Abstract
Background
1q43-q44 deletion syndrome is a well-defined chromosomal disorder which is characterized by moderate to severe mental retardation, and variable but characteristic facial features determined by the size of the segment and the number of genes involved. However, patients with 1q43-q44 duplication with a clinical phenotype comparable to that of 1q43-q44 deletion are rarely reported. Moreover, pure 1q43-q44 deletions and duplications derived from balanced insertional translocation within the same family with precisely identified breakpoints have not been reported.
Case presentation
The proband is a 6-year-old girl with profound developmental delay, mental retardation, microcephaly, epilepsy, agenesis of the corpus callosum and hearing impairment. Her younger brother is a 3-month-old boy with macrocephaly and mild developmental delay in gross motor functions. G-banding analysis of the subjects at the 400-band level did not reveal any subtle structural changes in their karyotypes. However, single-nucleotide polymorphism (SNP) array analysis showed a deletion and a duplication of approximately 6.0 Mb at 1q43-q44 in the proband and her younger brother, respectively. The Levicare analysis pipeline of whole-genome sequencing (WGS) further demonstrated that a segment of 1q43-q44 was inserted at 14q23.1 in the unaffected mother, which indicated that the mother was a carrier of a 46,XX,ins(14;1)(q23.1;q43q44) insertional translocation. Moreover, Sanger sequencing was used to assist the mapping of the breakpoints and the final validation of those breakpoints. The breakpoint on chromosome 1 disrupted the EFCAB2 gene in the first intron, and the breakpoint on chromosome 14 disrupted the PRKCH gene within the 12th intron. In addition, fluorescence in situ hybridization (FISH) further confirmed that the unaffected older sister of the proband carried the same karyotype as the mother.
Conclusion
Here, we describe a rare family exhibiting pure 1q43-q44 deletion and duplication in two siblings caused by a maternal balanced insertional translocation. Our study demonstrates that WGS with a carefully designed analysis pipeline is a powerful tool for identifying cryptic genomic balanced translocations and mapping the breakpoints at the nucleotide level and could be an effective method for explaining the relationship between karyotype and phenotype.
Electronic supplementary material
The online version of this article (10.1186/s13039-018-0371-7) contains supplementary material, which is available to authorized users.
Keywords: Pure 1q43-q44 deletion/duplication, Insertional translocation, Congenital anomaly, Whole-genome sequencing
Background
The clinical phenotype of 1q43-q44 deletion or duplication is highly variable, due to the size of the segment and the number of genes involved. The phenotypic features of patients with 1q43-44 deletion include moderate to severe mental retardation, development retardation, microcephaly, corpus callosum dysplasia, epilepsy and dysmorphic features. In individuals with 1q43-q44 duplication, the most recognizable features are macrocephaly, mental retardation, epilepsy and mild malformation. Interstitial deletions of the long arm of chromosome 1 involving only the 1q43-q44 region have been reported in more than 80 patients, with most of these patients arising de novo [1–7]. A few individuals exhibiting pure 1q43-q44 interstitial duplication have been reported [8–15]. However, both pure 1q43-q44 deletion and duplication occurring in a family have not been reported.
Several cytogenetic and molecular techniques have been applied to detect the deletion or duplication of pathogenic copies, such as G-banding, fluorescence in situ hybridization (FISH) and chromosomal microarrays (CMAs). However, these techniques present individual limitations and can often be technically challenging. Recent studies have shown that whole-genome sequencing (WGS) with a carefully designed data analysis pipeline is a more powerful tool for detecting chromosomal abnormalities due to its higher resolution and the ability to detect balanced translocations and small imbalances that cannot be detected with CMAs [16].
Insertional translocations are complex chromosomal rearrangements that require at least three breakpoints in the involved chromosome, with an incidence of 1:80,000 in live births [17]. Insertional translocations can be divided into simple intrachromosomal or interchromosomal insertional translocations and complex chromosomal insertional translocations [18]. Nowakowska et al. [19] found that 2.1% of de novo copy number variations (CNVs) are actually inherited from a parental balanced insertional translocation. However, this percentage may represent an underestimate because not all parental data may be collected in these studies, and due to technical limitations, some small imbalances have not yet to be discovered. However, WGS can identify nearly all cryptic chromosomal abnormalities or complex rearrangements present in the genome, in addition to characterizing translocation breakpoints at the nucleotide level.
Herein, we present a rare family in which two siblings presented with congenital anomalies. These two individuals harboured an approximately 6.0-Mb deletion or duplication of 1q43-q44 inherited from their mother, a carrier of a cryptic balanced insertional translocation. We further precisely identified the corresponding breakpoints via WGS and Sanger sequencing. This is the first report of the detection of an insertional translocation associated with 1q43-q44 deletion and duplication using WGS.
Case presentation
The proband (III-3, Fig. 1a) is the third child of a non-consanguineous, healthy couple. She is a 6-year-old Chinese girl with profound developmental delay, microcephaly, agenesis of the corpus callosum, epilepsy, language delayed and hearing impairment. She was born at full term after an uncomplicated spontaneous vaginal delivery with a normal birth weight (3400 g). She experienced seizures four times at 3 months of age, with spontaneous remission occurring after more than 10 s. At 7 months of age, she began turning over but could not grasp and sit without support. Intellectual evaluation with the Gesell Development Schedule (GDS) showed that her developmental quotient at 7 months of age was equivalent to that of a 10-week-old infant, indicating significant growth retardation [20]. The detailed data are shown in Additional file 1: Table S1. Brain magnetic resonance imaging (MRI) indicated absence of the corpus callosum and enlargement of the posterior horn of all three ventricles bilaterally. The results of brainstem auditory evoked potential (BAEP) analysis indicated bilateral hearing impairment. At 6 years of age, the proband presented with microcephaly (47.2 cm, <− 2 SD) and began learning to walk but could not speak (Fig. 1b).
Fig. 1.
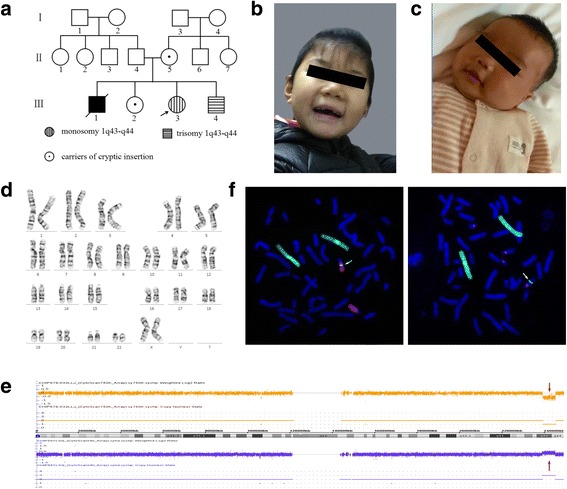
a Three-generation pedigree of the family with the proband (III-3) indicated by an arrow. Affected individuals are indicated with black, horizontal or vertical lines in the symbols, including III-3: monosomy 1q43-q44 and III-4: trisomy 1q43-q4; carriers of the cryptic insertion (II-5, III-2) are indicated with dots in the symbols. G-banding analysis was not performed for the elder brother (III-1) due to a lack of sample. b and c Facial profiles of the proband and younger brother at 6 years and 3 months, respectively. The proband presented with microcephaly, and the younger brother presented with macrocephaly. d G-banding analysis of the proband (III-3) at a band resolution of ∼400 showed no visual abnormal karyotype. e The results of SNP-array analysis. The proband (III-3) harbuored an interstitial 1q43-q44 deletion (upper), and the younger brother (III-4) carried an interstitial 1q43-q44 duplication (down). The deletion and duplication regions are indicated by red arrows. f The FISH results for the mother (II-5) with WCP1/14 (left) and WCP1/CEP14 (right) probes. Chromosome 1 is shown in green, and the chromosome 14 is shown in red (left); WCP1 and CEP14 are shown in green and red, respectively (right). The inserted segment is indicated by the white arrows
The elder brother (III-1, Fig. 1a) presented similar features to the proband, such as developmental delay, cerebral palsy and intracranial haemorrhage after birth, and died at 5 years of age. The elder sister (III-2, Fig. 1a) has a normal phenotype. The younger brother (III-4, Fig. 1a) is the fourth child. At 34 weeks of gestation, an MRI scan of the foetal head and ultrasonography revealed no obvious abnormalities. He was born at term via caesarean section after an uneventful pregnancy and exhibited a normal birth weight (3000 g). At the age of 3 months, he presented macrocephaly (Fig. 1c) and his head circumference was 44.5 cm (> + 2 SD). His developmental quotient was equivalent to that of an 11-week-old infant, with testing demonstrating a borderline full-scale developmental quotient (85), and he exhibited developmental delay in gross motor functions (Additional file 1: Table S1).
Materials and methods
G-banding at a band resolution of ∼400 was performed on metaphase peripheral blood lymphocytes obtained from the proband (III-3) and four other family members (II-4, II-5, III-2 and III-4) according to the laboratory’s protocols. DNA was isolated from their peripheral blood lymphocytes using the QIAamp® DNA blood midi kit (QIAGEN, Hilden, Germany). Single-nucleotide polymorphism (SNP) array analysis was performed using Cytoscan 750 K chips (Affymetrix, Santa Clara, CA, USA) as we described in a previous report [21]. The data were analysed using ChAS chromosome analysis software (Affymetrix, Santa Clara, CA, USA). To confirm the chromosomal imbalances of the patients and determine whether they were de novo or inherited from the parents, the parental DNA was evaluated by whole-genome low-coverage sequencing. Briefly, a non-size selected mate-pair library was prepared using ~ 3 μg of genomic DNA and then subjected to 50-bp-end multiplex sequencing on the Illumina HiSeq™ X10 platform. After automatically removing adaptor sequences and low-quality reads, high quality paired-end reads were aligned to the NCBI human reference genome (GRCh37/hg19) by SOAP2. Uniquely mapped reads were selected for subsequent analysis as previously described in detail [22]. After the bioinformatics analysis, we obtained the candidate breakpoint regions. The precise breakpoints were further confirmed by PCR and Sanger sequencing, and the genomic locations of the breakpoints were analysed according to the February 2009 (GRCh37/hg19) assembly in the UCSC Genome Browser (http://genome.ucsc.edu). Primers targeting the flanking sequences of the candidate breakpoints of chromosomes 1 and 14 were designed with Primer 5 software and are listed in Additional file 2: Table S2. To validate the abnormal karyotype, FISH was performed on metaphase chromosomes of peripheral blood lymphocytes using whole chromosome probes (WCPs) of chromosomes 1and 14 and a centromere probe (CEP) of chromosome 14 (CytoTrend, HK, China) following the manufacturer’s instructions. The chromosomes 14 and 22 with homologous regions in the centromeres were distinguished based on their different lengths.
Results
G-banding analysis at a band resolution of ∼400 revealed no karyotype abnormalities in the proband (Fig. 1d) or the four other family members. However, further SNP array analysis indicated pathological CNVs in the proband and her younger brother: arr[hg19]1q43q44 (239,019,924–245,142,519) × 1 and arr[hg19]1q43q44(239,033,439–245,142,567) × 3, respectively (Fig. 1e).
WGS analysis of the parents (II-4 and II-5) revealed a normal karyotype for the father but misalignment ~ 3.78 million reads for the mother. Further analysis showed that two records were highly credible (p < 0.001), roughly described as chr14-chr1:62,011,989–245,138,646 and chr14-chr1:62,006,695–239,045,980. These abnormal records indicated insertion of the 1q43-q44 segment into 14q23.1 in the mother’s genome, which was confirmed via FISH using the WCP1/14 and WCP1/CEP14 probes (Fig. 1f). The combination of WGS and FISH analyses revealed that the mother exhibited a 46,XX,ins(14;1)(q23.1;q43q44) karyotype. The three breakpoints were further determined by PCR and Sanger sequencing. Sanger sequencing further confirmed that the first breakpoint on chromosome 1 was located at chr1:239,045,641–239,046,656, the second breakpoint on chromosome 1 was located at chr1:245,145,720–245,145,726, and the breakpoint on chromosome 14 was located at chr14:62,011,535–62,011,546. There were no genes around the 1st breakpoint on chromosome 1. By contrast, the EFCAB2 gene was disrupted in the first intron by the 2nd breakpoint on chromosome 1, and the breakpoint on chromosome 14 disrupted the PRKCH gene within the 12th intron (Fig. 2a and b). Moreover, some small imbalances and microhomology sequences were also observed near these breakpoint sites (Fig. 2c-e).
Fig. 2.
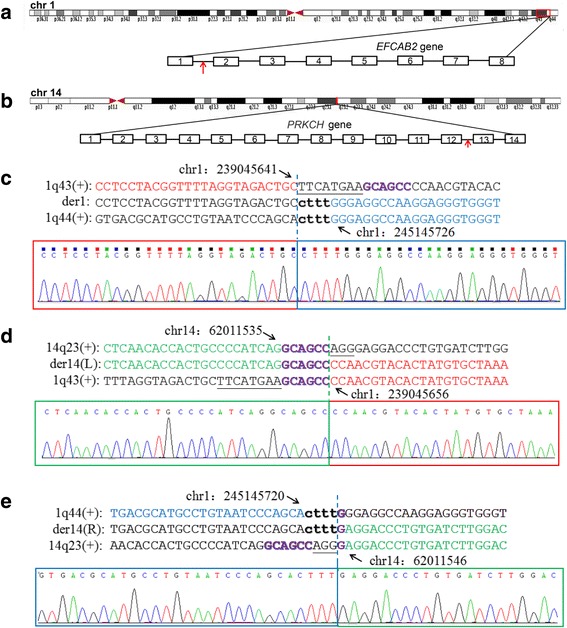
The chromosome breakpoints and disrupted genes within the insertional translocation t(1:14). a and b The disrupted genes at the breakpoints are indicated by red arrows. The breakpoint on chromosome 1q44 disrupts the EFCAB2 gene in intron 1, and the breakpoint on chromosome 14 disrupts the PRKCH gene in intron 12. c-e The breakpoints mapped at the base-pair level by Sanger sequencing. Translocation junction sequences (middle line) and matching reference sequences (top and bottom lines) are shown with different colours depending on the involved chromosome region (1q43-red, 1q44-blue, 14q23-green). The microhomology observed at the translocation breakpoint sites are indicated in purple letters in bold, deleted sequences are underlined, and duplicate sequences are shown in lower-case letters in bold. Der14(L) indicates the breakpoint sequence near the centromere, and der14(R) indicates the breakpoint sequence near telomere
Further family analysis by G-banding and FISH confirmed that the elder sister carries the same balanced insertional translocation as the mother.
Discussion
Here, we report a rare family in which two siblings exhibit 1q43-q44 deletion or duplication, respectively. The combination of SNP array, WGS and FISH analyses showed that both the deletion and duplication resulted from a 6.0-Mb cryptic balanced insertion of material from 1q43-q44 inserted into 14q23.1.
Chromosome 1q43-q44 deletion syndrome (OMIM: #612377) is characterized by moderate to severe mental retardation, limited or no speech, and variable but characteristic facial features, including a round face, prominent forehead, flat nasal bridge, hypertelorism, epicanthal folds, and low-set ears. Other characteristics may include developmental retardation, microcephaly, agenesis of the corpus callosum, and seizures [2]. Compared with 1q43-q44 deletion, the clinical manifestations of patients with 1q43-q44 duplication may be mild and mainly include macrocephaly, mental retardation and mild malformation [8, 23]. The clinical features of previously described patients with deletion or duplication of chromosome 1 overlapping q34-q44 are shown in Table 1 for a comparison of phenotypic differences. In the present study, the proband exhibiting 1q43-q44 deletion was found to show some characteristic features, such as profound developmental delay, microcephaly, agenesis of the corpus callosum, epilepsy, and unusual hearing impairment. The younger brother carrying this duplicated region presented with macrocephaly and mild developmental delay in gross motor functions. We speculate that some symptoms have not yet emerged in this child because he is very young or that other symptoms might be very mild.
Table 1.
Features presented in patients with 1q43-q44 deletion and duplication
| Clinical features | Part or all 1q43-q44 deletion [23] | This paper (proband) | Part or all 1q43-q44 duplication [8–14] | This paper (younger brother) |
|---|---|---|---|---|
| Microcephaly | 61/81 | + | 1/8 | – |
| macrocephaly | – | – | 5/8 | + |
| Intellectual disability | 63/81 | + | 7/8 | + |
| Corpus colllosum abnormalities | 48/81 | + | – | – |
| Seizures | 52/81 | + | 4/8 | – |
| Round face | 16/81 | – | – | – |
| Hypertelorism | 14/81 | – | 5/8 | – |
| Prominent forhead | 10/81 | – | 6/8 | – |
| Up or downward palpebral fissures | 18/81 | – | 4/8 | – |
| Long philtrum | 18/81 | – | 3/8 | – |
| Abnormal ear shape | 29/81 | – | 6/8 | – |
| Caradiac abnormalities | 23/81 | – | 4/8 | – |
| Abnormal hand or feet | 25/81 | – | 6/8 | – |
| Micrognathia | 21/81 | – | 3/8 | – |
| Hypotonia | 43/81 | – | 2/8 | – |
“+”: present; “-”: absent
Many patients with 1q43-q44 deletion or duplication including part or all of the regions identified in our patients have been reported. However, the identification of well-detailed genotype-phenotype correlations is hindered by inaccurate mapping of the detailed breakpoints, due to the use of karyotyping or FISH analyses, before the era of high-resolution cytogenetics and the fact that those patients exhibit affected 1q43-q44 regions that differ in size and location. We identified a cryptic chromosomal rearrangement in the mother (II-5) via WGS and confirmed it via FISH. Moreover, we accurately mapped the breakpoints with a combination of a carefully designed data analysis pipeline and Sanger sequencing. The combination of these molecular and cytogenetics techniques characterized the breakpoints at the base-pair level and identified two intron-disrupted genes, EFCAB2 and PRKCH, that have observable clinical phenotypes.
In our study, the two patients with 1q43-q44 deletion or duplication presented congenital anomalies, and the proband exhibited a more serious phenotype than her younger brother. Thus, dosage effects or pathogenic variants of some genes within 1q43-q44 likely contribute to their phenotypes. There are 20 known genes that lie within the 6.0-Mb genomic region, 9 of which are indicated to be disease genes in the Online Mendelian Inheritance in Man (OMIM) database according to NCBI Map Viewer (https://www.ncbi.nlm.nih.gov/mapview). The details of these 9 OMIM disease genes and their clinical characteristics and inheritances are shown in Table 2. We analysed the dominant genes for potential dose-effect phenotypes. The AKT3 gene encodes a serine-threonine kinase belonging to the protein kinase B family that is highly expressed in the brain tissue of humans and rodents [24]. The expression of this gene is significantly decreased in the brain and corpus callosum of AKT3-null mice [25], and some studies in humans and mice have demonstrated that AKT3 plays an important role in controlling the sizes of cells and organs [26, 27]. Boland et al. [5] reported a patient with a 46,XY,t(1;13)(q44;q32) translocation who presented postnatal microcephaly and agenesis of the corpus callosum and demonstrated that AKT3 was a candidate gene for these phenotypes. Another study showed that a critical region comprising CEP170, SDCCAG8 and AKT3 was associated with microcephaly [4]. However, among patients with 1q43-q44 duplication, macrocephaly is observed in patients exhibiting AKT3 gene duplication [8]. The ZBTB18 gene encodes a protein that acts as a transcriptional repressor of key pro-neurogenic genes. Xiang et al. [28] found that conditional knockout of the ZBTB18 gene in the central nervous system resulted in microcephaly, reduced thickness of the cortex, agenesis of the corpus callosum, and cerebellar hypoplasia. Thus, ZBTB18 was proposed as the most likely candidate gene for corpus callosum abnormalities [2, 29]. These studies support pathological roles of AKT3 and ZBTB18 in the 1q43-q44 region. Furthermore, our findings support the notion that ATK3 is a dosage-effect gene that may explain microcephaly or macrocephaly in patients with 1q43-q44 deletion or duplication, including our proband and her younger brother. Some studies have indicated that HNRNPU plays an important role in the regulation of embryonic brain development, and genetic mutation of HNRNPU might cause epileptic encephalopathy and intellectual disability [30–33]. Therefore, the HNRNPU gene may contribute to the seizure phenotypes of patients harbouring 1q43-q44 microdeletions. Furthermore, Bhatti et al. [34] found that homozygosity of 1q43-q44 deletion might cause non-syndromic hearing impairment and deemed a region containing CHLM, OPN3 and MAP1LC3C a new autosomal recessive non-syndromic hearing impairment locus. In the present study, the proband also showed bilateral hearing impairment, but it may have been caused by genes of unknown function or other pathogenic factors. The elder brother presented a similar phenotype to that of the proband, and we cannot rule out the possibility that he might have exhibited the same karyotype as the proband. Analysis of families harbouring translocations via WGS and the associated analysis strategy can help us to gain a better understanding of the relationship between phenotype and karyotype, in addition to providing evidence for genetic and reproductive counselling, which may be especially important for the unaffected mother and sister, who are carriers of the insertional translocation.
Table 2.
OMIM genes deleted or duplicated in our patients, with related phenotypes and model of inheritance
| Gene | OMIM | Phenotype | Inheritance |
|---|---|---|---|
| CHRM3 | 118,494 | Prune belly syndrome | AR |
| FMN2 | 606,373 | Mental retardation, autosomal recessive 47 | AR |
| GREM2 | 608,832 | Tooth agenesis, selective, 9 | AD |
| FH | 136,850 | Fumarase deficiency Leiomyomatosis and renal cell cancer |
AR AD |
| SDCCAG8 | 613,524 | Bardet-Biedl syndrome 16 Senior-Loken syndrome 7 |
AR - |
| AKT3 | 611,223 | Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome 2 | AD |
| ZBTB18 | 608,433 | Mental retardation, autosomal dominant 22 | AD |
| COX20 | 614,698 | Mitochondrial complex IV deficiency | AR |
| HNRNPU | 602,869 | Epileptic encephalopathy, early infantile, 54 | AD |
AD Autosomal dominant, AR Autosomal recessive
Accurate breakpoint mapping not only facilitates the elucidation of the relationship between phenotype and karyotype but also offers insights into the possible mechanisms involved in the generation of balanced translocations. In this study, the molecular characterization of the breakpoints showed that they occurred in homologous regions between two non-homologous chromosomes, in addition to demonstrating the presence of small imbalances around the breakpoint site. These findings suggest that the translocation was likely generated through microhomology-mediated repair (MHMR) of double-strand breaks.
Conclusion
In summary, we report a rare family in which two siblings exhibit pure 1q43-q44 deletion or duplication caused by a maternal balanced insertional translocation. Our study demonstrated that WGS is a powerful tool that allows rapid and accurate mapping of translocation breakpoints at the nucleotide level and could provide useful information for genetic and reproductive counselling for balanced translocation carriers. In addition, the results may help us to better understand detailed karyotype-phenotype correlations, and investigate the possible mechanisms underlying the generation of translocations.
Additional files
Table S1. The Gesell Development Scale results of the patients. (DOCX 16 kb)
Table S2. The list of primers for PCR amplification of chromosomal breakpoint regions. (DOCX 16 kb)
Acknowledgements
We would like to thank the patients and their family members who kindly agreed to participate in this study.
Funding
This study was supported by two grants from the National Natural Science Foundation of China (No. 81771645 and 81471432 to Y.T.).
Availability of data and materials
The authors declare that all relevant data are included in the article and the additional files.
Abbreviations
- CNV
Copy number variation
- FISH
Fluorescence in situ hybridization
- PCR
Polymerase chain reaction
- SNP
Single-nucleotide polymorphism
- WGS
Whole-genome sequencing
Authors’ contributions
LA collected patient data and drafted the initial manuscript. CD designed the primers, performed the FISH experiments and interpreted the results. YS helped draft the initial manuscript. LH performed PCR and Sanger sequencing. DJ provided the instruments and critically reviewed the manuscript. ZY and YC reviewed all laboratory results. LG provided the instruments and approved the final manuscript as submitted. ZW and TY conceived the study, participated in its design and coordination, and helped draft the initial manuscript and write the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Institutional Ethics Committee of the Reproductive and Genetic Hospital of Citic-Xiangya, and written informed consent was obtained from all participants prior to genetic analysis.
Consent for publication
Written informed consent for publication of the images was obtained from the mother of these patients.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13039-018-0371-7) contains supplementary material, which is available to authorized users.
Contributor Information
Aixiang Luo, Email: luoaixiang@csu.edu.cn.
Dehua Cheng, Email: 18631538@qq.com.
Shimin Yuan, Email: 132711015@csu.edu.cn.
Haiyu Li, Email: 166511023@csu.edu.cn.
Juan Du, Email: 1006540087@qq.com.
Yang Zhang, Email: zy0781@hku.hk.
Chuanchun Yang, Email: yangchuanchun@cheerlandgroup.com.
Ge Lin, Email: linggf@hotmail.com.
Wenyong Zhang, Email: zhangwy@sustc.edu.cn.
Yue-Qiu Tan, Phone: 86-731-82355301, Email: tanyueqiu@csu.edu.cn.
References
- 1.Shimojima K, Okamoto N, Suzuki Y, Saito M, Mori M, Yamagata T, et al. Subtelomeric deletions of 1q43q44 and severe brain impairment associated with delayed myelination. J Hum Genet. 2012;57(9):593–600. doi: 10.1038/jhg.2012.77. [DOI] [PubMed] [Google Scholar]
- 2.Ballif BC, Rosenfeld JA, Traylor R, Theisen A, Bader PI, Ladda RL, et al. High-resolution array CGH defines critical regions and candidate genes for microcephaly, abnormalities of the corpus callosum, and seizure phenotypes in patients with microdeletions of 1q43q44. Hum Genet. 2012;131(1):145–156. doi: 10.1007/s00439-011-1073-y. [DOI] [PubMed] [Google Scholar]
- 3.Nagamani SC, Erez A, Bay C, Pettigrew A, Lalani SR, Herman K, et al. Delineation of a deletion region critical for corpus callosal abnormalities in chromosome 1q43-q44. Eur J Hum Genet. 2012;20(2):176–179. doi: 10.1038/ejhg.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill AD, Chang BS, Hill RS, Garraway LA, Bodell A, Sellers WR, et al. A 2-Mb critical region implicated in the microcephaly associated with terminal 1q deletion syndrome. Am J Med Genet A. 2007;143A(15):1692–1698. doi: 10.1002/ajmg.a.31776. [DOI] [PubMed] [Google Scholar]
- 5.Boland E, Clayton-Smith J, Woo VG, McKee S, Manson FD, Medne L, et al. Mapping of deletion and translocation breakpoints in 1q44 implicates the serine/threonine kinase AKT3 in postnatal microcephaly and agenesis of the corpus callosum. Am J Hum Genet. 2007;81(2):292–303. doi: 10.1086/519999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Bon BW, Koolen DA, Borgatti R, Magee A, Garcia-Minaur S, Rooms L, et al. Clinical and molecular characteristics of 1qter microdeletion syndrome: delineating a critical region for corpus callosum agenesis/hypogenesis. J Med Genet. 2008;45(6):346–354. doi: 10.1136/jmg.2007.055830. [DOI] [PubMed] [Google Scholar]
- 7.Gai D, Haan E, Scholar M, Nicholl J, Yu S. Phenotypes of AKT3 deletion: a case report and literature review. Am J Med Genet A. 2015;167A(1):174–179. doi: 10.1002/ajmg.a.36710. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Zeesman S, Tarnopolsky MA, Nowaczyk MJ. Duplication of AKT3 as a cause of macrocephaly in duplication 1q43q44. Am J Med Genet A. 2013;161A(8):2016–2019. doi: 10.1002/ajmg.a.35999. [DOI] [PubMed] [Google Scholar]
- 9.Flatz S, Fonatsch C. Partial trisomy 1q due to tandem duplication. Clin Genet. 1979;15(6):541–542. doi: 10.1111/j.1399-0004.1979.tb00839.x. [DOI] [PubMed] [Google Scholar]
- 10.Clark BJ, Lowther GW, Lee WR. Congenital ocular defects associated with an abnormality of the human chromosome 1: trisomy 1q32-qter. J Pediatr Ophthalmol Strabismus. 1994;31(1):41–45. doi: 10.3928/0191-3913-19940101-09. [DOI] [PubMed] [Google Scholar]
- 11.Duba HC, Erdel M, Loffler J, Bereuther L, Fischer H, Utermann B, et al. Detection of a de novo duplication of 1q32-qter by fluorescence in situ hybridisation in a boy with multiple malformations: further delineation of the trisomy 1q syndrome. J Med Genet. 1997;34(4):309–313. doi: 10.1136/jmg.34.4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cocce MC, Villa O, Obregon MG, Salido M, Barreiro C, Sole F, et al. Duplication dup(1)(q41q44) defined by fluorescence in situ hybridization: delineation of the ‘trisomy 1q42-->qter syndrome’. Cytogenet Genome Res. 2007;118(1):84–86. doi: 10.1159/000106446. [DOI] [PubMed] [Google Scholar]
- 13.Kulikowski LD, Bellucco FT, Nogueira SI, Christofolini DM, Smith Mde A, de Mello CB, et al. Pure duplication 1q41-qter: further delineation of trisomy 1q syndromes. Am J Med Genet A. 2008;146A(20):2663–2667. doi: 10.1002/ajmg.a.32510. [DOI] [PubMed] [Google Scholar]
- 14.Nowaczyk MJ, Bayani J, Freeman V, Watts J, Squire J, Xu J. De novo 1q32q44 duplication and distal 1q trisomy syndrome. Am J Med Genet A. 2003;120A(2):229–233. doi: 10.1002/ajmg.a.20028. [DOI] [PubMed] [Google Scholar]
- 15.Polityko A, Starke H, Rumyantseva N, Claussen U, Liehr T, Raskin S. Three cases with rare interstitial rearrangements of chromosome 1 characterized by multicolor banding. Cytogenet Genome Res. 2005;111(2):171–174. doi: 10.1159/000086388. [DOI] [PubMed] [Google Scholar]
- 16.Aristidou C, Koufaris C, Theodosiou A, Bak M, Mehrjouy MM, Behjati F, et al. Accurate breakpoint mapping in apparently balanced translocation families with discordant phenotypes using whole genome mate-pair sequencing. PLoS One. 2017;12(1):e0169935. doi: 10.1371/journal.pone.0169935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Hemel JO, Eussen HJ. Interchromosomal insertions. Identification of five cases and a review. Hum Genet. 2000;107(5):415–432. doi: 10.1007/s004390000398. [DOI] [PubMed] [Google Scholar]
- 18.Kang SH, Shaw C, Ou Z, Eng PA, Cooper ML, Pursley AN, et al. Insertional translocation detected using FISH confirmation of array-comparative genomic hybridization (aCGH) results. Am J Med Genet A. 2010;152A(5):1111–1126. doi: 10.1002/ajmg.a.33278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nowakowska BA, de Leeuw N, Ruivenkamp CA, Sikkema-Raddatz B, Crolla JA, Thoelen R, et al. Parental insertional balanced translocations are an important cause of apparently de novo CNVs in patients with developmental anomalies. Eur J Hum Genet. 2012;20(2):166–170. doi: 10.1038/ejhg.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng XF, Chen JJ. Evaluation of intellectual development level of 300 children with language disorder. Zhongguo Dang Dai Er Ke Za Zhi. 2016;18(11):1115–1118. doi: 10.7499/j.issn.1008-8830.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan YQ, Tan K, Zhang SP, Gong F, Cheng DH, Xiong B, et al. Single-nucleotide polymorphism microarray-based preimplantation genetic diagnosis is likely to improve the clinical outcome for translocation carriers. Hum Reprod. 2013;28(9):2581–2592. doi: 10.1093/humrep/det271. [DOI] [PubMed] [Google Scholar]
- 22.Dong Z, Jiang L, Yang C, Hu H, Wang X, Chen H, et al. A robust approach for blind detection of balanced chromosomal rearrangements with whole-genome low-coverage sequencing. Hum Mutat. 2014;35(5):625–636. doi: 10.1002/humu.22541. [DOI] [PubMed] [Google Scholar]
- 23.Hemming IA, Forrest AR, Shipman P, Woodward KJ, Walsh P, Ravine DG, et al. Reinforcing the association between distal 1q CNVs and structural brain disorder: a case of a complex 1q43-q44 CNV and a review of the literature. Am J Med Genet B Neuropsychiatr Genet. 2016;171B(3):458–467. doi: 10.1002/ajmg.b.32427. [DOI] [PubMed] [Google Scholar]
- 24.Nakatani K, Sakaue H, Thompson DA, Weigel RJ, Roth RA. Identification of a human Akt3 (protein kinase B gamma) which contains the regulatory serine phosphorylation site. Biochem Biophys Res Commun. 1999;257(3):906–910. doi: 10.1006/bbrc.1999.0559. [DOI] [PubMed] [Google Scholar]
- 25.Easton RM, Cho H, Roovers K, Shineman DW, Mizrahi M, Forman MS, et al. Role for Akt3/protein kinase Bgamma in attainment of normal brain size. Mol Cell Biol. 2005;25(5):1869–1878. doi: 10.1128/MCB.25.5.1869-1878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potter CJ, Pedraza LG, Huang H, Xu T. The tuberous sclerosis complex (TSC) pathway and mechanism of size control. Biochem Soc Trans. 2003;31(Pt 3):584–586. doi: 10.1042/bst0310584. [DOI] [PubMed] [Google Scholar]
- 27.Shioi T, McMullen JR, Kang PM, Douglas PS, Obata T, Franke TF, et al. Akt/protein kinase B promotes organ growth in transgenic mice. Mol Cell Biol. 2002;22(8):2799–2809. doi: 10.1128/MCB.22.8.2799-2809.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang C, Baubet V, Pal S, Holderbaum L, Tatard V, Jiang P, et al. RP58/ZNF238 directly modulates proneurogenic gene levels and is required for neuronal differentiation and brain expansion. Cell Death Differ. 2012;19(4):692–702. doi: 10.1038/cdd.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perlman SJ, Kulkarni S, Manwaring L, Shinawi M. Haploinsufficiency of ZNF238 is associated with corpus callosum abnormalities in 1q44 deletions. Am J Med Genet A. 2013;161A(4):711–716. doi: 10.1002/ajmg.a.35779. [DOI] [PubMed] [Google Scholar]
- 30.Carvill GL, Heavin SB, Yendle SC, McMahon JM, O'Roak BJ, Cook J, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013;45(7):825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Kovel CG, Brilstra EH, van Kempen MJ, Van’t Slot R, Nijman IJ, Afawi Z, et al. Targeted sequencing of 351 candidate genes for epileptic encephalopathy in a large cohort of patients. Mol Genet Genomic Med. 2016;4(5):568–580. doi: 10.1002/mgg3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamdan FF, Srour M, Capo-Chichi JM, Daoud H, Nassif C, Patry L, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10(10):e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roshon MJ, Ruley HE. Hypomorphic mutation in hnRNP U results in post-implantation lethality. Transgenic Res. 2005;14(2):179–192. doi: 10.1007/s11248-004-8147-8. [DOI] [PubMed] [Google Scholar]
- 34.Bhatti A, Lee K, McDonald ML, Hassan MJ, Gutala R, Ansar M, Ahmad W, Leal SM. Mapping of a new autosomal recessive non-syndromic hearing impairment locus (DFNB45) to chromosome 1q43-q44. Clin Genet. 2008;73(4):395–398. doi: 10.1111/j.1399-0004.2008.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The Gesell Development Scale results of the patients. (DOCX 16 kb)
Table S2. The list of primers for PCR amplification of chromosomal breakpoint regions. (DOCX 16 kb)
Data Availability Statement
The authors declare that all relevant data are included in the article and the additional files.


