Abstract
The T4 lysozyme L99A mutant is often used as a model system to study small-molecule binding to proteins, but pathways for ligand entry and exit from the buried binding site and the associated protein conformational changes have not been fully resolved. Here, molecular dynamics simulations were employed to model benzene exit from its binding cavity using the weighted ensemble (WE) approach to enhance sampling of low-probability unbinding trajectories. Independent WE simulations revealed four pathways for benzene exit, which correspond to transient tunnels spontaneously formed in previous simulations of apo T4 lysozyme. Thus, benzene unbinding occurs through multiple pathways partially created by intrinsic protein structural fluctuations. Motions of several α-helices and side chains were involved in ligand escape from metastable microstates. WE simulations also provided preliminary estimates of rate constants for each exit pathway. These results complement previous works and provide a semiquantitative characterization of pathway heterogeneity for binding of small molecules to proteins.
Introduction
The binding of small molecules to proteins, a fundamental process in cellular metabolism, is also exploited pharmacologically to treat a large number of diseases. Drug development strategies usually try to improve ligand affinity to a target protein based on the structure and interactions observed in bound ligand-protein complexes. But binding kinetics and ligand residence time may also determine the physiological response and efficacy of a drug (1, 2, 3, 4).
Kinetic, affinity, and structural information on protein-ligand association may be obtained from the ensemble of transition pathways (5, 6, 7, 8) for the binding process. Each pathway describes the evolution of conformational and orientational degrees of freedom of ligand and protein that leads from an unbound to a bound configuration. As expected from path-ensemble symmetry (9), the reverse bound-to-unbound process should be described by the same pathway ensemble.
Computer simulations have been of great help in revealing possible pathways and kinetic information of protein-ligand binding (10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34). For example, Kubas and co-workers (29) simulated pathways for molecular O2 binding to a hydrogenase and proposed mutations along the binding protein tunnels that slowed down the access of small molecules, thus reducing the hydrogenase inhibition caused by O2. Casasnovas and co-workers (30) simulated dissociation of a potent pyrazol inhibitor from p38 MAP kinase and proposed congeneric ligands that could spend longer residence times when bound to the same kinase.
However, sampling of pathway ensembles for ligand-protein (un)binding is difficult because these are rare events on the timescales usually reached in molecular dynamics (MD) simulations. Methods such as weighted ensemble (WE) (35, 36), milestoning (37, 38), and transition interface sampling (39) are valuable because they enhance the sampling of pathways by increasing computational time spent in conformational regions with low visiting probability without introducing bias into the simulated potential energy (40, 41). The WE method has been applied to study pathways and kinetics for protein conformational transitions (42, 43, 44), host-guest association (45), protein-peptide association (46), and protein-ligand dissociation (4, 24, 33).
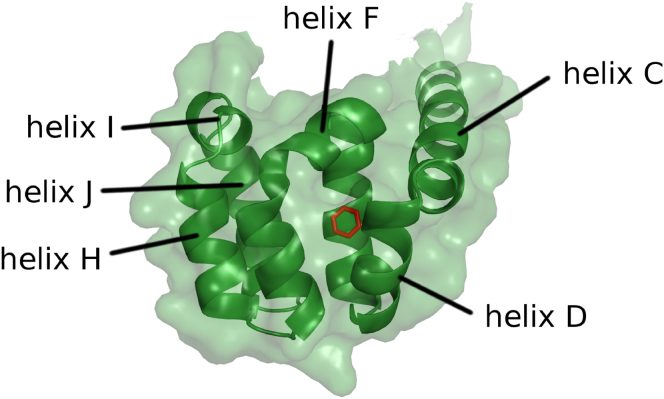
One of the proteins most often used to study association with small molecules is the T4 lysozyme (T4L) (47, 48, 49), the structure of which is shown in Fig. 1. The T4L L99A mutation creates a hydrophobic cavity in the protein C-terminal domain with a volume of ∼150 Å3. This site may accommodate gases such as xenon or O2 and small molecules such as benzene and its nonpolar derivatives (50, 51, 52, 53). The engineered cavity is dehydrated and shows little structural variation upon ligand complexation. Because of this simplicity and ease of experimental manipulation, more than 700 T4L complexes with small molecules have been characterized and used extensively to study structure-affinity relationships (49) and to compare with computational predictions of binding pose and affinity (54, 55, 56, 57).
Figure 1.
Crystal structure of the C-terminal domain of T4 lysozyme L99A mutant bound to benzene (red). The protein is represented with its molecular surface (green transparency), showing the ligand is fully buried. To see this figure in color, go online.
Crystal structures of T4L L99A mutant bound to benzene and a congeneric series of ligands (47, 50) show that the binding cavity is buried, without any clear tunnel or channel connecting to the protein surface (Fig. 1). Thus, T4L must undergo structural fluctuations or conformational “breathing” to allow ligand excursion into the binding cavity. The kinetics of benzene entry and exit from T4L L99A mutant has been estimated from NMR spectroscopy (= 950 s–1 for benzene at 303 K) (58), implying that the conformational motions involved in benzene escape should be observed in a millisecond timescale.
Several studies have addressed the possible pathways and the structural transitions used by small molecules to escape from inside T4L L99A. A weakly populated (or “excited”) state involving motion of the flexible T4L helix F (Fig. 1) was observed from relaxation-dispersion NMR measurements and suggested to be responsible for benzene exit (59). Later structural determination of this weakly populated state showed that the binding cavity remains inaccessible (60), but a recent metadynamics simulation showed that the conformational transition from the crystal structure to the weakly populated state forms a transient protein tunnel along which benzene can exit from T4L (61). An accelerated MD simulation was able to identify a different route for benzene entry (62), and two recent conventional MD simulations identified three other pathways for O2 unbinding (53) and five putative tunnels formed transiently in apo T4L L99A (63) that might accommodate small molecule excursions to the binding cavity.
To resolve the number and nature of ligand exit pathways and the conformational transitions involved in T4L L99A mutant, the WE approach is employed here as a discovery tool to sample trajectories for benzene exit. The ensemble of unbinding pathways obtained shows that benzene escapes via four different protein tunnels, all of which formed spontaneously in apo T4L L99A in a previous study that did not sample explicit ligand binding or unbinding (63). We find that motions in helix F, as previously suggested (49, 58, 59), are involved in ligand escape, but more importantly, so are those in helices C, H, and J, and side chains of several residues. Unbinding rates were estimated for each pathway and allowed a preliminary quantification of their contribution to the overall rate constant for benzene dissociation.
Methods
Structural model and molecular dynamics simulations
The structure of T4L L99A mutant bound to benzene was obtained from the Protein Data Bank structure 1L83 (47) after removal of water and crystallization molecules. Hydrogen atoms were constructed using the GROMACS Protein Data Bank parser (64).
Interactions were described by the CHARMM36 force field (65, 66, 67). All simulations employed implicit solvation in the generalized Born surface area form (68). This should be a reasonable approximation, as the binding cavity in T4L L99A is dehydrated, although any phenomena involving specific water interactions may not be modeled accurately. The OBC method was used to estimate Born radii (69) with a dielectric constant 80, and the nonpolar contribution was calculated as in Schaefer et al. (70), with a surface tension of 5.4 cal mol–1 Å–2 for all atoms.
Molecular dynamics trajectory segments in WE simulations were run using GROMACS 4.5 (64). Dynamics were carried out at 300 or at 400 K, with a 2 fs time-step, using a leapfrog stochastic dynamics integrator with a collision frequency of 10 ps–1. Covalent bonds between hydrogens and heavy atoms were constrained with LINCS (71). Structures were saved every 2 ps for analysis. The total aggregate simulation time was ∼29 μs.
WE simulations and rate estimation
The WE algorithm (35, 36) was used to enhance sampling of benzene unbinding events. Briefly, in the WE method, a progress coordinate is defined and divided into bins to describe the process of interest. A group of trajectories of the simulated system is propagated from an initial state with initially equal probabilities or weights. At fixed time intervals of length τ, the occupancy of each coordinate bin is reevaluated. Trajectories may be replicated or merged with a proper weight attribution to keep a given number (M) of trajectories per bin as follows. If there are more than M trajectories in one bin, trajectories to be terminated will be selected according to their weight, and the weight of the terminated trajectories will be shared among the remaining ones in the bin. If a trajectory reaches an unvisited empty bin, it will be replicated in M trajectories, and each daughter trajectory will receive 1/M of the weight of the mother trajectory. Based on this unbiased allocation of trajectory weights, a properly weighted trajectory ensemble is generated (72). Maintaining M trajectories per bin evenly distributes computing resources in configuration space in contrast to an ordinary simulation. The number of trajectories per bin and the binning scheme to divide the progress coordinate do not change the results of simulations but affect the efficiency of the WE method in sampling rare events (72).
The WESTPA software package (73) was used to manage trajectory splitting and merging. The initial state was defined as benzene bound to T4L in the crystallographic position, and the target or final state was reached when benzene had a solvent-accessible surface area (SASA) higher than 60% of its maximal SASA. This definition was enough to see benzene reaching the protein surface. But, as discussed below, this definition is not equivalent to complete unbinding as measured in experiments. Resampling intervals (τ) of 10 ps (for simulations run at 400 K) or 2 ps (for runs at 300 K) were used. The number of trajectories per bin varied from four to five according to the progress coordinate used. WE simulations were run for 4000 iterations, resulting in a maximum trajectory length of 8 ns. More details about the WE method are given in Table S2 (see Supporting Material).
WE simulations were run in a nonequilibrium steady-state scheme with trajectories being recycled back to the initial state once they reached the target state. This scheme allows estimation of transition rates from the initial to the target state (see below) (74). Estimated rate constants were expected to increase transiently in the beginning of a WE simulation after bin occupancy and later remain constant around the steady-state value after convergence was reached.
Time-windowed rate constants () were estimated from the nonequilibrium steady-state set of trajectories obtained from WE simulations (74):
| (1) |
where is the sum of probabilities or weights of the trajectories that reached the target state in the time interval to t. Transition rates from the bound to the unbound state (dissociation rate constants) were calculated using a of 2 ns. Errors for average rates were obtained from 90% confidence intervals from bootstrapping (75), calculated by resampling the rates obtained from six independent WE simulations.
Definition of progress coordinates for WE simulations
Two sets of WE simulations with different progress coordinates were carried out: an exploratory set to find possible routes for benzene unbinding and a production set to estimate rate constants and determine the protein conformational transitions involved in ligand escape for each unbinding route found.
In the exploratory simulations, two one-dimensional progress coordinates were used: the distance between benzene and binding site center-of-mass and the root mean-squared deviation (RMSD) between the current benzene position and the bound position found in the crystal structure. The binding site was defined by the Cα atoms of L84, V87, R95, A98, A99, V111, L118, N122, A129, and L133 in the artificial binding cavity. Only benzene carbon atoms were considered for RMSD calculations, and structures were previously aligned by superimposing the binding site Cαs just defined. The 12 possible symmetric images of benzene in each configuration were compared to the reference crystal structure, and the lowest RMSD value was adopted.
The exploratory progress coordinates were partitioned into small bins along coordinate regions where benzene movements were more restricted, as observed in initial simulations. Larger bins were employed along regions where benzene could diffuse more freely. Unbound states were defined by a distance or RMSD higher than 2.0 nm, in agreement with the SASA criteria described above. Simulations were run at high temperature (400 K) for 150 iterations, resulting in a maximum trajectory length of 1.5 ns. Bin boundaries and further details on the progress coordinates are given in Table S2.
Based on initial pathways found in the exploratory simulations, production simulations at 300 K were set up by using Voronoi bins (24, 33) to study each of four pathways separately. In a Voronoi mapping, a set of centers is specified and all points in configurational space are attributed to bins for which center they are closest to according to a given distance criteria. Here, a Voronoi center was manually selected as the structure of a ligand-protein complex along unbinding trajectories obtained in the exploratory set of simulations. A total of 25, 23, 24, and 26 Voronoi centers were used to sample trajectories for the paths denoted below by the colors blue, orange, pink, and cyan, respectively.
In the production set of simulations, a three-dimensional progress coordinate was employed for a subset of Voronoi bins based on two atomic pair distances. The distances were chosen to delineate small protein conformational transitions observed in the exploratory simulations, as shown in Table S1 (see Supporting Material). To avoid a combinatorial increase in the total number of bins and WE trajectories simulated, the distance dimensions were included in a nested way to only two Voronoi bins for each of the four paths.
Conformational transitions involved in ligand escape from metastable or long-lived microstates along the unbinding pathways were obtained from analysis of the transition structure observed immediately after the Voronoi bin with highest lifetime in a trajectory was unoccupied. Further details for this analysis are described in the Supporting Material.
Results
Identification of pathways for ligand escape
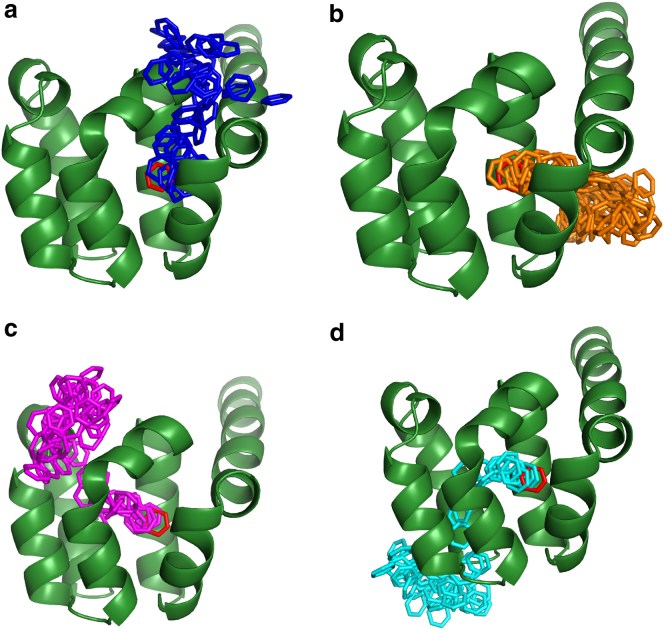
In the exploratory set of simulations run at 400 K, four exit pathways were found for benzene unbinding from T4L, as shown in Fig. 2 and denoted by color-coding. In the blue pathway, the ligand exits from the buried binding site by passing through helices C and F. Benzene mainly transits through helices C and D in the orange pathway, through helices F and I in the pink pathway, and through helices H and J in the cyan pathway.
Figure 2.
Four pathways were found for benzene exit from the buried binding site in T4 lysozyme. Blue (a), orange (b), pink (c), and cyan (d) colors show benzene positions sampled during one WE simulation. Each color represents a different exit pathway. The benzene position in the crystal structure is shown in red sticks. Only the T4L C-terminal domain is shown, but the complete protein was used in all simulations. To see this figure in color, go online.
Table 1 shows that four pathways were found when RMSD was used as the progress coordinate, whereas three of these paths were visited when the distance from the binding site was used as the progress coordinate. The four pathways found at 400 K could also be sampled at simulations performed at 300 K using a Voronoi mapping of the unbinding process.
Table 1.
Sampling of Exit Pathways by WE Simulations Performed with Distance to Cavity Center-of-Mass, Ligand RMSD, and Voronoi Bins Progress Coordinates in Different Temperatures
| Pathway | 400 K |
300 K |
||||||
|---|---|---|---|---|---|---|---|---|
| Dist | RMSD | Vblue |
Vorange |
Vpink | Vcyan | |||
| Beforea | After | Before | After | |||||
| Blue | 4 | 2 | 6 | 6 | 6 | 0 | 0 | 0 |
| Orange | 3 | 2 | 6 | 0 | 6 | 6 | 0 | 0 |
| Pink | 2 | 5 | 0 | 0 | 0 | 0 | 6 | 0 |
| Cyan | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 6 |
| Total | 4 | 5 | 6 | 6 | 6 | 6 | ||
The total refers to the total amount of WE simulations. Dist, distance to the center-of-mass; V, Voronoi bin.
Before and after post-production reassignment step, as explained in the main text.
The Voronoi bin map used as progress coordinate in the production set of simulations allowed sampling of trajectories for a specific pathway, thus facilitating the estimation of rate constants and identification of protein conformational transitions involved in ligand unbinding. Six independent WE simulations were carried out for each set of Voronoi bins defined (Table 1), leading to a total of 11,237, 16,062, 15,777, and 12,642 unbinding events through the blue, orange, pink, and cyan pathways, respectively. These events are correlated within a single WE run because of the WE procedure, and hence uncertainties are estimated based on independent WE runs.
Table 1 shows that WE simulations with a progress coordinate defined from Voronoi bins for the blue pathway also sampled unbinding events for the orange pathway and vice versa. This overlap for two different Voronoi mappings is due to a similar route followed by the ligand in the first half of the orange and blue pathways. Unbinding trajectories were reassigned to the correct pathway (blue or orange) before further analysis by adding two new Voronoi centers to the blue pathway (total of 27 centers) and one new center to the orange pathway (total of 24 centers), as detailed in the Supporting Material. Thus, the correct estimation of rate constants and detection of conformational transitions was possible for each pathway.
Protein structural transitions in long-lived microstates
Considering that small-molecule ligands such as benzene are clearly buried inside a hydrophobic cavity in T4L L99A holo crystal structures (47, 52) (Fig. 1), conformational transitions or some “breathing” of the protein structure are expected to allow ligand escape to the protein surface.
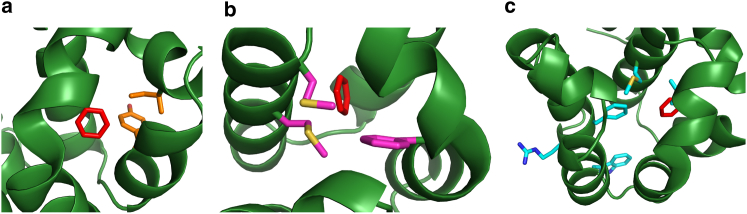
Transition structures involved in ligand escape from metastable or long-lived microstates along the unbinding pathways were analyzed. Table 2 shows fractions of unbinding trajectories with transition structures that displayed side chain motions involved in ligand unbinding. Different reference structures were used to show the calculated fractions have a small dependency on the reference. Motions in Y88 and I78 side chains are the most often observed for the blue and orange pathways. These side chains move away from the binding site, allowing the ligand to depart from the buried cavity. Motions of W126, R154, and V111 side chains in the cyan pathway are related to motions in helices H, J, and F, respectively. Motions of the other side chains listed on Table 2 allow the ligand to exit the bound state and reach the protein surface. Fig. 3 shows some of the side chains involved in exit pathways.
Table 2.
Fraction of Trajectories in WE Simulations Showing Residue Side Chain Motion Associated with Benzene Transit from the Microstate with Longest Lifetime along an Unbinding Event
| Blue |
Orange |
||||
|---|---|---|---|---|---|
| Side Chain | 100 ps | 10 ps | Side Chain | 100 ps | 10 ps |
| I78 | 0.78 | 0.67 | Y88 | 0.55 | 0.69 |
| Y88 | 0.52 | 0.48 | I78 | 0.57 | 0.49 |
| Pink |
Cyan |
||||
|---|---|---|---|---|---|
| Side Chain | 100 ps | 10 ps | Side Chain | 100 ps | 10 ps |
| F114 | 0.43 | 0.50 | M102 | 0.36 | 0.45 |
| M102 | 0.42 | 0.25 | F153 | 0.59 | 0.35 |
| M106 | 0.35 | 0.25 | R154 | 0.29 | 0.31 |
| W126 | 0.26 | 0.23 | |||
| V111 | 0.26 | 0.22 | |||
Reference structures were collected either 100 or 10 ps before the transition structure. Data obtained from six WE simulations for each pathway.
Figure 3.
Residue side chains involved in benzene unbinding. (a) Y88 and I78 (orange sticks) contribute to the blue and orange pathways. (b) F114, M102, and M106 (pink) contribute to the pink pathway. (c) M102, F153, R154, W126, and V111 (cyan) contribute to the cyan pathway. The benzene position in the crystal structure is shown in red sticks. The structure was rotated in each panel to allow a better view. To see this figure in color, go online.
Backbone fluctuation is also involved in benzene unbinding. Significant displacements in helix C in transition structures were observed in fractions of 0.35 and 0.65 of unbinding trajectories via blue and orange pathways, respectively. This helix moves away from the binding site and facilitates ligand exit from the buried cavity. The orange pathway also involved displacements of helix D in one-fifth of the transition structures. One-third of trajectories in the cyan pathway presented significant fluctuations in the backbone of helices H and J, increasing the distance between these helices and allowing benzene passage. On the other hand, only 15 and 12% of trajectories for the blue and pink pathways, respectively, presented transition structures with displacements in helix F. Fluctuations along this helix F were previously suggested as essential displacements for small molecule unbinding from T4L (58, 60, 61).
Preliminary rate constants for each unbinding pathway
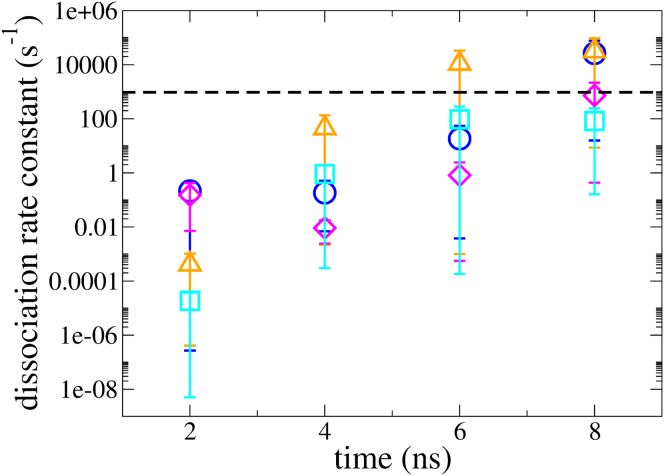
Preliminary estimates for dissociation rate constants were calculated as a function of simulation time for each exit pathway to quantify their relative contributions to the overall ligand escape rate constant, as shown in Fig. 4. However, the relative contributions of each pathway cannot be clearly distinguished, as their rank order changes over simulation time. Note that the probability fluxes, upon which the rates are estimated, are expected to increase with simulation time until they reach steady values. Additionally, these estimates are dominated by only one or two individual WE simulations (Fig. S1). Thus, further sampling appears to be required to enable confident distinctions among the pathways.
Figure 4.
Average rate constant estimated for each pathway of benzene unbinding from T4L. Symbols: blue pathway circle; orange pathway triangle; pink pathway diamond; cyan pathway square. The bars indicate a 90% confidence interval from bootstrapping. The dotted line indicates the experimental rate constant. Rates were averaged every 2 ns for 6 WE simulations. To see this figure in color, go online.
The experimental rate constant for benzene dissociation () from T4L L99A mutant has been determined (58) by lineshape analysis of protein NMR as 950 s–1. The experimental value is close to the rate calculated for the pink and cyan pathways (at 8 ns in Fig. 4) but lower than the rates estimated for the blue and orange pathways (Fig. 4). Rates calculated here from WE simulations are expected to be higher than the experimental value because a less strict definition of unbound state was used (60% of the ligand maximum SASA; see WE Simulations and Rate Estimation). The calculated rates should be corrected by the committor probability for full ligand unbinding (equivalent to 100% ligand SASA) from the unbound state defined here (6).
Discussion
Exit pathways and comparison to previous T4L studies
Four pathways were found here for benzene exit from the binding cavity buried in T4L L99A mutant. Each escape route was characterized by a preliminary rate constant and a different set of protein structural transitions.
Interestingly, a previous long timescale (30 μs) conventional MD simulation of apo T4L L99A, without sampling explicit ligand entry or exit, identified five protein tunnels that could be used by small molecules to access the buried cavity (63). Four of these tunnels, named D/F/G (tunnel through helices D, F, and G), C/D, F/G/H, and H/J, correspond respectively to the blue, orange, pink, and cyan pathways found here. An additional D/G tunnel was also identified, but the authors suggested that this D/G and the H/J tunnels would be too narrow to accommodate the passage of benzene (63). The WE simulations shown here were not able to find benzene escape through tunnel D/G but identified benzene transit via tunnel H/J (cyan pathway) with a slower rate and indeed a narrower tunnel than escape through the other pathways.
Other previous simulations also studied small-molecule (un)binding to T4L (53, 61, 62). Three routes were found for molecular O2 dissociation in conventional MD simulations (53). One route matches the cyan pathway (H/J tunnel) found here for benzene transit. The other two O2 routes correspond to the tunnel D/G mentioned above and to another tunnel found between helices D, E, G, H, and J. Both these two last tunnels were not observed here for benzene transit. Two MD simulations combined with enhanced sampling methods separately identified benzene transit from the binding site through the blue (62) and pink (61) pathways, with similar protein structural transitions to the ones observed here. To our knowledge, ligand excursion through the orange pathway was observed for the first time here.
The two previous simulations showing benzene (un)binding to T4L were able to find only one ligand transit pathway each (61, 62), probably because of the enhanced sampling procedure employed. The biasing boost potential used by Miao et al. (62) may not efficiently increase sampling of the heterogeneous processes exhibited by the full set of pathways found in our study. Moreover, sampling the binding (rather than unbinding) paths may be intrinsically more difficult under the conditions used. Wang et al. used metadynamics (76) with a path progress coordinate designed to sample the protein transitions involved in displacements of T4L residue F114 and helix F (60, 61). These transitions are associated with benzene unbinding via the pink pathway, which was thus the only ligand exit pathway they observed (61).
Taken together, these results show that small molecules such as benzene unbind from T4L L99A through multiple and kinetically competitive pathways that correspond to ligand passage through transient tunnels spontaneously opened by intrinsic fluctuations of the protein structure observed in the absence of any ligand. This is reminiscent of a conformational selection mechanism (77, 78) but for protein tunnels permitting small molecule transit. Our results also show that there is a degree of conformational diversity even within each of the four observed pathways.
It should be noted the results shown here are the first to our knowledge to find together and consistently the four pathways for small molecule exit from T4L by explicitly collecting exit trajectories instead of observing protein structural fluctuations that may lead to putative tunnels. The exploratory set of simulations presented here was carried out in 2016 without any knowledge of the pathways explored in the pink, orange, and cyan pathways. At that time, previous works (53, 61, 63) were not yet published or known to us. We were only aware of the blue path (62), although no information about it was used during our exploratory simulations. Remarkably, model composition (solvation, for instance) and force field descriptions differ significantly between our and previous studies (53, 61, 62, 63). Nevertheless, a consistent set of ligand exit tunnels and pathways was found.
Pathways for ligand entry and exit have also been investigated in other proteins containing binding sites with variable surface exposure. Multiple protein tunnels are usually available for small gaseous molecules to diffuse into binding sites. For instance, hydrogenases may react with molecular O2, and three entry routes for gaseous molecules have been characterized (12, 14, 29). On the other hand, a dominant route usually emerges for proteins with exposed binding sites and bulky ligands. This is the case for unbinding of a drug-like inhibitor from p38 MAP kinase (30). Binding to T4L lies somewhere in between and is similar to cytochrome P450, which binds organic molecules larger than gases but smaller than bulky drug-like ligands in a buried cavity via three or four different tunnels (10, 18, 19, 23, 28, 31).
Structural analysis of exit pathways
The analysis of transition structures presented in Protein Structural Transitions in Long-Lived Microstates will only find protein movements related to benzene escape from metastable microstates (or long-lived Voronoi bins). Thus, conformational transitions involved in short-lived microstates have not been analyzed. It is noticeable, however, that no specific conformational gate such as a single side chain or helix displacement is clearly dominant for each pathway. Instead, benzene exit depends on a combination of intrinsic protein motions, and these motions exhibited variability even within each (single color) pathway. Nonetheless, some side chains such as in residues Y88 and I78 (blue and orange pathways) or F114 (pink pathway) were often found to be involved in benzene escape from long-lived microstates. Thus, point mutations that exchange these residues for a less bulky side chain such as alanine could speed up benzene exit by allowing faster transit through the respective protein tunnels and dissociation pathways.
Crystallographic (47, 48, 52) and NMR (59, 60) data has shown that helix F is the most flexible element near the artificial cavity in T4L mutants. Thus, it has been suggested (49, 58, 79) that displacements along this helix are involved in ligand unbinding. The WE simulations suggest that, although observed, movements on helix F are not determinant or ubiquitous among unbinding pathways. Instead, transitions in helices C, H, and J appear to be more important for ligand transit. These motions might lead to conformational states which are too weakly populated to be detected experimentally.
Methodological limitations
Overall WE sampling led to detailed pathway information of a millisecond process based on a total aggregate simulation time of ∼30 μs. However, the large variance of rate estimates for each individual pathway (based on multiple WE runs) precluded discrimination among rates for different paths. This large variance may have been caused by high correlation among trajectories belonging to the same WE simulation, which is a consequence of the splitting and merging scheme of WE simulations to enhance sampling (36, 41). Because of correlation among trajectories, slow coordinates orthogonal to the progress coordinate may not be fully sampled, leading to different rate estimates for one single path.
The choice of coordinates and bins is always a concern in WE simulation. Based on our results, it appears that initial use of ligand RMSD as a progress coordinate in combination with high-temperature MD simulation is an efficient and reliable procedure to explore ligand exit pathways from buried cavities in proteins. Ligand distance to the binding site center-of-mass lacks information on ligand orientation and proved a less reliable progress coordinate, as one pathway was not found when this metric was used. It also should be noted that high-temperature MD simulations may generate pathways that differ from those at normal temperatures (80). The pathways found here were ultimately sampled in production runs at 300 K and are in agreement with other MD simulations performed at normal temperature and with different potential energy models (53, 61, 62).
Although WE simulation is guaranteed to yield unbiased values of the fluxes (and hence rate constants) with sufficient sampling, the relatively short (8 ns) trajectory lengths may prevent good sampling of certain types of unbinding events. All the WE unbinding trajectories are observed with a maximum length of 8 ns in our WE simulations, which is significantly shorter than the overall time that would be required in direct MD simulations. This overall time generally consists of a waiting period before a transition and the duration of the transition event itself (41). The WE approach enhances sampling in part by reducing the waiting time for the start of transitions, which is corrected by the weights. The event durations seen in WE, although short, may not be unreasonable in light of repeated observations of opening of protein tunnels during the 30 μs MD simulation of apo T4L (63).
Conclusions
Pathways for benzene exit from the buried binding cavity of T4L L99A mutant and the associated protein conformational changes were characterized here with the WE simulation method. For the first time to our knowledge, four separate ligand unbinding processes from T4L were observed in a single study. The four pathways found here are also in agreement with ligand transit routes observed in previous holo T4L simulations (53, 61, 62). Conformational transitions in several side chains, most notably Y88, I78, and F114, as well as displacements in helices C, D, F, H, and J, are involved in benzene unbinding. Our study also provided preliminary estimates of the dissociation rates for the different pathways, although further sampling is required to narrow statistical uncertainties.
This study used the WE method in two stages: an exploratory or discovery mode using naive progress coordinates to find possible pathways, followed by a production stage with more focused sampling. Combining the WE method with an RMSD progress coordinate and high temperature MD simulations appears to be a plausible approach to find multiple (un)binding pathways for small molecules. Production simulations using Voronoi bins may then be obtained to better characterize the dissociation rates and the conformational transitions involved in ligand transit.
The four protein pathways found for benzene exit may also be used for larger binders such as meta-xylene and N-butylbenzene (49). Similarly, it is also worth investigating if hydrophobic molecules such as 1,3,5-trimethylbenzene, which could still fit inside the binding cavity, experimentally do not bind T4L L99A mutant (49) because of a very slow entry kinetics (57). Such bulkier molecules may require wider protein tunnels to access the binding cavity, which WE simulation could help to rule out if extremely low rates were found.
Author Contributions
A.N.-A. performed research and wrote the manuscript. D.M.Z. designed research, analyzed data, and revised the manuscript. G.M.A. designed research, analyzed data, and wrote the manuscript.
Acknowledgments
A.N.-A. acknowledges Lillian Chong, Adam Pratt, Rory Donovan, and Ernesto Suarez for discussions and technical help during a recent visit to the University of Pittsburgh, and Lillian Chong and Murilo Teixeira (Universidade De São Paulo) for suggestions on the manuscript.
Grants from São Paulo Research Foundation (FAPESP projects 2014/17008-7, 2014/21900-2, 2015/19912-5, and 2016/24096-5) and from the National Institutes of Health (GM115805) are gratefully acknowledged. Computing resources were in part provided by the University of Pittsburgh Center for Research Computing.
Editor: Markus Buehler.
Footnotes
Supporting Materials and Methods, three figures, two tables, and one data file are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30140-1.
Contributor Information
Daniel M. Zuckerman, Email: zuckermd@ohsu.edu.
Guilherme Menegon Arantes, Email: garantes@iq.usp.br.
Supporting Citations
Reference (81) appears in the Supporting Material.
Supporting Material
References
- 1.Copeland R.A., Pompliano D.L., Meek T.D. Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 2.Copeland R.A. The drug-target residence time model: a 10-year retrospective. Nat. Rev. Drug Discov. 2016;15:87–95. doi: 10.1038/nrd.2015.18. [DOI] [PubMed] [Google Scholar]
- 3.Schuetz D.A., de Witte W.E.A., Ecker G.F. Kinetics for drug discovery: an industry-driven effort to target drug residence time. Drug Discov. Today. 2017;22:896–911. doi: 10.1016/j.drudis.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Tang Z., Roberts C.C., Chang C.A. Understanding ligand-receptor non-covalent binding kinetics using molecular modeling. Front. Biosci. 2017;22:960–981. doi: 10.2741/4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dellago C., Bolhuis P.G., Chandler D. Efficient transition path sampling: application to Lennard-Jones cluster rearrangements. J. Chem. Phys. 1998;108:9236–9245. [Google Scholar]
- 6.Bolhuis P.G., Chandler D., Geissler P.L. Transition path sampling: throwing ropes over rough mountain passes, in the dark. Annu. Rev. Phys. Chem. 2002;53:291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 7.Singhal N., Snow C.D., Pande V.S. Using path sampling to build better Markovian state models: predicting the folding rate and mechanism of a tryptophan zipper beta hairpin. J. Chem. Phys. 2004;121:415–425. doi: 10.1063/1.1738647. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B.W., Jasnow D., Zuckerman D.M. Efficient and verified simulation of a path ensemble for conformational change in a united-residue model of calmodulin. Proc. Natl. Acad. Sci. USA. 2007;104:18043–18048. doi: 10.1073/pnas.0706349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatt D., Zuckerman D.M. Beyond microscopic reversibility: are observable non-equilibrium processes precisely reversible? J. Chem. Theory Comput. 2011;7:2520–2527. doi: 10.1021/ct200086k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winn P.J., Lüdemann S.K., Wade R.C. Comparison of the dynamics of substrate access channels in three cytochrome P450s reveals different opening mechanisms and a novel functional role for a buried arginine. Proc. Natl. Acad. Sci. USA. 2002;99:5361–5366. doi: 10.1073/pnas.082522999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martínez L., Sonoda M.T., Polikarpov I. Molecular dynamics simulations reveal multiple pathways of ligand dissociation from thyroid hormone receptors. Biophys. J. 2005;89:2011–2023. doi: 10.1529/biophysj.105.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen J., Kim K., Schulten K. Finding gas diffusion pathways in proteins: application to O2 and H2 transport in CpI [FeFe]-hydrogenase and the role of packing defects. Structure. 2005;13:1321–1329. doi: 10.1016/j.str.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Buch I., Giorgino T., De Fabritiis G. Complete reconstruction of an enzyme-inhibitor binding process by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2011;108:10184–10189. doi: 10.1073/pnas.1103547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P.H., Best R.B., Blumberger J. Multiscale simulation reveals multiple pathways for H2 and O2 transport in a [NiFe]-hydrogenase. J. Am. Chem. Soc. 2011;133:3548–3556. doi: 10.1021/ja109712q. [DOI] [PubMed] [Google Scholar]
- 15.Shan Y., Kim E.T., Shaw D.E. How does a drug molecule find its target binding site? J. Am. Chem. Soc. 2011;133:9181–9183. doi: 10.1021/ja202726y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang D., Caflisch A. The free energy landscape of small molecule unbinding. PLoS Comput. Biol. 2011;7:e1002002. doi: 10.1371/journal.pcbi.1002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang M., Roberts C., Chang C.E. Gating and intermolecular interactions in ligand-protein association: coarse-grained modeling of HIV-1 protease. J. Chem. Theory Comput. 2011;7:3438–3446. doi: 10.1021/ct2004885. [DOI] [PubMed] [Google Scholar]
- 18.Cojocaru V., Winn P.J., Wade R.C. Multiple, ligand-dependent routes from the active site of cytochrome P450 2C9. Curr. Drug Metab. 2012;13:143–154. doi: 10.2174/138920012798918462. [DOI] [PubMed] [Google Scholar]
- 19.Yu X., Cojocaru V., Wade R.C. Conformational diversity and ligand tunnels of mammalian cytochrome P450s. Biotechnol. Appl. Biochem. 2013;60:134–145. doi: 10.1002/bab.1074. [DOI] [PubMed] [Google Scholar]
- 20.Bisha I., Rodriguez A., Magistrato A. Metadynamics simulations reveal a Na+ independent exiting path of galactose for the inward-facing conformation of vSGLT. PLoS Comput. Biol. 2014;10:e1004017. doi: 10.1371/journal.pcbi.1004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y.M., Kang M., Chang C.E. Switches of hydrogen bonds during ligand-protein association processes determine binding kinetics. J. Mol. Recognit. 2014;27:537–548. doi: 10.1002/jmr.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiwary P., Limongelli V., Parrinello M. Kinetics of protein-ligand unbinding: predicting pathways, rates, and rate-limiting steps. Proc. Natl. Acad. Sci. USA. 2015;112:E386–E391. doi: 10.1073/pnas.1424461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu X., Nandekar P., Wade R.C. Ligand tunnels in T. brucei and human CYP51: insights for parasite-specific drug design. Biochim. Biophys. Acta. 2016;1860:67–78. doi: 10.1016/j.bbagen.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickson A., Lotz S.D. Ligand release pathways obtained with WExplore: residence times and mechanisms. J. Phys. Chem. B. 2016;120:5377–5385. doi: 10.1021/acs.jpcb.6b04012. [DOI] [PubMed] [Google Scholar]
- 25.Teo I., Mayne C.G., Lelièvre T. Adaptive multilevel splitting method for molecular dynamics calculation of benzamidine-trypsin dissociation time. J. Chem. Theory Comput. 2016;12:2983–2989. doi: 10.1021/acs.jctc.6b00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paloncýová M., Navrátilová V., Otyepka M. Role of enzyme flexibility in ligand access and egress to active site: bias-exchange metadynamics study of 1,3,7-trimethyluric acid in cytochrome P450 3A4. J. Chem. Theory Comput. 2016;12:2101–2109. doi: 10.1021/acs.jctc.6b00075. [DOI] [PubMed] [Google Scholar]
- 27.Rydzewski J., Nowak W. Ligand diffusion in proteins via enhanced sampling in molecular dynamics. Phys. Life Rev. 2017;22–23:58–74. doi: 10.1016/j.plrev.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Rydzewski J., Nowak W. Thermodynamics of camphor migration in cytochrome P450cam by atomistic simulations. Sci. Rep. 2017;7:7736. doi: 10.1038/s41598-017-07993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubas A., Orain C., Léger C. Mechanism of O2 diffusion and reduction in FeFe hydrogenases. Nat. Chem. 2017;9:88–95. doi: 10.1038/nchem.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casasnovas R., Limongelli V., Parrinello M. Unbinding kinetics of a p38 MAP kinase type II inhibitor from metadynamics simulations. J. Am. Chem. Soc. 2017;139:4780–4788. doi: 10.1021/jacs.6b12950. [DOI] [PubMed] [Google Scholar]
- 31.Magistrato A., Sgrignani J., Cavalli A. Single or multiple access channels to the CYP450s active site? An answer from free energy simulations of the human aromatase enzyme. J. Phys. Chem. Lett. 2017;8:2036–2042. doi: 10.1021/acs.jpclett.7b00697. [DOI] [PubMed] [Google Scholar]
- 32.Tiwary P., Mondal J., Berne B.J. How and when does an anticancer drug leave its binding site? Sci. Adv. 2017;3:e1700014. doi: 10.1126/sciadv.1700014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickson A., Lotz S.D. Multiple ligand unbinding pathways and ligand-induced destabilization revealed by WExplore. Biophys. J. 2017;112:620–629. doi: 10.1016/j.bpj.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y.M., Raymundo M.A.V., Chang C.A. Mechanism of the association pathways for a pair of fast and slow binding ligands of HIV-1 protease. Biochemistry. 2017;56:1311–1323. doi: 10.1021/acs.biochem.6b01112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huber G.A., Kim S. Weighted-ensemble Brownian dynamics simulations for protein association reactions. Biophys. J. 1996;70:97–110. doi: 10.1016/S0006-3495(96)79552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuckerman D.M., Chong L.T. Weighted ensemble simulation: review of methodology, applications, and software. Annu. Rev. Biophys. 2017;46:43–57. doi: 10.1146/annurev-biophys-070816-033834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faradjian A.K., Elber R. Computing time scales from reaction coordinates by milestoning. J. Chem. Phys. 2004;120:10880–10889. doi: 10.1063/1.1738640. [DOI] [PubMed] [Google Scholar]
- 38.Votapka L.W., Amaro R.E. Multiscale estimation of binding kinetics using Brownian dynamics, molecular dynamics and milestoning. PLoS Comput. Biol. 2015;11:e1004381. doi: 10.1371/journal.pcbi.1004381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Erp T.S., Moroni D., Bolhuis P.G. A novel path sampling method for the calculation of rate constants. J. Chem. Phys. 2003;118:7762–7774. [Google Scholar]
- 40.Zuckerman D.M. Equilibrium sampling in biomolecular simulations. Annu. Rev. Biophys. 2011;40:41–62. doi: 10.1146/annurev-biophys-042910-155255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chong L.T., Saglam A.S., Zuckerman D.M. Path-sampling strategies for simulating rare events in biomolecular systems. Curr. Opin. Struct. Biol. 2017;43:88–94. doi: 10.1016/j.sbi.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhatt D., Zuckerman D.M. Heterogeneous path ensembles for conformational transitions in semi-atomistic models of adenylate kinase. J. Chem. Theory Comput. 2010;6:3527–3539. doi: 10.1021/ct100406t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adelman J.L., Dale A.L., Grabe M. Simulations of the alternating access mechanism of the sodium symporter Mhp1. Biophys. J. 2011;101:2399–2407. doi: 10.1016/j.bpj.2011.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suárez E., Lettieri S., Zuckerman D.M. Simultaneous computation of dynamical and equilibrium information using a weighted ensemble of trajectories. J. Chem. Theory Comput. 2014;10:2658–2667. doi: 10.1021/ct401065r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zwier M.C., Kaus J.W., Chong L.T. Efficient explicit–solvent molecular dynamics simulations of molecular association kinetics: methane/methane, Na(+)/Cl(-), methane/benzene, and K(+)/18–crown–6 ether. J. Chem. Theory Comput. 2011;7:1189–1197. doi: 10.1021/ct100626x. [DOI] [PubMed] [Google Scholar]
- 46.Zwier M.C., Pratt A.J., Chong L.T. Efficient atomistic simulation of pathways and calculation of rate constants for a protein-peptide binding process: application to the MDM2 protein and an intrinsically disordered p53 peptide. J. Phys. Chem. Lett. 2016;7:3440–3445. doi: 10.1021/acs.jpclett.6b01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eriksson A.E., Baase W.A., Matthews B.W. A cavity-containing mutant of T4 lysozyme is stabilized by buried benzene. Nature. 1992;355:371–373. doi: 10.1038/355371a0. [DOI] [PubMed] [Google Scholar]
- 48.Morton A., Matthews B.W. Specificity of ligand binding in a buried nonpolar cavity of T4 lysozyme: linkage of dynamics and structural plasticity. Biochemistry. 1995;34:8576–8588. doi: 10.1021/bi00027a007. [DOI] [PubMed] [Google Scholar]
- 49.Baase W.A., Liu L., Matthews B.W. Lessons from the lysozyme of phage T4. Protein Sci. 2010;19:631–641. doi: 10.1002/pro.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morton A., Baase W.A., Matthews B.W. Energetic origins of specificity of ligand binding in an interior nonpolar cavity of T4 lysozyme. Biochemistry. 1995;34:8564–8575. doi: 10.1021/bi00027a006. [DOI] [PubMed] [Google Scholar]
- 51.Mulder F.A.A., Hon B., Kay L.E. Flexibility and ligand exchange in a buried cavity mutant of T4 lysozyme studied by multinuclear NMR. Biochemistry. 2000;39:12614–12622. doi: 10.1021/bi001351t. [DOI] [PubMed] [Google Scholar]
- 52.Merski M., Fischer M., Shoichet B.K. Homologous ligands accommodated by discrete conformations of a buried cavity. Proc. Natl. Acad. Sci. USA. 2015;112:5039–5044. doi: 10.1073/pnas.1500806112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitahara R., Yoshimura Y., Mulder F.A. Detecting O2 binding sites in protein cavities. Sci. Rep. 2016;6:20534. doi: 10.1038/srep20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei B.Q., Baase W.A., Shoichet B.K. A model binding site for testing scoring functions in molecular docking. J. Mol. Biol. 2002;322:339–355. doi: 10.1016/s0022-2836(02)00777-5. [DOI] [PubMed] [Google Scholar]
- 55.Mobley D.L., Graves A.P., Dill K.A. Predicting absolute ligand binding free energies to a simple model site. J. Mol. Biol. 2007;371:1118–1134. doi: 10.1016/j.jmb.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang K., Chodera J.D., Shirts M.R. Identifying ligand binding sites and poses using GPU-accelerated Hamiltonian replica exchange molecular dynamics. J. Comput. Aided Mol. Des. 2013;27:989–1007. doi: 10.1007/s10822-013-9689-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nunes-Alves A., Arantes G.M. Ligand-receptor affinities computed by an adapted linear interaction model for continuum electrostatics and by protein conformational averaging. J. Chem. Inf. Model. 2014;54:2309–2319. doi: 10.1021/ci500301s. [DOI] [PubMed] [Google Scholar]
- 58.Feher V.A., Baldwin E.P., Dahlquist F.W. Access of ligands to cavities within the core of a protein is rapid. Nat. Struct. Biol. 1996;3:516–521. doi: 10.1038/nsb0696-516. [DOI] [PubMed] [Google Scholar]
- 59.Mulder F.A.A., Mittermaier A., Kay L.E. Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 2001;8:932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- 60.Bouvignies G., Vallurupalli P., Kay L.E. Solution structure of a minor and transiently formed state of a T4 lysozyme mutant. Nature. 2011;477:111–114. doi: 10.1038/nature10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y., Papaleo E., Lindorff-Larsen K. Mapping transiently formed and sparsely populated conformations on a complex energy landscape. eLife. 2016;5:e17505. doi: 10.7554/eLife.17505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miao Y., Feher V.A., McCammon J.A. Gaussian accelerated molecular dynamics: unconstrained enhanced sampling and free energy calculation. J. Chem. Theory Comput. 2015;11:3584–3595. doi: 10.1021/acs.jctc.5b00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schiffer J.M., Feher V.A., Amaro R.E. Capturing invisible motions in the transition from ground to rare excited states of T4 lysozyme L99A. Biophys. J. 2016;111:1631–1640. doi: 10.1016/j.bpj.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 66.MacKerell A.D., Jr., Feig M., Brooks C.L., III Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 67.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Still W.C., Tempczyk A., Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 1990;112:6127–6129. [Google Scholar]
- 69.Onufriev A., Bashford D., Case D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 70.Schaefer M., Bartels C., Karplus M. Solution conformations and thermodynamics of structured peptides: molecular dynamics simulation with an implicit solvation model. J. Mol. Biol. 1998;284:835–848. doi: 10.1006/jmbi.1998.2172. [DOI] [PubMed] [Google Scholar]
- 71.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 72.Zhang B.W., Jasnow D., Zuckerman D.M. The “weighted ensemble” path sampling method is statistically exact for a broad class of stochastic processes and binning procedures. J. Chem. Phys. 2010;132:054107. doi: 10.1063/1.3306345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zwier M.C., Adelman J.L., Chong L.T. WESTPA: an interoperable, highly scalable software package for weighted ensemble simulation and analysis. J. Chem. Theory Comput. 2015;11:800–809. doi: 10.1021/ct5010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhatt D., Zhang B.W., Zuckerman D.M. Steady-state simulations using weighted ensemble path sampling. J. Chem. Phys. 2010;133:014110. doi: 10.1063/1.3456985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Efron B., Tibshirani R.J. First Edition. Chapman & Hall/CRC; Boca Raton, Florida: 1993. An Introduction to the Bootstrap. [Google Scholar]
- 76.Laio A., Parrinello M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weikl T.R., Paul F. Conformational selection in protein binding and function. Protein Sci. 2014;23:1508–1518. doi: 10.1002/pro.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gianni S., Dogan J., Jemth P. Distinguishing induced fit from conformational selection. Biophys. Chem. 2014;189:33–39. doi: 10.1016/j.bpc.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 79.Vallurupalli P., Chakrabarti N., Kay L.E. Atomistic picture of conformational exchange in a T4 lysozyme cavity mutant: an experiment-guided molecular dynamics study. Chem. Sci. 2016;7:3602–3613. doi: 10.1039/c5sc03886c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang, B. W.; Jasnow, D.; Zuckerman, D. M. 2009, Weighted ensemble path sampling for multiple reaction channels. arXiv, arXiv:0902.2772, https://arxiv.org/abs/0902.2772.
- 81.Roux B. The calculation of the potential of mean force using computer simulations. Comput. Phys. Commun. 1995;91:275–282. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.