Abstract
Hepatitis B virus (HBV) targets the liver and is a major driver for liver cancer. Clinical data suggest that HBV infection is associated with reduced response to treatment with the multi-kinase inhibitor sorafenib, the first available molecularly targeted anti-hepatocellular carcinoma (HCC) drug. Given that Raf is one of the major targets of sorafenib, we investigated the activation state of the Raf-Mek-Erk pathway in the presence of HBV and in response to sorafenib. Here we show that hepatoma cells with replicating HBV are less susceptible to sorafenib inhibitory effect as compared to cells in which HBV expression is suppressed. However, although HBV replication is associated with increased level of pErk, its blockade only modestly augments sorafenib effect. In contrast, the phosphorylated form of the pro-oncogenic Mitogen-Activated Protein Kinase 14 (pMAPK14), a protein kinase that was recently linked to sorafenib resistance, is induced in sorafenib-treated hepatoma cells in association with HBV X protein expression. Knocking down pMAPK14 results in augmentation of the therapeutic efficacy of sorafenib and largely alleviates resistance to sorafenib in the presence of HBV. Thus, this study suggests that HBV promotes HCC resistance to sorafenib. Combining pMAPK14 inhibitors with sorafenib may be beneficial in patients with HBV-associated HCC.
Introduction
Hepatocellular carcinoma (HCC) is the fifth most prevalent tumor type and the third leading cause of cancer-related deaths worldwide [1]. Risk factors for HCC include chronic viral hepatitis, metabolic liver diseases such as non-alcoholic steatohepatitis (NASH) as well as cirrhosis from any cause. Chronic infection with hepatitis B virus (HBV), a small DNA virus that targets the liver, is a leading worldwide risk factor for HCC [2]. The risk of HBV infected patient to develop HCC is 5-100 times as high as the risk of healthy individual [3]. However, although extensively studied, the mechanism(s) by which HBV promotes liver carcinogenesis is still largely obscure [4].
Patients with early stage HCC can be treated with curative modalities such as tumor resection, liver transplantation or radio-frequency ablation [5]. Available options for patients with advanced disease are much more limited, since conventional systemic chemotherapy is usually ineffective in HCC. Therefore, the introduction of the first chemotherapy agent for patients with advanced HCC, the multi-kinase inhibitor sorafenib that blocks vascular endothelial growth factor receptor (VEGFR), platelet-derived growth factor receptor (PDGFR) and Raf family kinases [6], was accompanied by a great enthusiasm. However, early studies suggested only modest survival benefit for sorafenib at the cost of often substantial side effects [7].
Clinical trials performed in HBV endemic areas, investigating the efficacy of sorafenib in patients with HCC, suggest that the response rate to sorafenib among HBV infected patients is lower as compared to that observed in patients with HCC associated with other etiologies [8]. This observation raises the hypothesis that HBV might antagonize sorafenib effect, mandating investigation into alternative or additive therapies for patients with HBV-associated HCC.
In this study, we aimed to investigate whether HBV is implicated in resistance to sorafenib effect in hepatoma cell model systems. We further investigated the mechanism by which HBV confers resistance to sorafenib and explored for potential alternative pathways that can be targeted in order to overcome HBV-associated resistance to sorafenib.
Material and Methods
Cell Lines, Transfection, Transduction and Treatments
Human hepatoma cell lines HepG2 and the HepG2.2.15 cells [9] were cultured in high glucose Dulbecco’s Modified Eagle Medium (DMEM; Biological Industries, Beit Haemek, ISRAEL) supplemented with 5% fetal bovine serum (FBS; Biological Industries, Beit Haemek, ISRAEL). All cell lines were maintained at 37°C in humidified atmosphere with 5% CO2. For drug testing, cells were treated either with sorafenib (7-12μM) (BAY 43-9006, Enzo Life Science), with FR180204 (70μM) (Sigma) or with corresponding combinations. The compounds were dissolved in DMSO and treatment was performed one day after plating.
Recombinant lentiviral vectors (pLENTI4-HAX; pLENTI4-GFP) and lentiviral vectors encoding for shRNA targeting the MAPK14 gene or non-targeting (control) shRNA (GE Dharmacon) were produced by co-transfection of HEK-293T cells with lentiviral expression plasmids and packaging plasmids (2nd generation packaging plasmids Gag/Pol/Rev/Tat and VSV-G) using PEI (Linear polyethylenimines; Polyscience) transfection reagent. Supernatants were collected after 48 hours and passed through a 0.22 μm filter. The viral supernatant was added to target cells with 8 μg/ml Polybrene. Following 24h incubation, media was removed and replaced with fresh media. Cells were collected after 72 hours for further analyses.
Crystal Violet Cytotoxicity Assay
Cells were plated in 24 well plates and were stained with 0.1% crystal violet solution. Cell viability was detected using the crystal violet staining protocol, solubilization of the dye adsorbed by cell nuclei during staining of drug- and DMSO-treated cells was performed after 48 h of treatment and quantified using spectrophotometry.
XTT Cell Viability Assay
After verifying cell viability using trypan blue dye exclusion test, cells were seeded at approximately 1x10^4 cell/well in a final volume of 200 μl in 96-well fat-bottom microtiter plates. After an overnight incubation, cells were treated with increasing concentrations of sorafenib and DMSO as a control. Plates were incubated at 37°C in a 5% CO2 incubator for the indicated time periods. At the end of incubation, 100 μl of XTT (2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide) (Biological Industries, Israel) was added to each well, after which plates were incubated at 37°C for additional 4 hours. Absorbance was measured at 450 nm against a reference wavelength at 650 nm using a microplate reader. The mean of triplicate experiments for each dose was used to calculate cytotoxicity values.
Western Blot
Cells were washed in PBS and then lysed in RIPA lysis buffer containing protease and phosphatase inhibitors. 40 μg of protein was separated by 12% SDS-polyacrylamide electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (GE Healthcare). Membranes were blocked with 5% non-fat milk in PBST (containing 0.1% Tween-20) for 1 h, and incubated with antibodies specific to Phospho-p38 MAPK (Thr180/Tyr182) (1:1000; Cell Signaling), pERK (1:200; Santa Cruz), ERK2 (1:200; Santa Cruz), Phospho-HSP27 (Ser82) (1:1000; Cell Signaling), β-actin (1:5000; Abcam) and GAPDH (1:2500; Abcam) for ON at 4°C. Membranes were washed with PBS-T for three times followed by 1h incubation with appropriate secondary antibodies, and then visualized using the odyssey imaging system (LI-COR).
Immunohistochemistry
Cells were extensively washed with PBS and fixed with 4% paraformaldehyde in Phosphate Buffer (PB) for 30 min in room temperature and washed 3 times with PBS. Fixed cells were permeabilized with 0.5% Triton X-100 in PBS for 25 min at room temp, washed with PBS containing 0.2% Tween 20 (PBS-T), and blocked with FCS containing 10% (v/v) skin milk and 0.2% (v/v) tween 20 for 45 min. Cells were then incubated with Phospho-p38 MAPK (Thr180/Tyr182) (1:500; Cell Signaling) in PBS-T containing 10% skim milk for ON at 4°C. The Next day, cells were washed with PBS-T, and detection was performed using the fluorescent secondary antibody Alexa Fluor® 568 goat anti-rabbit IgG (1:500; Molecular Probes) for 45 min at room temperature. Finally, cells were washed with PBS-T and the coverslips were mounted in DAPI Fluoromount G (SouthenBiotech). Microscopic images were obtained using Olympus BX52 Fluorescence Microscope.
Quantitative mRNA Analysis
mRNA was isolated from whole cells by EZ-10 Total RNA miniprep-kit (Bio Basic). DNase treatment was performed using TURBO DNA-free™ Kit (Ambion). cDNA synthesis was done with qScript cDNA Synthesis Kit (Quanta Biosciences). Quantitative qPCR was performed with PerfeCTa SYBR Green FastMix (Quanta Biosciences). Values were normalized to RPS11 house-keeping gene.
HBV Viral Load Titer Expression and Quantitative PCR Analysis
HBV DNA quantification was done as previously described [10]. Briefly, viral DNA was isolated from whole cells via QIAamp DNA Blood mini kit (Qiagene). Quantitative qPCR was performed with TaqMan® Fast Advanced Master Mix (Applied Biosystems). Values were normalized to a standard curve performed with known concentrations of HBV DNA.
Signal Quantification
Fluorescence intensity of pMAPK14 expression was quantified using ImageJ software. For all western blots, quantification of differences relative to total protein levels of samples was made using Image Studio software.
Statistics
All statistical analyses were performed using Graph Pad Prism 5 software, if not otherwise stated. Results were expressed as mean ± SD. Comparisons between groups were assessed by two-tailed Student’s t test. P < .05 was considered to indicate statistical significance.
Results
HBV is Associated with Reduced Susceptibility of Hepatoma Cells to Sorafenib
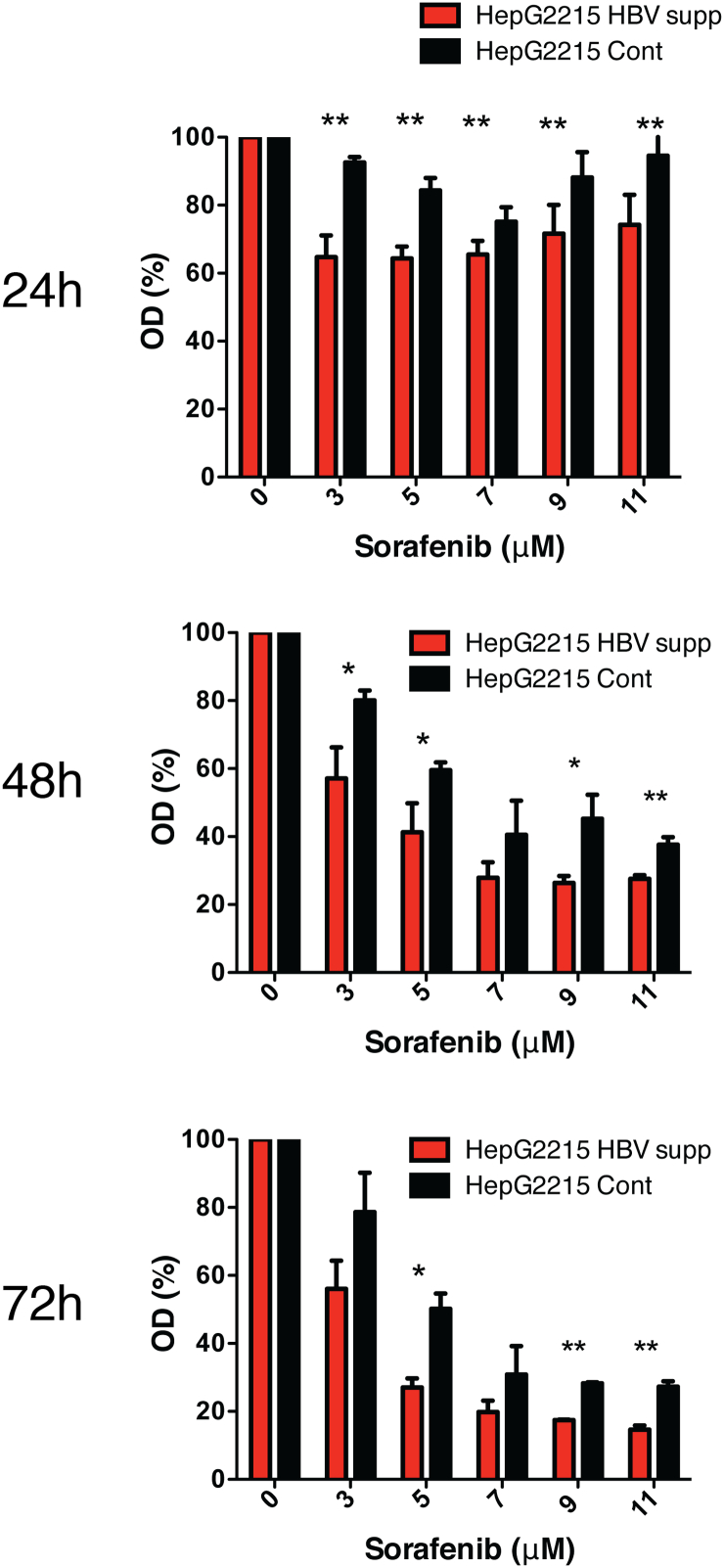
We hypothesized that hepatoma cells with replicating HBV are less susceptible to sorafenib treatment. To investigate this, we took advantage of the HBV-replicating HepG2.2.15 cell line [9] in which HBV expression was suppressed by the CRISPR-Cas9 technology [11] (HepG2.2.15 HBV supp.). The production of HBV particles, as well as the expression level of major viral transcripts in this cell line is suppressed as compared to the parental HepG2.2.15 cell line that was transduced with CRISPR and an inactive Cas-9 (HepG2.2.15 Cont.) (Supp. Figure 1). When both HepG2.2.15 Cont. cells and HepG2.2.15 HBV supp. cells were treated with sorafenib, a significantly higher cell viability was detected in HepG2.2.15 Cont. cells as compared to cells in which HBV replication was down regulated (Figure 1 and Supp. Figure 2). Notably, cell viability assays have shown a much higher difference in sorafenib effect between HepG2.2.15 cells and naive HepG2 cells that completely lack any HBV expression (Supp. Figure 2). Overall, these results suggest that in the presence of HBV, hepatoma cells are less susceptible to the inhibitory effect of sorafenib.
Figure 1.
HBV expression confers resistance to sorafenib treatment in vitro. HepG2.2.15 in which HBV expression was suppressed using the CRISPR/Cas9 system (HepG2.2.15 HBV supp.) and HepG2.2.15 cells transduced with CRISPR and an inactive form of Cas9 (HepG2.2.15 Cont.) were plated in 96-well plates and were incubated with various concentrations of sorafenib (0 to 11 μM). After 24, 48 and 72 hours, cell viability was assessed by XTT assay and quantified after additional 4 hours’ incubation at 37°C using spectrophotometry. Results are expressed as percentage of optical density (OD) reading (OD obtained with 0μM sorafenib considered as 100%) and represent average ± SDEV of biochemical triplicates. The asterisk denotes statistically significance (*P < .05, **P < .01).
HBV Induces Activation of the RAF-MEK-ERK Pathway
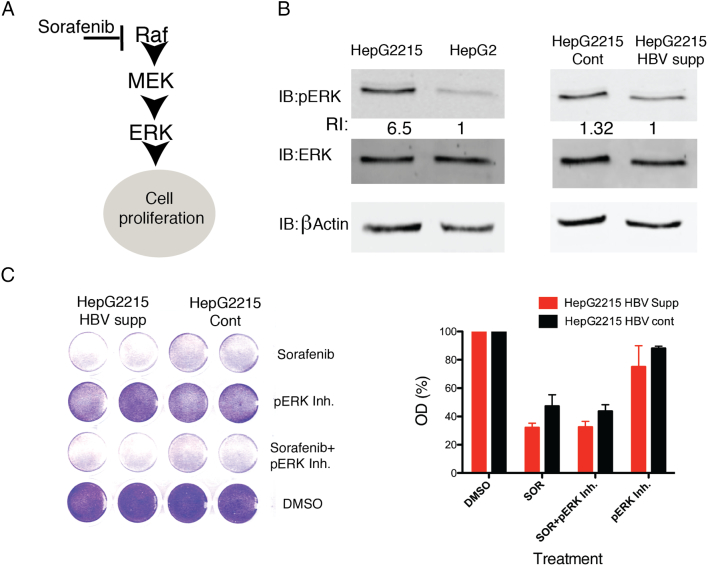
Sorafenib extracts much of its effect by blocking Raf, thereby largely neutralizing the RAF-MEK-ERK pro-oncogenic pathway (Figure 2A) [6]. We therefore hypothesized that HBV might directly or indirectly intervene with this pathway. To investigate this, we analyzed the level of pERK, a downstream kinase activated by Raf, in the presence or in the absence of HBV. As shown in Figure 2B, the level of pERK in HBV harboring hepatoma cells was much higher as compared to pERK level observed in HBV-null HepG2 cells. A higher expression of pERK was also observed in HBV positive (HepG2.2.15 Cont.) as compared to cells in which HBV expression was suppressed by the CRISPR-Cas9 system (HepG2.2.15 HBV supp.), although the difference was modest as compared to that observed in HepG2 versus HepG2.2.15 cells (Figure 2B). We concluded that baseline pERK level is higher in hepatoma cells with HBV. This finding may suggest that pERK induction is involved in HBV-associated resistance to sorafenib treatment.
Figure 2.
HBV is associated with enhanced pERK activation in hepatoma cells. (A) A scheme of the Raf-Mek-Erk signaling pathway and the role of the multi-kinase inhibitor sorafenib in blocking this oncogenic pathway. (B) HepG2, HepG2.2.15, HepG2.2.15 cont. and HepG2.2.15 HBV supp. cells were seeded and collected after 48 h, protein was extracted and analyzed by Western blot for the expression of ERK, pERK and β-actin proteins. The relative intensities (RI) of pERK normalized to β Actin are shown(C) HepG2.2.15 cont. and HepG2.2.15 HBV supp. cells were plated in 24-well plates, and were treated by sorafenib (9 μM) with or without the pERK inhibitor FR180204 (70 μM). Cell viability was assessed by crystal violet staining (left) after 48 h of treatment and quantified using spectrophotometry (right). Results of quantification are expressed as percentage of optical density (OD) reading and represent an average ± SDEV of biological duplicates.
Inhibition of pERK Does Not Alleviate HBV-Associated Resistance to Sorafenib
Since pERK level is induced in hepatoma cells with replicating HBV as compared to HBV-null cells, we next investigated whether pERK inhibition results in alleviation of HBV-associated resistance to sorafenib. For this, we pharmacologically inhibited pERK with a pERK inhibitor (FR180204) in sorafenib treated hepatoma cells (Supp. Figure 3) and tested cell viability under these conditions. As shown in Figure 2C, pERK inhibition by itself was not sufficient to induce cell death in either HBV replicating HepG2.2.15 cont. or HepG2.2.15 HBV supp. cells. Moreover, the combination of pERK inhibition with sorafenib treatment in HBV replicating HepG2.2.15 cont. cells had a very modest additive effect on cell death as compared to sorafenib treatment alone. A similar trend was observed when a combination of pERK inhibition with sorafenib treatment was employed in HepG2 versus HepG2.2.15 cells (Supp. Figure 4). Therefore, we concluded that although pERK is induced in HBV replicating hepatoma cells, its blockade was not sufficient to overcome the resistance to sorafenib treatment.
The Pro-Oncogenic Mitogen-Activated Protein Kinase 14 (MAPK14) is Induced in HBV Positive Hepatoma Cells
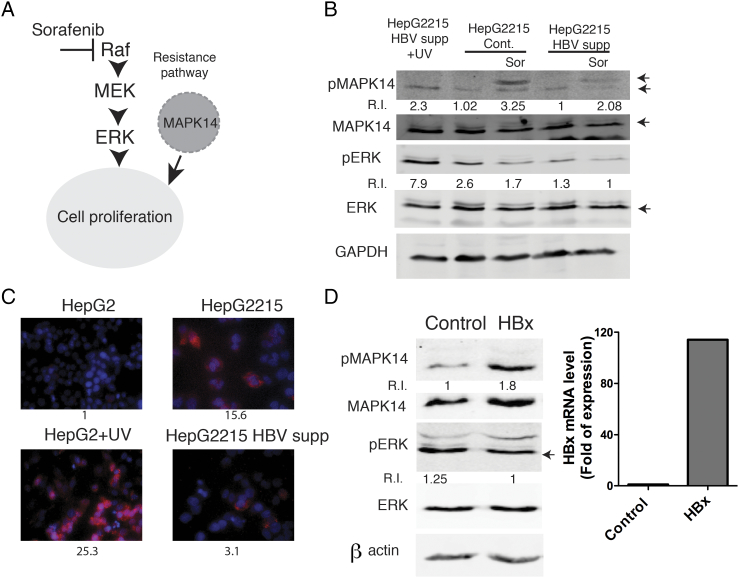
Recently, it was shown that the pMAPK14 (phosphorylated p38 alpha MAPK) protein is induced in a fraction of liver samples from patients with HCC, thereby conferring resistance to treatment with sorafenib by circumventing Raf to directly promote cell proliferation (Figure 3A) [12]. We therefore speculated that pMAPK14 might be involved in HBV associated resistance to sorafenib. To investigate this, we first analyzed the level of pMAPK14 in hepatoma cells in response to sorafenib. As shown in Figure 3B, pMAPK14 was robustly induced in sorafenib-treated HBV replicating hepatoma cells whereas this induction was attenuated in HepG2.2.15 cells in which HBV was largely suppressed.
Figure 3.
The oncogenic protein pMAPK14 is induced in the presence of HBV, possibly through HBx. (A) A scheme illustrating the possible role of Mapk14 in mediating sorafenib resistance in hepatoma cells. (B) HepG2215 Cont. and HepG2.2.15 HBV supp. cells were seeded and either treated with sorafenib (10 μM) or left untreated 24 h post seeding. Cells were collected at 12 h following treatment and protein was analyzed by WB, using the indicated antibodies. Cells treated with UV were used as a positive control for pERK and pMAPK14 activation. GAPDH protein expression is used for equal loading control. Relative intensities (R.I.) of the corresponding bands (marked by arrows) (pERK normalized to total ERK and pMAPK14 normalized to total MAPK14) are shown. (C) The indicated cells were analyzed by immunofluorescence for pMAPK14 expression (red). Cells nuclei were stained with DAPI (blue). The relative fluorescence intensities are indicated below for each panel (UV; ultraviolet) (D) HepG2 cells were transduced with pLENTI4-HA-X or pLENTI4-GFP plasmids. 72 h post transduction cells were lysed and analyzed by Western blot for the expression of the indicated proteins. Relative intensities of the corresponding bands (marked by arrows) (pERK normalized to total ERK and pMAPK14 normalized to total MAPK14) are shown. A q RT-PCR for HBx expression is shown in the right panel.
Consistent with these results, immunostaining for pMAPK14 showed a higher basal expression of pMAPK14 in HepG2.2.15 cells as compared to both, HBV-null HepG2 cells and HepG2.2.15 cells in which HBV was suppressed by the CRISPR-Cas9 system (Figure 3C).
HBV X protein (HBx) is a transcriptional trans-activator that has been implicated in liver carcinogenesis [13], [14]. We speculated that HBx might be involved in pMAPK14 induction in hepatoma cells and thereby contribute to their resistance to sorafenib treatment. To investigate this, hepatoma cells were transduced with either HBx expressing or non-coding control lentiviruses, and pMAPK14 expression was analyzed in cells following transduction. As shown in Figure 3D, the presence of HBx results in an increased expression of pMAPK14, but not in total MAPK14, pERK or total Erk, suggesting that HBx is involved in the induction of pMAPK14 in HBV harboring hepatoma cells.
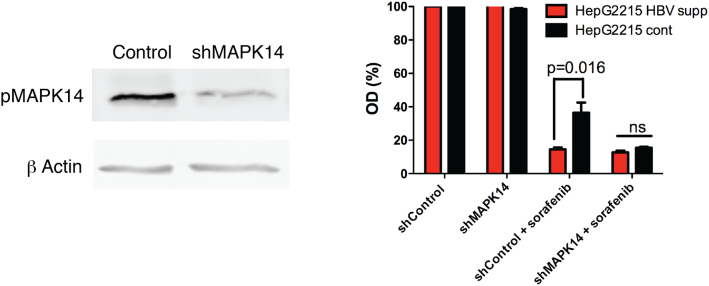
To better define the role of pMAPK14 in conferring resistance to sorafenib in the context of HBV infection, we knocked down pMAPK14 with specific shRNAs (Figure 4 left panel). In contrast to cells in which pMAPK14 expression was intact, the inhibitory effect of sorafenib in the context of pMAPK14 knock down was almost comparable between HBV expressing HepG2.2.15 cells (HepG2.2.15 Cont.) and cells in which HBV was suppressed (Figure 4 right panel). This strongly argues for a central role of HBV-associated pMAPK14 induction in resistance to sorafenib.
Figure 4.
Inhibition of MAPK14 largely reverses HBV-induced resistance to sorafenib. (Left) HepG2 cells were transduced with shMAPK14 or shControl-expressing lentiviruses. Seventy-two hours post transduction cells were treated with UV. After 24 h cells were collected and protein was extracted and analyzed by Western blot for the expression of pMAPK14 and β Actin. (Right) HepG2215 Cont. and HepG2.2.15 HBV supp. cells were transduced with shMAPK14 or shControl-expressing lentiviruses. 72 h post transduction, cells were treated with sorafenib (9 μM). After 48 h cell viability was analyzed using the XTT assay (see Methods section) and quantified using spectrophotometry. Results are expressed as percentage of optical density (OD) reading and represent average ± SDEV of biochemical triplicates (ns, non-significant).
Collectively, these results suggest that pMAPK14 is induced in an HBx-dependent manner and that blockage of the pMAPK14 pathway largely alleviates the resistance of HBV harboring hepatoma cells to sorafenib treatment.
Discussion
In this study, we show that hepatoma cells with replicating HBV are less susceptible to the inhibitory effect of the multi-kinase inhibitor sorafenib as compared to HBV-null cells. Our finding that HBV replication in hepatoma cells is associated with the induction of the Raf-MAPK pathway, a major target of sorafenib, is in line with other previous studies [3], [15]. However, in contrast to our prediction, inhibition of the downstream kinase pERK did not alleviate the resistance to sorafenib in HBV replicating cells. This was the rational for exploring alternative oncogenic pathway that might be induced in the presence of HBV.
Indeed, our results show that in the presence of HBV, treatment with sorafenib results in induction of pMAPK14. MAPK14 is the major isoform of the stress-activated kinases and its activity controls the expression of inflammatory mediators, cytokines as well as survival and antioxidant genes. Initially considered to have mainly a tumor suppressor role, recent evidence suggests that MAPK14 also shares a tumor promoter activity and might confer tumor cells with resistance to chemotherapy [16]. A recent in vivo RNAi screen has revealed that pMAPK14 is over-expressed in hepatoma samples from patients who are resistant to sorafenib treatment. Knocking down pMAPK14 expression, or inhibiting its kinase activity, markedly increased the therapeutic activity of sorafenib in this in vivo model of hepatocarcinogenesis [12]. Given the molecular heterogeneity of HCC, this important observation suggests that pMAPK14 could mediate sorafenib resistance across the wide spectrum of HCC, possibly in an etiology-dependent manner.
A key observation of our study concerns the induction of pMAPK14 in HBV positive hepatoma cells. This induction is associated with the expression of HBx, a viral protein that is heavily implicated in liver carcinogenesis [17], [18]. Importantly, our results suggest that pMAPK14 induction largely underlies HBV-associated resistance to sorafenib, since its inhibition by RNAi almost completely reverses the resistance phenotype. Therefore, our study suggests that in patients with HBV-associated HCC, the addition of pMAPK14 inhibitor to the conventional treatment with sorafenib may dramatically increase its inhibitory effect, as demonstrated in our in vitro experiments.
Hepatocellular carcinoma is a deadly cancer and to date, there are no good therapeutic options for patients who are diagnosed at an advanced stage of their disease. The high number of reported genes and pathways involved in this neoplasm [3] is one of the major obstacles in finding a “one size fits all” systemic therapy. This was the driving force for using the multi-kinase inhibitor sorafenib, which blocks several pathways possibly involved in hepatocarcinogenesis, for treating HCC [19]. However, clinical trials as well as real world data suggest only modest survival benefit for sorafenib treated patients [7], [8], [15], [20], [21]. Therefore, one can speculate that future anti-HCC therapies will not only combine several inhibitors of relevant oncogenic pathways but will also heavily rely on pre-specified etiology-related altered genes or pathways that can potentially be targeted [22], [23]. Indeed, our proof-of-principle work exemplifies the importance of defining etiology-specific molecular pathways in liver cancer that are also drug targetable, in order to achieve a much more efficient elimination of the cancerous cells.
Our study joins other previous publications, suggesting various molecular mechanisms for sorafenib resistance in the context of HBV-associated HCC; for example, the expression of myeloid cell leukemia-1 (Mcl-1) protein has been found to be elevated in HBV positive HCC cells and its down-regulation by over expressing miR193b restored the pro-apoptotic effect of sorafenib in these cells [24]. On the other hand, another study suggests that sorafenib destabilizes HBx and suppresses HBV replication, thereby maintaining the drug’s pro-apoptotic effect even in the presence of HBV [25]. These conflicting results might derive from inherent differences in experimental systems and conditions used in those studies.
Our in vitro experimental system is based on integrated HBV DNA that drives viral replication but do not recapitulate the full-cycle in the context of infection. However, we believe that the introduction of the CRISPR/Cas-9 system enabled us to achieve a more reliable comparison between HBV replicating cells and the very same cell line in which HBV replication is largely suppressed. Nevertheless, we are well aware that this system is not entirely clean, since a low level of HBV replication is still maintained even in cells transduced with HBV-targeting CRISPR/Cas9. This may explain the relatively small differences in pERK and pMAPK14 expression between HBV positive and HBV suppressed cells as compared to the differences observed between HBV-null HepG2 cells and HBV replicating HepG2.2.15 cell line.
Based on the results presented in this work, further studies should define the mechanism by which HBx induces pMAPK14 and whether this induction also underlies de novo HBV associated carcinogenesis. Indeed, a large variety of stimuli, such as oxidative stress, DNA damage, pro-inflammatory cytokines and growth factors can induce pMAPK14 [26]. Therefore, it is still possible that pX, and possibly other factors implicated in HBV gene expression and replication, promote changes in the cellular milieu that are ultimately responsible for MAPK14 induction through its phosphorylation. Further studies should better define these putative mechanisms.
Obviously, our results obtained in in vitro systems should be further validated in in vivo models to prove the utility of combined sorafenib and pMAPK14 inhibition in treating HBV-associated HCC. Those studies will also test possible non-specific toxicity effect, a mandatory requirement prior to application of combination therapies in humans.
In conclusion, we believe that our study has a high translational potential, suggesting that the addition of pMAPK14 inhibitor to sorafenib may have an additive effect over sorafenib mono therapy among HBV-infected individuals. This study represents the evolving concept of etiology-tailored therapy for HCC, a deadly cancer for which highly effective therapy is still unavailable.
Footnotes
Acknowledgements: This work was supported by the Israel Science Foundation (ISF) physician-scientist grant (A.S), the Binational Science Foundation (BSF) grant (A.S and R.Z) and by young physician grant from Beilinson hospital (M. B A).
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2018.02.015.
Appendix A. Supplementary Data
Supplementary figures
References
- 1.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. doi: 10.1056/NEJMra1001683. [Epub 2011/10/14. PubMed PMID: 21992124] [DOI] [PubMed] [Google Scholar]
- 2.Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350(11):1118–1129. doi: 10.1056/NEJMra031087. [Epub 2004/03/12. 350/11/1118 [pii]. PubMed PMID: 15014185] [DOI] [PubMed] [Google Scholar]
- 3.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6(9):674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 4.Shlomai A, de Jong YP, Rice CM. Virus associated malignancies: The role of viral hepatitis in hepatocellular carcinoma. Semin Cancer Biol. 2014;26:78–88. doi: 10.1016/j.semcancer.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruix J, Sherman M. Management of hepatocellular carcinoma: An update. Hepatology. 2011;53(3):1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, Schwartz B, Simantov R, Kelley S. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5(10):835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 7.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc J-F, de Oliveira AC, Santoro A, Raoul J-L, Forner A. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 8.Cheng A-L, Guan Z, Chen Z, Tsao C-J, Qin S, Kim JS, Yang T-S, Tak WY, Pan H, Yu S. Efficacy and safety of sorafenib in patients with advanced hepatocellular carcinoma according to baseline status: Subset analyses of the phase III Sorafenib Asia–Pacific trial. Eur J Cancer. 2012;48(10):1452–1465. doi: 10.1016/j.ejca.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Sells MA, Chen M-L, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. PNAS. 1987;84(4):1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shlomai A, Schwartz RE, Ramanan V, Bhatta A, de Jong YP, Bhatia SN, Rice CM. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci U S A. 2014;111(33):12193–12198. doi: 10.1073/pnas.1412631111. [doi: papers3://publication/doi/10.1073/pnas.1412631111] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramanan V, Shlomai A, Cox DBT, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833. doi: 10.1038/srep10833. [PubMed PMID: PMC4649911] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudalska R, Dauch D, Longerich T, McJunkin K, Wuestefeld T, Kang T-W, Hohmeyer A, Pesic M, Leibold J, von Thun A. In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat Med. 2014;20(10):1138–1146. doi: 10.1038/nm.3679. [ http://www.nature.com/nm/journal/v20/n10/abs/nm.3679.html - supplementary-information] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koike K. Hepatitis B virus X gene is implicated in liver carcinogenesis. Cancer Lett. 2009;286(1):60–68. doi: 10.1016/j.canlet.2009.04.010. [Epub 2009/05/26. PubMed PMID: 19464104] [DOI] [PubMed] [Google Scholar]
- 14.Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol. 2001;36(10):651–660. doi: 10.1007/s005350170027. [Epub 2001/11/01. PubMed PMID: 11686474] [DOI] [PubMed] [Google Scholar]
- 15.Di Marco V, De Vita F, Koskinas J, Semela D, Toniutto P, Verslype C. Sorafenib: from literature to clinical practice. Ann Oncol. 2013;24(Suppl. 2):ii30–ii37. doi: 10.1093/annonc/mdt055. [DOI] [PubMed] [Google Scholar]
- 16.Igea A, Nebreda AR. The stress kinase p38α as a target for cancer therapy. Cancer Res. 2015;75(19):3997–4002. doi: 10.1158/0008-5472.CAN-15-0173. [DOI] [PubMed] [Google Scholar]
- 17.Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26:144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- 18.Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene. 2006;25(27):3823–3833. doi: 10.1038/sj.onc.1209559. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D, Wilhelm S, Lynch M, Carter C. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66(24):11851–11858. doi: 10.1158/0008-5472.CAN-06-1377. [DOI] [PubMed] [Google Scholar]
- 20.Lencioni R, Marrero J, Venook A, Ye SL, Kudo M. Design and rationale for the non-interventional Global Investigation of therapeutic DEcisions in hepatocellular carcinoma and Of its treatment with sorafeNib (GIDEON) study. Int J Clin Pract. 2010;64(8):1034–1041. doi: 10.1111/j.1742-1241.2010.02414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iavarone M, Cabibbo G, Piscaglia F, Zavaglia C, Grieco A, Villa E, Cammà C, Colombo M, on behalf of the Ssg Field-practice study of sorafenib therapy for hepatocellular carcinoma: A prospective multicenter study in Italy. Hepatology. 2011;54(6):2055–2063. doi: 10.1002/hep.24644. [DOI] [PubMed] [Google Scholar]
- 22.Goossens N, Hoshida Y. Personalized management of hepatocellular carcinoma based on molecular information: future prospects. Clin Liver Dis. 2015;5(6):132–135. doi: 10.1002/cld.483. [PubMed PMID: PMC4512174] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miki D, Ochi H, Hayes CN, Aikata H, Chayama K. Hepatocellular carcinoma: Towards personalized medicine. Cancer Sci. 2012;103(5):846–850. doi: 10.1111/j.1349-7006.2012.02242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao K, Zhang J, He C, Xu K, Liu J, Sun J, Wu G, Tan C, Zeng Y, Wang J. Restoration of miR-193b sensitizes Hepatitis B virus-associated hepatocellular carcinoma to sorafenib. Cancer Lett. 2014;352(2):245–252. doi: 10.1016/j.canlet.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Kim HY, Jung HU, Yoo SH, Yoo KS, Cheong J, Park BS, Yun I, Yoo YH. Sorafenib overcomes the chemoresistance in HBx-expressing hepatocellular carcinoma cells through down-regulation of HBx protein stability and suppresses HBV gene expression. Cancer Lett. 2014;355(1):61–69. doi: 10.1016/j.canlet.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Cuadrado A, Nebreda Angel R. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429(3):403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures