms751 cell lines compared to the negative control group (P<0.05).
Expression of lncRNA RP11-396F22.1 in cervical tissues
We next assessed RP11-396F22.1 expression in cervical tissues via in situ hybridization. The data showed that RP11-396F22.1 was highly expressed in 24% cancerous tissues. We also found that the areas, in which RP11-396F22.1 overexpression was observed, were consistent with the cancer nests, while almost negative signals were found in interstitial, blood vessels, muscle layer and normal epithelial morphology. Furthermore, RP11-396F22.1 was highly expressed in 25% of 141 patients with CC according to ISH scoring criterion (Figure 3).
Figure 3.
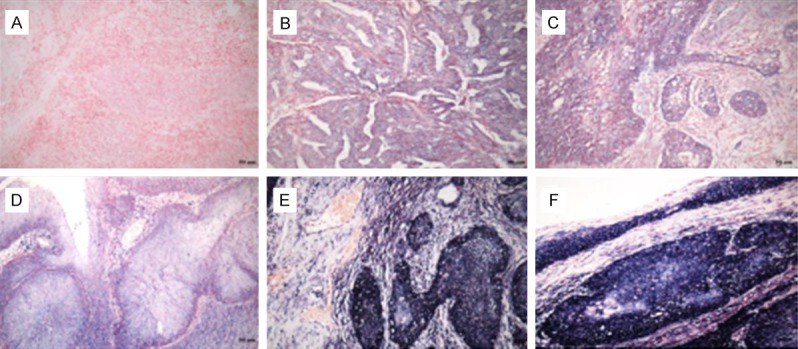
RP11-396F22.1 expression level in cervical tissues via in situ hybridization (×200). A: Negative expression in cervical squamous cell carcinomas nests and interstitial, ISH=0; B: Cervical adenocarcinoma, glandular epithelium shows shallow blue, more than 50% cell were stained positive, ISH=3; C: Cervical squamous-cell carcinoma, 10-50% cell was stained positive, ISH=4; D: Cervical squamous-cell carcinoma, more than 50% cell were stained positive, ISH=6; E, F: Abundant expression in cervical cancer nest, ISH=9; Cases with ISH≥6 was defined as LncRNA RP11-396F22.1 high expression group, while ISH<6 was defined as RP11-396F22.1 low expression group.
Correlations between expression of RP11-396F22.1 and clinical pathological parameters
We examined the correlation of RP11-396F22.1 expression level in cervical tissues via in situ hybridization with CC patients’ clinical features according to ISH scoring criterion, such as gender, age, smoking, chewing areca, pathological stage, tumor size (T stage), lymph-vascular invasion (N stage), invasion muscles of tongue, and relapse. As shown in Table 1, the expression of RP11-396F22.1 was correlated with clinical stage (P=0.0012), tumor differentiation (P=0.0031), tumor size (P=0.0031), invasion degree (P=0.036) and lymph node metastasis (P<0.001).
Table 1.
Correlations between the expression of RP11-396F22.1 and clinical pathological parameters
| RP11-396F22.1 (%) | χ2 | P | ||
|---|---|---|---|---|
|
|
||||
| Clinicopathologic parameters | Low expression | High expression | ||
| Age (Year) | ||||
| ≤35 | 24 (88.9) | 3 (11.1) | 3.365 | 0.067 |
| >35 | 82 (71.9) | 32 (28.1) | ||
| Pathological grade | ||||
| Ia | 9 (90.0) | 1 (10.0) | 15.941 | 0.0012* |
| Ib1 IIa1 | 82 (82.0) | 18 (18.0) | ||
| Ib2 IIa2 | 9 (45.0) | 11 (55.0) | ||
| IIb | 6 (54.6) | 5 (45. 5) | ||
| Histological type | ||||
| Squamous carcinoma | 76 (70.4) | 32 (29.6) | 5.939 | 0.051 |
| Adenocarcinoma | 25 (92.6) | 2 (7.4) | ||
| Adeno-squamous carcinoma | 5 (83.3) | 1 (16.7) | ||
| Differentiation status | ||||
| Well | 15 (100.0) | 0 (0.0) | 6.921 | 0.0031* |
| Moderate | 44 (77.2) | 13 (22.8) | ||
| Poor | 47 (68.1) | 22 (31.9) | ||
| Tumor Size (cm) | ||||
| <4 | 91 (80.5) | 22 (19.5) | 8.739 | 0.0031* |
| ≥.0 | 15 (53.6) | 13 (46.4) | ||
| Invasion depth | ||||
| <1/2 | 58 (82.9) | 12 (17.1) | 4.394 | 0.036* |
| ≥.03 | 48 (67.6) | 23 (32.4) | ||
| Lymphatic vascular invasion | ||||
| Negative | 95 (77.2) | 28 (22.8) | 2.188 | 0.139 |
| Positive | 11 (61.1) | 7 (38.8) | ||
| Lymphatic Metastasis | ||||
| Negative | 95 (84.1) | 18 (15.9) | 24.118 | <0.001* |
| Positive | 11 (39.3) | 17 (60.7) | ||
| Squamous cell carcinoma antigen SCC | ||||
| ≤q | 58 (85.3) | 10 (14.7) | 3.787 | 0.0516 |
| >4 | 6 (60.0) | 4 (40.0) | ||
| HPV16/18 | ||||
| Negative | 11 (57.9) | 8 (42.1) | 0.258 | 0.612 |
| Positive | 63 (51.6) | 59 (48.4) | ||
indicate there is a statistical difference.
LncRNA RP11-396F22.1 is an independent risk factor for prognosis of CC
Logistic regression analysis results indicated that lymph node metastasis (Table 2, OR=8.156, P<0.001), locally advanced CC (tumor stage Ib2+IIa2, OR=10.996, P=0.036), tumor size >40 mm (OR=3.585, P=0.004) and deep myometrial invasion (OR=2.361, P=0.039) were the high risks of overexpression of RP11-396F22.1. Multiple logistic regression analysis showed that lymph node metastasis (OR=7.25, P=0.004) was closely related to high expression of RP11-396F22.1. Kaplan-Meier survival analysis showed that RP11-396F22.1 overexpression was associated with reduced overall survival time and tumor-free survival time, compared to underexpression of RP11-396F22.1 (P<0.001, Figure 4). Multivariate COX regression analysis showed that RP11-396F22.1 (P=0.001), adenocarcinoma (P=0.017), squamous cell carcinoma (P<0.001), and lymph node metastasis (P=0.02) were independent prognostic factors for the prognosis of CC (Table 3). These results suggested that RP11-396F22.1 might be the molecular marker for predicting lymph node metastasis and evaluating prognosis in early CC.
Table 2.
Logistic regression analysis of RP11-396F22.1 expression and variable data
| Variable | Estimate | StdErr | χ2 | P | OR | LOR | UOR |
|---|---|---|---|---|---|---|---|
| Age | 1.138 | 0.647 | 3.097 | 0.078 | 3.121 | 0.879 | 11.091 |
| Ib1 IIa1 | 0.681 | 1.086 | 0.393 | 0.531 | 1.975 | 0.235 | 16.581 |
| Ib2 IIa2 | 2.398 | 1.146 | 4.379 | 0.036* | 10.996 | 1.164 | 103.876 |
| IIb | 2.015 | 1.216 | 2.747 | 0.097 | 7.497 | 0.692 | 81.196 |
| Adenocarcinoma vs squamous carcinoma | -1.661 | 0.765 | 4.719 | 0.030* | 0.190 | 0.042 | 0.850 |
| Adenosquamous carcinoma vs squamous carcinoma | -0.744 | 1.116 | 0.445 | 0.505 | 0.475 | 0.053 | 4.229 |
| Differentiation status | |||||||
| Moderate vs Well | 12.352 | 228.600 | 0.003 | 0.957 | >999.999 | <0.001 | >999.999 |
| Poor vs Well | 12.813 | 228.600 | 0.003 | 0.955 | >999.999 | <0.001 | >999.999 |
| Tumor Size | 1.277 | 0.447 | 8.148 | 0.004* | 3.585 | 1.492 | 8.613 |
| Stromal invasive depth | 0.840 | 0.406 | 4.278 | 0.039* | 2.316 | 1.045 | 5.133 |
| Lymphatic vascular invasion | 0.770 | 0.529 | 2.116 | 0.146 | 2.159 | 0.765 | 6.091 |
| Lymphatic Metastasis | 2.099 | 0.465 | 20.410 | <.0001* | 8.156 | 3.281 | 20.273 |
| SCC | 1.352 | 0.731 | 3.426 | 0.064 | 3.867 | 0.923 | 16.192 |
Figure 4.
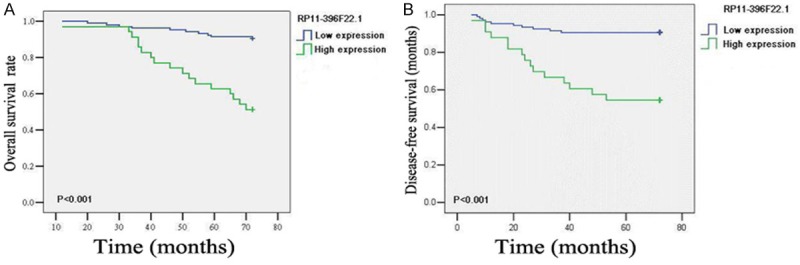
Survival curves of RP11-396F22.1 high expression group and RP11-396F22.1 low expression group. A: Overall survival; B: Disease-free survival. Log-rank test, P<0.001.
Table 3.
Filtering results from multivariate COX regression analysis
| B | SE | Wald | P | HR | Lower HR | Upper HR | |
|---|---|---|---|---|---|---|---|
| Pathological type (2 vs 1) | 2.101 | 0.877 | 5.743 | 0.017 | 8.173 | 1.466 | 45.559 |
| Pathological type (3 vs 1) | 4.298 | 0.907 | 22.446 | <0.001 | 73.577 | 12.43 | 435.52 |
| Lymphatic Metastasis | 1.533 | 0.657 | 5.441 | 0.02 | 4.631 | 1.278 | 16.791 |
| RP11-396F22.1 | 0.461 | 0.133 | 12.037 | 0.001 | 1.586 | 1.222 | 2.057 |
Note: Pathological type: 1, squamous carcinoma; 2, adenocarcinoma; 3, adenosquamous carcinoma.
Knockdown of lncRNA RP11-396F22.1 inhibited cell biological behavior
RP11-396F22.1 knockdown cells were established by siRNA transfection. The efficiencies of transfection were 70% and 86% in Hela cells treated with 24 h and 48 h, and those were 62% and 70% in Siha cells with the same treatment (Figure 5). The MTT assay results showed that there was no significant difference in cell viability among groups (Figure 6). FCM results showed that G2/M cell cycle arrest was observed both in Hela cells and Siha cells treated with siRNA transfection (Figure 6). Furthermore, the early apoptotic rate increased significantly in Hela cells treated with siRNA transfection (P<0.001, Figure 7), as well as the late apoptotic rate in Siha cells (P=0.026). The results of scratch and transwell test showed that the migration ability decreased remarkably in siRNA transfected group (P<0.001, Figure 8). Moreover, the expression of Cpne8 raised significantly in RP11-396F22.1 knockdown cells after 48 h (P<0.05, Figure 9A and 9B). Bioinformatics analysis using miRcode (http://www.mircode.org/) and DIANA TOOL (http://diana.imis.athena-innovation.gr/DianaTools/index.php) suggested that RP11-396F22.1 was associated with both Cpne8 and miRNA-32. Base on this prediction, we speculated that RP11-396F22.1 might regulate Cpne8 expression via miRNA-32. Indeed, we found that knockdown of RP11-396F22.1 significantly downregulated miRNA-32 expression (P<0.01, Figure 9C). Moreover, treatment with miRNA-32 mimics increased Cpne8 expression, whereas inhibition of miRNA-32 reduced Cpne8 expression (P<0.01, Figure 9D and 9E). To further explore the role of Cpne8 in RP11-396F22.1-induced cell migration, siRNA-Cpne8 was transfect into RP11-396F22.1 knockdown cells. The result showed that depletion of Cpne8 restored the migration ability of Hela cells after RP11-396F22.1 knockdown (P<0.01, Figure 9F). This suggested that knockdown of RP11-396F22.1 inhibited proliferation and migration of CC cells and increased G2/M cell cycle arrest and apoptotic rate, which was consistent to clinical researches.
Figure 5.
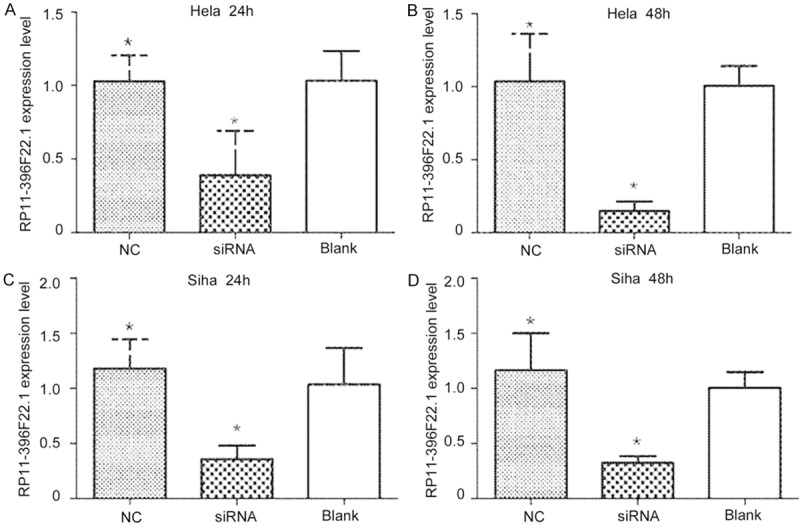
The efficiency of RP11-396F22.1 knockdown by siRNA treatment. After transfection for 24 to 48 hours, RP11-396F22.1 expression was effectively knocked-down in CC cells in vitro. A: Transfection for 24 h in Hela cells; B: Transfection for 48 h in Hela cells; C: Transfection for 24 h in Siha cells; D: Transfection for 48 h in Siha cells. *P<0.05 vs Blank group.
Figure 6.
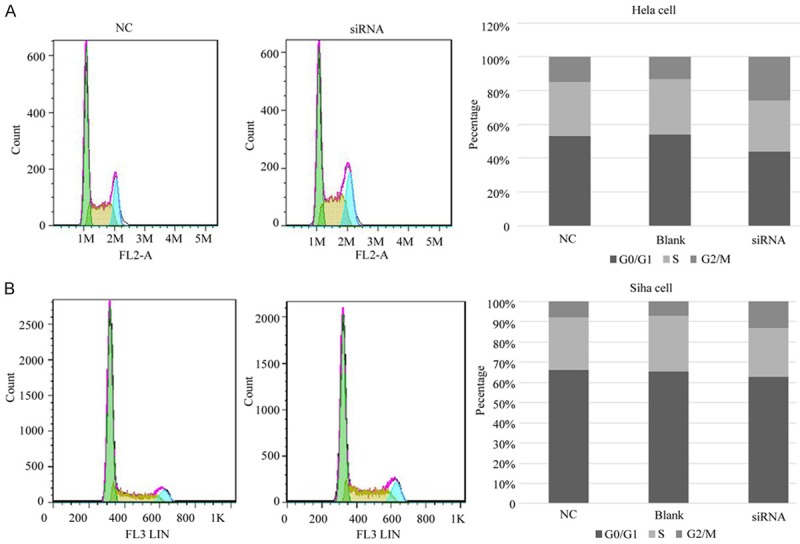
The effect of RP11-396F22.1 knockdown on cell cycle via FCM. A: G2/M phase cells increased in Hela cells in the transfection group; B: G2/M phase cells increased in Siha cells in the transfection group.
Figure 7.
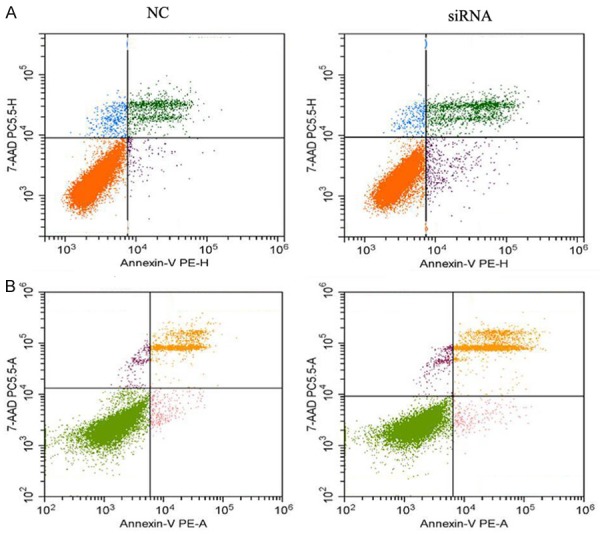
The effect of RP11-396F22.1 knockdown after 48 h on apoptosis. A: Early apoptosis rate increased significantly after RP11-396F22.1 knockdown in Hela cells (P=0.005); B: Late apoptosis rate increased significantly after RP11-396F22.1 knockdown in Siha cells (P=0.026).
Figure 8.
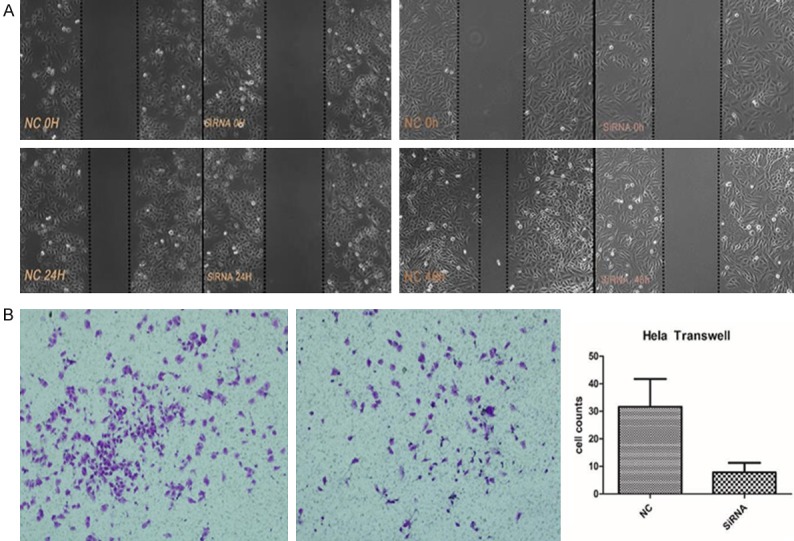
The effect of RP11-396F22.1 knockdown on cell migration. A: After transfection for 48 hours, cell migration ability was significantly decreased in Hela cells (left); After transfection for 72 hours, cell migration ability was significantly decreased in Siha cells (right). B: The effect of RP11-396F22.1 knockdown on migration of Hela cells. RP11-396F22.1 knockdown can significantly reduce the migration ability compared with the NC group (P<0.001).
Figure 9.
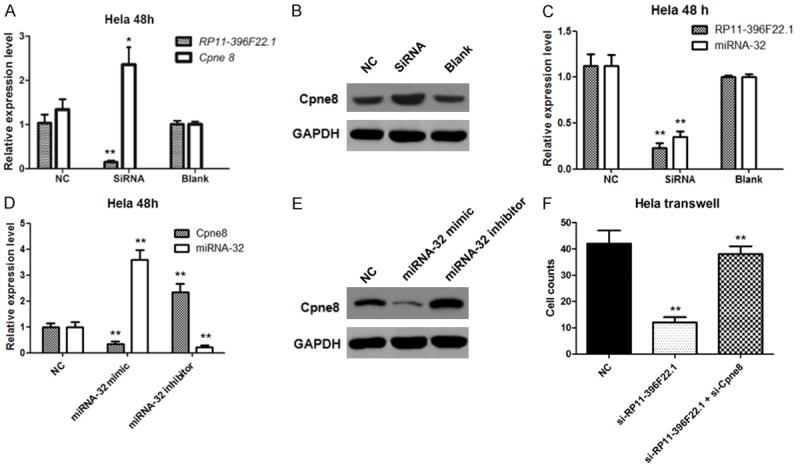
The effect of RP11-396F22.1 downregulation on Cpne8 expression. The effect of RP11-396F22.1 downregulation on Cpne8 expression was detected by qRT-PCR (A) and Western blotting (B). Interference group showed decreased expression of RP11-396F22.1 and increased levels of Cpne8 mRNA and protein expression. (C) The effect of RP11-396F22.1 downregulation on miRNA-32 expression was detected by qRT-PCR. Effects of overexpression and downregulation of miRNA-32 on Cpne8 expression were detected by qRT-PCR (D) and Western blotting (E). (F) Downregulation of Cpne8 restored the migration ability of Hela cells after RP11-396F22.1 knockdown. *P<0.05 vs NC and **P<0.01 vs NC.
Discussions
The progress of CC includes 4 processes: HPV infection, HPV persistent infection, canceration of infected epithelial cells and invasion through the basement membrane [12]. The tumor markers such as squamous cell carcinoma (SCC), carbohydrate antigen 125 (CA125) and ccarcinoembryonic antigen (CEA) raise in patients with terminal CC, which are associated with disease activity, inflammation and other physiological and pathological processes [13-15]. Long non-encoding RNAs (LncRNAs) have functions of regulation similar to proto oncogene or tumor suppressor gene, which are associated with biological behavior of malignant tumors. Zhang et al. [16] indicated that high expression of lncRNA MALAT-1 in cancer tissues was linked to tumor cell differentiation, lymph node metastasis and prognosis. Kim et al. [17] pointed out that overexpression of lncRNA HOTAIR in CC cells upregulated VEGF, MMP-9 and genes associated with epithelial mesenchymal transition for enhancement of tumor invasion. Moreover, Naemura et al. showed that lncRNA ANRIL promoted the proliferation of Hela cells by regulating the expression of P15 [18]. Other researches [19-21] suggested that lncRNA LET downregulation was connected with prognosis of CC and inhibited the progression of carcinoma of urinary bladder.
LncRNA RP11-396F22.1 (GenBank: HG504668.1) located on chromosome 12 is a 526 bp-long lncRNA that is transcribed in antisense orientation from intron. This study, for the first time, reported that RP11-396F22.1 expression significantly increased in CC tissues and decreased in adjacent tissues (cervical stroma and muscular layer). In this study, the expression of RP11-396F22.1 was correlated with clinical stage (P=0.0012), tumor differentiation (P=0.0031), tumor size (P=0.0031), invasion degree (P=0.036) and lymph node metastasis (P<0.001). We also found that RP11-396F22.1 overexpression reduced overall survival time and tumor-free survival time, compared to underexpression of RP11-396F22.1, which suggested that RP11-396F22.1 might be the molecular marker for predicting lymph node metastasis and evaluating prognosis in early CC. Moreover, in vitro researches indicated that knockdown of RP11-396F22.1 inhibited proliferation and migration of CC cells and increased G2/M cell cycle arrest and apoptotic rate.
Cpne8 is a member of the Copine proteins, which are phospholipid-binding proteins and play a critical role in development and mitogenesis [22,23] The function of Cpne8 is not well characterized. It has been reported that Cpne8 is involved in prion disease and parkinsonian neurodegeneration [24,25]. A recent study suggested that Cpne8 might play a role in tumor invasion [26]. Interestingly, in this study, we found that in RP11-396F22.1 knockdown CC cells, Cpne8 expression significantly increased, indicating that RP11-396F22.1 might enhance tumor aggressiveness via negatively regulating Cpne8.
In conclusion, high expression level of RP11-396F22.1 is independently associated with lymph node tumor metastasis, which serves as a novel candidate biomarker for unfavorable prognosis. RP11-396F22.1 served as an onco-lncRNA to execute its functions probably through Cpne8.
Acknowledgements
This study was supported by Grants from Natural Science Foundation of Guangdong Province, China (No. 2015A030313073); Science and Technology Program of Guangzhou, China (No. 201510010289/1563000183) and National Natural Science Foundation of China (No. 81672561).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Wang SM, Li J, Qiao YL. HPV prevalence and genotyping in the cervix of Chinese women. Front Med China. 2010;4:259–263. doi: 10.1007/s11684-010-0095-5. [DOI] [PubMed] [Google Scholar]
- 5.Willoughby BJ, Faulkner K, Stamp EC, Whitaker CJ. A descriptive study of the decline in cervical screening coverage rates in the North East and Yorkshire and the Humber regions of the UK from 1995 to 2005. J Public Health (Oxf) 2006;28:355–360. doi: 10.1093/pubmed/fdl062. [DOI] [PubMed] [Google Scholar]
- 6.Quinn M, Babb P, Jones J, Allen E. Effect of screening on incidence of and mortality from cancer of cervix in England: evaluation based on routinely collected statistics. BMJ. 1999;318:904–908. doi: 10.1136/bmj.318.7188.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: from pathogenesis to therapy. Tumour Biol. 2014;35:5083–5093. doi: 10.1007/s13277-014-1737-z. [DOI] [PubMed] [Google Scholar]
- 8.Fan Z, Cui H, Xu X, Lin Z, Zhang X, Kang L, Han B, Meng J, Yan Z, Yan X, Jiao S. MiR-125a suppresses tumor growth, invasion and metastasis in cervical cancer by targeting STAT3. Oncotarget. 2015;6:25266–25280. doi: 10.18632/oncotarget.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu Y, Chen T, Li G, Yu X, Lu Y, Wang H, Teng L. LncRNAs: emerging biomarkers in gastric cancer. Future Oncol. 2015;11:2427–2441. doi: 10.2217/fon.15.175. [DOI] [PubMed] [Google Scholar]
- 10.Deng K, Guo X, Wang H, Xia J. The lncRNA-MYC regulatory network in cancer. Tumour Biol. 2014;35:9497–9503. doi: 10.1007/s13277-014-2511-y. [DOI] [PubMed] [Google Scholar]
- 11.Lin ZY, Huang YQ, Zhang YQ, Han ZD, He HC, Ling XH, Fu X, Dai QS, Cai C, Chen JH, Liang YX, Jiang FN, Zhong WD, Wang F, Wu CL. MicroRNA-224 inhibits progression of human prostate cancer by downregulating TRIB1. Int J Cancer. 2014;135:541–550. doi: 10.1002/ijc.28707. [DOI] [PubMed] [Google Scholar]
- 12.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 13.Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 2006;110:525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- 14.Gocze PM, Vahrson HW, Freeman DA. Serum levels of squamous cell carcinoma antigen and ovarian carcinoma antigen (CA 125) in patients with benign and malignant diseases of the uterine cervix. Oncology. 1994;51:430–434. doi: 10.1159/000227378. [DOI] [PubMed] [Google Scholar]
- 15.Tomas C, Risteli J, Risteli L, Vuori J, Kauppila A. Use of various epithelial tumor markers and a stromal marker in the assessment of cervical carcinoma. Obstet Gynecol. 1991;77:566–572. [PubMed] [Google Scholar]
- 16.Zhang Y, Wang T, Huang HQ, Li W, Cheng XL, Yang J. Human MALAT-1 long non-coding RNA is overexpressed in cervical cancer metastasis and promotes cell proliferation, invasion and migration. J BUON. 2015;20:1497–1503. [PubMed] [Google Scholar]
- 17.Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naemura M, Murasaki C, Inoue Y, Okamoto H, Kotake Y. Long noncoding RNA ANRIL regulates proliferation of non-small cell lung cancer and cervical cancer cells. Anticancer Res. 2015;35:5377–5382. [PubMed] [Google Scholar]
- 19.Jiang S, Wang HL, Yang J. Low expression of long non-coding RNA LET inhibits carcinogenesis of cervical cancer. Int J Clin Exp Pathol. 2015;8:806–811. [PMC free article] [PubMed] [Google Scholar]
- 20.Ma MZ, Kong X, Weng MZ, Zhang MD, Qin YY, Gong W, Zhang WJ, Quan ZW. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol Carcinog. 2015;54:1397–1406. doi: 10.1002/mc.22215. [DOI] [PubMed] [Google Scholar]
- 21.Sanders I, Holdenrieder S, Walgenbach-Brunagel G, von Ruecker A, Kristiansen G, Muller SC, Ellinger J. Evaluation of reference genes for the analysis of serum miRNA in patients with prostate cancer, bladder cancer and renal cell carcinoma. Int J Urol. 2012;19:1017–1025. doi: 10.1111/j.1442-2042.2012.03082.x. [DOI] [PubMed] [Google Scholar]
- 22.Greene ND, Leung KY, Wait R, Begum S, Dunn MJ, Copp AJ. Differential protein expression at the stage of neural tube closure in the mouse embryo. J Biol Chem. 2002;277:41645–41651. doi: 10.1074/jbc.M203607200. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama T, Yaoi T, Kuwajima G, Yoshie O, Sakata T. Ca2(+)-dependent interaction of N-copine, a member of the two C2 domain protein family, with OS-9, the product of a gene frequently amplified in osteosarcoma. FEBS Lett. 1999;453:77–80. doi: 10.1016/s0014-5793(99)00700-0. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd SE, Maytham EG, Grizenkova J, Hummerich H, Collinge J. A Copine family member, Cpne8, is a candidate quantitative trait gene for prion disease incubation time in mouse. Neurogenetics. 2010;11:185–191. doi: 10.1007/s10048-009-0219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinhardt P, Schmid B, Burbulla LF, Schondorf DC, Wagner L, Glatza M, Hoing S, Hargus G, Heck SA, Dhingra A, Wu G, Muller S, Brockmann K, Kluba T, Maisel M, Kruger R, Berg D, Tsytsyura Y, Thiel CS, Psathaki OE, Klingauf J, Kuhlmann T, Klewin M, Muller H, Gasser T, Scholer HR, Sterneckert J. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12:354–367. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Dang TT, Westcott JM, Maine EA, Kanchwala M, Xing C, Pearson GW. DeltaNp63alpha induces the expression of FAT2 and Slug to promote tumor invasion. Oncotarget. 2016;7:28592–28611. doi: 10.18632/oncotarget.8696. [DOI] [PMC free article] [PubMed] [Google Scholar]


