Abstract
Plasma membrane proteins organize into structures named compartments, microdomains, rafts, phases, crowds, or clusters. These structures are often smaller than 100 nm in diameter. Despite their importance in many cellular functions, little is known about their inner organization. For instance, how densely are molecules packed? Being aware of the protein compaction may contribute to our general understanding of why such structures exist and how they execute their functions. In this study, we have investigated plasma membrane crowds formed by the amyloid precursor protein (APP), a protein well known for its involvement in Alzheimer’s disease. By combining biochemical experiments with conventional and super-resolution stimulated emission depletion microscopy, we quantitatively determined the protein packing density within APP crowds. We found that crowds occurring with reasonable frequency contain between 20 and 30 molecules occupying a spherical area with a diameter between 65 and 85 nm. Additionally, we found the vast majority of plasmalemmal APP residing in these crowds. The model suggests a high molecular density of protein material within plasmalemmal APP crowds. This should affect the protein’s biochemical accessibility and processing by nonpathological α-secretases. As clustering of APP is a prerequisite for endocytic entry into the pathological processing pathway, elucidation of the packing density also provides a deeper understanding of this part of APP’s life cycle.
Introduction
Biological membranes are flat, two-dimensional sheets with a hydrophobic core and a hydrophilic surface. Membrane-populating proteins often feature transmembrane segments (TMSs) that displace lipids from the hydrophobic core and bulky domains that decorate the hydrophilic surface (1). The area occupied by proteins is difficult to determine, and as a consequence numbers are available for only a few systems. For instance, a study employing a strong electric field to drag proteins to one end of a mitochondrial inner membrane surface shows that membrane proteins occupy 40–50% of the total surface area (2). This range is in line with a study presenting a membrane model of a synaptic vesicle, which proposes that the bulky parts of the proteins cover half of the membrane surfaces (3). Moreover, the aforementioned study and a further investigation of red blood cells determined that the protein’s TMSs occupy up to a quarter of the hydrophobic membrane core (4).
Given the large variety of protein types, when random protein distribution occurs, two proteins of the same type must be clearly separated from each other. However, experiments show the contrary, leading to two explanations. Firstly, the plasma membrane is composed of protein-poor and protein-rich areas (5, 6). In one case, protein-rich areas were so dense that they covered the complete membrane surface (7). This phenomenon generates areas into which proteins are preconcentrated. Secondly, within these areas, specific segregation mechanisms bring proteins of one type close together. These structures are known as membrane compartments, microdomains, rafts, phases, crowds, or clusters, and are often smaller than 100 nm in diameter (8, 9, 10, 11). This nanoscale organization is functionally relevant, as a plethora of biological processes depend on the localization, aggregation, clustering, and/or oligomerization of membrane proteins (12, 13, 14, 15, 16).
One aspect not yet fully understood is the protein packing density in such structures. Studying this aspect should be possible by employing super-resolution microscopy techniques like photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy. In fact, these methods have been used to directly count the number of molecules present. To give just one example, studying T cell receptor (TCR) clustering by PALM suggests that a TCR microcluster is composed of 7–30 TCR molecules occupying a circular area with a 35–70 nm radius (17). However, in this study the TCR was visualized by overexpression of CD3 fused to a fluorescent protein, which excludes the nonfluorescent endogenous CD3 from the count. Indeed, employing GFP labeling without downregulation of the endogenous, unlabeled protein is common practice when studying clusters by PALM (for an exception in Escherichia coli, see Greenfield et al. (18)). Additionally, one can never be certain that every protein has been counted due to uncontrollable complications such as misfolding, denaturation, or premature activation and bleaching of the expressed fusion protein (19, 20). These complications lead to a systematic underestimation of the packing density. Moreover, the fusion protein may not behave like the wild-type protein, particularly when the protein being studied is small. In this case, the bulky label may not allow packing as densely as the unlabeled variant. For these reasons, PALM is a suitable technique for studying relative changes in molecule numbers per cluster. However, it is not as useful for investigating the actual number of molecules, which is a prerequisite for elucidating the protein packing density. Stochastic optical reconstruction microscopy, on the other hand, requires fluorescent dyes to be introduced by affinity reagents, such as antibodies. For instance, activated B cell receptor clusters were visualized with directly labeled Fab fragments, revealing 30–120 copies of immunoglobulin D in a spherical radius of 60–80 nm (21). In this approach, three considerations make it difficult to obtain reliable estimates of the number of molecules per cluster. Firstly, after one imaging cycle the fluorophore is typically not bleached and may be resensitized and recounted in a following cycle, potentially resulting in overestimation. Secondly, the affinity reagents that are used often possess a variable amount of dye molecules, making calibration difficult and overestimation even more likely. Finally, affinity reagents generally do not reach every epitope in a densely packed cluster (22, 23, 24). In fact, labeling efficiency should correlate inversely with packing density. Such epitope shielding leads to a systematic underestimation of the molecule number.
To circumvent the challenge of stoichiometric 1:1 labeling and 100% counting efficiency, modeling based on combined imaging and biochemical data has also been applied. It was estimated that the SNARE protein syntaxin 1A is concentrated in small nanoclusters of 60 nm diameter, containing ∼75 molecules (25). This number is close to values from two other studies that determined the syntaxin 1A copy number more directly, either by comparing the intensity of single GFP molecules to the intensity of a cluster (yielding 50–70 molecules per cluster, taking endogenous proteins into account) (19) or by PALM (30–40 molecules per cluster, not counting endogenous proteins) (26).
To date, we have just begun to gather information on the packing density of protein clusters and to understand its biological relevance. Here, we address the question of the molecular packing density of amyloid precursor protein (APP) crowds. APP is involved in Alzheimer’s disease (AD), as its processing releases a neurotoxic peptide. Knowing more about APP crowds would be of interest because it has been speculated that the clustering process may modulate APP processing and endocytosis (27, 28).
APP has a canonical life cycle that can be described as follows. Nascent APP traffics from the Golgi apparatus to the plasma membrane. Here, APP may be processed by plasmalemmal α-secretases that shed the large N-terminal ectodomain. Subsequently, γ-secretase cleaves within the membrane anchoring region of the remaining APP stub, producing a small, harmless peptide that eventually becomes released into the extracellular environment. This cleavage sequence is referred to as the nonamyloidogenic processing of APP (29, 30). However, instead of being processed by α-secretases at the plasma membrane, APP may be endocytosed into acidifying vesicles. Here, β-secretases are activated in the acidic lumen of the endocytic pathway (31, 32). They cleave APP, thereby generating an N-terminal ectodomain that is a few amino acids shorter when compared to the α-secretase cleavage product. As a result, subsequent cleavage by γ-secretase generates a peptide that is a few amino acids longer, namely the AD causative neurotoxic Aβ-peptide (this pathway is referred to as amyloidogenic processing) (33). At present, it remains an enigma how APP can efficiently escape from α-processing to produce enough neurotoxic peptides for the development of AD.
One reason may be that plasmalemmal APP is so densely packed in clusters that the cleavage site is no longer accessible for α-secretases. As an initial step to shed light on this issue, and to learn more about the packing density of protein clusters in general, we set out to study the distribution of APP molecules in the plasma membrane in greater detail. For this purpose, we employed an established model for research on neurodegenerative diseases, the human neuronal SH-SY5Y cell line.
Materials and Methods
Cloning, expression, and purification of His6-APP695
A bacterial expression vector for human APP695 (National Center for Biotechnology Information reference sequence NM_201414) was cloned starting from a construct described previously (28). APP was amplified by PCR using primers introducing N-terminal NdeI and C-terminal BlpI restriction sites flanking the coding sequence of APP for ligation into the multiple cloning site of the pET-15b vector (Novagen, #69661-3; Merck, Kenilworth, NJ). The resulting construct had an N-terminal His6-tag, followed by a thrombin cleavage site and the APP coding sequence. ArcticExpress (DE3)RP competent cells (#230194; Agilent Technologies, Santa Clara, CA) were transformed with the expression vector, plated on agar plates, and the next day a starter culture was inoculated with bacteria from a single colony. The starter culture was incubated overnight at 30°C. The next day, the culture was diluted with lysogeny broth medium to OD600 = 0.1. 2 L lysogeny broth medium were grown at 30°C to OD600 = 0.6. The culture was cooled down to 12°C and protein expression was induced by the addition of 0.25 mM IPTG (#BP1755100; Fisher Scientific, Hampton, NH). After 16 h of incubation at 12°C, the cells were pelleted by centrifugation at 4000 × g for 10 min at 4°C, followed by a wash with precooled phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4, pH 7.4). The pellet was resuspended in ice-cold lysis buffer (50 mM NaH2PO4, 500 mM NaCl, 1% Tween-20, 10 mM imidazole, 15 mM β-mercaptoethanol, 1 mM PMSF, 1 mg/mL lysozyme and protease inhibitors (cOmplete Protease Inhibitor Cocktail, #11697498001; Sigma-Aldrich, St. Louis, MO), pH 8) using 5 mL lysis buffer per gram wet weight. The solution was incubated for 15 min with agitation at 4°C before adding DNase I (#M0303; NEB, Ipswich, MA) and RNase H (#M0297; NEB), followed by incubation for 15 min at 4°C. Then, the solution was sonicated for 1 min on ice at 40% power (100% cycle; using a Bandelin Sonoplus HD2070 (Bandelin Sonoplus, Berlin, Germany) with a MS 73 probe) and then for 5 min at 40% power (10% cycle); after a short pause, the last step was repeated. From the almost clear solution, insoluble material was removed by centrifugation at 10,000 × g for 30 min at 4°C. To isolate the His6-tagged APP695, Ni-NTA beads (50% slurry) (Protino, #745400; Macherey-Nagel, North Rhine-Westphalia, Germany) were washed three times with washing buffer (50 mM NaH2PO4, 500 mM NaCl, 1% Tween20, pH 8), added to the solution (1 mL of beads per 10 mL of cleared lysate), and the mixture was incubated overnight at 4°C with agitation. The beads were harvested by centrifugation at 500 × g for 5 min at 4°C, and the bead pellet was washed four times in an ice-cold washing buffer, containing increasingly higher concentrations of imidazole (20, 30, 40, and 50 mM) with each washing step. Beads in 50 mM imidazole containing washing buffer were transferred into a column. The bound protein was eluted from the beads with 15 mL elution buffer (50 mM NaH2PO4, 500 mM NaCl, 300 mM imidazole, pH 8), collecting 1 mL fractions. The fraction’s protein content was measured using the Pierce BCA Protein Assay kit (#23225; Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions. The three fractions containing the highest protein content were identified and pooled. For dialysis, the solution was transferred into a 10 K molecular weight cutoff Slide-A-Lyzer Dialysis Cassette (#66380; Thermo Fisher Scientific). Dialysis was performed in 500 mL PBS with stirring at 4°C for 8 h, replacing the buffer every 2 h. After supplementing the protein solution with 50% vol glycerol and 0.05% vol sodium azide, it was stored at −20°C. To determine the APP protein concentration, the sample was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (see Fig. S2 for details). Please note that bacterially expressed APP695 is N-terminally His6-tagged and nonglycosylated. Therefore, in Western blot analysis it runs slightly higher than cellular nonglycosylated APP695 (lowest band).
Cultivation, harvesting, and counting of SH-SY5Y cells
SH-SY5Y cells were acquired at passage 26 (#CRL-2266; ATCC, Manassas, VA). They were cultured in DMEM:F12 (#P04-41500; PAN Biotech, Aidenbach, Germany), supplemented with 10% heat-inactivated fetal bovine serum (#S0615; Biochrom AG, Berlin, Germany) and 1% penicillin-streptomycin, at 37°C in a 5% CO2 atmosphere. The medium was replaced every 3–4 days and the cells were passaged by trypsination and subsequent reseeding (usually diluting them 1:4, but never exceeding a dilution of 1:10). Cells were never used past total passage 40 and were regularly tested for mycoplasma contamination (SKU#B50400400; GATC mycoplasma check, Konstanz, Germany). For harvesting, cells were initially washed three times in Dulbecco’s phosphate-buffered saline (DPBS) (#P04-36500; PAN-Biotech) precooled to 4°C. Then, cells were mechanically detached using a cell scraper, followed by collection in ice-cold DPBS. Cells were pelleted, resuspended in 10 mL of precooled DPBS, and quantified without further dilution by counting the cells in four 1 mm2-squares of a Neubauer chamber. Alternatively, they were diluted 1:30 (in duplo) in a cell counting buffer (2% vol fetal bovine serum and 1 mM EDTA in DPBS) and analyzed employing a flow cytometer (Guava easyCyte 5; MilliporeSigma, Burlington, MA), collecting samples with at least 10,000 whole cells.
Next, we determined the protein content of the cell suspensions. To this end, cells were pelleted, resuspended in a radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors (#sc-24948, Santa Cruz, Dallas, TX), precooled to 4°C, and rigorously vortexed. The samples were rotated for 30 min at 4°C, followed by 10 min sonication in an ice-cold sonication bath. After centrifugation at 14,000 × g for 10 min at 4°C, the supernatant was collected and stored at −20°C for further analysis. Protein content was analyzed using the Pierce BCA Protein Assay kit (#23225; Thermo Fisher Scientific) following the manufacturer’s instructions. To obtain more accurate values, each lysate was analyzed three times and values were averaged. For each lysate, the amount of protein per cell was determined by dividing the protein concentration by the cell concentration.
Protein quantification by SDS-PAGE in-gel staining and Western blotting
SDS-PAGE in-gel staining
SDS-PAGE in-gel staining was used to determine the concentration of purified His6-APP695 in relation to a known standard. Nonneutralized 98.4% pure Albumin Fraction V (#2834; Carl Roth, Karlsruhe, Germany) was dissolved in PBS at 200 μg bovine serum albumin (BSA) per mL and a series of 1:2 dilutions was prepared, down to a concentration of 6.25 μg/mL. The BSA dilution series and three dilutions of the purified APP were prepared for SDS-PAGE analysis. The samples were mixed with 4× Lämmli buffer and agitated at 95°C for 10 min. Samples were analyzed using a 10% polyacrylamide running gel with a 4% stacking gel, mounted in a Mini-PROTEAN Tetra Cell (Bio-Rad, Hercules, CA), in SDS running buffer (25 mM Tris, 0.1% w/v SDS, 192 mM glycine, pH 8.3 in ddH2O). When the samples had left the stacking gel, the voltage was raised from initially 70 to 100 V.
After the run, gels were washed three times in ddH2O for 10 min with agitation. Then they were stained overnight either with colloidal Coomassie (0.02% w/v Coomassie Brilliant Blue G-250 (#20279; Thermo Fisher Scientific) in ddH2O, supplemented with 5% w/v Al2(SO4)3∗xH2O (x ∼14−18), 10% vol EtOH, and 2% vol orthophosphoric acid) or with Fast Green (0.001% w/v Fast Green FCF (#F7252; Sigma-Aldrich) in ddH2O, supplemented with 30% vol MeOH and 7% vol acetic acid). The gels were washed three times for 5 min in ddH2O and destained for 1 h with agitation in destaining solution (10% vol EtOH with 2% vol orthophosphoric acid in ddH2O for colloidal Coomassie gels, and 30% vol MeOH with 7% vol acetic acid in ddH2O for Fast Green gels). Finally, the gels were washed again twice for 5 min in ddH2O and imaged using the 700 nm channel of a Li-Cor Odyssey Classic Imaging System (Li-Cor Biotechnology, Lincoln, NE), with settings recommended for imaging of in-gel stainings (resolution: 338.983 μm; quality: highest, focus offset: 0.5, intensity: 5 (at 700 nm)). Single bands were quantified using the “Gel Analyzer” functionality of the ImageJ software. The APP concentration was calculated with reference to a standard curve generated from the band intensities of the BSA dilution series.
Western blotting
To determine the amount of APP per SH-SY5Y cell, we employed Western blot analysis. Cell lysates were calibrated to their cell concentration and a determined number of cells was analyzed together with an APP dilution series (24–3.2 fmol of purified His6-APP695). Preparation of samples and SDS-PAGE were performed as described above.
After the run, the gels were washed once in ddH2O for 5 min and subjected to ice-cold Towbin buffer (25 mM Tris, 192 mM glycine, 20% vol MeOH, pH 8.3 in ddH2O) for 10 min with agitation. Nitrocellulose membranes (0.2 μm pore-size, Roti-NC, #HP40.1; Carl Roth) were also equilibrated in ice-cold Towbin buffer for 30 min. Transfer was performed in a Bio-Rad Mini-PROTEAN Tetra Cell with a Mini Trans-Blot Module in Towbin buffer under constant agitation and cooling at 100 V for 2 h. After protein transfer, the membranes were washed twice with PBS for 5 min with agitation. Then, they were blocked with a 1:1 mixture of PBS and Odyssey Blocking Buffer (#927-40000; Li-Cor, Lincoln, NE) for 1 h with agitation. Membranes were incubated with an anti-APP (C1/6.1) primary antibody (#802801; BioLegend, San Diego, CA) diluted 1:3000 in 5 mL 1:1 PBS:Odyssey Blocking Buffer containing 0.1% Tween-20 for either 2–3 h at room temperature or overnight at 4°C (in both cases with agitation). Next, membranes were washed five times in PBS-T (PBS containing 0.1% Tween-20) for 10 min with agitation. For secondary antibody labeling, membranes were incubated with goat anti-mouse IRDye 800CW (#926-32210; Li-Cor) diluted 1:10,000 in 5 mL 1:1 PBS:Odyssey Blocking Buffer containing 0.1% Tween-20 for 1 h at room temperature with agitation. Finally, membranes were washed three times with PBS-T and twice with PBS for 5 min with agitation. Bands were detected using the 700 and 800 nm channels of a Li-Cor Odyssey Classic Imaging System with recommended settings for imaging of Western blot membranes (resolution: 84.674; quality: medium; offset: 0; intensity: 3 (at 700 nm) and 6–7.5 (at 800 nm)). Single bands were quantified as described above. The APP amount was calculated with reference to a standard curve generated from the band intensities of the His6-APP695 serial dilution. The amount of APP per cell was calculated by dividing the amount of APP in the lysate by the number of cells in the lysate.
Each biological replicate was analyzed at least twice on separate Western blot membranes, and the average was calculated. Moreover, from each Western blot membrane, 2–3 lysate values were obtained, which were averaged.
Protein biotinylation
To identify the plasmalemmal APP fraction, the Pierce Cell Surface Protein Isolation kit (#10230104; Thermo Fisher Scientific) was employed following the manufacturer’s instructions (using the following buffers from the kit: “Quenching Solution,” “Lysis Buffer,” “Wash Buffer” and “Elution Buffer”). Briefly, four T75 cell culture flasks of SH-SY5Y cells grown to 90% confluency were washed twice with DPBS precooled to 4°C. Then, each flask was incubated with 10 mL precooled DPBS supplemented with 250 μg/mL Sulfo-NHS-SS-Biotin. The flasks were gently agitated on a rocking platform for 30 min at 4°C. Afterwards, the reaction was stopped by the addition of 500 μL “Quenching Solution.” The biotinylated cells were mechanically detached using a cell scraper, pelleted by centrifugation (1000 × g for 3 min), washed once with 5 mL precooled TBS, and pelleted again. The cell pellet was resuspended in 566 μL precooled “Lysis Buffer” with protease inhibitors added (Inhibitor Cocktail Plus, #3751.1; Roth). For lysis, the cells were incubated on ice over a period of 15 min, vortexing every 5 min for 5 s. Then another 15 min incubation period on ice followed, this time applying a 1 s sonication burst at 5% power (Bandelin Sonoplus HD2070, with a MS 73 probe) every 5 min. The insoluble material was removed by centrifugation at 10,000 × g for 2 min at 4°C. For isolation of the biotinylated material, 500 μL of NeutrAvidin Agarose beads (50% slurry) were transferred into a spin column. The beads were washed three times by the addition of 500 μL “Wash Buffer,” followed by centrifugation at 1000 × g for 1 min. Then, the lysate was incubated with the NeutrAvidin Agarose beads for at least 1 h at room temperature with end-over-end rotation. Next, the column was centrifuged at 1000 × g for 1 min and the flow-through collected. The column was washed four times as described above, but using “Wash Buffer” supplemented with protease inhibitors (Inhibitor Cocktail Plus, #3751.1; Roth). The flow-throughs from the washes were collected. To elute the bound protein, the column was incubated with 500 μL “Elution Buffer” containing 66.67 mM DTT for 1 h with end-over-end rotation. The eluate was collected by centrifugation at 1000 × g for 1 min. A second elution step was performed overnight to ensure all bound protein was collected. All fractions were stored at −20°C until analysis by Western blotting using the anti-APP (C1/6.1) primary and anti-mouse IRDye 800CW secondary antibodies as described above. The plasmalemmal APP percentage is defined as: 100% x “Eluate”/(“Eluate” + “Flow-through” + “Wash 1”). Each biological replicate was analyzed at least twice on separate Western blots, and the individual values were averaged.
Plasma membrane surface area measurements
SH-SY5Y cells were seeded at low density (5000 cells per cm2) on cover slips coated with poly-L-lysine (#P1524; Sigma-Aldrich) and cultured for 24 h. They were washed twice with ice-cold DPBS supplemented with 1 mM CaCl2 and 1 mM MgCl2 (DPBS++) and fixed in ice-cold 1% paraformaldehyde (PFA) in DPBS++ for 30 min at 4°C. Fixation was stopped by rinsing once with 50 mM NH4Cl in PBS followed by a 20 min incubation step under gentle agitation with 50 mM NH4Cl in PBS. Then cells were washed twice with PBS for 5 min with agitation. The cover slips were mounted in a microscopy chamber containing imaging buffer prepared by diluting a saturated TMA-DPH (#T204; Thermo Fisher Scientific, Invitrogen, Carlsbad, CA) solution 1:10 in PBS. For optical sectioning, a Zeiss AXIO Observer Z1 inverted microscope with an LSM 710 confocal system and a LCI Plan-Neofluar 1.3 NA 63× Imm Corr DIC M27 water objective was employed. The cells were imaged by exciting fluorescence at 405 nm at 10% laser power and detecting fluorescence from 415 to 464 nm, employing a pixel depth of eight-bit, a lateral pixel size of 260 nm, axial sectioning steps of 240 nm, 2× line averaging, and a pinhole size of 45 μm. Initially, the z-image stacks were processed with ImageJ; i.e., single cells were cropped to remove residual signals followed by image smoothing (employing the “Smooth” filter). Using the Imaris three-dimensional (3D) modeling software (34), 3D cell models were initially created from the optical sections without thresholding. As a result, pixel interpolation and creation of voxels occurred. Then, the voxel size was calibrated with reference to the imaging settings (pixel size and axial steps during sectioning, see above). Using the “Surface Creation Tool,” each cell was thresholded manually to find the highest threshold resulting in a surface model in which neurites remain intact and no holes or disruptions in the cell surface occur. From such models the values of the cell surface areas were extracted.
Preparation of membrane sheets
All analyzed membrane sheets were prepared from nonoverexpressing cells, with the exception of the experiment shown in Fig. 3 and Fig. S5, for which APP-GFP was overexpressed. For overexpression, the pcDNA6.2-APP695-emGFP vector (28) was transfected using the Neon Transfection System (Thermo Fisher Scientific, Invitrogen), with 100 μL tips containing 1,000,000 cells mixed with 10 μg DNA. A single pulse of 1100 V with a 50 ms width was applied. The electroporated cells were transferred into cell culture medium without antibiotics, and cells were seeded at a density of 80,000 cells per cm2 by pipetting 500 μL menisci onto poly-L-lysine coated cover slips. Nonoverexpressing cells were seeded using normal cell culture medium at a density of 60,000 cells per cm2. Cells were allowed to settle down for 1 h before the additional normal culture medium with antibiotics was added, followed by an incubation of 21–23 h. For membrane sheet generation, cover slips were washed twice with ice-cold DPBS++ and transferred to the middle of a glass petri dish filled with ice-cold DPBS++. A sonicator tip was positioned directly above the cover slip at a 5 mm distance, and a 100 ms sonication pulse was applied (15% power; Bandelin Sonoplus HD2070, with a MS 73 probe) that removes the upper parts of the cells, leaving behind the intact basal plasma membranes. The plasma membrane sheets were washed once more with ice-cold DPBS++ before being fixed for 30 min at room temperature with ice-cold periodate-lysine-paraformaldehyde (PLP) fixative (10 mM sodium-metaperiodate, 1% w/v PFA, 75 mM L-lysine, pH 7.4 in DPBS++). For Fig. S4, we also fixed with 1 or 4% PFA at 4°C, or with 1% PFA at room temperature. To stop fixation, the cover slips were rinsed once with 50 mM NH4Cl in PBS and then incubated for 20 min with agitation in 50 mM NH4Cl in PBS. The membrane sheets were washed twice with PBS and blocked with 2.5% BSA in PBS for 1 h with agitation. Then, the cover slips were incubated with a 1:200 dilution of the anti-APP (C1/6.1) primary antibody in 1% BSA in PBS overnight at 4°C. For stochastic labeling, the antibody was diluted 1:8000, and was omitted for the background control. The cover slips were washed five times with 0.5% BSA in PBS for 10 min with agitation. Next, the cover slips were incubated with secondary antibody diluted 1:200 in 1% BSA in PBS for 1 h at room temperature. For stimulated emission depletion (STED) microscopy, donkey anti-mouse Alexa Fluor 594 (#A21203; Thermo Fisher Scientific) was used. For epifluorescence microscopy, donkey anti-mouse Alexa Fluor 647 (#A31571; Thermo Fisher Scientific) was used. Afterwards, the cover slips were washed twice with PBS. For epifluorescence microscopy, they were imaged immediately. For STED microscopy, the membranes were counterstained by incubation with 0.5 μM Fast-DiO (#D3898; Thermo Fisher Scientific) in PBS for 10 min with agitation, followed by washing three times in PBS for 5 min and mounting on microscopy slides with ∼15 μL ProLong Gold (#P36930; Thermo Fisher Scientific).
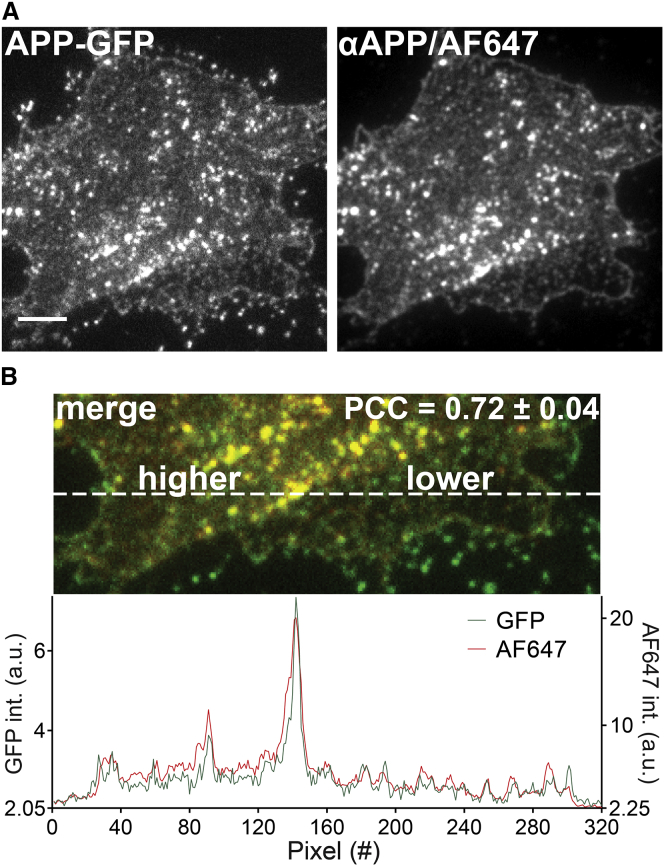
Figure 3.
Overlap between APP antibody staining and overexpressed APP-GFP. (A) On the left, GFP-signal was detected on plasma membranes generated from APP695-GFP overexpressing cells; on the right, the corresponding immunostaining fluorescence obtained by APP antibody staining is shown. Scale bar, 4 μm. (B) A zoomed-in view of an overlay from the bottom parts of the images shown in (A) is shown, with a line scan (dashed white line) illustrating the high similarity between the GFP- and antibody-staining signals. Overlap was quantified by calculation of the Pearson correlation coefficient (PCC). The PCC value is displayed in the image as mean ± SD (n = 3 independent experiments with at least 22 sheets per experiment). To see this figure in color, go online.
Epifluorescence microscopy on membrane sheets
Cover slips with membrane sheets were mounted in a microscopy chamber containing imaging buffer with TMA-DPH (see above). Epifluorescence microscopy was performed using an Olympus IX-81 inverted microscope equipped with an ImagEM C9100-13 16-bit EM CCD camera (Hamamatsu Photonics, Shizuoka, Japan), a MT20E illumination system (Olympus, Tokyo, Japan), an Apochromat NA 1.49 60× oil objective (Olympus), and filter sets (from AHF Analysentechnik) for TMA-DPH (F36-500 DAPI), GFP (F36-525 EGFP), and Alexa Fluor 647 (F46-009 Cy5 ET). Moreover, we recorded with additional 1.6× and 2× magnification lenses, yielding a pixel size of 83.33 nm. The lamp power was set to 23.13% and the acquisition time was 100 ms for both the GFP and Alexa Fluor 647 channels.
Membrane sheets were analyzed using the software ImageJ. Regions of interest (ROI) were placed in the membrane counterstain images assuring unbiased selection of the analyzed areas.
For the analysis of the Pearson correlation coefficient (PCC), the ROI’s positions were laterally aligned in the green and far red channels, and the PCC was quantified using an ImageJ macro. The PCC values were averaged for each experimental day, and the average of all days was calculated for the final PCC.
For Fig. 3, a single line scan was positioned in the merged images and the raw intensity profile was plotted for both channels. For both channels a linear scale for the intensity axes was used, but offset and scaling were adjusted to enable direct comparison.
For each membrane sheet the standard deviation (SD) of pixel intensities in the ROI was divided by the background corrected mean pixel intensity of the ROI, yielding the relative SD. The individual values, collected from 107 membrane sheets recorded from three independent preparations, were then plotted against the average intensity of the respective membrane sheet (Fig. S5 B).
To determine the level of APP overexpression above the endogenous level (Fig. S5 A), overexpressing cells were compared to nonoverexpressing cells by calculating the background corrected mean intensities from ROIs, averaging per condition and experimental day.
For the analysis of local maxima, membrane sheets were imaged with additional 1.6× and 4× magnification lenses, yielding a pixel size of 41.67 nm. The lamp power was set to 57.36%, and from each field of view 10 images were recorded, each with 100 ms acquisition time. The 10 images were averaged using ImageJ before the detection of maxima (for details see below).
STED microscopy
Cover slips with membrane sheets mounted on microscopy slides were imaged with a four-channel super-resolution STED microscope (Abberior Instruments, Goettingen, Germany) at the Life and Medical Sciences imaging facility (University of Bonn). The microscope was equipped with an easy 3D module, single-photon counting avalanche photodiodes, and an Olympus IX83 confocal microscope body (Olympus, Tokyo, Japan) with an UPlanSApo 100× (1.4 NA) objective (Olympus, Tokyo, Japan). For imaging of Fast-DiO, we used a pulsed 485 nm laser at 60 μW power with detection of fluorescence at 500–520 nm. For Alexa Fluor 594, we used a pulsed 561 nm laser at 60 μW power with detection of fluorescence at 580–630 nm. For de-excitation of Alexa Fluor 594, a pulsed 775 nm depletion laser was employed at 700 mW in two-dimensional mode.
For all images, the pixel size was 15 nm and the pinhole size was 60 μm. Confocal images were recorded with time-gated detection with 78.13 ps delay and 8 ns gate width. STED micrographs were recorded with 10 line accumulations and time-gated detection with 1.25 ns delay and 8 ns gate width.
Analysis of maxima using ImageJ
As outlined above, for an unbiased analysis, ROIs were placed in the plasma membrane counterstain images. Turning to the respective APP channel, a background area devoid of any signal was defined within the ROI. Then, single APP maxima were analyzed in the ROIs using a custom ImageJ macro. The macro recognizes and counts maxima based on the “Find Maxima” algorithm. A threshold was chosen to eliminate instrument noise. The threshold was three intensity counts for all STED recordings (the dynamic signal range was roughly from 0 to 200 counts) and 30 intensity counts for all epifluorescence images (here the dynamics ranged roughly from 2150 to 11,000 counts). Once maxima were identified, circular ROIs with a diameter of five pixels were centered on the detected maxima, measuring the maxima’s mean intensities. Maxima intensities were corrected for the value of the background area (see above). Additionally, the intensity profiles of the maxima were determined by line scan analysis. For each maximum, a horizontal and a vertical line scan with a length of 31 pixels for STED microscopy or 15 pixels for epifluorescence microscopy and a width of three pixels (which were averaged) were placed through the maximum. Intensity profiles of both line scans (horizontal and vertical) were fitted with a Gaussian function, choosing the horizontal or the vertical line scan based on fit quality to extract the full width at half maximum (FWHM). Maxima with fits with an R-squared value < 0.90 were excluded from the analysis, which accounted for less than 2% of all maxima in all samples.
For the intensity histograms of maxima in epifluorescence images, maxima obtained from all membrane sheets recorded from three experimental days were pooled. The number of maxima was normalized to the total plasma membrane area, yielding maxima per μm2. The intensity values were grouped into defined bins with a width of 75 intensity counts, and for each bin the maxima per μm2 was determined. Intensity ranges for background were subtracted from the anti-APP antibody-dependent signals (Fig. 4 B). Within the histogram shown in Fig. 4 B, the intensity range of single-labeled structures was estimated by subtracting from the stochastic labeling the background signals (Fig. S8).
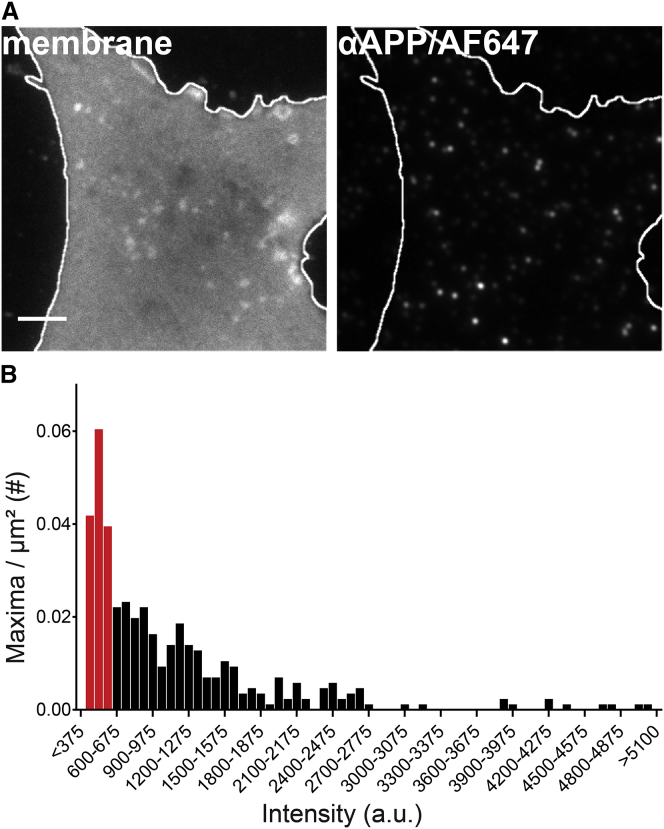
Figure 4.
Imaging of endogenous APP in the plasma membrane. (A) The plasma membrane from a nonoverexpressing cell is shown. The left image shows membrane morphology visualized by TMA-DPH staining. The right image shows APP staining from which an algorithm identified local maxima with very high sensitivity. Scale bar, 2 μm. (B) The histogram shows the intensity distribution of individual APP maxima obtained after background signal subtraction. From the intensity range of maxima arising from labeling by a single primary antibody, we estimate that 34% of the maxima represent noncrowded APP molecules (red bars). a.u., arbitrary units. To see this figure in color, go online.
Additionally, for epifluorescence and STED microscopy, the number of maxima per μm2 and the average intensity of the maxima was averaged per experimental day, from which the mean ± SD was calculated.
For the STED microscopy experiment, the values of the FWHM were used for a histogram showing the size distribution of crowds. Because the dynamic intensity range in STED micrographs is very small, the intensity range of single-labeled structures cannot be resolved. For this reason, after correcting for the background signal, the fraction comprising single-labeled structures was extracted from the epifluorescence microscopy experiment.
Molecular model of an APP crowd
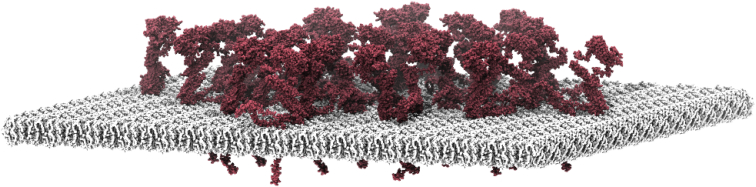
The model of full-length APP was generated by linking the following APP fragment structures (listed from N- to C-terminus) available in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB): 35XC (35) (chain B, 2.1 Å x-ray structure) (36), 2LOH (chain A, NMR model 1) (37), 1IYT (NMR model 1) (38), 3NYL (2.8 Å x-ray structure) (39), 1AAP (chain A, 1.5 Å x-ray structure) (40), 3KTM (chain A, 2.7 Å x-ray structure) (41). Gaps between the folded fragments were modeled by adding the nonresolved regions in a random coil conformation using PyMOL 1.7 (42). Two additional amino acids on each modeled linker were used to apply PyMOL’s iterative structural fitting routine based on short sequence overlaps (these additional amino acids were removed after the fit). The overall orientation of the protein structure is based on a POPC membrane-inserted model of the combined fragments of 2LOH and 1IYT, which was automatically rotated according to its hydrophobic belt using the program LAMBADA (43). Based on the modeled full-length structure, 27 proteins were placed on a POPC lipid bilayer patch (44) using the GROMACS 5.0.1 molecular modeling package (45). In order to mimic the 77 nm large protein arrangement in a model, the protein structure was centered in a box with the dimensions 12.3 × 12.3 nm along the membrane plane (xy). Seven individual layers i with a different number n of copies of the protein along the x-vector were generated (ni = 2, 4, 5, 7, 5, 3, 1). Each APP structure was randomly rotated by an angle of maximally ±10° around the respective vectors of the membrane plane (x, y) and maximally ±360° around a vector parallel to the membrane normal vector (z). The approximately circular arrangement of proteins was achieved by combining and shifting the individual layers along both membrane plane vectors (x, y). The POPC model bilayer patch was enlarged by copies along the vectors x and y to a dimension of 90.8 × 90.8 nm and shifted along the membrane-normal z axis according to the hydrophobic belt of the transmembrane regions of the APP models. VMD 1.9.2 was used for visualization (46).
Results
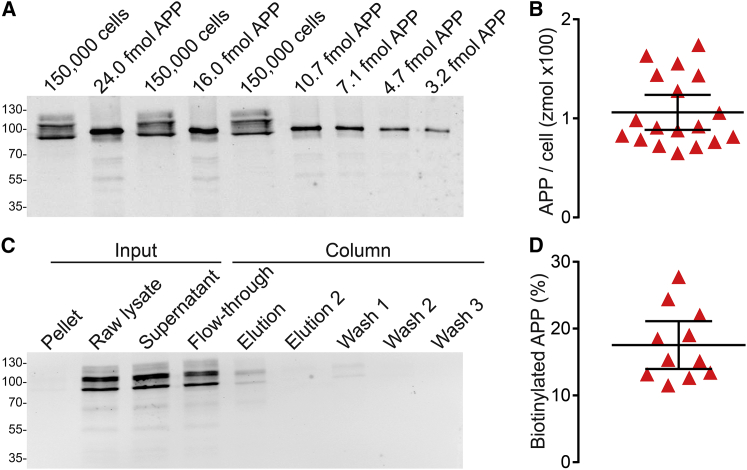
At first, we wished to clarify what the surface concentration of APP in the plasma membrane might be. We began by estimating the copy number of APP molecules in SH-SY5Y whole-cell lysates by quantitative Western blotting. SH-SY5Y cells were nonenzymatically harvested to avoid proteolysis of extracellular protein domains, which would affect the large ectodomain of plasmalemmal APP. The harvested cells were counted by two different methods, using either a hemocytometer or an automated cell counter based on flow cytometry. Cells were lysed in a radioimmunoprecipitation assay buffer, followed by a standardized bicinchoninic acid assay to determine the protein concentration. Independent of the counting method, the lysate’s protein content linearly correlated with the counted number of cells over a wide range (Fig. S1 A). The amount of total protein per SH-SY5Y cell was 76 pg (Fig. S1 B), providing a reference for calculating the number of loaded cells in Western blot analysis from a lysate’s protein concentration. Together with SH-SY5Y cell lysate, we loaded an APP protein standard using dilutions of recombinant human APP695 (Fig. S2). On the membranes, APP was detected by a widely used antibody that recognizes APP’s C-terminal domain. The C-terminal domain is distal to all of the glycosylation sites and is identical in all APP isoforms and the APP protein standard. Hence, the antibody detects all isoforms of APP at all maturation stages. Bands are expected to be detected for immature and/or mature APP695, APP751 and APP770. The band pattern we observed (Fig. 1 A) was in line with previously published data (47, 48, 49). The antibody did not detect any prominent bands that would indicate APP cleavage products (Fig. 1 A and Fig. S3, A and C). This concurs with previous reports suggesting that the sequential processing of APP is fast, meaning that intermediate products reside in the cell membrane only very briefly (50). Including all detected isoforms for quantification, we estimate that an average cell accommodates 106.1 zmol or ∼64,000 APP molecules (Fig. 1 B). The APP copy number per cell was comparable over the range of cell passages used in these experiments (population doubling level was at most 16; Fig. S3 B) and was thus unaffected by cellular aging. This seems to be different in a human fibroblast cell line where after roughly 20 population doublings the APP level was decreased (51).
Figure 1.
Copy number of APP molecules on the cell surface. (A and B) APP molecules per cell. (A) Western blot analysis of a His6-APP695 dilution series together with technical triplicates of a SH-SY5Y cell lysate are shown. The two major bands in the lysate correspond to the isoforms APP695 and APP751/770; weaker bands are differentially glycosylated APP species. For the entire blot see Fig. S3A. (B) Using the APP standard as reference, the copy number of APP molecules per SH-SY5Y cell was estimated to be 106.1 zmol. Error bars indicate the 95% confidence interval (n = 18 biological replicates). (C and D) Quantification of the plasmalemmal APP fraction. (C) Western blot analysis of the fractions of the cell surface biotinylation experiment is shown. Cells were biotinylated at 4°C for 30 min, lysed (“Raw lysate”), and the lysate was centrifuged, yielding “Pellet” and “Supernatant”. The biotinylated material in the “Supernatant” was bound to NeutrAvidin Agarose, collecting the “Flow-through,” followed by four washing steps (“Wash 1,” “Wash 2,” and “Wash 3”; the last wash is not shown) and two elution steps with DTT (“Elution” + “Elution 2”). During the experimental procedure, virtually no APP was lost (Fig. S3D; for the entire blot see Fig. S3C). (D) The percentage of plasmalemmal APP was 100% × “Elution”/(“Elution” + “Flow-through” + “Wash 1”), yielding 17.54%. Error bars indicate the 95% confidence interval (n = 11 biological replicates). To see this figure in color, go online.
In steady state, the bulk of APP is not localized to the plasma membrane (52). Hence, the next step required is to determine the fraction of plasmalemmal APP. APP is a single-pass transmembrane protein with more than 90% of its amino acids exposed to the extracellular milieu. This makes it ideally suited for extracellular biotinylation to elucidate the fraction of APP protein residing in the plasma membrane. Cells were biotinylated on ice and lysed, followed by separation of biotinylated- and nonbiotinylated proteins. Using Western blot analysis, the percentage of biotinylated APP was determined (Fig. 1 C). The plasmalemmal fraction constituted 17.5% (Fig. 1 D), indicating that from a total of 106.1 zmol per cell, roughly 11,180 molecules populate the cell membrane.
The next parameter required to obtain the cell surface concentration of APP was the average plasma membrane surface area of a SH-SY5Y cell. To this end, cells were fixed with ice-cold 1% PFA, a concentration that prevents blebbing and shrinking artifacts during the fixation process (53). Cellular membranes were visualized using TMA-DPH. This dye only fluoresces when it is integrated in a lipid membrane (54) and its insertion is reversible. Thus, imaging can be carried out with nonfluorescent TMA-DPH in the imaging buffer and bleached dye molecules are replenished by unbleached ones during the imaging procedure. This ensures a relatively constant labeling intensity over the entire procedure, which involves recording up to 51 images per stack. It should be noted that TMA-DPH is not plasma membrane specific, but increases its fluorescence in the presence of cholesterol, which is found in the plasma membrane (55). However, the vast majority of membranous material is located inside of the cell, meaning that in the images the cell periphery is defined by the weaker fluorescent signals (Fig. 2 A).
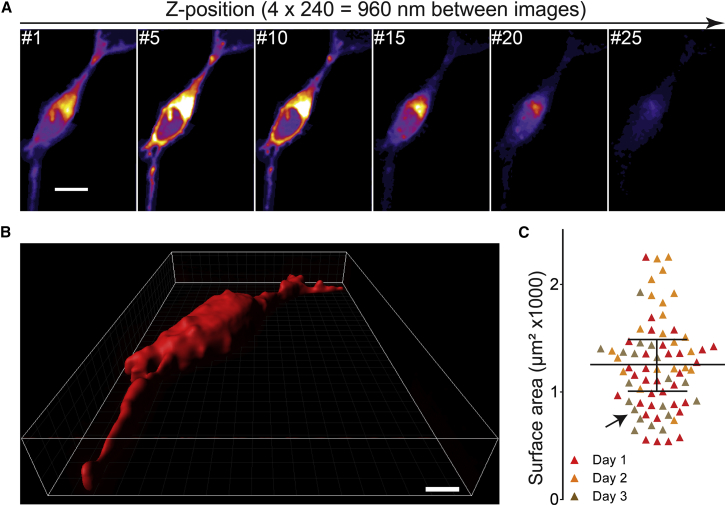
Figure 2.
SH-SY5Y cell surface area. (A) Optical sections of a fixed cell visualized by TMA-DPH are shown. For clarity, only every fifth image of a stack of 28 images is shown. Scale bar, 10 μm. (B) 3D cell morphology reconstructed from the entire image stack. Scale bar, 5 μm. (C) From cells reconstructed as illustrated in (B), we determined the average imaged cell surface area to be 1256 ± 254 μm2 (value is given as mean ± SD; n = 3 independent experimental days with at least 19 cells per experiment). The black arrow in (C) marks the cell shown in (B). To see this figure in color, go online.
After fluorescent labeling, optical sections from the cells were taken by confocal microscopy (Fig. 2 A). These sections were digitally aligned, stacked, thresholded manually for the cell periphery, and interpolated to acquire 3D models of the cell morphology (Fig. 2 B). The surface area could be obtained directly from these models. We found that on average the reconstructed cell surface area was 1256 μm2 (Fig. 2 C). This value is potentially an overestimate as light effects arising from strongly labeled intracellular parts tend to increase the apparent cell size in the images slightly.
We refer to the microscopically measured membrane areas as the “imaged” areas, since the methodology generates a flat projection of the real membrane area, which may be larger where it is curved or ruffled. 11,180 molecules on a surface of 1256 μm2 yield an average plasma membrane surface concentration of 9 APP molecules per imaged μm2. Please note that the error produced by ignoring membrane curvature (x = ignored membrane area in μm2) cancels out later when the number of molecules per crowd is calculated, because the molecule density given in per μm2 is divided by the crowd density in per μm2, and both times the unit is actually per (μm2 + x).
Having found the average surface concentration, we proceeded to study the structures in which these proteins appear. Fluorescence microscopy is the standard technique for studying the distribution of membrane proteins. Visualization can be achieved by targeting the protein of interest with affinity reagents (such as antibodies or derivatives thereof, aptamers, or protein-specific ligands) or genetically by overexpressing the protein fused to a fluorescent protein. However, overexpression of fusion constructs excludes the detection of the endogenous proteins. Furthermore, it may alter the behavior of the protein (e.g., resulting in less efficient clustering). As we are specifically interested in the unperturbed situation and need to correlate microscopic to biochemical data with as few additional corrections as possible (like normalizing for transfection efficiency), overexpression of a fluorescent APP fusion construct is not ideal. Thus, we opted to visualize our protein of interest via conventional primary-secondary antibody labeling, using the same primary antibody as for the quantitative Western blotting experiments. To evaluate whether the antibody immunostaining correctly reflects the distribution of APP molecules, we compared the antibody staining pattern to that of a C-terminally GFP-labeled APP695 fusion protein. If the antibody efficiently and specifically recognizes APP molecules, the patterns of the GFP and the immunostaining signals should look similar. For these experiments we performed staining and imaging on plasma membrane sheets that do not require the use of detergent and allow for a better signal-to-noise ratio during imaging. Moreover, to avoid postfixation movement of molecules, which for some molecules might still occur after standard 4% paraformaldehyde fixation (56), we used PLP fixative with a concentration of 1% paraformaldehyde. The periodate in the PLP fixative oxidizes carbohydrates and/or glycoproteins that interact with polymeric complexes of lysine and formaldehyde (57, 58). The lysine cross-links may not only immobilize APP, which is in part glycosylated, but also indirectly reduce APP mobility by cross-linking other plasma membrane glycoproteins. When compared to paraformaldehyde fixation alone, more intensity maxima were detected by immunostaining after PLP fixation, presumably because of reduced antibody-induced protein patching (Fig. S4).
Fig. 3 A shows both the APP-GFP fluorescence and the signal arising from its antibody labeling. As a measure of the similarity between the two signal distributions, we used the PCC and obtained a value of 0.72 (Fig. 3 B). Considering the technical limitations of these experiments, this PCC is very high and suggests almost complete conformity between the signals (please note that previously for a GFP and a myc double-tagged protein a lower value of 0.63 was obtained (25)). The strong similarity between these two signals can be appreciated when viewing the two merged channels or when comparing the two signal traces measured by a line scan (Fig. 3 B). In conclusion, the antibody is very well suited for the detection and visualization of APP.
It should be noted that the levels of overexpressed APP695-GFP exceeded the amount of endogenous APP on average by 27-fold (Fig. S5 A). Independent from the expression level, staining was always spotty with a strong variability in the intensities of single spots in both the GFP and the antibody channels. In particular at high expression levels spots appear on top of a uniform background signal that gets brighter with increasing expression levels (Fig. S5 B). This can also be seen in the line scan signal traces in Fig. 3 B, crossing two distinct membrane areas. In the left area, APP-GFP is present at a higher level and the aforementioned uniform background signal is readily visible, in contrast to the right area with less APP in which the uniform background is less prominent. This suggests that the weakest spots in the lower right membrane area may be single APP molecules (or very small entities such as dimers) only resolvable at low expression levels.
Hence, under nonoverexpression conditions, at a molecular density of 9 molecules per μm2 (see above) and a fraction of these nine molecules in crowds, the density of noncrowded molecules is so low that they can be resolved as single objects, even at diffraction limited resolution. However, at higher expression levels, noncrowded molecules are more abundant, come closer to each other, and become unresolvable, forming a faint uniform fluorescent background signal. Conversely, this means that the brighter spots must represent several APP molecules irresolvably close to each other (i.e., crowded APP).
Additionally, because of the variability of the intensities of these protein spots, it can be concluded that the number of molecules in such a protein crowd spans a considerable range.
Next, we performed a detailed analysis of the APP signals on plasma membranes from nonoverexpressing cells. PLP fixed membrane sheets were labeled for APP and imaged. To reduce random noise, we took 10 images in quick succession that were averaged to one image (Fig. 4). For analysis we employed an ImageJ algorithm that, after removing residual instrument noise by thresholding, identifies maxima and determines their intensity (for maxima distribution see Fig. S6 A). Aside from the standard immunostaining (“αAPP”), we prepared two additional samples. Firstly, to characterize signals that are produced by the secondary antibody alone (the condition “background”; through unspecific binding to the sample or the glass cover slip), we performed the same staining in the absence of primary antibody. This revealed surprisingly many (Figs. S6 C and S7 A), yet significantly dimmer maxima (Fig. S7 B). In the following text, these signals are referred to as background maxima or background signals. Secondly, we investigated the intensity of maxima arising from a single APP antibody. To this end, we labeled APP with highly diluted primary antibody. Though a crowd provides many epitopes for antibody binding, in most cases only one antibody molecule will bind to it. The subset of labeled epitopes is chosen randomly; therefore, we refer to this condition as “stochastic labeling” of epitopes (Fig. S6 B). As expected, these maxima were dimmer than in normally stained samples but brighter when compared to the condition omitting the primary antibody.
Therefore, we have three types of intensity maxima in the imaging data. Firstly, some instrument noise. Secondly, there are primary antibody-independent background maxima. Thirdly, we have the APP antibody-dependent signals over a broad intensity range.
Fig. 4 shows the intensity histogram corrected for the APP antibody-independent maxima. Moreover, subtracting the background maxima from the stochastic labeling, we identify an intensity range between 300 and 600 counts (Fig. S8; Fig. 4 B, red bins in the histogram) for singly labeled APP moieties. This range in Fig. 4 B comprises 34% of all maxima.
Some of the detected maxima may comprise several nonresolved maxima, though we assume that this number is very low because of the very low signal density in general. However, the diffraction limited resolution, here in the range of 250 nm, will also increase the size of the objects. The accuracy of the size measurement is critical as the area covered by a crowd is, next to the number of molecules populating it, one of the two key parameters in determining its packing density. Thus, to address both the number of maxima per surface area and the size of said maxima properly, we turned to higher resolving super-resolution imaging and employed pulsed STED microscopy with time-gated detection (Fig. 5 A). Images were analyzed by applying the same algorithm and methodology for background correction as for epifluorescence microscopy. After subtraction of the background maxima, a value of 0.5 APP maxima per imaged μm2 was obtained (Fig. 5 B).
Figure 5.
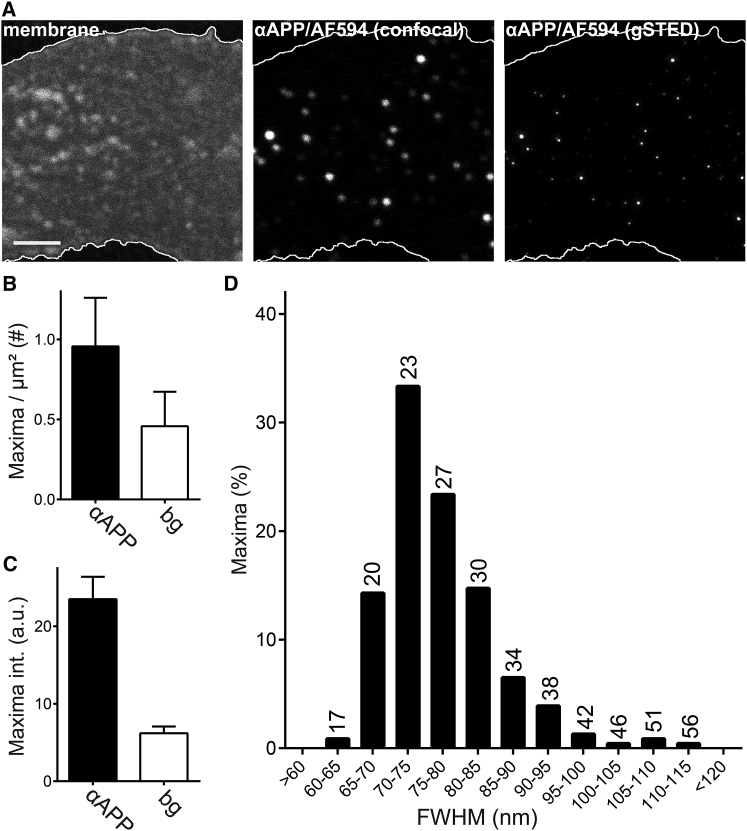
Density and size of APP crowds. (A) Antibody labeled APP structures in membrane sheets imaged with super-resolution microscopy are shown. On the left, a Fast-DiO stain for illustrating the membrane is shown. In the middle and on the right, APP imaged at confocal and STED resolution are shown, respectively. Scale bar, 2 μm. (B) Maxima per μm2 in the presence (“αAPP”) and absence (“bg”) of primary antibody are shown. Subtraction of the background density yields 0.5 APP maxima per imaged μm2. (C) Background maxima were on average fourfold dimmer than αAPP maxima (which also include background maxima). Values are given as means ± SD (n = 3 independent experiments with at least 20 membrane sheets per experiment). (D) For each experimental day, the αAPP maxima were corrected for the density of the background signal by eliminating a respective fraction of dimmest maxima. The background corrected maxima were pooled and corrected for the signal fraction expected for noncrowded APP (see Fig. 4B), subtracting the dimmest 34% of the maxima. Based on the FWHM of these maxima, the size distribution of crowded APP is shown. The average size is 77 ± 8 nm (mean ± SD; n = 231). The numbers above the bins give a speculative estimate on the molecules per crowd in the respective size ranges. To obtain these numbers, with reference to the average crowd number per cell (surface area × crowd density = 414 crowds), we calculated how many crowds of each size were present. Assuming spherical shape and a uniform distribution of molecules within a crowd, we calculated the surface area occupied by crowds for each size range and from this what fraction of molecules they would contain. For each bin, dividing the molecule fractions by the number of crowds yielded the number of molecules per crowd. a.u., arbitrary units; bg, background.
When background intensity was not corrected for, ∼70% more maxima were detected in epifluorescence images than in STED images. This suggests that epifluorescence microscopy, as expected, records with a higher sensitivity. After background correction, ∼20% more maxima were found in STED images than in epifluorescence images, probably due to the higher resolution of STED microscopy. In any case, the number of background corrected, APP antibody-dependent maxima quantified by the two different imaging techniques was remarkably similar.
The density of 0.5 maxima per imaged μm2, and the fact that 34% of the maxima are from noncrowded APP (see above), indicates that 0.17 maxima per μm2 are not from APP crowds. Therefore, 0.17 molecules/μm2 × 1256 μm2/cell surface = 214 molecules/cell surface are not in crowds (which equates to ∼2%) or conversely, ∼98% are present in crowds. Moreover, the average is 27 APP molecules per crowd (for details see Fig. S9). The number of molecules per APP crowd is not very sensitive to the estimate of free molecules (see Fig. S9). Additionally, should APP always be present as dimers, as has been suggested (59, 60, 61), the total fraction of crowded APP decreases only marginally to ∼96%. This is associated with a decrease of 0.5 molecules per crowd.
To determine the size distribution of the APP crowds, initially background maxima and the estimated noncrowded maxima fraction were eliminated (see legend of Fig. 5). For the remaining maxima the FWHM was plotted as a histogram (Fig. 5 D). As can be seen in the size distribution of the crowds, most of them are between 65 and 85 nm, although a few significantly larger entities were also present (Fig. 5 D). From these maxima, an average crowd size of 77 nm in diameter was calculated. Assuming an average of 27 molecules per crowd and equal spacing between all molecules means that every APP TMS is roughly 15 nm away from its nearest neighbor (Fig. 6). At the height of the extracellular bulky domains, the space between molecules must be much smaller.
Figure 6.
Molecular crowding in the APP crowds. To exemplify the packing density in an average APP crowd, we used a coarse-grain molecular dynamics model from APP and placed 27 full-length APP molecules evenly spaced in a circular lipid bilayer patch of 77 nm in diameter. Full-length APP695 was modeled based on various available x-ray and NMR structures (see Materials and Methods for details). The upper part of the image corresponds to the extracellular side. To see this figure in color, go online.
Using the size distribution in the histogram (Fig. 5 D), and assuming that molecule density is independent from the crowd size, the smallest APP crowds occurring with frequency contain 20 molecules, the largest 30 molecules, and crowds with 60 or more molecules are not detected. This is a relatively narrow range.
Discussion
Here we describe the anatomy of APP crowds in the plasma membrane of SH-SY5Y cells. SH-SY5Y cells are human cells with neuron-like characteristics and a widely used model for studying APP processing.
Roughly 20–30 APP molecules concentrate in a circular area with a diameter of 65–85 nm. For an average crowd, this yields a packing density suggesting mean distances between molecules in the nanometer range (see Fig. 6). However, for the following reasons we believe that molecular crowding is even denser and that direct contacts between APP molecules also occur, perhaps yielding very tight APP clusters. Firstly, the cell surface is likely an overestimate, whereas the cell surface molecule number is likely an underestimate, because not all molecules may become biotinylated. In addition, the average diameter of ∼75 nm is probably an overestimate, as we had to rely on sequential primary and/or secondary antibody labeling. On these spatial scales, the size of antibodies is no longer negligible and tends to increase the size of the labeled structure (62). Each of these uncertainties contributes to the underestimation of the packing density. Secondly, APP has been reported to form dimers or to cluster via a small region within the Aβ domain (28, 59, 60, 61). These interactions would most likely be retained in the crowd. Thirdly, other membrane-associated proteins, such as the amyloid precursor-like proteins (52), the α2A-adrenergic receptor (63) and several members of the low-density lipoprotein receptor-related proteins (64) have been shown to bind to or colocalize with APP. These proteins are likely filling empty space in the crowd. Finally, it has been reported that sections directly adjacent to the TMS form loops and may even be embedded in the plasma membrane (65). Such a conformational arrangement compresses the protein backbone in the axial dimension, which in turn increases the area occupied near the membrane.
Altogether, the data suggest that the APP crowd is a relatively large and dense structure, occupying a volume that nearly matches that of a small organelle, such as a synaptic vesicle.
What might the biological relevance of APP crowding be? At present, several ideas on the general role of protein clustering are discussed (12, 13, 14, 15, 16). The most obvious purpose of a crowd is to provide a local concentration of components for augmented activity (66, 67). On the contrary, shielding of active protein domains has been also suggested (12, 68, 69).
With respect to APP crowding, it has previously been shown that APP aggregation is a prerequisite for endocytosis and amyloidogenic processing (27, 28). More efficient crowding promotes internalization and consequently diminishes the possibility of APP processing by α-secretases. Our model proposes that almost all APP is crowded, possibly prepared for endocytosis. Hence, APP crowding contributes to the efficiency of internalization and thereby indirectly drives amyloidogenic processing. However, efficient endocytosis may not be the only reason why crowding favors the amyloidogenic pathway. Tight crowding should also decrease the accessibility for α-secretases significantly.
As crowding could influence APP processing by two different mechanisms, it is tempting to speculate that the efficiency of APP crowding in the plasma membrane is a key regulatory step in preventing APP from being processed by α-secretases. As has been pointed out, the APP clustering mechanism opens a new avenue for fighting AD, such as dissolving clusters by drugs applied from the extracellular environment, which should direct APP toward the nonamyloidogenic processing pathway (28).
The notion of crowding as a key regulatory step is also supported by genetic studies. Many mutations that cause familiar AD (70, 71, 72, 73, 74), as well as the only mutation known to protect against AD (75, 76), are located exactly in the region required for APP clustering (28). However, at present it is unclear whether these mutations affect crowding, processing, or trafficking of the molecule.
The plasmalemmal concentration of endogenous APP, 9 molecules per imaged μm2, is very low when compared to other proteins for which cell membrane concentrations are known (2000 and 7500 molecules per μm2 have been reported for syntaxin 1A and SNAP25, respectively (77)). However, it should also be noted that SNAREs are abundant proteins, and, for example, signaling proteins and receptors presumably are present at much lower concentrations (78, 79). We believe that this low endogenous concentration, as opposed to highly efficient overexpression of APP, explains why we observe a 27-fold increase of APP levels upon overexpression. This could affect the interpretation of studies on APP processing in which overexpression is employed, because at these concentrations APP tends to be less crowded and might be less efficiently endocytosed. Moreover, such high concentrations may saturate the binding sites of α-secretases in the plasma membrane, altering the kinetics of cleavage.
In conclusion, we suggest that the APP crowding is an important aspect for understanding the life cycle of APP and should be considered when studying the mechanisms involved in APP processing and the development of AD.
Author Contributions
T.L. was responsible for the planning, conceptual layout, and supervision of the project. D.d.C. was responsible for carrying out the experiments and collecting and presenting the data. T.H.S. generated the structural molecular model. J.-G.S. developed the ImageJ macro. T.L. and D.d.C. took equal parts in writing the work. All authors were involved in reviewing and editing of the manuscript.
Acknowledgments
We thank Caroline Sayers and Dr. Michael Pankratz (University of Bonn) for language editing of the manuscript.
This project was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB645 to T.L.).
Editor: Andreas Engel.
Footnotes
Nine figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30106-1.
Supporting Material
References
- 1.Engelman D.M. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 2.Sowers A.E., Hackenbrock C.R. Rate of lateral diffusion of intramembrane particles: measurement by electrophoretic displacement and rerandomization. Proc. Natl. Acad. Sci. USA. 1981;78:6246–6250. doi: 10.1073/pnas.78.10.6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takamori S., Holt M., Jahn R. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 4.Dupuy A.D., Engelman D.M. Protein area occupancy at the center of the red blood cell membrane. Proc. Natl. Acad. Sci. USA. 2008;105:2848–2852. doi: 10.1073/pnas.0712379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lillemeier B.F., Pfeiffer J.R., Davis M.M. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. USA. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saka S.K., Honigmann A., Rizzoli S.O. Multi-protein assemblies underlie the mesoscale organization of the plasma membrane. Nat. Commun. 2014;5:4509. doi: 10.1038/ncomms5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao W., Tian Y., Wang H. Studying the nucleated mammalian cell membrane by single molecule approaches. PLoS One. 2014;9:e91595. doi: 10.1371/journal.pone.0091595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 9.Kusumi A., Suzuki K.G.N., Fujiwara T.K. Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem. Sci. 2011;36:604–615. doi: 10.1016/j.tibs.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Nicolson G.L. The fluid-mosaic model of membrane structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim. Biophys. Acta. 2014;1838:1451–1466. doi: 10.1016/j.bbamem.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 11.Goyette J., Gaus K. Mechanisms of protein nanoscale clustering. Curr. Opin. Cell Biol. 2017;44:86–92. doi: 10.1016/j.ceb.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Lang T., Rizzoli S.O. Membrane protein clusters at nanoscale resolution: more than pretty pictures. Physiology (Bethesda) 2010;25:116–124. doi: 10.1152/physiol.00044.2009. [DOI] [PubMed] [Google Scholar]
- 13.Cebecauer M., Spitaler M., Magee A.I. Signalling complexes and clusters: functional advantages and methodological hurdles. J. Cell Sci. 2010;123:309–320. doi: 10.1242/jcs.061739. [DOI] [PubMed] [Google Scholar]
- 14.Matthews J.M., Sunde M. Dimers, oligomers, everywhere. Adv. Exp. Med. Biol. 2012;747:1–18. doi: 10.1007/978-1-4614-3229-6_1. [DOI] [PubMed] [Google Scholar]
- 15.Nussinov R. The spatial structure of cell signaling systems. Phys. Biol. 2013;10:045004. doi: 10.1088/1478-3975/10/4/045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Parajo M.F., Cambi A., Jacobson K. Nanoclustering as a dominant feature of plasma membrane organization. J. Cell Sci. 2014;127:4995–5005. doi: 10.1242/jcs.146340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lillemeier B.F., Mörtelmaier M.A., Davis M.M. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenfield D., McEvoy A.L., Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knowles M.K., Barg S., Almers W. Single secretory granules of live cells recruit syntaxin-1 and synaptosomal associated protein 25 (SNAP-25) in large copy numbers. Proc. Natl. Acad. Sci. USA. 2010;107:20810–20815. doi: 10.1073/pnas.1014840107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costantini L.M., Baloban M., Snapp E.L. A palette of fluorescent proteins optimized for diverse cellular environments. Nat. Commun. 2015;6:7670. doi: 10.1038/ncomms8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattila P.K., Feest C., Batista F.D. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 2013;38:461–474. doi: 10.1016/j.immuni.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 22.Zilly F.E., Halemani N.D., Lang T. Ca2+ induces clustering of membrane proteins in the plasma membrane via electrostatic interactions. EMBO J. 2011;30:1209–1220. doi: 10.1038/emboj.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merklinger E., Schloetel J.-G., Lang T. The packing density of a supramolecular membrane protein cluster is controlled by cytoplasmic interactions. Elife. 2017;6:e20705. doi: 10.7554/eLife.20705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batoulis H., Schmidt T.H., Lang T. Concentration dependent ion-protein interaction patterns underlying protein oligomerization behaviours. Sci. Rep. 2016;6:24131. doi: 10.1038/srep24131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sieber J.J., Willig K.I., Lang T. The SNARE motif is essential for the formation of syntaxin clusters in the plasma membrane. Biophys. J. 2006;90:2843–2851. doi: 10.1529/biophysj.105.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rickman C., Medine C.N., Duncan R.R. t-SNARE protein conformations patterned by the lipid microenvironment. J. Biol. Chem. 2010;285:13535–13541. doi: 10.1074/jbc.M109.091058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider A., Rajendran L., Simons M. Flotillin-dependent clustering of the amyloid precursor protein regulates its endocytosis and amyloidogenic processing in neurons. J. Neurosci. 2008;28:2874–2882. doi: 10.1523/JNEUROSCI.5345-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber A., Fischer S., Lang T. The amyloid precursor protein forms plasmalemmal clusters via its pathogenic amyloid-β domain. Biophys. J. 2012;102:1411–1417. doi: 10.1016/j.bpj.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parvathy S., Hussain I., Hooper N.M. Cleavage of Alzheimer’s amyloid precursor protein by alpha-secretase occurs at the surface of neuronal cells. Biochemistry. 1999;38:9728–9734. doi: 10.1021/bi9906827. [DOI] [PubMed] [Google Scholar]
- 30.Asai M., Hattori C., Ishiura S. Putative function of ADAM9, ADAM10, and ADAM17 as APP alpha-secretase. Biochem. Biophys. Res. Commun. 2003;301:231–235. doi: 10.1016/s0006-291x(02)02999-6. [DOI] [PubMed] [Google Scholar]
- 31.Koo E.H., Squazzo S.L. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J. Biol. Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- 32.Kinoshita A., Fukumoto H., Hyman B.T. Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J. Cell Sci. 2003;116:3339–3346. doi: 10.1242/jcs.00643. [DOI] [PubMed] [Google Scholar]
- 33.Fukumori A., Okochi M., Takeda M. Presenilin-dependent gamma-secretase on plasma membrane and endosomes is functionally distinct. Biochemistry. 2006;45:4907–4914. doi: 10.1021/bi052412w. [DOI] [PubMed] [Google Scholar]
- 34.Imaris (Bitplane AG, Zurich, Switzerland).
- 35.Berman H.M., Westbrook J., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radzimanowski J., Simon B., Wild K. Structure of the intracellular domain of the amyloid precursor protein in complex with Fe65-PTB2. EMBO Rep. 2008;9:1134–1140. doi: 10.1038/embor.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nadezhdin K.D., Bocharova O.V., Arseniev A.S. Dimeric structure of transmembrane domain of amyloid precursor protein in micellar environment. FEBS Lett. 2012;586:1687–1692. doi: 10.1016/j.febslet.2012.04.062. [DOI] [PubMed] [Google Scholar]
- 38.Crescenzi O., Tomaselli S., Picone D. Solution structure of the Alzheimer amyloid beta-peptide (1-42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur. J. Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Ha Y. The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain. Mol. Cell. 2004;15:343–353. doi: 10.1016/j.molcel.2004.06.037. [DOI] [PubMed] [Google Scholar]
- 40.Hynes T.R., Randal M., Kossiakoff A.A. X-ray crystal structure of the protease inhibitor domain of Alzheimer’s amyloid beta-protein precursor. Biochemistry. 1990;29:10018–10022. doi: 10.1021/bi00495a002. [DOI] [PubMed] [Google Scholar]
- 41.Dahms S.O., Hoefgen S., Than M.E. Structure and biochemical analysis of the heparin-induced E1 dimer of the amyloid precursor protein. Proc. Natl. Acad. Sci. USA. 2010;107:5381–5386. doi: 10.1073/pnas.0911326107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The PyMOL Molecular Graphics System, Version 1.3 r1. (Schrödinger, LLC).
- 43.Schmidt T.H., Kandt C. LAMBADA and InflateGRO2: efficient membrane alignment and insertion of membrane proteins for molecular dynamics simulations. J. Chem. Inf. Model. 2012;52:2657–2669. doi: 10.1021/ci3000453. [DOI] [PubMed] [Google Scholar]
- 44.Poger D., Mark A.E. On the validation of molecular dynamics simulations of saturated and cis-monounsaturated phosphatidylcholine lipid bilayers: a comparison with experiment. J. Chem. Theory Comput. 2010;6:325–336. doi: 10.1021/ct900487a. [DOI] [PubMed] [Google Scholar]
- 45.Van Der Spoel D., Lindahl E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 46.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 47.Lewis V., Whitehouse I.J., Hooper N.M. Cellular prion protein expression is not regulated by the Alzheimer’s amyloid precursor protein intracellular domain. PLoS One. 2012;7:e31754. doi: 10.1371/journal.pone.0031754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suh J., Lyckman A., Guénette S.Y. FE65 proteins regulate NMDA receptor activation-induced amyloid precursor protein processing. J. Neurochem. 2011;119:377–388. doi: 10.1111/j.1471-4159.2011.07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang Y., Mullaney K.A., Nixon R.A. Alzheimer’s-related endosome dysfunction in Down syndrome is Abeta-independent but requires APP and is reversed by BACE-1 inhibition. Proc. Natl. Acad. Sci. USA. 2010;107:1630–1635. doi: 10.1073/pnas.0908953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koo E.H., Squazzo S.L., Koo C.H. Trafficking of cell-surface amyloid beta-protein precursor. I. Secretion, endocytosis and recycling as detected by labeled monoclonal antibody. J. Cell Sci. 1996;109:991–998. doi: 10.1242/jcs.109.5.991. [DOI] [PubMed] [Google Scholar]
- 51.Kern A., Roempp B., Behl C. Down-regulation of endogenous amyloid precursor protein processing due to cellular aging. J. Biol. Chem. 2006;281:2405–2413. doi: 10.1074/jbc.M505625200. [DOI] [PubMed] [Google Scholar]
- 52.Kaden D., Voigt P., Multhaup G. Subcellular localization and dimerization of APLP1 are strikingly different from APP and APLP2. J. Cell Sci. 2009;122:368–377. doi: 10.1242/jcs.034058. [DOI] [PubMed] [Google Scholar]
- 53.Zeng F., Yang W., Chen Y. Determination of the lowest concentrations of aldehyde fixatives for completely fixing various cellular structures by real-time imaging and quantification. Histochem. Cell Biol. 2013;139:735–749. doi: 10.1007/s00418-012-1058-5. [DOI] [PubMed] [Google Scholar]
- 54.Illinger D., Duportail G., Kuhry J.G. A comparison of the fluorescence properties of TMA-DPH as a probe for plasma membrane and for endocytic membrane. Biochim. Biophys. Acta. 1995;1239:58–66. doi: 10.1016/0005-2736(95)00135-p. [DOI] [PubMed] [Google Scholar]
- 55.Sinha M., Mishra S., Joshi P.G. Liquid-ordered microdomains in lipid rafts and plasma membrane of U-87 MG cells: a time-resolved fluorescence study. Eur. Biophys. J. 2003;32:381–391. doi: 10.1007/s00249-003-0281-3. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka K.A., Suzuki K.G., Kusumi A. Membrane molecules mobile even after chemical fixation. Nat. Methods. 2010;7:865–866. doi: 10.1038/nmeth.f.314. [DOI] [PubMed] [Google Scholar]
- 57.McLean I.W., Nakane P.K. Periodate-lysine-paraformaldehyde fixative. A new fixation for immunoelectron microscopy. J. Histochem. Cytochem. 1974;22:1077–1083. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- 58.Hixson D.C., Yep J.M., Walborg E.F., Jr. Evaluation of periodate/lysine/paraformaldehyde fixation as a method for cross-linking plasma membrane glycoproteins. J. Histochem. Cytochem. 1981;29:561–566. doi: 10.1177/29.4.6265547. [DOI] [PubMed] [Google Scholar]
- 59.Scheuermann S., Hambsch B., Multhaup G. Homodimerization of amyloid precursor protein and its implication in the amyloidogenic pathway of Alzheimer’s disease. J. Biol. Chem. 2001;276:33923–33929. doi: 10.1074/jbc.M105410200. [DOI] [PubMed] [Google Scholar]
- 60.Gorman P.M., Kim S., Chakrabartty A. Dimerization of the transmembrane domain of amyloid precursor proteins and familial Alzheimer’s disease mutants. BMC Neurosci. 2008;9:17. doi: 10.1186/1471-2202-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fogel H., Frere S., Slutsky I. APP homodimers transduce an amyloid-β-mediated increase in release probability at excitatory synapses. Cell Reports. 2014;7:1560–1576. doi: 10.1016/j.celrep.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 62.Ziomkiewicz I., Sporring J., Schulz A. Novel approach to measure the size of plasma-membrane nanodomains in single molecule localization microscopy. Cytometry A. 2015;87:868–877. doi: 10.1002/cyto.a.22708. [DOI] [PubMed] [Google Scholar]
- 63.Zhang F., Gannon M., Wang Q. The amyloid precursor protein modulates α2A-adrenergic receptor endocytosis and signaling through disrupting arrestin 3 recruitment. FASEB J. 2017;31:4434–4446. doi: 10.1096/fj.201700346R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pohlkamp T., Wasser C.R., Herz J. Functional roles of the interaction of APP and lipoprotein receptors. Front. Mol. Neurosci. 2017;10:54. doi: 10.3389/fnmol.2017.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song Y., Mittendorf K.F., Sanders C.R. Impact of bilayer lipid composition on the structure and topology of the transmembrane amyloid precursor C99 protein. J. Am. Chem. Soc. 2014;136:4093–4096. doi: 10.1021/ja4114374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meyer A.C., Frank T., Moser T. Tuning of synapse number, structure and function in the cochlea. Nat. Neurosci. 2009;12:444–453. doi: 10.1038/nn.2293. [DOI] [PubMed] [Google Scholar]
- 67.Schmid A., Hallermann S., Sigrist S.J. Activity-dependent site-specific changes of glutamate receptor composition in vivo. Nat. Neurosci. 2008;11:659–666. doi: 10.1038/nn.2122. [DOI] [PubMed] [Google Scholar]
- 68.Bar-On D., Gutman M., Nachliel E. Evaluation of the heterogeneous reactivity of the syntaxin molecules on the inner leaflet of the plasma membrane. J. Neurosci. 2009;29:12292–12301. doi: 10.1523/JNEUROSCI.0710-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bethani I., Werner A., Rizzoli S.O. Endosomal fusion upon SNARE knockdown is maintained by residual SNARE activity and enhanced docking. Traffic. 2009;10:1543–1559. doi: 10.1111/j.1600-0854.2009.00959.x. [DOI] [PubMed] [Google Scholar]
- 70.Selkoe D.J. Alzheimer’s disease: genes, proteins, and therapy. Physiol. Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 71.Wakutani Y., Watanabe K., Nakashima K. Novel amyloid precursor protein gene missense mutation (D678N) in probable familial Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry. 2004;75:1039–1042. doi: 10.1136/jnnp.2003.010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di Fede G., Catania M., Tagliavini F. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science. 2009;323:1473–1477. doi: 10.1126/science.1168979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou L., Brouwers N., De Strooper B. Amyloid precursor protein mutation E682K at the alternative β-secretase cleavage β′-site increases Aβ generation. EMBO Mol. Med. 2011;3:291–302. doi: 10.1002/emmm.201100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lan M.-Y., Liu J.-S., Chang Y.-Y. A novel APP mutation (D678H) in a Taiwanese patient exhibiting dementia and cerebral microvasculopathy. J. Clin. Neurosci. 2014;21:513–515. doi: 10.1016/j.jocn.2013.03.038. [DOI] [PubMed] [Google Scholar]
- 75.Jonsson T., Atwal J.K., Stefansson K. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 76.Maloney J.A., Bainbridge T., Atwal J.K. Molecular mechanisms of Alzheimer disease protection by the A673T allele of amyloid precursor protein. J. Biol. Chem. 2014;289:30990–31000. doi: 10.1074/jbc.M114.589069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Destainville N., Schmidt T.H., Lang T. Where biology meets physics--a converging view on membrane microdomain dynamics. Curr. Top. Membr. 2016;77:27–65. doi: 10.1016/bs.ctm.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 78.Tan C.W., Gardiner B.S., Burgess A.W. Wnt signalling pathway parameters for mammalian cells. PLoS One. 2012;7:e31882. doi: 10.1371/journal.pone.0031882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Bakker B.I., de Lange F., Garcia-Parajo M.F. Nanoscale organization of the pathogen receptor DC-SIGN mapped by single-molecule high-resolution fluorescence microscopy. ChemPhysChem. 2007;8:1473–1480. doi: 10.1002/cphc.200700169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.