Abstract
BACKGROUND: Liquid biopsy is emerging as an important approach for tumor genotyping in non-small cell lung cancer, ddPCR and SuperARMS are both methods with high sensitivity and specificity for detecting EGFR mutation in plasma. We aimed to compare ddPCR and SuperARMS to detect plasma EGFR status in a cohort of advanced NSCLC patients. METHOD: A total of 79 tumor tissues and paired plasma samples were collected. The EGFR mutation status in tissue was tested by ADx-ARMS, matched plasma was detected by ddPCR and SuperARMS, respectively. RESULTS: The EGFR mutation rates were identified as 64.6% (tissue, ARMS), 55.7% (plasma, ddPCR), and 49.4% (plasma, Super ARMS), respectively. The sensitivity of ddPCR was similar with Super-ARMS in plasma EGFR detection (80.4% vs 76.5%), as well as the specificity (89.3% vs 100%). And the McNemar’s test showed there was no significant difference (P = .125). The concordance rate between SuperARMS and ddPCR was 91.1%. A significant interaction was observed between cfDNA EGFR mutation status and EGFR-TKIs treatment tested by both methods. CONCLUSION: Super-ARMS and ddPCR share the similar accuracy for EGFR mutation detection in plasma biopsy; both methods predicted well the efficacy of EGFR-TKIs by detecting plasma EGFR status.
Introduction
In recent years, although tumor tissue is still quoted as the “gold standard” for tumor genotyping, bloodstream-based genetic analyses—termed as “liquid biopsy” is emerging as an important approach [1]. For lung cancer, epidermal growth factor receptor (EGFR) mutation was regarded as the strongest predictor of effectiveness of treatment with EGFR tyrosine kinase inhibitors (TKIs), such as gefitinib and erlotinib [2], [3]. Many methods have been reported for detecting EGFR mutation in plasma cell-free DNA (cfDNA) of lung cancer patients, including Amplification Refractory Mutation System (ARMS) [4], [5], PNA-clamp [6], denaturing high-performance liquid chromatography (DHPLC) [7], and next-generation sequencing (NGS) [8]. The sensitivity and specificity of these technologies varied significantly among different studies. Droplet digital PCR (ddPCR) was reported with the sensitivity and specificity of 81.82% and 98.44% [9], respectively. Due the high sensitivity and specificity, ddPCR was favored by many researchers for plasma EGFR mutation detecting. Recently, a novel technique called SuperARMS, which was reported with high sensitive and specific, provides a promising approach for cfDNA detection [10].
In our pilot study, we detected 79 pairs plasma samples of non-small cell lung cancer patients used ADx-ARMS and SuperARMS, resulted showed the sensitivity of SuperARMS was higher than ARMS. Few studies were performed to compare SuperARMS and ddPCR for genotyping. In this reason, we use ddPCR and SuperARMS to detect plasma EGFR status in a cohort of advanced NSCLC patients.
Patients and Methods
Patients
A total of 79 patients were enrolled in this study, which were collected from Jan 1, 2012, to Jun 1, 2013, at The First People's Hospital of Foshan. All patients were histologically confirmed lung adenocarcinoma (include two adenosquamous carcinoma). Sufficient tumor tissue specimens and plasma specimens were sampled. The study was approved by the ethics committee of The First People's Hospital of Foshan. All patients had provided written information consents before the study procedures. Patients were followed up every month by the investigators.
Sample Collection and DNA Extraction
Tumor tissue sample and matched plasma with an interval less than 1 week was collected for each patient. All tumor tissue samples were formalin-fixed and paraffin-embedded (FFPE), and underwent pathologic evaluation to ensure the tumor content of at least 20%.
Tumor tissue DNA from FFPE was extracted by using an FFPE DNA Kit (Amoy Diagnostics Co, Xiamen, China) according to the standard protocols. Plasma was separated from 10 ml of peripheral blood from each patient before EGFR-TKI treatment. Then, cfDNA was isolated using a plasma/serum cell-free DNA kit (Amoy Diagnostics Co, Xiamen, China) according to the manufacturer’s instructions.
EGFR Mutation Detection
The EGFR mutations of the extracted DNA from the tissue were assessed with the amplification refractory mutation system (ARMS) by using an EGFR mutations detection Kit (Amoy Diagnostics, Xiamen, China) according to the manufacturer’s instructions. The method was reported previously [11].
Plasma DNA was analyzed by ddPCR and SuperARMS, respectively. The ddPCR was performed according to the manufacturer’s instructions and this method was reported previously [9].
The SuperARMS assay (Amoy Diagnostics, Xiamen, China), which is newly designed based on ARMS with indicative detection limit of 0.2% as the kit specification claims. This kit adopts both ARMS and real-time PCR technologies to identify 41 of the most common hotspots in EGFR exons 18-21 in plasma ctDNA samples. So far, the kit is for research use only. All samples were carried out following the manufacturer’s protocol. Briefly, the ADx-SuperARMS PCR was implemented in a final 80 μL reaction mixture consisting of 15 μL DNA templates as well as novel proprietary primers and probes, dNTPs, buffer, Mg2+ and Taq DNA polymerases. PCR reactions were carried out on Mx3000P PCR system using the cycling conditions as follows: 1 cycle at 95°C for 10 min for incubation, followed by 15 cycles at 95°C for 40 seconds, 64°C for 40 seconds and 72°C for 30 seconds, and then 28 cycles at 93°C for 40 seconds, 60°C for 45 seconds and 72°C for 30 seconds. The raw data were analyzed using the MxPro-Mx3000P v4.10 included with the instrument. ∆Ct was calculated as: mutant Ct value—internal control Ct value. If ∆Ct is less than the Cut-off value, the result is determined as positive. Otherwise, the result is determined as negative or beyond the limit of detection.
Statistical Analysis
Statistical analyses were performed with the IBM SPSS Statistics version 20. McNemar test was used to analysis the concordance of EGFR mutation between matched samples. The progression-free survival (PFS) according to EGFR mutation status was estimated by Kaplan–Meier method and the difference in survival was compared using log-rank test. P < .05 was deemed to be of statistical significance. All P values and 95% confidence intervals (95%CI) were two-sided.
Results
Patients Clinical Characteristics
A total of 79 patients were enrolled and 79 pairs of tumor tissue and plasma samples were collected. All tissue samples were tested EGFR mutation status by ADx-ARMS successfully. There were 51 (64.6%) patients harbor EGFR-activating mutations, among whom 26 were exon 21 L858R mutation and 25 were exon 19 deletion. 28 (35.4%) harbored wild-type EGFR. The EGFR mutation status in tumor tissue by ADx-ARMS was regarded as “gold standard” in the study. The clinical characteristics of patients were shown in Table 1.
Table 1.
Baseline demographic and clinical characteristics.
| Characteristics | No. of patients (%) |
|---|---|
| Sex | |
| Male | 36 (45.6%) |
| Female | 43(54.4%) |
| Age | |
| Median | 59 (30-75) |
| <60 | 43 (54.4%) |
| ≧60 | 36 (45.6%) |
| Smoking status | |
| Never-smoker | 54 (68.4%) |
| Current/ever-smoker | 25(31.6%) |
| ECOG PS | |
| PS 0-1 | 72 (91.1%) |
| PS 2 | 7 (8.9%) |
| EGFR mutation in tissue | |
| Mutation+ | 51 (64.6%) |
| Wild-type | 28 (35.4%) |
| Histology | |
| adenocarcinoma | 77 (97.5%) |
| adenosquamous | 2 (2.5%) |
EGFR Mutation in Plasma by SuperARMS and ddPCR
All 79 plasma samples were detected EGFR mutation status by ddPCR and SuperARMS successfully. When tested by ddPCR, 44 of 79 (55.7%) patients were identified to be EGFR mutation, including three patients who were detected to be wild-type in tumor tissue, which was considered as false positive. The sensitivity, specificity, positive predictive value and negative predictive value was 80.4%, 89.4%, 93.2%, 71.4%, respectively. The concordance of EGFR mutations between plasma and tissue was 83.5% by ddPCR.
When using SuperARMS to detect plasma EGFR, 39 of 79 (49.4%) patients were demonstrated to be EGFR mutation. Twelve patients were EGFR mutation in tumor tissue but EGFR wild-type in plasma. The sensitivity and negative predictive value of SuperARMS was 76.5% and 70.0%, respectively. The specificity and positive predictive value were both 100%. The concordance of EGFR mutations between plasma and tissue was 84.8%. The result of EGFR mutation status by ddPCR and SuperARMS was shown in Table 2.
Table 2.
EGFR mutation status by ddPCR and SuperARMS.
| Plasma cfDNA | Tumor tissue | Results | ||
|---|---|---|---|---|
| SuperARMS | M+ | M- | Total | Sensitivity = 76.5% |
| M+ | 39 | 0 | 39 | Specificity = 100% |
| M- | 12 | 28 | 40 | Positive predictive value = 100% |
| Total | 51 | 28 | 79 | Negative predictive value = 70.0% Concordance = 84.8% |
| ddPCR | M+ | M- | Total | Sensitivity = 80.4% |
| M+ | 41 | 3 | 44 | Specificity = 89.3% |
| M- | 10 | 25 | 35 | Positive predictive value = 93.2% |
| Total | 51 | 28 | 79 | Negative predictive value = 71.4% Concordance = 83.5% |
Compare SuperARMS with ADx-ARMS in Plasma EGFR Detection
The sensitivity of ddPCR was similar with Super-ARMS in plasma EGFR detection (80.4% vs. 76.5%), as well as the specificity (89.3% vs 100%). The McNemar’s test showed there was no significant difference (P = .125). And the concordance rate of EGFR mutation status between SuperARMS and ddPCR was 91.1% (72/79).
EGFR Mutation Status and Prediction of EGFR-TKI Efficacy
There were 51 patients who received EGFR-TKI treatment. The overall objective response rate (ORR) was 56.9% (29/51) for EGFR-TKI treatment.
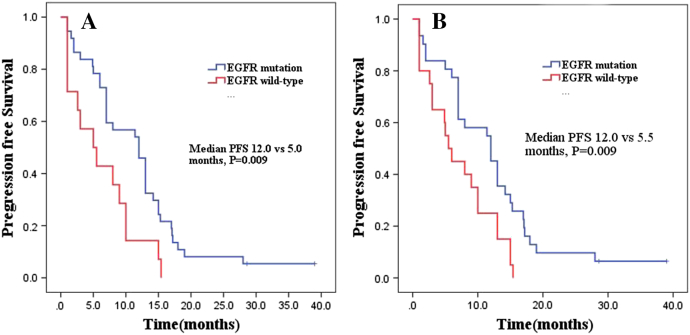
For plasma samples by ddPCR, the ORR was 67.6% (25/37) and 28.6% (4/14) respectively in patients with or without EGFR mutation, P = .025. Median PFS was significantly prolonged in EGFR mutation patients compared with EGFR wild-type patients (12.0months vs 5.0 months, P = .009) (Figure 1A).
Figure 1.
PFS in EGFR mutation patients compared with EGFR wild-type patients.
(A) Plasma samples by ddPCR. (B) Plasma samples by SuperARMS.
For plasma samples by SurperARMS, the ORR in EGFR mutation or wild-type patients was 74.2% (23/31) and 30.0% (6/20), respectively, P = .002. EGFR mutation patients tended to have longer PFS than those with EGFR wild-type patients (12.0months vs 5.5 months, P = .009) (Figure 1B).
These data suggested that EGFR mutation status in blood detected by ddPCR and SuperARMS can be predictive of tumor response to EGFR-TKIs and survival results.
Discussion
Nowadays, liquid biopsy represents a promising approach for noninvasive assessment of cancer gene profile. In this study, we performed a comparison of ddPCR and SuperARMS for the detection of the EGFR mutation in plasma cfDNA from NSCLC patients. All plasma samples were successfully processed for DNA extraction and genotyping. Our results suggested that both SuperARMS and ddPCR have high sensitivity and specificity for EGFR detection in plasma samples, and are good predictors of the efficacy of EGFR-TKIs.
Various methods had been reported to detect EGFR mutation in cfDNA of lung cancer patients. The ddPCR was one with high sensitivity and specificity that was widely used. Zhu et al had optimized the ddPCR assays to reach the sensitivity of 0.04%.[9] In his study, EGFR Exon19-Del mutation was detected with the sensitivity of 81.82% and the specificity of 98.44%, while EGFR L858R mutation was tested with sensitivity of 80.00% and the specificity of 95.77%, compared with the paired tumor tissues. Oxnard et al. reported the ddPCR assays for detecting EGFR with high sensitivity and 100% specificity and ddPCR was demonstrated valuable in monitoring resistance mutations which may offer broader clinical application [12]. The results were consisted with that in our study. The SuperARMS is a novel technique which was modified on the basis of ARMS by optimizing the primers designation, PCR amplification reaction-system, which makes it much more sensitive. Preciously, Li et al. used SuperARMS to test plasma EGFR mutation of 109 patients, results showed the sensitivity of 82% and specificity of 100%, and the concordance rate between plasma and tissue was 89.9%[10]. The results were concordant with our study, with sensitivity of 76.5% and specificity of 100%, the concordance between plasma and tissue was 84.8%. The sensitivity of ddPCR was similar with Super-ARMS (80.4% vs 76.5%), while the specificity tended to be lower (89.3% vs 100%), but there was no significant difference (P = .125). One great advantage of ddPCR is the quantification of DNA; SuperARMS cannot quantify but it is fast and of low dependency upon the laboratory equipment, and more importantly, it covers multiple abnormal positions of 41 EGFR mutation hotspots within one run.
About 12% of cases by ddPCR and 15% by SuperAMRS were false-negative in this study. The major reason may be the low abundance of cfDNA in plasma. Nonetheless, cancers are heterogeneous, with intra-tumor or inter-tumor showing different genetic profiles. In Table 2, we can see three patients were EGFR negative in tumor tissue, but positive in plasma by ddPCR. This may be due to tumor heterogeneity between tumor tissue and plasma. One important advantage of liquid biopsy is potentially fewer heterogeneity issues than tumor tissue biopsy [13].
Many previous studies demonstrated that there was a significant interaction between cfDNA EGFR mutation status and treatment for PFS [4], [5], [14], [15]. In accordance with these studies, our study revealed that patients with plasma EGFR mutations had longer PFS as compared to patients without plasma EGFR mutations detected by both ddPCR and SuperARMS, which means both methods predicted well the efficacy of EGFR-TKI by detecting plasma EGFR status.
As we know, although NSCLC patients with EGFR mutation show dramatic responses to EGFR-TKIs, resistance eventually develops in most patients. T790 M mutation is the most common mechanism of EGFR TKI resistance [16]. Liquid biopsy plays an important role in real-time molecular monitoring of resistance and personalized cancer therapy [17], [18]. Both ddPCR and SuperARMS were reported to detect T790M successfully [12], [19], [20]. One main limitation of this study was that we failed to track the dynamic changes of EGFR mutations during treatment. That's what we're going to do in the future works.
Conclusion
In conclusion, Super-ARMS and ddPCR share the similar accuracy for EGFR mutation detection in plasma, both methods predicted well the efficacy of EGFR-TKIs.
Disclosure
The authors report no potential conflicts of interest in this work.
Acknowledgements
We would like to give our appreciation to Amoy Diagnostics Co, Xiamen, China, for providing the technical guidance for us. This work was supported by the Special fund project for technology innovation of Foshan City (No. 2014AG10003), and Wu Jieping Medical Foundation (No. 320.6750.15241).
References
- 1.Buder A, Tomuta C, Filipits M. The potential of liquid biopsies. Curr Opin Oncol. 2016;28(2):130–134. doi: 10.1097/CCO.0000000000000267. [DOI] [PubMed] [Google Scholar]
- 2.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 3.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 4.Duan H, Lu J, Lu T, Gao J, Zhang J, Xu Y, Wang M, Wu H, Liang Z, Liu T. Comparison of EGFR mutation status between plasma and tumor tissue in non-small cell lung cancer using the Scorpion ARMS method and the possible prognostic significance of plasma EGFR mutation status. Int J Clin Exp Pathol. 2015;8(10):13136–13145. [PMC free article] [PubMed] [Google Scholar]
- 5.Goto K, Ichinose Y, Ohe Y, Yamamoto N, Negoro S, Nishio K, Itoh Y, Jiang H, Duffield E, McCormack R. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol. 2012;7(1):115–121. doi: 10.1097/JTO.0b013e3182307f98. [DOI] [PubMed] [Google Scholar]
- 6.Pasquale R, Fenizia F, Esposito A, Sacco A, Esposito C, Forgione L, Rachiglio AM, Bevilacqua S, Montanino A, Franco R. Assessment of high-sensitive methods for the detection of EGFR mutations in circulating free tumor DNA from NSCLC patients. Pharmacogenomics. 2015;16(10):1135–1148. doi: 10.2217/pgs.15.45. [DOI] [PubMed] [Google Scholar]
- 7.Bai H, Mao L, Wang HS, Zhao J, Yang L, An TT, Wang X, Duan CJ, Wu NM, Guo ZQ. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol. 2009;27(16):2653–2659. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- 8.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, Bentley D. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 9.Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y, McCormack R, Gu Y, Liu X. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA from Patients with Advanced Non-Small Cell Lung Cancer. J Mol Diagn. 2015;17(3):265–272. doi: 10.1016/j.jmoldx.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Xu H, Su S, Ye J, Chen J, Jin X, Lin Q, Zhang D, Ye C, Chen C. Clinical validation of a highly sensitive assay to detect EGFR mutations in plasma cell-free DNA from patients with advanced lung adenocarcinoma. PLoS One. 2017;12(8):e0183331. doi: 10.1371/journal.pone.0183331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Lu Y, Zhu G, Lei Y, Zheng L, Qin H, Tang C, Ellison G, McCormack R, Ji Q. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol. 2013;66(12):1065–1069. doi: 10.1136/jclinpath-2013-201728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O'Connell A, Messineo MM, Luke JJ, Butaney M, Kirschmeier P, Jackman DM. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20(6):1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz LJ, Bardelli A. Liquid biopsies: genotyping circulating tumoe DNA. J Clin Oncol. 2014;32(6):579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Zhuo M, Ye X, Bai H, Wang Z, Sun Y, Zhao J, An T, Duan J, Wu M. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget. 2016;7(15):20810–20824. doi: 10.18632/oncotarget.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu YJ, Zhang HB, Liu YH, Zhu YZ, Chen J, Li Y, Bai JP, Liu LR, Qu YC, Qu X. Association of mutant EGFR L858R and exon 19 concentration in circulating cell-free DNA using droplet digital PCR with response to EGFR-TKIs in NSCLC. Oncol Lett. 2017;14(2):2573–2579. doi: 10.3892/ol.2017.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16(9):e447–e459. doi: 10.1016/S1470-2045(15)00246-6. [DOI] [PubMed] [Google Scholar]
- 17.Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, Ladrera G, Thongprasert S, Srimuninnimit V, Liao M. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res. 2015;21(14):3196–3203. doi: 10.1158/1078-0432.CCR-14-2594. [DOI] [PubMed] [Google Scholar]
- 18.Oellerich M, Schutz E, Beck J, Kanzow P, Plowman PN, Weiss GJ, Walson PD. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit Rev Clin Lab Sci. 2017;54(3):205–218. doi: 10.1080/10408363.2017.1299683. [DOI] [PubMed] [Google Scholar]
- 19.Zhu L, Zhang S, Xun Y, Jiang Y, Xia B, Chen X, Wang L, Jiang H, Ma S. Comparison of the Amplification Refractory Mutation System, Super Amplification Refractory Mutation System, and Droplet Digital PCR for T790 M Mutation Detection in non-small cell lung cancer after failure of tyrosine kinase inhibitor treatment. Pathol Oncol Res. 2017;2017 doi: 10.1007/s12253-017-0286-3. [DOI] [PubMed] [Google Scholar]
- 20.Takahama T, Sakai K, Takeda M, Azuma K, Hida T, Hirabayashi M, Oguri T, Tanaka H, Ebi N, Sawa T. Detection of the T790M mutation of EGFR in plasma of advanced non-small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors (West Japan oncology group 8014LTR study) Oncotarget. 2016;7(36):58492–58499. doi: 10.18632/oncotarget.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]