Abstract
Metabolic engineering offers an exquisite capacity to produce new molecules in a renewable manner. However, most industrial applications have focused on only a small subset of elements from the periodic table, centered around carbon biochemistry. This review aims to illustrate the expanse of chemical elements that can currently (and potentially) be integrated into useful products using cellular systems. Specifically, we describe recent advances in expanding the cellular scope to include the halogens, selenium and the metalloids, and a variety of metal incorporations. These examples range from small molecules, heteroatom-linked uncommon elements, and natural products to biomining and nanotechnology applications. Collectively, this review covers the promise of an expanded range of elemental incorporations and the future impacts it may have on biotechnology.
Keywords: Metabolic engineering, Synthetic biology, Unnatural metabolism, Halogenation, Cellular factories, Non-canonical elements
1. Introduction
Metabolic engineering expands the chemical palate of cells to yield industrially-relevant products that compete with traditional chemical methods, especially with respect to product specificity and mild process conditions [1], [2], [3]. These advances range from traditional examples of products such as alcohols, citric acid, and penicillin to more recent examples including artemisinin [4], lycopene [5], plant alkaloids [6], energy dense fatty acids [7], among others. However, the chemical space of cellular production has not yet been fully harnessed in most applications. In this regard, the field of metabolic engineering is currently accessing and competing with a subset of molecules accessed through traditional synthetic organic chemistry and primarily limited to combinations of only six common elements (carbon, hydrogen, oxygen, nitrogen, phosphorus and sulfur) comprising most compounds [8]. Synthetic biology has begun to enable access to “unnatural” combinations of these common elements through composite pathways leading to molecules such as muconic acid, 1,4 butanediol, unnatural alcohols and amino acids, and rare heteroatom-heteroatom bonded compounds [9], [10], [11], [12]. However, new synthetic tools coupled with examination of native genetic diversity have uncovered unique chemistries that enable assimilation of uncommon elements. Such discoveries provide an exciting glimpse into how classic cellular biochemistry can transcend into cellular bioorganic and bioinorganic chemistry.
Indeed, there is a cellular advantage for incorporating uncommon elements. Inherently, each element has its own unique characteristics that have both biological and biotechnological advantages. Prevailing examples include halogenated molecules with enhanced biological activities used in pharmaceutics and agriculture [13], boronated antibiotics and signaling molecules [14], and transition metal nanoparticles used for optics and medical applications [15]. Likewise, uncommon metals themselves may be inherent products in biomining applications where organisms can concentrate valuable metals, such as lithium and platinum. These applications begin to expand the subset of “bioreachable” products beyond the traditional six elements of canonical biochemistry.
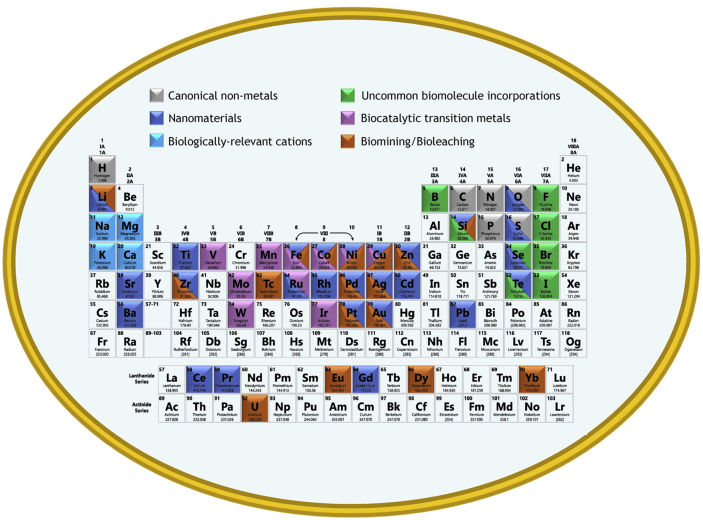
In this review, we provide an up-to-date synopsis on the expanding “cellular periodic table” (Fig. 1). Specifically, we focus on compounds and structures containing halogens, other non-metals/metalloids (such as selenium and boron) and a collection of transition and rare earth metals. Through this overview, we aim to highlight previous and nascent engineering efforts aimed at incorporating uncommon elements into cellular metabolism to produce interesting products.
Fig. 1.
Cellular Periodic Table - Visual summary of the elemental interactions of microorganisms addressed in this review. As a note, this figure does not consider every possible interaction with the elements, especially rarer metals, but instead focuses on some of the most promising current developments.
2. Cellular destinations for uncommon elements
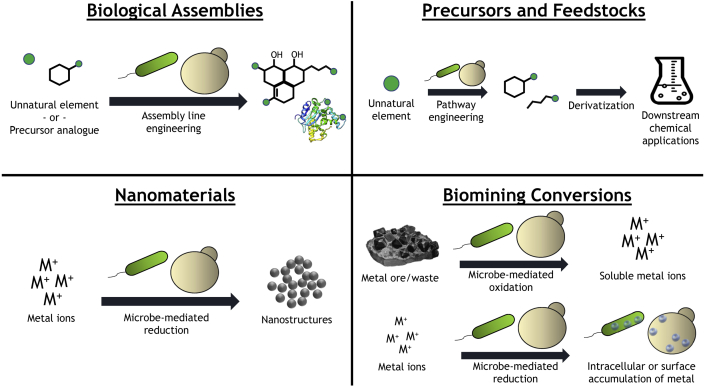
For the sake of this review, we examine four major destinations for uncommon elements. These comprise: biological assemblies, precursors/feedstocks, biomining, and nanomaterials (Fig. 2). We briefly describe each of these four here with respect to product examples and industrial applications before discussing the types of uncommon elements in more depth.
Fig. 2.
Cellular Destinations for Uncommon Elements - An overview of the four major destinations for biologically-uncommon elements discussed in this review.
2.1. Biological assemblies
A great deal of structural and biochemical diversity can be found in products stemming from biological assembly lines such as polyketides, non-ribosomal peptides, and proteins. Not surprisingly, engineering these components has led to therapeutically relevant natural products [16]. Likewise, these compounds and proteins are targets for uncommon elements with several case studies leading to generation and incorporation of non-canonical amino acids [17], [18], [19] and molecules with improved biological function [20]. Molecules like polyketides are both natural and synthetic substrates for halogenation [21], [22]. Precise incorporation of uncommon elements into these end-products is still an ongoing challenge assisted by advances in synthetic biology.
2.2. Precursors and feedstocks
As described above, metabolic engineering provides a unique opportunity for replacing traditional chemical synthesis of small molecules. However, many industrial compounds produced with toxic chemical syntheses also have biochemically uncommon elements. To this end, microorganisms capable of producing a variety of precursor molecules have the potential to be expanded in their use to include rare-element analogues which can be further utilized in downstream chemical applications [3]. Such transformations include small molecules containing silicon and halogens, for example, that are suitable for reactions like cross-couplings [23], electrophilic cleavage [24], and polymerizations [25]. These efforts involve both protein engineering and synthetic biology along with metabolic engineering to expand the chemical reach of cellular metabolism.
2.3. Biomining
Microorganisms adapt to challenging environments, especially through mechanisms of detoxification. In the case of biomining, organisms use both reductive and oxidative mechanisms to convert and sequester metals of potential interest. As an example, industrial biotechnology has utilized microorganisms for biomining of copper and other more common elements for decades and has begun to expand its elemental ambit to include transformations of rarer and more valuable metals including lithium, palladium, and platinum [26]. This field shows great promise especially as we deplete rare-earth metals and generate substantial electronic waste.
2.4. Nanomaterials
As a final destination for uncommon elements, cellular systems are capable of serving biocatalytic roles in forming nanomaterials. The recent explosion of nanotechnology has yielded industrially-relevant products for a wide range of applications such as biosensing, drug delivery, catalysis, optics, and agriculture [27]. As with chemical production, microorganisms can provide precise nanoparticle functionalization and low-toxicity. Although not mutually exclusive from biomining, nanoparticle synthesis optimization requires distinct modifications of strain characteristics. Thus, the rewiring of biology's native capacity to reduce metallic salts into nanostructures has resulted in many breakthroughs in this emerging field.
3. Halogenation
Among the many uncommon elements that can find their way into biological molecules, halogens are particularly interesting as halogenated organic compounds play a substantial role in the pharmaceutical, diagnostic, agricultural, and materials industries. While halogenation of organic compounds using synthetic chemistry is well-established, these procedures are often characterized by highly toxic chemicals and poor atom economy [28]. Additionally, these processes typically suffer from low yields due to difficulty of separation and purification of enantiomeric substances [29]. Thus, metabolic engineering can offer a more efficient and environmentally friendly method for producing halogenated molecules.
The field of halogenation biochemistry has grown immensely over the past few decades, evidenced by the discovery of over 5000 halogenated natural products to date [30]. Halogenated secondary metabolites have been found in nearly all kingdoms of microbial life, with the majority concentrated around marine life and particularly in algae and cyanobacteria [31]. Significant biochemical analysis has been done to discover the underlying pathways for halogen incorporation. The first discovered halogenases were the electrophile-directing haloperoxidases [32], a class of enzymes with high turnover rates but lacking any specificity due to a release of free reactive halogen species into solution—a non-ideal trait for most immediate metabolic engineering approaches. Since these initial findings, an array of other halogenases with divergent mechanisms and substrates have been discovered [29]. These new enzymes span broad substrate ranges including small-molecule aromatics, olefins, aliphatics, and polyketides. The resulting products are attractive chemically and have been used to improve protein stabilization and functionality, modulate bioactivity, and enable functionalizable handles for further chemical modifications via cross-coupling reactions [33], [34], [35], [36]. In the subsections below, we describe prominent examples of halogenation incorporation organized by halogen type (Table 1).
Table 1.
Non-metal and metalloid metabolic products - Select organic compounds with rare element incorporations (including halogens, non-metals, and metalloids) generated in vivo, both natural and engineered, are provided in order of their appearance. (*) Denotes completely unnatural products.
| Element | Highlighted Compounds | Organism(s) | Engineered vs. Natural | Titer | References |
|---|---|---|---|---|---|
| Fluorine | Fluorosalinosporamide* | Salinispora tropica | Engineered | 1.5 mg/L | Eustaquio and Moore [167] |
| Nucleocidin | Streptomyces calvus | Natural | N/A | Zhu et al. [42] | |
| Fluorinated rapamycin analogues* | Streptomyces hygroscopicus | Engineered, precursor directed | 5-28 mg/L | Goss et al. [44] | |
| Fluorinated tetraketide lactones* | E. coli | Engineered, precursor directed | N/A | Walker et al. [45] | |
| Chlorine | 2-chloro-resveratrol* | E. coli | Engineered | 7.0 mg/L | Wang et al. [61] |
| 5-chloro daurichromenicacid* | Aspergillus oryzae | Engineered | 2.06 mg/L | Okada et al. [55] | |
| 8-chloro-7-hydroxycoumarin* | E. coli | Engineered | 1.1 mg/L | Menon et al. [64] | |
| Chloramphenicol | Streptomyces avermitilis | Engineered | 250 mg/L | Komatsu et al. [70] | |
| Bromine | Bromoalterochromide A | E. coli | Engineered | N/A | Ross et al. [72] |
| Bromophenols, bromocatechols, and related molecules | E. coli | Engineered | N/A | Agarwal et al. [73] | |
| 7-bromo-tryptophan and bromo-pacidamycin D* | E. coli | Engineered | N/A | Sharma et al. [74] | |
| Iodine | Methyl iodide | E. coli, S. cerevisiae | Engineered | Bayer et al. [80] | |
| Iodocionin | Ciona edwardsii | Natural | N/A | Aiello et al. [78] | |
| Selenium | Dimethyldiselenide | E. coli | Engineered | N/A | Swearingen et al. [168] |
| 2-selenouridine | E. coli, Methanococcus vannielii, Clostridium sticklandii, etc. | Natural | N/A | Sun et al. [101] | |
| Selenobiotin | Phycomyces blakesleeanus | Natural | N/A | Sum bui et al. [169] | |
| Boron | Borophycin and structurally related compounds | Nostoc spongiaeforme, Teredinibacter turnerae, etc. | Natural | N/A | Dembitsky et al. [170] Elshahawi et al. [171] |
| Borolithochromes | Solenopora jurassica | Natural | N/A | Wolkenstein et al. [109] | |
| Silicon | Acetyldimethylphenylsilane* | Kloeckera cortices | Natural, precursor directed | N/A | Frampton and Zelisko [113] |
| Dimethylphenylsilane diol products* | E. coli | Engineered, precursor directed | >10g scale | Smith et al. [118] | |
| α-hydroxy silanes* | S. cerevisiae | Natural, precursor directed | N/A | Zani [117] | |
| Ethyl 2-((4-aminophenyl)dimethylsilyl)propanoate* | E. coli | Engineered, precursor directed | N/A | Kan et al. [119] | |
| Tellurium | Methanetellurol | E. coli | Engineered | N/A | Swearingen et al. [121] |
| Dimethyl tellurenyl sulfide | E. coli | Engineered | N/A | Swearingen et al. [121] |
3.1. Fluorine
While fluorine is the most abundant halogen in the Earth's crust, it is the rarest found in natural products and is only known to be incorporated into a handful of molecules. This reality can be attributed to a multitude of factors including fluorine's high electronegativity, low bioavailability, and high heat of solvation [37]. Moreover, a major drawback of fluorinated natural products is the toxicity associated with certain metabolites, notably fluoroacetate and its downstream metabolite fluorocitrate, which inhibits aconitase in the citrate cycle. This toxicity is an issue since most known fluorinated metabolites are closely related to these compounds and originate from a single pathway, the well-studied fluorinase pathway, first discovered in the bacterium Streptomyces cattleya [38]. Recent studies by Ma et al. [39] found that pathway intermediate 5-fluoro-5-deoxy-d-ribose was converted into 5-fluoro-2,3,4-trihydroxypentanoic acid, an undiscovered fluorinated metabolite. The reaction is catalyzed via the novel enzyme FdrC in the organism Streptomyces sp. MA37 and thus reveals new possibilities for the fluorinase pathway. Likewise, higher efficiency fluorinases (including one isolated from the marine bacterium Streptomyces xinghaiensis) can show specificity increases by over an order of a magnitude, thus opening the door to more efficient fluorination [40]. Beyond these initial pathways, a few slightly more complex fluorometabolites have been discovered, including ω-fluorooleic acids from the plant Dichapetalum toxicarium and the antibiotic nucleocidin from Streptomyces calvus [41]. Nucleocidin is unique in that the fluorine is bonded at the C-4 position of a ribose ring, while every other recognized metabolite has originated from a fluoroacyl molecule [42]. Further details of fluorinated natural products and the fluorinase enzyme have been published in other reviews [41].
Synthetic incorporation of fluorine has been achieved through precursor-directed incorporation aimed to mitigate the inherent toxicity of fluorinated anti-metabolites. This approach consists of determining desirable locations for incorporation and engineering promiscuity of existing enzymes and pathways to preferentially react with halogenated substrates. Several successful examples of incorporating pre-fluorinated precursors into metabolites can be found in the polyketide pathway. As examples, fluorine, along with the other halogens, have been incorporated into pacidamycin analogues via precursor directed biosynthesis [43]. Another group produced six fluorinated rapamycin analogous via precursor feeding in the bacterium S. hygroscopicus [44].
The Chang group has worked extensively to create fluorinated polyketides and has identified locations in the polyketide synthesis machinery that are amenable to fluorine incorporation through the addition of fluorinated precursors [45]. Through this work in E. coli, it was possible to convert fluoromalonate to fluoromalonyl-CoA and subsequently incorporate this molecule into novel fluorinated derivatives of tetraketide lactones. Further efforts have been made to enable loading of fluorinated extender units into this synthetic machinery, albeit at lower rates than native substrates [46].
Beyond small molecules, fluorinated precursors have been used to insert fluorine into amino acids and polypeptide chains, an approach that shows great promise for protein engineering and the production of bioactive peptide compounds [47], [48]. Aromatic fluorinated amino acids are most commonly used for protein incorporation and biorthogonal fluorinated amino acids have utility as probes for molecular interactions [49]. More specifically, incorporation of fluorinated amino acids has shown great capacity for improving specific properties of proteins, such as refoldability, thermoactivity, and overall protein stability [50], [51], [52]. An excellent review on fluorinated amino acids and protein engineering was recently published for more details (Odar et al. [47]). More recently, the biosynthesis of fluorinated peptaibols was enabled by using a site-direction building block incorporation approach in the fungus T. arundinaceum [53]. Overall, the incorporation of fluorine remains a challenge for synthetic biology primarily due to toxicity and low enzyme function and specificity.
3.2. Chlorine
Chlorine is the most prevalent halogen with respect to bioavailability, especially given the abundance of halogenating organisms in marine environments. As a result, natural chlorinated molecules are highly diverse particularly in bioactive natural products such as rebeccamycin, chloramphenicol, radicicol, and vancomycin [21]. Here, we highlight several notable examples of chlorine incorporation with a primary focus on synthetic approaches and promising routes for future metabolic engineering.
Based on their bioactivity, the first major contributions in the field were natural products such as the in vivo combinatorial synthesis efforts by Sanchez et al. [54]. In this work, the authors reconstituted the pathway for the chlorinated bioactive molecule rebeccamycin and created novel pathway derivatives by co-expressing the tryptophan halogenases PyrH and ThaI in Streptomyces albus. These two enzymes uniquely bind the chlorine atom to tryptophan's aromatic ring with different regioselectivities. The resulting chlorinated derivatives have potential applications as antibiotics and antitumor agents. A similar approach was taken by Okada et al. to produce chlorinated derivatives of daurichromenic acid with higher antibacterial activity via the halogenase AscD [55]. Introducing a similar tryptophan halogenase into the plant C. roseus yielded novel chlorinated alkaloids, which have potential as powerful bioactive pharmaceuticals [56]. Likewise, PrnA was introduced into Streptomyces coeruleorubidus to yield chlorinated pacidamycin [35], a molecule that can be modified via cross-coupling reactions to produce further unnatural pacidamycin derivatives. As can be assumed from these efforts, halogenated tryptophan plays a central role for halogenation pathways. Likewise, most engineering efforts on halogenases have been performed on tryptophan halogenases, notably RebH [57], [58], and demonstrate the potential to switch regioselectivity [59].
Chlorine incorporation has also been seen in pathways related to phenolics and flavonoids. For example, introduction of fungal flavin-dependent halogenase Rdc2 can be used to drive radicicol biosynthesis in S. cerevisiae leading to a new halogenated analogue, 6-chloro, 7,8-dehydrozearalenol [60]. Production of 2-chloro-resveratrol in E. coli was also accomplished by incorporating Rdc2, which is known to have broad substrate specificity, into a heterologously expressed resveratrol pathway at an initial titer of 7.0 mg/L [61]. The same Rdc2 gene can also be used to convert fed hydroxyquinolines into a chlorinated derivative in vivo [62], [63]. The RadH enzyme, an ortholog of Rdc2, has been investigated in E. coli for the production of a novel chlorometabolite, 8-chloro-7-hydroxycoumarin [64]. To accomplish this feat, site-saturated mutagenesis coupled with a fluorescence screening assay was employed to isolate a RadH variant with an improved affinity for 7-hydroxycoumarin.
Beyond aromatics, chlorine can be incorporated into different chemical scaffolds. As an example, enzymes such as WelO5 [65], AmbO5 [66], and AioQ [67] have been well characterized for their ability to charge a chlorine atom to an aliphatic, non-activated carbon atom – a conversion that cannot currently be performed through chemical means. Among these enzymes, AmbO5 is an especially promising candidate for enzyme engineering approaches due to its promiscuous nature. Regarding substrate diversity, the recent discovery of the enzyme CylC displays the potential for chlorination of fatty acid residues [68].
Metabolic engineering strategies have been coupled with some of these incorporation approaches to increase titers of chlorinated biomolecules. For example, chlortetracycline biosynthesis in Streptomyces aureofaciens was increased nearly 2-fold in titer to 25.9 g/L [69]. In another example, the entire gene cluster for chloramphenicol was imported into the industrial microorganism Streptomyces avermitilis [70]. Resulting chloramphenicol titers were initially 6-fold higher than the original host (S. venezuelae) and further shikimate pathway modifications increased titers over 12-fold, reaching a titer of 250 mg/L. These initial metabolic efforts coupled with chlorine's moderate properties and biological accessibility are quite impactful for further expanding bioprocesses for chlorinated molecule generation.
3.3. Bromine
Bromine and chlorine are nearly identical in both mechanism of incorporation and possible bonding locations. More often than not, chlorinase enzymes also incorporate bromine. However, brominases have a more difficult time accepting chlorine due to differences in the oxidation potentials of intermediates and dehydration penalties, i.e. addition of chlorine requires more energy and enzymatic effectiveness [71]. As a result, brominated natural metabolites are more common and are found in a greater variety of molecular positions. Despite its prominence, the biological activity modulation imparted by bromine is not easily predictable and varies by the molecule [30].
In contrast to chlorinated molecules, brominated molecules and pathways have not been as extensively engineered. Initial efforts involved simply replacing chloride with bromide in solution. In doing so, Sanchez et al. were also able to produce brominated indolocarbazole derivatives through combinatorial biosynthesis in a similar fashion to how they produced chlorinated derivatives [54]. Using a transformation-associated recombination strategy, Ross et al. heterologously expressed an entire alterochromide lipopeptide pathway from Pseudoalteromonas piscicida in E. coli and obtained unusual brominated lipopeptides, originating from tyrosine, where bromine is added early during the NRPS assembly via the enzyme AltN [72].
Additional brominated small molecules have been explored in heterologous hosts as well. As examples, Agarwal and Moore engineered genes from the bmp gene locus of a marine γ-proteobacteria to produce a variety of brominated phenolic molecules in E. coli [73]. A very recent effort produced 7-bromo-tryptophan in E. coli and subsequently modified downstream peptide molecules in vivo using Suzuki-Miyaura cross-coupling reactions [74]. Beyond these initial efforts, there is great potential for expanding cellular bromination pathways into structures such as terpenoids, nonterpenoid C15-acetogenins (ACGs), indoles, and phenols/aromatics [75]. Thus, future bioprospecting and engineering efforts will continue to expand this unnatural element incorporation.
3.4. Iodine
Iodine plays an important role in thyroid health and hormone production, but is not ubiquitously found in biology mainly due to its very large atomic radius and electron cloud. Additionally, reports have found that the ability to produce iodinated compounds is more dependent on an organism's ability to concentrate the element, rather than on the activity of its halogenases [76]. Despite this, several biological routes in natural products suggest incorporation via haloperoxidase action. This mechanism is consistent with the human thyroid hormone triiodothyronine and prohormone thyroxine produced using peroxidase enzymes. Beyond human hormones, other medical roles for iodinated molecules include contrast agents (with multiple iodine atoms attached to an aromatic ring [77]) and natural products, including iodocionin [78], that show efficacy in cancer treatments.
Most iodine-based metabolic engineering efforts have been pursued in plants as a means of crop fortification for iodine supplementation in humans [79]. In microbes, Bayer, et al. achieved the production of methyl halides using a methyl halide transferase, an S-adenosyl-l-methionine-dependent halogenase [80] and was able to produce methyl iodide. The other recognized enzymes, such as iodoperoxidases, are particularly nonspecific and thus impractical for the production of precise chemical structures [29]. Nevertheless, there remains the possibility of identifying novel pathway enzymes for expanding iodine incorporation in microorganisms [81].
3.5. Halogenation outlook
Collectively, these examples demonstrate the capacity for halogen atoms to be integrated into a wide variety of metabolic destinations in vivo in both native and nonnative hosts. Through rational protein design, directed evolution, and metabolic engineering, the underlying reaction mechanisms and associated pathways can be altered and improved, thereby enabling further advancements. Although promising, notable challenges exist for halogen biochemistry including off-target effects of toxic products, poor enzymatic turnover rates, low-throughput analytical methods, and a lack of information on related pathways and enzymes compared to halogenated natural product discoveries. Continued efforts have been made to improve substrate recognition [82], turnover rates [57], and often necessary carrier proteins [83] to expand the scope and yield of halogenated metabolites. To address the low-throughput nature of analytical analyses for these metabolites (NMR, HPLC, or LC-MS), high-throughput screening techniques for halogenated molecules are continually being developed that accelerate the process of protein and metabolic engineering [64], [84], [85], [86], [87]. Nevertheless, halogenated metabolites remain a fertile and promising area of expansion for metabolic engineering, especially for the case of natural products and secondary metabolites. Advances in large-scale genome mining and further discovery and engineering of halogenases will continue to unlock new reaction possibilities, moving us one step closer to highly robust cellular factories.
4. Selenium and the metalloids
Various non-metals and metalloids can be incorporated into cells both through covalent bonds to metabolites (as in the case with halogens described above) and as nanoparticle assemblies. We highlight here advances in the incorporation of selenium, boron, silicon, and tellurium as examples of the expanding periodic table for cells (Fig. 1, Table 1).
4.1. Selenium
Perhaps the most ubiquitous and well-characterized unnatural incorporation in biology is selenium in the form of the 21st proteinogenic amino acid – selenocysteine [88]. Beyond selenocysteine, selenium metabolism [89] can also lead to bioactive natural products [90] and potentially complex pharmaceuticals including the synthetic drug Ebselen that is being investigated as a treatment for hearing loss and many mental disorders [91]. Additionally, simple metabolites such as Se-methyl-selenocysteine and methylseleninic acid have been shown to be potent chemopreventative compounds [92]. Finally, given the trace element dietary requirement of selenium, enriched yeast provide an economical means to supplement selenium for human nutrition and cancer prevention [93]. The selenium metabolic pathway in yeast and bacteria has been mapped to identify key selenium metabolites with selenomethionine being the most dominant and accounting for 60–85% of selenium in yeast [94]. Further selenium incorporation beyond this local pathway has not yet been fully explored, but various microorganisms and plants contain compounds like 3-Butenyl isoselenocyanate, selenobiotin, and selenosinigrins [95]. Additionally, the volatile selenium compounds dimethylselenide and dimethyldiselenide were produced in both bacteria and yeast as part of selenium metabolism, however these compounds have limited known applications [96], [97]. Overall incorporation of selenium from selenite in strains was increased by Pusztahelyi et al. through increased in glutathione concentration and glutathione reductase activity [98].
Current research in the field focuses on examining the interplay between sulfur and selenium metabolism [99]. For example, replacing the sulfur with a selenium atom in biologically active molecules like the plant alkaloid camalexin, could potentially improve functionality [100]. Enzymes also exist that can selectively exchange sulfur for selenium, including a selenouridine synthase that coverts 2-thiouridine to 2-selenouridine, found in the wobble position of certain bacterial tRNAs [101]. Further investigation into similar enzymes could potentially open new doors for selenium incorporation.
Beyond metabolite compounds, a common destination for selenium in a variety of microorganisms are selenium nanoparticles [102]. Cellular systems can provide an advantage to the field with better control of size, polydispersity, and morphology of these nanoparticles. For example, Dobias et al. investigated the role of different proteins in controlling nanoparticle size in E. coli [103]. Specifically, four proteins were identified that bound tightly to SeNPs resulting in narrower size distributions and more spherical nanoparticles with further investigations also being reported recently [104], [105]. Expression of a metal-resistant variant of reductase enzyme cytochrome b5 in Pichia pastoris led to selenium nanoparticles that were far less cytotoxic to human cells than conventional nanoparticles [106]. With regard to applications, selenium nanospheres have the ability to sequester elemental mercury for bioremediation applications [107].
Selenium metabolism and pathways within cells can be expanded both with respect to metabolites and nanomaterials, but will require fine tailoring to balance toxicity and selenium incorporation. Future metabolic engineering opportunities exist to balance two competing pathways: selenium metabolite incorporation vs. selenium reduction into nanoparticles. Once realized, selenium metabolism could establish a new path for bioactive molecules and materials.
4.2. Boron
Despite being an essential micronutrient, boron is rarely found in natural products and instead is commonly found in the cell walls of plants in complexes with polysaccharides and other structural features. A small handful of boronated natural products exist, all of which share the similar structure of a tetracoordinate boron-oxygen complex. These molecules include borophycin, boromycin, aplasmomycin, the tartrolons, autoinducer AI-2, and the recently discovered but extinct borolithochromes [108], [109]. Evidence from Chen et al. suggests that the boron atom of these naturally-occurring molecules is incorporated into the macrocyclic ring in the final synthesis step without enzymatic catalysis required [110]. The bacteria-produced Autoinducer AI-2 is a quorum sensing compound produced from the reaction of naturally occurring boric acid with 1-deoxy-3-dehydro-d-ribulose [111]. Other non-natural boron-containing molecules can be altered using promiscuous enzymes, however a heteroatom boronating enzyme is yet to be found [108].
Although not yet a mature field, boron chemistry has carved a space in modern medicinal chemistry and drug discovery. A recent review on the versatility of boron's bioactivity provides mechanistic and structural insights into the development of primarily synthetic boron molecules designed to be bioactive [112]. Yet, the recent discovery of a complex boronated polyketide compound in a prehistoric fossil displays the potential for boronated complexes to act as bridges between complex backbones to create larger scale unique molecules [109]. Future efforts in the field may enhance the ability to form functional boronated molecules using microorganisms.
4.3. Silicon
Silicon makes up nearly 28% of the Earth's crust and plays a vital role in modern chemistry as well as in elastomer production, microelectronics, and in many biotechnology applications. Standard chemical production of many enantioselective silicon molecules relies on multi-step chemical syntheses and separations, but biocatalysis has seen a slow yet steady adoption in the industry for certain reactions [113]. In this regard, silicon biotechnology is still a fairly new field. While not discussed here, additional applications in the area of nanotechnology are possible with the formation of complex silica structures [114], [115].
Until recently, the only known silicon-related bond formations that occurred in biological systems were silicon-oxygen bonds (such as the elaborate silica structures formed by diatoms [114]). Research performed in the yeast Kloeckera cortices demonstrated that enzymes can promiscuously react with silicon-bearing analogues of their native substrates, a form of precursor directed biosynthesis [116]. This method has been applied to other microorganisms as well to produce a handful of enantioselective silanes, with the oxidoreductase systems thought to serve as the catalytic machinery. Notably, Saccharomyces cerevisiae could reduce a variety of acylsilane substrates with high efficiency, but had trouble reducing phenyl-functionalized silicon molecules [117]. Other organisms, including Pichia pijperi, have shown to be more efficient at reducing these and expression of toluene dioxygenase in E. coli results in the dioxygenation of several phenyl silanes [118]. Free enzymes in non-aqueous solvents can perform more complex reactions such as polymerizations and generation of siloxane-phospholipids, but these conditions are not immediately compatible with in vivo use [24].
Silicon's chemical similarity to carbon has always hinted that microorganisms and biocatalysts may have a greater capacity for novel silicon-heteroatom bonds than what has been discovered previously. To this end, the first instance of an enzyme directly catalyzing the formation of a carbon-silicon bond was shown very recently in the Arnold lab using an engineered cytochrome C [119]. This evolved system was capable of catalyzing multiple substrates to siliconated products with high enantioselectivity and in vivo experiments demonstrated the feasibility for in vivo silicon-carbon bond formations. This example demonstrates the potential for broadening the production scope of enantiomerically pure, complex heteroatom organosilicon compounds.
4.4. Tellurium
Tellurium is most often found in various metal ores with below detectable limits in seawater [95]. Tellurium is assimilated by microorganisms and reduced into forms similar to that of sulfur and selenium. The tellurium-containing amino acids, telluro-cysteine and telluro-methionine can be detected in microorganisms when sulfur is replaced with tellurium, and much like selenium, can be incorporated into proteins where it has some benefits for crystallographic structure determination and other applications [120].
Analogous to selenium, engineering efforts with tellurium have enabled E. coli to produce volatile organotellurium compounds through the expression of a gene cluster from the organism Geobacillus stearothermophilus V [121]. These molecules include dimethyl telluride, dimethyl ditelluride, methanetellurol, and dimethyl tellurenyl sulfide, but have yet to be found useful for downstream applications. Much like selenium, certain tellurium compounds possess chemopreventative, anticancer, and antibiotic properties but extensive research is needed due to the toxicity of tellurium compounds, especially relative to human health [122], [123].
A potentially more promising aspect of tellurium metabolism is the reduction of tellurium salts into nanostructures. Mechanistically similar to selenium, microorganisms can produce elemental tellurium nanoparticles in various shapes, including spheres and rods [124], [125], [126]. These nanoparticles showed efficacious antimicrobial activity, especially in biofilm eradication [127]. Tellurium also plays a promising role in semiconductor nanoparticle composites (as discussed below for other metals). As with certain other elements described above, tellurium metabolism is rather rare and limited by toxicity, but holds promise in certain product applications.
4.5. Selenium and the metalloids outlook
The profound biological function of molecules containing selenium and the metalloids discussed give rise to a variety of unique biotechnological applications. Moreover, microbial-based synthesis of metalloid compounds can produce highly enantioselective products that are unable to be easily synthesized using conventional methods. Further investigation into the design of heteroatom-forming enzymes and the identification of promising metabolic routes will provide insight into the production of novel metabolites and nanomaterials with more fascinating properties.
5. Metals
Microorganisms have been exposed to trace elements and minerals for billions of years and have evolved uses for these as both critical catalytic components and important co-factors as is the case with elements like calcium, cobalt, copper, iron, magnesium, manganese, molybdenum, potassium, sodium, and zinc [128]. Likewise, disparate organisms show the potential to assimilate and reduce nearly every metal in bioremediation applications [129]. Many extensive reviews cover the fundamentals of biomining, bioremediation, and nanomaterial structure synthesis for these metals [26], [130], [131]. Instead, this review will focus primarily on biologically-uncommon metals that possess value for modern applications in biomining and nanomaterials (Table 2, Table 3).
Table 2.
Cellular metal incorporations - Microorganisms can interact with a large variety of metals. This table presents select examples of metal incorporation and applications, organized by element.
| Element | Description | Application | Organism(s) | References |
|---|---|---|---|---|
| Cobalt | Recovery from laterite tailings | Bioleaching | Acidithiobacillus thiooxidans and Acidithiobacillus ferrooxidans | Marrero et al. [172] |
| Intracellular, 550 nm average, flakes | Biomining and nanomaterials | Pseudomonas aeruginosa | Srivastava and Constanti [173] | |
| Copper | Copper bioleaching performed industrially | Bioleaching | Consortium of bacteria, archea, mesophiles, and thermophiles. | Gentina and Acevedo [174] |
| Extracellular nanoparticles, 3–10 nm, spherical | Nanomaterials - Antifungal | Stereum hirsutum | Cuevas et al. [175] | |
| Dysprosium | Intracellular accumulation of Dy | Biomining and bioremediation | Penidiella sp. T9 | Horiike and Yamashita [176] |
| Europium | Accumulation on cell surface | Biomining and bioremediation | Chlorella vulgaris | Ozaki et al. [177] |
| Gold | Ultra-efficient recovery from acidic leachate obtained from jewelry waste | Biomining | E. coli, Desulfovibrio desulfuricans | Deplanche and Macaskie [151] |
| Nanoclusters of various sizes and shapes depending on conditions | Nanomaterials – catalytic and medicinal | Shewanella haliotis | Zhu et al. [153] | |
| Iron | Recovery of iron from iron-containing minerals | Bioleaching | Acidithiobacillus thiooxidans | Marrero et al. [178] |
| Extracellular, 20 nm average, flakes | Nanomaterials | Pseudomonas aeruginosa | Srivastava and Constanti [173] | |
| Lithium | Lithium solubilization from various ores | Bioleaching | Aspergillus niger and Rhodotorula rubra | Marcincakova et al. [138], [139] |
| Lithium nanoparticles formed intracellularly, 750 nm average size | Biomining and nanomaterials | Pseudomonas aeruginosa | Srivastava and Constanti [173] | |
| Nickel | Recovery from laterite tailings | Bioleaching | Acidithiobacillus thiooxidans and Acidithiobacillus ferrooxidans | Marrero et al. [172] |
| Extracellular, 3 nm average, dense polygons | Nanomaterials | Pseudomonas aeruginosa | Srivastava and Constanti [173] | |
| Palladium | Monodisperse, small (4–5 nm) nanoparticles were observed | Nanomaterials - catalytic | E. coli | Zhu et al. [149] |
| Intracellular accumulation of palladium nanoparticles | Biomining | Desulfovibrio desulfuricans, Bacillus benzeovorans | Omajali et al. [150] | |
| Platinum | Extracellular nanoparticles, 5–30 nm | Nanomaterials - catalytic | Fusarium oxysporum | Syed and Ahmad [145] |
| Intracellular accumulation of platinum nanoparticles | Biomining | Acinetobacter calcoaceticus | Gaidhani et al. [147] | |
| Rhodium | Extracellular, 10 nm average, spherical | Nanomaterials – catalytic | Pseudomonas aeruginosa | Srivastava and Constanti [173] |
| Ruthenium | Extracellular, 3 nm average, dense polygons | Nanomaterials – catalytic | Pseudomonas aeruginosa | Srivastava and Constanti [173] |
| Selenium | Extracellular, rod-shaped Se nanoparticles, average size 17 nm | Nanomaterials | Streptomyces bikiniensis | Ahmad et al. [179] |
| Silver | Extracellular nanoparticles, 10–100 nm, protein functionalized | Nanomaterials – catalytic and medicinal | Cladosporium cladosporioides | Balaji et al. [180] |
| Silver uptake capabilities of up to 153 mg/L were observed | Biomining | Trichoderma harzianum | Cecchi et al. [181] | |
| Technetium | Reduction of Tc(VII) to Tc(IV) via various reducing agents | Biomining and bioremediation | Fe(III)-reducing, sulfate-reducing, fermentative, aerobic, and anaerobic bacteria | Chernyh et al. [182] |
| Tellurium | Intracellular, rod-shaped Te nanoparticles, 20 × 180 nm | Nanomaterials and biomining | Bacillus sp. | Zare et al. [124] |
| Uranium | Uranium bioprecipitation engineered for different cellular loci | Biomining | Deinococcus radiodurans, E. coli | Kulkarni et al. [142] |
| Ytterbium | Accumulation on cell surface | Biomining and bioremediation | S. cerevisiae | Jiang et al. [183] |
| Zinc | A 75% Zn extraction was obtained from Zn-plant leach residues under optimized conditions | Bioleaching | Acidithiobacillus thiooxidans | Sethurajan et al. [184] |
Table 3.
Cellular composite nanomaterials - This table displays examples of composite nanomaterials produced through interactions with cellular systems.
| Composition | Description | Application | Organism(s) | Reference |
|---|---|---|---|---|
| AuPd | Intracellular, 2–4 nm, gold-palladium core-shell particles | Nanomaterials | E. coli | Deplanche et al. [185] |
| BaTiO3 | Extracellular, 4–5 nm | Nanomaterials - ferroelectric | Fusarium oxysporum | Bansal et al. [161] |
| CdSe | Intracellular, 7–13 nm, uniform size | Nanomaterials - semiconductor | E. coli | Yan et al. [186] |
| CdSeZnTe, AuCdSeZn, SrGd, PrGd | Various sizes and shapes | Nanomaterials | E. coli | Park et al. [163] |
| CdTe | Extracellular, 2–4 nm, uniform size | Nanomaterials - semiconductor | S. cerevisiae | Bao et al. [187] |
| Cerium oxides | Extracellular formation of CeO2 nanoparticles containing Ce (III) and Ce (IV) mixed oxidation states | Nanomaterials | Humicola sp. | Khan and Ahmad [188] |
| CoFe2O4 | Extracellular, 3–15 nm, spherical | Nanomaterials - magnetic | S. cerevisiae | Jha and Prasad [189] |
| Copper oxides | Extracellular, various size and shape, mechanism investigated | Nanomaterials | E. coli | Singh et al. [190] |
| NiO | Hollow Cylinder NiO Nanostructured Material | Nanomaterials | Sporosarcina pasteurii | Vaidyanathan et al. [191] |
| PbS | Intracellular, 2–5 nm, cubic structure | Nanomaterials - semiconductor | Rhodosporidium diobovatum | Seshadri et al. [192] |
| SiO2 | Extracellular, 15 nm average size, spherical | Nanomaterials | Thermophilic bacterium (BKH1) | Show et al. [193] |
| TiO2 | Extracellular, 50–100 nm, spherical | Nanomaterials | Bacillus subtilis | Kirthi et al. [194] |
| Zircon sand (Zirconia and silica) | Selective leaching of silica to form SiO2 nanoparticles and enrich zirconia content | Nanomaterials and bioleaching | Fusarium oxysporum | Bansal et al. [195] |
| Zirconia | Extracellular, quasi-spherical, 3–11 nm | Nanomaterials | Fusarium oxysporum | Bansal et al. [196] |
The foundation of bionanotechnology as a field is still young, but many proteins and other biomolecules have been discovered that interact with an assortment of metals. There are two major industrial outcomes of this field with two distinct modes of production. For nanoparticle formation, extracellular formation is desired, whereas with biomining applications, it is advantageous for the metal to accumulate within the cells to facilitate facile downstream separations. Biomining may also be used to solubilize insoluble forms of a metal to enable simpler extraction, a process called bioleaching. Favorable aspects of these bioconversions include lower capital expenditures, simpler operation, and a lower overall carbon footprint than traditional reaction-based processes.
Metabolic engineering approaches are beginning to improve upon the relatively slow reaction rates inherent in metal conversion processes [132]. Beyond these applications, another area where biological systems are interacting with uncommon elements is metalloenzyme engineering. In one instance, the iron in heme proteins was replaced with iridium to catalyze novel reactions [133]. Another study achieved artificial ring-closing metathesis in vivo using a biotin-ruthenium (Biot-Ru) catalyst complex localized in the periplasmic space to improve activity [134]. Thus, the chemical palette has expanded beyond metabolism into biocatalysis, a topic of significant interest [135]. The sections that follow outline advances in prevalent biological metal incorporations while other elements that are not discussed in depth are highlighted in Table 2, Table 3
5.1. Lithium
Lithium has become pervasive in our modern-day society primarily for its use in materials and batteries. As a result, lithium mining operations have continued to expand and although the global availability is currently stable, sustainable recycling and extraction are major concerns of the future due to a growing global market [136]. Unsurprisingly, traditional lithium extractions are energy and reagent intensive and incur high capital expenditures. In this regard, biomining via microorganisms may be an attractive avenue for lithium extraction and recycling.
Lithium is present in many minerals with varying degrees of resistance to traditional chemical leaching. Among these minerals, the best studied are spodumene (6.9% Li2O) and lepidolite (3.8% Li2O). Lithium extraction from spodumene has been investigated using Penicillium purpurogenum, Aspergillus niger, and Rhodotorula rubra [137]. Similarly, A. niger and R. rubra were also shown to extract lithium from lepidolite [138], [139]. Although the maximum lithium yield was 413 μg/g biomass, these processes show potential as ecofriendly methods for lithium extraction. Lithium accumulation in these systems occurs via two mechanisms: (1) adherence to the outer surface of the cell via exopolymer interactions and (2) uptake of lithium into the cytoplasm through an unknown transport mechanism. The initial step of the bioleaching process, the solubilization of lithium from the ore, occurs via a reaction with biogenic organic acids. Thus, organic acid overproducing strains could lead to improved lithium extraction in the future. While still in its early stages, cellular systems along with metabolic engineering can lead to a more sustainable recycling of lithium from batteries and traditional mining-resistant minerals.
5.2. Uranium
Microorganisms have the potential to sequester and convert uranium into a useable source. Accordingly, uranium biomining through bioleaching has been industrially used for nearly 6 decades, beginning in the 1960s [140]. Several efforts to understand this process have been conducted including modeling bioleaching efficiency [141] and evaluating the influence of phosphatase enzymes localization in the cell for Deinococcus radiodurans and E. coli [142]. A further report discovered that non-crystalline U(IV) from biogenic sources is the predominant form of uranium in characterized ore deposits [143]. This report supports the idea that biogenic processes are very important to uranium ore genesis and further characterization of ore and microorganism composition could guide future research involving biomining efforts.
5.3. Platinum
Platinum's intrinsic low reactivity allows it to perform as an excellent catalyst and surface coating material, especially when formulated into nanoparticles [144]. This area has been investigated with cellular systems through the secretion of proteins by the fungus F. oxysporum that can stabilize and organize platinum nanoparticles [145]. Likewise, sulfate-reducing bacteria are a platform for studying the bioreductive mechanism behind platinum nanoparticle formation [146]. This process likely involves a two-step reduction, each with separate enzymes, reducing the initial Pt(IV) to Pt(II) and then to Pt(0) before the metal precipitates into nanocrystals. Finally, intracellular precipitation and formation of smaller nanocrystals has been observed, opening the future door to biomining [147].
5.4. Palladium
Palladium, a widely-used platinum group transition metal, is also highly sought after for its catalytic capabilities. Biological reductions and particle formation is possible in cells fed with palladium sources. For example, using E. coli mutant strains, Deplanche et al. determined that [NiFe] hydrogenases are responsible for accelerating the reduction of Pd(II) to Pd(0) [148]. E. coli could also be used as a vehicle to make biogenic Pd nanoparticle catalysts [149]. These catalysts provided lower cis-trans isomerization during hydrogenation of alkynes and alkenes, an advantage over conventionally produced Pd-NPs. Similar Pd-NPs were also produced in the sulfate-reducing bacteria Desulfovibrio desulfuricans, which created smaller and thus more effective Pd catalyst particles. Biomining efforts can also be envisioned based on observations of intracellular accumulation of palladium in Desulfovibrio desulfuricans, Bacillus benzeovorans, and other microorganisms [150].
5.5. Gold
Gold has historically been one of the most valuable elements, especially given its inherent oxidation resistance, malleability, and medicinal properties. Biorecovery of gold has been studied in both Escherichia coli and Desulfovibrio desulfuricans as an alternative to conventional reclaiming methods [151]. Gold nanoparticle synthesis using the fungus Penicillium brevicompactum has been explored and tested for cytotoxicity effects on mouse mayo blast cancer cells [152]. More recently, a 99% biorecovery efficiency of gold into small, spherical nanoparticles and subsequent catalytic reduction activity was shown in Shewanella haliotis [153]. As with other metals, several other organisms have been explored to both sequester gold and to produce nanostructures with exquisite size and shape control [154].
5.6. Silver
As the most conductive of all elements, silver has a multitude of applications, especially in the fields of catalysis and material coatings. Once again, cellular systems provide an excellent platform for both nanoparticle synthesis and biomining applications. As an example, Fusarium oxysporum is capable of efficiently producing size-controlled silver nanoparticles [155]. The industrially-relevant yeast Yarrowia lipolytica and fungus Neurospora intermedia can also mediate the synthesis of Ag nanoparticles [156]. Given the antimicrobial properties of silver nanoparticles, each of these studies demonstrated significant activity with biogenically created structures. Regarding silver nanoparticles, biological synthesis has been shown to proceed faster than chemical or physical processes, providing a potential advantage. To this end, monodispersed silver nanoparticles (5–25 nm) were produced extracellularly by the fungus Aspergillus fumigatus in only 10 min [157].
5.7. Nanoparticle composites
Nanotechnology has demonstrated the importance of nanoparticle composites for functionality, especially in the areas of magnetic nanoparticles, semiconductor materials, and quantum dots. Among the biologically-produced quantum dot and semiconductor materials are PbSe, CdSe, CdTe, and PbS [158], [159], [160]. Ferroelectric materials are also capable of being produced within microorganisms, evidenced by the synthesis of barium titanate in Fusarium oxysporum [161]. In addition, Au-Ag alloy nanoparticles were produced by S. cerevisiae and displayed anti-microbial properties [162]. These efforts were taken further by Park et al., who used recombinant E. coli for the in vivo biosynthesis of a wide variety of nanoparticle composites [163]. Their results show that cellular reduction mechanisms can reduce a wide range of metallic elements (Table 3). Moreover, this system can produce nanoassemblies which have not yet been possible through chemical means, such as CdSeZnTe and AuCdSeZn. Further applications of coupling microfluidic devices to optimize process conditions resulted in homogeneous nanocluster production by E. coli [164].
5.8. Metalloid and metal metabolism outlook
As described in the sections above, nanoparticle synthesis in cells is an interesting interaction between metabolism, protein expression, and cell capacity. Mechanisms for formation are beginning to be elucidated, paving the way for the tailoring of size, shape, and other properties [165]. Certainly, there are newfound possibilities to interface nanomaterials with synthetic biology and metabolic engineering. Although not mutually exclusive from nanoparticle synthesis, biomining developments will also require disparate optimization of strain characteristics and process specifics. In this regard, motivated by organisms naturally capable of metal reduction and oxidation, model organisms and microbial functional communities can be engineered to selectively capture an assortment of valuable metals [166]. Ideally, advances in these areas will parallel those of metabolic engineering, where elucidation of mechanisms and subsequent process optimization will enable microorganisms to perform desired traits beyond the levels currently seen in Nature.
6. Conclusions and future prospects
Metabolic engineering and cellular rewiring have the capability of greatly expanding the chemical diversity within cells (Fig. 1). Through this review, we seek to demonstrate this premise and provide insight into the various avenues of further innovation to incorporate uncommon elemental groups. Products containing heteroatom-bonded, conjugated, and nanoparticle-sequestered uncommon elements are important in all fields and industries. Although many challenges exist in the field of alternative element incorporation, especially with regard to toxicity, insight gained from the remarkable chemical feats of nonconventional organisms can be used to engineer more capable model organisms. Thus, the expanding array of synthetic tools will play an important role in future developments. Ultimately, enabling novel elemental chemistries within cellular systems will be a major contribution of the biotechnology era that expands the scope of renewable chemistries performed by microorganisms.
Acknowledgements
We acknowledge support from The Camille and Henry Dreyfus Teacher Scholar Program, The Welch Foundation under Grant F-1753, and the Air Force Office of Scientific Research under Award No. FA9550-14-1-0089.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Curran K.A., Alper H.S. Expanding the chemical palate of cells by combining systems biology and metabolic engineering. Metab Eng. 2012;14:289–297. doi: 10.1016/j.ymben.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Sun J., Alper H.S. Metabolic engineering of strains: from industrial-scale to lab-scale chemical production. J Ind Microbiol Biotechnol. 2015;42:423–436. doi: 10.1007/s10295-014-1539-8. [DOI] [PubMed] [Google Scholar]
- 3.Markham K.A., Alper H.S. Synthetic biology for specialty chemicals. Annu Rev Chem Biomol Eng. 2015;6:35–52. doi: 10.1146/annurev-chembioeng-061114-123303. [DOI] [PubMed] [Google Scholar]
- 4.Ro D.-K., Paradise E.M., Ouellet M., Fisher K.J., Newman K.L., Ndungu J.M. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 5.Alper H., Miyaoku K., Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol. 2005;23:612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 6.Ehrenworth A.M., Peralta-Yahya P. Accelerating the semisynthesis of alkaloid-based drugs through metabolic engineering. Nat Chem Biol. 2017;13:249–258. doi: 10.1038/nchembio.2308. [DOI] [PubMed] [Google Scholar]
- 7.Blazeck J., Hill A., Liu L., Knight R., Miller J., Pan A. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat Commun. 2014;5 doi: 10.1038/ncomms4131. ncomms4131. [DOI] [PubMed] [Google Scholar]
- 8.King J.R., Edgar S., Qiao K., Stephanopoulos G. Accessing Nature's diversity through metabolic engineering and synthetic biology. F1000Research. 2016:5. doi: 10.12688/f1000research.7311.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang K., Sawaya M.R., Eisenberg D.S., Liao J.C. Expanding metabolism for biosynthesis of nonnatural alcohols. Proc Natl Acad Sci. 2008;105:20653–20658. doi: 10.1073/pnas.0807157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K., Li H., Cho K.M., Liao J.C. Expanding metabolism for total biosynthesis of the nonnatural amino acid L-homoalanine. Proc Natl Acad Sci. 2010;107:6234–6239. doi: 10.1073/pnas.0912903107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldman A.J., Ng T.L., Wang P., Balskus E.P. Heteroatom–heteroatom bond formation in natural product biosynthesis. Chem Rev. 2017;117:5784–5863. doi: 10.1021/acs.chemrev.6b00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yim H., Haselbeck R., Niu W., Pujol-Baxley C., Burgard A., Boldt J. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 13.Latham J., Brandenburger E., Shepherd S.A., Menon B.R.K., Micklefield J. Development of halogenase enzymes for use in synthesis. Chem Rev. 2017 doi: 10.1021/acs.chemrev.7b00032. [DOI] [PubMed] [Google Scholar]
- 14.Breydo D.L. Boron, biologically active compounds. In: Kretsinger R.H., Uversky V.N., Permyakov E.A., editors. Encycl. Met. Springer; New York: 2013. pp. 295–299. [Google Scholar]
- 15.Pereira L., Mehboob F., Stams A.J.M., Mota M.M., Rijnaarts H.H.M., Alves M.M. Metallic nanoparticles: microbial synthesis and unique properties for biotechnological applications, bioavailability and biotransformation. Crit Rev Biotechnol. 2015;35:114–128. doi: 10.3109/07388551.2013.819484. [DOI] [PubMed] [Google Scholar]
- 16.Ladner C.C., Williams G.J. Harnessing natural product assembly lines: structure, promiscuity, and engineering. J Ind Microbiol Biotechnol. 2016;43:371–387. doi: 10.1007/s10295-015-1704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravikumar Y., Nadarajan S.P., Hyeon Yoo T., Lee C., Yun H. Unnatural amino acid mutagenesis-based enzyme engineering. Trends Biotechnol. 2015;33:462–470. doi: 10.1016/j.tibtech.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Rovner A.J., Haimovich A.D., Katz S.R., Li Z., Grome M.W., Gassaway B.M. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sardar D., Tianero M.D., Schmidt E.W. Chapter one - directing biosynthesis: practical supply of natural and unnatural cyanobactins. In: O'Connor S.E., editor. vol. 575. Academic Press; 2016. pp. 1–20. (Methods enzymol). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumann T., Nickling J.H., Bartholomae M., Buivydas A., Kuipers O.P., Budisa N. Prospects of in vivo incorporation of non-canonical amino acids for the chemical diversification of antimicrobial peptides. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner C., El Omari M., König G.M. Biohalogenation: nature's way to synthesize halogenated metabolites. J Nat Prod. 2009;72:540–553. doi: 10.1021/np800651m. [DOI] [PubMed] [Google Scholar]
- 22.Brown S., O'Connor S.E. Halogenase engineering for the generation of new natural product analogues. ChemBioChem. 2015;16:2129–2135. doi: 10.1002/cbic.201500338. [DOI] [PubMed] [Google Scholar]
- 23.Jedinák L., Zátopková R., Zemánková H., Šustková A., Cankař P. The suzuki–Miyaura cross-coupling reaction of halogenated aminopyrazoles: method development, scope, and mechanism of dehalogenation side reaction. J Org Chem. 2017;82:157–169. doi: 10.1021/acs.joc.6b02306. [DOI] [PubMed] [Google Scholar]
- 24.Frampton M.B., Zelisko P.M. Biocatalysis in silicon chemistry. Chem Asian J. 2017;12:1153–1167. doi: 10.1002/asia.201700214. [DOI] [PubMed] [Google Scholar]
- 25.Adkins J., Pugh S., McKenna R., Nielsen D.R. Engineering microbial chemical factories to produce renewable “biomonomers.”. Front Microbiol. 2012;3 doi: 10.3389/fmicb.2012.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schippers A., Hedrich S., Vasters J., Drobe M., Sand W., Willscher S. Geobiotechnology I. Springer; Berlin, Heidelberg: 2013. Biomining: metal recovery from ores with microorganisms; pp. 1–47. [DOI] [PubMed] [Google Scholar]
- 27.Mishra S., Keswani C., Abhilash P.C., Fraceto L.F., Singh H.B. Integrated approach of agri-nanotechnology: challenges and future trends. Front Plant Sci. 2017;8 doi: 10.3389/fpls.2017.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dicks A., Hent A. Springer; 2014. Green chemistry metrics: a guide to determining and evaluating process greenness. [Google Scholar]
- 29.Agarwal V., Miles Z.D., Winter J.M., Eustáquio A.S., El Gamal A.A., Moore B.S. Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem Rev. 2017;117:5619–5674. doi: 10.1021/acs.chemrev.6b00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gribble G.W. Biological activity of recently discovered halogenated marine natural products. Mar Drugs. 2015;13:4044–4136. doi: 10.3390/md13074044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gribble G.W. Springer Science & Business Media; 2009. Naturally occurring organohalogen compounds - a comprehensive update. [Google Scholar]
- 32.Littlechild J. Haloperoxidases and their role in biotransformation reactions. Curr Opin Chem Biol. 1999;3:28–34. doi: 10.1016/s1367-5931(99)80006-4. [DOI] [PubMed] [Google Scholar]
- 33.Borozan S.Z. Stojanović SĐ. Halogen bonding in complexes of proteins and non-natural amino acids. Comput Biol Chem. 2013;47:231–239. doi: 10.1016/j.compbiolchem.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Ohtake K., Yamaguchi A., Mukai T., Kashimura H., Hirano N., Haruki M. Protein stabilization utilizing a redefined codon. Sci Rep. 2015:5. doi: 10.1038/srep09762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deb Roy A., Grüschow S., Cairns N., Goss R.J.M. Gene expression enabling synthetic diversification of natural products: chemogenetic generation of pacidamycin analogs. J Am Chem Soc. 2010;132:12243–12245. doi: 10.1021/ja1060406. [DOI] [PubMed] [Google Scholar]
- 36.Salah Ayoup M., Cordes D.B., Slawin A.M., O'Hagan D. Fluorine containing amino acids: synthesis and peptide coupling of amino acids containing the all- cis tetrafluorocyclohexyl motif. Org Biomol Chem. 2015;13:5621–5624. doi: 10.1039/c5ob00650c. [DOI] [PubMed] [Google Scholar]
- 37.O'Hagan D., Deng H. Enzymatic fluorination and biotechnological developments of the fluorinase. Chem Rev. 2015;115:634–649. doi: 10.1021/cr500209t. [DOI] [PubMed] [Google Scholar]
- 38.Deng H., O'Hagan D., Schaffrath C. Fluorometabolite biosynthesis and the fluorinase from Streptomyces cattleya. Nat Prod Rep. 2004;21:773–784. doi: 10.1039/b415087m. [DOI] [PubMed] [Google Scholar]
- 39.Ma L., Bartholome A., Tong M.H., Qin Z., Yu Y., Shepherd T. Identification of a fluorometabolite from Streptomyces sp. MA37: (2R3S4S)-5-fluoro-2,3,4-trihydroxypentanoic acid. Chem Sci. 2015;6:1414–1419. doi: 10.1039/c4sc03540b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma L., Li Y., Meng L., Deng H., Li Y., Zhang Q. Biological fluorination from the sea: discovery of a SAM-dependent nucleophilic fluorinating enzyme from the marine-derived bacterium Streptomyces xinghaiensis NRRL B24674. RSC Adv. 2016;6:27047–27051. [Google Scholar]
- 41.Carvalho M.F., Oliveira R.S. Natural production of fluorinated compounds and biotechnological prospects of the fluorinase enzyme. Crit Rev Biotechnol. 2017;0:1–18. doi: 10.1080/07388551.2016.1267109. [DOI] [PubMed] [Google Scholar]
- 42.Zhu X.M., Hackl S., Thaker M.N., Kalan L., Weber C., Urgast D.S. Biosynthesis of the fluorinated natural product nucleocidin in Streptomyces calvus is dependent on the blda-specified Leu-tRNAUUA molecule. ChemBioChem. 2015;16:2498–2506. doi: 10.1002/cbic.201500402. [DOI] [PubMed] [Google Scholar]
- 43.Grüschow S., Rackham E.J., Elkins B., Newill P.L.A., Hill L.M., Goss R.J.M. New pacidamycin antibiotics through precursor-directed biosynthesis. Chembiochem Eur J Chem Biol. 2009;10:355–360. doi: 10.1002/cbic.200800575. [DOI] [PubMed] [Google Scholar]
- 44.Goss R.J.M., Lanceron S., Deb Roy A., Sprague S., Nur-e-Alam M., Hughes D.L. An expeditious route to fluorinated rapamycin analogues by utilising mutasynthesis. ChemBioChem. 2010;11:698–702. doi: 10.1002/cbic.200900723. [DOI] [PubMed] [Google Scholar]
- 45.Walker M.C., Thuronyi B.W., Charkoudian L.K., Lowry B., Khosla C., Chang M.C.Y. Expanding the fluorine chemistry of living systems using engineered polyketide synthase pathways. Science. 2013;341:1089–1094. doi: 10.1126/science.1242345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ad O., Thuronyi B.W., Chang M.C.Y. Elucidating the mechanism of fluorinated extender unit loading for improved production of fluorine-containing polyketides. Proc Natl Acad Sci. 2017;114:E660–E668. doi: 10.1073/pnas.1614196114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Odar C., Winkler M., Wiltschi B. Fluoro amino acids: a rarity in nature, yet a prospect for protein engineering. Biotechnol J. 2015;10:427–446. doi: 10.1002/biot.201400587. [DOI] [PubMed] [Google Scholar]
- 48.Yu A.C.-S., Yim A.K.-Y., Mat W.-K., Tong A.H.-Y., Lok S., Xue H. Mutations enabling displacement of tryptophan by 4-fluorotryptophan as a canonical amino acid of the genetic code. Genome Biol Evol. 2014;6:629–641. doi: 10.1093/gbe/evu044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arntson K.E., Pomerantz W.C.K. Protein-observed fluorine NMR: a bioorthogonal approach for small molecule discovery. J Med Chem. 2016;59:5158–5171. doi: 10.1021/acs.jmedchem.5b01447. [DOI] [PubMed] [Google Scholar]
- 50.Montclare J.K., Son S., Clark G.A., Kumar K., Tirrell D.A. Biosynthesis and stability of coiled-coil peptides containing (2S,4R)-5,5,5-trifluoroleucine and (2S,4S)-5,5,5-trifluoroleucine. ChemBioChem. 2009;10:84–86. doi: 10.1002/cbic.200800164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker P.J., Montclare J.K. Enhanced refoldability and thermoactivity of fluorinated phosphotriesterase. ChemBioChem. 2011;12:1845–1848. doi: 10.1002/cbic.201100221. [DOI] [PubMed] [Google Scholar]
- 52.Merkel L., Budisa N. Organic fluorine as a polypeptide building element: in vivo expression of fluorinated peptides, proteins and proteomes. Org Biomol Chem. 2012;10:7241–7261. doi: 10.1039/c2ob06922a. [DOI] [PubMed] [Google Scholar]
- 53.Rivera-Chávez J., Raja H.A., Graf T.N., Burdette J.E., Pearce C.J., Oberlies N.H. Biosynthesis of fluorinated peptaibols using a site-directed building block incorporation approach. J Nat Prod. 2017;80:1883–1892. doi: 10.1021/acs.jnatprod.7b00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sánchez C., Zhu L., Braña A.F., Salas A.P., Rohr J., Méndez C. Combinatorial biosynthesis of antitumor indolocarbazole compounds. Proc Natl Acad Sci U S A. 2005;102:461–466. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okada M., Saito K., Wong C.P., Li C., Wang D., Iijima M. Combinatorial biosynthesis of (+)-Daurichromenic acid and its halogenated analogue. Org Lett. 2017;19:3183–3186. doi: 10.1021/acs.orglett.7b01288. [DOI] [PubMed] [Google Scholar]
- 56.Glenn W.S., Nims E., O'Connor S.E. Reengineering a tryptophan halogenase to preferentially chlorinate a direct alkaloid precursor. J Am Chem Soc. 2011;133:19346–19349. doi: 10.1021/ja2089348. [DOI] [PubMed] [Google Scholar]
- 57.Payne J.T., Poor C.B., Lewis J.C. Directed evolution of RebH for site-selective halogenation of large biologically active molecules. Angew Chem Int Ed. 2015;54:4226–4230. doi: 10.1002/anie.201411901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andorfer M.C., June Park H., Vergara-Coll J., Lewis J.C. Directed evolution of RebH for catalyst-controlled halogenation of indole C–H bonds. Chem Sci. 2016;7:3720–3729. doi: 10.1039/c5sc04680g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shepherd S.A., Menon B.R.K., Fisk H., Struck A., Levy C., Leys D. A structure-guided switch in the regioselectivity of a tryptophan halogenase. Chembiochem. 2016;17:821–824. doi: 10.1002/cbic.201600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou H., Qiao K., Gao Z., Vederas J.C., Tang Y. Insights into radicicol biosynthesis via heterologous synthesis of intermediates and analogs. J Biol Chem. 2010;285:41412–41421. doi: 10.1074/jbc.M110.183574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang S., Zhang S., Xiao A., Rasmussen M., Skidmore C., Zhan J. Metabolic engineering of Escherichia coli for the biosynthesis of various phenylpropanoid derivatives. Metab Eng. 2015;29:153–159. doi: 10.1016/j.ymben.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 62.Xu F., Merkley A., Yu D., Zhan J. Selective biochlorination of hydroxyquinolines by a flavin-dependent halogenase. Tetrahedron Lett. 2016;57:5262–5265. [Google Scholar]
- 63.Zeng J., Lytle A.K., Gage D., Johnson S.J., Zhan J. Specific chlorination of isoquinolines by a fungal flavin-dependent halogenase. Bioorg Med Chem Lett. 2013;23:1001–1003. doi: 10.1016/j.bmcl.2012.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Menon B, Brandenburger E, Sharif H, Klemstein U, Shepherd S, Greaney M, et al. RadH a versatile halogenase for integration into synthetic pathways. Angew Chem Int Ed n.d.:n/a-n/a. doi:10.1002/anie.201706342. [DOI] [PMC free article] [PubMed]
- 65.Mitchell A.J., Zhu Q., Maggiolo A.O., Ananth N.R., Hillwig M.L., Liu X. Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5. Nat Chem Biol. 2016;12:636–640. doi: 10.1038/nchembio.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hillwig M.L., Zhu Q., Ittiamornkul K., Liu X. Discovery of a promiscuous non-heme iron halogenase in ambiguine alkaloid biogenesis: implication for an evolvable enzyme family for Late-stage halogenation of aliphatic carbons in small molecules. Angew Chem Int Ed. 2016;55:5780–5784. doi: 10.1002/anie.201601447. [DOI] [PubMed] [Google Scholar]
- 67.Chankhamjon P., Tsunematsu Y., Ishida-Ito M., Sasa Y., Meyer F., Boettger-Schmidt D. Regioselective dichlorination of a non-activated aliphatic carbon atom and phenolic bismethylation by a multifunctional fungal flavoenzyme. Angew Chem Int Ed. 2016;55:11955–11959. doi: 10.1002/anie.201604516. [DOI] [PubMed] [Google Scholar]
- 68.Nakamura H., Schultz E.E., Balskus E.P. A new strategy for aromatic ring alkylation in cylindrocyclophane biosynthesis. Nat Chem Biol. 2017 doi: 10.1038/nchembio.2421. advance online publication. [DOI] [PubMed] [Google Scholar]
- 69.Zhu T., Cheng X., Liu Y., Deng Z., You D. Deciphering and engineering of the final step halogenase for improved chlortetracycline biosynthesis in industrial Streptomyces aureofaciens. Metab Eng. 2013;19:69–78. doi: 10.1016/j.ymben.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 70.Komatsu M., Komatsu K., Koiwai H., Yamada Y., Kozone I., Izumikawa M. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol. 2013;2:384–396. doi: 10.1021/sb3001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teufel R., Agarwal V., Moore B.S. Unusual flavoenzyme catalysis in marine bacteria. Curr Opin Chem Biol. 2016;31:31–39. doi: 10.1016/j.cbpa.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ross A.C., Gulland L.E.S., Dorrestein P.C., Moore B.S. Targeted capture and heterologous expression of the Pseudoalteromonas alterochromide gene cluster in Escherichia coli represents a promising natural product exploratory platform. ACS Synth Biol. 2015;4:414–420. doi: 10.1021/sb500280q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agarwal V., Moore B.S. Enzymatic synthesis of polybrominated dioxins from the marine environment. ACS Chem Biol. 2014;9:1980–1984. doi: 10.1021/cb5004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharma S.V., Tong X., Pubill-Ulldemolins C., Cartmell C., Bogosyan E.J.A., Rackham E.J. Living GenoChemetics by hyphenating synthetic biology and synthetic chemistry in vivo. Nat Commun. 2017;8:229. doi: 10.1038/s41467-017-00194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang B.-G., Gloer J.B., Ji N.-Y., Zhao J.-C. Halogenated organic molecules of rhodomelaceae origin: chemistry and biology. Chem Rev. 2013;113:3632–3685. doi: 10.1021/cr9002215. [DOI] [PubMed] [Google Scholar]
- 76.Wang L., Zhou X., Fredimoses M., Liao S., Liu Y. Naturally occurring organoiodines. RSC Adv. 2014;4:57350–57376. [Google Scholar]
- 77.Pasternak J.J., Williamson E.E. Clinical pharmacology, uses, and adverse reactions of iodinated contrast agents: a primer for the non-radiologist. Mayo Clin Proc. 2012;87:390–402. doi: 10.1016/j.mayocp.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aiello A., Fattorusso E., Imperatore C., Menna M., Müller W.E.G. Iodocionin, a cytotoxic iodinated metabolite from the mediterranean ascidian ciona edwardsii. Mar Drugs. 2010;8:285–291. doi: 10.3390/md8020285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gonzali S., Kiferle C., Perata P. Iodine biofortification of crops: agronomic biofortification, metabolic engineering and iodine bioavailability. Curr Opin Biotechnol. 2017;44:16–26. doi: 10.1016/j.copbio.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 80.Bayer T.S., Widmaier D.M., Temme K., Mirsky E.A., Santi D.V., Voigt C.A. Synthesis of methyl halides from biomass using engineered microbes. J Am Chem Soc. 2009;131:6508–6515. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 81.Agarwal V., Li J., Rahman I., Borgen M., Aluwihare L.I., Biggs J.S. Complexity of naturally produced polybrominated diphenyl ethers revealed via mass spectrometry. Environ Sci Technol. 2015;49:1339–1346. doi: 10.1021/es505440j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Andorfer M.C., Grob J.E., Hajdin C.E., Chael J.R., Siuti P., Lilly J. Understanding flavin-dependent halogenase reactivity via substrate activity profiling. ACS Catal. 2017;7:1897–1904. doi: 10.1021/acscatal.6b02707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin S., Huang T., Shen B. Tailoring enzymes acting on carrier protein-tethered substrates in natural product biosynthesis. Meth Enzymol. 2012;516:321–343. doi: 10.1016/B978-0-12-394291-3.00008-3. [DOI] [PubMed] [Google Scholar]
- 84.Schnepel C., Minges H., Frese M., Sewald N. A high-throughput fluorescence assay to determine the activity of tryptophan halogenases. Angew Chem Int Ed. 2016;55:14159–14163. doi: 10.1002/anie.201605635. [DOI] [PubMed] [Google Scholar]
- 85.Hosford J., Shepherd S.A., Micklefield J., Wong L.S. A high-throughput assay for arylamine halogenation based on a peroxidase-mediated quinone–amine coupling with applications in the screening of enzymatic halogenations. Chem Weinh Bergstr Ger. 2014;20:16759–16763. doi: 10.1002/chem.201403953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin J.-L., Wagner J.M., Alper H.S. Enabling tools for high-throughput detection of metabolites: metabolic engineering and directed evolution applications. Biotechnol Adv. 2017 doi: 10.1016/j.biotechadv.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 87.Abatemarco J., Hill A., Alper H.S. Expanding the metabolic engineering toolbox with directed evolution. Biotechnol J. 2013;8:1397–1410. doi: 10.1002/biot.201300021. [DOI] [PubMed] [Google Scholar]
- 88.Böck A., Forchhammer K., Heider J., Leinfelder W., Sawers G., Veprek B. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 89.Kieliszek M., Błażejak S., Gientka I., Bzducha-Wróbel A. Accumulation and metabolism of selenium by yeast cells. Appl Microbiol Biotechnol. 2015;99:5373–5382. doi: 10.1007/s00253-015-6650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Organoselenium compounds in biology and medicine. 2017. [Google Scholar]
- 91.Azad G.K., Tomar R.S. Ebselen, a promising antioxidant drug: mechanisms of action and targets of biological pathways. Mol Biol Rep. 2014;41:4865–4879. doi: 10.1007/s11033-014-3417-x. [DOI] [PubMed] [Google Scholar]
- 92.Geng X., Liu L., Tsai K.-J., Liu Z. Essent. Non-essent. Met. Humana Press; Cham: 2017. Selenium: roles in cancer prevention and therapies; pp. 39–68. [Google Scholar]
- 93.Herrero E., Wellinger R.E. Yeast as a model system to study metabolic impact of selenium compounds. Microb Cell. 2015;2:139–149. doi: 10.15698/mic2015.05.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rao Y., McCooeye M., Windust A., Bramanti E., D'Ulivo A., Mester Z. Mapping of selenium metabolic pathway in yeast by Liquid Chromatography−Orbitrap mass spectrometry. Anal Chem. 2010;82:8121–8130. doi: 10.1021/ac1011798. [DOI] [PubMed] [Google Scholar]
- 95.Řezanka T., Sigler K. Biologically active compounds of semi-metals. Stud Nat Prod Chem. 2008;35:835–921. doi: 10.1016/j.phytochem.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 96.Zhou X., Yuan Y., Yang Y., Rutzke M., Thannhauser T.W., Kochian L.V. Involvement of a broccoli COQ5 methyltransferase in the production of volatile selenium compounds. Plant Physiol. 2009;151:528–540. doi: 10.1104/pp.109.142521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ranjard L., Prigent-Combaret C., Nazaret S., Cournoyer B. Methylation of inorganic and organic selenium by the bacterial thiopurine methyltransferase. J Bacteriol. 2002;184:3146–3149. doi: 10.1128/JB.184.11.3146-3149.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pusztahelyi T., Kovács S., Pócsi I., Prokisch J. Selenite-stress selected mutant strains of probiotic bacteria for Se source production. J Trace Elem Med Biol. 2015;30:96–101. doi: 10.1016/j.jtemb.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 99.Mapelli V., Hillestrøm P.R., Patil K., Larsen E.H., Olsson L. The interplay between sulphur and selenium metabolism in yeast influences the intracellular redox balance. FEMS Yeast Res. 2012;12:20–32. doi: 10.1111/j.1567-1364.2011.00757.x. [DOI] [PubMed] [Google Scholar]
- 100.Burow M., Wittstock U., Gershenzon J. Sulfur metab. Phototrophic Org., Springer; Dordrecht: 2008. Sulfur-containing secondary metabolites and their role in plant defense; pp. 201–222. [Google Scholar]
- 101.Sun H., Jiang S., Caton-Williams J., Liu H., Huang Z. 2-Selenouridine triphosphate synthesis and Se-RNA transcription. RNA N Y N. 2013;19:1309–1314. doi: 10.1261/rna.038075.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chaudhary S., Umar A., Mehta S.K. Selenium nanomaterials: an overview of recent developments in synthesis, properties and potential applications. Prog Mater Sci. 2016;83:270–329. [Google Scholar]
- 103.Dobias J., Suvorova E.I., Bernier-Latmani R. Role of proteins in controlling selenium nanoparticle size. Nanotechnology. 2011;22:195605. doi: 10.1088/0957-4484/22/19/195605. [DOI] [PubMed] [Google Scholar]
- 104.Tugarova A.V., Kamnev A.A. Proteins in microbial synthesis of selenium nanoparticles. Talanta. 2017;174:539–547. doi: 10.1016/j.talanta.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 105.Lenz M., Kolvenbach B., Gygax B., Moes S., Corvini P.F.X. Shedding light on selenium biomineralization: proteins associated with bionanominerals. Appl Environ Microbiol. 2011 doi: 10.1128/AEM.01713-10. AEM.01713–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Elahian F., Reiisi S., Shahidi A., Mirzaei S.A. High-throughput bioaccumulation, biotransformation, and production of silver and selenium nanoparticles using genetically engineered Pichia pastoris. Nanomed Nanotechnol Biol Med. 2017;13:853–861. doi: 10.1016/j.nano.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 107.Fellowes J.W., Pattrick R.A.D., Green D.I., Dent A., Lloyd J.R., Pearce C.I. Use of biogenic and abiotic elemental selenium nanospheres to sequester elemental mercury released from mercury contaminated museum specimens. J Hazard Mater. 2011;189:660–669. doi: 10.1016/j.jhazmat.2011.01.079. [DOI] [PubMed] [Google Scholar]
- 108.Dembitsky V.M., Smoum R., Al-Quntar A.A., Ali H.A., Pergament I., Srebnik M. Natural occurrence of boron-containing compounds in plants, algae and microorganisms. Plant Sci. 2002;163:931–942. [Google Scholar]
- 109.Wolkenstein K., Sun H., Falk H., Griesinger C. Structure and absolute configuration of jurassic polyketide-derived spiroborate pigments obtained from microgram quantities. J Am Chem Soc. 2015;137:13460–13463. doi: 10.1021/jacs.5b08191. [DOI] [PubMed] [Google Scholar]
- 110.Chen T.S.S., Chang C.-J., Floss H.G. Biosynthesis of the boron-containing macrolide antibiotic aplasmomycin by Streptomyces griseus. J Am Chem Soc. 1981;103:4565–4568. [Google Scholar]
- 111.Galloway W.R.J.D., Hodgkinson J.T., Bowden S.D., Welch M., Spring D.R. Quorum sensing in gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem Rev. 2011;111:28–67. doi: 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]
- 112.Diaz D.B., Yudin A.K. The versatility of boron in biological target engagement. Nat Chem. 2017;9:731–742. doi: 10.1038/nchem.2814. [DOI] [PubMed] [Google Scholar]
- 113.Frampton M.B., Zelisko P.M. Organosilicon biotechnology. Siliconindia. 2009;1:147–163. [Google Scholar]
- 114.Fernandes F.M., Coradin T., Aimé C. Self-assembly in biosilicification and biotemplated silica materials. Nanomaterials. 2014;4:792–812. doi: 10.3390/nano4030792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jain K.K. Humana Press; New York, NY: 2017. Nanotechnologies. Handb. Nanomedicine; pp. 11–71. [Google Scholar]
- 116.Tacke R., Linoh H., Stumpf B., Abraham W., Kieslich K., Ernst L. Microbiological Transformation of Silicon Compounds: enantioselective Reduction of Trimethylsilylalkyl Acetoacetates and their Carba-Analogues. Zeitschrift für Naturforschung B. 1983 https://www.degruyter.com/view/j/znb.1983.38.issue-5/znb-1983-0516/znb-1983-0516.xml [Google Scholar]
- 117.Zani P. Biotransformations of organosilicon compounds: enantioselective reduction of acyl silanes by means of baker's yeast. J Mol Catal B Enzym. 2001;11:279–285. [Google Scholar]
- 118.Smith W.C., Whited G.M., Lane T.H., Sanford K., McAuliffe J.C. vol. 999. American Chemical Society; 2008. Enzymatic dihydroxylation of aryl silanes; pp. 434–459. (Polym. Biocatal. Biomater. II). [Google Scholar]
- 119.Kan S.B.J., Lewis R.D., Chen K., Arnold F.H. Directed evolution of cytochrome c for carbon–silicon bond formation: bringing silicon to life. Science. 2016;354:1048–1051. doi: 10.1126/science.aah6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kaur M., Rob A., Caton-Williams J., Huang Z. vol. 1152. American Chemical Society; 2013. Biochemistry of nucleic acids functionalized with sulfur, selenium, and tellurium: roles of the single-atom substitution; pp. 89–126. (Biochalcogen chem. Biol. Chem. Sulfur selenium tellurium). [Google Scholar]
- 121.Swearingen J.W., Araya M.A., Plishker M.F., Saavedra C.P., Vásquez C.C., Chasteen T.G. Identification of biogenic organotellurides in Escherichia coli K-12 headspace gases using solid-phase microextraction and gas chromatography. Anal Biochem. 2004;331:106–114. doi: 10.1016/j.ab.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 122.Turner R.J., Borghese R., Zannoni D. Microbial processing of tellurium as a tool in biotechnology. Biotechnol Adv. 2012;30:954–963. doi: 10.1016/j.biotechadv.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 123.Silberman A., Kalechman Y., Hirsch S., Erlich Z., Sredni B., Albeck A. The anticancer activity of organotelluranes: potential role in integrin inactivation. ChemBioChem. 2016;17:918–927. doi: 10.1002/cbic.201500614. [DOI] [PubMed] [Google Scholar]
- 124.Zare B., Faramarzi M.A., Sepehrizadeh Z., Shakibaie M., Rezaie S., Shahverdi A.R. Biosynthesis and recovery of rod-shaped tellurium nanoparticles and their bactericidal activities. Mater Res Bull. 2012;47:3719–3725. [Google Scholar]
- 125.Borghese R., Brucale M., Fortunato G., Lanzi M., Mezzi A., Valle F. Extracellular production of tellurium nanoparticles by the photosynthetic bacterium Rhodobacter capsulatus. J Hazard Mater. 2016;309:202–209. doi: 10.1016/j.jhazmat.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 126.Espinosa-Ortiz E.J., Rene E.R., Guyot F., van Hullebusch E.D., Lens P.N.L. Biomineralization of tellurium and selenium-tellurium nanoparticles by the white-rot fungus Phanerochaete chrysosporium. Int Biodeterior Biodegrad. 2017 [Google Scholar]
- 127.Zonaro E., Lampis S., Turner R.J., Qazi S.J.S., Vallini G. Biogenic selenium and tellurium nanoparticles synthesized by environmental microbial isolates efficaciously inhibit bacterial planktonic cultures and biofilms. Front Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barton L.L., Goulhen F., Bruschi M., Woodards N.A., Plunkett R.M., Rietmeijer F.J.M. The bacterial metallome: composition and stability with specific reference to the anaerobic bacterium Desulfovibrio desulfuricans. BioMetals. 2007;20:291–302. doi: 10.1007/s10534-006-9059-2. [DOI] [PubMed] [Google Scholar]
- 129.Nancharaiah Y.V., Mohan S.V., Lens P.N.L. Biological and bioelectrochemical recovery of critical and scarce metals. Trends Biotechnol. 2016;34:137–155. doi: 10.1016/j.tibtech.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 130.Hulkoti N.I., Taranath T.C. Biosynthesis of nanoparticles using microbes—a review. Colloids Surf B Biointerfaces. 2014;121:474–483. doi: 10.1016/j.colsurfb.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 131.de Lorenzo V., Marlière P., Solé R. Bioremediation at a global scale: from the test tube to planet Earth. Microb Biotechnol. 2016;9:618–625. doi: 10.1111/1751-7915.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Giebner F., Kaschabek S., Schopf S., Schlömann M. Three adapted methods to quantify biomass and activity of microbial leaching cultures. Miner Eng. 2015;79:169–175. [Google Scholar]
- 133.Key H.M., Dydio P., Clark D.S., Hartwig J.F. Abiological catalysis by artificial haem proteins containing noble metals in place of iron. Nature. 2016;534:534–537. doi: 10.1038/nature17968. [DOI] [PubMed] [Google Scholar]
- 134.Jeschek M., Reuter R., Heinisch T., Trindler C., Klehr J., Panke S. Directed evolution of artificial metalloenzymes for in vivo metathesis. Nature. 2016;537:661–665. doi: 10.1038/nature19114. [DOI] [PubMed] [Google Scholar]
- 135.Lewis J.C. Artificial metalloenzymes and metallopeptide catalysts for organic synthesis. ACS Catal. 2013;3:2954–2975. [Google Scholar]
- 136.Swain B. Recovery and recycling of lithium: a review. Sep Purif Technol. 2017;172:388–403. [Google Scholar]
- 137.Rezza I., Salinas E., Calvente V., Benuzzi D., Tosetti M.I. Extraction of lithium from spodumene by bioleaching. Lett Appl Microbiol. 1997;25:172–176. doi: 10.1046/j.1472-765x.1997.00199.x. [DOI] [PubMed] [Google Scholar]
- 138.Marcincakova R., Kadukova J., Mrazikova A., Velgosova O., Vojtko M. Lithium bioleaching from lepidolite using the yeast Rhodotorula Rubra. Inż Miner. 2015;16 nr 1. [Google Scholar]
- 139.Marcincakova R., Mrazikova A., Velgosova O., Kadukova J., Vojtko M., Holub M. The influence of spore age of Aspergillus Niger on lithium dissolution from lepidolite. Inż Miner. 2014;15 nr 2. [Google Scholar]
- 140.Hefnawy M.A., El-Said M., Hussein M., Amin M.A. Fungal leaching of uranium from its geological ores in alloga area, west central sinai. Egypt. J Biol Sci. 2002;2:346–350. [Google Scholar]
- 141.Rashidi A., Safdari J., Roosta-Azad R., Zokaei-Kadijani S. Modeling of uranium bioleaching by Acidithiobacillus ferrooxidans. Ann Nucl Energy. 2012;43:13–18. [Google Scholar]
- 142.Kulkarni S., Misra C.S., Gupta A., Ballal A., Apte S.K. Interaction of uranium with bacterial cell surfaces: inferences from phosphatase-mediated uranium precipitation. Appl Environ Microbiol. 2016;82:4965–4974. doi: 10.1128/AEM.00728-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bhattacharyya A., Campbell K.M., Kelly S.D., Roebbert Y., Weyer S., Bernier-Latmani R. Biogenic non-crystalline U(IV) revealed as major component in uranium ore deposits. Nat Commun. 2017;8 doi: 10.1038/ncomms15538. ncomms15538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bendale Y. Green synthesis, characterization and anticancer potential of platinum nanoparticles Bioplatin. J Chin Integr Med. 2012;10:681–689. doi: 10.3736/jcim20120613. [DOI] [PubMed] [Google Scholar]
- 145.Syed A., Ahmad A. Extracellular biosynthesis of platinum nanoparticles using the fungus Fusarium oxysporum. Colloids Surf B Biointerfaces. 2012;97:27–31. doi: 10.1016/j.colsurfb.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 146.Riddin T.L., Govender Y., Gericke M., Whiteley C.G. Two different hydrogenase enzymes from sulphate-reducing bacteria are responsible for the bioreductive mechanism of platinum into nanoparticles. Enzym Microb Technol. 2009;45:267–273. [Google Scholar]
- 147.Gaidhani S.V., Yeshvekar R.K., Shedbalkar U.U., Bellare J.H., Chopade B.A. Bio-reduction of hexachloroplatinic acid to platinum nanoparticles employing Acinetobacter calcoaceticus. Process Biochem. 2014;49:2313–2319. [Google Scholar]
- 148.Deplanche K., Caldelari I., Mikheenko I.P., Sargent F., Macaskie L.E. Involvement of hydrogenases in the formation of highly catalytic Pd(0) nanoparticles by bioreduction of Pd(II) using Escherichia coli mutant strains. Microbiology. 2010;156:2630–2640. doi: 10.1099/mic.0.036681-0. [DOI] [PubMed] [Google Scholar]
- 149.Zhu J., Wood J., Deplanche K., Mikheenko I., Macaskie L.E. Selective hydrogenation using palladium bioinorganic catalyst. Appl Catal B Environ. 2016;199:108–122. [Google Scholar]
- 150.Omajali J.B., Mikheenko I.P., Merroun M.L., Wood J., Macaskie L.E. Characterization of intracellular palladium nanoparticles synthesized by Desulfovibrio desulfuricans and Bacillus benzeovorans. J Nanoparticle Res. 2015;17:264. doi: 10.1007/s11051-015-3067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Deplanche K., Macaskie L. Biorecovery of gold by Escherichia coli and Desulfovibrio desulfuricans. Biotechnol Bioeng. 2008;99:1055–1064. doi: 10.1002/bit.21688. [DOI] [PubMed] [Google Scholar]
- 152.Mishra A., Tripathy S.K., Wahab R., Jeong S.-H., Hwang I., Yang Y.-B. Microbial synthesis of gold nanoparticles using the fungus Penicillium brevicompactum and their cytotoxic effects against mouse mayo blast cancer C2C12 cells. Appl Microbiol Biotechnol. 2011;92:617–630. doi: 10.1007/s00253-011-3556-0. [DOI] [PubMed] [Google Scholar]
- 153.Zhu N., Cao Y., Shi C., Wu P., Ma H. Biorecovery of gold as nanoparticles and its catalytic activities for p-nitrophenol degradation. Environ Sci Pollut Res. 2016;23:7627–7638. doi: 10.1007/s11356-015-6033-y. [DOI] [PubMed] [Google Scholar]
- 154.Gericke M., Pinches A. Microbial production of gold nanoparticles. Gold Bull. 2006;39:22–28. [Google Scholar]
- 155.Husseiny S.M., Salah T.A., Anter H.A. Biosynthesis of size controlled silver nanoparticles by Fusarium oxysporum, their antibacterial and antitumor activities. Beni-Suef Univ J Basic Appl Sci. 2015;4:225–231. [Google Scholar]
- 156.Apte M., Sambre D., Gaikawad S., Joshi S., Bankar A., Kumar A.R. Psychrotrophic yeast Yarrowia lipolytica NCYC 789 mediates the synthesis of antimicrobial silver nanoparticles via cell-associated melanin. AMB Express. 2013;3:32. doi: 10.1186/2191-0855-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bhainsa K.C., D'Souza S.F. Extracellular biosynthesis of silver nanoparticles using the fungus Aspergillus fumigatus. Colloids Surf B Biointerfaces. 2006;47:160–164. doi: 10.1016/j.colsurfb.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 158.U P, Gowda K.M.A., M G E, Teja B.S., N N, Mohan B.R. Biologically synthesized PbS nanoparticles for the detection of arsenic in water. Int Biodeterior Biodegrad. 2017;119:78–86. [Google Scholar]
- 159.Bao H., Lu Z., Cui X., Qiao Y., Guo J., Anderson J.M. Extracellular microbial synthesis of biocompatible CdTe quantum dots. Acta Biomater. 2010;6:3534–3541. doi: 10.1016/j.actbio.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 160.Jacob J.M., Lens P.N.L., Balakrishnan R.M. Microbial synthesis of chalcogenide semiconductor nanoparticles: a review. Microb Biotechnol. 2016;9:11–21. doi: 10.1111/1751-7915.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bansal V., Poddar P., Ahmad A., Sastry M. Room-Temperature biosynthesis of ferroelectric barium titanate nanoparticles. J Am Chem Soc. 2006;128:11958–11963. doi: 10.1021/ja063011m. [DOI] [PubMed] [Google Scholar]
- 162.Zheng D., Hu C., Gan T., Dang X., Hu S. Preparation and application of a novel vanillin sensor based on biosynthesis of Au–Ag alloy nanoparticles. Sens Actuators B Chem. 2010;148:247–252. [Google Scholar]
- 163.Park T.J., Lee S.Y., Heo N.S., Seo T.S. In vivo synthesis of diverse metal nanoparticles by recombinant Escherichia coli. Angew Chem Int Ed. 2010;49:7019–7024. doi: 10.1002/anie.201001524. [DOI] [PubMed] [Google Scholar]
- 164.Park T.J., Lee K.G., Lee S.Y. Advances in microbial biosynthesis of metal nanoparticles. Appl Microbiol Biotechnol. 2016;100:521–534. doi: 10.1007/s00253-015-6904-7. [DOI] [PubMed] [Google Scholar]
- 165.Singla R., Guliani A., Kumari A., Yadav S.K. vol. 2. Springer; Cham: 2017. Role of bacteria in nanocompound formation and their application in medical; pp. 3–37. (Microb. Appl). [Google Scholar]
- 166.Butu A., Rodino S., Dobre A., Butu M. Potential of microbial functional communities for high-tech critical metals recovery. Stud Univ Vasile Goldis Ser Stiintele Vietii Life Sci Ser. 2016;26:287–292. [Google Scholar]
- 167.Eustáquio A.S., Moore B.S. Mutasynthesis of fluorosalinosporamide, a potent and reversible inhibitor of the proteasome. Angew Chem Int Ed. 2008;47:3936–3938. doi: 10.1002/anie.200800177. [DOI] [PubMed] [Google Scholar]
- 168.Swearingen J.W., Fuentes D.E., Araya M.A., Plishker M.F., Saavedra C.P., Chasteen T.G. Expression of the ubiE gene of Geobacillus stearothermophilus V in Escherichia coli K-12 mediates the evolution of selenium compounds into the headspace of selenite- and selenate-amended cultures. Appl Environ Microbiol. 2006;72:963–967. doi: 10.1128/AEM.72.1.963-967.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tse Sum Bui B., Mattioli T.A., Florentin D., Bolbach G., Marquet A. Escherichia coli biotin synthase produces selenobiotin. Further evidence of the involvement of the [2Fe-2S]2+ cluster in the sulfur insertion step. Biochemistry (Mosc) 2006;45:3824–3834. doi: 10.1021/bi052388m. [DOI] [PubMed] [Google Scholar]
- 170.Dembitsky V.M., Al Quntar A.A.A., Srebnik M. Natural and synthetic small boron-containing molecules as potential inhibitors of bacterial and fungal quorum sensing. Chem Rev. 2011;111:209–237. doi: 10.1021/cr100093b. [DOI] [PubMed] [Google Scholar]
- 171.Elshahawi S.I., Trindade-Silva A.E., Hanora A., Han A.W., Flores M.S., Vizzoni V. Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills. Proc Natl Acad Sci. 2013;110:E295–E304. doi: 10.1073/pnas.1213892110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Marrero J., Coto O., Goldmann S., Graupner T., Schippers A. Recovery of nickel and cobalt from laterite tailings by reductive dissolution under aerobic conditions using acidithiobacillus species. Environ Sci Technol. 2015;49:6674–6682. doi: 10.1021/acs.est.5b00944. [DOI] [PubMed] [Google Scholar]
- 173.Srivastava S.K., Constanti M. Room temperature biogenic synthesis of multiple nanoparticles (Ag, Pd, Fe, Rh, Ni, Ru, Pt, Co, and Li) by Pseudomonas aeruginosa SM1. J Nanoparticle Res. 2012;14:831. [Google Scholar]
- 174.Acevedo F., Gentina J.C. Application of bioleaching to copper mining in Chile. Electron J Biotechnol. 2013:16. [Google Scholar]
- 175.Cuevas R., Durá N., Diez M.C., Tortella G.R., Rubilar O. Extracellular biosynthesis of copper and copper oxide nanoparticles by Stereum hirsutum, a native white-rot fungus from chilean forests. J Nanomater. 2015 [Google Scholar]
- 176.Horiike T., Yamashita M. A new fungal isolate, Penidiella sp. Strain T9, accumulates the rare earth element dysprosium. Appl Environ Microbiol. 2015;81:3062–3068. doi: 10.1128/AEM.00300-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ozaki T., Kimura T., Ohnuki T., Yoshida Z., Gillow J.B., Francis A.J. Sorption behavior of europium(III) and curium(III) on the cell surfaces of microorganisms. Radiochim Acta. 2004;92:741–748. [Google Scholar]
- 178.Marrero J., Coto O., Schippers A. Anaerobic and aerobic reductive dissolutions of iron-rich nickel laterite overburden by Acidithiobacillus. Hydrometallurgy. 2017;168:49–55. [Google Scholar]
- 179.Ahmad M.S., Yasser M.M., Sholkamy E.N., Ali A.M., Mehanni M.M. Anticancer activity of biostabilized selenium nanorods synthesized by Streptomyces bikiniensis strain Ess_amA-1. Int J Nanomedicine. 2015;10:3389–3401. doi: 10.2147/IJN.S82707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Balaji D.S., Basavaraja S., Deshpande R., Mahesh D.B., Prabhakar B.K., Venkataraman A. Extracellular biosynthesis of functionalized silver nanoparticles by strains of Cladosporium cladosporioides fungus. Colloids Surf B Biointerfaces. 2009;68:88–92. doi: 10.1016/j.colsurfb.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 181.Cecchi G., Marescotti P., Piazza S.D., Zotti M. Native fungi as metal remediators: silver myco-accumulation from metal contaminated waste-rock dumps (Libiola Mine, Italy) J Environ Sci Health Part B. 2017;52:191–195. doi: 10.1080/03601234.2017.1261549. [DOI] [PubMed] [Google Scholar]
- 182.Chernyh N.A., Gavrilov S.N., Sorokin V.V., German K.E., Sergeant C., Simonoff M. Characterization of technetium(vII) reduction by cell suspensions of thermophilic bacteria and archaea. Appl Microbiol Biotechnol. 2007;76:467–472. doi: 10.1007/s00253-007-1034-5. [DOI] [PubMed] [Google Scholar]
- 183.Jiang M., Ohnuki T., Tanaka K., Kozai N., Kamiishi E., Utsunomiya S. Post-adsorption process of Yb phosphate nano-particle formation by Saccharomyces cerevisiae. Geochem Cosmochim Acta. 2012;93:30–46. [Google Scholar]
- 184.Sethurajan M., Lens P.N., Rene E.R., van de Vossenberg J., Huguenot D., Horn H.A. Bioleaching and selective biorecovery of zinc from zinc metallurgical leach residues from the Três Marias zinc plant (Minas Gerais, Brazil) J Chem Technol Biotechnol. 2017;92:512–521. [Google Scholar]
- 185.Deplanche K., Merroun M.L., Casadesus M., Tran D.T., Mikheenko I.P., Bennett J.A. Microbial synthesis of core/shell gold/palladium nanoparticles for applications in green chemistry. J R Soc Interface. 2012;9:1705–1712. doi: 10.1098/rsif.2012.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yan Z., Qian J., Gu Y., Su Y., Ai X., Wu S. Green biosynthesis of biocompatible CdSe quantum dots in living Escherichia coli cells. Mater Res Express. 2014;1 [Google Scholar]
- 187.Bao H., Hao N., Yang Y., Zhao D. Biosynthesis of biocompatible cadmium telluride quantum dots using yeast cells. Nano Rev. 2010;3:481–489. [Google Scholar]
- 188.Khan S.A., Ahmad A. Fungus mediated synthesis of biomedically important cerium oxide nanoparticles. Mater Res Bull. 2013;48:4134–4138. [Google Scholar]
- 189.Jha A.K., Prasad K. Biological synthesis of cobalt ferrite nanoparticles. Nanotechnol Dev. 2012;2:9. [Google Scholar]
- 190.Ajay V., Singh, Patil R., Anand Atul, Milani Paolo, Gade W.N. Biological synthesis of copper oxide nano particles using Escherichia coli. Curr Nanosci. 2010;6:365–369. [Google Scholar]
- 191.Vaidyanathan S., Cherng J.-Y., Sun A.-C., Chen C.-Y. Bacteria-templated NiO nanoparticles/microstructure for an enzymeless glucose sensor. Int J Mol Sci. 2016;17:1104. doi: 10.3390/ijms17071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Seshadri S., Saranya K., Kowshik M. Green synthesis of lead sulfide nanoparticles by the lead resistant marine yeast, Rhodosporidium diobovatum. Biotechnol Prog. 2011;27:1464–1469. doi: 10.1002/btpr.651. [DOI] [PubMed] [Google Scholar]
- 193.Show S., Tamang A., Chowdhury T., Mandal D., Chattopadhyay B. Bacterial (BKH1) assisted silica nanoparticles from silica rich substrates: a facile and green approach for biotechnological applications. Colloids Surf B Biointerfaces. 2015;126:245–250. doi: 10.1016/j.colsurfb.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 194.Kirthi A.V., Rahuman A.A., Rajakumar G., Marimuthu S., Santhoshkumar T., Jayaseelan C. Biosynthesis of titanium dioxide nanoparticles using bacterium Bacillus subtilis. Mater Lett. 2011;65:2745–2747. [Google Scholar]
- 195.Bansal V., Syed A., Bhargava S.K., Ahmad A., Sastry M. Zirconia enrichment in zircon sand by selective fungus-mediated bioleaching of silica. Langmuir. 2007;23:4993–4998. doi: 10.1021/la062535x. [DOI] [PubMed] [Google Scholar]
- 196.Bansal V., Rautaray D., Ahmad A., Sastry M. Biosynthesis of zirconia nanoparticles using the fungus Fusarium oxysporum. J Mater Chem. 2004;14:3303–3305. [Google Scholar]