Abstract
The staphylococcal pathogenicity islands (SaPIs) are highly mobile 15 kb genomic islands that carry superantigen genes and other virulence factors and are mobilized by helper phages. Helper phages counteract the SaPI repressor to induce the SaPI replication cycle, resulting in encapsidation in phage like particles, enabling high frequency transfer. The SaPIs split from a protophage lineage in the distant past, have evolved a variety of novel and salient features, and have become an invaluable component of the staphylococcal genome. This review focuses on recent studies describing 3 different mechanisms of SaPI interference with helper phage reproduction and other studies demonstrating that helper phage mutations to resistance against this interference impact phage evolution. Also described are recent results showing SaPIs contribute in a major way to lateral transfer of host genes as well as enabling their own transfer. SaPI-like elements, readily identifiable in the bacterial genome, are widespread throughout the Gram-positive cocci, though functionality has thus far been demonstrated for only a single one of these.
Introduction
The staphylococcal pathogenicity islands (SaPIs) are highly mobile chromosomal islands that exploit specific bacteriophages for their mobility. They contain a characteristic set of genes required for their distinctive life style and typically carry accessory genes, including those for toxic shock syndrome toxin (TSST-1), enterotoxin B (SEB), and other superantigens. These are expressed during the island's integrated state and contribute to the host pathotype. The SaPIs are the sole genetic carriers of TSST-1 and SEB and are responsible for the staphylococcal toxic shock syndrome, including necrotizing fasciitis [1]. The SaPIs are extremely common in S. aureus, with an average of one per strain, some strains having 3 or more. They are also common in non-aureus staphylococci and similar elements have been identified by sequence analysis in streptococci and lactococci [2], though these elements have not been found to carry virulence genes, nor has their functionality been demonstrated as yet.
Recent literature on the SaPIs reflects the evolution of our thinking about these fascinating mobile chromosomal islands (see recent reviews: [3] [4]. Originally, we focused on their cargo, then their life cycle and interaction with particular bacteriophages. Later, we understood that these islands play a more significant role for the staphylococci than mere high-speed couriers of fancy virulence genes. Although they are clearly derived from prophages, they long ago diverged, forming a highly distinctive and entirely separate lineage. They mediate transfer of unlinked chromosomal genes [5], interfere with phage reproduction [6], increase host cell survival of phage infection [7] and determine animal host specificity [8]. Consequently, the SaPIs appear to be critical for the evolution and adaptation of the organism.
The Basics
Like prophages, the SaPIs reside quiescently at specific chromosomal attachment (att) sites under the control of their master repressors [9]. However, unlike the classical phage repressors, SaPI repressors are not SOS-inactivated. Following infection by a helper phage or SOS induction of a resident helper prophage, an antirepressor protein [9] is produced by the phage, counteracting the SaPI repressor (Stl). Different SaPIs are derepressed by different phage-coded anti-repressors [3]. At present, the phage coded anti-repressors have been identified for only a very few SaPIs: dUTPase for SaPIs 2 and bov1, Sri for SaPI1 and 80α GP15 for bov2. 80α encodes all of these, Φ11 encodes only Sri. Following derepression, int and xis are expressed, leading to SaPI excision (by the Campbell mechanism [10]), expression of SaPI-coded replication initiation genes and commencement of extensive SaPI DNA replication, using the host replication system for DNA polymerization [11]. In addition to forming phage procapsids phage virion proteins are directed by SaPI genes cpmA & B to form small procapsids commensurate with the 15 kb SaPI genome [7, 12, 13]. SaPI replication, like phage replication, results in the formation of concatemers [7]. Post-replicative Phage and SaPI DNA's are packaged by the headful mechanism [7, 11] [14], with specificity determined by the respective TerS proteins of phage and SaPI, which recognize phage and SaPI pac sites respectively, and which promote packaging, indiscriminately, into procapsids, of either size [15]. Since only 1/3 of the phage genome will fit in the SaPI-sized capsids, this is a dead-end for the phage and generally results in a 10-fold reduction in infectious phage particles compared with phage production by a non-SaPI containing strain [16]. SaPI particles released by phage-induced lysis infect other staphylococci with a high degree of promiscuity.
To effect this carefully orchestrated life cycle, the SaPIs contain a set of functional genes that encode a master repressor (analogous to λc1) that maintains the SaPI genome in its integrated state, an integrase, required for SaPI genome integration into a specific att site, an excisionase, non-specifically required in conjunction with the integrase, for SaPI excision from its att site, a primase homolog (not known to be an actual primase), a replication initiator protein that catalyzes melting and unwinding of the origin DNA [11], and usually a SaPI-specific homolog of the phage terminase small subunit (TerS). TerS recognizes and binds to a specific site, the pac site, at which DNA packaging is initiated. The TerS-DNA complex binds to the large terminase subunit, TerL, which introduces the pac site cleavage that initiates headful packaging [14]. These genes, are organized in functional modules and form a pair of divergent transcription units reminiscent of the standard pro phage genome organization (for typical examples, see [17]. A critical feature of all SaPIs is their interference with phage packaging. Genes required for this interference are located directly upstream of terS and are transcribed from the major rightward promoter. terS and most of these genes also form a separate operon regulated by LexA [18]. Accessory genes are located either in the leftward transcript between the repressor and integrase genes or distal to terS and the rightward transcript. The accessory genes are expressed and regulated independently of the functional genes and the SaPI life cycle. Several SaPIs (SaPI1 [7, 16], SaPI2 [19], SaPIbov1 [12] and SaPIbov5 [20] have been analyzed in detail. The map of a typical SaPI, SaPI2, is shown in Fig.1.
Fig. 1. SaPI2 genome.
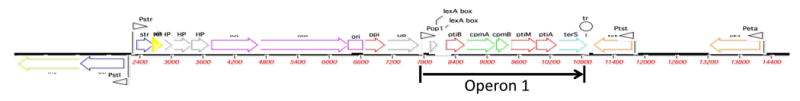
Color scheme: yellow – int/xis; blue – regulation; gray – HP; purple – replication; red – interference; green – capsid morphogenesis, also interference; aqua – terS; orange – accesssory genes; small pink arrows – promoters. Scale is in base pairs.
Abbreviations: int – integrase; xis – excision; stl – repressor of leftward SaPI transcription; str – repressor of rightward SaPI transcription; pri – primase homolog; rep – initiator protein; ori – replication origin; ppi – phage packaging inhibition; ptiB phage transcription inhibitor independent of ptiA; cpm – capsid morphogenesis; ptiA – phage transcription inhibitor; ptiM - modulator of ptiA; terS – terminase small subunit; HP – hypothetical protein.
Host specificity
SaPIs, being packaged in phage-like particles, would be expected a priori to have the same host range as their helper phages. Since temperate staphylococcal phages [21] (Siphoviridae) attach rather nonspecifically to the riboteichoic acid of the cell wall, they have a very broad potential host range. A limitation to this host range is the variation in teichoic acid structure in ST395 S. aureus and most non-aureus staphylococci. These have glycerol phosphate instead of ribose in their wall teichoic acid [22]. In practice, however, individual phages have quite limited infectivity ranges as measured by plaque formation – which gave rise to the once widely used but now obsolete phage typing system [23]. Host range is well-known to be determined by post-adsorption factors that determine the ability to replicate and form plaques – such as prophage immunity, restriction-modification, host factors required for phage reproduction, phage-specific inhibitory factors, etc. [24]. Since the SaPIs need only to integrate into a chromosomal att site, their host range is much wider than that of the phages, being limited only by the availability of an att site, and, perhaps by DNA restriction. This has been dramatically illustrated by the ability of SaPIs to stably infect L. monocytogenes [25], a species in which no known staphylococcal phage can replicate, although many can attach to and infect the organism. Although L. monocytogenes does not contain any of the native SaPI att sites, infecting SaPIs insert into secondary sites similar to those identified in S. aureus [25]. There are, however, many strains of L. monocytogenes that SaPIs do not infect, presumably owing to differences in teichoic acid structure.
Phage interference
A remarkable feature of the SaPIs is that they universally interfere with reproduction of their helper phages, and they do so by several unrelated mechanisms, all of which cause partial but never complete blockage of the targeted phage function. The first of these interference mechanisms that was identified was simply their ability to divert the vast majority of the phage virion proteins to the formation of small (SaPI-sized) heads commensurate with the 15 kb SaPI genome [7, 12, 13, 26], using SaPI genes cpmA & B (capsid morphogenesis) or ccm (cos capsid morphogenesis). Concatemeric post-replicative phage and SaPI DNAs are packaged by the headful mechanism [7, 11] with packaging specificity being determined by the respective TerS proteins of phage and SaPI, which recognize phage and SaPI pac sites respectively. Consequently, packaging is non-specific with respect to head size [15]. Since packaging specificity is based on TerS recognition of pac sites and not on prohead size, small heads cause the phage DNA to be wasted since only 1/3 of the phage genome can be accommodated. The second known interference mechanism also blocks phage DNA packaging, but by an entirely different mechanism. Here, interference is mediated by a single SaPI gene, ppi (phage packaging interference), which binds the phage TerS protein and blocks its function [26] by mechanism that has yet to be determined. Ppi is widely conserved among the SaPIs and exists as several distinct SaPI-specific variants [26]. The third interference mechanism thus far identified is the pti (phage transcription inhibitor) system, which consists of 3 genes: ptiA, ptiB and ptiM directed toward interference with transcription of the late phage region [27], which encodes the virion and lytic proteins [17]. Late phage gene transcription among the staphylococcal Siphoviridae is driven by a single promoter, which is activated by the regulatory protein, LtrC (late transcriptional regulator). This protein, first thought to interfere with integrase transcription, was initially designated RinA (regulation of integrase) [28, 29]. Transcriptional activation by LtrC is absolutely essential for SaPI as well as for phage reproduction [6]; therefore as PtiA binds directly to LtrC and blocks activation of the phage late promoter, it must be modulated in order to avoid complete blockage of late transcription. This modulation is accomplished by PtiM, which binds to PtiA and diminishes the PtiA-mediated inhibition of LtrC [27]. PtiB also inhibits late phage transcription; however, it is also highly toxic for the host cell [27], which has complicated its analysis so that its precise mechanism of phage interference has yet to be determined. It is remarkable that at least 3 unrelated interference mechanisms have been developed by the SaPIs, and are all located in a small region of the SaPI genome. This convergent evolution suggests that phage interference is of major importance for SaPI biology. Although all known SaPIs possess one or more interference systems, different SaPIs possess different combinations of these. One might imagine that the SaPIs would benefit from interference by increasing their reproduction at the expense of the phage. Surprisingly, however, this is not the case: deletion of any of the 3 systems does not decrease SaPI particle production, even though phage production is always increased. Even more remarkably, deletion of cpmA&B increases SaPI particle production as well as phage reproduction [27]. It is assumed that in phage-infected cells, phage products are generally in vast excess and are not exhausted during the lytic cycle, even if there is a co-replicating SaPI. Therefore, the reason for increase in production of viable phage particles seen with ΔcpmAB is probably that phage DNA is no longer being wasted by being packaged in small capsids. We are left, however, with the question of the biological rationale for interference. Our best guess is that it helps the SaPI to keep pace with its helper phage during successive infection cycles.
Although phage interference is seen only when a SaPI is induced by a helper phage, it exerts strong purifying selection for phage mutants that have lost SaPI-inducing activity through mutations in either the gene targeted by the interference [26] or in the phage-coded SaPI de-repressor genes [30], since interference is expressed only when the SaPI has been induced. Indeed, helper phages are rather scarce – it is quite unusual to find a naturally occurring SaPI-helper phage combination. Why, then, are any helper phages still in existence? One simple and obvious reason is that the derepressor genes are perfectly stable so long as there is no responsive SaPI around. Moreover, the anti-repressor genes, while not essential, are not entirely dispensable for phage biology, so that the de-repressor mutants are somewhat disadvantaged; additionally, there may be some selective value in the SaPI-helper phage combination. Although the SaPIs could respond to the scarcity of helper phages by developing inactivating mutations in their repressors, this would be very unlikely in nature, since de-repressed SaPIs are rather toxic to their host cells and are rapidly lost [12].
SaPI-mediated generalized transduction
Given that SaPIs use the same headful packaging mechanism as do many Siphoviridae, it was not a surprise to find that they mediate generalized transduction, based on mis-packaging of host DNA, identically to the phages [5]. As with the phages, homologs of the SaPI pac site (pseudo-pac sites) are scattered around the host genome and have dramatically variable efficiencies, resulting in transduction of some genes at very high frequency, others at very low [5]. Remarkably, many of the pseudo-pac sites are adjacent or very near to certain classes of genes, especially those involved in iron metabolism and other important adaptivity genes that may impact the virulence of the host organism [5]; more remarkably, SaPI operon 1, within which is located the SaPI terS, is SOS-induced [18] – this was a highly mysterious feature of the SaPI genome until it was realized that SOS induction of terS in a lysogenic strain enables the mis-packaging of host DNA via the SaPI pseudo-pac sites although the SaPI itself is not SOS induced and is not transferred specifically following SOS induction. Any resident prophage, whether it is a helper or not, is thus enabled to expand its transductional repertoire via any co-resident SaPI [5].
Variations on a theme: The SaPIbov5 family
A widespread group of highly atypical SaPIs harbored by many S. aureus strains of domestic animals [20, 31], were presumably derived by recombinational replacement of most of operon 1 by a DNA segment containing phage derived cos and pac sites and a gene encoding a homolog of the phage major capsid protein. They lack any functional terS but have a C-terminal terS remnant, presumably marking one end of the recombinational event. They are packaged by either pac or cos phages using the respective phage TerS's, reportedly in small capsids [32]. The Cap protein homolog (Ccm) causes severe phage interference and seems to be required for the formation of small SaPI particles. Many of these SaPIs carry a homolog of the von Willebrand factor binding protein, which is involved in the determination of animal host specificity [8], and also the phage-derived Scin (staphylococcal complement interference) protein. These genes are also carried by a more typical SaPI from an equine strain (SaPIeq1), consistent with the recombinational origin of SaPIbov5, et al.
Evolution
A glance at the SaPI genome organization and the list of SaPI genes indicates an obvious relation to the prophage genome and genes (see references [17, 33] and, especially [4] for comparative diagrams– indeed, SaPIs have generally been annotated as defective prophages and their genes as phage genes. Although there is clearly an ancestral relation, we consider it very distant for the following reasons: i) the SaPIs are a coherent family, within which the similarities between SaPI genes are much greater than similarities between SaPI and phage genes or genes of other genetic elements [2, 4]; ii) they possess genes that are shared among SaPIs but are not found elsewhere, especially the hypothetical genes, and the interference genes, of which the latter are all located in the same region of the island, a region that has probably been imported [2, 4]; iii) they share a rare (though not absolutely unique) biological functionality – the property of being induced by prophage anti-repressor genes; iv) they occupy unique SaPI-specific att sites that are conserved throughout the staphylococci and are never occupied by other elements; v) they carry certain accessory genes that are not found on other genetic elements.
Other species
Elements that are clearly within the SaPI family can readily be identified in various non-aureus staphylococci by performing a BLAST search of the staphylococcus database using the TerS protein of e.g., SaPI2 as query. Sequence similarities of 50% or greater represent SaPI-like elements. Below 50% similarity, phages begin to appear in the orthology lists. With these criteria, such elements are readily identified in most of the available genomes of the non-aureus staphylococci. Three typical examples are: a 14 kb element in S. pasteuri SP1 at nt 1580572, an 11 kb element in S. warneri SG1 at nt 2395104, and a 15 kb element in S. lugdunensis HKU09-01 at nt 2072768. None of these SaPI-like elements, however, has been shown to be inducible or transferred at high frequency. Some carry identifiable accessory genes, especially fusidic acid resistance in S. epidermidis [34]. Similarly, large families of SaPI-like elements have been identified in the genomes of lactococci, pneumococci, and streptococci [2, 4] [35]; functionality has yet to be demonstrated for any of those in lactococci and pneumococci. Enterococcus faecalis strain V583 possesses the only SaPI-like-like element, EfCIV583, outside of the staphylococci whose SaPI-like functionality has been demonstrated [33] [2]: it is induced by a helper phage (Φ1) whose xis gene is the de-repressor, is packaged in small particles, is transferred at high frequency, and interferes with its helper phage. However, as it lacks both an identifiable terS and identifiable cpm genes, key aspects of its life cycle remain to be determined. E. faecalis does not possess a family of such elements so its place among the SaPI-like elements is unclear. One family of SaPI--like elements in Streptococcus pyogenes, with an att site between mutS and mutL, blocking transcription of mutL, has the remarkable property of reversible excision during the exponential growth of its host organism, which restores mutL transcription. It is not known whether or not any bacteriophage is involved in this remarkable phenomenon, nor is it known whether this element is transferrable. SaPI-like elements in other locations are more like those seen in other Gram-positive cocci [36-38].
Conclusions
The SaPIs are a family of mobile genetic elements that we believe to have split from an ancestral protophage, retaining the basic prophage replicon and genomic organization and being efficiently packaged in infectious phage-like particles. Following this split, they embarked on an independent evolutionary pathway, acquiring or developing the following key features that provide them with a unique biological identity, endear them to their host bacteria, and guarantee their continued survival: They possess unique att site-integrase combinations and DNA packaging specificity; they exploit their helper phages by commandeering specific phage-encoded proteins as de-repressors; they engage in (necessarily incomplete) interference with the reproduction of their helper phages, enabled by their unique 3 kb interference operon; they contribute materially to the horizontal transfer of host genes; and they are the sole carriers and transmitters of several superantigen genes, especially that for TSST-1.
We note an interesting contrast between the genomes of Gram-positive cocci and those of enteric bacteria. The latter are littered with remnants and fragments of prophages, which are largely incidental findings of genome analysis. – this is readily understandable since prophages, being dispensable, are subject to every imaginable type of DNA rearrangement. Prophage remnants, however, are quite infrequent in the genomes of Gram-positive cocci despite the near universality of prophages in these organisms – instead, their genomes are replete with SaPIs, defective SaPIs, and SaPI-like elements. Perhaps the basis of this contrast lies in the primary role of SaPIs and their ancestral prophages in the performance of horizontal gene transfer (HGT), which could well be selective, in contrast to the primacy of conjugation and DNA-mediated transformation in the Gram-negative bacteria. This would also be consistent with the rarity in staphylococci of CRISPR/Cas systems, which prevent HGT.
Fig. 2. Interference mechanisms.
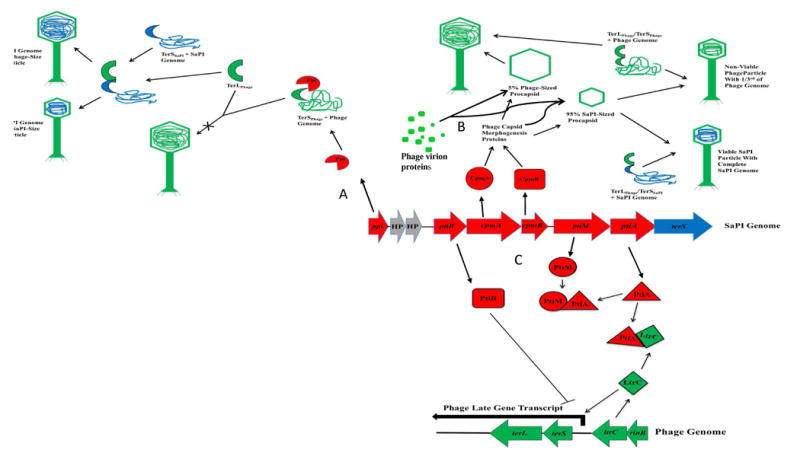
Upper left, SaPI terS (blue crescent) binds to SaPI DNA then complexes with phage TerL (green semicircle, enabling packaging of SaPI DNA in capsids of either size. A. Ppi (red ¾ circle) complexes with phage terS (green crescent) blocking its function in phage DNA packaging. B. Virion proteins are diverted by Cpm A & B to form mostly small capsids, which are filled with SaPI DNA, using SaPI TerS, or with phage DNA using phage TerS. C. Pti interference with transcription of late phage genes. PtiA, at right, complexes with LtrC, blocking activation of late phage gene transcription. PtiA is partially countered by PtiM, preventing the complete blockage of late phage gene expression. PtiB blocks late phage gene expression independently, by an unknown mechanism.
Table 1. The SaPI family.
| Element (strain) | Size, Kb, (comment) | att | genes | Phage interference genes | Orient. | References | |
|---|---|---|---|---|---|---|---|
| SaPI4 (I) (MRSA 252) | 15.1 | Endogenous prophage | AAAGAAGAACAATA ATAT (∼8′) | none | + | [39] | |
| SaPI1028 (I)(NY940) | 15.6 | Endogenous prophage | AAAGAAGAACAATA ATAT (∼8′) | none | + | [40] | |
| SaPIbov1 (II)(RF122) | 15.8, (excised spontaneously) | Φ11 Φ1477, 80α | TAATTATTCCC ACTCAAT (∼9′) | tst, sel, sek | cpmAB ppi, ptiAB | + | [41] [42] |
| SaPIbov2 (II)(V329) | 27, (excised spontaneously) | 80α | TAATTATTCCC ACTCGAT (∼9′) | bap | ppi | + | [42] |
| SaPIbov5 (II)(JP5338) | 13.5 (cos&pac packaging) | Φ12 | GAGTGGGAATAATT ATATATA (∼9′) | ada, scin, VWbp | ppi | [8] | |
| SaPIm4 (III) (mu50: | 14.4 | Endogenous prophage | TCCCGCCGTCT CCAT (∼18′) | fhuD | + | [43] | |
| SaPImw2 (III)(mw2) | 14.4 | Endogenous prophage | TCCCGCCGTCT CCAT (∼18′) | ear, se2l, sec4 | + | [43] | |
| ShPI2 (III) (S. haemolyticus) | 16.6 | TCCCGCCGTCT CCAT (48′)* | none | - | [44] | ||
| SaPI1 (IV) (RN4282) | 15.2 | 80α, Φ13 | TTATTTAGCAGGAA TAA (∼19′) | ear, tst, sek seq | cpmAB ppi | + | [16] |
| SaPI3 (IV)(COL) | 15.6, | 29 (?) | TTATTTAGCAGGAA TAA (∼19′) | ear, seb sel sek | + | [45] | |
| SaPI5 (IV) (USA300) | 14.0 | TTATTTAGCAGGAA TAA (∼19′) | ear, sek, seq | + | [46] | ||
| SaPI(IV)-OC3 | 14.6 | TTATTTAGCAGGAA TAA (∼19′) | sek, seq, FeHyR1 | cpmAB ppi, ptiAB | + | [47] | |
| SaPIeq1(V) (DL:584) | 15.0 | ATGCCAGGAATGAT GTAAAA)(44′) | ada, scin, VWbp | ppi, ptiAB | [8] | ||
| SaPIn1 (V) (n315) SaPIm1(mu50) | 15.0 (SaPIs n1 and m1 are identical) | GTTTTACCATC ATTCCCGGCAT (∼44′) | tst, sel, sec3 | - | [48] | ||
| SaPI2 (V) (RN3984) | 14.7 | 80 | ATTTTACATCATTC CTGGCAT (∼44′) | tst, eta | cpmAB ppi, ptiAB | - | [7]; |
| SaPI122 (V) (RF122) | 17.9 | Endogenous prophage | GTTTTACATCATTC CTGGCAT (∼44′) | mdr | - | GenBa nk NC 00 | |
| SaPI6Δ (V)(many) | (identical) 3.14 (excised spontaneously) | GTTTTACCATCATT CCCGGCAT, GTTTTACATCATTC CTGGCAT (∼44′) | none | - | [43] | ||
| SsPI15305 (S. saprophyticus 15305 | aad, fosB | [49] | |||||
| SaPI2R(V) (OC3) | 14.8 | TTTTACATCA TTCCTGGCAT (∼44′) | eta, tst | cpmAB ppi, ptiAB | - | [47] |
ShPI2 is located 180° away from the other SaPIs having the same att core sequence, owing to the major chromosomal inversion that has been documented in the S. haemolyticus genome [Takeuchi, 2005 #8070].
No homolog detected.
Highlights.
SaPIs are helper phage inducible, highly mobile elements that carry toxin genes
SaPIs have a distinctive life style that is highly beneficial to their host bacteria
SaPIs in animal strains determine the animal specificity of their host bacteria
SaPIs engage in lateral host gene transfer independently of their own transfer
SaPIs have independently evolved 3 different ways of interfering with helper phages
Phage mutations reveal interference targets and drive phage evolution
Acknowledgments
The research in the authors' lab described in this review was supported by grant R01-AI22159 from the National Institutes of Health and by a Grant-in-aid from NYU School of Medicine
Footnotes
Conflict of interest statement: We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schlievert PM, et al. Secreted virulence factor comparison between methicillin-resistant and methicillin-sensitive Staphylococcus aureus, and its relevance to atopic dermatitis. J Allergy Clin Immunol. 2010;125(1):39–49. doi: 10.1016/j.jaci.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2*.Martinez-Rubio R, et al. Phage-inducible islands in the Gram-positive cocci. ISME J. 2016 doi: 10.1038/ismej.2016.163. Describes families of SaPI-like elements, (referred to as phage-inducible chromosomal islands (PICIs) in sttreptococci, pneumococci and lactococci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novick RP, et al. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol. 2010;8:541–51. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4**.Novick RP, Ram G. The Floating (Pathogenicity) Island: A Genomic Dessert. Trends Genet. 2016;32(2):114–26. doi: 10.1016/j.tig.2015.11.005. This review compares the SaPIs with other mobile genomic islands, including the integrative and conjugative elements (ICEs) and the gene transfer agents (GTAs). It highlights the uniqueness of the SaPIs as a close-knit family sharply distinct from their ancestral bacteriophages, and suggests an interesting contrast between the evolution of the SaPIs and of the GTAs. It also demonstrates the cohesiveness of a large family of SaPI-like elements in Lactococcus lactis. A key feature of this analysis is the demonstration that ORFs encoding hypothetical proteins are nearly always co-inherited by these islands and largely excluded from other gemomic elements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, et al. Pathogenicity island-directed transfer of unlinked chromosomal virulence genes. Mol Cell. 2015;57(1):138–49. doi: 10.1016/j.molcel.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ram G, Ross HF, Novick RP. Precisely modulated SaPI interference with late phage gene transcription. Proc Natl Acad Sci, USA. 2014;111(40):4536–41. doi: 10.1073/pnas.1406749111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruzin A, et al. Molecular genetics of SaPI1 - a mobile pathogenicity island in Staphylococcus aureus. Mol Microbiol. 2001;41(2):365–77. doi: 10.1046/j.1365-2958.2001.02488.x. [DOI] [PubMed] [Google Scholar]
- 8*.Viana D, et al. Adaptation of Staphylococcus aureus to ruminant and equine hosts Demonstration of the possible determination of staphylococcal specificity for animal hosts by a SaPI-carried gene. involves SaPI-carried variants of von Willebrand factor-binding protein. Mol Microbiol. 2010;77(6):1583–94. doi: 10.1111/j.1365-2958.2010.07312.x. [DOI] [PubMed] [Google Scholar]
- 9*.Tormo-Mas MA, et al. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature. 2010;465(7299):779–82. doi: 10.1038/nature09065. Identification of dUTPase as a dual-funtion protein that serves as the helper phage encoded SaPIbov1 de-repressor in addition to its classical enzymatic activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell AM. Episomes. Adv Genet. 1962;11:101–145. [Google Scholar]
- 11.Ubeda C, et al. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proc Nat Acad Sci, USA. 2007;104:14182–88. doi: 10.1073/pnas.0705994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Ubeda C, et al. Characterization of mutations defining SaPI functions and enabling autonomous replication in the absence of helper phage. Mol Microbiol. 2008;67:493–503. doi: 10.1111/j.1365-2958.2007.06027.x. Gene-byh-gene analysis of the entire set of ORFs in SaPIbov1. [DOI] [PubMed] [Google Scholar]
- 13.Poliakov A, et al. Capsid size determination by Staphylococcus aureus pathogenicity island SaPI1 involves specific incorporation of SaPI1 proteins into procapsids. J Mol Biol. 2008;380(3):465–75. doi: 10.1016/j.jmb.2008.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, et al. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol Microbiol. 2002;45(6):1631–46. doi: 10.1046/j.1365-2958.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- 15*.Ubeda C, et al. Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations. Mol Microbiol. 2009;72(1):98–108. doi: 10.1111/j.1365-2958.2009.06634.x. Shows that packaging specificty is determined solely by TerS and not by the size of the procapsid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Lindsay JA, et al. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. Discovery of the SaPIs. [DOI] [PubMed] [Google Scholar]
- 17.Christie GE, et al. The complete genomes of Staphylococcus aureus bacteriophages 80 and 80alpha--implications for the specificity of SaPI mobilization. Virology. 2010;407(2):381–90. doi: 10.1016/j.virol.2010.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ubeda C, et al. SaPI operon I is required for SaPI packaging and is controlled by LexA. Mol Microbiol. 2007;65(1):41–50. doi: 10.1111/j.1365-2958.2007.05758.x. [DOI] [PubMed] [Google Scholar]
- 19.Subedi A, et al. Sequence analysis reveals genetic exchanges and intraspecific spread of SaPI2, a pathogenicity island involved in menstrual toxic shock. Microbiology. 2007;153:3235–45. doi: 10.1099/mic.0.2007/006932-0. [DOI] [PubMed] [Google Scholar]
- 20*.Quiles-Puchalt N, et al. Staphylococcal pathogenicity island DNA packaging system involving cos-site packaging and phage-encoded HNH endonucleases. Proc Natl Acad Sci U S A. 2014;111(16):6016–21. doi: 10.1073/pnas.1320538111. A subgroup of SaPIs, widespread among S. aureus strains isolated from domestic animals, has acquired, presumably by one or more recombination events, a phage cos site, a phage pac site, and a homolog of the major phage capsid protein. These elements lack any terS homolog and can be packaged by either pac or cos phages.** [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deghorain M, Van Melderen L. The Staphylococci phages family: an overview. Viruses. 2012;4(12):3316–35. doi: 10.3390/v4123316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winstel V, et al. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat Commun. 2013;4:2345. doi: 10.1038/ncomms3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hood AM. Phage typing of Staphylococcus aureus. J Hyg (Lond) 1953;51(1):1–15. doi: 10.1017/s0022172400015448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Labrie SJ, et al. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–27. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 25**.Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009;323(5910):139–41. doi: 10.1126/science.1164783. Demonstration that SaPIs can be transferred to Listeria monocytogenes, and that it occurs at remarkably high frequencies. [DOI] [PubMed] [Google Scholar]
- 26.Ram G, et al. Staphylococcal pathogenicity island interference with helper phage reproduction is a paradigm of molecular parasitism. Proc Natl Acad Sci U S A. 2012;109(40):16300–5. doi: 10.1073/pnas.1204615109. Discovery of a highly conserved SaPI gene dedicated explicitly to the blockage of phage DNA packaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ram G, et al. Precisely modulated pathogenicity island interference with late phage gene transcription. Proc Natl Acad Sci U S A. 2014;111(40):14536–41. doi: 10.1073/pnas.1406749111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye ZH, Lee CY. Cloning, sequencing, and genetic characterization of regulatory genes, rinA and rinB, required for the activation of staphylococcal phage phi 11 int expression. J Bacteriol. 1993;175(4):1095–102. doi: 10.1128/jb.175.4.1095-1102.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrer MD, et al. RinA controls phage-mediated packaging and transfer of virulence genes in Gram-positive bacteria. Nucleic Acids Res. 2011;39(14):5866–78. doi: 10.1093/nar/gkr158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Frigols B, et al. Virus Satellites Drive Viral Evolution and Ecology. PLoS Genet. 2015;11(10):e1005609. doi: 10.1371/journal.pgen.1005609. Thereason that SaPI helper phages are so infrequent is that helper phages mutate spontaneously to decommission their SaPI de-repressor proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, et al. Intra- and inter-generic transfer of pathogenicity island-encoded virulence genes by cos phages. ISME J. 2015;9(5):1260–3. doi: 10.1038/ismej.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carpena N, et al. Convergent evolution of pathogenicity islands in helper cos phage interference. Philos Trans R Soc Lond B Biol Sci. 2016;371(1707) doi: 10.1098/rstb.2015.0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Matos RC, et al. Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 2013;9(6):e1003539. doi: 10.1371/journal.pgen.1003539. First demonstration of a functional SaPI-like element outside of the staphylococci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen HJ, et al. Identification of fusB-mediated fusidic acid resistance islands in Staphylococcus epidermidis isolates. Antimicrob Agents Chemother. 2011;55(12):5842–9. doi: 10.1128/AAC.00592-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35*.Martinez-Rubio R, et al. Phage-inducible islands in the Gram-positive cocci. ISME J. 2017;11(4):1029–1042. doi: 10.1038/ismej.2016.163. Describes families of SaPI-like elements, (referred to as phage-inducible chromosomal islands (PICIs) in sttreptococci, pneumococci and lactococci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott J, et al. Phage-associated mutator phenotype in group A streptococcus. J Bacteriol. 2008;190(19):6290–301. doi: 10.1128/JB.01569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott J, et al. Phage-Like Streptococcus pyogenes Chromosomal Islands (SpyCI) and Mutator Phenotypes: Control by Growth State and Rescue by a SpyCI-Encoded Promoter. Front Microbiol. 2012;3:317. doi: 10.3389/fmicb.2012.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Nguyen SV, McShan WM. Chromosomal islands of Streptococcus pyogenes and related streptococci: molecular switches for survival and virulence. Front Cell Infect Microbiol. 2014;4:109. doi: 10.3389/fcimb.2014.00109. Along with references 36 and 37, presents evidence for two types of SaPI-like elements in streptococci - one type involved in HGT, the other involved in local genetic regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holden MT, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101(26):9786–91. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwan T, et al. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A. 2005;102(14):5174–9. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fitzgerald JR, et al. Characterization of a putative pathogenicity island from bovine staphylococcus aureus encoding multiple superantigens [In Process Citation] J Bacteriol. 2001;183(1):63–70. doi: 10.1128/JB.183.1.63-70.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ubeda C, et al. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol Microbiol. 2003;49(1):193–210. doi: 10.1046/j.1365-2958.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- 43.Baba T, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359(9320):1819–27. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi F, et al. Whole-genome sequencing of staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol. 2005;187(21):7292–308. doi: 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novick RP, et al. Pathogenicity and resistance islands of staphylococci. Microbes Infect. 2001;3(7):585–94. doi: 10.1016/s1286-4579(01)01414-9. [DOI] [PubMed] [Google Scholar]
- 46.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367(9512):731–9. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 47.Khokhlova OE, et al. Healthcare- and Community-Associated Methicillin-Resistant Staphylococcus aureus (MRSA) and Fatal Pneumonia with Pediatric Deaths in Krasnoyarsk, Siberian Russia: Unique MRSA's Multiple Virulence Factors, Genome, and Stepwise Evolution. PLoS One. 2015;10(6):e0128017. doi: 10.1371/journal.pone.0128017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuroda M, et al. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet. 2001;357(9264):1225–40. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 49.Kuroda M, et al. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc Natl Acad Sci U S A. 2005;102(37):13272–7. doi: 10.1073/pnas.0502950102. [DOI] [PMC free article] [PubMed] [Google Scholar]


