Abstract
Traditional informatics in comparative genomics work only with static representations of biomolecules (i.e., sequence and structure), thereby ignoring the molecular dynamics (MD) of proteins that define function in the cell. A comparative approach applied to MD would connect this very short timescale process, defined in femtoseconds, to one of the longest in the universe: molecular evolution measured in millions of years. Here, we leverage advances in graphics-processing-unit-accelerated MD simulation software to develop a comparative method of MD analysis and visualization that can be applied to any two homologous Protein Data Bank structures. Our open-source pipeline, DROIDS (Detecting Relative Outlier Impacts in Dynamic Simulations), works in conjunction with existing molecular modeling software to convert any Linux gaming personal computer into a “comparative computational microscope” for observing the biophysical effects of mutations and other chemical changes in proteins. DROIDS implements structural alignment and Benjamini-Hochberg-corrected Kolmogorov-Smirnov statistics to compare nanosecond-scale atom bond fluctuations on the protein backbone, color mapping the significant differences identified in protein MD with single-amino-acid resolution. DROIDS is simple to use, incorporating graphical user interface control for Amber16 MD simulations, cpptraj analysis, and the final statistical and visual representations in R graphics and UCSF Chimera. We demonstrate that DROIDS can be utilized to visually investigate molecular evolution and disease-related functional changes in MD due to genetic mutation and epigenetic modification. DROIDS can also be used to potentially investigate binding interactions of pharmaceuticals, toxins, or other biomolecules in a functional evolutionary context as well.
Introduction
In recent decades, many advances in our understanding of biology at the level of the molecular genotype have been facilitated by biologists working in the fields of molecular evolution and comparative genomics. Most comparative methods in bioinformatics rely primarily upon symbolic character representation of nucleic acid and/or amino acid sequences or else static three-dimensional models of molecular structure. Subsequently, the many dynamic aspects of biomolecules are explicitly ignored in comparative genomics, often with the tacit assumption that the most important “information” regarding function and evolution can be abstracted from the physical details of molecular behavior itself (1, 2, 3). Currently, researchers in genomics and structural biology have spent enormous effort to more efficiently generate, process, and analyze sequence and structural data with a variety of heuristic, probabilistic, and now machine-learning-based methods. However, we currently lack statistical methods and user-friendly comparative bioinformatics tools that biologists can apply to biomolecular dynamics despite a wealth of homologous structures from the Protein Data Bank (PDB) that could be data mined for this purpose. This leaves the comparative impact of genetic mutation, epigenetic modification, and binding interaction largely unknown at the level of protein dynamics.
The rapid expansion of entries in the PDB as well as the rapid development of computer programs accelerated by graphics processing units (GPUs) (4, 5) to simulate the molecular dynamics on PDB structures now provide an opening to develop theoretically sound comparative methods and tools that can enable both functional and evolutionary comparisons rooted in how macromolecular polymers dynamically move. We can also examine how they ultimately self-organize to form functionally stable structures and cross-interact with each other to control molecular processes in the cell. A computationally expensive but direct way of studying molecular interaction is provided by the field of molecular dynamics (MD) simulation. The technique of MD simulation often begins with a molecular structure file (.pdb) and an empirically parameterized force field representing atom interactions between fixed-point charges or polarizable masses that treat covalent chemical bond stretching, torsion, and angular motion as simple harmonic functions. Solvent can be added either explicitly or implicitly, then initial velocities are applied to individual atoms randomly according to the Boltzmann distribution for a given temperature, and finally Newtonian mechanics are numerically approximated over very short femtosecond-scale time steps because of the rapid vibration of chemical bonds. Energy transfer via nonbonded interactions between molecules in the simulation is typically approximated using Coulomb’s law, sometimes with a distance constraint to minimize calculations in very large systems. Despite a relatively enormous computational expense, if set up carefully and sampled for a long enough time frame on stably equilibrated systems, current GPU technology applied to MD simulation potentially provides tremendous accuracy and resolution for the biophysical behaviors of protein-based systems (4, 5). In the last several years, the demand from the personal computer gaming community for GPUs with thousands of computing cores now allows desktop computers to run enough MD simulations to adequately sample transition states of long-term molecular biological processes such as protein folding that play out at nanosecond-to-microsecond timescales (6). A less explored potential application of this new technology is the sampling of mutational space through the lens of comparative protein dynamics. This is enabled by the recent discovery that in very-short-timescale dynamics, the atom fluctuations that play out over picosecond timescales are also intrinsic drivers of longer-timescale functional dynamics through general hierarchical properties of protein energy landscapes ((7, 8); Fig. 1 A). Theoretically, this association between short- and long-timescale protein dynamics could allow for the biologically meaningful comparative statistical analysis of two “sets” of multiple short-timescale runs (picoseconds to nanoseconds) representing differences between homologous dynamic conditions related to long-timescale protein functioning (Fig. 1, B and C). Luckily, this magnitude and scale of computation is currently within the performance capability of MD simulation on modern GPUs. Software enabling comparative protein dynamics could be particularly useful for investigating the molecular evolution of protein stability, disease malfunction, and gene regulatory binding interaction.
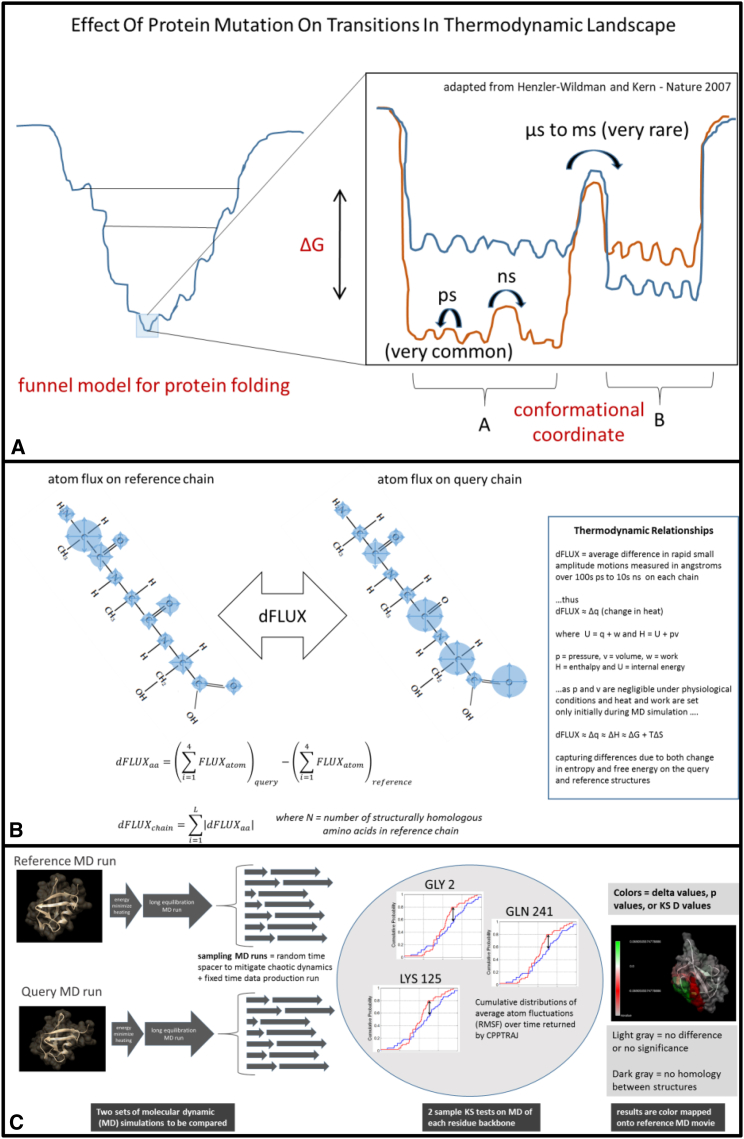
Figure 1.
(A) A hypothetical representation of the effect of protein mutation on the thermodynamic landscape of a protein (adapted from (8)). In the inset image, the mutation destabilizes the original thermodynamic landscape, shown in orange, to the state shown in blue. Under functional conservation of the protein function, many mutations will likely have little effect on the free-energy landscape, whereas only a very few may have more devastating impacts, as shown here. (B) dFLUX can be visualized here as the hypothetical differences in atom fluctuation (blue circles) on two homologous protein chains. In the DROIDS color mapping, dFLUX is averaged over the four backbone atoms of each amino acid. Global dFLUX for the whole chain is simply the sum of absolute dFLUX over the length of the polypeptide chain. (C) A schematic representation of DROIDS comparative molecular dynamic analysis software is shown. DROIDS 1.2 is a software tool for multiple-test-corrected pairwise comparison of molecular dynamics of two comparable PDB structures at the amino acid level. The three main phases of analysis include MD sampling runs and vector trajectory analysis, statistical comparison via multiple-test-corrected KS tests, and visualization results on static and moving images. RMSF, root mean-square fluctuation.
We introduce DROIDS 1.20, a graphical user interface (GUI) pipeline for Amber16 MD simulations and subsequent statistical analyses and visual representations capable of directly addressing functional and evolutionary changes in MD (digital object identifier: 10.5281/zenodo.1001755). It is available at https://github.com/gbabbitt/DROIDS-1.0. DROIDS (Detecting Relative Outlier Impacts in Dynamic Simulations) implements multiple-test-corrected Kolmogorov-Smirnov statistics at single-amino-acid resolution along the polypeptide backbone to identify significant functional differences in protein dynamics due to differences in nonsynonymous genetic mutation, epigenetic covalent modifications, or chemical binding interactions affected by the presence or absence of different natural or drug ligands or toxins. The DROIDS statistics are applied to samples of atom fluctuation (i.e., the cpptraj atom fluctuation (FLUX) calculation) to address local or global effects on thermodynamic stability and conformational rigidity that may affect longer-timescale functioning of proteins. The DROIDS software combines GPU-accelerated Amber16 and AmberTools17 MD simulation software by utilizing the pmemd.cuda executable (4, 5), cpptraj program for vector trajectory analysis (9), R software for statistical calculation and graphics, and the UCSF Chimera extensible molecular modeling software for structural comparison and visualization (10) to create a seamless GUI-based experience that allows users to identify significant differences in how proteins move. DROIDS automates the repeated random sampling of MD to allow for proper statistical comparison of two sets of simulations run on any pair of homologous queries and reference PDB files that can be superimposed and structurally aligned with a high degree of overlap. DROIDS then produces molecular visualizations in which significant changes in MD are color mapped to static images or movies according to quantitative differences (i.e., delta values in angstroms for atom fluctuations, or “dFLUX”), and it also indicates statistically significant p-values of the Kolmogorov-Smirnov (KS) test colored against gray-scaled nonsignificant values and nonhomologous regions.
Materials and Methods
The DROIDS 1.0 pipeline
The DROIDS pipeline is run as a series of three perl-tk scripts that are initiated at the Linux command line and controlled via a pop-up GUI interface. The Quick Start Guide walks the user through the steps shown schematically in Fig. 1 C. We also offer a more detailed user manual and installation guide with the download from GitHub. The user starts the pipeline by placing the two PDB files to be compared in the DROIDS main folder, opening a terminal, and typing “perl GUI_START_DROIDS.pl.” This GUI interface is designed to control and run all stages of the MD simulations of both the query and reference PDB structures that will be needed for later DROIDS analysis. The start menu begins with the construction of a structure-based sequence alignment in Chimera. This is followed by typical teLeap setup of the PDB files and a single energy minimization, heating, and equilibration run on each PDB file. These single MD runs are followed by N number of sampling runs, with N specified by the user and starting at the end of equilibration. For adequate estimation of average dFLUX, we generally recommend 50 or more sampling runs on each protein conducted at 0.5 ns per sample (see Fig. S1 for the effect of sample size on dFLUX estimation and Table S1 for observed false discovery rates in null comparisons on ubiquitin). Random spacer runs precede each sampling run to minimize the impact of initial conditions on the MD sampling. This is important, as it is well known that MD simulations are often chaotic in their behavior, and the randomization of the start of sampling during each run helps to average out any effects of chaotic dynamics on the statistical comparisons to be made. Afterwards, a GUI for vector trajectory analysis will pop up. This leads the user through typical cpptraj commands to collect atom fluctuation for each sampling run. Lastly, this GUI leads the user through data preparation for later DROIDS statistical analysis. In this step, the user is offered a choice of “strict” versus “loose” homology (which determines the amino acids to which the DROIDS statistics will be applied). See the user manual for more details regarding this step. The third and final GUI allows users to run the statistical comparisons and choose the method of multiple-test correction (11), followed by color and graphics options to be applied to the static and moving images of the reference PDB. The statistical test is a KS test applied specifically to the collective backbone MD of each amino acid residue (i.e., atoms N, CA, C, and O masked during cpptraj). Note that many more specific details of the MD can be modified by the user by editing the lines in the perl scripts that write the control files within GUI_START_DROIDS.pl.
MD simulation protocols for case examples
All MD simulations were tested on an Intel Xeon E5 board (Intel, Santa Clara, CA) mounting a GTX Titan X GPU Maxwell accelerator or a smaller graphics workstation mounting a pair of GTX 1080 Founder’s Edition GPU Pascal accelerators (Nvidia, Santa Clara, CA) running pmemd.cuda and cpptraj released with Amber16 and AmberTools17 (4, 5, 9). The force field we chose was protein.ff14SB. Each structure underwent initial energy minimization. The structures were then heated from 100 to 300 K under Langevin dynamics over 0.5 ns. The production MDs for most of the case examples we posted to YouTube were performed under the Hawkins, Cramer, and Truhlar pairwise Generalized Born model for implicit solvation (12); 50 sampling simulation runs lasted for 0.5 ns (0.5E9 steps), with an integration time step of 2.0 fs and an infinite cutoff. Before sampling the MD on each structure, we equilibrated for 10 ns. After equilibration, 50 sampling runs at 0.5 ns each were taken using a random starting time to average out any small differences caused by the sensitivity of MD to initial conditions set at the end of the equilibration stage. A Langevin dynamics thermostat was used to control for our constant temperature simulations (300 K) at a collision frequency of 10 ps−1. Bond length constraints were applied to all hydrogen bonds using the SHAKE algorithm. Center-of-mass motion was removed every 1000 steps, and slowly varying forces were evaluated every two steps. The minimal distance calculated for the effective Born radii was 25.0 Å. Simulation data were output every 500 steps to a binary NetCDF trajectory for cpptraj analysis.
Results and Discussion
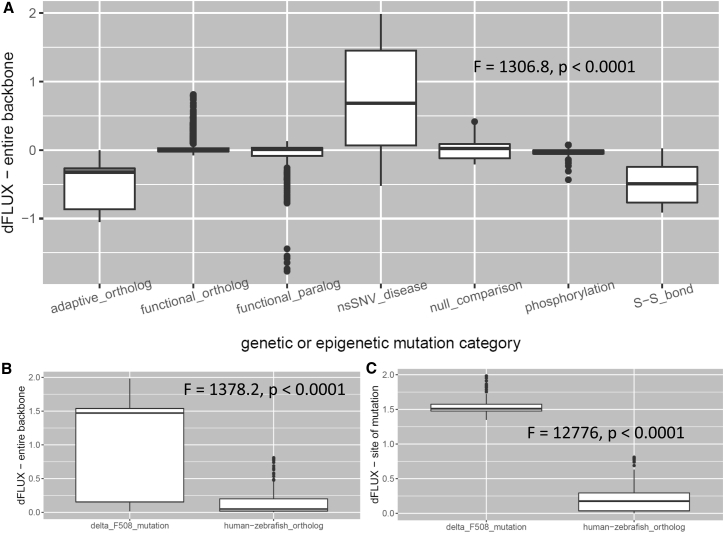
A summary of dFLUX values sampled across various types of mutation and epimutation is shown in Fig. 2. Summary details of each protein comparison conducted here are also presented in Table 1. DROIDS clearly demonstrates that most orthologous and paralogous evolutionary changes in functionally conserved proteins have little effect on MD (Fig. 2 A). Most changes in dynamics observed under purifying selection are similar in magnitude to differences in dynamics observed across separate runs on the same protein. Adaptive genetic changes occurring under a regime of positive selection promoting thermostability demonstrate consistent dampening of atom fluctuation of roughly the same magnitude as that observed for disulfide bridging in natural signaling proteins and bioengineered enzymes. Most interestingly, the several disease-related mutations we have initially analyzed with DROIDS can be observed to globally destabilize proteins (i.e., increase atom fluctuation), confirming recent general hypotheses about the role of general biophysical malfunction in disease (13, 14). In our modest sampling of a few disease mutations, we found this global destabilization is particularly notable in the case of the impact of the ΔF508 mutation and other single mutations engineered at this site on the CFTR protein, as it is the cause of over 90% of cases of human cystic fibrosis (15, 16). Mutations at this site have a much larger singular effect on both global and local protein dynamics even when compared to the cumulative effect of many orthologous changes that have arisen since the divergence of vertebrates (Fig. 2, B and C). We also observed a global destabilization of the B-RAF kinase caused by the V600E mutation (17), which is associated with roughly 50% of human melanoma cases (see Table 1). Together, these results demonstrate a potential for DROIDS to verify the underlying molecular mechanics of regions of low mutational tolerance in at least some proteins. Despite being much faster than MD, current sequence- and structure-based machine learning classifiers of mutational tolerance do not currently provide any biophysical details as to the root cause of low mutational tolerances involved in any disease. DROIDS analysis might provide detailed additional biophysical information regarding low mutational tolerance of some proteins identified in genomic scans using current sequence-based methods (e.g., SIFT, PolyPhen, or MegaMD (18, 19, 20, 21)).
Figure 2.
(A) Comparison of global dFLUX distribution (entire protein) across different mutation and epimutation categories in all DROIDS analyses listed in Table 1. (B) Here, a comparison is shown of global dFLUX distribution in the nucleotide-binding domain of CFTR caused by changes at the site of cystic fibrosis mutation (ΔF508) and orthologous changes in CFTR that have occurred since the divergence of vertebrates (human-zebrafish). (C) Local dFLUX only at sites of mutations is also shown for contrast.
Table 1.
Comparative Protein Dynamics Analyses Conducted with DROIDS
| Category | Protein | PDB ID | Species | Sequence Similarity (%) | Grantham Distance (Average) | Dynamic Similarity (Average dFLUX) |
|---|---|---|---|---|---|---|
| Adaptive ortholog | p450 cytochrome | 1f4t-1phd | Sulfolobus solfactaricus | 15.95 | 79.93 | 0.28 |
| Pseudomonas putida | ||||||
| Adaptive ortholog | p450 cytochrome | 1n97-1phd | Thermus thermophilus | 10.88 | 82.10 | 0.25 |
| P. putida | ||||||
| Adaptive ortholog | p450 cytochrome | 1t2b-1phd | Citrobacter braakii | 23.68 | 76.63 | 0.27 |
| P. putida | ||||||
| Adaptive ortholog | DNA polymerase | 4n56-1kfd | Thermus aquaticus | 29.1 | 77.86 | 0.89 |
| E. coli | ||||||
| Functional ortholog | alcohol dehydrogenase | 1htb-6adh | Homo sapiens | 86.91 | 66.47 | 0.02 |
| Equus caballus | ||||||
| Functional ortholog | NBD1 domain of CFTR | 1xf9-2pze | Mus musculus | 83.12 | 52.58 | 0.29 |
| H. sapiens | ||||||
| Functional ortholog | L-Dap aminotransferase | 3ei7-3qgu | Arabidopsis thaliana | |||
| Chalmydomonas reinhardtii | ||||||
| Functional ortholog | ATP-free CFTR | 5uar-5uak | Danio rerio | 68.61 | 53.26 | 0.04 |
| H. sapiens | ||||||
| Functional paralog | lyase (DCoH2 and DCoH) | 1ru0-1dch | M. musculus | 66.68 | 52.10 | 0.02 |
| Rattus norvegicus | ||||||
| Functional paralog | serine proteases (Trypsin and pancreatic elastase) | 2ptn-3est | Bos taurus | 35.43 | 75.96 | 0.06 |
| Sus scrofa | ||||||
| Functional paralog | interferon regulatory factors (IRF-5 and IRF-3) | 3dsh-1j2f | H. sapiens | 23.90 | 84.27 | 0.29 |
| nsSNV disease | V599E mutant of B-RAF kinase (melanoma) | 1uwj-1uwh | H. sapiens | 99.29 | 113.50 | 0.73 |
| nsSNV disease | F508R disease and F508S nondisease mutant of NBD1 domain of CFTR (cystic fibrosis) | 1xfa-1xf9 | H. sapiens | 99.65 | 110.00 | 1.28 |
| nsSNV disease | F508R disease and wild-type NBD1 domain of CFTR (cystic fibrosis) | 1xfa-2pze | H. sapiens | 93.12 | 51.05 | 1.54 |
| nsSNV disease | ΔF508 disease and wild-type NBD1 domain of CFTR (cystic fibrosis) | 2pzf-2pze | H. sapiens | 99.57 | NA | 0.51 |
| nsSNV disease | Htt36Q3H and Htt17Q (Huntington’s disease) | 4fec-3iou | H. sapiens | 97.37 | 76.27 | 0.05 |
| nsSNV disease | PrP226 and human prion protein (prion-based amyloid disease) | 5l6r-1qlx | H. sapiens | 96.24 | 99.75 | 0.09 |
| Null comparison | collagen | 1bkv | H. sapiens | 100 | 0% | 0.12 |
| Null comparison | collagen-like peptide | 1cag | H. sapiens | 100 | 0% | 0.17 |
| Null comparison | DNA polymerase (Klenow fragment) | 1kfd | E. coli | 100 | 0% | 0.13 |
| Null comparison | T4 lysozyme | 1lyd | E. virus T4 | 100 | 0% | 0.15 |
| Null comparison | ubiquitin | 1ubq | H. sapiens | 100 | 0% | 0.04 |
| Null comparison | pancreatic elastase | 3est | S. scrofa | 100 | 0% | 0.06 |
| Phosphorylation | phosphorylated IRF-3 and dephosphorylated IRF-3 | 3a77-1j2f | H. sapiens | 98.68 | 0% | 0.02 |
| Phosphorylation | phosphorylated DesR (inactive) and dephosphorylated DesR (active) | 4le1-4le2 | Bacillus subtilis | 100 | 0% | 0.06 |
| Disulfide (S-S) | proinsulin (with and without three disulfide bonds) | 2kqp | H. sapiens | 100 | 0% | 0.63 |
| Disulfide (S-S) | T4 lysozyme with and without a bioengineered disulfide bond) | 1l35-1lyd | E. virus T4 | 100 | 0% | 0.08 (0.47) at site of S-S |
The impacts of genetic mutation on each protein are compared at the levels of amino acid sequence and chemical distance, as well as MD. This data is summarized in Figure 2. ID, identifier; NA, not applicable.
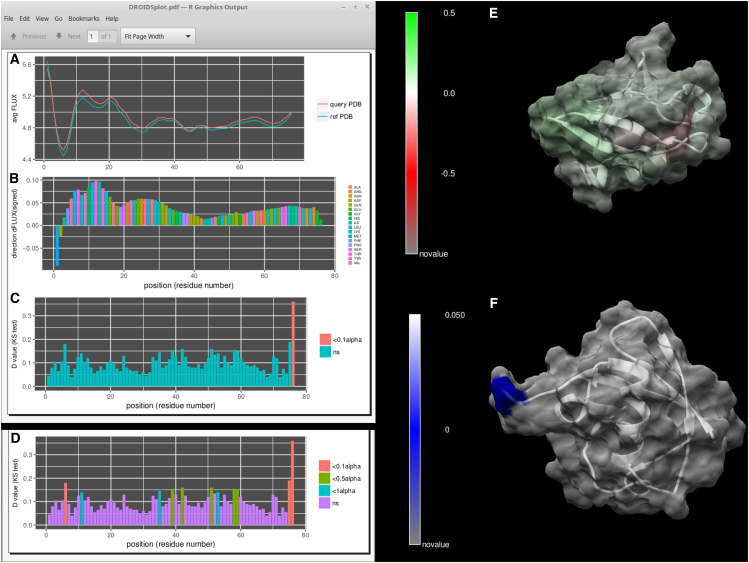
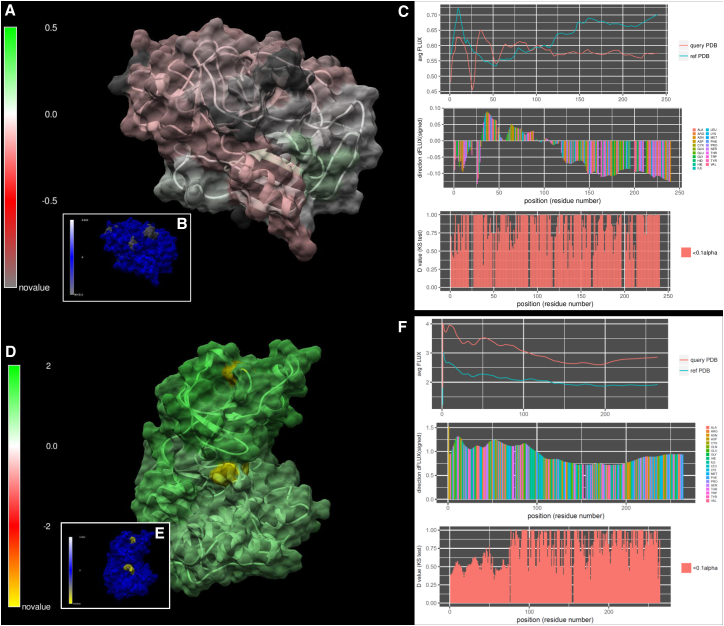
In addition to this general summary (Fig. 2; Table 1), we provide images from several case examples below that highlight the color-mapped visualization of DROIDS. Here, one can see some of the options for how movies rendered with DROIDS can actually look. In the remaining figures, we show some image output from a few selected MD runs in Table 1. Many of the others are available for viewing on our YouTube channel at https://www.youtube.com/channel/UCJTBqGq01pBCMDQikn566Kw. The first example is a simple null comparison on ubiquitin (Fig. 3), a very small and stable protein. The difference in the MD profile for atom fluctuation was very small, indicating reproducibility of the average FLUX values in the MD profile (Fig. 3, A, B, and E). This was essentially implemented as a “sanity check” for the method of multiple-test correction (i.e., Benjamini-Hochberg method), and it was deemed effective in eliminating most of the false-positive results from the comparison most of the time. Table S1 shows a more detailed analysis of false discovery rates at various settings and sampling regimes. The correction eliminated all false-positive results except one significant p-value at the C-terminal end of the protein (Fig. 3, C, D, and F). We suspect that the MD here might actually be significantly different, as the C-terminal end of ubiquitin, which was used to “tag” many other proteins for proteolysis, is relatively disordered and does not reside in a relatively stable potential energy state.
Figure 3.
A null comparison of molecular dynamics on a small protein (PDB: 1ubq – 1ubq). (A) The profiles in average FLUX as a function of position are nearly identical. (B) The differences (i.e., dFLUX) that are colored as a function of amino acid type are (C) almost entirely nonsignificant except at the terminal end of the protein. (D) Results without correction for false discovery rate are also shown for comparison. (E) dFLUX and (F) p-values of the KS tests are shown color mapped to PDB: 1ubq. ns, not significant.
For our next example, we conducted a series of comparisons of thermostable enzymes to normal homologs (Fig. 4). We chose thermostable p450 orthologs identified by (22) (Fig. 4, A and B) as well as Taq polymerase compared to the Klenow fragment in Escherichia coli (E. coli) (Fig. 4 C). We found that a similar mechanism of thermostability evolved in all cases. When the N- or C-terminal end of the protein lies near the surface and is lacking secondary structure, the sequence divergence observed conveys a great deal of local thermostability, as shown by the darker red regions of dampened atom fluctuation. We also included an example of a thermostabilizing effect of a bioengineered disulfide bridge in a phage T4 lysozyme (23) (Fig. 4 D). Our result seems to contradict the original results of this study, showing a large region of dampened atom fluctuation near the disulfide bond. As this original study only examined crystallographic temperature factors and not MD, and because it is well known that disulfide bridges stabilize protein structure, our finding is perhaps not surprising. We show a similar stabilization result using proinsulin as an example on our YouTube channel.
Figure 4.
Comparison of protein dynamics of several thermostable and wild-type enzymes. (A) Sulfolobus p450 cytochrome is compared to Pseudomonas (PDB: 1t2b and 1phd), and (B) Citrobacter p450 cytochrome is compared to Pseudomonas (PDB: 1f4t – 1phd). (C) Taq DNA polymerase and an E. coli Klenow fragment are shown here, and (D) the effect of a bioengineered disulfide bond in lysozyme is shown here. The differences in atom fluctuation (i.e., dFLUX) are color scaled: green indicates amplified motion, and red indicates dampened motion. (A)–(C) exhibit strongest thermostability (i.e., darkest red) near the terminal ends of the protein, especially when these regions lack secondary structure and are near the protein surface. For more details about (A) and (B), see (22).
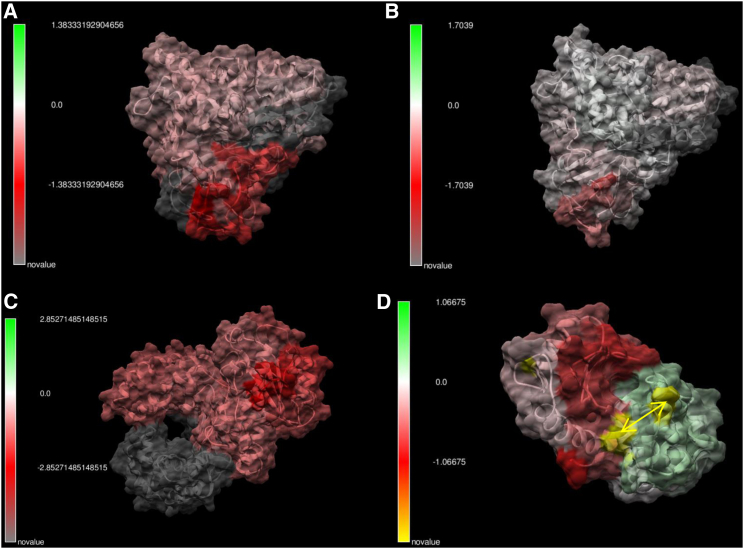
For our last example, we examined the functional biophysical consequences of mutation in serine protease paralogs (i.e., gene duplication events, Fig. 5, A–C) and a well-studied cancer mutation (V600E) on the B-RAF kinase oncogene (17, 24) (Fig. 5, D–F). The serine proteases we compared—trypsin and pancreatic elastase—exhibit large sequence divergence and also relatively small but significant changes in functional divergence in terms of atom fluctuation (dFLUX). In contrast, the functional changes to the B-RAF kinase are much more considerable even though the only sequence differences are due to the well-known valine-to-glutamine change in the activation loop of the kinase domain and an additional alanine-to-lysine mutation elsewhere on the chain. Because of their locations and amino acid characteristics, both of these mutations might be expected to shift the hydrophobicity of the protein. Our results indicate a relatively large global destabilization of the B-RAF kinase, indicated by the large increase of atom fluctuation over the whole structure. The original study reported that whereas the growth pathway in which B-RAF was involved is effectively less regulated, the B-RAF protein is often rendered less functional (17). Our analysis indicating the destabilization of the kinase would be congruent with previous results. We ran this analysis three times on two different machines and confirmed the same MD result each time. It seems that large protein destabilizations may be a common cause of low mutational tolerances in proteins with disease-causing malfunctions because of nonsynonymous mutation, as originally theorized by Linus Pauling many years ago (25). We have observed this significant global protein destabilization of atom fluctuation as a common underlying effect of the few disease-related mutations we have analyzed thus far (i.e., with cystic fibrosis and melanoma in particular but also to a lesser effect with Huntington’s disease and amyloid-related prion disease; see Table 1). We hope our software will allow medical researchers to examine this possibility more easily, directly, and frequently in relation to the many specific molecular details of disease.
Figure 5.
Comparison of evolutionary effects on protein dynamics. (A) The change in atom fluctuation (dFLUX) due to gene duplication in serine proteases (compares trypsin PDB: 2ptn to pancreatic elastase PDB: 3est) is shown. (B) Shown are the p-values of the KS tests, with blue indicating significant change. (C) Shown is the R graphical output revealing the average FLUX (top), dFLUX (middle), and KS statistics (bottom) as a function of amino acid position. (D)–(F) show the same results for destabilization of a B-RAF kinase by two cancer mutations (yellow).
Current and future uses for DROIDS
The potential uses for making protein comparisons with DROIDS are many. Some ideas we have imagined during its development include not only the visualization of the functional effects of natural mutation at the protein sequence level but also the potential cost-saving computational screening of the effects of site-directed mutagenesis by the pharmaceutical industry. Further investigation into human population variation associated with disease-related malfunction (i.e., nonsynonymous single-nucleotide variants (nsSNVs)) might also be analyzed, particularly with respect to determining molecular phenomena that drive personal genomic differences in mutational tolerance (20) and drug response in clinical trials. The functional impacts of many more posttranslational modifications (e.g., disulfide bridging or phosphorylation) and epigenetic modifications to chromatin (e.g., histone acetylation and methylation) will also be of considerable interest when run on larger computers that can handle larger molecular systems. We have demonstrated here that the functional consequences of natural evolutionary divergences created through the processes of speciation, gene duplication, and genetic drift and/or genomic decay can be compared in a pairwise manner. However, future releases might include methods of distinguishing selection from drift based upon randomization tests (26, 27, 28). The study of functional binding interactions (protein-ligand, protein-DNA, and protein-protein) may also be possible upon future versioning that can simultaneously analyze larger multichain systems.
We have shown that null comparisons are also potentially useful. These are when the exact duplicate copies of the same PDB files are compared through DROIDS. Because MD can diverge wherever the system does not settle into potential energy wells, a null comparison on a single structure using DROIDS can show users where the MD is potentially failing to replicate reproducible biophysics. This can be particularly useful for demonstrating and testing the efficacy of newly developed force fields and for identifying where on a given structure the scientific inferences made with existing force fields are most sound.
We hope to engage students at our home institutions as well as the open-source development community on GitHub to design future editions of DROIDS that are more specific to particular areas of interest in molecular evolutionary biology (e.g., chromatin dynamics, transcription factor binding function, and ATP and/or GTP protein activation). The immediate future development of DROIDS will incorporate GPU-accelerated MD conducted using the open-source OpenMM libraries (29) as well as Amber16 and AmberTools17. We openly invite the open-source community and gaming enthusiasts to be creative with our code repository and work toward leveraging the enormous computing resources collectively held by the personal computer gaming community toward our future goal of simulation-based comparative “microscopy” that can be managed by the everyday computer user.
Author Contributions
G.A.B. conceived of the project and wrote the article. G.A.B., J.S.M., and E.E.C. did the bulk of the main code development. L.E.A. wrote the Python code for the DROIDS movie player. J.K.L. wrote the Python code to color map our statistics on static Chimera images.
Acknowledgments
We thank the College of Science at the Rochester Institute of Technology for supporting this work with an internal grant. We gratefully acknowledge the support of Nvidia Corporation for the donation of GPU hardware used for this research.
Editor: Nathan Baker.
Footnotes
One figure and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30146-2.
Supporting Material
References
- 1.Wilke C.O. Bringing molecules back into molecular evolution. PLOS Comput. Biol. 2012;8:e1002572. doi: 10.1371/journal.pcbi.1002572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danchin E., Pocheville A. Inheritance is where physiology meets evolution. J. Physiol. 2014;592:2307–2317. doi: 10.1113/jphysiol.2014.272096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babbitt G.A., Coppola E.E., Hudson A.O. Can all heritable biology really be reduced to a single dimension? Gene. 2016;578:162–168. doi: 10.1016/j.gene.2015.12.043. [DOI] [PubMed] [Google Scholar]
- 4.Götz A.W., Williamson M.J., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salomon-Ferrer R., Götz A.W., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 6.Pande V.S., Beauchamp K., Bowman G.R. Everything you wanted to know about Markov state models but were afraid to ask. Methods. 2010;52:99–105. doi: 10.1016/j.ymeth.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henzler-Wildman K.A., Lei M., Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 8.Henzler-Wildman K., Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 9.Roe D.R., Cheatham T.E., 3rd PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 10.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 11.Noble W.S. How does multiple testing correction work? Nat. Biotechnol. 2009;27:1135–1137. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawkins G.D., Cramer C.J., Truhlar D.G. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J. Phys. Chem. 1996;100:19824–19839. [Google Scholar]
- 13.Redler R.L., Das J., Dokholyan N.V. Protein destabilization as a common factor in diverse inherited disorders. J. Mol. Evol. 2016;82:11–16. doi: 10.1007/s00239-015-9717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yue P., Li Z., Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J. Mol. Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z., Liu F., Chen J. Conformational changes of CFTR upon phosphorylation and ATP binding. Cell. 2017;170:483–491.e8. doi: 10.1016/j.cell.2017.06.041. [DOI] [PubMed] [Google Scholar]
- 16.Liu F., Zhang Z., Chen J. Molecular structure of the human CFTR ion channel. Cell. 2017;169:85–95.e8. doi: 10.1016/j.cell.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 17.Wan P.T., Garnett M.J., Marais R., Cancer Genome Project Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 18.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;20:1–7. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adzhubei I.A., Schmidt S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng P.C., Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerek N.Z., Liu L., Kumar S. Evolutionary diagnosis of non-synonymous variants involved in differential drug response. BMC Med. Genomics. 2015;8(Suppl 1):S6. doi: 10.1186/1755-8794-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meharenna Y.T., Poulos T.L. Using molecular dynamics to probe the structural basis for enhanced stability in thermal stable cytochromes P450. Biochemistry. 2010;49:6680–6686. doi: 10.1021/bi100929x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pjura P.E., Matsumura M., Matthews B.W. Structure of a thermostable disulfide-bridge mutant of phage T4 lysozyme shows that an engineered cross-link in a flexible region does not increase the rigidity of the folded protein. Biochemistry. 1990;29:2592–2598. doi: 10.1021/bi00462a023. [DOI] [PubMed] [Google Scholar]
- 24.Hubbard S.R. Oncogenic mutations in B-Raf: some losses yield gains. Cell. 2004;116:764–766. doi: 10.1016/s0092-8674(04)00256-9. [DOI] [PubMed] [Google Scholar]
- 25.Eaton W.A. Linus Pauling and sickle cell disease. Biophys. Chem. 2003;100:109–116. doi: 10.1016/s0301-4622(02)00269-7. [DOI] [PubMed] [Google Scholar]
- 26.Babbitt G.A., Kim Y. Inferring natural selection on fine-scale chromatin organization in yeast. Mol. Biol. Evol. 2008;25:1714–1727. doi: 10.1093/molbev/msn127. [DOI] [PubMed] [Google Scholar]
- 27.Babbitt G.A., Tolstorukov M.Y., Kim Y. The molecular evolution of nucleosome positioning through sequence-dependent deformation of the DNA polymer. J. Biomol. Struct. Dyn. 2010;27:765–780. doi: 10.1080/07391102.2010.10508584. [DOI] [PubMed] [Google Scholar]
- 28.Babbitt G.A., Alawad M.A., Hudson A.O. Synonymous codon bias and functional constraint on GC3-related DNA backbone dynamics in the prokaryotic nucleoid. Nucleic Acids Res. 2014;42:10915–10926. doi: 10.1093/nar/gku811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eastman P., Swails J., Pande V.S. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLOS Comput. Biol. 2017;13:e1005659. doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.