Abstract
Lipopolysaccharides (LPS) are a main constituent of the outer membrane of Gram-negative bacteria. Salmonella enterica, like many other bacterial species, are able to chemically modify the structure of their LPS molecules through the PhoPQ pathway as a defense mechanism against the host immune response. These modifications make the outer membrane more resistant to antimicrobial peptides (AMPs), large lipophilic drugs, and cation depletion, and are crucial for survival within a host organism. It is believed that these LPS modifications prevent the penetration of large molecules and AMPs through a strengthening of lateral interactions between neighboring LPS molecules. Here, we performed a series of long-timescale molecular dynamics simulations to study how each of three key S. enterica lipid A modifications affect bilayer properties, with a focus on membrane structural characteristics, lateral interactions, and the divalent cation bridging network. Our results discern the unique impact each modification has on strengthening the bacterial outer membrane through effects such as increased hydrogen bonding and tighter lipid packing. Additionally, one of the modifications studied shifts Ca2+ from the lipid A region, replacing it as a major cross-linking agent between adjacent lipids and potentially making bacteria less susceptible to AMPs that competitively displace cations from the membrane surface. These results further improve our understanding of outer membrane chemical properties and help elucidate how outer membrane modification systems, such as PhoPQ in S. enterica, are able to alter bacterial virulence.
Introduction
Pathogenic bacteria are a significant cause of human disease, with Gram-negative bacterial infections being both more difficult to treat and more likely to become resistant to antibiotics than their Gram-positive counterparts (1, 2, 3). Gram-negative bacteria contain a thin peptidoglycan cell wall surrounding their plasma membrane and a second, outer membrane. This outer membrane is distinct from typical plasma membranes both in its structure and its lipid constituents. Although most bilayers contain phospholipids in both leaflets, the outer membrane is highly asymmetric, with an external leaflet composed almost entirely of lipopolysaccharides (LPS) and an inner leaflet rich in the more prototypical phosphatidylethanolamine lipids (4, 5, 6). The structure of LPS is different from that of phospholipids in that it contains multiple hydrophobic tails covalently linked to a large, highly charged polysaccharide (6, 7). Because of its unique structure, LPS is central to the organization and function of the bacterial outer membrane and is implicated in its low permeability and antibiotic resistance (8, 9, 10).
LPS from different bacterial species have the same general structure, consisting of three distinct regions: lipid A, a hydrophobic anchor that contains a variety of gel-like lipid chains depending on the species (6); the core region, containing a collection of branched oligosaccharides and several negatively charged moieties; and the O-antigen, a polymer of repeating saccharide subunits. Because of their large number of phosphoryl and carboxyl groups, LPS membranes must be stabilized by a network of divalent cations bridging these moieties (11, 12, 13, 14). This unique structure leads to a gel state of very low fluidity at the membrane center that acts as a barrier against polar solutes, whereas the hydrated core region with its strong charge interactions impedes the movement of hydrophobic molecules (15), making the outer membrane a highly effective barrier to translocation (6, 9, 16).
Salmonella enterica have evolved a defense mechanism in which they remodel the LPS in their outer membrane through the PhoPQ two-component regulatory system (17). This pathway is triggered by sensed changes in the environment that occur during host infection (6), such as low divalent ion concentration (18), acidic conditions (19), hyperosmotic stress (20), and the presence of antimicrobial peptides (AMP) (21), and is critical both for attaining full virulence as well as survival within the host macrophages (22). When activated, the PhoPQ pathway increases transcription of genes responsible for LPS modifications, resulting in three key additions to lipid A (Fig. 1): a palmitoyl chain, a hydroxyl group, and a positively charged aminoarabinose sugar (23). These modifications lead to an increased resistance to host cationic AMPs (24), a reduced permeability to large lipophilic agents (10), and a decreased ability to activate an immune response through TLR4 (25), making them essential to the pathogen’s ability to survive in a host.
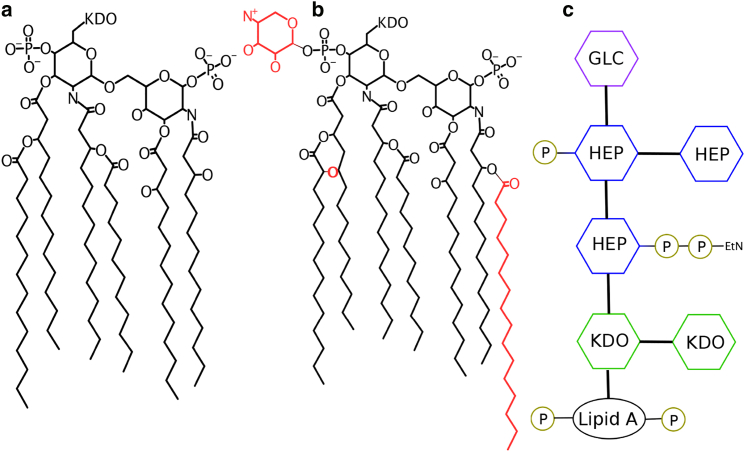
Figure 1.
Structure of LPS used in this study. (A) Shown here is the structure of unmodified lipid A. (B) Modification by the PhoPQ regulatory system results in three key additions to the lipid A structure, shown in red. (C) Shown here is a pictorial representation of Rc LPS in S. enterica, with lipid A shown in black and the sugars of the core region denoted by colored hexagons. EtN, ethanolamine; Glc, glucose; Hep, heptose; KDO, 2-keto-3-deoxyoctulosonic acid; P, phosphate. To see this figure in color, go online.
To date, much experimental work has been done to understand the role of PhoPQ-mediated LPS modification on the structure and function of the outer membrane. S. enterica that constitutively express the PhoPQ system were observed to have a twofold decrease in susceptibility to the antibiotic drug novobiocin compared to the null mutant, showing that LPS modification through the PhoPQ system results in a stronger permeability barrier (10). Additionally, recent experiments on Langmuir monolayers of pure LPS or pure modified LPS have shown that LPS modifications help to stabilize the monolayer and prevent the penetration of novobiocin into the hydrocarbon tail region, as well as to reduce the influx of the hydrophobic fluorescent dye ethidium bromide in vivo (23). One proposed mechanism for this barrier strengthening is that these modifications increase the lateral interactions between neighboring LPS molecules, thus preventing the penetration of large molecules.
Though the role of the PhoPQ system is well studied, it is still unclear whether these LPS modifications do in fact affect permeability through increased lateral interactions and what roles specific modifications play in the structure and dynamics of these bilayers. Here, we present results from a series of molecular dynamics (MD) simulations designed to examine in more detail the effects of each modification. Long-timescale atomistic simulations of symmetric bilayers of eight distinct variations of LPS were performed (Table 1), with variations ranging from the unmodified form to modified LPS containing all three additions (mLPS). Results show that both the aminoarabinose and 2-hydroxyl groups function to strengthen interactions between neighboring LPS molecules through increased hydrogen bonding. Additionally, the incorporation of aminoarabinose onto the lipid A headgroup displaces Ca2+ ions and their associated water molecules, with aminoarabinose bridging similar functional groups to those that Ca2+ cross-links. Palmitoylation slightly weakens the hydrogen bonding network between LPS; however, this is offset by a thickening of the bilayer and more tightly packed, ordered lipid tails. Finally, for many of the properties studied here, the effects of LPS modifications appear to be cooperative, as modification effects are typically not strictly additive.
Table 1.
Descriptions of the 10 LPS Bilayer Systems Simulated on Anton 2
| System Description | |
|---|---|
| LPS | LPS |
| Asymmetric LPS | LPS leaflet + POPE leaflet |
| Partially modified LPS | LPS + OH |
| LPS + AAB | |
| LPS + palmitoyl | |
| LPS + OH + AAB | |
| LPS + OH + palmitoyl | |
| LPS + AAB + palmitoyl | |
| Modified LPS | LPS + OH + AAB + palmitoyl |
| Asymmetric mLPS | mLPS leaflet + POPE leaflet |
OH refers to the added hydroxyl group, AAB refers to the aminoarabinose sugar, and palmitoyl refers to the extra palmitoyl group. All systems are symmetric bilayers unless otherwise indicated.
Materials and Methods
Symmetric LPS and mLPS system preparation
Initial coordinates and structure for a single Escherichia coli LPS, consisting of lipid A and the R1 core, were obtained from the force field parameters of Wu et al. (26). To model the Rc chemotype derived from (27), all outer core sugars past Glc I were removed. An mLPS molecule was constructed by adding a 2-hydroxyl, a palmitoyl, and aminoarabinose groups to match the lipid A structure of S. enterica serovar typhimurium that constitutively express the PhoPQ system (17). The structure of lipid A, both before and after modification by the PhoPQ regulatory system, as well as the GalE mutant core region, are shown in Fig. 2.
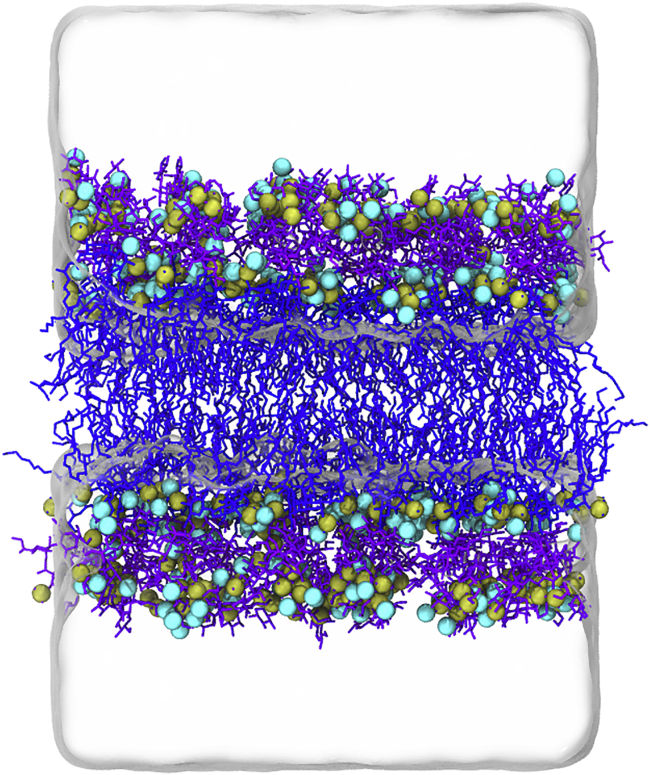
Figure 2.
The equilibrated symmetric LPS simulation box. Lipid A and the Rc core region are shown in blue and purple, respectively. LPS phosphate groups are shown in the tan sphere representation, calcium ions are displayed as cyan spheres, and water molecules are depicted as a transparent box. Hydrogen atoms, Na+, and Cl− have been removed for clarity. To see this figure in color, go online.
Initial standard and modified LPS monolayers were constructed by placing nine LPS molecules in a 3 × 3 grid, then replicating this grid in the +x, +y, and +xy directions to create a leaflet with 36 LPS molecules. Symmetric bilayers of were built by rotating a second copy of the monolayer 90° about the x axis, then aligning the two. Experimental data exist for LPS with both Mg2+ or Ca2+, as well as more exotic polyvalent cations (28, 29); Ca2+ was used as the divalent ion of choice in these simulations, consistent with past LPS simulation work (26, 30, 31, 32, 33). These Ca2+ ions were added to the core region of each leaflet in the amount needed for charge neutralization, then 20 Å water with 0.15 M NaCl was added in the ±z dimensions. All systems utilized the LPS parameter set of Wu et al. (26), modified Lennard-Jones parameters for sodium ion interactions with certain lipid oxygens (34), and TIP3P water (35). Parameters for the aminoarabinose sugar and the additional hydroxyl group of modified LPS were assigned by analogy to existing CHARMM 36 (C36) lipid (36, 37) and carbohydrate (38, 39, 40, 41) parameters; no additional parameters were needed for the palmitoyl group.
Systems were converted to AMBER-compatible format using chamber (42) in ParmEd. Both bilayers were minimized for 20,000 steps, using both the steepest descent and conjugate gradient algorithms, with 10 kcal/(molÅ2) restraints on all carbon and phosphorus atoms of the lipid A GlcN residues. Systems were heated to 310 K over 60 ps in the NVT ensemble, then simulated for 5 ns in the NPT ensemble with 10 kcal/(molÅ2) restraints to equilibrate the water density. Both bilayers were then simulated for 5.5 μs in the NPT ensemble to allow the bilayers to fully equilibrate in both area and lipid orientation. The pressure was maintained at 1.0 bar with semiisotropic coordinate scaling, utilizing the Berendsen barostat (43) and a relaxation time of 1.0 ps, and the temperature was controlled at 310 K using Langevin dynamics with a collision frequency of 1.0 ps−1. All hydrogen bonds were constrained using the SHAKE algorithm (44). A hard cutoff of 10.0 Å, which has been shown to suitably reproduce C36 lipid properties, was used (45). Long-range electrostatics were treated with the particle mesh Ewald method (46) with a grid spacing of 1.0 Å. Long-timescale equilibrations for all systems were performed locally with the GPU-accelerated version of pmemd in AMBER 16 (Amber Software, San Francisco, CA) (47). Final frames from these simulations (Fig. 2), referred to as the equilibrated LPS and mLPS bilayers, were used to construct two asymmetric LPS/palmitoyloleoyl phosphatidylethanolamine (POPE) and mLPS/POPE bilayers (Section S1.1 in the Supporting Material) and the six partially modified LPS systems described below.
Partially modified LPS system preparation
The final frame of the mLPS bilayer equilibration simulation was used to construct six partially modified LPS systems, representing each possible permutation of the three modifications studied here (Table 1). Systems were built by removing the appropriate moieties from the mLPS system. As before, neutralizing Ca2+ ions were added to the core region of each leaflet, then 20 Å water with 0.15 M NaCl was added in the ±z dimensions. These systems were minimized, heated, and density equilibrated as described above, followed by 1000 ns of long-timescale equilibration per system before being moved to Anton 2 (D.E. Shaw Research, Pittsburgh, PA) (48) for production MD.
MD simulations on Anton 2
In all, 10 distinct LPS bilayer systems were simulated on Anton 2 (D. E. Shaw Research) (Table 1): one symmetric LPS bilayer, one asymmetric LPS bilayer, one symmetric mLPS bilayer, one asymmetric mLPS bilayer, and six symmetric partially modified LPS bilayers. The asymmetric bilayer simulations were performed to determine whether the opposing leaflet identity affected properties of the LPS leaflet. In all analyses performed, we observed almost no discernible differences between symmetric and asymmetric LPS or mLPS leaflets (Section S1.2 in the Supporting Material). Therefore, only simulations of symmetric bilayers were performed for the partially modified LPS systems, as this effectively doubled the sampling. All simulations performed on Anton 2 (D. E. Shaw Research) were in the NPT ensemble, utilizing the Multigrator framework (49), with the Nosé-Hoover thermostat and semiisotropic MTK barostat. The temperature and pressure were controlled at 310 K and 1 bar, and a 2.0 fs time step was used. The cutoff varied by simulation for best performance, as is standard protocol for simulations on Anton 2 (D. E. Shaw Research) (48, 50), but was always between 10.0 and 13.3 Å; long-range electrostatics were evaluated with the u-series method (48). All systems were simulated for 7.0 μs. The first 2.0 μs were removed as equilibration, as this was when the largest changes to lipid area (Fig. 3) and volume (Fig. S4) were observed.
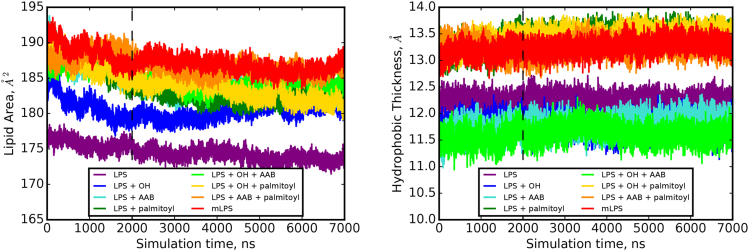
Figure 3.
Time series of lipid areas (left panel) and leaflet thicknesses (right panel) for all symmetric systems. The dashed horizontal line at 2.0 μs indicates the portion of the trajectory that was removed as equilibration. To see this figure in color, go online.
Simulation analysis
Trajectory analysis was performed for the final 5.0 μs of each simulation. Lipid area, hydrogen bonds, carbon-deuterium order parameters, and electron density profiles along the bilayer normal were calculated using CPPTRAJ (51) from AmberTools 17 (Amber Software) (52). Hydrogen bond calculations utilized a distance cutoff of 3.0 Å and an angle cutoff of 135°. All carbon-deuterium order parameters are reported as . Ca2+ coordinating groups were determined using a distance-based cutoff of 2.5 Å between Ca2+ and all LPS heavy atoms, calculated in Visual Molecular Dynamics (VMD) (Theoretical and Computational Biophysics Group, Urbana-Champaign, IL) (53) and updated every 20 frames. VMD was also utilized to calculate leaflet hydrophobic thicknesses by measuring the distance along the z-dimension between the first carbon atoms of the four acyl chains attached directly to the glucosamine sugars (C11, C21, C31, and C51) and the terminal carbons of all acyl chains. The area per lipid tail was calculated from the area per lipid divided by the number of acyl chains in each lipid. Ca2+ radial distribution functions (RDFs) were calculated using LOOS (Grossfield Lab, Rochester, NY) (54), and coordination numbers were determined by integration of the RDF through the first peak (2.5 Å). The Diffusion Coefficient Tool (55) plugin for VMD was used to determine two-dimensional mean-squared displacement (MSD) curves; the MSD was averaged over all lipids and multiple time origins in 500 to 2500 ns blocks with time origins separated by 100 ns, and the center of mass drift was subtracted for each leaflet. In all systems, however, the MSD was not linear with time (Figs. S5 and S6), instead displaying subdiffusive properties, so reliable diffusion coefficients could not be calculated from these data. Conformations of all lipid A and core oligosaccharide pyranose rings were monitored by calculating the C2-C3-C4-C5 dihedral; for all cases, the chair conformation predominated (Fig. S7).
Results
Gross bilayer properties
To distinguish the effects of LPS modification on the overall bilayer structure, the area per lipid and leaflet hydrophobic thicknesses were calculated and compared for all systems. The average lipid area (Fig. 3), per-leaflet hydrophobic thickness, and area per lipid tail for each system are given in Table 2. All three chemical modifications led to an increased lipid area (Fig. 4), with the most dramatic increase occurring with the added aminoarabinose group. For all three modification types, the largest contribution to area increase occurred upon adding that single modification to LPS, whereas subsequent chemical modification of an already partially modified system had a lesser effect (Fig. 4). Finally, we note that although the addition of either aminoarabinose or the palmitoyl tail led to an area increase to ∼183 Å2, both modifications together increased the area further to ∼186 Å2, indicating that these two modifications do not occupy the same space in the xy-plane.
Table 2.
Mesoscopic Bilayer Properties for All Eight Symmetric LPS Bilayer Systems
| System | Lipid Area (Å2) | Hydrophobic Thickness, per Leaflet (Å) | Area per Lipid Tail (Å2) |
|---|---|---|---|
| LPS | 174.0 0.3 | 12.2 0.1 | 29.0 0.1 |
| LPS + OH | 180.2 0.5 | 11.9 0.1 | 30.0 0.1 |
| LPS + AAB | 183.2 0.2 | 11.8 0.1 | 30.5 0.1 |
| LPS + palmitoyl | 182.4 0.3 | 13.5 0.1 | 26.1 0.1 |
| LPS + OH + AAB | 184.2 0.3 | 11.6 0.1 | 30.7 0.1 |
| LPS + OH + palmitoyl | 183.1 0.7 | 13.5 0.1 | 26.2 0.1 |
| LPS + AAB + palmitoyl | 186.2 0.2 | 13.3 0.1 | 26.6 0.1 |
| mLPS | 186.8 0.4 | 13.3 0.1 | 26.7 0.1 |
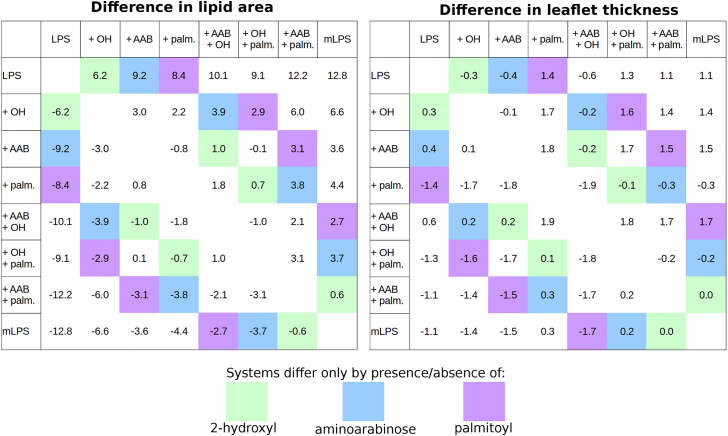
Figure 4.
Difference matrix for average lipid area (left panel) and leaflet hydrophobic thickness (right panel). Systems that differ only by the presence/absence of one modification are highlighted in green for 2-hydroxyl, blue for aminoarabinose, and purple for the palmitoyl tail. To see this figure in color, go online.
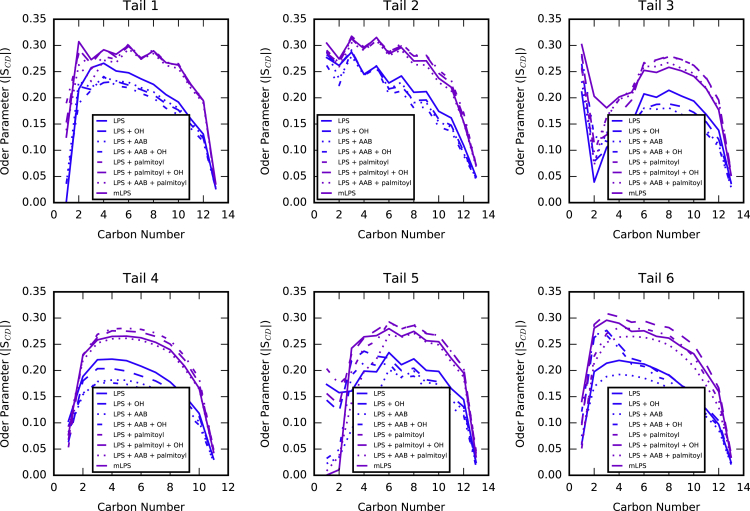
The bilayer thickness was directly affected by the addition of a palmitoyl tail: addition of the palmitoyl tail led to an increased hydrophobic thickness of ∼1.5 Å (Fig. 4, purple boxes) to compensate for the longer lipid tail, whereas inclusion of 2-hydroxyl (green boxes) or aminoarabinose (blue boxes) led to a slight thinning of the bilayer’s hydrophobic thickness, likely because of increased spreading of the lipid tails to accommodate the lipid area increase that accompanies these modifications. In addition to bilayer thickening, palmitoylation also resulted in a decreased area per lipid tail and a related increase in lipid tail ordering. This trend was observed in all six lipid tails common to all eight LPS species (Fig. 5) and indicates that palmitoylation may serve to decrease bilayer fluidity.
Figure 5.
Lipid tail order parameters () for all six lipid tails. Data for systems containing the palmitoyl group are shown in purple, and those without are shown in blue. To see this figure in color, go online.
Modifications alter lateral interactions
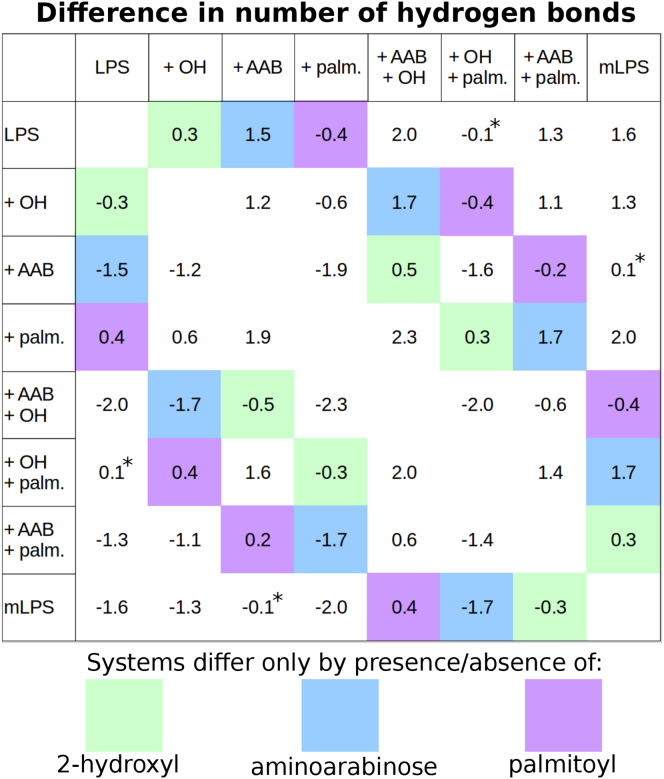
To examine how modifications affect interactions between neighboring LPS molecules, the average number of interlipid A hydrogen bonds was calculated for each of the eight systems (Table 3). The addition of the palmitoyl tail (Fig. 6, purple boxes) decreased the average number of hydrogen bonds that a lipid can make, likely a result of the increased lipid area and therefore interlipid distance. As hypothesized, the addition of the hydroxyl group (green boxes) led to a moderate increase in the number of hydrogen bonds, whereas the incorporation of aminoarabinose (blue boxes) had the much larger effect of increasing hydrogen bonding by 1.5–1.7 bonds per lipid. Finally, we observe that the effects of each addition are in some cases influenced by the presence of other modifications, though not to the extent observed in the lipid area or bilayer thickness discussed above.
Table 3.
Interlipid A Hydrogen Bonding for All Eight Symmetric LPS Bilayer Systems
| System | Interlipid A Hydrogen Bonds |
|---|---|
| LPS | 0.75 0.02 |
| LPS + OH | 1.03 0.01 |
| LPS + AAB | 2.24 0.02 |
| LPS + palmitoyl | 0.39 0.01 |
| LPS + OH + AAB | 2.71 0.03 |
| LPS + OH + palmitoyl | 0.67 0.01 |
| LPS + AAB + palmitoyl | 2.08 0.02 |
| mLPS | 2.34 0.02 |
Values are reported as the number of hydrogen bonds per lipid, and errors represent the standard error of the mean.
Figure 6.
Difference matrix for interlipid A hydrogen bonding. Systems that differ only by the presence/absence of one modification are highlighted in green for 2-hydroxyl, blue for aminoarabinose, and purple for the palmitoyl tail. All differences are significant at p < 0.0005, except those marked by an asterisk, which are significant only to the p < 0.01 level. To see this figure in color, go online.
Calcium and water interactions
LPS bilayers contain a network of divalent cations in the core region, which recent simulations have demonstrated are crucial for the bilayer to maintain a lamellar structure (56). Previous simulations of unmodified LPS have shown that these cations (typically Ca2+) are hexacoordinated, interacting primarily with water molecules and the phosphate groups (26, 32). To study how the addition of the positively charged aminoarabinose group alters Ca2+ localization, coordination numbers were calculated for the calcium ions and different charged groups of interest (Table 4). In all systems, the calcium cations were found to be primarily hexacoordinated, with the Ca2+-heavy atom radial distribution functions nearly identical between systems (Fig. S8). Additionally, in all eight systems the Ca2+ remained well hydrated, with each Ca2+ ion coordinated to three to four water molecules. There was only slight binding of the Ca2+ ions to the carboxyl groups of the inner core saccharides; the Ca2+ ions interacted primarily with the phosphate groups of the lipid A and core regions (Figs. S9–S11), with each ion coordinated to a total of ∼2 phosphate groups.
Table 4.
Coordination Numbers for Ca2+ in the Eight LPS Bilayer Systems, Corresponding to the Integration up through the First Peak of the Respective Radial Distribution Functions
| System | Ca2+-Wat | Ca2+-PLipA | Ca2+-PCore | Ca2+-CCore |
|---|---|---|---|---|
| LPS | 3.5 | 1.0 | 1.0 | 0.2 |
| LPS + OH | 3.5 | 1.0 | 1.1 | 0.2 |
| LPS + AAB | 3.5 | 0.6 | 1.3 | 0.3 |
| LPS + palmitoyl | 3.5 | 1.0 | 1.0 | 0.3 |
| LPS + OH + AAB | 3.6 | 0.6 | 1.3 | 0.3 |
| LPS + OH + palmitoyl | 3.5 | 1.0 | 1.1 | 0.2 |
| LPS + AAB + palmitoyl | 3.6 | 0.7 | 1.3 | 0.3 |
| mLPS | 3.6 | 0.6 | 1.3 | 0.3 |
PLipA and PCore correspond to the phosphorus atoms in the lipid A head group and in the core oligosaccharide region, respectively; CCore corresponds to the carboxyl carbons in the inner core.
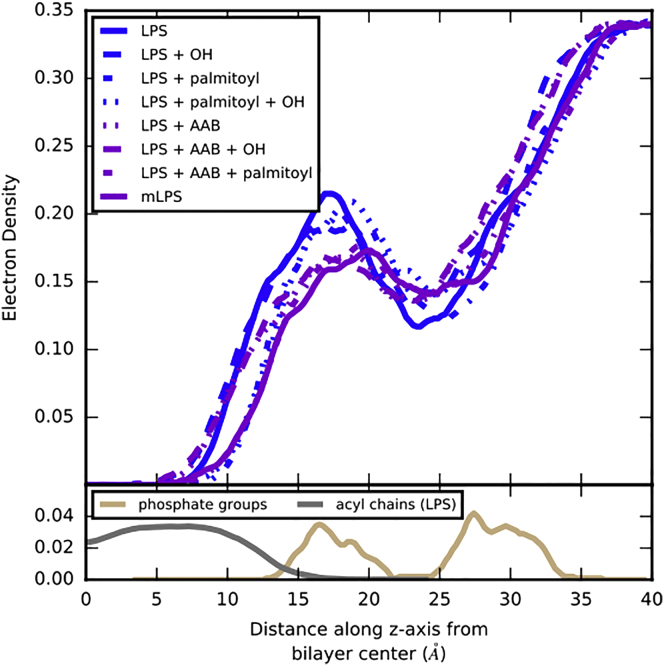
Though the total number of Ca2+-phosphate interactions were the same, the addition of the positively charged aminoarabinose group to one of the lipid A phosphate groups caused a shift in Ca2+ location: instead of a roughly equal distribution between the lipid A and core phosphate groups, Ca2+ ions were more likely to be found in the core region than the lipid A region in systems with the aminoarabinose modification. Additionally, electron density plots of the water distribution along the z axis (Fig. 7) show that the presence of aminoarabinose also affected the localization of water. Systems without aminoarabinose showed a greater density of water around ± ∼18 Å (where the lipid A phosphate groups are located) than systems with aminoarabinose present. This effect is associated with the Ca2+ ion localization, as Ca2+ ions in these systems are heavily hydrated.
Figure 7.
Water electron density along the z axis, highlighting the effect that aminoarabinose has on water distribution. Data for systems containing aminoarabinose are shown in purple, and those without are shown in blue. For reference, electron densities of the acyl chains (gray) and lipid A and core phosphate groups (tan) from the LPS system are plotted below. Data for the acyl chains have been scaled by a factor of 0.1. To see this figure in color, go online.
Calcium and aminoarabinose coordinate similar functional groups
RDFs, calcium coordination, and number density plots all revealed decreased Ca2+ interactions in the lipid A region in systems containing aminoarabinose. Additionally, these systems also displayed greater interlipid hydrogen bonding as a direct result of aminoarabinose presence. To determine whether aminoarabinose is bridging similar functional groups in this region as Ca2+, a detailed analysis was performed of all Ca2+ bridging interactions as well as a breakdown of all hydrogen bonds that aminoarabinose formed.
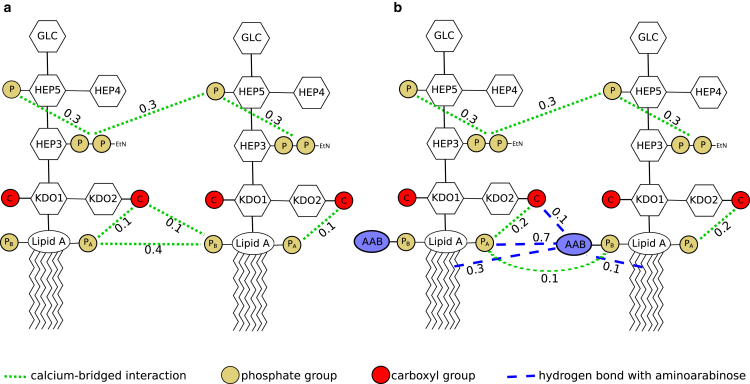
The dominant hydrogen bonds formed by the aminoarabinose amine and hydroxyl groups were with the lipid A PA phosphate group and carbonyl oxygens in the neighboring lipid tails (Fig. 8; Table S4). To a lesser extent, hydrogen bonding was also observed with carbonyl groups within the same molecule, as well as the KDO2 carboxyl group of neighboring molecules. In this way, aminoarabinose increases interactions both between and within the lipid A region of the bilayer.
Figure 8.
Dominant bridging interactions in systems (a) without and (b) with aminoarabinose present. Moieties bridged by Ca2+ are indicated in dotted green lines, and hydrogen bonds with aminoarabinose are depicted by dashed blue lines. Phosphate groups and the KDO carboxyl groups are represented as tan and red circles, respectively. Lines pointing to lipid tails indicate hydrogen bonds with tail carbonyl groups. Values listed are the probability that calcium is bridging these groups or that the hydrogen bond exists and were calculated from the LPS and LPS + AAB systems. Values for all other systems are given in Tables S4 and S5. To see this figure in color, go online.
Calcium ions are known to serve a similar cross-linking purpose in LPS (11, 13, 14). In our simulations, most Ca2+ ions coordinated two or three functional groups at a time, with some coordinating up to five functional groups; this was unchanged by the presence or absence of aminoarabinose in the system (Fig. S12). However, distinct differences were observed in the types of bridging interactions present as well as their likelihood (Fig. 8; Table S5). In systems without aminoarabinose, the dominant Ca2+-mediated interactions observed were between the PA and PB phosphate groups of neighboring molecules, as has been observed in previous simulations of lipid A bilayers (56), as well as both inter- and intralipid bridging of the heptose 3 and heptose 5 phosphate groups. Additionally, bridging of the PA (intramolecular) and PB (intermolecular) phosphates to the KDO2 carbonyl group was present to a lesser degree.
Aminoarabinose presence greatly perturbed Ca2+ interactions in the lipid A region but left bridging interactions in the core unaffected. PA-PB bridges between neighbors were greatly reduced, whereas the intermolecular PB-KDO2 bridges were entirely eliminated. Instead, the dominant interlipid links in the lipid A region were mediated by aminoarabinose to the same PA and KDO2 carboxyl groups that Ca2+ bridged in nonaminoarabinose systems, as well as to a new acceptor in the lipid tails.
Discussion
Each modification studied here has a distinct effect on both the larger scale structure of the bilayer as well as the interaction between neighboring lipids and ions. Of the three, 2-hydroxylation has the least prominent effect, resulting in a slightly larger lipid area as well as a modest increase to interlipid hydrogen bonding. It is interesting to note that the number of additional hydrogen bonds hydroxyl forms (0.3–0.5 per lipid) closely matches number of hydrogen bonds lost upon palmitoyl incorporation (0.2–0.4 per lipid). In this way, hydroxylation may not serve to function on its own, per se, so much as to compensate for the slight loss in lateral electrostatic interactions that palmitoylation confers.
Palmitoylation leads to an increased hydrophobic thickness and decreased fluidity of the lipid tails, which may help to offset this detrimental weakening of interlipid hydrogen bonding. Additionally, the added palmitoyl tail is longer than the other six fatty acids in lipid A, which may increase hydrophobic and van der Waals interactions between adjacent lipids, thus strengthening the leaflet (6). Indeed, pentaacylated Salmonella display significant growth defects, especially in environments with low ionic strength (57), indicating a high dependence on the number of acyl chains; it is thought that with one fewer lipid tail, the hydrophobic interactions are no longer sufficient to counterbalance electrostatic repulsions within the lipid A headgroup (6). This increased stabilization of the hydrophobic region could explain why pagP-mediated acylation is, by itself, sufficient to render Salmonella less susceptible to cationic AMPs (24).
In vitro, LPS adopts a gel-like state with a nearly crystalline arrangement of lipid tails (6). Indeed, Langmuir monolayers of LPS and mLPS produce coherent scattering patterns in grazing-incidence x-ray diffraction experiments; these experiments reveal ordered domains within the monolayer, with an average lateral coherence length of 216 Å for LPS and 164 Å for mLPS (23). In these diffraction experiments, Nobre et al. observed that introduction of novobiocin increased the size of these crystalline domains, with a much more dramatic effect for LPS than mLPS; they suggest that this crystalline growth could be responsible for the ability of novobiocin to penetrate through the LPS leaflet. Jefferies et al. (58) recently proposed a similar mechanism for the AMP polymyxin B (PMB). They performed a series of coarse-grained simulations of six PMB molecules interacting with symmetric LPS bilayers and found that PMB induces the formation of crystalline patches within the bilayer (58). Atomistic simulations of PMB with symmetric and asymmetric bilayers of E. coli lipid A revealed a decrease in the lateral diffusion coefficients of the membrane lipids (59), further supporting this notion of increased membrane ordering upon peptide binding. Additionally, a number of AMPs are known to induce phase separation in more traditional phospholipid bilayers (60). The bilayer thickening and concomitant increase in lipid tail ordering we observed because of palmitoylation could make the differences between lipids in the liquid-ordered and liquid-disordered states less pronounced in mLPS than in LPS, potentially making the bilayer to be less susceptible to permeation at these boundaries.
Incorporation of aminoarabinose onto the phosphate group radically alters the localization of Ca2+ and water near the lipid A head group, as well as decreasing the net charge of the molecule and increasing interlipid hydrogen bonding by 1.5–1.7 bonds per lipid. Not only are these hydrogen bonds strengthening interactions between adjacent LPS, they also bridge many of the same key functional groups as Ca2+ within the lipid A headgroup, rendering Ca2+ less essential for bilayer stability. In addition, the decreased net charge conferred by aminoarabinose may help to protect bacteria from cationic AMPs that rely on favorable long-range electrostatic interactions to induce cellular association (61).
Additionally, some AMPs are known to competitively displace divalent cations from the LPS core (62, 63, 64), which is presumed to facilitate outer membrane diffusion through surface defects. In fact, this divalent ion displacement is the proposed mechanism by which the PhoQ sensor domain detects AMP presence (65). Lam et al. (66) used a charge binding site model of LPS to show that cationic AMPS and EDTA are able to displace Mg2+ from the LPS layer and weaken the bilayer. Furthermore, recent MD simulations of Pseudomonas lipid A also revealed Ca2+ displacement by the AMP polymyxin B, along with increased membrane curvature, with the effect more pronounced in the native pentaacylated systems than in hexaacylated forms (33). The authors also performed simulations of the pentaacylated form with aminoarabinose bound to both phosphate groups and observed that, although polymyxin surface binding occurred, it did not induce the membrane curvature seen in other systems (33); we note, however, that no Ca2+ ions were included in this system, so it is unclear from that work how aminoarabinose presence would affect this polymyxin-induced ion displacement. Given the decreased Ca2+ density around the lipid A headgroup and the fact that aminoarabinose is instead mediating these contacts, we hypothesize that the addition of aminoarabinose makes bacteria less dependent on Ca2+, thus protecting them from AMPs that induce defects through divalent ion displacement.
Although these modifications clearly serve to protect bacteria against host defenses, they also function to modulate the host immune response. The recognition of LPS through TLR4 is highly dependent on the number of acyl chains present; lipid IVa, a lipid A precursor, is able to bind to the TLR4–MD-2 complex, but does not induce the oligomerization necessary to activate the innate immune response (67). Crystal structures of the TLR4–MD-2 dimer indicate that the lipid A phosphate groups play a critical role in stabilizing the dimerization interface (68); these phosphate groups are exposed when lipid A is bound to MD-2, but lipid IVa binds in a different orientation, leaving its phosphate groups buried (68, 69). Recent simulations of MD-2 demonstrate a remarkable plasticity in the size of the hydrophobic cavity, which contracts when antagonists such as lipid IVa are bound, increasing flexibility of the gating loop that stabilizes the TLR4–MD-2 complex (70, 71); these results indicate that MD-2 binding is primarily hydrophobically driven, whereas changes to the headgroup structure are more likely to manifest through changes to the TLR4–MD-2 complex affinity (71). Both pentaacylated and heptaacylated forms of LPS are significantly less potent inducers of TLR4-mediated signaling (25, 57, 72), with six acyl chains of 12–14 carbons in length being the optimum for TLR4 activation in humans (73, 74). Aside from palmitoylation, the addition of aminoarabinose may also factor into the decreased immune response to PhoPQ-modified LPS (17), given the important role of the lipid A phosphates in the dimerization interface.
Although this work focused on PhoPQ-mediated modification in S. enterica, many other Gram-negative bacteria exhibit similar modification systems. PagP homologs have been discovered in Bordetella (75), Legionella (76), and Yersinia (77), among others (74, 78, 79), for which they play similar roles of increasing virulence, reducing AMP sensitivity, and decreasing TLR4 activation. Hydroxylation of lipid A acyl chains is also prevalent in Bordetella, Pseudomonas, and Legionella (78, 79). Finally, adornment of the lipid A phosphate groups with aminoarabinose or phosphoethanolamine are some of the most common LPS modifications, occurring in E. coli, P. aeruginosa, Y. pestis, V. cholerae, and others (74, 78, 79). Although less common than lipid A remodeling, modification of moieties within the core and O-antigen are known to occur; additional work must be done to characterize how changes within the core or O-antigen influence the properties studied here. Alterations to lipid A, like those studied here, directly impact the virulence of Gram-negative bacteria through direct bilayer strengthening and increased resistance to AMPs and lipophilic drugs; future studies of outer membrane modification systems will further elucidate the link between the multitude of LPS structural variations and their roles in pathogenesis.
Author Contributions
A.R. and J.W. designed the simulations. A.R. performed and analyzed the simulations. A.R. and J.W. analyzed the results and wrote the manuscript.
Acknowledgments
The authors thank Prof. David Gidalevitz and Dr. Michael Martynowycz for valuable discussions concerning the work presented here.
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health (grant 1R35GM119647). Anton 2 computer time was provided by the Pittsburgh Supercomputing Center through grant R01GM116961 from the National Institutes of Health. The Anton 2 machine at the Pittsburg Supercomputing Center was generously made available by D. E. Shaw Research.
Editor: D. Peter Tieleman.
Footnotes
Supporting Materials and Methods, twelve figures, five tables, and one data file are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30209-1.
Supporting Material
References
- 1.Delcour A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta. 2009;1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blair J.M., Webber M.A., Piddock L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 3.Miller S.I. Antibiotic resistance and regulation of the gram-negative bacterial outer membrane barrier by host innate immune molecules. MBio. 2016;7:e01541-16. doi: 10.1128/mBio.01541-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smit J., Kamio Y., Nikaido H. Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studies with lipopolysaccharide mutants. J. Bacteriol. 1975;124:942–958. doi: 10.1128/jb.124.2.942-958.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamio Y., Nikaido H. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 6.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raetz C.R. Bacterial endotoxins: extraordinary lipids that activate eucaryotic signal transduction. J. Bacteriol. 1993;175:5745–5753. doi: 10.1128/jb.175.18.5745-5753.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob. Agents Chemother. 1989;33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papo N., Shai Y. A molecular mechanism for lipopolysaccharide protection of Gram-negative bacteria from antimicrobial peptides. J. Biol. Chem. 2005;280:10378–10387. doi: 10.1074/jbc.M412865200. [DOI] [PubMed] [Google Scholar]
- 10.Murata T., Tseng W., Nikaido H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J. Bacteriol. 2007;189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock R.E. Alterations in outer membrane permeability. Annu. Rev. Microbiol. 1984;38:237–264. doi: 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- 12.Rietschel E.T., Kirikae T., Brade H. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 13.Kucerka N., Papp-Szabo E., Katsaras J. Effect of cations on the structure of bilayers formed by lipopolysaccharides isolated from Pseudomonas aeruginosa PAO1. J. Phys. Chem. B. 2008;112:8057–8062. doi: 10.1021/jp8027963. [DOI] [PubMed] [Google Scholar]
- 14.Nascimento A., Jr., Pontes F.J., Soares T.A. Hydration, ionic valence and cross-linking propensities of cations determine the stability of lipopolysaccharide (LPS) membranes. Chem. Commun. (Camb.). 2014;50:231–233. doi: 10.1039/c3cc46918b. [DOI] [PubMed] [Google Scholar]
- 15.Carpenter T.S., Parkin J., Khalid S. The free energy of small solute permeation through the Escherichia coli outer membrane has a distinctly asymmetric profile. J. Phys. Chem. Lett. 2016;7:3446–3451. doi: 10.1021/acs.jpclett.6b01399. [DOI] [PubMed] [Google Scholar]
- 16.Allende D., McIntosh T.J. Lipopolysaccharides in bacterial membranes act like cholesterol in eukaryotic plasma membranes in providing protection against melittin-induced bilayer lysis. Biochemistry. 2003;42:1101–1108. doi: 10.1021/bi026932s. [DOI] [PubMed] [Google Scholar]
- 17.Guo L., Lim K.B., Miller S.I. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 18.García Véscovi E., Soncini F.C., Groisman E.A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 19.Prost L.R., Daley M.E., Miller S.I. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Yuan J., Jin F., Sourjik V. Osmosensing by the bacterial PhoQ/PhoP two-component system. Proc. Natl. Acad. Sci. USA. 2017;114:E10792–E10798. doi: 10.1073/pnas.1717272114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bader M.W., Navarre W.W., Miller S.I. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 2003;50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- 22.Miller S.I., Mekalanos J.J. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J. Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nobre T.M., Martynowycz M.W., Gidalevitz D. Modification of Salmonella Lipopolysaccharides prevents the outer membrane penetration of novobiocin. Biophys. J. 2015;109:2537–2545. doi: 10.1016/j.bpj.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo L., Lim K.B., Miller S.I. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 25.Kawasaki K., Ernst R.K., Miller S.I. 3-O-deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through Toll-like receptor 4. J. Biol. Chem. 2004;279:20044–20048. doi: 10.1074/jbc.M401275200. [DOI] [PubMed] [Google Scholar]
- 26.Wu E.L., Engström O., Im W. Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys. J. 2013;105:1444–1455. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devyatyarova-Johnson M., Rees I.H., Jack D.L. The lipopolysaccharide structures of Salmonella enterica serovar Typhimurium and Neisseria gonorrhoeae determine the attachment of human mannose-binding lectin to intact organisms. Infect. Immun. 2000;68:3894–3899. doi: 10.1128/iai.68.7.3894-3899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snyder S., Kim D., McIntosh T.J. Lipopolysaccharide bilayer structure: effect of chemotype, core mutations, divalent cations, and temperature. Biochemistry. 1999;38:10758–10767. doi: 10.1021/bi990867d. [DOI] [PubMed] [Google Scholar]
- 29.Garidel P., Rappolt M., Brandenburg K. Divalent cations affect chain mobility and aggregate structure of lipopolysaccharide from Salmonella minnesota reflected in a decrease of its biological activity. Biochim. Biophys. Acta. 2005;1715:122–131. doi: 10.1016/j.bbamem.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Kirschner K.N., Lins R.D., Soares T.A. A glycam-based force field for simulations of lipopolysaccharide membranes: parametrization and validation. J. Chem. Theory Comput. 2012;8:4719–4731. doi: 10.1021/ct300534j. [DOI] [PubMed] [Google Scholar]
- 31.Wu E.L., Fleming P.J., Im W. E. coli outer membrane and interactions with OmpLA. Biophys. J. 2014;106:2493–2502. doi: 10.1016/j.bpj.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dias R.P., da Hora G.C., Soares T.A. Outer membrane remodeling: the structural dynamics and electrostatics of rough lipopolysaccharide chemotypes. J. Chem. Theory Comput. 2014;10:2488–2497. doi: 10.1021/ct500075h. [DOI] [PubMed] [Google Scholar]
- 33.Santos D.E.S., Pol-Fachin L., Soares T.A. Polymyxin binding to the bacterial outer membrane reveals cation displacement and increasing membrane curvature in susceptible but not in resistant lipopolysaccharide chemotypes. J. Chem. Inf. Model. 2017;57:2181–2193. doi: 10.1021/acs.jcim.7b00271. [DOI] [PubMed] [Google Scholar]
- 34.Venable R.M., Luo Y., Pastor R.W. Simulations of anionic lipid membranes: development of interaction-specific ion parameters and validation using NMR data. J. Phys. Chem. B. 2013;117:10183–10192. doi: 10.1021/jp401512z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jorgensen W., Chandrasekhar J., Klein M. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 36.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pastor R.W., Mackerell A.D., Jr. Development of the CHARMM force field for lipids. J. Phys. Chem. Lett. 2011;2:1526–1532. doi: 10.1021/jz200167q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guvench O., Greene S.N., Mackerell A.D., Jr. Additive empirical force field for hexopyranose monosaccharides. J. Comput. Chem. 2008;29:2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guvench O., Hatcher E.R., Mackerell A.D. CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J. Chem. Theory Comput. 2009;5:2353–2370. doi: 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guvench O., Mallajosyula S.S., Mackerell A.D., Jr. CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comput. 2011;7:3162–3180. doi: 10.1021/ct200328p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mallajosyula S.S., Guvench O., Mackerell A.D., Jr. CHARMM additive all-atom force field for phosphate and sulfate linked to carbohydrates. J. Chem. Theory Comput. 2012;8:759–776. doi: 10.1021/ct200792v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crowley M., Williamson M., Walker R. CHAMBER: comprehensive support for CHARMM force fields within the AMBER software. Int. J. Quantum Chem. 2009;109:3767–3772. [Google Scholar]
- 43.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 44.Krautler V., Van Gunsteren W.F., Hunenberger P.H. A fast SHAKE: algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001;22:501–508. [Google Scholar]
- 45.Lee J., Cheng X., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darden T., York D., Pedersen L. Particle mesh Ewald - an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 47.Case D.A., Cheatham T.E., 3rd, Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw D.E., Grossman J.P., Young C. SC 14 Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis. IEEE Press; 2014. Anton 2: raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer; pp. 41–53. [Google Scholar]
- 49.Lippert R.A., Predescu C., Shaw D.E. Accurate and efficient integration for molecular dynamics simulations at constant temperature and pressure. J. Chem. Phys. 2013;139:164106. doi: 10.1063/1.4825247. [DOI] [PubMed] [Google Scholar]
- 50.Shaw D.E., Dror R.O., Towles B. SC 09 Proceedings of the Conference on High Performance Computing Networking, Storage and Analysis. Vol. 65. ACM; 2009. Millisecond-scale molecular dynamics simulations on Anton. 65:1–65:11. [Google Scholar]
- 51.Roe D.R., Cheatham T.E., 3rd PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 52.Case D.A., Cerutti D.S., Kollman P. University of California; San Francisco: 2017. AMBER 2017. [Google Scholar]
- 53.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 54.Romo T.D., Grossfield A. LOOS: an extensible platform for the structural analysis of simulations. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2009;2009:2332–2335. doi: 10.1109/IEMBS.2009.5335065. [DOI] [PubMed] [Google Scholar]
- 55.Giorgino, T. 2015. Diffusion Coefficient Tool. GitHub: https://github.com/tonigi/vmd_diffusion_coefficient/.
- 56.Pontes F.J., Rusu V.H., Lins R.D. The effect of temperature, cations, and number of Acyl chains on the lamellar to non-lamellar transition in lipid-A membranes: A microscopic view. J. Chem. Theory Comput. 2012;8:3830–3838. doi: 10.1021/ct300084v. [DOI] [PubMed] [Google Scholar]
- 57.Murray S.R., Bermudes D., Low K.B. Extragenic suppressors of growth defects in msbB Salmonella. J. Bacteriol. 2001;183:5554–5561. doi: 10.1128/JB.183.19.5554-5561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jefferies D., Hsu P.C., Khalid S. Through the lipopolysaccharide glass: a potent antimicrobial peptide induces phase changes in membranes. Biochemistry. 2017;56:1672–1679. doi: 10.1021/acs.biochem.6b01063. [DOI] [PubMed] [Google Scholar]
- 59.Berglund N.A., Piggot T.J., Khalid S. Interaction of the antimicrobial peptide polymyxin B1 with both membranes of E. coli: a molecular dynamics study. PLoS Comput. Biol. 2015;11:e1004180. doi: 10.1371/journal.pcbi.1004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Epand R.M., Epand R.F. Bacterial membrane lipids in the action of antimicrobial agents. J. Pept. Sci. 2011;17:298–305. doi: 10.1002/psc.1319. [DOI] [PubMed] [Google Scholar]
- 61.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 62.Hancock R.E., Chapple D.S. Peptide antibiotics. Antimicrob. Agents Chemother. 1999;43:1317–1323. doi: 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hancock R.E., Scott M.G. The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. USA. 2000;97:8856–8861. doi: 10.1073/pnas.97.16.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hagge S.O., Hammer M.U., Gutsmann T. Calcium adsorption and displacement: characterization of lipid monolayers and their interaction with membrane-active peptides/proteins. BMC Biochem. 2006;7:15. doi: 10.1186/1471-2091-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bader M.W., Sanowar S., Miller S.I. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 66.Lam N.H., Ma Z., Ha B.Y. Electrostatic modification of the lipopolysaccharide layer: competing effects of divalent cations and polycationic or polyanionic molecules. Soft Matter. 2014;10:7528–7544. doi: 10.1039/c4sm01262c. [DOI] [PubMed] [Google Scholar]
- 67.Saitoh S., Akashi S., Miyake K. Lipid A antagonist, lipid IVa, is distinct from lipid A in interaction with Toll-like receptor 4 (TLR4)-MD-2 and ligand-induced TLR4 oligomerization. Int. Immunol. 2004;16:961–969. doi: 10.1093/intimm/dxh097. [DOI] [PubMed] [Google Scholar]
- 68.Park B.S., Song D.H., Lee J.O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 69.Garate J.A., Oostenbrink C. Lipid A from lipopolysaccharide recognition: structure, dynamics and cooperativity by molecular dynamics simulations. Proteins. 2013;81:658–674. doi: 10.1002/prot.24223. [DOI] [PubMed] [Google Scholar]
- 70.Paramo T., Piggot T.J., Bond P.J. The structural basis for endotoxin-induced allosteric regulation of the Toll-like receptor 4 (TLR4) innate immune receptor. J. Biol. Chem. 2013;288:36215–36225. doi: 10.1074/jbc.M113.501957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paramo T., Tomasio S.M., Bond P.J. Energetics of endotoxin recognition in the toll-like receptor 4 innate immune response. Sci. Rep. 2015;5:17997. doi: 10.1038/srep17997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Low K.B., Ittensohn M., Bermudes D. Lipid A mutant Salmonella with suppressed virulence and TNFalpha induction retain tumor-targeting in vivo. Nat. Biotechnol. 1999;17:37–41. doi: 10.1038/5205. [DOI] [PubMed] [Google Scholar]
- 73.Stöver A.G., Da Silva Correia J., Hershberg R.M. Structure-activity relationship of synthetic toll-like receptor 4 agonists. J. Biol. Chem. 2004;279:4440–4449. doi: 10.1074/jbc.M310760200. [DOI] [PubMed] [Google Scholar]
- 74.Miller S.I., Ernst R.K., Bader M.W. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 75.Preston A., Maxim E., Maskell D.J. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol. Microbiol. 2003;48:725–736. doi: 10.1046/j.1365-2958.2003.03484.x. [DOI] [PubMed] [Google Scholar]
- 76.Robey M., O’Connell W., Cianciotto N.P. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 2001;69:4276–4286. doi: 10.1128/IAI.69.7.4276-4286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bishop R.E., Gibbons H.S., Raetz C.R. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raetz C.R., Reynolds C.M., Bishop R.E. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Needham B.D., Trent M.S. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 2013;11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.