Abstract
It has long been a puzzle in cancer treatment that despite the initial appearance of apoptosis, the process could be reversed in some cancer cells and often results in more aggressive tumors and metastasis. The mechanism for this recurrence is yet unknown. Here we report that human mammary carcinoma cells induced to undergo apoptosis could recover with increased tumorigenicity in vitro and in vivo, and induced lymph node metastasis. Specifically, the reversed cells underwent epithelial-to-mesenchymal transitions in the primary tumors in situ, and mesenchymal-to-epithelial transitions in the metastatic cells. Flow cytometry confirmed an elevated percentage of cells carrying cancer stem cells (CSCs) markers (CD44+/CD24-) in the reversed breast cancer cell population, with hypomethylated CD44 promoters and hypermethylated CD24 promoters. More importantly, CSCs were generated anew from non-stem cancer cells after apoptosis reversal possibly through epigenetic modifications. The results from this study can open doors to discovering more effective cancer treatments by suppressing apoptosis reversal.
Introduction
Cancer is among the leading causes of mortality worldwide [1]. The greatest therapeutic challenge it poses is its cellular heterogeneity, probably the product of clonal evolution [2] and/or cancer stem cells (CSC) [3]. CSCs are more resistant to conventional cancer therapies than non-CSC populations [4], [5], [6] and more likely to cause cancer recurrences [7]. Current cancer treatments including chemotherapy, γ-irradiation and immunotherapy eliminate cancer cells mostly through activating apoptosis pathways [8], [9], [10]. However, recent studies demonstrated that cancer cell lines could survive apoptosis induction and the reversed cancer cells possessed some features of CSCs and became more aggressive [11], [12], [13].
Here, we showed that breast cancer cells undergoing reversal of apoptosis acquired increased tumorigenicity both in vitro and in vivo and there was a correlated elevation of the percentage of cells with CSC markers in the reversed cells comparing with the untreated ones. Importantly, this reversal process triggered a transition from non-stem cancer cells (NSCCs) to CSCs, which could be suppressed by pre-treatment with epigenetic modulators. These results suggest that targeting the epigenetic regulation could be a promising strategy for decreasing CSCs and hence tumorigenicity and/or metastasis.
Materials and Methods
Cell Culture and Apoptosis Induction
The culture of MCF-7, T47D and MDA-MB-231 (American Type Culture Collection, Manassas, VA, USA) were done as described in the Supplementary data. 2.5 μM staurosporine (STS) (Sigma-Aldrich, St. Louis, MO, USA) and 5 μM paclitaxel (Sigma-Aldrich) were applied to MCF-7 cells for 6 hours and to T47D cells for 10 hours, respectively, to induce apoptosis when cell density reached 70% confluency. Solvent controls of STS and paclitaxel were 0.25% v/v and 0.05% v/v dimethyl sulfoxide (DMSO) (Sigma-Aldrich) respectively.
CellEvent™ Staining and Fluorescence-Activated Cell Sorting (FACS)
Apoptotic MCF-7 and T47D cells were stained with 3 μM CellEvent™ (Invitrogen, Carlsbad, CA, USA) at a cell concentration of 106 cells/mL in the dark for 30 minutes at 37°C. Cells were then filtered through a 40-μm nylon mesh (BD Biosciences, Bedford, MA, USA) before running on the Bio-Rad S3 sorter (Bio-Rad, Hercules, CA, USA) (refer to Supplementary data for details on cell sorting and the subsequent procedure for culturing of cells undergoing apoptosis reversal).
Propidium Iodide (PI) Staining for Live/Dead Cell Detection
Cells were collected, washed twice with phosphate buffered saline (PBS) and stained with 2 μg/mL PI (Thermo Fisher Scientific, Waltham, MA, USA) just before running on the BD FACSVerse flow cytometer (BD Biosciences).
In Vivo Tumorigenicity Assay
The following experimental procedures were approved by the Animal Ethics Committee, the Chinese University of Hong Kong and conformed to the Guide for the Care and Use of Laboratory Animals published by the United States National Institutes of Health (NIH Publication, 8th Edition, 2011). Single MCF-7 cells were resuspended in 50 μL serum-free RPMI 1640 and mixed with Growth Factor Reduced Matrigel Matrix (BD Biosciences) at 1:1 ratio (v:v) and injected subcutaneously into the mammary fat pad of anesthetized 6-week old female nude mice (refer to Supplementary data for details on animal surgery). The control group received only serum-free RPMI 1640 medium and Matrigel mixture (1:1). Tumor volumes were determined every 3 days. Tumor volume (mm3) = 0.52 × width (mm)2 × length (mm).
Mammosphere Culture
Single cell suspension was plated in an ultra-low attachment 6-well plate (Corning, NY, USA) at a density of 4 × 104 cells/mL in the primary or 104 cells/mL in the secondary and tertiary mammosphere formation experiments. The culture condition for mammosphere formation was described in the Supplementary data. Mammosphere formation efficiency (MFE) (%) = (number of mammospheres per well / number of cells seeded per well) × 100. Only mammospheres with a diameter of ≥60 μm were counted at the end of each passage (day 7).
Immunohistochemical Analysis
Tissues and xenograft tumors were fixed and sectioned for analysis (refer to Supplementary data for details on slice preparation). For detection of CSC markers, fluorochrome-conjugated antibodies against human CD44 (PERCP-CY5.5, BD Biosciences) and CD24 (PE, BD Biosciences) were added at 1:40 and 1:10 dilution, respectively. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) at 1:5000 dilution. For EMT markers, sections were incubated with mouse anti-E-Cadherin (BD Biosciences) and rabbit anti-N-Cadherin antibodies (Abcam, Cambridge, UK) at 4°C overnight, followed by goat anti-mouse Alexa Fluor® 488- and goat anti-rabbit Alexa Fluor® 594-labeled secondary antibodies (Thermo Fisher Scientific).
Immunostaining Followed by Flow Cytometric and Confocal Analysis
For preparing cells for flow cytometry, cells were blocked with 1:50 diluted Fc block (Miltenyi Biotec, Auburn, CA, USA) for 20 minutes. For preparing cells for confocal microscopy, cells were fixed, permeabilized and blocked with 1% BSA and 5% normal goat serum (Invitrogen) for 1 hour. In both cases, antibodies against human CD44 (PERCP-CY5.5, BD Biosciences) and CD24 (PE, BD Biosciences) were added at 1:40 and 1:10 dilution, respectively, and incubated at 4°C in dark. For flow cytometry, cells were analyzed on FACSVerse equipped with FACSuite software (BD Biosciences). For confocal analysis, images were acquired using the Olympus FluoView FV1000 confocal laser scanning microscope with a 60X objective and analyzed using the FV1000 software (Olympus, Tokyo, Japan) (refer to Supplementary data for details).
FACS of NSCCs
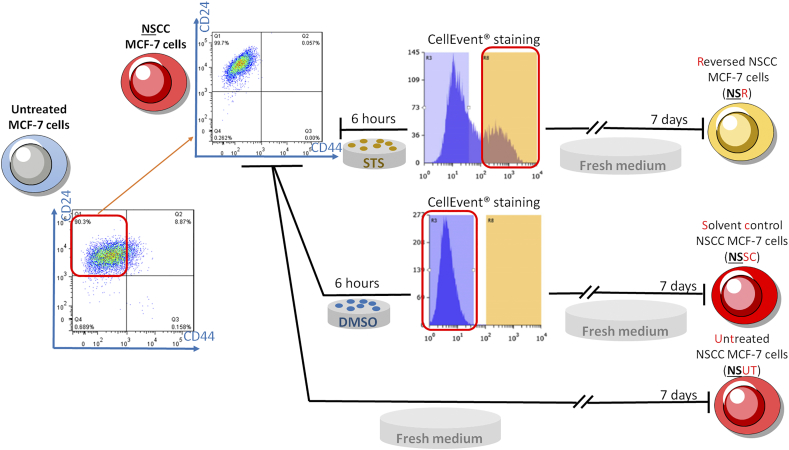
CSCs in breast cancer were defined as CD44+/CD24- cells, and NSCCs were defined as CD44-/CD24+ cells. Cells were immunostained under identical conditions as described above and CSCs and NSCCs were collected using Bio-Rad S3 sorter. Sorted NSCCs were cultured in the collection medium for 7 days before STS (or solvent control) treatment as described in the 'basic procedure for apoptosis induction and reversal of apoptosis' in Supplementary data.
Isolation of Limited Numbers of CSCs by Magnetic-Activated Cell Sorting (MACS) Using Dynabeads®
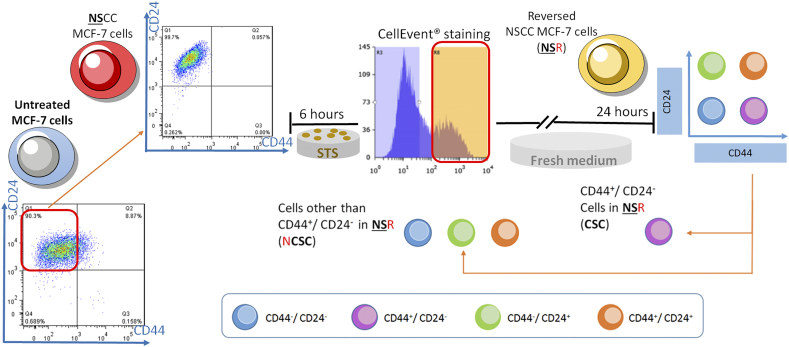
NSCCs in MCF-7 were obtained through FACS and reversal of apoptosis was done as described above. Reversed NSCC (NSR) cells were harvested at day 7. Then, MACS using Dynabeads® (Invitrogen) was first applied for CD24 negative selection to collect CD24- cells, followed by a CD44 positive selection to eventually obtain the CD44+/CD24- (CSC) population. Cells remaining after this sorting procedure were named NCSCs.
Reverse Transcription Followed by Real-time PCR (RT-qPCR)
mRNA was extracted and reverse-transcribed to cDNA using SuperScript® III 1st Strand Synthesis System (Invitrogen). Real-time PCR was performed as described in the Supplementary data.
Bisulfite Conversion and Methylation-Specific PCR (MSP)
Genomic DNA was extracted and bisulfite conversion of DNA was performed using the EpiTect Bisulfite Kit (Qiagen, Duesseldorf, Germany). For methylation analyses of the promoter regions of CD44 and CD24, two rounds of end-point PCR were performed (refer to Supplementary data for details).
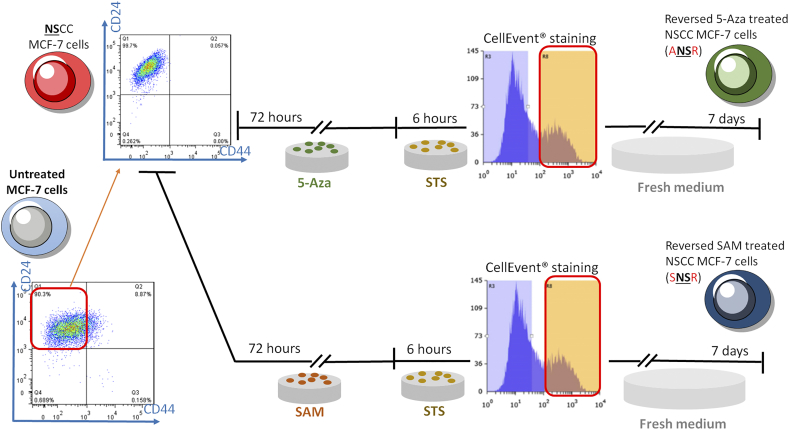
Epigenetic Modulator Experiments
FACS-sorted NSCCs were treated with 5 μM 5-Azacytidine (5-Aza) (Sigma-Aldrich) or 0.5 mM S-(5′-Adenosyl)-L-methionine chloride dihydrochloride (SAM) (Sigma-Aldrich). Solvent was used as control. The media containing the drugs were replaced every 24 hours and the cells were treated for a total of 72 hours before procedure for apoptosis reversal (refer to Supplementary data for details).
Live Cell Imaging
Cells were stained sequentially with CellEvent™, Mitotracker Red CMXRos (Invitrogen) and Hoechst 33342 (Invitrogen) (refer to Supplementary data for details). Cell morphology was observed using the Olympus FluoView FV1000 confocal laser scanning microscope with a 60X objective and further analyzed using the FV1000 software.
Western Blotting
Total proteins were extracted from cells and analyzed by Western blotting. Immunodetection were performed using anti-PARP at 1:1000 dilution(Cell Signaling, Danvers, MA, USA) and anti-β-tubulin (1:1000 dilution) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) (refer to Supplementary data for details).
Results
Apoptosis Reversal Increased Tumorigenicity of Breast Cancer Cells In Vitro and In Vivo
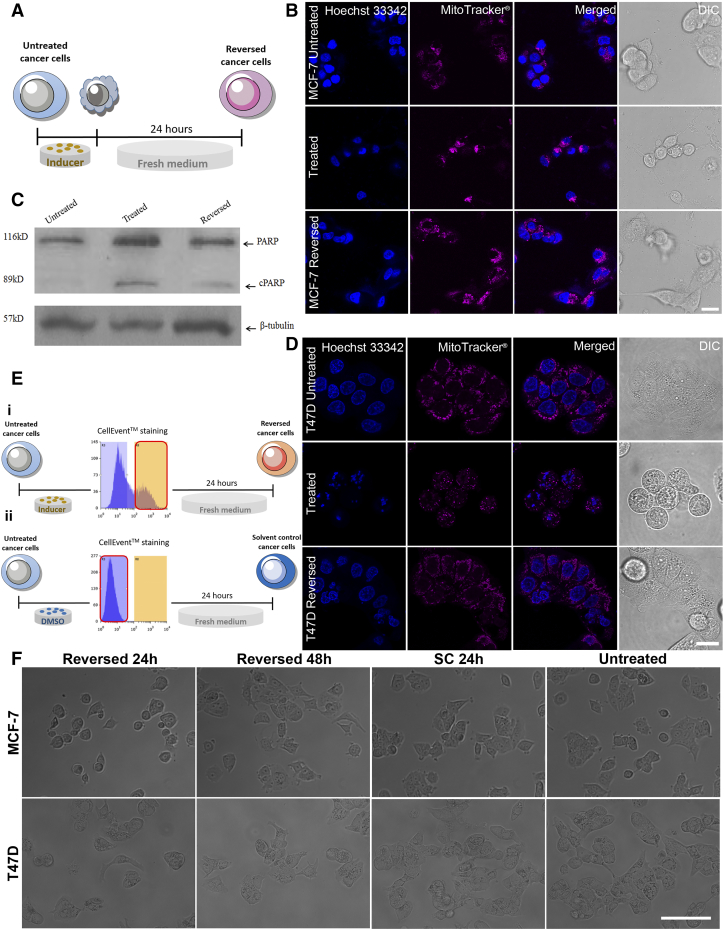
First, to show how apoptosis reversal affects tumorigenesis, we used the MCF-7 cell line and apoptotic inducer, STS [14], to build an apoptosis reversal model (Supplementary Figure 1A). Typical apoptotic morphological changes, including cell shrinkage, membrane blebbing, mitochondrial fragmentation and nuclear condensation, were observed 6 hours after STS treatment. Twenty-four hours after replacing STS with fresh medium, cells appeared to recover (Supplementary Figure 1B). Western blotting revealed that poly (ADP-ribose) polymerase (PARP) was cleaved after STS treatment, confirming the activation of caspase, a biochemical marker of apoptosis (Supplementary Figure 1C). To show that apoptosis reversal induced transformation of NSCCs to CSCs is not restricted to a particular cell line, another breast cancer cell line T47D was treated with the chemotherapy drug paclitaxel. Chemotherapy so far has been the mainstay in breast cancer treatment and paclitaxel is one of the clinically used drug for breast cancer [15], [16]. The mechanism of action of paclitaxel involves its interference with microtubule assembly during mitosis [17], [18] and its inhibition on the anti-apoptosis protein Bcl-2 to induce apoptosis in cancer cells [19]. Similar to the treatment of MCF-7 with STS, after 10 hours of paclitaxel induction, characteristic changes in cell morphology were also observed in T47D and cells regained their normal morphology after drug removal and medium replenishment for 24 hours (Supplementary Figure 1D). We then labeled caspase-activated cells using CellEvent™ and isolated them through fluorescence-activated cell sorting (FACS) (Supplementary Figure 1E). In parallel, DMSO-treated cells were also stained with CellEvent™, and CellEvent™-negative cells were collected as solvent control (Supplementary Figure 1E). Both MCF-7 and T47D cells that recovered from apoptosis were viable, able to proliferate and had morphology similar to untreated and solvent-treated cells (Supplementary Figure 1F).
Supplementary Figure 1.
Breast cancer cells were able to recover from apoptosis. (A) Schematics of cell generation for confocal imaging and western blotting to confirm the reversal of apoptosis. (B, D) Confocal images showing cellular morphological changes in MCF-7 cells (B) after STS induction (treated) and after the withdrawal of STS (reversed) and in T47D cells (D) after Paclitaxel induction (treated) and after withdrawal (reversed). Nuclei were stained by Hoechst 33342 (blue) while mitochondria were stained by MitoTracker® Red CMXRos (red). A series of typical apoptotic morphologies were observed in the treated group including cell shrinkage, membrane blebbing, mitochondrial fragmentation and nuclear condensation. Scale bars: 20 μm. (C) Western blot analysis of poly (ADP-ribose) polymerase (PARP). Cleaved PARP (cPARP) was detected in MCF-7 cells treated with STS for 6 hours, confirming the activation of caspase. β-tubulin was used as a loading control. (E) Schematics of the sorting of treated, caspase-activated cancer cells (i) and solvent-treated and non-caspase-activated cancer cells (ii) by CellEvent™ staining. (F) Inverted microscopy showing both MCF-7 (upper panel) and T47D (lower panel) cells that recovered from apoptosis (reversed 24h and 48h) were viable and were able to proliferate. Solvent control (SC) had similar morphology as untreated cells. Scale bar: 100 μm.
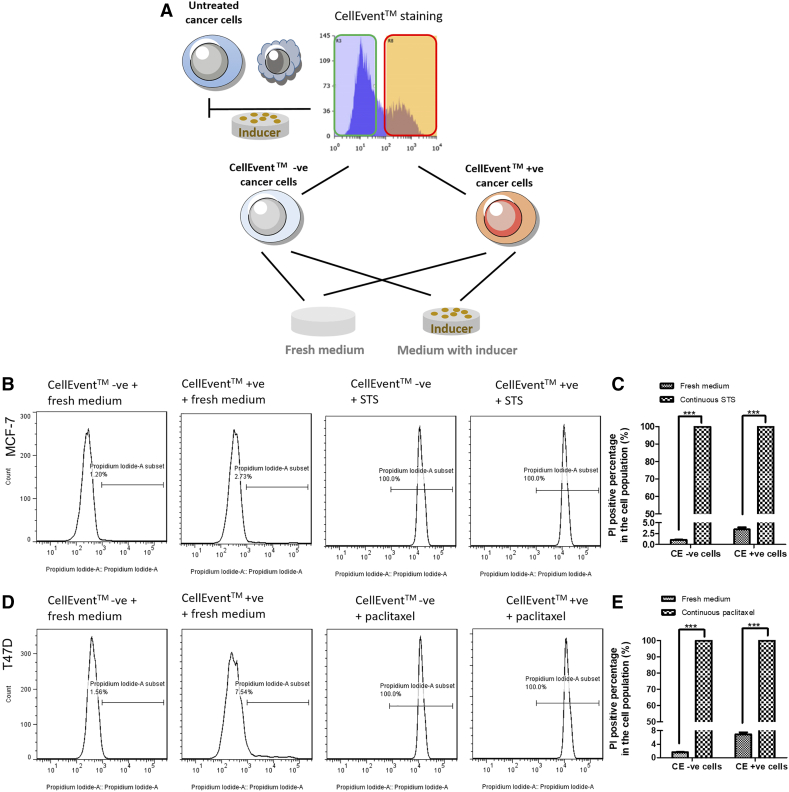
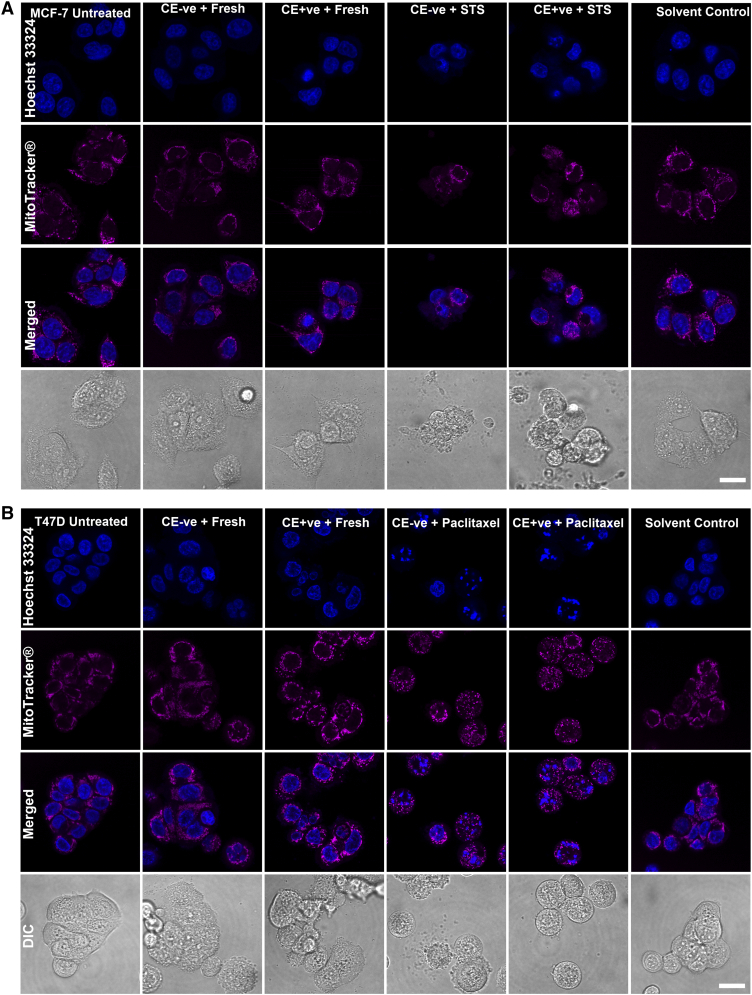
In order to demonstrate that reversal of apoptosis was not a process to select drug-resistant cells, we prolonged the induction of apoptosis to reveal the fate of both CellEvent™-positive and CellEvent™-negative cells. Cells after STS or paclitaxel treatment were stained with CellEvent™ and FACS-sorted. Unlike the reversal steps, the sorted cells were cultured back in the medium with previous inducers for another 24 hours before PI staining and microscopy detection. In parallel, some sorted cells were put into fresh medium without drug (Supplementary Figure 2A). Without the reversal steps, both CellEvent™-positive and CellEvent™-negative cells were kept dying in the continuous apoptotic induction (Supplementary Figures 2B-E and 3A and B), which exclude the possibility that drug-resistant cells were preferably selected during apoptosis reversal. In contrast, under the reversal protocol, attached cells that were put back into fresh medium owned normal morphology with intact cell membrane function (Supplementary Figures 2B-E and 3A and B).
Supplementary Figure 2.
Continuous apoptotic treatment on sorted CellEvent™ positive cancer cell led to cell death. (A) Schematics of cell generation for flow cytometry and confocal imaging to confirm cell death after continuous inducers treatment. (B, D) Distributions of PI staining in CellEvent™ sorted MCF-7 cells in fresh medium or STS group (B) and in CellEvent™ sorted T47D cells in fresh medium or Paclitaxel group (D). Cells were treated with 2.5 μM STS for 6 hours (MCF-7) or with 5 μM Paclitaxel for 10 hours (T47D) then sorted into CellEvent™ positive and negative groups. Each group was further separated and put into either fresh medium or medium with its original inducers for another 24 hours. All CellEvent™ positive and negative cells under additional 24 hours’ induction had 100% PI positive staining. (C, E) Percentage of cells with PI positive staining. CE +ve: sorted CellEvent™ positive cells. CE –ve: sorted CellEvent™ negative cells. Data represent mean±SEM. (n = 3). ***P < .001. n.d.: not detected.
Supplementary Figure 3.
Confocal imaging on the morphological changes in the continuous treated CellEvent™ positive and negative cells. Confocal images showing cellular morphological changes in both CellEvent™ positive and negative cells after continuous STS (MCF-7 cells) (A) or paclitaxel (T47D cells) (B) induction or putting into fresh medium for recovery. Nuclei were stained by Hoechst 33342 (blue) while mitochondria were stained by MitoTracker® Red CMXRos (red). A series of typical apoptotic morphologies were observed in the continuous treated groups including cell shrinkage, membrane blebbing, mitochondrial fragmentation and nuclear condensation. Scale bars: 20 μm.
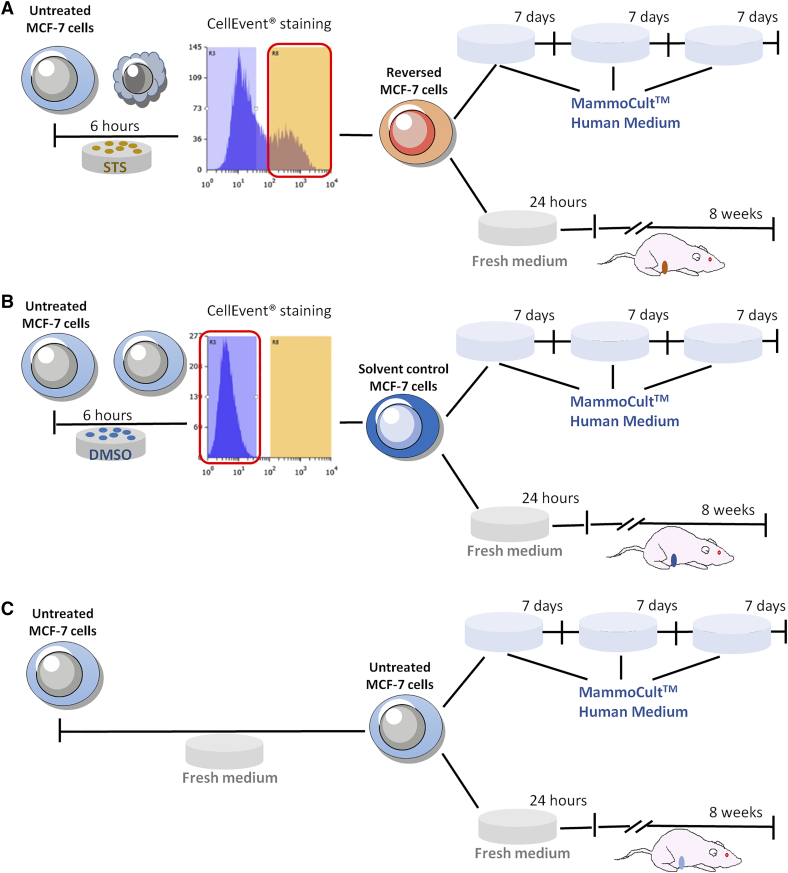
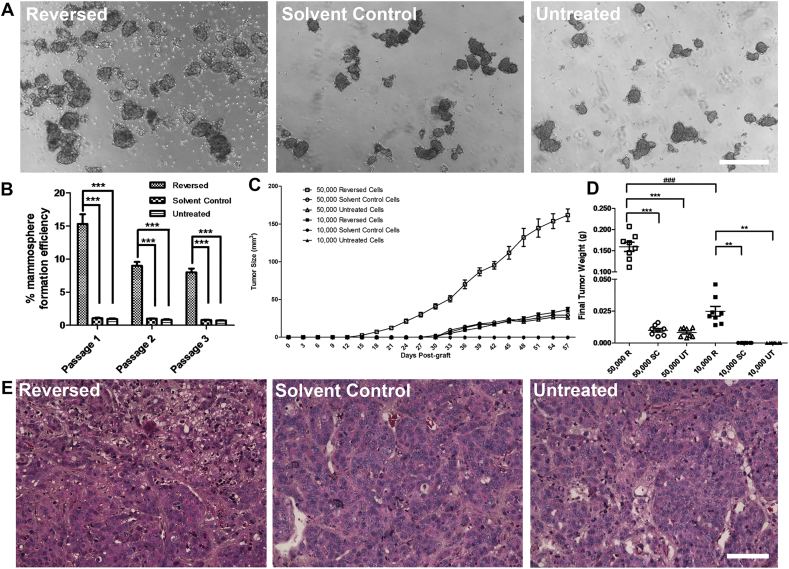
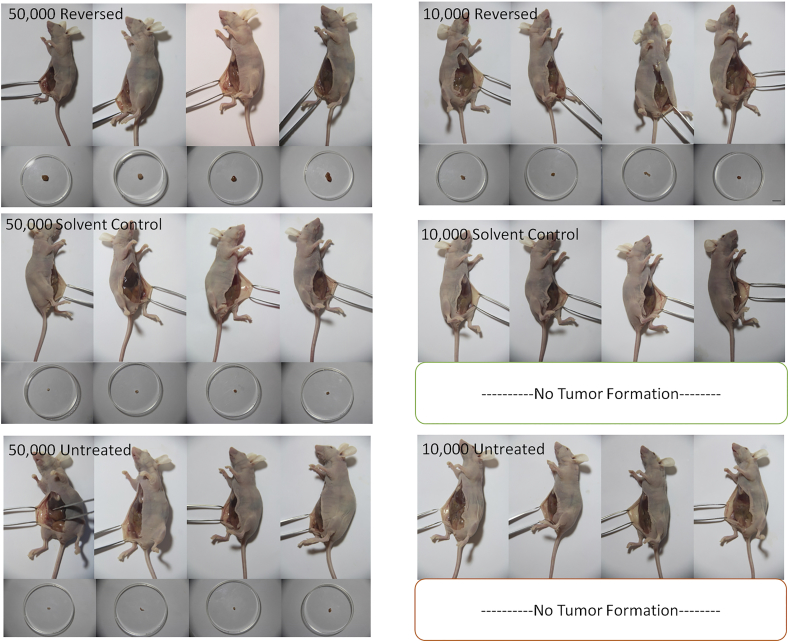
FACS-sorted MCF-7 cells were then used for in-vitro and in-vivo tumorigenicity analyses (Supplementary Figure 4). All sorted cells retained their tumorsphere-forming ability [20] (Figure 1A), which was significantly enhanced in the reversed cells after one week of culture compared to the solvent control or untreated cells. The ability to maintain mammospheres in serial suspension passage is related to the stem cells' self-renewal ability [21], [22], thus, mammospheres were usually digested and passed for more passages [23], [24]. In our experiment, the ability of the reversed cells to form mammospheres was observed to persist through two subsequent serial passages (Figure 1B). On the other hand, it is now realized that tumor-initiating cells have plasticity and their “stemness” could be affected by extrinsic factors in the microenvironment [25], [26], which were not always supplemented in the in vitro culture condition. Thus, the tumor transplantation in immunodeficient mice is the current “gold standard” for identifying CSCs [18], [26]. Tumorigenicity of the reversed cells was assessed in vivo by injecting them into the mammary fat pads of 6-week-old female nude mice (Supplementary Figure 4). Among the 50,000-injected-cell groups, transplanted reversed cells led to rapid tumor development, with visible tumors emerging at 2 weeks after injection while visible tumors did not appear until after 4 weeks for the solvent-control and untreated groups (Figure 1C). The mean final tumor size (Figure 1C and Supplementary Figure 5) and mean final tumor weight (Figure 1D) was significantly greater in the reversed group than in the control groups. Among the 10,000-injected-cell groups, tumors were first detected at about 4 weeks after injection of the reversed cells while no tumor was found in the control groups throughout the experiment (Figure 1C and Supplementary Figure 5). Tumors derived from all 3 cell types possessed similar morphologies (Figure 1E).
Supplementary Figure 4.
Schematics of in vitro and in vivo tumorigenicity assays using mammosphere culture and xenograft transplantation. (A) apoptosis-reversed MCF-7 cells; (B) solvent control; (C) untreated cells. Upper panel in each case: mammosphere culture; lower panel: xenograft transplantation.
Figure 1.
Reversed MCF-7 cells showed increased tumorigenicity in vitro and in vivo.
(A) Mammospheres formed by reversed, solvent control and untreated cells. Scale bar: 500 μm. (B) Mammosphere formation efficiency of the three passages of reversed, solvent control and untreated cells. Data represent mean±SEM. (n = 3). ***P < .001. (C) Kinetics of the in-vivo xenograft assay. Data represent mean±SEM. (n = 8). (D) Final tumor weight of xenografts. R: reversed cells; SC: solvent control; UT: untreated. Horizontal bar in the middle of each data cluster represents mean±SEM. ***/###P < .001; **P < .01. (E) H&E-stained xenografts from the initial injection of 50,000 cells in reversed, solvent control and untreated groups. Scale bar: 200 μm.
Supplementary Figure 5.
MCF-7 cells reversed from apoptosis showed increased tumorigenicity in vivo as determined using a xenograft model in nude mice. The tumors harvested from the mammary fat pads of nude mice in the reversed group were generally larger than those in the solvent control and the untreated groups, regardless of the number of MCF-7 cells used in the injection. The tumors formed in the 50,000-reversed cells group were also larger than those in the 10,000-reversed cells group. No visible tumor was formed in the groups injected with 10,000 solvent-treated cells or 10,000 untreated cells.
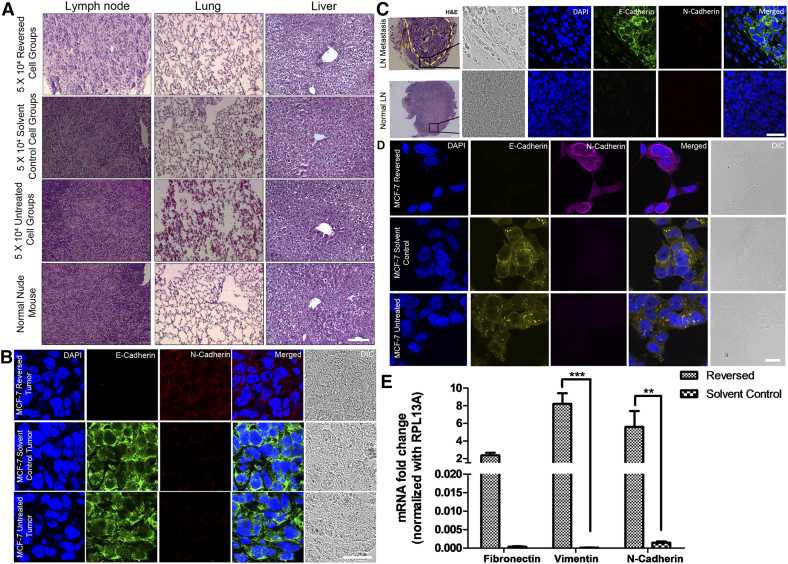
Autopsy revealed that in the 50,000 reversed-cell group, lymph node metastasis was detected in 4 out of 8 mice (Figure 2A). Immunostaining of epithelial-mesenchymal transition (EMT) markers revealed that primary tumors formed from both solvent control and untreated cells expressed the epithelial marker E-cadherin [27] while those from reversed cells expressed the mesenchymal marker N-cadherin (Figure 2B). Interestingly, the metastasis detected in the lymph node re-expressed E-cadherin (Figure 2C). The same epithelial/mesenchymal expression patterns were also found in cells undergoing apoptosis reversal in vitro (Figure 2D). Transcript levels of N-cadherin and other mesenchymal markers including fibronectin and vimentin [28] in cultured reversed cells were all elevated when compared to those in solvent control cells (Figure 2E).
Figure 2.
EMT occurred during apoptosis reversal both in vitro and in vivo.
(A) H&E-stained sectioned organs from tumor-bearing mice in the 50,000-injected-cell groups versus normal mice. Scale bar: 100 μm. (B) Immunofluorescence images of E-cadherin (green) and N-cadherin (red) in reversed (top), solvent control (middle) and untreated (bottom) MCF-7 xenografts. Scale bar: 25 μm. (C) H&E-stained and immunofluorescence images showing a tumor (T) in the lymph node (LN) from the reversed MCF-7 group (top) and a normal lymph node (bottom). Dash line indicates the LN-T boundary. Scale bar: 50 μm (H&E); 25 μm (confocal). (D) Immunofluorescence images showing the switch from epithelial (E-cadherin, green) to mesenchymal marker (N-cadherin, magenta) in reversed cells. Scale bar: 25 μm. (B-D) Nuclei were stained with DAPI. (E) Relative expressions of mesenchymal markers in the reversed and solvent control cells. Data represent mean±SEM (n = 3). **P < .01; ***P < .001.
CD44+/CD24- CSC-Like Cells Were Transited from NSCCs During Apoptosis Reversal
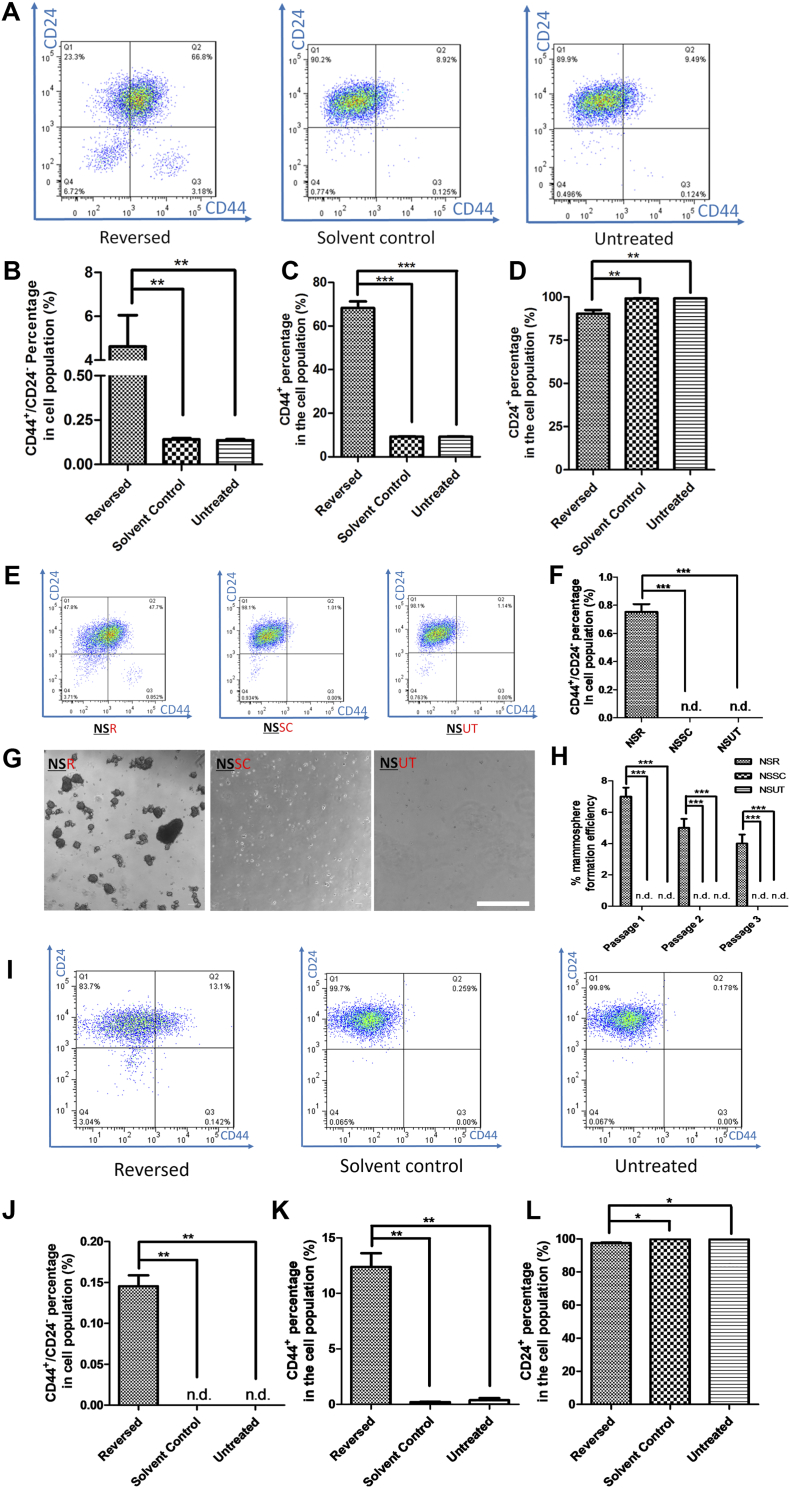
Next, we used flow cytometry to evaluate the percentage of tumor-initiating or CSC-like cells before and after apoptosis reversal, based on the expression of two cell-surface markers, CD44 and CD24 [8]. Cells with the CD44-/CD24+ configuration were designated as NSCCs, while those with CD44+/CD24- were designated as CSC-like cells [29]. We found that, as in previous studies [30], <2% (~0.1% in our case) of untreated MCF-7 cells were CD44+/CD24- (Figure 3A), whereas the percentage of reversed cells that were CD44+/CD24- was significantly elevated (Figure 3A and B). Higher CD44 and lower CD24 expressions were separately observed in the reversed MCF-7 population than in the control groups (Figure 3C and D). Further confirmation was provided by immunostaining of these two markers in cultured MCF-7 cells (Supplementary Figure 6A), and in the corresponding tumor sections (Supplementary Figure 6B). Although the increased percentage of CSC-like cells in the reversed population could be attributed to the enrichment of pre-existing CSCs present in the cell population before treatment [31], an alternative explanation could be that some of the original NSCCs were transformed into new CSCs by apoptosis reversal. To test the latter hypothesis, we induced apoptosis in NSCCs from untreated cells. CellEvent™ staining and FACS were used to isolate caspase-activated cells. Solvent-treated NSCC controls were sorted in parallel (Supplementary Figure 7). We were able to show that new CSCs could originate from NSCCs after apoptosis reversal (NSR) since no CSC was observed among NSCCs treated with solvent only (NSSC) or untreated NSCCs (NSUT) (Figure 3E and F). Moreover, these NSRs formed stable spheroids in the in-vitro mammosphere assay while NSSCs and NSUTs failed to yield any mammosphere (Figure 3G and H).
Figure 3.
Reversed breast cancer cells have higher percentage of cells with CSC markers (CD44+/CD24-).
(A) Distributions of MCF-7 cells expressing CD44 and CD24 in reversed, solvent control or untreated group. More cells had CSC markers (CD44+/CD24-) (Q3) after apoptosis reversal. (B-D) Percentage of MCF-7 cells expressing CD44+/CD24- (B), CD44+ (C) and CD24+ (D). Data represent mean±SEM. (n = 6). **P < .01; ***P < .001. (E) Expressions of CD44 and CD24 in reversed NSCC (NSR), solvent control NSCC (NSSC) and untreated NSCC (NSUT) cells by FACS analyses. (F) Percentages of CD44+/CD24- cells in all three groups. Data represent mean±SEM (n = 3). ***P < .001. n.d.: not detected. (G) Inverted microscopic images of mammospheres formed by NSRs, NSSCs and NSUTs. Scale bar: 500 μm. (H) Mammosphere formation efficiencies of the three passages of NSRs, NSSCs and NSUTs. Data represent mean±SEM (n = 3). ***P < .001. n.d.: not detected. (I) Distributions of T47D cells expressing CD44 and CD24 in reversed, solvent control or untreated group. Some cells possessed CSC markers (CD44+/CD24-) (Q3) after apoptosis reversal. (J-L) Percentage of T47D cells expressing CD44+/CD24- (J), CD44+ (K) and CD24+ (L). Data represent mean±SEM. (n = 3). *P < .05; **P < .01.
Supplementary Figure 6.
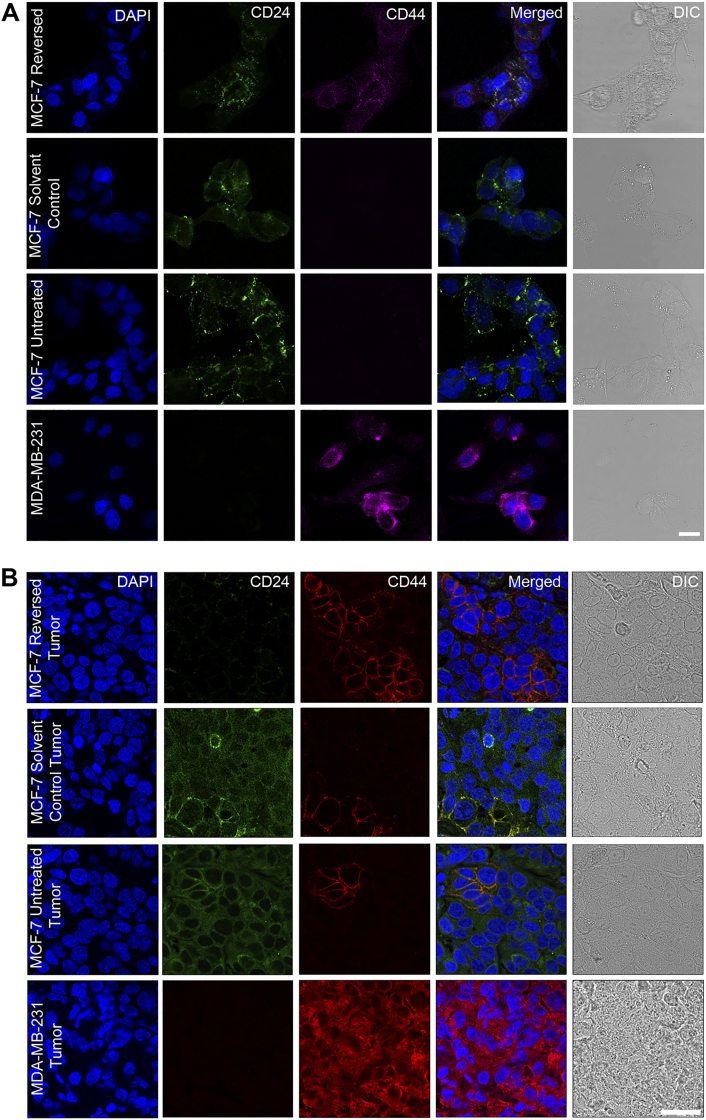
Confocal images showing immunofluorescence detection of CD44 and CD24 in reversed, solvent control and untreated MCF-7 cells (A) and xenografts (B). Reversal of apoptosis increased the percentage of cells expressing CD44 and decreased the percentage of cells expressing CD24. MDA-MB-231 was used as a positive control for CD44 staining. DAPI was used to stain the nuclei. Scale bar: 50 μm.
Supplementary Figure 7.
Schematics of fluorescence-activated cell sorting (FACS) of non-stem cancer cells (NSCCs) for the subsequent apoptosis reversal experiments.
The discovery on NSCC to CSC transition during apoptosis reversal prompted us to investigate whether this phenomenon was repeatable in different apoptotic induction condition and in other cancer cell line. Unlike MCF-7 cells, there was no CSCs detectable in untreated T47D population (Figure 3I). Results from T47D-paclitaxel model showed that after reversal of apoptosis, CD44+/CD24- cells appeared in the population, which could not be detected in the solvent control group (Figure 3I and J). Consistently, the increase of CD44 and decrease of CD24 expressions were significant in the reversed T47D cells when compared to untreated and solvent control groups (Figure 3K and L). These results suggested that reversal of apoptosis assists the transformation of NSCCs to CSCs and this is a general phenomenon in breast cancer cells.
The Expression of CSC Markers CD44 and CD24 Was Correlated with DNA Methylation Status at Their Promoters During Apoptosis Reversal
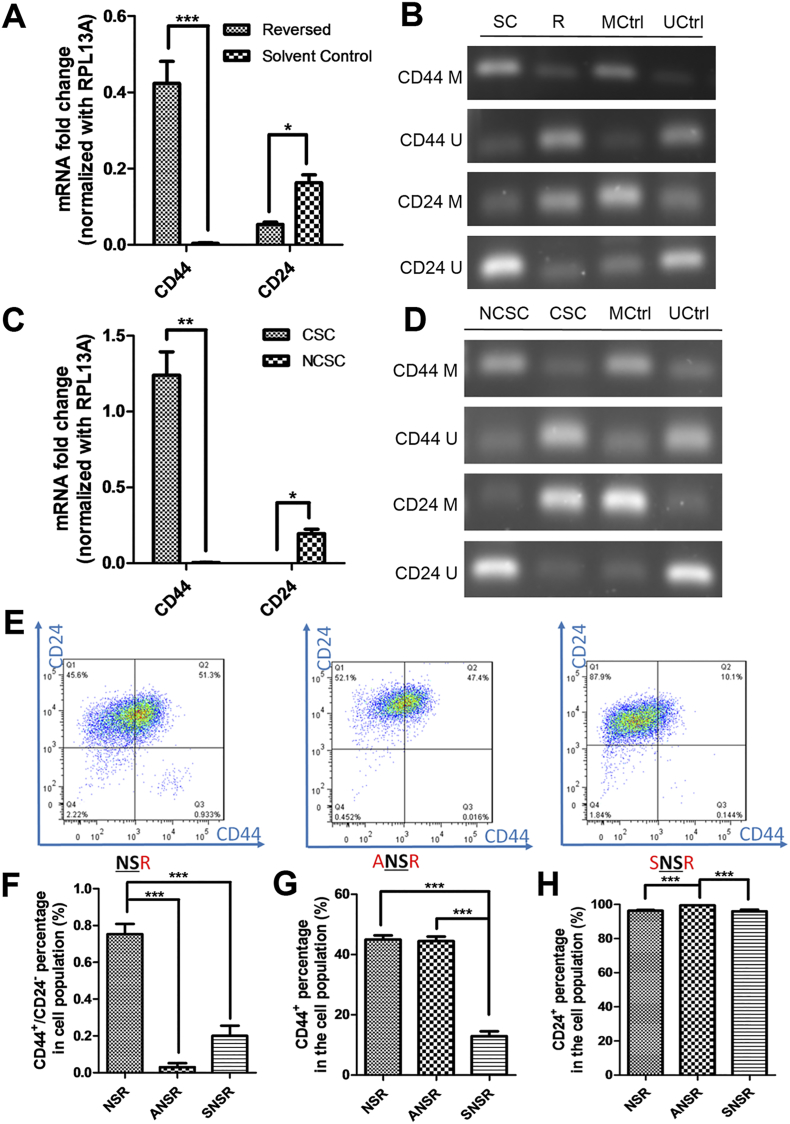
During cancer development, cells acquire capabilities for malignant growth through both genetic and epigenetic mechanisms [32]. However, genetic advantages take longer to accumulate under selective pressure whereas epigenetic changes are faster-acting. To test if the transformation from NSCCs to CSCs by apoptosis reversal involves methylation/demethylation of specific genes, we first investigated the transcript levels of CD44 and CD24 in the reversed versus control MCF-7 cells. Consistent with flow cytometry results, the mRNA level of CD44 was elevated while that of CD24 was decreased in the reversed cells compared to control (Figure 4A). The expression data were confirmed by the methylation statuses of CD44 and CD24 in these two groups using methylation-specific PCR (MSP). In control cells, CD44 were hypermethylated while CD24 were hypomethylated (Figure 4B). In contrast, cells having undergone apoptosis reversal showed hypomethylation of CD44 and hypermethylation of CD24 (Figure. 4B). We then induced apoptosis reversal on FACS-sorted NSCCs. Reversed cells from NSCCs (CellEvent™-positive) (NSRs) were further sorted into CSC-like cells (CSC) and non-CSC cells (NCSCs) (Supplementary Figure 8), and the mRNA levels of CD44 and CD24 in each group were consistent with their corresponding protein expressions (Figure 4C). In accordance, hypomethylation of CD44 and hypermethylation of CD24 were observed in CSCs when compared to NCSCs (Figure 4D). These results clearly indicated that methylation statuses of CD44 and CD24 determined the expression levels of these genes, and that apoptosis reversal drove the epigenetic modifications of CD44 and CD24 and gave rise to cells with CSC properties. The contribution by epigenetic regulations during the reversal of apoptosis and CSC generation was further confirmed by pre-treatment of NSCCs with the methylation inhibitor, 5-azacytidine (5-Aza) [33] (ANSRs), or the methyl group donor, S-(5'-adenosyl)-L-methionine chloride dihydrochloride (SAM) [34] (SNSRs), before the induction of apoptosis (Supplementary Figure 9). Both drugs suppressed the formation of CSC-like cells after apoptosis reversal, as indicated by the lower percentage of CD44+/CD24- cells in ANSRs and SNSRs (Figure 4E and F). Meanwhile, CD44 expression was suppressed in SNSRs and CD24 expression was up-regulated in ANSRs (Figure 4G and H). These results suggest that the NSCC-to-CSC transition during apoptosis reversal is at least partially attributable to epigenetic regulations.
Figure 4.
Dynamic regulation of CD44 and CD24 methylation statuses during apoptosis reversal.
(A and B) Relative expressions (A) and methylation statuses by methylation-specific PCR (B) of CD44 and CD24 in reversed versus solvent control cells. (C and D) Relative expressions (C) and methylation statuses (D) of CD44 and CD24 in CSCs and NCSCs sorted from NSRs. (B and D) MCtrl and UCtrl: methylated and unmethylated bisulfite-converted control human DNA; CD44 M and CD44 U: CD44 methylated- and unmethylated-specific primers; CD24 M and CD24 U: CD24 methylated- and unmethylated-specific primers. (E) FACS analyses of the expressions of CD44 and CD24 in NSR, ANSR (5-Aza-pre-treated NSR) and SNSR (SAM-pre-treated NSR). (F-H) Effects of 5-Aza or SAM pre-treatment on the percentage of CD44+/CD24- cells (F), CD44+ cells (G) and CD24+ cells (H) in NSR. For A, C, F, G and H, Data represent mean±SEM (n = 3). *P < .05; **P < .01; ***P < .001.
Supplementary Figure 8.
Schematics for the isolation of CSCs and NCSCs from NSRs.
Supplementary Figure 9.
Schematics for the pre-treatment of NSR with DNA-demethylating agent, 5-Aza, or methyl group donor, SAM, before the apoptosis reversal experiment.
Discussion
While previous studies showed that apoptosis reversal increased drug resistance, cell migration and invasion in vitro [13], our study is the first to demonstrate that cancer cells recovering from apoptosis reversal have higher tumorigenicity and metastatic potential in vivo, and that apoptosis reversal actually induced the formation of new CSCs from NSCCs in breast cancer cells. It has long been thought that cancer cell development is uni-directional: with CSCs at the apex, giving rise to progenitors and other differentiated cells [3]. Nevertheless, more recent studies have proposed the concept of CSC plasticity in which cells can transit between the non-CSC and the CSC states in both directions [35], [36], [37]. Our current study has clearly shown that the percentage of CSC-like cells (CD44+/CD24-) increased in the cell population which had undergone apoptosis reversal, and that these CSCs originated from NSCCs.
Besides, here we show that by inhibiting DNA methylation or demethylation before apoptosis induction, the CSC formation rate after apoptosis reversal is also reduced. Other epigenetic mechanisms [38] may also be involved. Therefore, it is critical to consider the involvement of chromatin remodeling during apoptosis reversal.
This study has clearly shown that through apoptosis reversal, NSCCs are transformed into CSCs, at least partly by the epigenetic modifications of CSC marker genes, and the resulting apoptosis-reversed cells undergo EMT [39] and possess enhanced tumorigenicity and metastatic properties in vivo. Thus when applying the commonly used tactic of eliminating cancer cells by triggering apoptosis [8], [9], [10], one needs to be mindful of at least ensuring cancer cells undergoing treatment have passed the point-of-no-return before terminating the treatment, in order to minimize the chances of relapse and/or metastasis.
The following are the supplementary data related to this article.
Supplementary material
Funding
This work was supported by the Innovative Technology Fund of Innovation Technology Commission: Funding Support to Partner State Key Laboratories in Hong Kong (to H.-M.L., M.-C.F., and S.-Y.T.), the Lo Kwee-Seong Biomedical Research Fund (to H.-M.L.), and Lee Hysan Foundation (to H.-M.L. and M.-C.F.).
Role of the Funding Source
All funding agencies play no role in the research design, or collection, analysis or interpretation of data or in the manuscript preparation, editing or review.
Acknowledgments
Acknowledgements
This work was supported by the Innovative Technology Fund of Innovation Technology Commission: Funding Support to Partner State Key Laboratories in Hong Kong, the Lo Kwee-Seong Biomedical Research Fund, and Lee Hysan Foundation. Y.X. was supported by the postgraduate studentship from the CUHK. We thank Ms. J-Y. Chu for copyediting the manuscript.
Conflict of Interest Statement
We declared no conflict of interest.
Contributor Information
Hon-Ming Lam, Email: honming@cuhk.edu.hk.
Ming-Chiu Fung, Email: fl9923mc@gmail.com.
Suk-Ying Tsang, Email: fayetsang@cuhk.edu.hk.
References
- 1.McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien CA, Kreso A, Dick JE. Cancer Stem Cells in Solid Tumors: An Overview. Semin Radiat Oncol. 2009;19:71–77. doi: 10.1016/j.semradonc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, La Noce M, Laino L, De Francesco F, Papaccio G. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB J. 2013;27:13–24. doi: 10.1096/fj.12-218222. [DOI] [PubMed] [Google Scholar]
- 6.Yu Y, Ramena G, Elble RC. The role of cancer stem cells in relapse of solid tumors. Front Biosci. 2012;4:1528–1541. doi: 10.2741/e478. [DOI] [PubMed] [Google Scholar]
- 7.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 8.Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25:4798–4811. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- 9.Wong RS. Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassan M, Watari H, AbuAlmaaty A, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014;2014:150845. doi: 10.1155/2014/150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Tang HL, Tang HM, Mak KH, Hu S, Wang SS, Wong KM, Wong CS, Wu HY, Law HT, Liu K. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol Biol Cell. 2012;23:2240–2252. doi: 10.1091/mbc.E11-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang HL, Yuen KL, Tang HM, Fung MC. Reversibility of apoptosis in cancer cells. Br J Cancer. 2009;100:118–122. doi: 10.1038/sj.bjc.6604802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang SS, Xie X, Wong CST, Choi PY, Fung MC. HepG2 cells recovered from apoptosis show altered drug responses and invasiveness. HBPD Int. 2014;13:293–300. doi: 10.1016/s1499-3872(14)60042-4. [DOI] [PubMed] [Google Scholar]
- 14.Grant S. Therapeutic Protein Kinase Inhibitors. Cell Mol Life Sci. 2009;66:1163–1177. doi: 10.1007/s00018-008-8539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes FA, Walters RS, Theriault RL, Forman AD, Newton LK, Raber MN, Buzdar AU, Frye DK, Hortobagyi GN. Phase II trial of taxol, an active drug in the treatment of metastatic breast cancer. J Natl Cancer Inst. 1991;83:1797–1805. doi: 10.1093/jnci/83.24.1797-a. [DOI] [PubMed] [Google Scholar]
- 16.Reichman BS, Seidman AD, Crown JPA, Heelan R, Hakes TB, Lebwohl DE, Gilewski TA, Surbone A, Currie V, Hudis CA. Paclitaxel and Recombinant Human Granulocyte-Colony-Stimulating Factor as Initial Chemotherapy for Metastatic Breast-Cancer. J Clin Oncol. 1993;11:1943–1951. doi: 10.1200/JCO.1993.11.10.1943. [DOI] [PubMed] [Google Scholar]
- 17.Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by taxol. Nature. 1979;277:665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- 18.Schiff PB, Horwitz SB. Taxol stabilizes microtubules in mouse fibroblast cells. Proc Natl Acad Sci U S A. 1980;77:1561–1565. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haldar S, Jena N, Croce CM. Inactivation of Bcl-2 by Phosphorylation. Proc Natl Acad Sci U S A. 1995;92:4507–4511. doi: 10.1073/pnas.92.10.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 21.Shaw FL, Harrison H, Spence K, Ablett MP, Simoes BM, Farnie G, Clarke RB. A Detailed Mammosphere Assay Protocol for the Quantification of Breast Stem Cell Activity. J Mammary Gland Biol Neoplasia. 2012;17:111–117. doi: 10.1007/s10911-012-9255-3. [DOI] [PubMed] [Google Scholar]
- 22.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponti D, Zaffaroni N, Capelli C, Daidone MG. Breast cancer stem cells: An overview. Eur J Cancer. 2006;42:1219–1224. doi: 10.1016/j.ejca.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 24.Vermeulen L, Melo FDSE, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 25.Plaks V, Kong NW, Werb Z. The Cancer Stem Cell Niche: How Essential Is the Niche in Regulating Stemness of Tumor Cells? Cell Stem Cell. 2015;16:225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 27.Lu L, Zhou D, Jiang X, Song K, Li K, Ding W. Loss of E-cadherin in multidrug resistant breast cancer cell line MCF-7/Adr: possible implication in the enhanced invasive ability. Eur Rev Med Pharmacol Sci. 2012;16:1271–1279. [PubMed] [Google Scholar]
- 28.Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147:631–643. doi: 10.1083/jcb.147.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin H, Glass J. The Phenotypic Radiation Resistance of CD44(+)/CD24(-or) (low) Breast Cancer Cells Is Mediated through the Enhanced Activation of ATM Signaling. PLoS One. 2011;6:e24080. doi: 10.1371/journal.pone.0024080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borst P. Cancer drug pan-resistance: pumps, cancer stem cells, quiescence, epithelial to mesenchymal transition, blocked cell death pathways, persisters or what? Open Biol. 2012;2:120066. doi: 10.1098/rsob.120066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadikovic B, Al-Romaih K, Squire JA, Zielenska M. Cause and Consequences of Genetic and Epigenetic Alterations in Human Cancer. Curr Genomics. 2008;9:394–408. doi: 10.2174/138920208785699580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christman JK. 5-Azacytidine and 5-aza-2 '-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 34.Laduron P. N-methylation of dopamine to epinine in brain tissue using N-methyltetrahydrofolic acid as the methyl donor. Nat New Biol. 1972;238:212–213. doi: 10.1038/newbio238212a0. [DOI] [PubMed] [Google Scholar]
- 35.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D'Alessio AC, Young RA, Weinberg RA. Poised Chromatin at the ZEB1 Promoter Enables Breast Cancer Cell Plasticity and Enhances Tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta PB, Fillmore CM, Jiang GZ, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic State Transitions Give Rise to Phenotypic Equilibrium in Populations of Cancer Cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 38.Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nat Struct Mol Biol. 2013;20:282–289. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- 39.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material