Abstract
Background
Marek’s disease virus (MDV) resides in the genus Mardivirus in the family Herpesviridae. MDV is a highly contagious virus that can cause neurological lesions, lymphocytic proliferation, immune suppression, and death in avian species, including Galliformes (chickens, quails, partridges, and pheasants), Strigiformes (owls), Anseriformes (ducks, geese, and swans), and Falconiformes (kestrels).
Case presentation
In 2015, two red-crowned cranes died in Nanjing (Jiangsu, China). It was determined that the birds were infected with Marek’s disease virus by histopathological examination, polymerase chain reaction (PCR), gene sequencing and sequence analysis of tissue samples from two cranes. Gross lesions included diffuse nodules in the skin, muscle, liver, spleen, kidney, gizzard and heart, along with liver enlargement and gizzard mucosa hemorrhage. Histopathological assay showed that infiltrative lymphocytes and mitotic figures existed in liver and heart. The presence of MDV was confirmed by PCR. The sequence analysis of the Meq gene showed 100% identity with Md5, while the VP22 gene showed the highest homology with CVI988. Furthermore, the phylogenetic analysis of the VP22 and Meq genes suggested that the MDV (from cranes) belongs to MDV serotype 1.
Conclusion
We describe the first molecular detection of Marek’s disease in red-crowned cranes based on the findings previously described. To our knowledge, this is also the first molecular identification of Marek’s disease virus in the order Gruiformes and represents detection of a novel MDV strain.
Keywords: Marek’s disease virus, Red-crowned crane, Clinical necropsy, PCR, Homology
Background
The red-crowned crane (Grus japonensis) is classified as an endangered species with a small global population of 1830 mature individuals [1]. The red-crowned crane breeds in south-eastern Russia, north-east China, Mongolia, and eastern Hokkaido, Japan [2]. The Russian and Chinese populations mainly migrate to the Yellow River Delta and the coast of Jiangsu province, China, and the demilitarized zone of North Korea/South Korea in winter [3]. The number of over wintering cranes in China is now only 8% of what it was in the 1980s due to habitat degradation [4]. This also leads to the overconcentration of cranes at a few sites, which therefore decrease the genetic diversity [5]. The small population and low genetic diversity make this species especially vulnerable to epidemic diseases.
Marek’s disease (MD) is a highly contagious disease, characterized by immunosuppression, neurological disorder, CD4+ T cells transformation and eventual tumor formation in peripheral nerves and visceral organs [6]. The causative agent of MD is Marek’s disease virus (MDV) which is a member of the genus Mardivirus belonging to the subfamily Alphaherpesvirinae of the family Herpesviridae. MD was first reported in chicken by Josef Marek in 1907 [7], and is prevalent throughout the world. Later, it was also reported in Galliformes (such as turkeys [8], quails [9] and pheasants [10]), Strigiformes (owls), Anseriformes (ducks, geese and swans) and Falconiformes (kestrels). The most important natural hosts for MDV are domestic and wild chicken, including game fowl [11], native breeds [12] and jungle fowl [13]. MDV contains three serotypes: MDV-1 (GaHV-2), MDV-2 (GaHV-3) and MDV-3 (MeHV-1) [14]. However, only MDV-1 can induce disease in chicken, whereas MDV-2 and MDV-3 are avirulent and are used as vaccines. Symptoms of MD depend on the age of the bird [15, 16]. For example, young chickens infected with virulent MDV strains may exhibit high mortality in 8–16 days post infection with early mortality syndrome [17]. MDV can be transmitted horizontally through aerosols and enter the host through the respiratory tract, but it cannot be transmitted vertically from chicken to eggs [18]. The infectious virus particles replicate in epithelial cells of the feather follicles, and then are shed to the environment with the skin dander [19]. MDV in the dust survives for at least several months at room temperature [20]. Thus, skin dander, house dust, feces, and saliva can be a source of infection. Because cross-species spreading of the virus is possible, this mode of transmission enables it to infect an even broader spectrum of hosts [21]. Despite several cases of herpesvirus infection in cranes that were previously reported [22, 23], there is a lack of molecular evidence on MDV infection and knowledge of which species are infected.
This article reports the molecular detection of a novel MDV isolate in the red-crowned crane. It is also the first molecular determination of MDV from individual red-crowned cranes (Grus japonensis), indicating that red-crowned cranes can be infected by MDV.
Case presentation
In August 2015, reduced feed consumption was observed in a 2-month-old red-crowned crane (crane A) that was bred and incubated in Hongshan Forest Zoo in Nanjing, China. After segregation and palliative treatment, it exhibited astasia and then died. Subcutaneous palpation examination revealed many diffuse soybean-sized nodules (Table 1). Red-crowned crane B showed obvious swelling of its left knee joint. After antibiotic treatment, although it showed an improved appetite and a decrease in symptoms, the crane died unexpectedly ten days later (Table 1). The dead cranes were not immunized with the MDV vaccine. Although there had been no previous reports of MDV-infected cranes, the cranes were fed in mesh cages that were accessible to wild birds.
Table 1.
Timeline table of the information in the case report
| Date | Clinical Observation | Intervention | Samples Collected |
|---|---|---|---|
| Aug 19-20 | Red-crowned crane Aa: emaciation, reduced appetite. Diagnostic check: Heart rate: 106 bp/min Body temperature: 39.7°C Weight: 1.6kg; Crane Aa: astasia. |
Rocephin (100 mg/kg) + Dexamethasone (0.25 mg/kg) + 0.9% NaCl 15 ml (intravenous infusion once daily for 3 days) Vitamin C (100 mg/kg) + Vitamin B1(15 mg/kg) + ATP (5 mg/kg) + Inosine (15 mg/kg) + coenzyme A (15 IU/kg) + 10% glucose 15 ml (intravenous infusion once daily for 3 days) 10% glucose + mixture of fish and corn, orally, for 3 days Calcium gluconate (25 mg/kg) + 10% glucose 15 ml (intravenous infusion once daily), for 1 day |
Crane Aa: •Histopathological examination of liver, heart, and spleen. Three repeated samples were collected from each tissue. The ratio of MDV-positive tissues: Heart: 67%, liver: 100%, spleen: 67%. •RNA extraction of feather, liver, spleen, kidney and muscle. Three of each. Positive ratio of all samples was 100%. •Genome extraction of muscle, feather, feather-pulp and spleen; three of each. The positive ratio of all samples was 100%. Crane Bb: same as crane A. |
| Aug 21 | Diagnostic check: Heart rate: 156 bp/min Body temperature: 39.8°C Crane Aa died. |
||
| Aug 22 | Clinical necropsy: Nodules found in skin, muscle, liver, heart, trachea, and gizzard. (MD suspected). |
||
| Sept 11 | Crane Bb: obvious swelling of its left knee joint. |
Musk analgesic aerosol, external use | |
| Sept 12-17 | Clinical improvement: Recovered appetite, left knee joint swelled | Musk analgesic aerosol + ichthammol ointment, external use Calcium tablets (10 mg/kg) + Vitamin D2(500 IU/kg) + Vitamin A (1500 IU/kg), orally |
|
| Sept 18-19 | Relapse, anorexia, depression | ||
| Sept 20 | Crane Bb died. Clinical necropsy: Inflammatory exudates; diffuse yellow-white nodules in muscle, liver, spleen, kidney, pancreas, gizzard, and heart. |
aCrane A: hatched on June 10, died on Aug 22, 2015. Body weight: 1.6 kg
bCrane B: hatched on June 9, died on Sep 22. Body weight: 2.4 kg
Clinical necropsy revealed diffuse yellow-white nodules from millet to soybean size in the skin, muscle, trachea, liver, gizzard, and heart in both cranes (Fig. 1a-f). Diffused yellow-white nodules were also observed in the vertical section of the liver (Fig. 1g). Several hemorrhage sites were observed in the gizzard mucosa (Fig. 1h). A large amount of inflammatory exudates oozed from a surface cut of the swollen joint (Fig. 1i). No other significant gross lesions were observed.
Fig. 1.
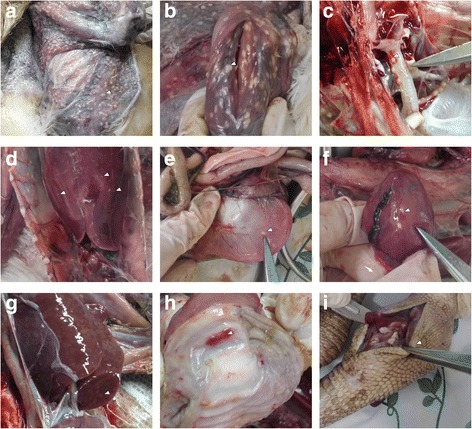
Clinical symptoms and pathological lesions. a-f, Nodules in the (a) skin, (b) muscle, (c) trachea, (d) liver, (e) gizzard, (f) heart; g, nodules in the vertical section of the liver; h, hemorrhage sites in the gizzard mucosa; i, left knee joint swelling
Representative tissue samples were collected during necropsy from the liver, spleen, kidney, muscle, heart, feather follicles, and skin, and these were used for pathological and virological investigations (Table 1). Two sets of tissue samples were collected for different purposes, and were processed in different ways: one set was used for virology analysis and stored at − 80 °C and the other was used for conventional histopathology analysis and fixed in 10% neutral formalin. Formalin-fixed samples were subsequently dehydrated through graded alcohols before being embedded in paraffin wax. Several 4 μm-thick sections were cut from each sample and stained using hematoxylin and eosin (H&E). The heart histology showed that numerous lymphoid cells had infiltrated between adjacent cardiac muscle fibers. We also found that there were various polymorphic lymphoid cells with mitotic figures (Fig. 2a-b). Similarly, the liver structure was disrupted, and a large number of variably sized lymphoblastic cells was dispersed or exhibited mass distribution in the hepatic lobules (Fig. 2c-d). These findings were consistent with the symptoms of MDV infection. However, bacterial infection cannot be excluded from this case, as swollen joints could be a consequence of bacterial infection. It is possible that the birds were infected with MDV, which is immunosuppressive, but succumbed to other conditions such as bacterial superinfection.
Fig. 2.
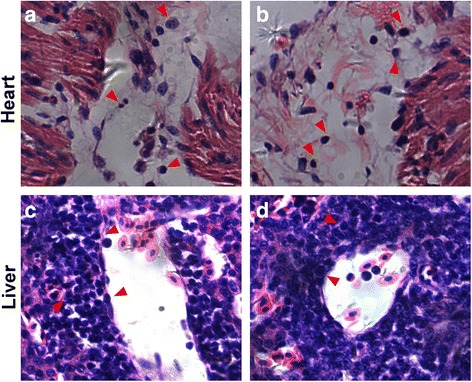
Histopathological section (H&E staining) of the liver and heart. a and b, lymphomatous infiltration in the liver; c and d, lymphomatous infiltration in the heart. The arrow shows a range of leukocytes, including large lymphocytes, small lymphocytes lymphoblasts, and malignant cells with mitotic figure.
For virological detection, frozen tissue samples of feather, liver, spleen, kidney, and muscle were homogenized using a dispersing homogenizer (IKA Ultra-Turrax, T10, Germany). Total RNA was extracted using TRIzol reagent (Sigma, USA) according to the manufacturer’s protocol. The cDNA was prepared using an iScript™ Reverse Transcription Supermix kit (Bio-Rad, USA). The PCR was carried out with 2× Taq Master Mix (Vazyme, China) with the following conditions: denaturation at 94 °C for 5 min; 28 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30s; final extension at 72 °C for 7 min; and maintenance at 4 °C. To optimize the PCR conditions, primers were modified on the basis of Yamaguchi et al. [24]. The Meq primer used herein amplified the DNA 10 bp longer than published primer, due to the forward primer being located 10 bp upstream from the published forward primer. For example, the sequence of the published primer is 5’-GGTCGACTTCGAGACGGAAA-3′, while here, we used 5’-CTTTCTCTCGGGTCGACTTC-3′. The underlined parts are the overlapped sequence. Our gB primers can amplify shorter sequences, but still within the same region compared with the published one. The amplified fragments were resolved by electrophoresis in 1.5% agarose gels (Sigma, USA). Here, RT-PCR was used to determine the viral gene expression. Meq mRNA was examined to measure general mRNA expression regardless of the phase of infection, and gB mRNA was examined to indicate virus lytic replication. Samples were analyzed for the presence of MDV and the reactions were positive for all sampled tissues when amplified with both Meq (Fig. 3a) and gB (Fig. 3b) primers [24] in both crane A (Fig. 3a, b) and crane B (Fig. 3c, d).
Fig. 3.
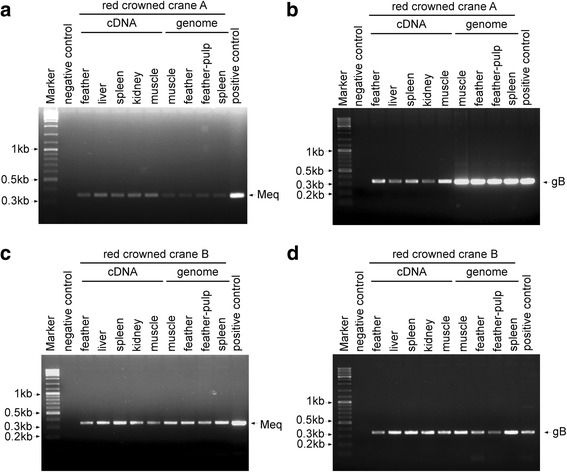
Agarose gel electrophoresis of MDV from RT-PCR of the Meq and gB genes. a and c, Meq (347 bp, partial) was amplified from cDNAs and genomes of different tissues. b and d, gB (338 bp, partial) was amplified from cDNAs and genomes of different tissues. The feather sample was a homogenate of ground feathers and skin that contained feather follicle epithelium. The entire cell genome from Md5-infected CEF cells was used as a positive control. The cDNA from CEF cells was used as a negative control
To further characterize the virus, samples from spleen, skin, muscle, feather and feather pulp were cut and digested overnight at 55 °C in lysis buffer (10 mM Tris, 100 mM EDTA, 0.5% SDS, and 0.2 mg/ml Proteinase K), and total cellular DNA was extracted with phenol-chloroform-isoamylalcohol (25:24:1), then precipitated with ethanol, and dissolved in TE buffer (10 mM Tris-HCl 1 mM EDTA pH 8.0). Total cellular DNA samples were used as templates for amplifying viral genes. The PCR was carried out with LA Taq DNA polymerase (Takara, Japan). Primers were designed to target the entire-length of the Meq and VP22 genes of Md5 (Table 2). The Meq gene was successfully amplified from the spleen tissue (Fig. 4a). The sequence alignment revealed that the analyzed fragment presented 100% sequence identity with reference MDV strains such as Md5 and Md11 (Fig. 4b and c). It also shared a high homology (97.9%) to the very virulent strain LMS from China (Fig. 4b). An additional PCR was carried out to amplify the VP22 gene, which is a major structural component of the virion [25] (Fig. 5a). The obtained band was sequenced and aligned using DNAMAN software. The sequence alignment showed that the VP22 protein shared the highest identity with the vaccine strain CVI988 as compared to other strains (Fig. 5b). Two substitutions at residues 152 (A → P), 155 (S → C) were found in CVI988, while 2–3 substitutions were found when aligned with other eight isolates. The same as CVI988, six amino acids (TKSERT) were deleted from residues 201 to 206 compared with other isolates (Fig. 5b). The deleted site was located in the domain (SKSERTTKSERT), which contained two copies of motif (KSERT) in virulent MDV strains. It has been shown that regardless of the deletion, the VP22 of the CVI988/Rispens vaccine strain still maintains an intercellular-trafficking function [26].
Table 2.
Primers used for amplification of MDV viral genes
| Name | Sequence | Length |
|---|---|---|
| Meq-RT-F | CTTTCTCTCGGGTCGACTTC | 347 bp |
| Meq-RT-R | GTAAGCAGTCCAAGGGTCAC | |
| gB-RT-F | CTTCACAGTTGGGTGGGAC | 338 bp |
| gB-RT-R | GAGCCAGGGATTTGGATAG | |
| MDV-Meq-F | AGAGATGTCTCAGGAGCCAGAGCC | 1020 bp |
| MDV-Meq-R | ATCATCAGGGTCTCCCGTCACCTG | |
| MDV-VP22-F | ATCGGATCCATGGGGGATTCTGAAAG | 750 bp |
| MDV-VP22-R | ACACTCGAGTTATTCGCTATCACTGC |
Fig. 4.
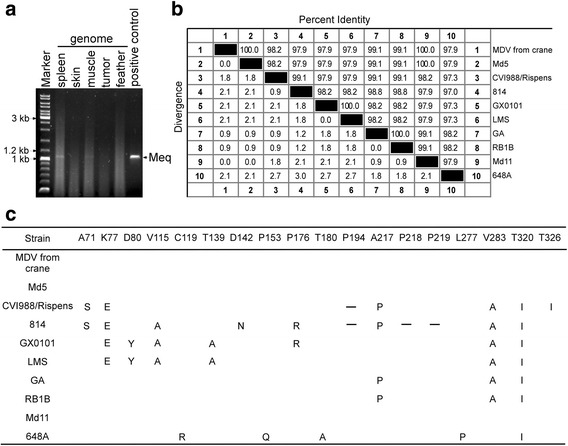
Amino acid sequence alignment of Meq (from crane). a, The full-length Meq gene (1020 bp) was amplified from the spleen genome. b, The amino acid (aa) sequences of Meq (339 aa) from cranes were aligned with 9 previously published MDV isolates (including CVI988, 814, GX0101, LMS, GA, RB-1B, Md11 and 648a). c, Substituted amino acids are listed, while deleted amino acids are denoted by strips in the alignment
Fig. 5.
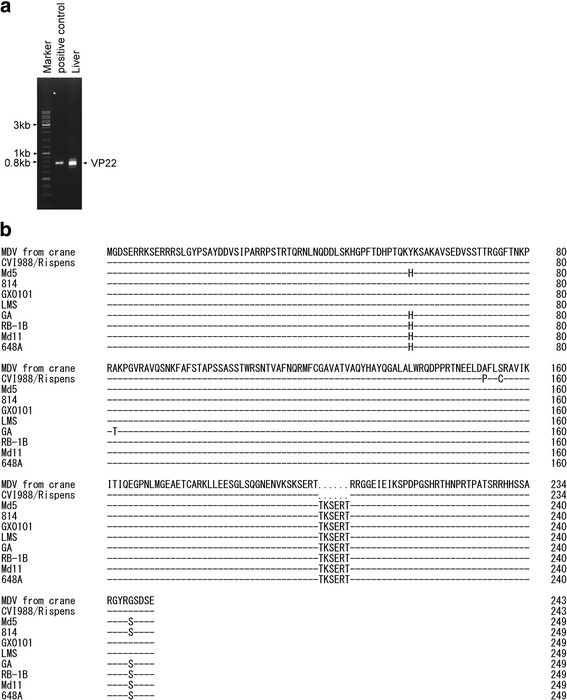
Amino acid sequence alignment of VP22 (from crane). a, The full-length VP22 gene (750 bp) was amplified from the liver genome. b, The amino acid sequence of VP22 (243 aa) from cranes was aligned with 9 previously published MDV isolates (including CVI988, 814, GX0101, LMS, GA, RB-1B, Md11 and 648a). Identical amino acids are denoted by strips in the alignment, and deleted amino acids are denoted by dots in the alignment
Phylogenetic analysis of the Meq gene showed that our MDV clustered with very virulent strain and was most closely related to Md5, Md11 and W (Fig. 6a). However, phylogenetic analysis of the VP22 gene showed that our MDV was closely related with attenuated virulent strain CVI988 (Fig. 6b).
Fig. 6.
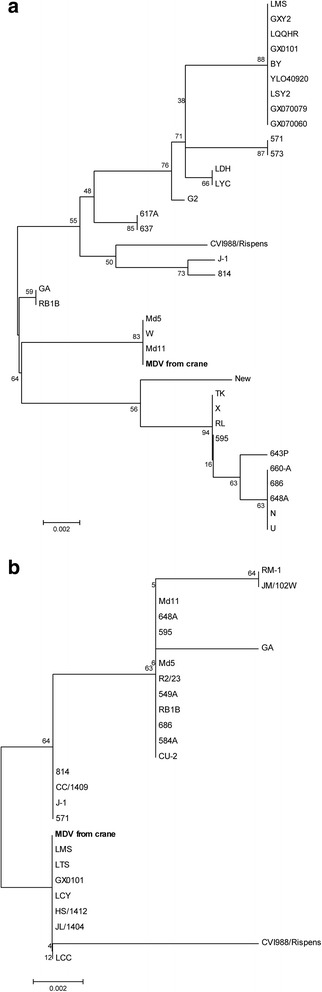
Phylogenetic profile showing the relationships among MDV isolates based on a comparison of the (a) Meq gene and (b) VP22 gene. Phylogenetic analysis of MDV based on (a) Meq and (b) VP22 amino acid sequences. The tree was constructed using the neighbor-joining (N-J) analysis method in the MEGA 5.0 program with bootstrapping (1000)
The Meq gene is only present in MDV1m and its gene product, MEQ, which is an MDV1-specific 339 amino acids protein with an N-terminal basic leucine zipper (bZIP) [27, 28]. L-Meq is a longer isoform of Meq that contains a 180 bp nucleotide insertion in the transactivation domain of Meq and is normally detected in non-oncogenic MDV1 vaccine strains such as CVI988 [29]. Interestingly, L-Meq was also amplified from crane spleen cDNA (Fig. 7a). Sequence alignment indicated that it shared 100% identity with the BC-1 strain (Fig. 7b). One substitution was found in the American strains GA and JM10 (Fig. 7c), and four substitutions were found in the JM102 strain (Fig. 7c).
Fig. 7.
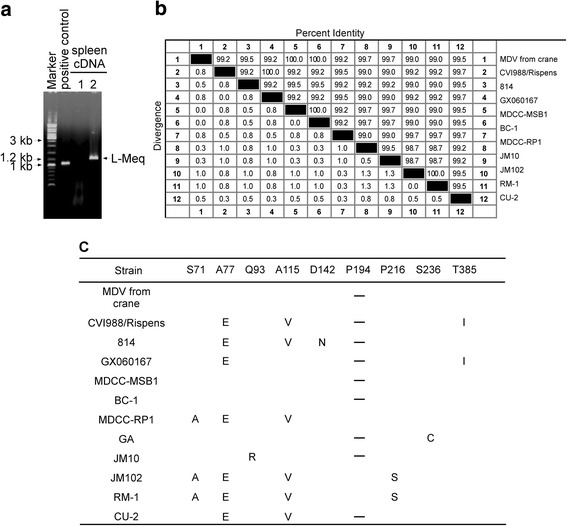
Amino acid sequence alignment of L-Meq (from crane). a, The full-length L-Meq gene (1197 bp) was amplified from spleen cDNA. b, The amino acid sequence of L-Meq (398 aa) from cranes was aligned with 11 previously published MDV isolates (including CVI988, 814, GX060167, MDCC-RP1, MDCC-MSB1, BC-1, GA, JM10, JM102, RM-1 and CU-2). Sequences were aligned using the DNAMAN software. c, Substituted amino acids are listed, while deleted amino acids are denoted by strips in the alignment
Discussion and conclusions
This article describes the first reported case of MD in red-crowned cranes and confirms that red-crowned cranes are susceptible to MDV infection. Previous research has suggested that multiple avian species can be infected by MDV such as Galliformes (turkeys [8], quails [9] and pheasants [10]), Strigiformes (owls), Anseriformes (ducks, geese, swans), and Falconiformes (kestrels). Therefore, this is also the first molecular detection of Marek’s disease virus in the order Gruiformes. One possible sources of infection for red-crowned cranes might be from migrating wild birds. Murata et al. tested feather-tip samples of wild geese from Japan and the Far East region of Russia by nested PCR, and 30% of analyzed white-fronted geese contained MDV [30], which suggested that migratory birds such as white-fronted geese could be MDV carriers during their migration. Previous research showed that MDV can be transmitted horizontally and across-species. Kenzy and Cho reported that a MDV-positive quail can transmit MD to monitor chicks by contact exposure [31]. Additionally, MDV is fairly resistant to environmental factors. Indeed, MDV can survive at least a few months in the dust at room temperature [32]. The infection source is also diverse, including chicken feather follicle, scurf, house dust, feces, and saliva. Many chickens with a normal appearance can be MDV carriers and spread infection. All of these provide a conducive environment for the spreading of MDV. Considering that the red-crowned cranes were fed in mesh cages that were accessible to wild birds and were never immunized with MDV vaccine before, it is possible that the MDV infection originated from migratory birds.
This case report describes the first detection and characterization of MDV from red-crowned cranes in a Chinese zoo. The molecular analysis of MDV genes suggested that the MDV (from crane) belongs to MDV serotype 1. The red-crowned cranes described in this study showed clinical signs of disease including decreased feed consumption, depression, paralysis, joint swelling, and eventual death. The results of necropsies of dead cranes revealed diffuse nodules in skin, muscle, liver, gizzard and heart. Histological investigation demonstrated lymphomatous infiltration into the affected tissue and neoplastic changes with evidence of mitosis. Additionally, a range of leukocytes was observed, including small/large lymphocytes, lymphoblasts, and occasional plasma cells. The demonstration of polymorphic leukocytes along with molecular detection is highly suggestive of Marek’s disease. RT-PCR targeting oncogenic gene Meq and glycoprotein B (gB) was performed to confirm the presence of MDV, with observation of expected amplification products throughout all tissue samples. Further phylogenetic analysis of the Meq and VP22 genes suggested that our MDV could belong to MDV-1, but it is a novel MDV that contains both Meq and L-Meq gene in its genome.
The detection of MDV in red-crowned cranes suggests that the virus may have a huge impact on the endangered crane’s survival in the wild. However, because this is the first verified instance of crane mortality resulting from MD, its true impact on the population is unknown. A possible solution for management of this species is to vaccinate all the red-crowed cranes to provide immunity against this disease. There are several available MDV vaccines including serotypes MDV-2 (SB-1), HVT, and attenuated MDV-1 (CVI988/Rispens), which are used singly or jointly [33]. MDV vaccination began in the late 1960s and made great contributions to protecting the poultry industry for more than 50 years. However, the current problem involves attenuated MDV vaccine that cannot provide effective protection against new virulent MDV strains. For example, previous reports have shown that molecular evolution of MDV genes could increase the pathogenicity of MDV virulent strains [34]. Infection of very virulent MDV (vvMDV) could also break post-vaccinal protection and cause the outbreak of MDV [35]. Therefore, the safety of this measurement needs to be studied in depth.
Acknowledgements
We thank Dr. Yingjun Lv for review of histological slides and constructive input for data analysis strategies.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant No. 31472218), the Natural Science Foundation of Jiangsu Province (Grant No. BK20140711), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), SRF for ROCS, SEM([2014]1685), and the Fundamental Research Funds for the Central Universities (Y0201300526; Y0201300527). The funders did not play any role in the design, conclusions or interpretation of the study.
Availability of data and materials
The datasets analysed during the current study are available in the TreeBASE repository. Our data are posted as NEXUS files in the following public domain resources: http://purl.org/phylo/treebase/phylows/study/TB2:S22355. The viral protein sequences of different MDV strains were deposited in GenBank under the accession number in Tables 3, 4 and 5.
Table 3.
GenBank accession numbers of the Meq amino acid sequences used in this study
| Isolate | Country of Origin | Accession Number | Pathotype |
|---|---|---|---|
| Md5 | USA | AAG14255.1 | vvMDV |
| Md11 | USA | AAS01627.1 | vvMDV |
| GA | USA | AAF66798.1 | vMDV |
| RB1B | USA | AY243332 | vvMDV |
| RL | USA | AAR13332.1 | vv + MDV |
| TK | USA | AAR13333.1 | vv + MDV |
| N | USA | AAR13330.1 | vv + MDV |
| New | USA | AAR13331.1 | vv + MDV |
| U | USA | AAR13334.1 | vv + MDV |
| W | USA | AAR13335.1 | vv + MDV |
| X | USA | AAR13336.1 | vv + MDV |
| 571 | USA | AAR13322.1 | vMDV |
| 573 | USA | AAR13323.1 | vMDV |
| 595 | USA | AAR13327.1 | vvMDV |
| 617A | USA | AAR13324.1 | vMDV |
| 637 | USA | AAR13325.1 | vMDV |
| 643P | USA | AAR13328.1 | vvMDV |
| 660-A | USA | AAR13338.1 | vv + MDV |
| 648A | USA | AAR13337.1 | vv + MDV |
| 686 | USA | AAR13339.1 | vv + MDV |
| CVI988/Rispens | Netherlands | AAP06940.1 | attMDV |
| 814 | China | AAL99997.1 | attMDV |
| J-1 | China | AEA06596.1 | N.A. |
| LQQHR | China | AEM63533.1 | N.A. |
| LSY2 | China | AEM63534.1 | N.A. |
| LMS | China | AEZ51745.1 | vvMDV |
| GX0101 | China | AFX97850.1 | vvMDV |
| GX070060 | China | ACA13267.1 | vvMDV |
| GX070079 | China | ACA13268.1 | N.A. |
| BY | China/Tibet | AEA06593.1 | N.A. |
| GXY2 | China | ABQ23868.1 | N.A. |
| LDH | China | AEM63523.1 | N.A. |
| LYC | China | AEM63536.1 | N.A. |
| YLO40920 | China | ABA54944.1 | N.A. |
| G2 | China | AAM00003.1 | vvMDV |
Abbreviation: Att attenuated, N.A not available, v virulent, vv very virulent, vv + very virulent plus
Table 4.
GenBank accession numbers of the L-meq amino acid sequences used in this study
| Isolate | Country of Origin | Accession Number | Pathotype |
|---|---|---|---|
| CVI988/Rispens | Netherlands | ABF72204.1 | attMDV |
| 814 | China | ADA83412.1 | attMDV |
| GX060167 | China | ACD75766.1 | N.A. |
| HNGS201 | China | CCO02736.1 | N.A. |
| MDCC-MSB1 | Japan | BAC57989.1 | N.A. |
| MDCC-RP1 | Japan | BAC57991.1 | N.A. |
| BC-1 | USA | AAR13319.1 | vMDV |
| GA | USA | AAC54628.1 | vMDV |
| JM10 | USA | AAP06937.1 | vMDV |
| JM/102 W | USA | ABG22905.1 | vMDV |
| RM-1 | USA | ABG22996.1 | attMDV |
| CU-2 | USA | ACF94907.1 | mMDV |
| 3004 | Russia | ABS84657.1 | attMDV |
Abbreviation: att attenuated; m mild, N.A not available, v virulent, vv very virulent, vv + very virulent plus
Table 5.
GenBank accession numbers of the vp22 amino acid sequences used in this study
| Isolate | Country of Origin | Accession Number | Pathotype |
|---|---|---|---|
| Md5 | USA | YP-001033978.1 | vvMDV |
| CVI988/Rispens | Netherlands | ABF72291.1 | attMDV |
| GA | USA | AAF66784.1 | vMDV |
| RB1B | USA | ABR13137.1 | vvMDV |
| Md11 | USA | AAS01692.1 | vvMDV |
| 648A | USA | AFM7426.1 | vv + MDV |
| 549A | USA | ABV31181.1 | vvMDV |
| 571 | USA | ABV31182.1 | vMDV |
| 584A | USA | ABV31183.1 | vv + MDV |
| 595 | USA | ABV31184.1 | vvMDV |
| 686 | USA | ABV31179.1 | vv + MDV |
| JW/102 W | USA | ABV31188.1 | vMDV |
| CU-2 | USA | ABV31189.1 | mMDV |
| R2/23 | USA | ABV31191.1 | N.A. |
| RM-1 | USA | ABV31180.2 | N.A. |
| CC/1409 | China | AQN78018.1 | N.A. |
| HS/1412 | China | AQN78190.1 | N.A. |
| J-1 | China | AQN77146.1 | N.A. |
| 814 | China | AEV55030.1 | attMDV |
| GX0101 | China | AFX97970.1 | vvMDV |
| LMS | China | AEZ51712.1 | vvMDV |
| JL/1404 | China | AQN77845.1 | N.A. |
| LCC | China | AQN77320.1 | N.A. |
| LCY | China | ARE59101.1 | N.A. |
| LTS | China | AQN77496.1 | N.A. |
Abbreviation: att attenuated, N.A not available, v virulent, vv very virulent, vv + very virulent plus
Abbreviations
- gB
Glycoprotein B
- MDV
Marek’s disease virus
- PCR
Polymerase chain reaction
Authors’ contributions
XL, XM, YSJ, and YJQ designed the research. XL, WC and XM performed laboratory experiments. XL, XM, JX, WC, XZ, HC, CD, YSJ, and YJQ analyzed the data. WC initiated the study and organized samples. XL, XM, YSJ, and YJQ wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the author’s institution (Nanjing Agricultural University ethics committee) and owner consent was obtained for the animals used for the post-mortem examinations.
Consent for publication
Consent was obtained from the owner of the animal for publication of this case report.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yong-Sam Jung, Email: ysjung@njau.edu.cn.
Yingjuan Qian, Email: yqian@njau.edu.cn.
References
- 1.Grus japonensis:The IUCN Red List of Threatened Species 2016. http://www.iucnredlist.org/details/full/22692167/0.
- 2.Collar NJ, Andreev A, Chan S, Crosby M, Subramanya S, Tobias J, Srinivasan C, Subramanya HS, Awatramani N, Subramanya HM. Threatened birds of Asia: the BirdLife international red data book. Cambridge (RU): Birdlife international; 2001. [Google Scholar]
- 3.Kanai Y, Ueta M, Germogenov N, Nagendran M, Mita N, Higuchi H. Migration routes and important resting areas of Siberian cranes (Grus leucogeranus) between northeastern Siberia and China as revealed by satellite tracking. Biol Conserv. 2002;106(3):339–346. doi: 10.1016/S0006-3207(01)00259-2. [DOI] [Google Scholar]
- 4.Su L, Zou H. Status, threats and conservation needs for the continental population of the red-crowned crane. Chinese Birds. 2012;3(3):147–164. doi: 10.5122/cbirds.2012.0030. [DOI] [Google Scholar]
- 5.Wang Q. Threats for red-crowned crane. China Crane News. 2008;12(2):7–12. [Google Scholar]
- 6.Calnek BW. Pathogenesis of Marek's disease virus infection. Curr Top Microbiol Immunol. 2001;255:25–55. doi: 10.1007/978-3-642-56863-3_2. [DOI] [PubMed] [Google Scholar]
- 7.Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S. Marek's disease virus: from miasma to model. Nat Rev Microbiol. 2006;4(4):283–294. doi: 10.1038/nrmicro1382. [DOI] [PubMed] [Google Scholar]
- 8.Davidson I, Borenstein R. Multiple infection of chickens and turkeys with avian oncogenic viruses: prevalence and molecular analysis. Acta Virol. 1999;43(2–3):136–142. [PubMed] [Google Scholar]
- 9.Kobayashi S, Kobayashi K, Mikami T. A study of Marek's disease in Japanese quails vaccinated with herpesvirus of turkeys. Avian Dis. 1986;30(4):816–819. doi: 10.2307/1590591. [DOI] [PubMed] [Google Scholar]
- 10.Lesnik F, Pauer T, Vrtiak OJ, Danihel M, Gdovinova A, Gergely M. transmission of Marek's disease to wild feathered game. Veterinarni medicina. 1981;26(10):623–630. [PubMed] [Google Scholar]
- 11.Kenzy S, Mclean G, Mathey W, Lee H. Preliminary observations of gamefowl neurolymphomatosis. J Nat Cancer Inst. 1964;17:121–130. [Google Scholar]
- 12.Grewal G, Singh B, Singh H. Epidemiology of Marek's disease: incidence of viral specific antigen in feather follicle epithelium of domestic fowl of Punjab. Indian journal of poultry science. 1977;
- 13.Weiss RA, Biggs PM. Leukosis and Marek's disease viruses of feral red jungle flow and domestic fowl in Malaya. J Natl Cancer Inst. 1972;49(6):1713–1725. doi: 10.1093/jnci/49.6.1713. [DOI] [PubMed] [Google Scholar]
- 14.Bulow VV, Biggs PM. Differentiation between strains of Marek's disease virus and Turkey herpesvirus by immunofluorescence assays. Avian pathology : journal of the WVPA. 1975;4(2):133–146. doi: 10.1080/03079457509353859. [DOI] [PubMed] [Google Scholar]
- 15.Swayne DE, Fletcher OJ, Schierman LW. Marek's disease virus-induced transient paralysis in chickens. 1. Time course association between clinical signs and histological brain lesions. Avian pathology : journal of the WVPA. 1989;18(3):385–396. doi: 10.1080/03079458908418613. [DOI] [PubMed] [Google Scholar]
- 16.Witter RL, Gimeno IM, Reed WM, Bacon LD. An acute form of transient paralysis induced by highly virulent strains of Marek's disease virus. Avian Dis. 1999;43(4):704–720. doi: 10.2307/1592740. [DOI] [PubMed] [Google Scholar]
- 17.Stephens EA, Witter RL, Nazerian K, Sharma JM. Development and characterization of a Marek's disease transplantable tumor in inbred line 72 chickens homozygous at the major (B) histocompatibility locus. Avian Dis. 1980;24(2):358–374. doi: 10.2307/1589703. [DOI] [PubMed] [Google Scholar]
- 18.Boodhoo N, Gurung A, Sharif S, Behboudi S. Marek's disease in chickens: a review with focus on immunology. Vet Res. 2016;47(1):119. doi: 10.1186/s13567-016-0404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nazerian K, Witter RL. Cell-free transmission and in vivo replication of Marek's disease virus. J Virol. 1970;5(3):388–397. doi: 10.1128/jvi.5.3.388-397.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calnek BW. Marek's disease--a model for herpesvirus oncology. Crit Rev Microbiol. 1986;12(4):293–320. doi: 10.3109/10408418509104432. [DOI] [PubMed] [Google Scholar]
- 21.Imai K, Yuasa N, Kobayashp S, Nakamura K, Tsukamoto K, Hihara H. Isolation of Marek's disease virus from Japanese quail with lymphoproliferative disease. Avian pathology : journal of the WVPA. 1990;19(1):119–129. doi: 10.1080/03079459008418661. [DOI] [PubMed] [Google Scholar]
- 22.Docherty DE, Henning DJ. The isolation of a herpesvirus from captive cranes with an inclusion body disease. Avian Dis. 1980;24(1):278–283. doi: 10.2307/1589787. [DOI] [Google Scholar]
- 23.Foerster S, Chastel C, Kaleta EF. Crane hepatitis herpesviruses. Zentralbl Veterinarmed B. 1989;36(6):433–441. doi: 10.1111/j.1439-0450.1989.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi T, Kaplan SL, Wakenell P, Schat KA. Transactivation of latent Marek's disease herpesvirus genes in QT35, a quail fibroblast cell line, by herpesvirus of turkeys. J Virol. 2000;74(21):10176–10186. doi: 10.1128/JVI.74.21.10176-10186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorange F, Tischer BK, Vautherot JF, Osterrieder N. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J Virol. 2002;76(4):1959–1970. doi: 10.1128/JVI.76.4.1959-1970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, Song C, Qin A, Zhang C. Expression and intercellular trafficking of the VP22 protein of CVI988/Rispens vaccine strain of Marek's disease virus. Sci China Ser C Life Sci. 2007;50(1):75–79. doi: 10.1007/s11427-007-2038-1. [DOI] [PubMed] [Google Scholar]
- 27.Liu JL, Ye Y, Lee LF, Kung HJ. Transforming potential of the herpesvirus oncoprotein MEQ: morphological transformation, serum-independent growth, and inhibition of apoptosis. J Virol. 1998;72(1):388–395. doi: 10.1128/jvi.72.1.388-395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu JL, Kung HJ. Marek's disease herpesvirus transforming protein MEQ: a c-Jun analogue with an alternative life style. Virus Genes. 2000;21(1–2):51–64. doi: 10.1023/A:1008132313289. [DOI] [PubMed] [Google Scholar]
- 29.Lee SI, Takagi M, Ohashi K, Sugimoto C, Onuma M. Difference in the meq gene between oncogenic and attenuated strains of Marek's disease virus serotype 1. J Vet Med Sci. 2000;62(3):287–292. doi: 10.1292/jvms.62.287. [DOI] [PubMed] [Google Scholar]
- 30.Murata S, Hayashi Y, Kato A, Isezaki M, Takasaki S, Onuma M, Osa Y, Asakawa M, Konnai S, Ohashi K. Surveillance of Marek's disease virus in migratory and sedentary birds in Hokkaido, Japan. Vet J. 2012;192(3):538–540. doi: 10.1016/j.tvjl.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Kenzy S, Cho B. Transmission of classical Marek's disease by affected and carrier birds. Avian Dis. 1969:211–4. [PubMed]
- 32.Calnek BW. Marek's disease virus and lymphoma. In: Rapp F, editor. Oncogenic Herpesviruses. Boca Raton: CRC Press; 1980. p. 103-143.
- 33.Witter RL. Increased virulence of Marek's disease virus field isolates. Avian Dis. 1997;41(1):149–163. doi: 10.2307/1592455. [DOI] [PubMed] [Google Scholar]
- 34.Wozniakowski G, Samorek-Salamonowicz AE. Molecular evolution of Marek's disease virus (MDV) field strains in a 40-year time period. Avian Dis. 2014;58(4):550–557. doi: 10.1637/10812-030614-Reg.1. [DOI] [PubMed] [Google Scholar]
- 35.Madej JP, Wozniakowski G, Gawel A. Morphology of immune organs after very virulent plus strain of Marek's disease virus infection in vaccinated hens. Pol J Vet Sci. 2016;19(2):325–335. doi: 10.1515/pjvs-2016-0040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analysed during the current study are available in the TreeBASE repository. Our data are posted as NEXUS files in the following public domain resources: http://purl.org/phylo/treebase/phylows/study/TB2:S22355. The viral protein sequences of different MDV strains were deposited in GenBank under the accession number in Tables 3, 4 and 5.
Table 3.
GenBank accession numbers of the Meq amino acid sequences used in this study
| Isolate | Country of Origin | Accession Number | Pathotype |
|---|---|---|---|
| Md5 | USA | AAG14255.1 | vvMDV |
| Md11 | USA | AAS01627.1 | vvMDV |
| GA | USA | AAF66798.1 | vMDV |
| RB1B | USA | AY243332 | vvMDV |
| RL | USA | AAR13332.1 | vv + MDV |
| TK | USA | AAR13333.1 | vv + MDV |
| N | USA | AAR13330.1 | vv + MDV |
| New | USA | AAR13331.1 | vv + MDV |
| U | USA | AAR13334.1 | vv + MDV |
| W | USA | AAR13335.1 | vv + MDV |
| X | USA | AAR13336.1 | vv + MDV |
| 571 | USA | AAR13322.1 | vMDV |
| 573 | USA | AAR13323.1 | vMDV |
| 595 | USA | AAR13327.1 | vvMDV |
| 617A | USA | AAR13324.1 | vMDV |
| 637 | USA | AAR13325.1 | vMDV |
| 643P | USA | AAR13328.1 | vvMDV |
| 660-A | USA | AAR13338.1 | vv + MDV |
| 648A | USA | AAR13337.1 | vv + MDV |
| 686 | USA | AAR13339.1 | vv + MDV |
| CVI988/Rispens | Netherlands | AAP06940.1 | attMDV |
| 814 | China | AAL99997.1 | attMDV |
| J-1 | China | AEA06596.1 | N.A. |
| LQQHR | China | AEM63533.1 | N.A. |
| LSY2 | China | AEM63534.1 | N.A. |
| LMS | China | AEZ51745.1 | vvMDV |
| GX0101 | China | AFX97850.1 | vvMDV |
| GX070060 | China | ACA13267.1 | vvMDV |
| GX070079 | China | ACA13268.1 | N.A. |
| BY | China/Tibet | AEA06593.1 | N.A. |
| GXY2 | China | ABQ23868.1 | N.A. |
| LDH | China | AEM63523.1 | N.A. |
| LYC | China | AEM63536.1 | N.A. |
| YLO40920 | China | ABA54944.1 | N.A. |
| G2 | China | AAM00003.1 | vvMDV |
Abbreviation: Att attenuated, N.A not available, v virulent, vv very virulent, vv + very virulent plus
Table 4.
GenBank accession numbers of the L-meq amino acid sequences used in this study
| Isolate | Country of Origin | Accession Number | Pathotype |
|---|---|---|---|
| CVI988/Rispens | Netherlands | ABF72204.1 | attMDV |
| 814 | China | ADA83412.1 | attMDV |
| GX060167 | China | ACD75766.1 | N.A. |
| HNGS201 | China | CCO02736.1 | N.A. |
| MDCC-MSB1 | Japan | BAC57989.1 | N.A. |
| MDCC-RP1 | Japan | BAC57991.1 | N.A. |
| BC-1 | USA | AAR13319.1 | vMDV |
| GA | USA | AAC54628.1 | vMDV |
| JM10 | USA | AAP06937.1 | vMDV |
| JM/102 W | USA | ABG22905.1 | vMDV |
| RM-1 | USA | ABG22996.1 | attMDV |
| CU-2 | USA | ACF94907.1 | mMDV |
| 3004 | Russia | ABS84657.1 | attMDV |
Abbreviation: att attenuated; m mild, N.A not available, v virulent, vv very virulent, vv + very virulent plus
Table 5.
GenBank accession numbers of the vp22 amino acid sequences used in this study
| Isolate | Country of Origin | Accession Number | Pathotype |
|---|---|---|---|
| Md5 | USA | YP-001033978.1 | vvMDV |
| CVI988/Rispens | Netherlands | ABF72291.1 | attMDV |
| GA | USA | AAF66784.1 | vMDV |
| RB1B | USA | ABR13137.1 | vvMDV |
| Md11 | USA | AAS01692.1 | vvMDV |
| 648A | USA | AFM7426.1 | vv + MDV |
| 549A | USA | ABV31181.1 | vvMDV |
| 571 | USA | ABV31182.1 | vMDV |
| 584A | USA | ABV31183.1 | vv + MDV |
| 595 | USA | ABV31184.1 | vvMDV |
| 686 | USA | ABV31179.1 | vv + MDV |
| JW/102 W | USA | ABV31188.1 | vMDV |
| CU-2 | USA | ABV31189.1 | mMDV |
| R2/23 | USA | ABV31191.1 | N.A. |
| RM-1 | USA | ABV31180.2 | N.A. |
| CC/1409 | China | AQN78018.1 | N.A. |
| HS/1412 | China | AQN78190.1 | N.A. |
| J-1 | China | AQN77146.1 | N.A. |
| 814 | China | AEV55030.1 | attMDV |
| GX0101 | China | AFX97970.1 | vvMDV |
| LMS | China | AEZ51712.1 | vvMDV |
| JL/1404 | China | AQN77845.1 | N.A. |
| LCC | China | AQN77320.1 | N.A. |
| LCY | China | ARE59101.1 | N.A. |
| LTS | China | AQN77496.1 | N.A. |
Abbreviation: att attenuated, N.A not available, v virulent, vv very virulent, vv + very virulent plus


