Abstract
The major categories of pancreatic neuroendocrine tumor (PanNET) are well-differentiated NET and poorly-differentiated neuroendocrine carcinoma. Sequencing of these tumors has identified multiple important genes in the pathogenesis of PanNETs, such as DAXX/ATRX, MEN1, TP53, RB and mTOR pathway genes. We identified a case of well-differentiated PanNET with high-grade progression with simultaneous low and high-grade histologic regions containing variable genomic profiles. We performed tumor microdissection and analyzed both regions using a 409-gene comprehensive cancer panel using next generation sequencing in addition to immunohistochemical and morphologic studies. The low-grade region showed a change in the DAXX gene as a copy number variant (CNV) deletion. The high-grade region showed CNV deletion changes in the DAXX gene as well as the MEN1 gene. We observed additional mutational changes in the PTEN gene and SMAD4 gene in the high-grade region. Our data supports that high-grade progression in PanNETs may be the result of the progressive accumulation of genetic changes (CNVs and point mutational changes) within the body of the tumor. NGS sequencing may provide pathologists and clinicians with ancillary information to accurately characterize and treat these tumors.
Keywords: pancreatic neuroendocrine tumor, high-grade progression, next-generation sequencing, proliferative index, chromosome instability, translational oncology
Introduction
Pancreatic neuroendocrine tumors (PanNETs) are relatively uncommon and account for roughly 1–2% of all pancreatic neoplasms. A grading system incorporating mitotic count and Ki-67-labelling index is employed by the World Health Organization (WHO) classification of neuroendocrine neoplasms of the digestive system (2010 guidelines).1 Well-differentiated neuroendocrine tumors (WD-NETs) are comprised of grade 1 (Ki-67 <3%, <2 mitotic figures/10 high-power fields (HPF)) and grade 2 (Ki-67 3–20%, 2–20 mitotic figures/HPF) lesions, and typically follow a far less aggressive clinical course than high-grade/poorly differentiated neuroendocrine carcinoma (PD-NEC) (grade 3; Ki-67 >20%, >20 mitotic figures/HPF), which are lethal. Grade 3 lesions are further divided into small and large cell types.1 Distinction between well-differentiated lesions and poorly-differentiated lesions is paramount, not only for prognosis, but also in terms of clinical response to chemotherapy regimens.2
It is recognized that not all grade 3 neuroendocrine neoplasms are morphologically poorly-differentiated (i.e. large cell or small cell type). Rather, despite Ki-67-labelling indices of >20%, some maintain well-differentiated neuroendocrine morphology.3–5 In addition, cases of WD-NET with apparent high-grade progression (grade 3) have been described previously.4,6 In these cases, diagnostic ambiguity is not easily resolved by traditional methods of analysis (histomorphology and proliferative index).
The literature currently supports that pure WD-NETs (G1 and G2) with increased Ki-67 indices (>20%), as well as WD-NETs with high-grade progression are molecularly distinct from pure PD-NECs (large cell or small cell carcinoma). Whereas PD-NECs have been proposed to be associated with mutations in TP53, RB1, and SMAD4, well-differentiated lesions are associated with mutations in genes such as DAXX, ATRX, and MEN1 in a mutually exclusive manner. Furthermore, grade 3 WD-NETs and WD-NETs with high-grade progression have been suggested to retain the latter mutation profile.4,6 As such, it is becoming increasingly apparent that the current WHO grade 3 category is comprised of two very distinct tumor types: (1) WD-NET with increased proliferation; and (2) PD-NEC, small or large cell types.1 Grouping them together does not accurately convey the diagnostic importance and distinction between these entities.3 While the prognostic and pathologic differences between WD-NETs and PD-NECs are well described, further study is required for these difficult to classify well-differentiated grade 3 lesions.
We identified a case of well-differentiated PanNET with high-grade progression, containing heterogeneous proliferative indices between low and high-grade histologic regions. The objective of this study was to investigate the differential histopathologic and molecular features in these two regions and paired non-neoplastic tissue using a 409-gene comprehensive cancer panel. The overall goal of this study was to identify differential mutational patterns in the heterogeneous low-grade vs high-grade regions of this tumor.
MATERIALS AND METHODS
Tumor Samples
The study was conducted according to the exempt rules for a single case specimen at the local institution. FFPE blocks of the specimen were sectioned to obtain unstained sections as well as a single Hematoxylin and Eosin stained slide. Three areas of interest relevant to the project were identified a) Non-neoplastic duodenal mucosa, b) Area corresponding to low-grade WD-NET, and c) Area corresponding to WD-NET with high-grade progression.
Tissue Microdissection and DNA Extraction
We used a modified tissue microdissection procedure to obtain the DNA from the relevant areas of interest in the case. Hematoxylin and Eosin (H and E) stains of the tissue of interest were reviewed by the pathologists (D.R.M, J.A.H., and R.R.G.). After marking the areas of interest on the H and E slides, FFPE sections on 5 μm-thick blank slides were pre-stained using the hematoxylin stain for four minutes. The corresponding areas of interest were then identified and carefully dissected under direct vision using an 80X stereomicroscope using a separate handled needle for each tissue sample. The tumor cell percentages in the microdissected regions were more than ~80%. Adequate care was taken to prevent any cross-contamination of tissue. Literature indicates the hematoxylin stain has minimal to no effect on the DNA sequencing protocol in a previous comprehensive study. The microdissected tissue was then cleaned using multiple rounds of washes in 100% ethanol to eliminate the hematoxylin stain and to remove any potential impurities. DNA extraction was then performed using the QIAamp DNA FFPE Tissue Kit (Cat #: 56404, Qiagen, Germantown, Md) according to the manufacturer’s instructions. The extracted DNA was quantified using the Qubit 2.0 DNA HS assay kit (Cat #: Q32854, Thermo Fisher Scientific, Waltham, Mass) and the Nanodrop 1000 Spectrophotometer (Thermo Fisher Scientific) to ensure availability of adequate DNA material.
Library Preparation for the 409 Cancer Gene Panel
Library construction of gDNA prepared from the microdissected FFPE samples was performed using the Ion AmpliSeq™ Library Kit 2.0 (Cat# 4480441, Thermo Fisher Scientific). A total of 10 ng gDNA per primer pool (4 primer pools for a total of ~40 ng) from FFPE samples was used as recommended in the product guide. Genomic DNA (gDNA) concentrations were measured prior to library preparation by qPCR quantitation using the TaqMan® by Life Technologies™ RNaseP Detection Reagents (FAM™) (Cat# 1512123, Life Technologies, Carlsbad, Calif). Briefly, samples were PCR amplified using the Ion AmpliSeqTM Comprehensive Cancer Panel (CCP, Cat# 4477685, Thermo Fisher Scientific) targeting 409 genes implicated in cancer research as potential cancer-causing genes. The Ion CCP panel contains 4 primer pools with a total of ~16,000 amplicons to enable amplification-based analysis and sequencing of the coding regions of 409 cancer relevant genes. Multiplexing of samples in an individual run was accomplished using the Ion Xpress barcode adapters 1-96 kit (Cat# 4474517, Life Technologies) according the manufacturer’s instructions.
Emulsion PCR and Sequencing
Equimolar library products from the four primer pools obtained after the library preparation were then purified using Agencourt©AMPure®XP magnetic beads (Cat# A63880, Beckman Coulter, Brea, Calif). The purified products were then quantitated using qPCR with either the Ion Library TaqMan™ Quantitation Kit (Cat. # 4468802, Thermo Fisher Scientific) or the KAPA Library Quantification Kits for Ion Torrent (Cat# KK4857, Kapa Biosystems, Wilmington, Mass). The multiplexed barcoded libraries were then enriched using emulsion PCR clonal amplification using the Ion PI™ Hi-Q™ OT2 kit (Cat# A26428, Thermo Fisher Scientific) and the Ion OneTouch 2 system for 16 cycles according to the manufacturer’s specifications. Following templating, samples were prepared for next generation sequencing using the Ion PI™ Hi-Q™ Sequencing 200 Reagents and the Ion PI™ sequencing nucleotides (Cat# A26431, A26432, Thermo Fisher Scientific) according to the manufacturer’s instructions. The final products were then loaded onto an Ion PI™ Chip v3 (Cat# QMB496W11, Thermo Fisher Scientific). Massively parallel next generation sequencing was performed using an Ion Torrent Proton semiconductor sequencing system (Thermo Fisher Scientific).
Bioinformatics Analysis of Ion Proton Sequencing Data
We adopted a hybrid approach to the analysis of the sequencing data. The primary analysis was performed using the Ion software followed by a confirmatory analysis using an open-source Linux-based pipeline to confirm the calls. The visualization of the sequencing data was provided by the open-source software, Integrated Genomics Viewer version 2.4.3 (Broad Institute, Cambridge, Mass).
Single Nucleotide Variant Pipeline
Primary alignment of the sequencing reads to the reference genome and subsequent base calling was performed using the instrument provided Torrent Suite software v.5.0.4 (Thermo Fisher Scientific). The Torrent Suite software pipeline was adapted in view of the highly optimized analysis parameters specific to Ion instrument data. The human genome build 19 was used for the alignment, calling and annotations of potential variant changes in the specimen. Subsequently, the generated BAM files were uploaded to the Ion Reporter Cloud. Detailed annotation of the individual variant calls was generated by the Ion Reporter v.5.2 (Thermo Fisher Scientific) software as shown previously.7 In addition to the Ion Reporter annotation, a custom built software pipeline composed of Linux bash scripts was used to confirm the somatic variants of single nucleotide variants (SNVs) and insertion-deletion (indel) changes called by Ion Reporter. For the purposes of variant calling, we set a coverage threshold of 200X reads for SNVs and 350X reads Indels at a variant allele fraction of 5%. Anything below these limits was not called.
Copy Number Variant Pipeline
For copy number changes (CNV) in the sample, the primary calling and annotation of the CNV calls was provided by the Ion Reporter software (v.5.2), which is based on a matched tumor-normal median of the absolute values of all pairwise differences (MAPD) algorithm. The confidence of the score is determined by the log likelihood that the called copy number state is not normal ploidy, i.e. 2 on autosomes (lower scores imply low confidence in the CNV calls). An orthogonal CNV analysis second pipeline was implemented to verify the CNV calls provided by Ion Reporter. The orthogonal CNV pipeline was implemented using the open-source software, CNVKit (available at https://github.com/etal/cnvkit).8 The CNVKit software uses a circular binary segmentation (CBS) algorithm to segment the targeted amplicon sequencing data to do the CNV calling.
Immunohistochemistry
The following immunohistochemical (IHC) stains were performed on the Benchmark Ultra (Ventana Medical Systems, Tucson, Ariz): Ki-67 (Ventana Medical Systems, Clone 30-9, prediluted), Synaptophysin (Thermo Scientific, Clone-SP11, dilution 1:100), Chromogranin (Ventana Medical Systems, Clone LK2H10, prediluted), and TP53 (Dako, Santa Clara, Calif, Clone DO-7, dilution 1:1600). IHC stains were performed on 4 μm-thick sections of formalin-fixed paraffin-embedded tissue. The sections were placed on the immunostainer and deparaffinized with EZ Prep solution. Antigen retrieval was performed using Ventana’s proprietary method (Protocol CC1) at pH 8 for all antibodies. After the primary antibody was applied, Ventana’s Opti-view DAB detection kit was used. The sections were counterstained with Hematoxylin. For the p-Akt IHC staining, 5 μm-thick sections were deparaffinized in xylene followed by heat-induced antigen retrieval in 10 mM Sodium Citrate buffer at 95°C. Slides were then incubated with clone D9E of Phospho-Akt (Ser473) antibody (1:8000, #4060, Cell Signaling Technology, Danvers, Mass) for 45 minutes at room temperature, followed by goat anti-rabbit IgG-Biotin secondary antibody (1:1000, sc-2040, Santa Cruz Biotechnology, Dallas, Texas). p-Akt was detected using the SignalStain® DAB Substrate Kit (#8059, Cell Signaling Technology) and counterstained with Hematoxylin.
RESULTS
Clinical and Histopathological Findings
A patient was found to have a 3 cm pancreatic head mass on a CT scan (Fig. 1), and subsequently underwent a pancreatoduodenectomy. Gross examination revealed a firm gray mass, 3.8 × 3.0 × 2.9 cm, centered in the pancreatic head, abutting the duodenum, and involving both the common bile duct and pancreatic duct.
FIGURE 1.
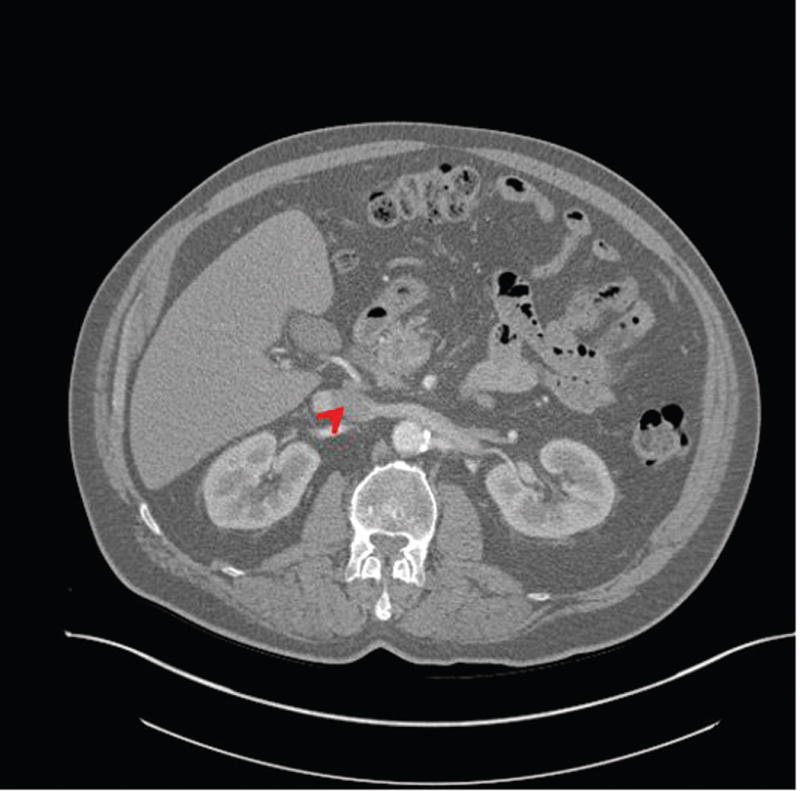
Cross sectional view of CT scan at the time of clinical presentation. A 3.8 × 3.0 × 2.9 cm mass was identified in the head of the pancreas (see arrow) of the patient.
Histologic examination showed a neuroendocrine tumor with areas of infiltrative growth pattern, invading the duodenum up to the level of submucosa as well as the peripancreatic tissue. Large vessel invasion was present and no lymph node metastases were identified (pT3N0).
The neoplasm contained both low-grade as well as high-grade regions. Low-grade areas contained well-circumscribed tumor nodules with trabecular (ribbon-like) and nested architecture admixed with areas of hyalinized, amyloid-like stroma (Fig. 2A). High-grade areas showed more overtly invasive features, associated with trabecular widening/solid areas, angulated tumor nests, as well as desmoplastic-type stroma (Fig. 2B). Mitotic figures were numerous in high-grade regions, and nuclear pleomorphism was more pronounced. Occasional foci with single cell necrosis were present (Fig. 2C). While some of the tumor nests contained features reminiscent of large cell carcinoma (e.g., prominent nucleoli), there was no geographic comedo-type necrosis. Focal osseous metaplasia was also present adjacent to high-grade foci. Interestingly, both low-grade and high-grade areas were associated with an invasive growth pattern and perineural invasion.
FIGURE 2.
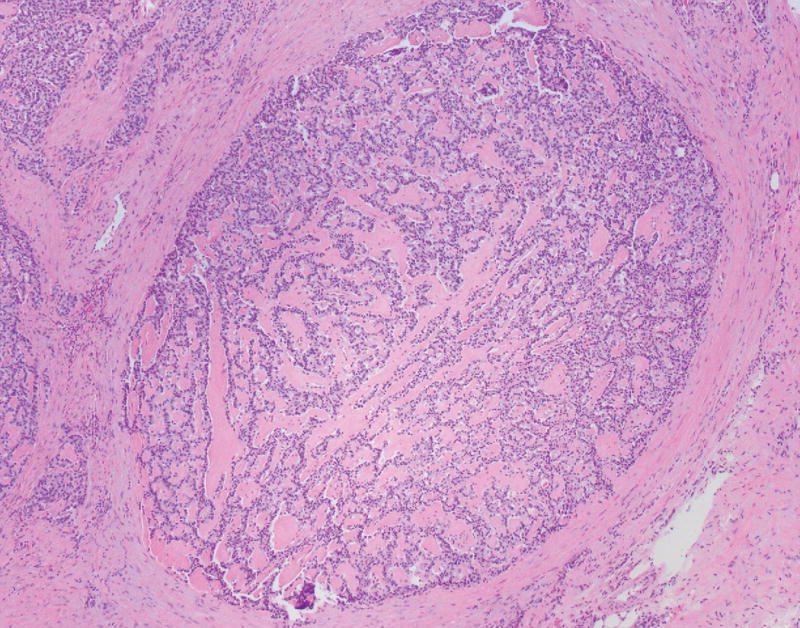
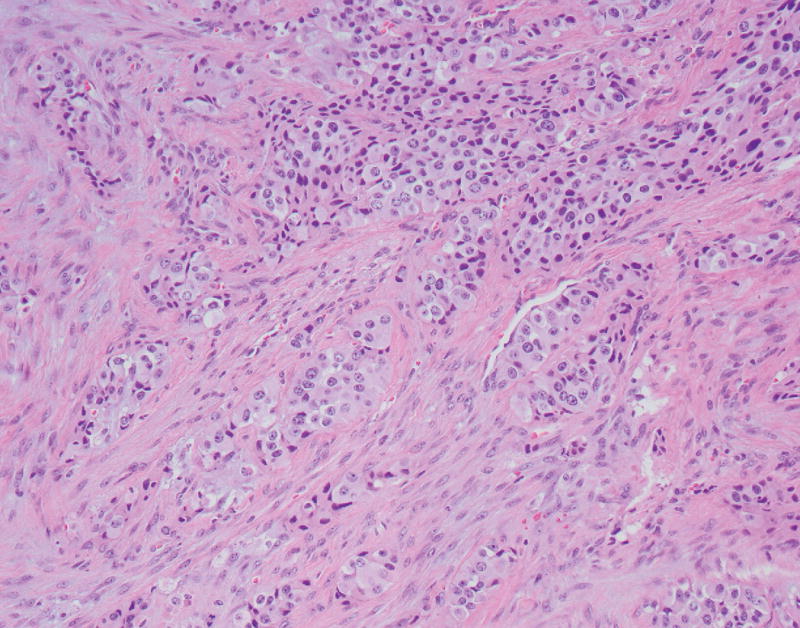
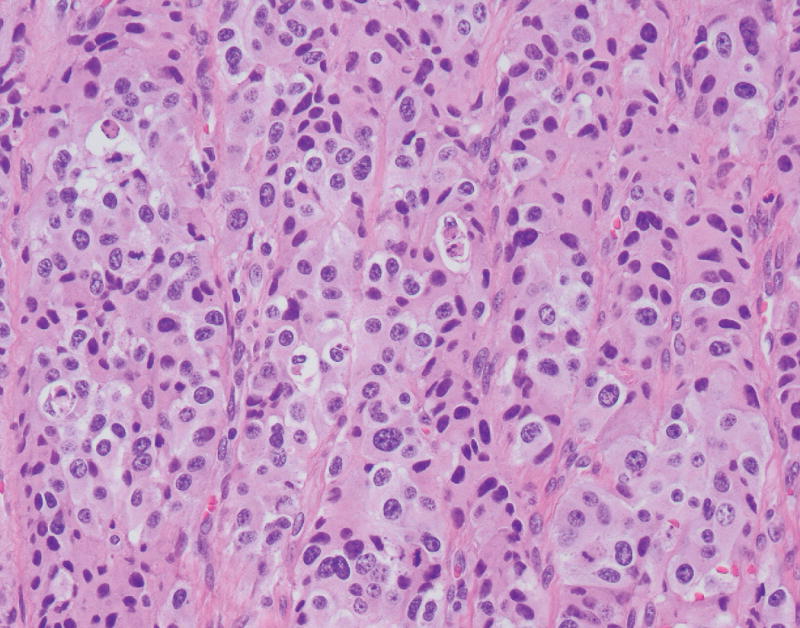
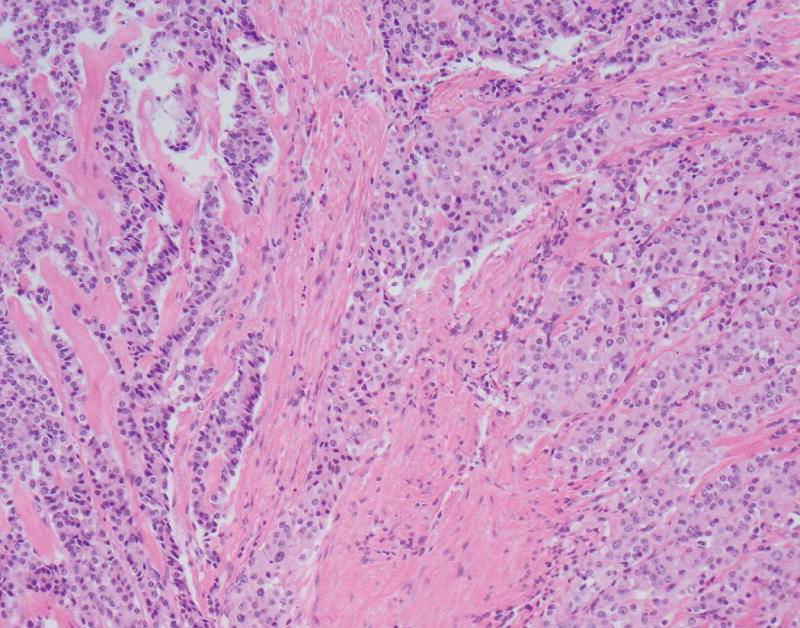
A, A low-power view of a low-grade focus with trabecular architecture and associated hyalinized, amyloid-like stroma. B, High-grade focus with invasive tumor nests associated with desmoplastic-type stroma. C, High-grade focus with nuclear pleomorphism and single cell necrosis. Mitotic figures are readily identified. D, Low-grade area is seen on the left, with adjacent progression to more intermediate/high-grade area.
Immunostains for synaptophysin, chromogranin, TP53, Ki-67, and p-Akt were performed on a section of tumor containing both low and high-grade regions. Synaptophysin and chromogranin were diffusely positive, although chromogranin was weaker in high-grade foci. The TP53 stain showed mild non-specific enhancement within the high-grade regions compared to low-grade regions. The Ki-67 stain highlighted a progression from low-grade to high-grade regions. In the histologically well-differentiated areas, the proliferative index was <5% (4.7% in a representative “hot spot” focus; mitotic rate 3/10 HPF) by manual count,9 while in the high-grade areas, the Ki-67 index approached 67% (66.3%; mitotic rate 26/10 HPF) (Figs. 3A, B). Interestingly, there were distinct zones between the low and high-grade regions with intermediate histomorphology (Fig. 2D) as well as proliferative index (proliferative index in a representative intermediate “hot spot” focus was 21.9%). The active p-Akt stain showed strong patchy expression in high-grade regions whereas low-grade regions were negative (Figs. 3C, D).
FIGURE 3.
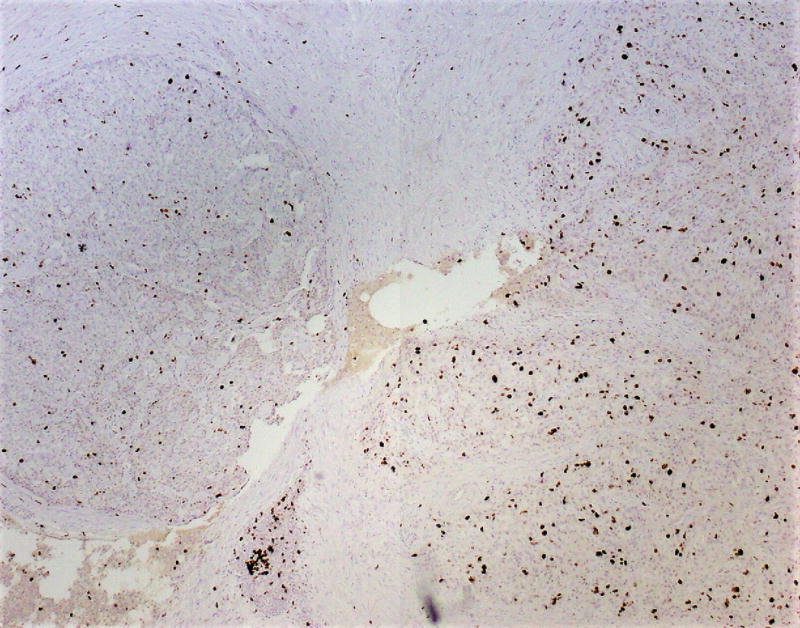
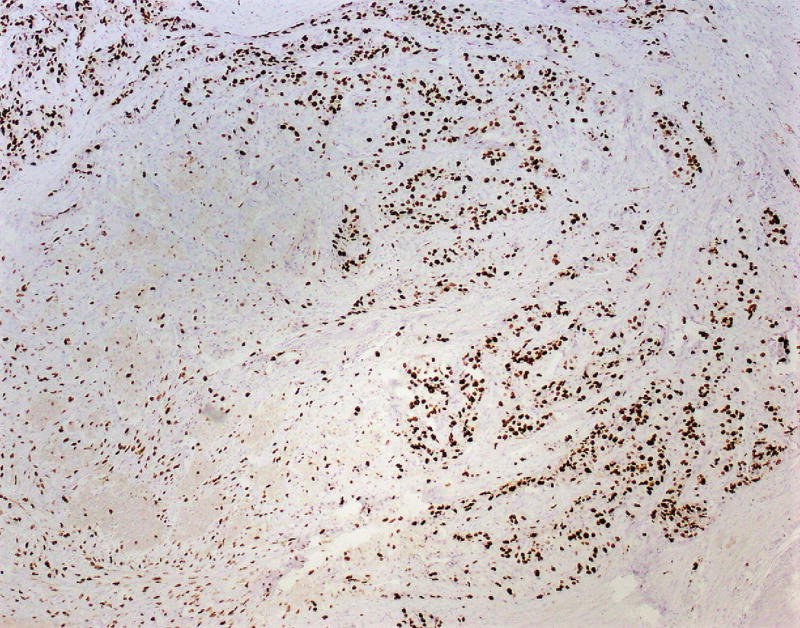
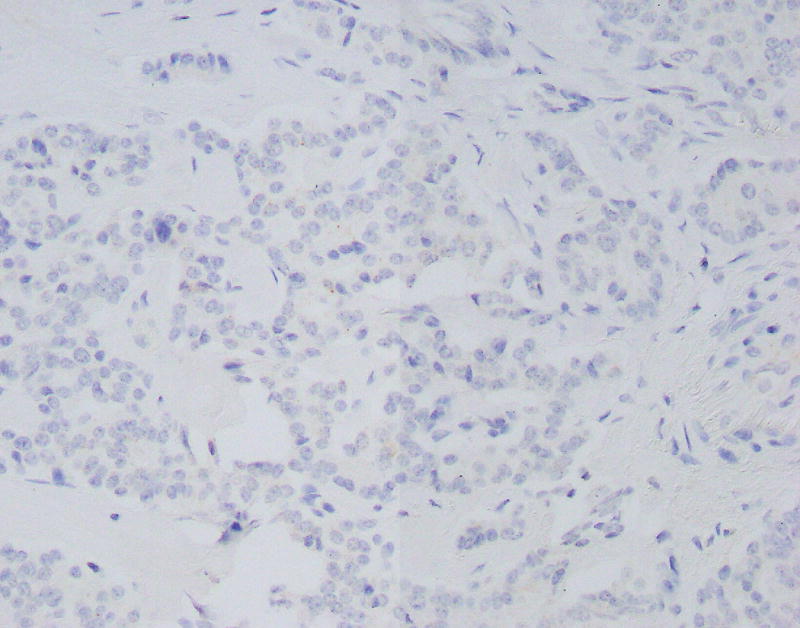
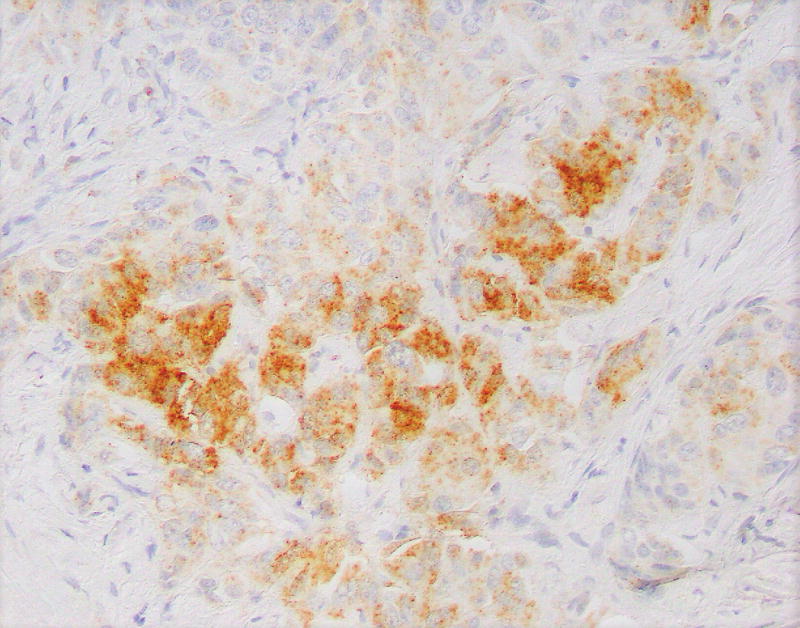
A, Ki-67 stain (low power), showing a low-grade area (left) and adjacent intermediate area (right). Proliferative indices in low power areas approached 5%, whereas the intermediate areas approached 22%. B, Ki-67 stain showing intermediate area (left) and adjacent high-grade area (right). Proliferative indices in high-grade areas approached 67%. C, Phosphorylated – Akt stain within a low-grade focus is mostly negative. D, Phosphorylated – Akt stain showing patchy strong expression in a high-grade focus of tumor.
Copy Number Changes
We examined the copy-number variation in the low-grade and high-grade regions of this PanNET case using a bioinformatics read-depth based approach. NGS based CNV approaches are more robustly developed for whole genome sequencing and whole exome sequencing. Applications of read-depth based approaches for targeted read sequencing (TRS) samples as in the current case are still evolving and increasingly useful. Multiple copy number variant changes were identified in the high-grade and low-grade regions of the PanNET sample (Table 1). We identified a 4.2 Kb copy number loss at the 6p21.32 location corresponding to the location of the DAXX gene in the low-grade tumor area. Interestingly, the CNV loss call for this location and the associated DAXX gene was the highest confidence call observed in both the Ion Reporter and CNVKit bioinformatics pipelines. A corresponding copy number loss at 6p21.32 was also identified in the high-grade tumor area sample area as well (Table 1). The ploidy call for this location was ×1, highly indicative of at least a single copy loss at this chromosomal location. It is important to note that the ploidy calls for any given location using the read-depth based approaches are a function of multiple factors, such as the normal-tumor contamination ratios, GC content, read-depth and other factors.10
TABLE 1.
Copy Number Losses
| ISCN Names | Size, kb | Ploidy | CNV Confidence Score | Genes in CNV region |
|---|---|---|---|---|
| PanNET high grade tumor area | ||||
| 6p21.32 | 4.2 | ×1 | 55.08 | DAXX |
| 11q13.1 | 6 | ×1 | 36.90 | MEN1 |
| PanNET low grade tumor area | ||||
| 6p21.32 | 4.2 | ×1 | 92.07 | DAXX |
Copy number losses identified in the high-grade area and the low-grade areas of tumor. Only the high confidence calls with the associated CNV confidence scores are shown over here.
A second, ~6Kb CNV deletion loss was identified in the 11q13.1 region which corresponds to the MEN1 gene location only in the high-grade region of the tumor. The MEN1 gene is of well-known significance in PanNET tumors. Similar to the DAXX gene, the call was made by both the bioinformatics pipelines and was a second highest significance CNV call. Pertinent to this case, the 11q13.1 deletion was not identified in the low-grade area. The ploidy call for the 11q13.1 location was ×1, indicative of at least a single copy loss at this region, similar to the DAXX gene. This CNV change was identified at a high significance in both the pipelines.
Read-depth based approaches for CNV calling for TRS are much more accurate for copy number losses than copy gains for statistical and bioinformatics reasons (for a more in-depth discussion, see 10). In the current case, we focus mainly on the CNV losses called in the two heterogeneous PanNET tumor fields under study. The top 10 high-confidence CNV gains identified by the Ion Reporter pipeline are listed in Supplementary Table 1 for the sake of completion. Interpretation of single copy gains in the context of cancer is still not well understood (as opposed to amplification e.g., HER2) and is not discussed further here.11 Of note, the CNV gains identified by Ion Reporter pipeline had a significant overlap with the orthogonal CNV bioinformatics pipeline, CNVKit.
Somatic Mutational Changes
We manually examined the sequencing data for the presence or absence of mutations in the DAXX/ATRX and MEN1 genes in both the high-grade/low-grade areas. We did not find any single nucleotide variants, indel variants or frameshift mutations within these genes in our sample. We identified a nonsense mutation, p. Tyr16 (Stop)*, in the PTEN gene in the high-grade tumor area at a ~18% variant allele frequency (see Table 2, Fig. 4). The PTEN (Phosphatase and tensin homolog) gene is a tumor suppressor gene that is highly mutated in multiple cancers. PTEN protein is a negative regulator of the PI3K/Akt/mTOR pathway. A nonsense mutation of the PTEN gene would thus be classified as pathogenic. Interestingly and pertinent to this case, this mutation was absent in the low-grade tumor region. We also identified a SMAD4 gene missense mutation, p.Gln455Pro, in the high-grade tumor area at a variant allele frequency of ~25.5% (Table 2, Fig. 5). A single SMAD4 mutation was identified by Yuan et al at the amino acid location 392 of the SMAD4 gene in 37 cases cohort they had studied.12 Changes to proline amino acid tend to create sharp dislocations in protein structure leading to loss of functionality and thus this change is highly likely to be pathogenic. Similar to the PTEN gene, we observed this mutational change only in the high-grade area and not within the low-grade area. SMAD4 is a transcription factor involved in the transforming growth factor-beta (TGF-beta) signaling pathway. TGF-beta is a pleomorphic cytokine involved in a wide variety of cell signaling activities. TGF-beta acts as a tumor suppressor as well as a tumor promoter in various contexts. The full functionality of TGF-beta is an active area of research at the current time.13 SMAD4 gene is present downstream of TGF-beta and loss of functionality of SMAD4 was associated with poor survival and was a negative prognostic indicator in patients with pancreatic adenocarcinomas.13
TABLE 2.
Somatic Mutational Changes
| Gene | Nucleotide (cDNA) | Amino Acid | Mutation Type | Pathogenicity |
|---|---|---|---|---|
| PanNET high grade tumor area | ||||
| PTEN | c.48T>A | p.Tyr16Ter | Nonsense | Pathogenic |
| SMAD4 | c.1364A>C | p.Gln455Pro | Missense | Pathogenic |
| WRN | c.2773G>A | p.Ala925Thr | Missense | VUS |
| CASC5 | c.4162G>A | p.Asp1388Asn | Missense | VUS |
| PanNET low grade tumor area | ||||
| None of significance | ||||
Somatic mutational changes identified in the high-grade and low-grade areas of tumor.
FIGURE 4.
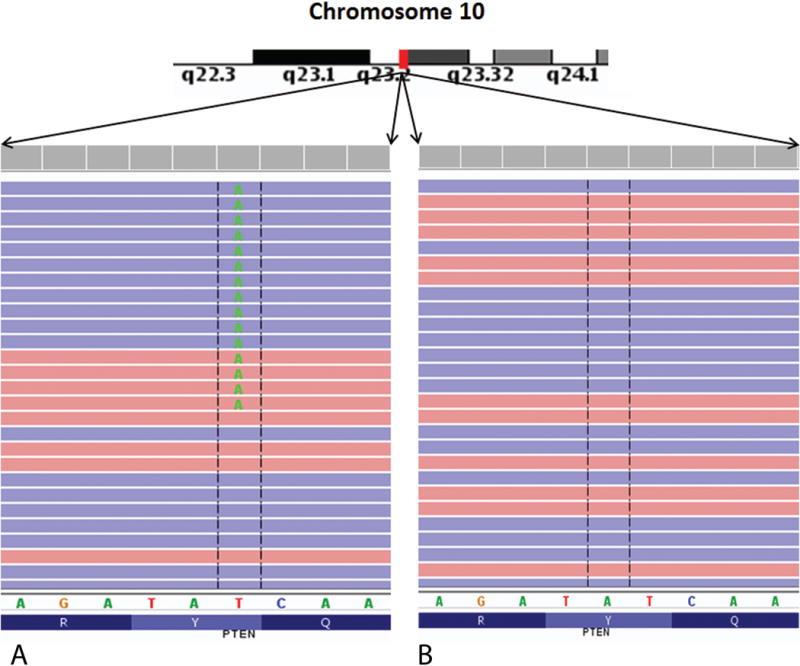
Integrated genome viewer (IGV) pileup of the PTEN mutation seen in the current sample. A nonsense mutation at p.Tyr16Stop was identified in this gene in the high grade tumor area, but not in the low grade tumor area. PTEN is a tumor suppressor gene of significant importance in carcinogenesis. A, High-grade area. B, Low-grade area.
FIGURE 5.
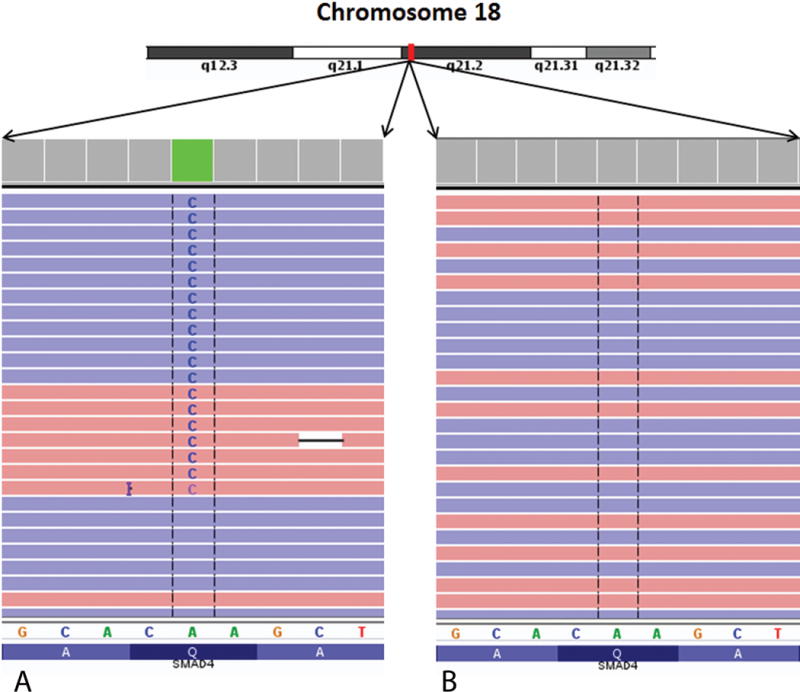
Integrated genome viewer (IGV) pileup of the SMAD4 gene mutation seen in the tumor. A pathogenic missense mutation change, p.Gln455Pro was identified in this gene in the high grade tumor area, but not in the low grade tumor area similar to the PTEN gene change. A, High-grade area. B, Low-grade area.
Additionally, we observed two variants of unknown significance only in the high-grade area (and not in the low-grade area). A p.Ala925Thr missense variant in the WRN gene was noted in the high-grade area at a variant allele frequency of ~10% (Table 2). WRN, gene is a member of the RecQ helicase family. The helicase families of proteins are involved in the DNA replication process as well as protein transcription. They are important to maintain the integrity of the normal DNA during the process of replication. In the current context, the loss of this protein function may indicate a loss of DNA replication control commonly associated with chromosomal instability (CIN). A p.Asp1388Asn variant of unknown significance was identified in CASC5 gene in the high-grade area of the tumor at a variant allele frequency of ~35%. CASC5 is involved in the attachment of the microtubules to the centromeres and activation of the spindle checkpoint during mitotic cell division.
DISCUSSION
The pathological classification of WD-NET versus PD-NEC carries major therapeutic and prognostic implications for patients with these neoplasms. One study showed that the median survival for patients with WD-NET versus patients with PD-NEC was 75 months and 11 months respectively, indicative of the diagnostic importance of properly classifying these tumors.14 Multiple studies have attempted to classify these tumors based on the mitotic rate as well as the Ki-67 proliferative index (Ki-67 proliferative index is used by the European Endocrine Tumor Society as a means to assess the grade of the tumor; part of 2010 WHO grading).1 Very little consensus was obtained in previous studies in a large proportion of cases (66%) based on histological features alone.4
The NORDIC NEC study showed that neuroendocrine tumors with Ki-67 < 55% were less responsive to platinum-based chemotherapy compared to those with indices > 55%.2,5 Perhaps in the future a cutoff point of 55% will be considered more clinically relevant than 20%. Furthermore, the literature supports better clinical outcome and prognosis for high-grade (grade 3) morphologically WD-NETs and low-grade (grade 1-2) WD-NETs, compared to highly aggressive PD-NECs.3 Indeed, our patient had an indolent clinical presentation that permitted an R0 resection, and was not treated with adjuvant chemotherapy and remained free of tumor recurrence on repeat CT imaging 5 months later.
A previous study investigated the role of copy number abnormalities and genomic instability in thirty-seven primary PanNETs and eleven metastastic PanNETs using an array-based comparative genomic hybridization approach.15 Chromosomal gains were most commonly seen at 6p22.2–p22.1 (27.1%), 17p13.1 (20.8%), 7p21.3–p21.2 (18.8%), and 9q34.11 (18.8%) in this study.15 The study identified genomic losses at 8q24.3 (6.3%) chromosomal locus. There was a much higher prevalence of chromosomal abnormalities in the metastases compared to the primary tumors. The authors concluded the presence of active chromosomal instability as a feature of PanNETs in this cohort of patients.15 Scarpa et al recently published a comprehensive and detailed analysis examining the genomic profile of ninety-eight PanNETs using whole genome sequencing (WGS) methods.16 This study identified recurrent genomic losses in the following chromosomal regions (pertinent gene in parenthesis): 11q13.1 (MEN1), 9q21.1 (CDKN2A), 8q13.1 (EYA1), 3p21.1 (FMBT1), 1q25.1 (RABGAP1L). Significant recurrent amplifications were seen at 19p13.3 (PSPN) and 12q24.33 (ULK1) gene locations.16 The differences between the Gebauer and Scarpa in the copy number variants may be due to the sample numbers involved. It is important to note the relative percentages of samples in which copy number changes are seen is low (~<30%, mostly <10%). However, chromosomal locations such as 11q13.1 representing the MEN1 gene were commonly identified in both the studies.15,16
Previous studies on sporadic PanNETs have focused on single nucleotide and insertion-deletion mutational changes in body of the tumor. Jiao et al performed whole exome sequencing in 10 tumor samples followed by targeted screening in 58 patients of sporadic PanNETs.17 Mutations were identified in this 68 patient PanNET cohort in the following genes: MEN1 gene (44%), DAXX/ATRX (43%), mTOR gene pathway (15%) and TTP53 (3%). A second study by Yuan et al examined the mutational patterns in a cohort of 37 Chinese patients. Similar to the Jiao et al study, mutations were observed in the following genes: MEN1 (35.14%), DAXX/ATRX (54.05%), mTOR pathway genes (54.05%), VHL (40.54%), TTP53 (13.5%), KRAS (10.8%) and SMAD/DPC4 (2.7%).12 Neither study examined the copy number status of the genes of the samples in the study. The latest study (March 2017) by Scarpa et al described the somatic mutational spectrum in that cohort.16 Similar to Jiao et al and Yuan et al, statistically significant mutations were identified in 16 genes. The chief genes with mutations were MEN1, DAXX, ATRX, PTEN and TTP53 genes. MEN1 gene was most frequently mutated (37% of the cases). Similar to other studies, mutations in the DAXX and ATRX genes were mutually exclusive and seen in 22 and 11 samples, respectively. TTP53 mutations were uncommon and seen only in 3 tumors.
In all of the above-mentioned studies, the sample was treated a single, homogeneous entity without accounting for tumor heterogeneity. In the current study, we present for the first time, a unique case of a patient with a mixed PanNET (morphologically and based on proliferative index) with comprehensive next generation sequencing targeting each region. Recently the literature has dealt with this seemingly heterogeneous and broad grade 3 category. According to 2010 WHO guidelines, neuroendocrine tumors with proliferative indices >20% should be categorized as PD-NEC. However, there seems to be greater appreciation that not all high-grade (Ki-67 >20%) neuroendocrine tumors should be classified as small or large cell type, and these proliferative/mitotically active WD-NETs should not be considered synonymous with PD-NEC. It can be confusing for surgical pathologists to accurately categorize neuroendocrine tumors that are morphologically well differentiated or morphologically ambiguous, but have proliferative indices of >20%.
Our findings implicate the importance of tumor heterogeneity in WD-NETs with high-grade progression. In our WD-NET case, we analyzed two tumor areas: low-grade WD-NET and WD-NET with high-grade progression. The proliferative index was approximately 5% in the low-grade areas and 66% in the high-grade areas. An intermediate area was also highlighted with the Ki-67 stain, showing an index of approximately 22%. While the Ki-67 count exceeded the 55% threshold in the high-grade area, the tumor did not meet criteria for large or small cell morphology. The high-grade areas had subtle “large cell-like” features with nuclear enlargement and focally prominent nucleoli with foci of single cell necrosis, yet we believe the histomorphology fell short of large cell carcinoma. The literature suggests that even when the high-grade foci morphologically resemble large cell carcinoma, the mutation profile of the high-grade component is more similar to the low-grade component than that of PD-NEC.6 Apparently these WD-NETs with high-grade progression may not show a long-term benefit from platinum-based chemotherapy either. It has been suggested that tumors such as these be described as WD-NETs with a high-grade component or WD-NET, grade 3.4,6
The definitive molecular signature of sporadic PanNETs is still under investigation.16 Unlike MEN1 syndrome-associated PanNETs, in which the role of the MEN1 gene is well known, the genetics of sporadic PanNETs were a mystery until 2011 when Jiao et al sequenced a case series of PanNETs. The DAXX/ATRX, MEN1 and mTOR pathway genes were mutated in nearly half of the cases sequenced.17 A similar study in a PanNET cohort of Chinese cases identified mutations within the same set of genes (DAXX/ATRX, MEN1, mTOR pathway) at similar frequencies as reported in the Jiao et al study.12 It is worthwhile to mention that neither of these studies focused on the role of copy number alterations. However, the latest study by Scarpa et al sheds light on PanNET genomics including simultaneous somatic and copy number profiling. Maxwell et al proposed a model for the likely genomic mechanisms of tumorigenesis in inherited as well as sporadic PanNETs.14 The main mechanisms proposed by Maxwell et al for genomic alteration in PanNETs include 1) Somatic homozygous mutation with loss of heterozygosity or 2) Somatic heterozygous mutation with epigenetic silencing.14
Marinoni et al provided significant evidence for the association of loss of DAXX/ATRX expression and chromosomal instability in PanNETs.18 DAXX functions as a histone chaperone protein while the ATRX protein, with an ATRX-DNMTT3-DNMT3L amino-terminal domain and a carboxy-terminal helicase domain function together for the CpG methylation and H3.3 incorporation into the telomeres of normal cells. The role of DAXX/ATRX mutations in PanNETs has been associated with opposite outcomes. Jiao et al showed a prolonged survival of patients with DAXX/ATRX mutations while Marinoni et al and Yuan et al showed the opposite effect on survival outcomes. Yuan et al speculated the reasons behind this discrepancy to be due to higher mutational rates of DAXX/ATRX, ethnicity based differences and location of mutations.12
In light of our current findings, we provide a likely alternative explanation. In this case we sought to examine the CNV and SNV genomic alterations in two histologically distinct regions of the tumor. To our knowledge, this is the first instance where the single mutation and copy number changes have been examined simultaneously in a PanNET. In line with previous studies, we identified changes in the DAXX gene. However, we did not identify any form of somatic mutations (SNV, Indel or frameshift) in the DAXX/ATRX genes. Instead, we identified a copy number alteration (i.e., high-confidence copy number deletion) in the DAXX gene in both the low-grade and high-grade regions of this tumor. We hypothesize that the alternate allele inactivation in this case may occur through epigenetic silencing mechanisms as has been suggested to occur in PanNETs.14 CNV deletion changes may also account for some of the cases that were point mutation negative in the DAXX/ATRX and MEN1 genes in the previous sequencing studies. We propose that the likely discrepancies observed in the survival outcomes in the prior studies may be due to the non-inclusion of the copy number variant data in the evaluation of the sequencing data.
A second unique finding in our study was the unique genomic alterations within different tumor areas examined. In the high-grade region, we identified a high-confidence copy number deletion in the MEN1 gene that was not identified in the low-grade region. Similar to the DAXX gene, we did not observe any single nucleotide, indel or frameshift mutations in the MEN1 gene either. MEN1 gene encodes the protein product menin, which is involved in the regulation of cell proliferation, genomic stability and apoptosis of normal cells. We found additional variants of unknown significance in the WRN and CASC5 genes, which are implicated in the process of cell repair and cell division respectively, lending support to the notion of existence of chromosomal instability as a fundamental feature of PanNETs.
The receptor tyrosine kinase (RTK)-PI3K-PTEN-Akt-TSC1/2-mTOR pathway plays a central role in cellular signaling, survival and differentiation in normal cells. This canonical pathway plays a central role in the progression of multiple cancers as well. PTEN is a tumor suppressor gene that inhibits the activation of Akt gene by PI3Kinase. Loss of PTEN tumor suppressor function leads to unobstructed activation of Akt that leads to downstream over activation of the mTOR pathway (Fig. 6). In this case, we identified a nonsense deactivating mutation in the PTEN gene in the high-grade area, which may potentially lead to over activity of p-Akt. Indeed, we confirm this hypothesis by the demonstration of increased immunohistochemical expression of phosphorylated-Akt protein in high-grade area of the tumor (figure 3C and D). As expected, over expression of p-Akt was not observed in the low-grade area, consistent with the absence of PTEN mutation in this area. This finding is of significance as there are multiple therapeutic trials currently available that target the mTOR pathway. A cartoon schematic of the mTOR pathway is shown in Figure 6.
FIGURE 6.
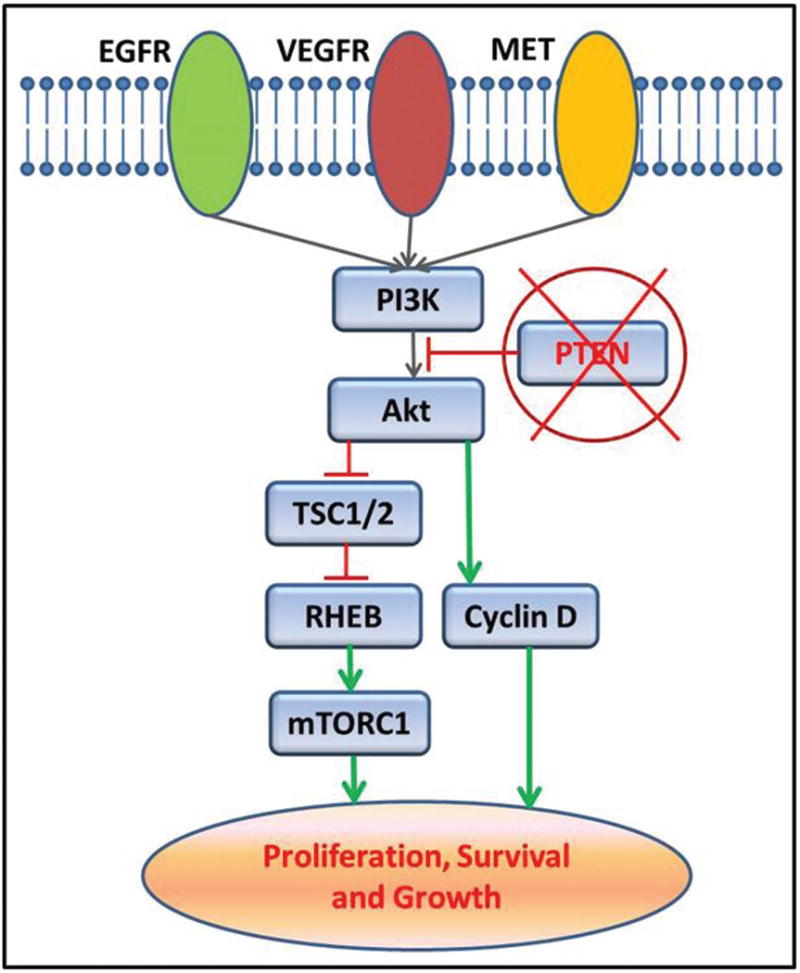
A cartoon schematic showing the mTOR signaling pathway in cells. PTEN acts as a negative regulator of the PI3Kinase to Akt signaling process. Loss of function mutations of PTEN gene leads to excessive activation of Akt and downstream mTOR pathway genes.
Tang et al proposed a diagnostic algorithm to classify WHO grade 3 WD-NET tumors from PD-NEC tumors in a thirty-three case cohort using IHC staining methods.4 The authors used IHC staining of DAXX/ATRX, TP53, Rb and SMAD4 proteins to evaluate their PanNET diagnostic algorithm.4 They concluded abnormal staining patterns of DAXX/ATRX genes are indicative of WD-NET tumors while altered IHC expression of TTP53, Rb and/or SMAD4 genes are suggestive of PD-NEC. However, altered IHC expression of SMAD4 (along with TP53) was reported in only 1 of 12 cases (~8%) classified as a PD-NEC.4 Of note, a component of ductal adenocarcinoma (commonly mutated for SMAD4) was also reported in this case. On the other hand, 11 of these cases classified as PD-NEC (92%) showed an alteration in TP53 and/or Rb genes.4 In our case, we identified a pathogenic SMAD4 gene mutation in the high-grade area of the tumor and no mutations in the TP53 or the Rb gene. In light of these findings, we believe SMAD4 mutations are clinically important, and likely contribute to high-grade transformation in WD-NETs. However, the presence of a SMAD4 mutation alone does not appear to be diagnostic of PD-NEC and we speculate somatic mutational changes/altered IHC expression of TP53 or Rb are likely to be a much stronger marker of PD-NEC.
From previous sequencing studies, mutual exclusivity of DAXX/ATRX gene mutations have been described in WD-PanNETs.12,17 However, mutations in either of these genes can co-exist with mutations of the MEN1 gene. We identified a similar pattern in our case as well (DAXX and MEN1 genes), though unlike previous studies, we identified copy number deletion changes instead of somatic mutations. Based on these findings, we hypothesize that inactivation of either the DAXX or ATRX gene (through somatic mutations or copy number deletions) is likely to be the preliminary step in WD-PanNET carcinogenesis. Due to the propensity for CIN, downstream changes occurring in genes like MEN1 may in turn lead to high-grade progression as a continuous spectrum. Additional mutations in genes such as PTEN, TTP53, VHL, Rb, SMAD4, TSC2 and KRAS as seen in other studies may represent the progressive changes in the evolution of this type of tumor.
Due to the substantial therapeutic and prognostic differences between WD-NETs and PD-NECs, it is important for clinicians to know the definitive diagnoses a priori to tailor treatment and follow-up.14 However, tumor categorization of grade 3 WD-PanNETs/WD-PanNETs with high-grade progression is not straightforward. In our case, we observe clear-cut areas of low-grade (grade 1–2) WD-PanNET with adjacent areas of high-grade transformation containing its own unique mutational profile (MEN1, PTEN, SMAD4, WRN, and CASC5) in addition to the DAXX mutation shared with the low-grade region. Based on these findings, we propose that for cases in which morphologic and immunohistochemical classification are not certain, clinical genomic testing may be useful to determine the possible biological behavior of the tumor. All known gene mutations in PanNETs (with the exception of DAXX/ATRX) are usually included in the commonly available solid tumor genomic testing panels available at the current time. In conclusion, we present an interesting case of WD-NET with high-grade progression and location-specific molecular changes. Additional studies are surely required to correlate these findings and further characterize the behavior of WD-PanNET with high-grade progression.
Supplementary Material
Acknowledgments
JMP, ELB and RRG were supported by the Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103451 and start-up funds from the UNM department of Pathology.
List of abbreviations
- PanNET
Pancreatic Neuroendocrine Tumor
- WD-NET
well-differentiated neuroendocrine tumor
- PD-NEC
poorly-differentiated neuroendocrine carcinoma
- CNV
Copy number variant
Footnotes
Authors’ contributions:
JMP and ELB performed the necessary experimental work to generate the data. DRM, VKC and JAH obtained the case and reviewed the slides. RRG performed the bioinformatics analysis. DRM and RRG wrote the paper. All authors read and approved the manuscript.
Conflict of Interests:
The authors declare that they have no competing interests.
References
- 1.Bosman FT, Carneiro F, Hruban RH, et al. WHO classification of tumours of the digestive system. World Health Organization; International Agency for Research on Cancer. Lyon, France: IARC Press; 2010. [Google Scholar]
- 2.Janson ET, Sorbye H, Welin S, et al. Nordic guidelines 2014 for diagnosis and treatment of gastroenteropancreatic neuroendocrine neoplasms. Acta Oncol. 2014;53:1284–1297. doi: 10.3109/0284186X.2014.941999. [DOI] [PubMed] [Google Scholar]
- 3.Basturk O, Yang Z, Tang LH, et al. The high-grade (WHO G3) pancreatic neuroendocrine tumor category is morphologically and biologically heterogenous and includes both well differentiated and poorly differentiated neoplasms. Am J Surg Pathol. 2015;39:683–690. doi: 10.1097/PAS.0000000000000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang LH, Basturk O, Sue JJ, Klimstra DS. A Practical Approach to the Classification of WHO Grade 3 (G3) Well-differentiated Neuroendocrine Tumor (WD-NET) and Poorly Differentiated Neuroendocrine Carcinoma (PD-NEC) of the Pancreas. Am J Surg Pathol. 2016;40:1192–1202. doi: 10.1097/PAS.0000000000000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorbye H, Welin S, Langer SW, et al. Predictive and prognostic factors for treatment and survival in 305 patients with advanced gastrointestinal neuroendocrine carcinoma (WHO G3): the NORDIC NEC study. Ann Oncol. 2013;24:152–160. doi: 10.1093/annonc/mds276. [DOI] [PubMed] [Google Scholar]
- 6.Tang LH, Untch BR, Reidy DL, et al. Well-Differentiated Neuroendocrine Tumors with a Morphologically Apparent High-Grade Component: A Pathway Distinct from Poorly Differentiated Neuroendocrine Carcinomas. Clin Cancer Res. 2016;22:1011–1017. doi: 10.1158/1078-0432.CCR-15-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh RR, Patel KP, Routbort MJ, et al. Clinical massively parallel next-generation sequencing analysis of 409 cancer-related genes for mutations and copy number variations in solid tumours. Br J Cancer. 2014;111:2014–2023. doi: 10.1038/bjc.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid MD, Bagci P, Ohike N, et al. Calculation of the Ki-67 index in pancreatic neuroendocrine tumors: a comparative analysis of four counting methodologies. Mod Pathol. 2015;28:686–694. doi: 10.1038/modpathol.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao M, Wang Q, Wang Q, et al. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics. 2013;14(Suppl 11):S1. doi: 10.1186/1471-2105-14-S11-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra S, Whetstine JR. Different Facets of Copy Number Changes: Permanent, Transient, and Adaptive. Mol Cell Biol. 2016;36:1050–1063. doi: 10.1128/MCB.00652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan F, Shi M, Ji J, et al. KRAS and DAXX/ATRX gene mutations are correlated with the clinicopathological features, advanced diseases, and poor prognosis in Chinese patients with pancreatic neuroendocrine tumors. Int J Biol Sci. 2014;10:957–965. doi: 10.7150/ijbs.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shugang X, Hongfa Y, Jianpeng L, et al. Prognostic Value of SMAD4 in Pancreatic Cancer: A Meta-Analysis. Transl Oncol. 2016;9:1–7. doi: 10.1016/j.tranon.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxwell JE, Sherman SK, Howe JR. Translational Diagnostics and Therapeutics in Pancreatic Neuroendocrine Tumors. Clin Cancer Res. 2016;22:5022–5029. doi: 10.1158/1078-0432.CCR-16-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebauer N, Schmidt-Werthern C, Bernard V, et al. Genomic landscape of pancreatic neuroendocrine tumors. World J Gastroenterol. 2014;20:17498–17506. doi: 10.3748/wjg.v20.i46.17498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scarpa A, Chang DK, Nones K, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543:65–71. doi: 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]
- 17.Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marinoni I, Kurrer AS, Vassella E, et al. Loss of DAXX and ATRX are associated with chromosome instability and reduced survival of patients with pancreatic neuroendocrine tumors. Gastroenterology. 2014;146:453–460.e5. doi: 10.1053/j.gastro.2013.10.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


