Abstract
Lung cancer is the leading cause of cancer-related mortality, and approximately 80% of cases are non-small cell lung cancer (NSCLC). Recently, the incidence of NSCLC has been quickly increasing, while the age of patients at diagnosis is decreasing. To date, it is still controversial whether younger patients have better or worse outcomes compared with their older counterparts. MicroRNAs (miRNAs) have been defined to play a key role in cancer pathogenesis, and their aberrant expression has been suggested as a potential biomarker of prognosis in lung adenocarcinoma. To understand the molecular features of young and old adenocarcinoma patients, we investigated the expression level of a panel of miRNAs selected after a mini-literature review. The expression analysis was performed by the nCounter System® (NanoString Technologies) directly on RNA, including small RNAs. The analysis revealed that 7 miRNAs (miR-25-3p, miR-29c-3p, miR-33a-5p, miR-144-3p, miR-153-3p, miR-342-5p and miR-485-3p) were differentially expressed in the two groups (P<0.05). All of these miRNAs showed higher expression levels in young compared to old patients, and their predicted targets included EGFR, MET, VEGF-A, TP53 and PDGFRa. miR-144-3p had an opposite influence on overall survival since its upregulation was associated with a worse prognosis in young patients (P=0.01) and with a better outcome in the older group (P=0.03). We observed that lung cancer in young and old patients may be influenced by different regulatory mechanisms. Moreover, one of the down-regulated miRNAs showed a different prognostic impact in the two groups, confirming that young and old patients deserve a specific clinical approach.
Keywords: miRNA, lung adenocarcinoma, young
Introduction
Lung cancer is the leading cause of cancer-related mortality in both men and women, and approximately 80% are non-small cell lung cancer (NSCLC) patients [1]. In the last decade, the incidence of NSCLC has been quickly increasing and patients’ age at diagnosis continues to decrease [2,3]. However, it remains controversial whether younger patients have better or worse outcomes compared with the older counterparts with lung cancer. In fact, several studies have indicated that lung cancer in young patients could represent a separate clinical pathological entity, suggesting that is more common in non-smoker and female patients with a predominance of adenocarcinoma, an advanced stage at diagnosis and thus a generally poor prognosis [4]. In contrast, some reports have shown that younger patients had a better outcome [5-7] or similar survival as older patients [8-10].
MicroRNAs (miRNAs) are a class of small noncoding RNA that regulate the expression of many target genes via mRNA degradation or translation inhibition [11]. By regulating a great diversity of mRNAs, miRNAs are involved in gene function during various biological processes, such as proliferation, apoptosis, and differentiation. Most importantly, several studies have demonstrated that aberrant expression of miRNAs appears to be causatively linked to the pathogenesis of cancer [12]. Thus, specific miRNA profiles could be useful in the diagnosis, subclassification and prognosis of NSCLC as well as the comprehension of mechanisms regulating the resistance or sensitivity of lung cancer to conventional chemotherapy (Figure 1) [13,14].
Figure 1.
Role of microRNAs as diagnostic, prognostic and predictive markers.
Despite the accumulating evidence linking miRNAs to lung carcinogenesis, very little is known about the expression level of these small RNAs in the young compared to old lung adenocarcinoma patients.
In this review we will give a brief summary of the most valuable deregulated miRNAs in lung cancer. Moreover, we will report our own experience regarding the evaluation of specific miRNA expression levels as prognostic markers in young and old ADC patients. In details, the analyzed miRNAs were selected on the basis of recent literature and their expression levels were correlated with patients’ clinical pathological and follow-up parameters.
Materials and methods
Data sources for mini-literature review
To collect studies of interest we searched on PubMed using the following keywords: microRNAs, lung cancer, lung adenocarcinoma, prognosis. Only English language publications were examined. We considered a total of 5 papers giving information about miRNAs as prognostic markers. We focused our attention at the studies that used the most innovative molecular techniques.
Patients and tumor characteristics
Regarding our own experience, a total of 88 NSCLC patients who consecutively underwent surgical resection at the Unit of Thoracic Surgery in the Department of Surgical, Medical, Molecular Pathology and Critical Area at Pisa University were retrospectively selected. The patients were divided into two groups according to the age at diagnosis: a young age group including patients who were 50 years or younger, and an old age group including patients who were older than 50 years. All samples were formalin fixed and paraffin embedded (FFPE) for microscopic examination, and the histological diagnoses were formulated according to the World Health Organization classifications [15,16]. The most representative paraffin blocks of the tumor tissues were selected for molecular analysis for each group. The clinical pathological characteristics and survival data were collected for all patients. Informed consent for the tissue collection and molecular analysis was obtained from each patient.
miRNA and DNA isolation
According to the manufacturer’s instructions, three to five paraffin sections with a thickness of 5-10 μm per sample were utilized to purify DNA and total RNA including miRNAs after standard deparaffinization and manual macrodissection of the neoplastic area, using the QIAmp DNA Mini Kit (QIAGEN Inc., Hilden, Germany) and the miRNeasy FFPE kit (QIAGEN Inc., Hilden, Germany), respectively. The samples, after quality and quantity evaluation using a NanoDrop ND-1000 (ThermoScientific, USA) spectrophotometer, were stored at -80/-20°C until used in the experiments.
Nanostring custom panel
The NanoString nCounter assay kit was used to test the miRNAs expression profile. The nCounter custom code set used in this study was designed and synthesized by NanoString Technologies (Seattle, WA, USA). It consists of reporter and capture probe pairs specific for 30 miRNAs reported in Table 1. In total, 150 ng of RNA was used for the nCounter miRNA sample preparation reactions. All sample preparations were performed in accordance with the manufacturer’s instructions (NanoString Technologies). The small RNA molecules were ligated with a specific DNA tag onto the 3’ end of each mature miRNA. After hybridization and the removal of excess probes, counts of digital reports were performed on the nCounter digital analyzer according to the manufacturer’s protocol. Raw NanoString counts were analyzed by Nanostring nSolver version 2.5 software. A background level of expression for each sample using the mean level of the negative controls plus two standard deviations of the mean was calculated. MiRNAs expressing less than two standard deviations from the mean were considered not expressed. Then, miRNAs were normalized using a scaling factor based on the top 5 miRNAs with the lower variability coefficients according to the manufacturer’s protocol.
Table 1.
miRNAs included in the Nanostring panel
| Official name | Accession |
|---|---|
| hsa-miR-25-3p | MIMAT0000081 |
| hsa-miR-29b-3p | MIMAT0000100 |
| hsa-miR-29c-3p | MIMAT0000681 |
| hsa-miR-30b-5p | MIMAT0000420 |
| hsa-miR-30c-5p | MIMAT0000244 |
| hsa-miR-33a-5p | MIMAT0000091 |
| hsa-miR-34a-5p | MIMAT0000255 |
| hsa-miR-34c-3p | MIMAT0004677 |
| hsa-miR-93-5p | MIMAT0000093 |
| hsa-miR-96-5p | MIMAT0000095 |
| hsa-miR-103a-3p | MIMAT0000101 |
| hsa-miR-125a-5p | MIMAT0000443 |
| hsa-miR-133a-3p | MIMAT0000427 |
| hsa-miR-133b | MIMAT0000770 |
| hsa-miR-137 | MIMAT0000429 |
| hsa-miR-138-5p | MIMAT0000430 |
| hsa-miR-145-5p | MIMAT0000437 |
| hsa-miR-144-3p | MIMAT0000436 |
| hsa-miR-153-3p | MIMAT0000439 |
| hsa-miR-182-5p | MIMAT0000259 |
| hsa-miR-183-5p | MIMAT0000261 |
| hsa-miR-191-5p | MIMAT0000440 |
| hsa-miR-210-3p | MIMAT0000267 |
| hsa-miR-214-3p | MIMAT0000271 |
| hsa-miR-221-3p | MIMAT0000278 |
| hsa-miR-299-3p | MIMAT0000687 |
| hsa-miR-342-3p | MIMAT0000753 |
| hsa-miR-342-5p | MIMAT0004694 |
| hsa-miR-425-5p | MIMAT0003393 |
| hsa-miR-485-5p | MIMAT0002175 |
Mutation detection
The status of EGFR exon 18-21 and KRas exon 2-3 was simultaneously evaluated using the MALDI-TOF method on a Sequenom (Agena Bioscience, San Diego, CA, USA) platform. In detail, we used the Myriapod Lung Status CE-IVD kit (Diatech Pharmacogenetics) that can also detect BRAF, NRAS, PIK3CA, ALK, ERBB2, DDR2, RET and MAP2K1 mutations.
Statistical analysis
Once miRNA raw data were normalized, differential expression was tested applying the Mann-Whitney U test with linearity correction using JMP10 software (SAS) in order to investigate the association between miRNAs expression and clinic-pathological parameters. Survival analyses were performed using the Kaplan-Meier method with log-rank test and the Cox proportional hazard model. A two-tailed P value <0.05 was considered significant.
MiRNA target prediction and pathway analysis
DIANA-mirPath v3.0 [17] was used to perform the enrichment analysis of predicted target genes by one or more miRNAs into known Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The software links miRNAs to experimentally validated target genes from Tarbase v7.0, microT-CDS v5.0, TargetScan.
Results
Mini-literature review
According to criteria described above, we reviewed the recent literature in order to deal with the usefulness of miRNAs as prognostic markers in non-small cell lung cancer.
Peng et al, performing a comprehensive analysis based on lung ADC miRNome profiling studies, found 4 miRNAs (miR-21-5p, miR-210-3p, miR-182-5p and miR-183-5p) up-regulated and 2 miRNAs (miR-126-3p and miR-218-5p) down-regulated in lung adenocarcinoma tissues. Moreover, they revelead the potential prognostic values of the six deregulated miRNAs in lung adenocarcinoma patients [18].
Also Sui et al employed an integrative computational method for the analysis of the available large-scale samples in The Cancer Genome Atlas (TCGA) database. They identified 15 aberrantly expressed miRNAs with respect to clinical features of lung adenocarcinoma. Among which, the low miR-378c and miR-221-5p expression levels were negatively correlated with overall survival, but the miR-142-3p overexpression had a better influence on overall survival [19].
A study conducted by Xu et al reported that miR-25 expression in tissue was found to be associated with lymph node metastasis and disease stage. In addition they found that high miR-25 expression was also associated with poorer overall survival of women with lung ADC [20]. A potential prognostic value of miR-138 expression levels and its target PDK1 was described by Han and collaborators. They found that patients with low levels of miR-138 combined with high levels of PDK1 had shortest overall survival [21].
Hou and collegues claimed that miR-125a-3p is a significant prognostic biomarker for patients with NSCLC since they found that patients treated with chemotherapy with high miR-125a-3p expression levels had a longer overall survival and disease free survival rate compared with untreated patients with low expression of miR-125a-3p [22].
Patients and tumor characteristics
Forty-four patients younger than 50 years (including 50) were enrolled as the younger group and forty-four patients older than 50 years were collected as the older group. In the young age group, females accounted for 47.7% (n=21), whereas in the older age group females accounted for 25% (n=11) (Chi-square test; P=0.02). Regarding the histological classification, the different subtypes of adenocarcinoma were defined according to the predominant component, as follows: 19 acinar (19/44, 43.18%), 13 lepidic (13/44, 29.55%), 9 solid (9/44, 20.45%), and 3 papillary (3/44, 6.82%) among the younger patients; 17 solid (17/44, 38.63%), 16 lepidic (16/44, 36.37%), 8 papillary (8/44, 18.19%), and 3 acinar (3/44, 6.82%) among the older group. Regarding tumor grading of the younger patients, 3 tumors were G1, 30 were G2 and 11 were G3. Among the old group, no tumors were classified as G1, whereas 28 tumors were G2 and 16 were graded G3. Features of patients and tumor characteristics are shown in Table 2. Follow-up data, in terms of disease-free interval (DFI) and overall survival (OS) were available for all patients.
Table 2.
Clinical pathological characteristics of young and old ADC patients
| Young (n) | Old (n) | Total (n) | |
|---|---|---|---|
| Sample size | 44 | 44 | 88 |
| Gender | |||
| Males | 23 | 33 | 56 |
| Females | 21 | 11 | 32 |
| ADC patterns | |||
| Lepidic | 13 | 16 | 29 |
| Acinar | 19 | 3 | 22 |
| Solid | 9 | 17 | 26 |
| Papillary | 3 | 8 | 11 |
| Tumor grading | |||
| G1 | 3 | 0 | 3 |
| G2 | 30 | 28 | 58 |
| G3 | 11 | 16 | 27 |
| Stage | |||
| IA | 8 | 9 | 17 |
| IB | 14 | 9 | 23 |
| IIA | 6 | 7 | 13 |
| IIB | 4 | 5 | 9 |
| IIIA | 10 | 13 | 23 |
| IIIB | 1 | 0 | 1 |
| IV | 1 | 1 | 2 |
ADC: adenocarcinoma.
miRNAs differentially expressed between young and old patients
To explore differentially expressed miRNAs between young and old lung cancer patients, we evaluated the expression profile of 30 miRNAs using the nCounter Nanostring platform. After raw data normalization, 3 samples were excluded from the statistical analysis because of the poor quality and quantity of the mRNA input. The Mann-Whitney U test revealed that 7 out of 30 miRNAs were differentially expressed between the two groups. Specifically, the miR-25-3p, miR-29c-3p, miR-33a-5p, miR-144-3p, miR-153-3p, miR-342-5p and miR-485-3p were found to be up-regulated in young compared to the old patients with a statistical significance of P<0.05.
microRNA target prediction and pathway analysis
In order to understand the potential involvement of miRNAs in the pathogenesis of NSCLC, potential targets were identified for the 7 differentially expressed miRNAs using the DIANA-mirPath v3.0 software on the gene targets predicted by Tarbase v7.0. Our analysis showed that 43 KEGG biological processes were significantly enriched (P<0.05, FDR corrected). As reported in Table 3, important pathways contributing to tumor aggressiveness were regulated by at least six of these miRNAs. Many of the predicted miRNAs targets are involved in critical pathways such as adherens junction (P=9.08e-06), the p53 signaling pathway (P=1.69e-05), cell cycle (P=1.88e-05), and the FoxO signaling pathway (P=2.0e-04).
Table 3.
Prediction target analysis using DIANA-mirPath v3.0 software
| KEGG Pathway | P-value | #Genes | #miRNAs |
|---|---|---|---|
| p53 signaling pathway | 1.69e-05 | 30 | 6 |
| Cell cycle | 1.88e-05 | 49 | 6 |
| Regulation of actin cytoskeleton | 8.0e-04 | 60 | 6 |
| FoxO signaling pathway | 2.0e-04 | 47 | 5 |
| Adherens junction | 9.08e-06 | 28 | 5 |
| Non small cell lung cancer | 0.01 | 17 | 5 |
Mutation analysis
Three EGFR mutations (3.4%) and 14 KRas mutations (15.9%) were identified in the younger group, while 6 EGFR mutations (6.8%) and 20 KRas mutations (22.7%) were found in the old patients. The molecular characterizations of the above-reported mutations are detailed in Table 4. No alterations in genes other than EGFR and KRas were detected. The difference in the frequency of oncogenic mutations between the two groups was not significant (Chi-square test; P=0.16).
Table 4.
EGFR and KRas status of young and old ADC patients
| Young (n, %) | Old (n, %) | Total (n, %) | |
|---|---|---|---|
| EGFR status | |||
| Wild-type | 41 (93.2%) | 38 (86.4%) | 79 (89.8%) |
| Mutated | 3 (6.8%) | 6 (13.6%) | 9 (10.2%) |
| KRas status | |||
| Wild-type | 30 (68.2%) | 24 (54.5%) | 54 (61.4%) |
| Mutated | 14 (31.8%) | 20 (45.5%) | 34 (38.6%) |
Survival analysis
Survival analysis using the Kaplan-Meier method with log-rank test and the Cox proportional hazard model showed no difference in the DFI and OS between young and old patients (data not shown). Then, we further investigated the prognostic impact of the analyzed miRNAs, and it was found that miR-144-3p had an opposite influence on overall survival between the two age-groups. Interestingly, high levels of miR-144-3p were associated with a worse outcome in young patients (P=0.01) and a better prognosis in the old group (P=0.03) (Figure 2).
Figure 2.
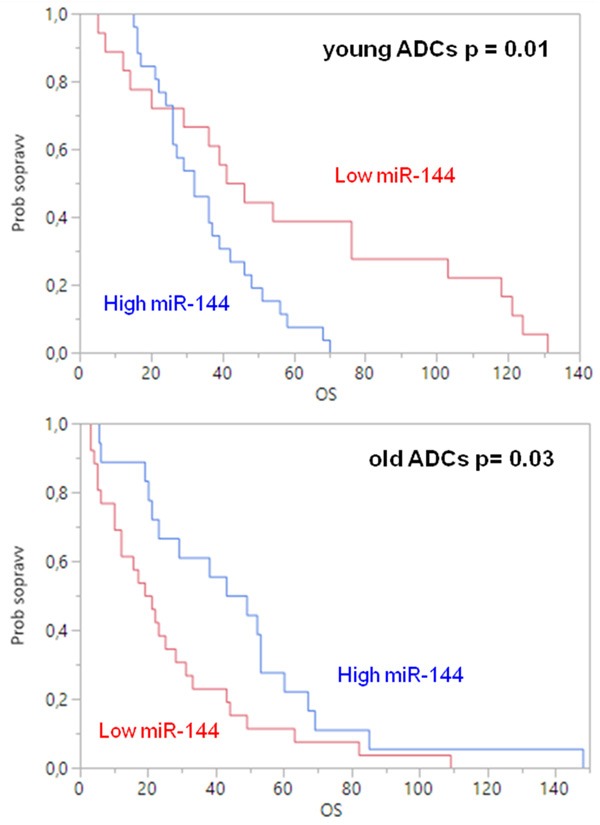
Overall survival for mir-144 in young and old adenocarcinoma patients (blue curve: high expression, red curve: low expression).
Discussion
In the last few years, several reports have shown trends of increasing incidence rates of lung cancer among young patients [23], with peculiar characteristics such as a higher incidence in females, a predominance of adenocarcinoma and an advanced stage at diagnosis [6,8,10]. Although there is no consensus about the specific cut-off age defining youthful lung cancer, an age of forty-fifty years is commonly used to separate the young patients from the old ones. To date, only few data are available about the molecular characteristics of young lung cancer patients compared to their old counterparts, and controversial results have been reported in this field. Moreover, no studies have been conducted on miRNAs expression profile in the two age-groups. A putative association between miRNAs and prognosis of lung adenocarcinoma, with the aim to find age-specific miRNAs, may highlight the molecular mechanisms and predict the clinical outcome.
The present study aimed to investigate the expression profile of 30 selected miRNAs between ADC patients younger and older than 50 years. We found that 7 miRNAs (miR-25-3p, miR-29c-3p, miR-33a-5p, miR-144-3p, miR-153-3p, miR-342-5p and miR-485-3p) were significantly up-regulated in the young patients.
A study of genome-wide assessment of 800 miRNAs conducted by Noren Hooten et al [24] in peripheral blood mononuclear cells from 14 young and 14 old healthy individuals by real-time RT-PCR analysis revealed that the expression of 9 miRNAs (miR-103, miR-107, miR-128, miR-130a, miR-155, miR-24, miR-221, miR-496, and miR-1538) was significantly lower in older individuals. Investigating the diseases, molecular functions and canonical pathways associated with each miRNA, it was determined that cancer was the most common disease associated with the differentially expressed miRNAs.
Furthermore, an age-related miRNA signature has been identified in breast cancer by Peña-Chilet et al [25]. They identified and validated a panel of six miRNAs differentially expressed in very young breast cancer patients, suggesting that breast cancer in young patients appears to be a different biological entity. The validated miRNAs noted pathways related to cell motility, invasion and proliferation.
To the best of our knowledge, our study is the first demonstrating the decrease of miRNA levels with age in lung adenocarcinoma patients. The biological role in lung cancer of miRNAs found as differentially expressed in our study has already been investigated. For example, the analysis of 81 NSCLCs patients showed that miR-25 promotes cell proliferation and inhibits apoptosis in NSCLC cells by negatively regulating the Modulator of apoptosis 1 expression [26]. A previous study demonstrates that miR-29c suppresses lung cancer cell adhesion to the extracellular matrix and metastasis by directly inhibiting integrin β1 and MMP2 expression [27]. Yang et al [28] reported that miR-33a regulates the epithelial-mesenchymal transition by targeting Twist1 in NSCLC cells and inhibits lung cancer metastasis. A report from Zhang et al [29] claimed that miR-144 promoted proliferation, migration, and invasion of nasopharyngeal carcinoma through repression of phosphatase and tensin homolog (PTEN). Although miR-153 has been shown to play an important role in various cancers, the function of miR-153 is not completely understood. miR-153 suppresses tumor growth in glioblastoma, epithelial cancer and leukemia [30]. In contrast, in prostate cancer, miR-153 promotes cell proliferation via down-regulation of the PTEN tumor suppressor gene. A study conducted by Mou and colleagues [31] found that the expression level of miR-485 was down-regulated in four lung adenocarcinoma cell lines and tissues and that the reduced miR-485 expression was associated with tumor metastasis. Luciferase assay revealed that Flot2 is a direct target of miR-485, while the expression levels of Flot2 were inversely correlated with the expression levels of miR-485 in lung adenocarcinoma tissues.
In addition, the analysis of the predicted targets regulated by these miRNAs and the relative pathway enrichment displayed the involvement of biological processes crucial for cancer such as adherens junction, the p53 signaling pathway, cell cycle and the FoxO signaling pathway.
The outcomes of young and old patients with lung cancer have been previously studied, but the results remain to be addressed. In our cohort of patients, overall survival time was similar between the younger and older groups as assessed in some reports [3,6,7]. However, some studies described a worse [4,5] and others a better overall survival [2,8] for the two age-groups. This controversial issue could be due to either the rare occurrence of lung cancer in young patients, the lack of a commonly accepted age cut-off, the different enrollment criteria of patients (not only surgically treated cases but also unresectable ones) or ethnic origin. While investigating the prognostic influence of each miRNA analyzed in our study, we found that miR-144-3p had an unexpected impact on overall survival between the two age-groups since its upregulation was associated with a worse prognosis in young patients and a better outcome in the old group. After further validations, these data could suggest that the expression level of miR-144 could be a novel indicator of prognosis depending on the age of lung adenocarcinoma patients.
In order to examine the presence of molecular alterations that frequently occur in the lung adenocarcinoma, we performed the analysis of exon 18-21 of EGFR and exon 2-3 of KRas. The EGFR mutation rate was 3.4% among young patients and 6.8% among old patients, whereas KRas mutations occurred in the 15.9% of young patients and 22.7% of old patients. The frequencies of EGFR and KRas mutations were not significantly different in relation to the age of patients, which is consistent with reported studies [32,33]. It should be noted that studying the relationship between age and genotype in young patients is challenging given the presence of multiple confounding factors including smoking status, sex and race.
In conclusion, our study provides new insights into the role of miRNAs in lung adenocarcinoma occurring in young patients. We observed that lung cancer in young and old patients may be influenced by different regulatory mechanisms since we found 7 miRNAs down-regulated in the older group, probably due to distinct age-related genetic and epigenetic alterations. Moreover, one of the dysregulated miRNAs showed a different prognostic impact in the two groups, thus confirming that young and old patients deserve a specific clinical approach. Further validations are needed to better define if an age-based genomic signature could be used as a prognostic marker in lung cancer.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Liu NS, Spitz MR, Kemp BL, Cooksley C, Fossella FV, Lee JS, Hong WK, Khuri FR. Adenocarcinoma of the lung in young patients: The M. D. Anderson experience. Cancer. 2000;88:1837–1841. doi: 10.1002/(sici)1097-0142(20000415)88:8<1837::aid-cncr12>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 3.Levi F, Bosetti C, Fernandez E, Hill C, Lucchini F, Negri E, La Vecchia C. Trends in lung cancer among young European women: the rising epidemic in France and Spain. Int J Cancer. 2007;121:462–465. doi: 10.1002/ijc.22694. [DOI] [PubMed] [Google Scholar]
- 4.Duan L, You Q, Chen X, Wang H, Zhang H, Xie D, Xu X, Jiang G. Outcome and prognosis for patients younger than thirty with primary lung cancer. Minerva Chir. 2013;68:175–182. [PubMed] [Google Scholar]
- 5.Subramanian J, Morgensztern D, Goodgame B, Baggstrom MQ, Gao F, Piccirillo J, Govindan R. Distinctive characteristics of non-small cell lung cancer (NSCLC) in the young: a surveillance, epidemiology, and end results (SEER) analysis. J Thorac Oncol. 2010;5:23–28. doi: 10.1097/JTO.0b013e3181c41e8d. [DOI] [PubMed] [Google Scholar]
- 6.Kuo C, Chen Y, Chao J, Tsai C, Perng R. Non-small cell lung cancer in very young and very old patients. Chest. 2000;117:354–357. doi: 10.1378/chest.117.2.354. [DOI] [PubMed] [Google Scholar]
- 7.Arnold BN, Thomas DC, Rosen JE, Salazar MC, Blasberg JD, Boffa DJ, Detterbeck FC, Kim AW. Lung cancer in the very young: treatment and survival in the national cancer data base. J Thorac Oncol. 2016;11:1121–1131. doi: 10.1016/j.jtho.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 8.Skarin AT, Herbst RS, Leong TL, Bailey A, Sugarbaker D. Lung cancer in patients under age 40. Lung Cancer. 2001;32:255–264. doi: 10.1016/s0169-5002(00)00233-6. [DOI] [PubMed] [Google Scholar]
- 9.Ak G, Metintas M, Metintas S, Yildirim H, Erginel S, Alatas F. Lung cancer in individuals less than 50 years of age. Lung. 2007;185:279–286. doi: 10.1007/s00408-007-9021-2. [DOI] [PubMed] [Google Scholar]
- 10.Blanco M, Garcia-Fontan E, Rivo JE, Repaaz JR, Obeso GA, Canizares MA. Bronchogenic carcinoma in patients under 50 years old. Clin Transl Oncol. 2009;11:322–325. doi: 10.1007/s12094-009-0361-7. [DOI] [PubMed] [Google Scholar]
- 11.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129–139. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 13.MacDonagh L, Gray SG, Finn SP, Cuffe S, O’Byrne KJ, Barr MP. The emerging role of microRNAs in resistance to lung cancer treatments. Cancer Treat Rev. 2015;41:160–169. doi: 10.1016/j.ctrv.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Sin TK, Wang F, Meng F, Wong SC, Cho WC, Siu PM, Chan LW, Yung BY. Implications of microRNAs in the treatment of gefitinib-resistant non-small cell lung cancer. Int J Mol Sci. 2016;17:237. doi: 10.3390/ijms17020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, Garg K, Austin JH, Asamura H, Rusch VW, Hirsch FR, Scagliotti G, Mitsudomi T, Huber RM, Ishikawa Y, Jett J, Sanchez-Cespedes M, Sculier JP, Takahashi T, Tsuboi M, Vansteenkiste J, Wistuba I, Yang PC, Aberle D, Brambilla C, Flieder D, Franklin W, Gazdar A, Gould M, Hasleton P, Henderson D, Johnson B, Johnson D, Kerr K, Kuriyama K, Lee JS, Miller VA, Petersen I, Roggli V, Rosell R, Saijo N, Thunnissen E, Tsao M, Yankelewitz D. International association for the study of lung cancer/American thoracic society/European respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Travis WD, Brambilla E, Noguchi M, Nicholson A, Geisinger K, Yatabe Y, Ishikawa Y, Wistuba I, Flieder DB, Franklin W, Gazdar A, Hasleton PS, Henderson DW, Kerr KM, Petersen I, Roggli V, Thunnissen E, Tsao M. Diagnosis of lung cancer in small biopsies and cytology: Implications of the 2011 international association for the study of lung cancer/American thoracic society/European respiratory society classification. Arch Pathol Lab Med. 2013;137:668–684. doi: 10.5858/arpa.2012-0263-RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–1993. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- 18.Peng Z, Pan L, Niu Z, Li W, Dang X, Wan L, Zhang R, Yang S. Identification of microRNAs as potential biomarkers for lung adenocarcinoma using integrating genomics analysis. Oncotarget. 2017;8:64143–64156. doi: 10.18632/oncotarget.19358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sui J, Yang RS, Xu SY, Zhang YQ, Li CY, Yang S, Yin LH, Pu YP, Liang GY. Comprehensive analysis of aberrantly expressed microRNA profiles reveals potential biomarkers of human lung adenocarcinoma progression. Oncol Rep. 2017;38:2453–2463. doi: 10.3892/or.2017.5880. [DOI] [PubMed] [Google Scholar]
- 20.Xu FX, Su YL, Zhang H, Kong JY, Yu H, Qian BY. Prognostic implications for high expression of MiR-25 in lung adenocarcinomas of female non-smokers. Asian Pac J Cancer Prev. 2014;15:1197–203. doi: 10.7314/apjcp.2014.15.3.1197. [DOI] [PubMed] [Google Scholar]
- 21.Han L, Zhang G, Zhang N, Li H, Liu Y, Fu A, Zheng Y. Prognostic potential of microRNA-138 and its target mRNA PDK1 in sera for patients with non-small cell lung cancer. Med Oncol. 2014;31:129. doi: 10.1007/s12032-014-0129-y. [DOI] [PubMed] [Google Scholar]
- 22.Hou L, Luo P, Ma Y, Jia C, Yu F, Lv Z, Wu C, Fu D. MicroRNA-125a-3p downregulation correlates with tumorigenesis and poor prognosis in patients with non-small cell lung cancer. Oncol Lett. 2017;14:4441–4448. doi: 10.3892/ol.2017.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marugame T, Yoshimi I, Kamo K, Imamura Y, Kaneko S, Mizuno S, Sobue T. Trends in lung cancer mortality among young adults in Japan. Jpn J Clin Oncol. 2005;35:177–180. doi: 10.1093/jjco/hyi054. [DOI] [PubMed] [Google Scholar]
- 24.Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. microRNA expression patterns reveal differential expression of target genes with age. PLoS One. 2010;5:e10724. doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peña-Chilet M, Martínez MT, Pérez-Fidalgo JA, Peiró-Chova L, Oltra SS, Tormo E, Alonso-Yuste E, Martinez-Delgado B, Eroles P, Climent J, Burgués O, Ferrer-Lozano J, Bosch A, Lluch A, Ribas G. MicroRNA profile in very young women with breast cancer. BMC Cancer. 2014;14:529. doi: 10.1186/1471-2407-14-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu T, Chen W, Kong D, Li X, Lu H, Liu S, Wang J, Du L, Kong Q, Huang X, Lu Z. miR-25 targets the modulator of apoptosis 1 gene in lung cancer. Carcinogenesis. 2015;36:925–935. doi: 10.1093/carcin/bgv068. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Zhu Y, Zhao M, Wu C, Zhang P, Tang L, Zhang H, Chen X, Yang Y, Liu G. miRNA-29c suppresses lung cancer cell adhesion to extracellular matrix and metastasis by targeting integrin β1 and matrix metalloproteinase2 (MMP2) PLoS One. 2013;8:e70192. doi: 10.1371/journal.pone.0070192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L, Yang J, Li J, Shen X, Le Y, Zhou C, Wang S, Zhang S, Xu D, Gong Z. MircoRNA-33a inhibits epithelial-to-mesenchymal transition and metastasis and could be a prognostic marker in non-small cell lung cancer. Sci Rep. 2015;5:13677. doi: 10.1038/srep13677. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Zhang LY, Ho-Fun Lee V, Wong AM, Kwong DL, Zhu YH, Dong SS, Kong KL, Chen J, Tsao SW, Guan XY, Fu L. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis. 2013;34:454–463. doi: 10.1093/carcin/bgs346. [DOI] [PubMed] [Google Scholar]
- 30.Yuan Y, Du W, Wang Y, Xu C, Wang J, Zhang Y, Wang H, Ju J, Zhao L, Wang Z, Lu Y, Cai B, Pan Z. Suppression of AKT expression by miR-153 produced anti-tumor activity in lung cancer. Int J Cancer. 2015;136:1333–1340. doi: 10.1002/ijc.29103. [DOI] [PubMed] [Google Scholar]
- 31.Mou X, Liu S. MiR-485 inhibits metastasis and EMT of lung adenocarcinoma by targeting Flot2. Biochem Biophys Res Commun. 2016;477:521–526. doi: 10.1016/j.bbrc.2016.04.043. [DOI] [PubMed] [Google Scholar]
- 32.Kim L, Kim KH, Yoon YH, Ryu JS, Choi SJ, Park IS, Han JY, Kim JM, Chu YC. Clinicopathologic and molecular characteristics of lung adenocarcinoma arising in young patients. J Korean Med Sci. 2012;27:1027–1036. doi: 10.3346/jkms.2012.27.9.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye T, Pan Y, Wang R, Hu H, Zhang Y, Li H, Wang L, Sun Y, Chen H. Analysis of the molecular and clinicopathologic features of surgically resected lung adenocarcinoma in patients under 40 years old. J Thorac Dis. 2014;6:1396–1402. doi: 10.3978/j.issn.2072-1439.2014.08.50. [DOI] [PMC free article] [PubMed] [Google Scholar]


