Abstract
BACKGROUND: Regional lymph node (LN) metastasis is a strong and well-established prognostic factor in colon cancer, and recent data suggest a prognostic value of detecting micrometastases and isolated tumor cells in regional LNs. The aim of the study was to investigate the clinical relevance of detecting sentinel lymph node (SLN) metastases in colon cancer patients by measuring the novel metastasis marker PHGR1 mRNA. METHODS: Using quantitative reverse-transcription polymerase chain reaction, we measured PHGR1 mRNA levels in SLNs and primary tumors from 206 patients surgically treated for stage I to III colon cancer and 52 normal LNs from patients undergoing surgery for benign colon diseases. The prognostic impact of these findings was evaluated by Kaplan-Meier analysis and Cox proportional-hazards regression. RESULTS: Compared to normal LNs, elevated PHGR1 mRNA levels were detected in SLNs from 56 (89%) of the 63 patients with pN+ disease. Furthermore, 68 (48%) of the 143 node-negative (pN0) patients had elevated PHGR1 mRNA levels in SLNs, suggesting occult metastases. With a median follow-up of 7.2 years, a significantly shorter recurrence-free (P=.005) and disease-specific (P=.02) survival was observed in patients with elevated PHGR1 mRNA levels in SLNs. Multivariable modeling showed that the SLN PHGR1 mRNA level was an independent prognostic factor. However, when the survival analyses were restricted to pN0 patients, no significant prognostic information was found. CONCLUSION: Measuring PHGR1 mRNA in SLNs provided independent prognostic information on operable colon cancer patients but not in the pN0 subgroup.
Introduction
Lymph node (LN) status is an important prognostic factor in patients with operable colon cancer and currently the single most essential indication for adjuvant systemic therapy [1]. Eventually, approximately 30% of patients without LN metastases (pN0) experience disease recurrence [2]. Thus, despite its widespread use, the tumor-node-metastasis (TNM) system is imperfect at best for prediction and prognosis [3], [4]. Many pN0 patients have occult LN metastases not detected by current diagnostic methods [5]. Although several studies have failed to demonstrate a prognostic significance of occult LN metastases in colorectal cancer (CRC), a recent meta-analysis concluded that patients with molecularly detected occult LN metastases have significantly shorter overall, disease-specific, and recurrence-free survival [5].
Several techniques have been utilized to improve the detection of LN metastasis, including ultrasectioning, immunohistochemistry, and polymerase chain reaction (PCR) [5]. Some of these methods are rather laborious and cost intensive. Thus, the idea of restricting analyses to a defined and relevant LN subgroup is attractive. Sentinel lymph node (SLN) mapping has been evaluated extensively as a tool for identifying the LNs most likely to harbor metastases [6], [7], [8]. Two meta-analyses have reported sensitivities of only 76% and 69% for routine histological analysis to detect LN metastases when the analysis was restricted to SLNs only [6], [7]. On the other hand, 15% to 19% of pN0 patients have been found to have occult LN disease by immunohistochemical or molecular analyses of SLNs [6], [7]. Thus, the clinical relevance of occult SLN metastasis in this clinical setting remains unclear, emphasizing the need for additional studies [9], [10], [11], [12], [13].
We recently reported the first functional characterization of the novel proline-, histidine-, and glycine-rich 1 (PHGR1) protein [14]. PHGR1 is primarily expressed in mature epithelial cells of the gastrointestinal tract and seems to be functionally related to transport and metabolic processes. We also obtained preliminary evidence that PHGR1 mRNA and protein may be promising novel markers for the detection of LN metastasis in colon cancer, based on high expression levels in colon tumors and extremely low levels in normal LNs. Here, we determine PHGR1 mRNA level in SLNs from patients with operable colon cancer and relate the findings to disease outcome.
Materials and Methods
Patients
Consecutive patients (n=278) undergoing elective surgery for primary colon cancer at Stavanger University Hospital were prospectively recruited for ex vivo SLN mapping between May 2003 and February 2010 as described previously [11]. The hospital serves a population of 350,000 as the only medical institution, which reduces potential referral bias. Patients with noninvasive tumors (pTis), distant metastases (M+) at the time of surgery, tumors in the lower rectosigmoideum, or deviations from the SLN mapping procedure were excluded. Patients treated preoperatively with temporary colon stents were also excluded.
Treatment and Follow-Up
All patients included in this study were treated surgically with curative intent followed by adjuvant chemotherapy administered according to the national guidelines at the time (i.e., offered to patients with stage III disease and up to 75 years of age) [15]. The patients were followed according to the national guidelines [16]. Follow-up data were obtained from the hospital's patient records in April 2016 and blinded for the results of molecular SLN analyses.
Ethics
The study was approved by the institutional review committee and regional medical ethical committee (REK 197.04). Informed consent was obtained from all patients.
Ex Vivo SLN Mapping
The ex vivo SLN mapping procedure was described in detail previously [17]. The SLNs were bisected; one half was formalin-fixed and paraffin-embedded for routine histology by hematoxylin, eosin, and safranin (HES) staining, and the other half was snap-frozen in liquid nitrogen and stored at −80°C for molecular analyses. Biopsies of 20 to 50 mm3 were taken from the outer rim of the tumors immediately after resection (<2 hours), snap-frozen in liquid nitrogen, and stored at −70°C until RNA extraction. In addition, 52 normal control LNs were removed from the resected specimens of 12 patients undergoing surgery for benign bowel diseases (6 with ulcerative colitis, 4 with Crohn’s disease, and 2 with diverticulitis).
RNA Isolation and Reverse Transcription
RNA isolation, integrity assessment, and reverse transcription were performed as described previously [17].
PCR Primers
The PCR primers were designed to bind to different exons in the proline-histidine-glycine-rich 1 gene (PHGR1; GenBank accession number NM_001145643) to avoid amplification of genomic DNA. We also amplified the two reference transcripts BCR (NM_004327) and HPRT1 (NM_000194). The primer sequences were as follows: PHGR1-F, 5′-CCCTGCTCTGCACTCTCAG-3′; PHGR1-R, 5′-CGCAGTGACCTGGAGGAT-3′; BCR-F, 5′-GCTCTATGGGTTTCTGAATG-3′; BCR-R, 5′-AAATACCCAAAGGAATCCAC-3′; HPRT1-F, 5′-TTCCTTGGTCAGGCAGTA-3′; HPRT1-R, 5′-TATCCAACACTTCGTGGG-3′. The amplicons for PHGR1, BCR, and HPRT1 mRNA were 109, 99, and 81 bp in length, respectively.
Real-Time PCR
PCR amplification was performed using the SYBR Green Core Kit (Eurogentec) according to the manufacturer's recommendations. Twenty nanograms of reverse-transcribed RNA was amplified in a total volume of 25 μl containing 1× reaction buffer, 0.2 mM dNTP, 0.15 μM forward and reverse primers, 0.75 μl 1:200 SYBR Green I diluted in DMSO, and MgCl2 at a concentration of 1.3, 2, and 2 mM for PHGR1, BCR, and HPRT1 reactions, respectively. Thermocycling and real-time fluorescence measurements were performed in an MX3000P real-time PCR instrument (Stratagene). The PCRs were incubated at 95°C for 10 minutes followed by 40 cycles of 30 seconds at 95°C and 60 seconds at 60°C. Finally, the PCR products were analyzed by melting curves, revealing well-defined peaks with the expected melting temperatures, confirming the specificity of the primers. The amplicon's identities were also confirmed by sequencing. No-template controls were included in every run to monitor potential contamination.
Computations
The amplification efficiency of each assay was determined by the standard curve method using four different samples in three repeated experiments. The mean amplification efficiencies for PHGR1, BCR, and HPRT1 amplicons were 1.96 ± 0.03, 2.03 ± 0.03, and 2.03 ± 0.03, respectively. The relative PHGR1 mRNA concentration was determined by normalization against the two reference transcripts, BCR and HPRT1, and a calibrator sample included in every run, as described previously [17]. The reproducibility of the assays was determined by quantifying a reference sample with low marker concentration (CV = 22%) in all runs and a reference sample with high marker concentration (CV = 11%) in the first 110 runs. The threshold for positivity for the PHGR1 mRNA marker (=6.5·10−4) was established from the highest levels in normal control LNs (n=52) by adding two standard deviations of the quantification of the reference sample with low concentration. Patients were considered positive for SLN PHGR1 mRNA if the marker level exceeded the threshold for positivity in at least one SLN. The real-time PCR quantifications were performed by a single person, blinded to the characteristics of the patients and the primary tumors.
Microsatellite Instability and Mutational Analysis
Primary tumor microsatellite instability (MSI) and the BRAF and KRAS mutation status were determined previously [11].
Cell Spiking Experiment
The LS174T human colon adenocarcinoma cell line was cultured according to the supplier's recommendations (ECACC, Salisbury, UK). Normal lymphocytes were isolated from normal whole blood by Lymphoprep density centrifugation (Axis-Shield, Dundee, UK). The cells were trypsinized and washed in PBS, and the cell density was counted in a hemocytometer. Increasing numbers of LS174T cells were then added to 107normal lymphocytes in three separate spiking series. Each spiking series was quantified in two separate runs by real-time quantitative reverse-transcription PCR as described above. We were able to detect as few as 10 LS174T cells, and the relationship between PHGR1 mRNA levels and the number of LS174T cells added was linear (Supplemental Figure S1).
Supplementary Figure S1.
Relative PHGR1 mRNA concentrations in dilutions of LS174T cells in 1E7 normal lymphocytes. Error bars indicate standard deviations.
Statistical Analysis
Most computations and all plots were achieved using the R software package (www.r-project.org) version 3.1.1. The normality of continuous variables was checked by the Shapiro-Wilks test. Due to the lack of normality, all continuous variables were compared using the nonparametric Mann-Whitney U test. Categorical variables were compared using the χ2 test or Fisher's exact test. Correlation coefficients were determined according to the Pearson method and tested for significance using Pearson's product moment correlation coefficient test. The whiskers in the boxplots (vertical lines extending from the boxes) extended to the most extreme data point, which was no more than 1.5 times the interquartile range from the box. A primary tumor location in the right colon including transversum was defined as proximal, whereas other locations were defined as distal.
Survival analyses were conducted using Kaplan-Meier survival estimates, log-rank tests, and Cox proportional-hazards regression. Clinical endpoints were disease recurrence (both locoregional and systemic), systemic recurrence, disease-specific death, and overall death [18]. Patients were censored at last follow-up or at death for reasons other than colon cancer. Cox proportional-hazards regression was used to model the impact of PHGR1 mRNA measurements and clinicopathological parameters on recurrence-free and disease-specific survival. In the multivariable analyses, both backward and forward stepwise selections of variables were used, and both resulted in the same final model. Variables with P<.25 from the univariate analysis were included in the initial model, but only variables with a significant effect (P<.05) were kept in the multivariable model. Only covariates that were significant after stepwise removal were included in tables. The proportional-hazards assumption was tested using the χ2 test on the Schoenfield residuals of the models and was rejected for age and disease-specific survival [19]. To adjust for age despite this observation, we used a two-step function for age, splitting the model at 2.5 years after surgery. The model showed that age provided significant prognostic information only in the first time step; thus, we only presented results for this step. Although some of the variables in the multivariable modeling were significantly associated, the variation inflation score was <2, which suggested that they could all be included in the multivariable analysis [20]. Test power calculations were using the SPSS Sample Power 2 software. Statistical tests were two-sided, and P values <.050 were considered significant. Reporting was performed according to the Reporting Recommendations for Tumor Marker Prognostic Studies [21].
Results
PHGR1 mRNA Measurements in SLNs
We previously reported successful SLN mapping in 206 (97%) of 213 operable colon cancer patients using an ex vivo approach (Figure 1) [11], [17]. Clinicopathological characteristics of the patients are provided in Table 1. Routine HES staining of one half of the bisected SLNs indicated that 42 of the 63 node-positive patients had SLN metastases (Figure 1). Thus, the diagnostic sensitivity of routine HES staining, when restricted to SLNs only, was only 67%. To improve the diagnostic sensitivity of SLN analysis and detect potential occult SLN metastases, the PHGR1 mRNA levels were measured in the other half of the isolated SLNs. Normal LNs from patients with benign colon disease were used as a reference group. Figure 2 shows the results in comparison to previously reported levels of PHGR1 mRNA in primary tumors from the same patients [14]. Compared to normal LNs, the PHGR1 mRNA levels were significantly higher in both HES-positive and HES-negative SLNs (P<.001 for both). The PHGR1 mRNA level was also significantly higher in HES-positive SLNs than in HES-negative SLNs (P<.001).
Figure 1.
Flowchart of patient inclusion, SLN mapping, and diagnostics.
Table 1.
Clinicopathological Characteristics of the Patients with Successful SLN Mapping Stratified According to SLN PHGR1 Status
| Patient Characteristics | ||||
|---|---|---|---|---|
| Parameter | All Patients | PHGR1− SLNs | PHGR1+ SLNs | PValue |
| Group size | 206 | 82 | 124 | |
| Median age [years] (range) | 76 (21-93) | 76 | 75 | .534 |
| Gender, no. (%) | .2 | |||
| Female | 115 (56) | 41 (50) | 74 (60) | |
| Male | 91 (44) | 41 (50) | 50 (40) | |
| Tumor stage, no. (%) | .4 | |||
| pT1 | 12 (6) | 5 (6) | 7 (6) | |
| pT2 | 34 (17) | 16 (20) | 18 (14) | |
| pT3 | 149 (72) | 57 (70) | 92 (74) | |
| pT4 | 11 (5) | 4 (5) | 7 (6) | |
| Median tumor diameter [mm] (range) | 50 (7-130) | 55 (13-120) | 50 (7-130) | .019 |
| Tumor grade, no. (%) | .356 | |||
| I | 10 (5) | 3 (4) | 7 (6) | |
| II | 145 (70) | 17 (21) | 34 (27) | |
| III | 51 (25) | 62 (76) | 83 (67) | |
| Tumor localization, no. (%) | .006 | |||
| Proximal | 140 (68) | 65 (79) | 75 (60) | |
| Distal | 66 (32) | 17 (21) | 49 (40) | |
| Tumor microsatellite status, no. (%) | .06 | |||
| MSSa | 121 (59) | 41 (50) | 80 (65) | |
| MSIa | 81 (39) | 39 (48) | 42 (34) | |
| Not analyzed | 4 (2) | 2 (2) | 2 (2) | |
| Tumor KRAS mutation,bno. (%) | .8 | |||
| Mutated | 76 (37) | 29 (35) | 47 (38) | |
| Wild-type | 129 (63) | 53 (65) | 76 (61) | |
| Not analyzed | 1 (0) | 1 (1) | ||
| Tumor BRAF mutation,cno. (%) | .9 | |||
| Mutated | 56 (27) | 23 (28) | 33 (27) | |
| Wild-type | 145 (70) | 57 (70) | 88 (71) | |
| Not analyzed | 5 (2) | 2 (2) | 3 (2) | |
| Median no. lymph nodes (range) | 14 (3-43) | 13.5 (5-43) | 14 (3-35) | .14 |
| Lymph node metastasis, no. (%) | <.001 | |||
| pN0 | 143 (69) | 75 (91) | 68 (55) | |
| pN1 | 48 (23) | 6 (7) | 42 (34) | |
| pN2 | 15 (7) | 1 (1) | 14 (11) | |
| Adjuvant chemotherapy, no. (%) | .03 | |||
| Adjuvant chemotherapy | 32 (16) | 7 (9) | 25 (20) | |
| No adjuvant chemotherapy | 174 (84) | 75 (91) | 99 (80) | |
Fisher’s exact test for categorical data, Mann-Whitney U test for continuous data.
Microsatelite stable/unstable.
Codon 12 and 13 mutations.
V600E mutation.
Figure 2.
Boxplot showing relative PHGR1 mRNA levels in primary colon tumors, normal lymph nodes (LNs), and HES-positive (HES+) and -negative (HES−) SLNs. The horizontal line shows the threshold used to characterize the SLNs as positive or negative for the marker.
By examining SLN PHGR1 mRNA status at the patient level, the sensitivity to detect pN+ patients (as defined by routine histopathological examination of SLNs and non-SLNs) was 89% (56/63). Furthermore, 68 (48%) of the 143 patients without known LN metastases (pN0) had elevated PHGR1 mRNA levels in SLNs, potentially reflecting the presence of occult metastases or micrometastases. Next, we tested for associations between the patients' SLN PHGR1 status and any of the clinicopathological characteristics in Table 1. Small tumor size, distal tumor localization, LN metastasis, and adjuvant chemotherapy were significantly associated with positive SLN PHGR1 mRNA status (Table 1).
We also compared primary tumor PHGR1 mRNA levels and the patients' clinicopathological characteristics. Patients with positive microsatellite instability score and those with BRAF V600E mutations both had significantly higher primary tumor PHGR1 mRNA levels than those who were negative for these tests (P<.001 for both).
Survival Analysis According to PHGR1 Levels
Within a median 7.2-year follow-up (maximum 12.7 years), 34 (17%) of the 206 patients with successful SLN mapping experienced disease recurrence (i.e., distant metastases and/or locoregional relapses). Distant metastases were found in 30 (88%) of the 34 patients with relapse. In addition, 100 (49%) of the patients died during follow-up, and 30 (15%) of the deaths were disease-specific.
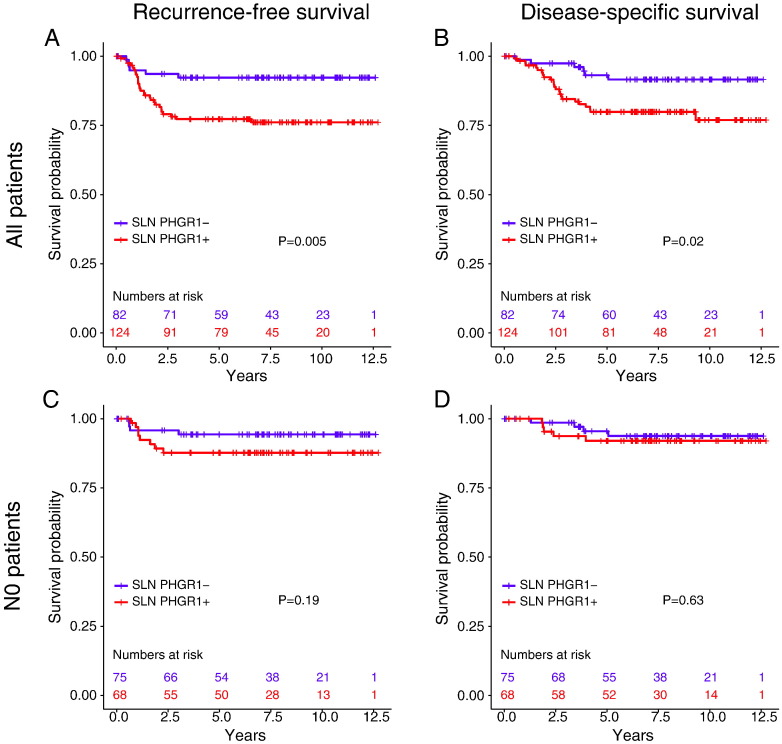
Kaplan-Meier survival analyses and log-rank tests showed that patients with positive SLN PHGR1 status had significantly shorter recurrence-free (P=.005), systemic recurrence-free (P=.006), and disease-specific (P=.02) survival in the overall patient group (Figure 3, A and B). No association with overall survival was observed (P=.7). However, when we stratified the patients according to pN stage (pN0 and pN+), SLN PHGR1 status was not a significant prognostic factor in either group for the investigated endpoints (Figure 3, C and D, and results not shown). Thus, the potential detection of occult metastases and micrometastases by PHGR1 mRNA quantification in the otherwise node-negative patients did not provide prognostic information.
Figure 3.
Kaplan-Meier survival analyses of recurrence-free and disease-specific survival according to SLN PHGR1 status. (A and C) Recurrence-free survival in all patients (A) and pN0 patients (C). (B and D) Disease-specific survival in all patients (B) and pN0 patients (D).
Recurrence-free and disease-specific survivals in the overall cohort were modeled by univariate (Table 2) and multivariable (Table 3) Cox proportional-hazards regression. In univariate analysis, only SLN PHGR1 mRNA status (categorical variable) and level (continuous variable), T and N stage, and adjuvant chemotherapy were significant predictors of recurrence-free survival. In addition, SLN PHGR1 mRNA status and level, age, N stage, and adjuvant chemotherapy were significant predictors of disease-specific survival. Stratified analyses according to N stage revealed that SLN PHGR1 mRNA levels, but not status, predicted recurrence-free and disease-specific survival in pN+ patients but not in pN0 patients. The primary tumor PHGR1 mRNA level did not have any prognostic value. In multivariable analysis, SLN PHGR1 mRNA level, N stage, and age were the only covariates retained in the model for both endpoints. According to the P values in the final model, SLN PHGR1 mRNA level was the strongest independent prognostic factor tested in this cohort when adjusting for other relevant covariates. When SLN PHGR1 mRNA status as categorical variable replaced PHGR1 mRNA level in the multivariable Cox regressions, it was not retained in the final model (results not shown).
Table 2.
Univariate Cox Proportional-Hazards Regression of Recurrence-Free and Disease-Specific Survival According to PHGR1 mRNA Measurements and Clinicopathological Parameters
| Recurrence-Free Survival |
Disease-Specific Survival |
|||
|---|---|---|---|---|
| Parameter | HR | 95% CI | HR | 95% CI |
| SLN PHGR1 status (pos. vs neg.) | 3.27 | 1.35-7.90 | 2.87 | 1.17-7.03 |
| PHGR1 mRNA level in SLNs | 1.41 | 1.22-1.62 | 1.56 | 1.34-1.82 |
| PHGR1 mRNA level in tumors | 1.03 | 0.97-1.09 | 1.03 | 0.96-1.09 |
| Age (continuous) | 1.03 | 1.00-1.07 | 1.09c | 1.03-1.16 |
| Gender (F vs M) | 1.53 | 0.76-3.10 | 1.42 | 0.67-2.98 |
| Tumor stage (T3/T4 vs T1/2) | 4.97 | 1.19-20.7 | 4.28 | 1.02-18.0 |
| Median tumor diameter [mm] | 1.00 | 0.99-1.02 | 1.00 | 0.99-1.02 |
| Tumor grade (III vs I/II) | 1.81 | 0.89-3.65 | 1.66 | 0.78-3.55 |
| Tumor localization (dist. vs prox.) | 1.54 | 0.78-3.04 | 1.41 | 0.68-2.94 |
| Tumor MSI status (MSS vs MSI) | 1.34 | 0.65-2.76 | 1.84 | 0.82-4.17 |
| Tumor KRAS mutationa (mut vs wt) | 0.81 | 0.40-1.67 | 0.72 | 0.33-1.57 |
| Tumor BRAF mutationb (mut vs wt) | 1.01 | 0.47-2.18 | 0.88 | 0.38-2.07 |
| Number of lymph nodes (> = 12) | 0.93 | 0.46-1.88 | 0.984 | 0.46-2.10 |
| Lymph node metastasis (N+ vs N0) | 4.7 | 2.33-9.51 | 6.04 | 2.77-13.2 |
| Adjuvant chemotherapy (yes vs no) | 2.36 | 1.13-4.95 | 2.63 | 1.23-5.62 |
Codon 12 and 13.
V600E.
Age coefficient modeled as a two-step function. Results for time <2.5 years are shown.
Table 3.
Multivariable Cox Proportional-Hazards Regression of Recurrence-Free and Disease-Specific Survival According to PHGR1 mRNA Measurements and Clinicopathological Parameters
| Recurrence-Free Survival |
Disease-Specific Survival |
|||
|---|---|---|---|---|
| Parameter | HR | 95% CI | HR | 95% CI |
| SLN PHGR1 mRNA level (continuous) | 1.21 | 1.03-1.43 | 1.35 | 1.12-1.61 |
| Lymph node metastasis (N+ vs N0) | 3.59 | 1.65-7.83 | 3.75 | 1.56-9.03 |
| Age (continuous) | 1.03 | 1.00-1.07 | 1.10a | 1.03-1.18 |
Age coefficient modeled as a two-step function. Results for time <2.5 years are shown.
Discussion
In the current study of stage I to III colon cancers, we found significantly shorter recurrence-free and disease-specific survival for patients with elevated SLN PHGR1 mRNA levels. However, when restricted to patients with pN0 disease, no significant prognostic information was found for elevated PHGR1 mRNA. Though the first finding demonstrates a role for PHGR1 mRNA assessment in determining prognosis, the latter does not allow us to conclude on its utility as a marker of occult disease.
The present conclusions regarding the clinical value of potential occult SLN disease are similar to what we previously reported for SLN KRT20 and MUC2 mRNA measurements in the same patient cohort [11]. In contrast, a large pooled analysis and a recent meta-analysis demonstrated that the detection of occult tumor cells in regional LNs has clear prognostic value [5], [22]. One of the reports recognized that several single studies have concluded differently, but this may be explained in part by small and underpowered study populations [5]. We originally designed the study to have an estimated log-rank test power of 80% using survival estimates based on previous Cancer in Norway reports [23]. However, the actual 5-year survival rate of stage I/II patients in our cohort (93%) was substantially better than prospectively estimated. When using the actual survival of our pN0 patients and hazard ratio (HR) estimates from the pooled analysis (HR=3.37), we estimated a test power of 67%, which suggests that our cohort of pN0 patients may have been too small to demonstrate a significant effect. This notion is especially interesting considering that a minor difference in recurrence-free survival was observed, although it was not significant. The high 5-year survival rate in our cohort could be related to the relatively high percentage of MSI and right-sided tumors [24]. Interestingly, stratification of pN0 patients by MSI status did not reveal any significant prognostic differences among the resulting subgroups (results not shown).
PHGR1 mRNA levels were downregulated in a subset of primary tumors (Figure 2), possibly reducing its potential for the detection of neoplastic cells from these tumors [25]. Interestingly, there was a tendency towards lower primary tumor PHGR1 mRNA levels in the seven patients with SLNs negative for PHGR1 mRNA despite known nodal disease. The disadvantage of such marker downregulation can be compensated for, to some extent, by using multiple markers [10], [25], [26]. However, occult SLN disease suggested by combinations of our three markers (PHGR1, KRT20, and MUC2 mRNA) in a multimarker panel did not reveal any further prognostic information (results not shown).
The SLN concept has been well established in breast and skin cancer, with advantages in terms of more focused LN diagnostics and a reduction in the extent of LN dissection for many patients [27], [28], [29]. Accordingly, there has been some focus on evaluating the SLN concept in CRC over the previous decade, with the aim of more accurate LN evaluation, not necessarily reduced surgical interventions for CRC [6], [7]. However, many individual studies and two large pooled analyses concluded that the number of false negatives, based on routine histological analysis, was too high to allow LN diagnostics to be restricted to SLNs only [6], [7]. On the other hand, a more focused examination of SLNs revealed occult LN disease of potential prognostic value [6], [7]. The fact that most studies have applied more focused examination techniques to SLNs and not other regional LNs precludes direct evaluation of the SLN concept per se [7], [30]. This objection is also relevant for our study, as we applied the molecular markers only to SLNs. Thus, we cannot preclude that the failure to show prognostic value of occult SLN disease is due to suboptimal selection of nodes for analysis by SLN mapping. Unfortunately, none of the studies presenting survival data after SLN analysis have analyzed both SLNs and non-SLNs using immunohistochemistry or molecular techniques [9], [10], [11], [12], [13], [31]. Furthermore, none of the studies reported a prognostic value of occult SLN disease in the group of otherwise node-negative patients. Thus, the present evidence does not support a clinical value of SLN mapping in CRC beyond the information retrieved by routine LN examination. Accordingly, the SLN concept per se does not seem to have the same validity and diagnostic potential in CRC as in breast cancer and melanoma.
SLN PHGR1 mRNA level was a stronger prognostic factor in univariate Cox regression than the corresponding categorical SLN PHGR1 status (Table 2), especially when the analyses were restricted to pN+ patients. This finding probably reflects very high PHGR1 mRNA levels, presumably corresponding to large tumor cell deposits, being associated with a worse prognosis than levels just above the threshold. This notion corresponds well with the established difference in prognostic value of macrometastases, micrometastases, and isolated tumor cells [32]. The observation that both pN stage and SLN PHGR1 mRNA level were retained in the multivariable regression model emphasizes that the magnitude of the PHGR1 mRNA level contributes to independent prognostic information beyond routine N staging. Accordingly, a recent meta-analysis demonstrated a prognostic value of LN micrometastases but not isolated tumor cells [22]. Therefore, we optimized the threshold of PHGR1 mRNA for positive test status in order to minimize the P value of log-rank tests for recurrence-free survival (results not shown). Interestingly, a considerably higher threshold resulted in a much lower P value for the overall patient group but not for the pN0 group.
PHGR1 mRNA level in an SLN reflects both the number of tumor cells present and the PHGR1 mRNA level per tumor cell. A higher marker level per tumor cell results in higher sensitivity for tumor cells. Interestingly, we found an association between primary tumor PHGR1 mRNA level and positive MSI and BRAF status. In spite of this, positive SLN PHGR1 mRNA status seemed to be inversely related to primary tumor MSI and associated proximal localization and large tumor diameter (Table 1). This apparent discrepancy might be due to other explanatory variables being stronger, for example the differences in overall lymph node yields between left and right colon [33].
Conclusions
We have shown that PHGR1 mRNA is a promising marker for the detection of tumor cells in SLNs from operable colon cancer patients, providing significant prognostic information in the overall patient group, but not patients who were node-negative upon routine examination. Low numbers of disease-related endpoints in the node-negative group in this study suggest that the prognostic impact of occult SLN disease should be investigated in larger cohorts in future studies.
The following are the supplementary data related to this article.
Acknowledgments
Acknowledgements
This work was supported by the Western Norway Regional Health Authorities, the Norwegian Cancer Society (grant number T-97452/001), and the Folke Hermansen Fund. We are very grateful to statistician Jan Terje Kvaløy, PhD, for advice regarding the survival analyses.
Conflict of Interest Statement
Declarations of interest: none.
References
- 1.Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol. 2015;33:1787–1796. doi: 10.1200/JCO.2014.60.0213. [DOI] [PubMed] [Google Scholar]
- 2.Figueredo A, Coombes ME, Mukherjee S. Adjuvant therapy for completely resected stage II colon cancer. Cochrane Database Syst Rev. 2008 doi: 10.1002/14651858.CD005390.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lea D, Håland S, Hagland HR, Søreide K. Accuracy of TNM staging in colorectal cancer: a review of current culprits, the modern role of morphology and stepping-stones for improvements in the molecular era. Scand J Gastroenterol. 2014;49:1153–1163. doi: 10.3109/00365521.2014.950692. [DOI] [PubMed] [Google Scholar]
- 4.Veen T, Nedrebø BS, Stormark K, Søreide JA, Kørner H, Søreide K. Qualitative and quantitative issues of lymph nodes as prognostic factor in colon cancer. Dig Surg. 2013;30:1–11. doi: 10.1159/000349923. [DOI] [PubMed] [Google Scholar]
- 5.Rahbari NN, Bork U, Motschall E, Thorlund K, Büchler MW, Koch M, Weitz J. Molecular detection of tumor cells in regional lymph nodes is associated with disease recurrence and poor survival in node-negative colorectal cancer: a systematic review and meta-analysis. J Clin Oncol. 2012;30:60–70. doi: 10.1200/JCO.2011.36.9504. [DOI] [PubMed] [Google Scholar]
- 6.van der Pas MH, Meijer S, Hoekstra OS, Riphagen II, de Vet HC, Knol DL, van Grieken NC, Meijerink WJ. Sentinel-lymph-node procedure in colon and rectal cancer: a systematic review and meta-analysis. Lancet Oncol. 2011;12:540–550. doi: 10.1016/S1470-2045(11)70075-4. [DOI] [PubMed] [Google Scholar]
- 7.van der Zaag ES, Bouma WH, Tanis PJ, Ubbink DT, Bemelman WA, Buskens CJ. Systematic review of sentinel lymph node mapping procedure in colorectal cancer. Ann Surg Oncol. 2012;19:3449–3459. doi: 10.1245/s10434-012-2417-0. [DOI] [PubMed] [Google Scholar]
- 8.Huynh KT, Bilchik AJ. Sentinel lymph node biopsy and nodal ultrastaging in colorectal cancer. Cancer J. 2015;21:11–16. doi: 10.1097/PPO.0000000000000093. [DOI] [PubMed] [Google Scholar]
- 9.Lim SJ, Feig BW, Wang H, Hunt KK, Rodriguez-Bigas MA, Skibber JM, Ellis V, Cleary K, Chang GJ. Sentinel lymph node evaluation does not improve staging accuracy in colon cancer. Ann Surg Oncol. 2008;15:46–51. doi: 10.1245/s10434-007-9629-8. [DOI] [PubMed] [Google Scholar]
- 10.Koyanagi K, Bilchik AJ, Saha S, Turner RR, Wiese D, McCarter M, Shen P, Deacon L, Elashoff D, Hoon DSB. Prognostic relevance of occult nodal micrometastases and circulating tumor cells in colorectal cancer in a prospective multicenter trial. Clin Cancer Res. 2008;14:7391–7396. doi: 10.1158/1078-0432.CCR-08-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordgård O, Oltedal S, Aasprong OG, Søreide JA, Søreide K, Tjensvoll K, Gilje B, Heikkilä R, Guriby M, Lothe RA. Prognostic relevance of occult metastases detected by cytokeratin 20 and mucin 2 mRNA levels in sentinel lymph nodes from colon cancer patients. Ann Surg Oncol. 2012;19:3719–3726. doi: 10.1245/s10434-012-2454-8. [DOI] [PubMed] [Google Scholar]
- 12.Nissan A, Protic M, Bilchik AJ, Howard RS, Peoples GE, Stojadinovic A. United States Military Cancer Institute Clinical Trials Group (USMCI GI-01) randomized controlled trial comparing targeted nodal assessment and ultrastaging with standard pathological evaluation for colon cancer. Ann Surg. 2012;256:412–427. doi: 10.1097/SLA.0b013e31826571c8. [DOI] [PubMed] [Google Scholar]
- 13.Braat AE, Pol RA, Oosterhuis JWA, de Vries JE, Mesker WE, Tollenaar RAEM. Excellent prognosis of node negative patients after sentinel node procedure in colon carcinoma: a 5-year follow-up study. Eur J Surg Oncol. 2014;40:747–755. doi: 10.1016/j.ejso.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Oltedal S, Skaland I, Maple-Grødem J, Tjensvoll K, Janssen EAM, Gilje B, Smaaland R, Heikkilä R, Nordgård O. Expression profiling and intracellular localization studies of the novel Proline-, Histidine-, and Glycine-rich protein 1 suggest an essential role in gastro-intestinal epithelium and a potential clinical application in colorectal cancer diagnostics. BMC Gastroenterology. 2018;18:26. doi: 10.1186/s12876-018-0752-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahl O. Adjuvant chemotherapy for colon cancer. Tidsskr Nor Laegeforen. 2007;127:3094–3096. [PubMed] [Google Scholar]
- 16.Guren MG, Vonen B, Edna TH, Kørner H, Dørum LMR. Helsedirektoratet; 2015. Nasjonalt handlingsprogram medretningslinjer for diagnostikk, behandling og oppfølging av kreft i tykktarm og endetarm. [Google Scholar]
- 17.Nordgård O, Oltedal S, Kørner H, Aasprong OG, Tjensvoll K, Gilje B, Heikkila R. Quantitative RT-PCR detection of tumor cells in sentinel lymph nodes isolated from colon cancer patients with an ex vivo approach. Ann Surg. 2009;249:602–607. doi: 10.1097/SLA.0b013e31819ec923. [DOI] [PubMed] [Google Scholar]
- 18.Punt CJA, Buyse M, Köhne C-H, Hohenberger P, Labianca R, Schmoll HJ, Påhlman L, Sobrero A, Douillard J-Y. Endpoints in adjuvant treatment trials: a systematic review of the literature in colon cancer and proposed definitions for future trials. J Natl Cancer Inst. 2007;99:998–1003. doi: 10.1093/jnci/djm024. [DOI] [PubMed] [Google Scholar]
- 19.Grambsch P, Therneau T. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81:515–526. [Google Scholar]
- 20.Dormann CF. Collinearity: A review of methods to eal with it and a simulation study evaluating their performance. Ecography. 2012;35:1–20. [Google Scholar]
- 21.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 22.Sloothaak DAM, Sahami S, van der Zaag-Loonen HJ, van der Zaag ES, Tanis PJ, Bemelman WA, Buskens CJ. The prognostic value of micrometastases and isolated tumour cells in histologically negative lymph nodes of patients with colorectal cancer: a systematic review and meta-analysis. Eur J Surg Oncol. 2014;40:263–269. doi: 10.1016/j.ejso.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Registry of Norway Cancer in Norway 2001. Cancer Registry of Norway; 2002. [Google Scholar]
- 24.Dienstmann R, Mason MJ, Sinicrope FA, Phipps AI, Tejpar S, Nesbakken A, Danielsen SA, Sveen A, Buchanan DD, Clendenning M. Prediction of overall survival in stage II and III colon cancer beyond TNM system: a retrospective, pooled biomarker study. Ann Oncol. 2017;28:1023–1031. doi: 10.1093/annonc/mdx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohlsson L, Hammarstrom ML, Israelsson A, Naslund L, Oberg A, Lindmark G, Hammarstrom S. Biomarker selection for detection of occult tumour cells in lymph nodes of colorectal cancer patients using real-time quantitative RT-PCR. Br J Cancer. 2006;95:218–225. doi: 10.1038/sj.bjc.6603206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tjensvoll K, Oltedal S, Farmen RK, Shammas FV, Heikkilä R, Kvaløy JT, Gilje B, Smaaland R, Nordgård O. Disseminated tumor cells in bone marrow assessed by TWIST1, cytokeratin 19, and mammaglobin A mRNA predict clinical outcome in operable breast cancer patients. Clin Breast Cancer. 2010;10:378–384. doi: 10.3816/CBC.2010.n.050. [DOI] [PubMed] [Google Scholar]
- 27.Krag DN, Anderson SJ, Julian TB, Brown AM, Harlow SP, Costantino JP, Ashikaga T, Weaver DL, Mamounas EP, Jalovec LM. Sentinel-lymph-node resection compared with conventional axillary-lymph-node dissection in clinically node-negative patients with breast cancer: overall survival findings from the NSABP B-32 randomised phase 3 trial. Lancet Oncol. 2010;11:927–933. doi: 10.1016/S1470-2045(10)70207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrelli F, Lonati V, Barni S. Axillary dissection compared to sentinel node biopsy for the treatment of pathologically node-negative breast cancer: a meta-analysis of four randomized trials with long-term follow up. Oncol Rev. 2012;6 doi: 10.4081/oncol.2012.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong SL, Balch CM, Hurley P, Agarwala SS, Akhurst TJ, Cochran A, Cormier JN, Gorman M, Kim TY, McMasters KM. Sentinel lymph node biopsy for melanoma: American Society of Clinical Oncology and Society of Surgical Oncology joint clinical practice guideline. J Clin Oncol. 2012;30:2912–2918. doi: 10.1200/JCO.2011.40.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nordgård O, Smaaland R. SLN mapping in colorectal cancer. Lancet Oncol. 2011;12:990. doi: 10.1016/S1470-2045(11)70176-0. [DOI] [PubMed] [Google Scholar]
- 31.Bilchik AJ, Hoon DSB, Saha S, Turner RR, Wiese D, DiNome M, Koyanagi K, McCarter M, Shen P, Iddings D. Prognostic impact of micrometastases in colon cancer: interim results of a prospective multicenter trial. Ann Surg. 2007;246:568–575. doi: 10.1097/SLA.0b013e318155a9c7. [DOI] [PubMed] [Google Scholar]
- 32.Edge S, Byrd D, Compton C, Fritz A, Greene F, Trotti A, editors. AJCC Cancer Staging Manual. Seventh ed. Springer; New York: 2010. [Google Scholar]
- 33.Berg M, Guriby M, Nordgård O, Nedrebø BS, Ahlquist TC, Smaaland R, Oltedal S, Søreide JA, Kørner H, Lothe RA. Influence of microsatellite instability and KRAS and BRAF mutations on lymph node harvest in stage I-III colon cancers. Mol Med. 2013;19:286–293. doi: 10.2119/molmed.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]