Abstract
Background
This study investigated the prevalence of HPVs in heterosexual South African men and the impact of HIV co-infection.
Methods
HPV was detected in penile swabs from 195 HIV-infected and 140 HIV-uninfected men using PCR with FAP59/64 primers and Roche Linear Array HPV genotyping (LA). Genotyping of FAP positive specimens was achieved by high-throughput sequencing of amplicons.
Results
HPV was detected by FAP PCR and LA in 79% (266/335) of the men. Men with HIV co-infection and men with HIV infected sexual partners had a significantly (p<0.0001) higher HPV infection risk (adjusted odds ratio 4.0 (2.1–8.2) and 3.7 (2.1–6.7), respectively). LA genotyping and 454 sequencing of 218 FAP positive specimens detected 45 known α-HPV types, 45 β-HPV types (34 known, 10 putative and 1 novel putative), and 91 γ-HPV types (26 known, 51 putative and 14 novel putative). Alpha, beta and gamma types were detected in 89.8%, 51.4% and 62.4% of the 218 men with HPV-62, HPV-5 and HPV-121 most common in each genus, respectively.
Conclusion
A great diversity of known and novel alpha, beta and gamma HPV types were detected with higher prevalence in HIV co-infected men and unknown associations, if any, with genital lesions and cancers.
Keywords: Alpha papillomavirus, Beta papillomavirus, Gamma papillomavirus, HIV, Penile HPV infection, High-throughput sequencing
1. Introduction
Human papillomaviruses (HPVs) are small non-enveloped circular dsDNA viruses of the Papillomaviridae family that infect the stratified epithelial layers of the mucosa and skin. HPVs represent a diverse group of over 200 recognised types. The majority of HPV types belong to one of three genera; Alpha (α), Beta (β) and Gamma (γ) [1]. To date 65 α-HPV, 52 β-HPV and 82 γ-HPV types have been fully characterised (http://www.hpvcenter.se/ accessed 08 Nov 2016). Classification of HPVs is based on nucleotide sequence identity within the ORF encoding the major capsid protein L1. HPVs of the same type share >90% identity, and those within a genus have >60% identity in the L1 coding sequence. Novel types have <90% identity in the L1 ORF to any other type [1].
Infections with HPVs are associated with a wide range of diseases, from benign lesions to invasive cancers. The α-HPV genus includes 12 carcinogenic types (group I) and 13 probable or possible carcinogenic types (group 2), as classified by the International Agency for Research on Cancer (IARC) [2]. These types have been associated with malignant anogenital cancers, including cervical [3], vaginal, vulvar and penile cancer, as well as oral cancers, reviewed in [4]. Members of the β HPV species 1, HPV-5 and −8, were found in epidermodysplasia verruciformis (EV) and have been classified as “possible carcinogenic” (IARC group 2B) exclusively in squamous cell carcinoma (SCC) of the skin in patients with this autosomal recessive disorder [2]. The β−1, −2 and −3 species as well as γ−1 species were associated with SCC of the skin in a recent systematic review [5]. Several new studies have also indicated that many niches on the healthy body, including the skin, mouth, nose and gut, are inhabited by diverse HPV types that may be considered commensal [6], [7], [8], [9], [10], [11].
The majority of studies of genital HPV infection in men have focused on α-HPVs only and few have examined the distribution of HPVs from the β and γ genera [12], [13], [14]. The impact of HIV-infection on genital β- and γ-HPV types is also understudied. In this study we describe the prevalence of HPV in penile samples from heterosexual South African men using the FAP59/64 primer system (FAP PCR) and Roche Linear Array (LA) HPV Genotyping of 37 α-HPV types. We assess the impact of concurrent human immunodeficiency virus (HIV) infection on HPV infection. The FAP59/64 primer set was designed by Forslund and co-workers [15] to preferentially detect cutaneous HPV types, but are able to detect a broad range of both cutaneous and mucosal HPV types from the α, β and γ genera [10], [15], [16]. We additionally assess the prevalence of α, β, γ HPV types in a subset of the FAP PCR positive penile samples using high throughput sequencing of FAP amplicons.
2. Materials and methods
2.1. Study population and sample collection
A total of 335 penile swabs samples from a study conducted on genital HPV transmission in heterosexual couples in Gugulethu, Cape Town, South Africa [17] were retrospectively analysed. Study protocols were approved by the research ethics committee of the University of Cape Town (reference: 258/2006) and all participants provided written informed consent. Penile samples were collected by dry swabbing of the penile shaft, glans and foreskin, if present, with a Digene swab. Swabs were stored in 1 mL Digene specimen transport medium (STM, Qiagen) at −80 °C.
2.2. DNA isolation and amplification
DNA was extracted from 400 µl of specimen using the MagNA Pure Compact System (Roche) and the MagNA Pure Compact Nucleic Acid Isolation kit (Roche) into a final volume of 200 µl.
2.3. HPV genotyping by Roche Linear Array
HPV genotyping was performed using the Roche LA HPV genotyping test, which identifies 37 α-HPV genotypes, on 50 µl of the extracted DNA according to the manufacturer's instructions. Oncogenic types include HPV-16, −18, −31, −33, −35, −39, −45, −51, −52, −56, −58, −59, −68, −73, and −82; probable oncogenic types include HPV-26, −53, and −66; and non-oncogenic HPV types include HPV-6, −11, −34 (previously classified as HPV-64), −40, 42, −44 (previously classified as HPV-55), −54, −61, −62, −67, −69, −70, −71, −72, −81, −83, −84, −82 (HPV-IS39) and −89 (HPV-CP6108). The Roche LA HPV genotyping test includes amplification of a human β-globin gene fragment as an internal control for sample adequacy. All samples had positive β-globin results.
2.4. FAP PCR
An approximately 480 bp region of the HPV L1 gene was amplified using the degenerate primers FAP59 and FAP64 [15]. PCR was performed using 5 units/µl TaKaRa Ex Taq HS (ClonTech Laboratories) with 2.5 mM each dNTP, TaKaRa Ex Taq buffer, 4 µM of each primer and 5 µl DNA. Cycling conditions were denaturation at 95 °C for 2 min followed by 32 cycles of 95 °C for 30 s, 50 °C for 1 min and 72 °C for 1 min.
2.5. FAP 454 sequencing
For 454 sequencing the FAP64 primer was fused to the 454 Lib-L Adaptor A and one of 20 different 10 base pair multiplex identifiers (MIDs) [18]. The FAP59 primer was fused to the 454 Lib-L Adaptor B. PCR was performed as described above, except the cycling conditions were 95 °C for 2 min followed by 32 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min. Amplicons were gel purified using the MinElute gel extraction kit (Qiagen) and quantified using the Quant-iT Picogreen dsDNA assay (ThermoFischer). Amplicons were pooled in equal amounts with 20 amplicons per library. Sequencing was done by Macrogen (Seoul, South Korea) on the 454 GS-FLX platform using Titanium chemistry on 1/8 region of the plate per library.
2.6. Data analysis
Sequences were processed with UPARSE [19]. Reads were quality filtered using a maximum expected error of 0.5. The median number of 454 reads per sample was 6738 (range 2334–14490). Reads were clustered at 97% identity using the cluster_otus command and then re-clustered at 90% identity with UCLUST [20]. Clustering in UPARSE includes removal of reads that are PCR chimeras of the most abundant reads. Clustering further allows for the reduction of noise due to PCR and sequencing errors. The centroid method was used for picking representative sequences for each cluster. Singletons were removed. Representative sequences were aligned to the L1 genes of all HPV types in the Papilloma Virus Episteme (PaVE) database (http://pave.niaid.nih.gov/) [21] and the 202 putative HPVs defined by Chouhy and co-workers (2013) [22]. Any unassigned reads were aligned to the NCBI nonredundant nucleotide database using blast for classification. Reads were assigned to an HPV genotype or isolate if they had >90% identity. Putative novel types (KY062999-KY063013) were defined as those that had <90% identity to any known or previously described putative HPV.
Sequences were aligned with Mafft v7 [23] and a maximum likelihood tree generated using PhyML 3.0 [24] with the GTR+I+R substitution model, as determined by jmodeltest [25]. Branch support was estimated using approximate likelihood ratio test (aLRT) [26].
2.7. Statistical analyses
Risk factors for penile HPV infection were summarised by frequency (percent) if discrete or median (interquartile range) if continuous. Effect sizes were estimated by odds ratio (95% confidence interval). P-values were calculated by Fisher's exact test or Wilcoxon rank sum test as appropriate. Univariable and multivariable logistic regression models were used to estimate effect sizes, and associated p-values, of risk factors stratified by HIV infection status. Variables were carried forward to multivariable analysis based on having a univariate p-value<0.1. Statistical significance was set at p<=0.05. Statistical analysis was carried out in R 3.2 (R Core Team 2016).
3. Results
3.1. HPV prevalence and associations with HIV infection, partner HIV infection and other factors
Penile samples from 335 men (195 HIV-uninfected and 140 HIV-infected) were screened for the presence of HPV using a PCR assay with FAP59/64 primers (FAP PCR) and Roche Linear Array HPV genotyping (LA HPV). Characteristics of the men are given in Supplementary Table 1 (FAP PCR/LA study).
Overall 79% (266/335) of the specimens tested positive for HPV, this included 17 specimens positive by LA HPV only, 59 positive by FAP PCR only and 190 specimens positive for both assays. The prevalence of HPV was significantly higher in HIV-infected men compared to HIV-uninfected men in an unadjusted analysis (91% compared to 71%; P<0.0001), as shown in Table 1. In HIV-infected men, HPV prevalence was similar in men with a CD4 count less than 350 cells/mm3 and men with a CD4 count greater than 350 cells/mm3 (93% compared with 90%, p=1.000). HPV prevalence was therefore not significantly associated with CD4 counts in HIV-positive men. The unadjusted analysis (Table 1) indicated that age, smoking and sexual behaviour variables (lifetime number of sexual partners and number of sex acts with their partner in the previous month). HPV infection was however found to be significantly different in men with HIV-infected female sexual partners compared to men with HIV-uninfected female sexual partners (86% compared to 65%, P<0.0001). Older age at sexual debut (greater than 18 years old) was also associated with significantly higher HPV infection than younger ages of sexual debut (Table 1).
Table 1.
HPV prevalence by Roche Linear Array HPV genotyping and FAP PCR in South African men and risk associations with HIV status and other clinical and demographic variables (univariate analysis)a.
| % HPV infected (n/N) | OR (95% CI) | P-value | |
|---|---|---|---|
| Total | 79 (266/335) | ||
| HIV status | |||
| Negative | 71 (139/195) | Ref | |
| Positive | 91 (127/140) | 3.9 (2.1–7.8) | <0.0001 |
| CD4 count (if HIV-positive) | |||
| ≤350 | 93 (63/69) | Ref | |
| >350 | 90 (61/67) | 1.0(0.3–3.2) | 1.0000 |
| Partner HIV status | |||
| Negative | 65 (69/106) | Ref | |
| Positive | 86 (197/229) | 3.3 (1.9–5.7) | <0.0001 |
| Partner CD4 count (if HIV positive) | |||
| ≤350 | 86 (96/111) | Ref | |
| >350 | 86 (99/115) | 1.0 (0.5–2.1) | 0.8679 |
| Age group | |||
| <30 | 84 (59/70) | Ref | |
| 30–40 | 79 (113/143) | 0.7 (0.3–1.4) | 0.3615 |
| >40 | 77 (94/122) | 0.6 (0.3–1.3) | 0.2328 |
| Age at sexual debut | |||
| <16 | 74 (70/95) | Ref | |
| 16–18 | 79 (138/174) | 1.4 (0.8–2.5) | 0.2932 |
| >18 | 89 (56/63) | 2.9 (1.2–7.6) | 0.0236 |
| Lifetime number of sexual partners | |||
| 1–2 | 77 (41/53) | Ref | |
| 3–5 | 79 (77/97) | 1.1 (0.5–2.5) | 0.7726 |
| 6–10 | 84 (84/100) | 1.5 (0.7–3.5) | 0.3141 |
| >10 | 75 (61/81) | 0.9 (0.4–2) | 0.7856 |
| Number of acts of sex with study partner in last month | |||
| <5 | 79 (145/183) | Ref | |
| ≥5 | 80 (115/144) | 1.0 (0.6–1.8) | 0.8892 |
| Smoker | |||
| Never | 78 (45/54) | Ref | |
| Ex | 83 (41/51) | 0.8 (0.3–2.2) | 0.6959 |
| Current | 79 (178/226) | 0.7 (0.3–1.6) | 0.4547 |
Abbreviations: OR, Odds Ratio. Ref, Reference. CI, Confidence Interval.
There was no information for the following: CD4 counts for 4 HIV-positive men, CD4 counts for HIV-positive partners of 3 men, age at sexual debut for 3 men, lifetime number of sexual partners for 4 men, number of acts of sex with study partner in last month for 8 men and smoker status for 4 men.
HIV-infected men were found in an adjusted assessment (Table 2) to be at a higher risk of HPV infection (adjusted odds ratio (aOR) 4.0 (95%CI 2.1–8.2, p<0.0001) compared to HIV-uninfected men. Men with female sexual partners infected with HIV had a greater risk of HPV infection (Table 2, aOR 3.7 (2.1–6.7)) than men with HIV-negative partners. Men with an age of sexual debut of over 18 years had a greater risk of HPV infection than men with sexual debut younger than 16 years of age (Table 2 aOR 2.9 (1.1–8.1), p=0.0315).
Table 2.
Risk factors for HPV infection in men (multivariate analysis).
| Adjusted OR (95% CI) | P-value | |
|---|---|---|
| HIV status | ||
| Negative | Ref | |
| Positive | 4.0 (2.1–8.2) | <0.0001 |
| Partner HIV status | ||
| Negative | Ref | |
| Positive | 3.7 (2.1–6.7) | <0.0001 |
| Age at sexual debut | ||
| <16 | Ref | |
| 16–18 | 1.3 (0.7–2.6) | 0.3755 |
| >18 | 2.9 (1.1–8.1) | 0.0315 |
Abbreviations: OR, Odds Ratio. Ref, Reference. CI, Confidence Interval.
3.2. Diversity of HPV genera and types in penile samples
To investigate the spectrum of HPV types in penile samples we used 454 sequencing of FAP amplicons (454-FAP). For the sequencing a subset of 218 of the 249 FAP-PCR positive samples were selected. These included penile samples from 104 HIV-negative and 114 HIV-positive men. The characteristics of the men included in the 454-FAP/LA sub-study are given in Supplementary Table 1.
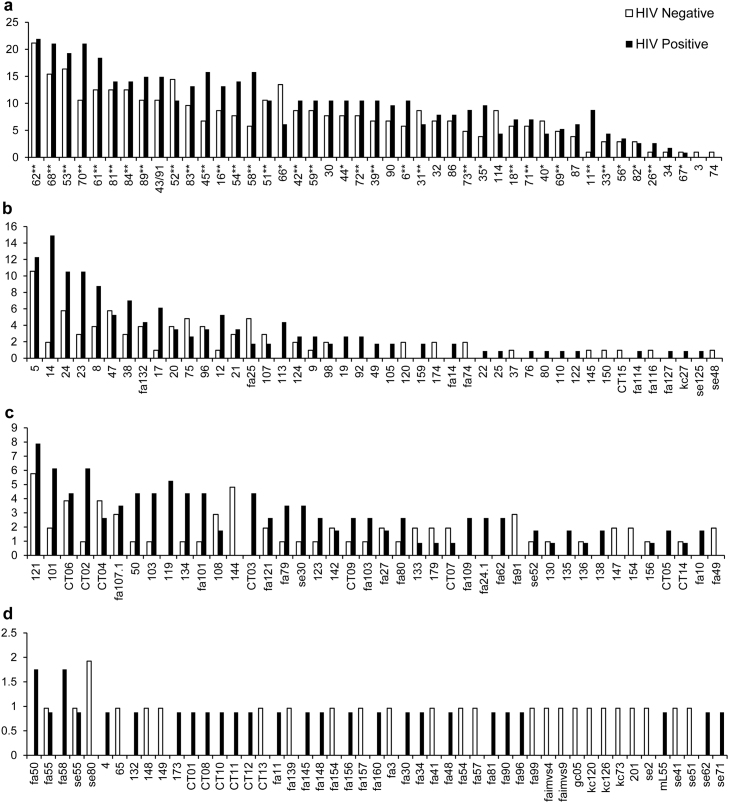
The prevalence of HPV genotypes detected in the 218 men are shown in Fig. 1. Samples were considered positive for a specific type if the type was detected by 454-FAP, LA HPV genotyping or both detection systems. Overall 181 unique HPV types were detected in 1330 infections in the 218 men. A total of 45 known α-HPV types, 45 β-HPV types (34 known, 10 putative and 1 novel putative), and 91 γ-HPV types (26 known, 51 putative and 14 novel putative) were detected (Fig. 1). Many HPV types (34.3%; 62/181) occurred only once in the penile samples, the majority of these belonged to the γ genus (44, 70.9%), followed by 16 (25.8%) β-HPV and 2 (3.2%) α-HPV types.
Fig. 1.
Prevalence (% of men, y-axis) of human papillomavirus (HPV) types from the α (a), β (b) and γ (c and d) genera in penile samples from 218 HPV positive men by human immunodeficiency virus (HIV) infection status. HPVs were detected by Roche Linear Array HPV genotyping and 454 sequencing of HPV FAP L1 amplicons. *Denotes HPV types detected by Roche Linear Array HPV genotyping only. **Denotes types detected by LA HPV genotyping and 454-FAP. All other types or putative types were detected by 454-FAP.
The most common α-HPV types were HPV-62, HPV-68 and HPV-53 with prevalence of 21.6%, 18.3% and 17.9%, respectively. In HIV-positive men the most prevalent α-HPV types were 62 (21.9%), 68 (21.1%), 70 (21.1%), 53 (19.3%), 61 (18.4%), 45 (15.8%) and 58 (15.8%). In HIV-negative men HPV types 62 (21.2%), 53 (16.3%), 68 (15.4%), 52 (14.4%) and 66 (13.5%) were the most frequently detected (Fig. 1). Of the 45 α-HPV types detected, 37 types had a higher prevalence in HIV-infected men compared to HIV-uninfected men. Only eight HPV types (3, 31, 40, 52, 66, 67, 74 and 114) were detected at a higher prevalence in HIV-uninfected men. The prevalence of IARC group 1 high-risk α-HPV types ranged from 3.2% to 12.4%, with HPV types 52 (12.4%), 45 (11.5%), 58 (11.0%), 16 (11.0%) and 51 (10.6%) most frequently detected.
The most prevalent β-HPV types were HPV-5 (11.5%), HPV-14 (8.7%) and HPV-24 (8.3%); all members of the β−1 species. In HIV-positive men β-HPV types 14 (14.9%), 5 (12.3%), 24 (10.5%) and 23 (10.5%) were the most frequently detected. The prevalence of HPV-14 was lower in HIV-negative men (1.9%), and HPV types 5 (10.6%), 24 (5.8%) and 47 (5.8%) were the most common. Of the 29 β-HPV types detected in more than one sample, 19 were more frequently detected in HIV-infected men.
The most frequently detected γ-HPVs were HPV-121 (6.9%), HPV-101 (4.1%) and a novel putative HPV type, CT06 (4.1%) (Fig. 1). In HIV-positive men types 121 (7.9%), 101 (6.1%) and CT02 (6.1%) and in HIV-negative men types 121 (5.8%), 144 (4.8%), CT06 (3.8%) and CT04 (3.8%) were the most prevalent types. Of the 47 types detected in more than one sample, 28 had higher prevalence in HIV-infected men.
The majority of the men (88.1%, 192/218) were co-infected with multiple HPV types. The median number of co-infecting types was six, with the number of co-infecting types ranging from two to 18. The distribution of HPV single and multiple infections stratified by HIV status is shown in Table 3. Multiple HPV infections were detected more frequently in HIV-infected men than HIV-uninfected men (p=0.002, Table 3). HIV-infected men had a median of six co-infecting HPV types and HIV-negative men four. Multiple infections were higher than single infections in both HIV-infected and HIV–uninfected men (p<0.0001, Table 3).
Table 3.
Prevalence of single and multiple infections with all, alpha, beta and gamma HPV types by HIV status among the men included in the 454 FAP/LA sub-study.
|
HIV-negative N=104 |
HIV-positive N=114 |
|||||
|---|---|---|---|---|---|---|
| n | Mean (95% CI) | n | Mean (95% CI) | P-valuea | ||
| All | Single | 20 | 19.2 (11.5–26.9) | 6 | 5.3 (1.1–9.4) | 0.002 |
| Multiple | 84 | 80.8 (73.1–88.5) | 108 | 94.7 (90.6–98.9) | 0.002 | |
| P-valueb | <0.0001 | <0.0001 | ||||
| Alpha | Any | 91 | 87.5 (81.0–94.1) | 105 | 92.1(87.1–97.1) | 0.262 |
| Single | 25 | 24.0 (15.7–32.4) | 14 | 12.3 (6.2–18.4) | 0.024 | |
| Multiple | 66 | 63.5 (54.1–72.9) | 91 | 79.8 (72.4–87.3) | 0.007 | |
| P-valueb | <0.0001 | <0.0001 | ||||
| Beta | Any | 45 | 43.3 (33.6–53.0) | 67 | 58.8 (49.6–68.0) | 0.023 |
| Single | 25 | 24.0 (15.7–32.4) | 29 | 25.4 (17.3–33.6) | 0.813 | |
| Multiple | 20 | 19.2 (11.5–26.9) | 38 | 33.3 (24.6–42.1) | 0.019 | |
| P-valueb | 0.402 | 0.192 | ||||
| Gamma | Any | 56 | 53.8 (44.1–63.6) | 80 | 70.2 (61.7–78.7) | 0.013 |
| Single | 56 | 36.5 (27.1–46.0) | 40 | 35.1 (26.2–44.0) | 0.825 | |
| Multiple | 18 | 17.3 (9.9–24.7) | 40 | 35.1 (26.2–44.0) | 0.003 | |
| P-valueb | 0.002 | 0.999 | ||||
Statistically significant values (<0.05) indicated in bold.
P-value for HIV-negative vs HIV-positive.
P-value for single vs multiple infections.
The prevalence of α-HPV infection detected by 454 FAP and Roche LA was 89.9% (196/218). Of the men with α-HPV infections, 80.1% (157/196) were infected with multiple α-HPV types (median four α-HPV types and range 2–15 types). HIV-infected men had a median of five α-HPV types (range 2–15) and HIV-uninfected men four α-HPV types (range 2–14). Infection with a single α-HPV type was significantly higher in HIV-negative men than HIV-positive men (p=0.024). Infection with multiple α-HPV types was significantly more common in HIV-infected men compared to uninfected men (p=0.007) (Table 3). Multiple infections with α-HPV types were higher than single infections in both HIV-infected and –uninfected men (p<0.0001, Table 3). The prevalence of β-HPV infection was lower than α-HPV infection at 51.4% (112/218) overall. A total of 51.7% (58/112) of the β-HPV positive men had multiple infections with a median of two β-HPV types (range 2–8). HIV-uninfected had a median of two β-HPV types (range 2–7) and HIV-infected three β-HPV types (range 2–8). Infection with a single β-HPV type was not significantly different between HIV-infected and uninfected men. Multiple infections with β-HPV types were significantly higher in HIV-infected men compared to uninfected men (p=0.019, Table 3). Multiple infections were not significantly higher than single infections in both HIV-infected and HIV–uninfected men (Table 3). The prevalence of γ-HPV types was 62.4% (136/218). Among the γ-HPV positive men 42.6% (58/136) were infected with multiple γ-HPV types, with a median of two types and range of 2–9 types. Infection with single γ-HPV types was similar in HIV-infected and HIV-uninfected men. A significantly higher number of HIV-infected men had multiple γ-HPV types compared to HIV-negative men (p=0.003). HIV-uninfected men were infected with a median of two γ-HPV types (range 2–4) and HIV-infected two γ-HPV types (range 2–9). Infection with a single γ-HPV type was higher than multiple γ-HPV infection in HIV-uninfected men (p=0.002), while single and multiple infections were not significantly different in HIV-infected men (Table 3).
Samples (N=218) for this sub-study (454-FAP) were selected chronologically from the 249 FAP-PCR positive specimens. There were no significant differences in the selected characteristics of the men in the sub-study, except the HIV-infected men in the 454-FAP sub-study had significantly less HIV-infected sexual partners (75% (78/104) vs 30% (3/10), p=0.0043), Supplementary Table 1). The HPV genotyping in the 104 HIV-infected men may therefore not accurately represent that in all the HIV-infected FAP positive men.
3.3. Putative novel HPVs
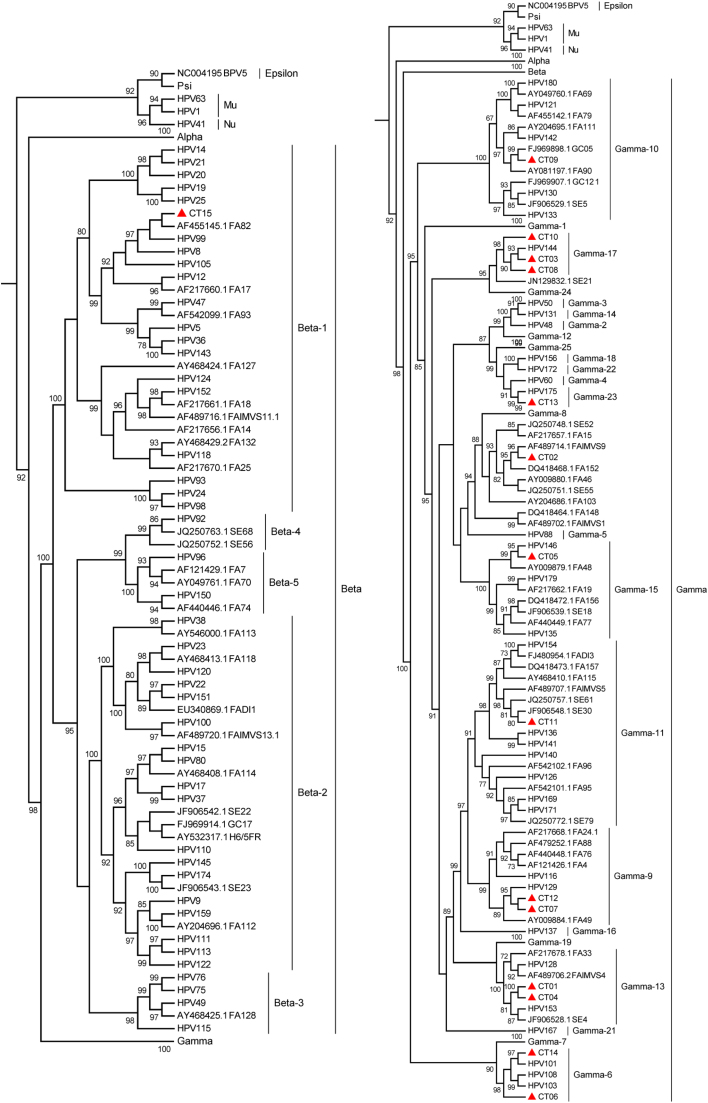
A total of fifteen novel putative HPV types, accounting for 47 infections, were detected based on the partial L1 sequences (Supplementary Table 2). These were named CT01-CT15. Complete L1 gene sequences for these putative types would be required to confirm that they are novel HPV types. The pairwise nucleotide identity based on the sequenced FAP region of the novel types to known and previously described putative HPV types is shown in Supplementary Table 2 and ranged from 69.1% to 89%. The prevalence of the novel types ranged from 0.5% (1/218) to 4.1% (9/218), with the most common being CT06 (4.1%, 9/218), CT02 (3.7%, 8/218) and CT04 (3.3%, 7/218). Most of the novel types were however rare, with 46.7% (7/15) occurring only once in the 218 men (Supplementary Table 2). Phylogenetic analysis of the putative novel viruses (Fig. 2) classified fourteen (CT01-CT14) as γ–HPV types and one (CT15) as a β-HPV type. No novel α-HPV types were detected.
Fig. 2.
Maximum likelihood tree inferred from the global alignment of L1 nucleotide sequences from known, putative and novel putative HPV types. The L1 nucleotide sequences from known HPV types were downloaded from the Papillomavirus Episteme and previously identified putative HPV types from GenBank. The GenBank accession numbers of the putative HPV types are indicated. The novel putative HPV types (CT01-CT15) are indicated with triangles. HPV groups were marked based on previously described phylogenetic relationships [22]. Branch support was measured using the approximate likelihood-ratio test (aLRT) and support greater than 70% is indicated.
4. Discussion
In this study we examined HPV infection in penile specimens from HIV-positive and HIV-negative South African heterosexual men. HPV typing by Roche LA HPV genotyping and next generation sequencing of FAP amplicons detected 45 α-HPV types, 45 β-HPV types, and 91 γ-HPV types in penile samples from 218 HPV-positive men. This represents an extremely broad range of HPV types to be reported in a single study and is the first study using this technology on penile samples from Africa.
A significant number of men (79%) were HPV-positive by FAP PCR and LA. There was an increased risk of HPV infection in HIV infected men. This risk was not associated with CD4 count levels. This is in agreement with other studies where HPV prevalence increased substantially after HIV infection and was therefore an early event associated with HIV seroconversion and not a drop in CD4 counts [27]. We also demonstrated that men with HIV-infected female sexual partners had a higher risk of HPV infection as detected by HPV FAP PCR and LA. This is consistent with a previous study on these couples using only Roche LA HPV genotyping of 37 α-HPV types where it was shown that both HIV-infected men and women had a higher prevalence of HPV, that the impact of decreasing CD4 count is greater in women than men, and that in men, in contrast to women, HPV infection risk may be influenced by the HIV status of their female partner [17]. HIV positive women have a higher prevalence of α HPV types, and in some cases higher HPV viral load, which impacts on transmission [28]. However, in the present study the FAP PCR used detects α, β and γ HPV types. The prevalence of β and γ HPV types in cervical specimens from the women in this couples cohort is not known.
A broad range of α, β and γ HPV types in penile specimens were identified using a combination of 454 sequencing of FAP amplicons and Roche LA HPV genotyping. Multiple HPV infections were more common than single infections (88.1 vs 11.9%), with the median number of co-infecting types being six (range 2–18).
α-HPV types were detected in 89.9% of the 218 FAP positive samples. Sequencing of FAP amplicons identified 38 α-HPV types. Of the 37 α-HPV types included in the LA HPV genotyping assay, 29 types (6, 11, 16, 18, 26, 31, 33, 39, 42, 45, 51, 52, 53, 54, 58, 59, 61, 62, 68, 69, 70, 71, 72, 73, 81, 82, 83, 84, 89) were detected by both 454-FAP and LA HPV genotyping, seven HPV types (35, 40, 55, 56, 66, 67 and 82) were detected by LA HPV genotyping only and one type, HPV-34 (previously classified as HPV-64), was detected by 454-FAP only (Fig. 1). Infection with any of the 12 h-HPV types occurred in 56% (122/218) of the men. The high burden of α-HPVs in this cohort is unexpected as the majority of the men were circumcised (91.2%, Supplementary Table 1). Removal of the mucosal epithelium during circumcision is thought to reduce mucosal HPV infection and previous studies have indicated that circumcised men are less likely to have α-HPVs than uncircumcised men [29].
β-HPVs were detected in 51.4% of the FAP positive penile samples. In concurrence with previous studies on men from USA, Mexico and Brazil [12], [13], [14] the most frequently detected β HPV species were β−1 and β−2. HPV types 5, 14, 24 and 8 of the β−1 species and HPV-23 of β−2 species were the most prevalent. Previous investigations of β-HPVs in genital specimens from men in the USA, Brazil and Mexico identified HPV-38 and −22 (β−2) as well as HPV-21 and −24 (β−1) [14], and HPV-107 and HPV-120 (β−2) [12] as most prevalent. Infection with any and multiple β-HPVs was significantly higher in HIV-infected men compared to uninfected men.
A total of 62.4% of the FAP positive penile samples were positive for γ-HPVs, with 91 different γ-HPV types detected. This study represents to our knowledge the most comprehensive investigation of genital γ-HPV infection in men. Only one other study examined genital infection with γ-HPV and reported ten γ-HPV types [12]. HPV-121, a member of the γ−10 species, was the most common γ-HPV in the South African penile specimens (6.9%). Previous studies of γ-HPV in the anal canal of men who have sex with men (MSM) identified HPV-121 and γ−10 species as the most prevalent γ-HPVs [30], [31]. Infection with any and multiple γ-HPVs was significantly higher in HIV-infected men compared to uninfected men. The γ-PV genus is a rapidly expanding group of diverse viruses that may be ubiquitous and have been found in both cutaneous and mucosal sites, including the skin, anal, nasal, oral and cervical sites [1], [5], [32].
5. Conclusion
To our knowledge this is the first report of the prevalence of genital α, β and γ HPVs in African men. Overall, the present study showed a strong association between HIV status and genital infection with α, β and γ HPVs in men. The potential contribution of β and γ HPVs to lesions and cancer in immunosuppressed and immunocompromised individuals is not yet understood and requires further investigation.
Footnote Page.
Declaration of interest statement
TLM: declares no commercial or other association that might pose a conflict of interest.
ZM: declares no commercial or other association that might pose a conflict of interest.
ML: declares no commercial or other association that might pose a conflict of interest.
DC: declares no commercial or other association that might pose a conflict of interest.
ALW: declares no commercial or other association that might pose a conflict of interest.
Funding
This study was supported by grants from Poliomyelitis Research Foundation [grant number 11/24] and South African Research Chairs Initiative of the Department of Science and Technology [grant number 64815]. TLM was supported by awards from the National Research Foundation South Africa and Clinical Infectious Diseases Research Initiative (Wellcome Trust).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.pvr.2017.05.001.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Bzhalava D., Eklund C., Dillner J. International standardization and classification of human papillomavirus types. Virology. 2015;476:341–344. doi: 10.1016/j.virol.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 2.Bouvard V., Baan R., Straif K., Grosse Y., Secretan B., El Ghissassi F., Benbrahim-Tallaa L., Guha N., Freeman C., Galichet L., Cogliano V. A review of human carcinogens--Part B: biological agents. Lancet Oncol. 2009;10:321–322. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 3.Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., Snijders P.J., Peto J., Meijer C.J., Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 4.Cubie H.A. Diseases associated with human papillomavirus infection. Virology. 2013;445:21–34. doi: 10.1016/j.virol.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Bzhalava D., Guan P., Franceschi S., Dillner J., Clifford G. A systematic review of the prevalence of mucosaland cutaneous human papillomavirus types. Virology. 2013;445:224–231. doi: 10.1016/j.virol.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Bzhalava D., Muhr L.S., Lagheden C., Ekstrom J., Forslund O., Dillner J., Hultin E. Deep sequencing extends the diversity of human papillomaviruses in human skin. Sci. Rep. 2014;4:5807. doi: 10.1038/srep05807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foulongne V., Sauvage V., Hebert C., Dereure O., Cheval J., Gouilh M.A., Pariente K., Segondy M., Burguiere A., Manuguerra J.C., Caro V., Eloit M. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonsson A., Forslund O., Ekberg H., Sterner G., Hansson B.G. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J. Virol. 2000;74:11636–11641. doi: 10.1128/jvi.74.24.11636-11641.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y., Madupu R., Karaoz U., Nossa C.W., Yang L., Yooseph S., Yachimski P.S., Brodie E.L., Nelson K.E., Pei Z. Human papillomavirus community in healthy persons, defined by metagenomics analysis of human microbiome project shotgun sequencing data sets. J. Virol. 2014;88:4786–4797. doi: 10.1128/JVI.00093-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forslund O., Johansson H., Madsen K.G., Kofoed K. The nasal mucosa contains a large spectrum of human papillomavirus types from the Betapapillomavirus and Gammapapillomavirus genera. J. Infect. Dis. 2013;208:1335–1341. doi: 10.1093/infdis/jit326. [DOI] [PubMed] [Google Scholar]
- 11.Bottalico D., Chen Z., Dunne A., Ostoloza J., McKinney S., Sun C., Schlecht N.F., Fatahzadeh M., Herrero R., Schiffman M., Burk R.D. The oral cavity contains abundant known and novel human papillomaviruses from the Betapapillomavirus and Gammapapillomavirus genera. J. Infect. Dis. 2011;204:787–792. doi: 10.1093/infdis/jir383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sichero L., Pierce Campbell C.M., Ferreira S., Sobrinho J.S., Luiza Baggio M., Galan L., Silva R.C., Lazcano-Ponce E., Giuliano A.R., Villa L.L., Broad HPV distribution in the genital region of men from the HPV infection in men (HIM) study. Virology. 2013;443:214–217. doi: 10.1016/j.virol.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sichero L., Pierce Campbell C.M., Fulp W., Ferreira S., Sobrinho J.S., Baggio M., Galan L., Silva R.C., Lazcano-Ponce E., Giuliano A.R., Villa L.L. High genital prevalence of cutaneous human papillomavirus DNA on male genital skin: the HPV infection in men study. BMC Infect. Dis. 2014;14:677. doi: 10.1186/s12879-014-0677-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nunes E.M., Sudenga S.L., Gheit T., Tommasino M., Baggio M.L., Ferreira S., Galan L., Silva R.C., Pierce Campbell C.M., Lazcano-Ponce E., Giuliano A.R., Villa L.L., Sichero L. Diversity of beta-papillomavirus at anogenital and oral anatomic sites of men: the HIM study. Virology. 2016;495:33–41. doi: 10.1016/j.virol.2016.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forslund O., Antonsson A., Nordin P., Stenquist B., Hansson B.G. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J. Gen. Virol. 1999;80(Pt 9):2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- 16.Ekström J., Bzhalava D., Svenback D., Forslund O., Dillner J. High throughput sequencing reveals diversity of Human Papillomaviruses in cutaneous lesions. Int. J. Cancer. 2011;129:2643–2650. doi: 10.1002/ijc.26204. [DOI] [PubMed] [Google Scholar]
- 17.Mbulawa Z.Z., Marais D.J., Johnson L.F., Boulle A., Coetzee D., Williamson A.L. Influence of human immunodeficiency virus and CD4 count on the prevalence of human papillomavirus in heterosexual couples. J. Gen. Virol. 2010;91:3023–3031. doi: 10.1099/vir.0.020669-0. [DOI] [PubMed] [Google Scholar]
- 18.J.C.H.M.P.D.G.W. Group Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One. 2012;7:e39315. doi: 10.1371/journal.pone.0039315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 20.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 21.Van Doorslaer K., Tan Q., Xirasagar S., Bandaru S., Gopalan V., Mohamoud Y., Huyen Y., McBride A.A. The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res. 2013;41:D571–D578. doi: 10.1093/nar/gks984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chouhy D., Bolatti E.M., Perez G.R., Giri A.A. Analysis of the genetic diversity and phylogenetic relationships of putative human papillomavirus types. J. Gen. Virol. 2013;94:2480–2488. doi: 10.1099/vir.0.055137-0. [DOI] [PubMed] [Google Scholar]
- 23.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 25.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anisimova M., Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 27.Wang C., Wright T.C., Denny L., Kuhn L. Rapid rise in detection of human papillomavirus (HPV) infection soon after incident HIV infection among South African women. J. Infect. Dis. 2011;203:479–486. doi: 10.1093/infdis/jiq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mbulawa Z.Z., Johnson L.F., Marais D.J., Gustavsson I., Moodley J.R., Coetzee D., Gyllensten U., Williamson A.L. Increased alpha-9 human papillomavirus species viral load in human immunodeficiency virus positive women. BMC Infect. Dis. 2014;14:51. doi: 10.1186/1471-2334-14-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castellsague X., Bosch F.X., Munoz N., Meijer C.J., Shah K.V., de Sanjose S., Eluf-Neto J., Ngelangel C.A., Chichareon S., Smith J.S., Herrero R., Moreno V., Franceschi S. Male circumcision, penile human papillomavirus infection, and cervical cancer in female partners. N. Engl. J. Med. 2002;346:1105–1112. doi: 10.1056/NEJMoa011688. [DOI] [PubMed] [Google Scholar]
- 30.Dona M.G., Gheit T., Latini A., Benevolo M., Torres M., Smelov V., McKay-Chopin S., Giglio A., Cristaudo A., Zaccarelli M., Tommasino M., Giuliani M. Alpha, beta and gamma Human Papillomaviruses in the anal canal of HIV-infected and uninfected men who have sex with men. J. Infect. 2015;71:74–84. doi: 10.1016/j.jinf.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Torres M., Gheit T., McKay-Chopin S., Rodriguez C., Romero J.D., Filotico R., Dona M.G., Ortiz M., Tommasino M. Prevalence of beta and gamma human papillomaviruses in the anal canal of men who have sex with men is influenced by HIV status. J. Clin. Virol. 2015;67:47–51. doi: 10.1016/j.jcv.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Ameur A., Meiring T.L., Bunikis I., Haggqvist S., Lindau C., Lindberg J.H., Gustavsson I., Mbulawa Z.Z., Williamson A.L., Gyllensten U. Comprehensive profiling of the vaginal microbiome in HIV positive women using massive parallel semiconductor sequencing. Sci. Rep. 2014;4:4398. doi: 10.1038/srep04398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material