Abstract
5-Aminolevulinic acid (ALA) is a precursor in the biosynthesis of tetrapyrroles including chlorophylls and heme. The formation of ALA involves two enzymatic steps which take place in the chloroplast in plants. The first enzyme, glutamyl-tRNA reductase, and the second enzyme, glutamate-1-semialdehyde-2,1-aminomutase, are encoded by the nuclear HEMA and GSA genes, respectively. To assess the significance of the HEMA gene for chlorophyll and heme synthesis, transgenic Arabidopsis plants that expressed antisense HEMA1 mRNA from the constitutive cauliflower mosaic virus 35S promoter were generated. These plants exhibited varying degrees of chlorophyll deficiency, ranging from patchy yellow to total yellow. Analysis indicated that these plants had decreased levels of chlorophyll, non-covalently bound hemes, and ALA; their levels were proportional to the level of glutamyl-tRNA reductase expression and were inversely related to the levels of antisense HEMA transcripts. Plants that lacked chlorophyll failed to survive under normal growth conditions, indicating that HEMA gene expression is essential for growth.
Porphyrin compounds such as chlorophyll and heme play vital roles in plant metabolism. The porphyrin ring system is derived from 5-aminolevulinic acid (ALA). In plants ALA is formed from the five-carbon skeleton of Glu in two steps known as the C5 pathway (Beale, 1978; von Wettstein et al., 1995; Kumar et al., 1996b). All of the components required for such a conversion are located in the chloroplasts. The initial metabolite of the C5 pathway is chloroplast-specific glutamyl-tRNAGlu (Schön et al., 1986). In the first step, glutamyl-tRNA reductase (GluTR) reduces the glutamyl moiety of Glu-tRNA to Glu-1-semialdehyde (GSA). In the subsequent reaction, GSA is converted to ALA by GSA-2,1-aminomutase (GSA-AM). GluTR and GSA-AM are encoded by the nuclear HEMA and GSA genes. In Arabidopsis and other plants two HEMA genes exist (Ilag et al., 1994; Kumar et al., 1996a; Tanaka et al., 1996; Grimm, 1998; Sangwan and O'Brian, 1999). The HEMA1 gene is regulated by light and expressed in all parts of the plant, while the expression of the HEMA2 gene was only found in roots and flowers in a light-independent fashion. The HEMA1 and HEMA2 genes are very similar (79% at the nucleotide level in the coding region and differ in the 5′- and 3′-untranslated regions), while the gene products also show high similarity (83%).
Although it is widely accepted that all ALA in plants is formed in the C5 pathway, the presence of the C4 pathway in plants has been implied (Beale, 1978). In this biosynthetic route, which operates in animal mitochondria and in yeast, ALA is generated by ALA synthase from Gly and succinyl coenzyme A (May et al., 1990). ALA synthase-like activity has been monitored in greening potato skin (Ramaswamy and Nair, 1973) and soybean callus (Meller and Gassman, 1982); however, the enzyme was never characterized from plants. Other lines of evidence, e.g. photodestruction of chloroplasts (Thomsen et al., 1993), mutations affecting the tRNAGlu (Hess et al., 1992), and inhibition of GSA-AT activity by gabaculin (Flint, 1984), also support the C4 pathway of ALA biosynthesis in plants. We describe studies of antisense HEMA1 Arabidopsis plants that unequivocally demonstrate the involvement of GluTR in ALA formation.
MATERIALS AND METHODS
Bacterial Strains
Escherichia coli strain DH5α was used for routine recombinant DNA work. Agrobacterium tumefaciens strains EHA101 (for hypocotyl infection) and GV 3101 (for vacuum infiltration) were used in the generation of Arabidopsis transgenic lines.
Construction of the HEMA Antisense Vector
A NcoI site was created by site-directed mutagenesis at the translation start site in HEMA1 cDNA (Sambrook et al., 1989). After confirming the sequence, the entire coding region of the HEMA1 cDNA was released by digestion with NcoI and SphI restriction endonucleases. This fragment was blunt-ended with Klenow DNA polymerase and ligated to NcoI/NruI-digested, blunt-ended pRTL2-GUS plasmid. Transformants were analyzed for the constructs containing the HEMA nucleotide sequence in the antisense orientation downstream of the cauliflower mosaic virus 35S promoter. From the confirmed construct, a DNA fragment containing the cauliflower mosaic virus 35S promoter, HEMA cDNA (in antisense orientation), and the terminator was isolated as a cassette and ligated to SalI-digested, blunt-ended binary vector pCIT20. Plasmids pRTL2-GUS and pCIT20 were from Dr. X.-W. Deng's laboratory (Yale University). The resultant construct was transformed into the A. tumefaciens strains EHA101 and GV3101.
Plant Growth and Transformation
Hypocotyl Infection
Arabidopsis (ecotype Columbia) plants were grown from surface-sterilized seeds on sterile Murashige and Skoog agar medium containing 0.1% (w/v) Suc and 0.1 g/L myo-inositol under standard conditions (22°C, 60% relative humidity with a regimen of 16 h of 90 μE m−2 s−1 white light and 8-h dark day cycle). Hypocotyls were isolated from 10-d-old seedlings and hand-prepared for A. tumefaciens infection as previously described (Akama et al., 1992). Growth conditions involved in generating well-differentiated plants from the infected hypocotyls using root- and growth-inducing media were as previously described (Akama et al., 1992).
Vacuum Infiltration
Arabidopsis seeds were grown in vermiculite under the standard growth conditions. When primary inflorescence shoots attained a reasonable height (2–3 cm), they were cut off to generate multiple secondary inflorescences (which took about 7–10 d). At this stage, plants were used for A. tumefaciens infection by vacuum infiltration as described previously (Berthold et al., 1993).
DNA Isolation and Analysis
Total DNA (5 μg) isolated from the 2-week-old transgenic and control Arabidopsis plants was digested with BamHI, and the restricted fragments were separated on a 0.8% (w/v) agarose gel. Transfer of restriction fragments to a nitrocellulose membrane, prehybridization, hybridization, and washing of the membrane were performed under stringent conditions (Sambrook et al., 1989). The probe (a 600-bp NheI fragment from the HEMA1 cDNA) was labeled with [α-32P]dATP as described previously (Sambrook et al., 1989).
RNA Extractions and Northern-Blot Analysis
Total RNA was isolated (Ilag et al., 1994) from 2-week-old transgenic and control Arabidopsis plants. RNA (20 μg) was heat-denatured (85°C for 2 min) in the presence of formamide (50%, w/v) and separated on a denaturing 1.2% (w/v) agarose gel as described previously (Sambrook et al., 1989). Equal loading of RNA in northern-blot analysis was confirmed by hybridization with a pea 18S rDNA fragment. Prehybridization, hybridization, and washing conditions of the membranes were described above. To monitor the expression levels of endogenous genes, respective DNA fragments were labeled with [α-32P]dATP using random hexanucleotide priming (Sambrook et al., 1989). We used strand-specific RNA probes prepared from HEMA cDNA clone using T3 and T7 RNA polymerase for the estimation of sense and antisense expression of the HEMA gene in these plants (Sambrook et al., 1989).
Protein Extractions and Western-Blot Analysis
Plants were ground in liquid nitrogen and suspended (100 μL/100 mg) in extraction buffer containing 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 50 mm β-mercaptoethanol, 0.5% (w/v) Triton X-100, 0.2 mm phenylmethylsulfonylfluoride, 1 μg/mL leupeptin, and 1 μg/mL apoprotein. After a brief vortexing, the suspension was centrifuged at 13,000g for 10 min and the supernatant collected. The protein in the supernatant was measured with the dye-binding assay (Bradford, 1976). Separation of proteins by SDS-PAGE and blotting was performed as described previously (Sambrook et al., 1989). Blots were incubated for 1 h with respective antibodies (1:1,000 dilution) in a solution containing 50 mm Tris-HCl (pH 10.2), 150 mm NaCl, 1% (w/v) bovine serum albumin, and 0.1% (w/v) Triton X-100. The blots were washed three times (for 5 min each) with the same solution without bovine serum albumin. The western-blot analysis of protochlorophyllide oxidoreductase (Por) proteins was performed as previously described (Armstrong et al., 1995).
Biochemical Analysis
Chlorophyll Estimation
Chlorophyll pigment was extracted into 80% (w/v) aqueous acetone and the total chlorophyll (chlorophyll a+b) was estimated as described previously (Lichtenthaler, 1987).
Quantitation of Heme
Non-covalently bound hemes (protoheme and heme a) were extracted by acid acetone and purified on a DEAE-Sepharose column as reported previously (Weinstein and Beale, 1984). Pyridine (final concentration, 25%) and NaOH (final concentration, 0.1 n) were added to the processed eluate (to convert the protoheme and heme a in the eluate to pyridine homochrome), followed by few crystals of sodium dithionite (to reduce the pyridine homochromes), as described previously (Stillman and Gassman, 1978). The absorption spectrum of the reduced pyridine homochrome was recorded between wavelengths of 400 to 600 nm with a UV/VIS spectrometer (Lambda 2, Perkin-Elmer, Foster City, CA).
ALA Determination
ALA was extracted from Arabidopsis plants, purified, and measured colorimetrically, as described previously (Weinstein and Beale, 1985). However, prior to the extraction, plants were incubated for 6 h in Murashige and Skoog medium containing levulinic acid (100 mm).
RESULTS
HEMA1 and HEMA2 Map to Chromosome I
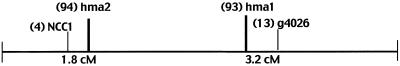
Several restriction enzymes were used to reveal the polymorphisms between pure Arabidopsis ecotypes Columbia and Landsberg erecta for HEMA1 and HEMA2 genes (Lister and Dean, 1993). The 3′-untranslated DNA fragments of HEMA1 and HEMA2 genes were used as gene-specific probes. After identifying the RFLP pattern for HEMA1 and HEMA2 genes, the genomic DNA isolated from 29 inbred lines derived from a cross of Columbia and Landsberg erecta ecotypes was digested with EcoRV (for HEMA1) and BglII (for HEMA2) and Southern-blot analysis was carried out with gene-specific probes as described above. The data on segregation of polymorphisms in these lines were analyzed with the Mapmaker program (Lander et al., 1987), which placed the HEMA1 and HEMA2 genes to chromosome I in Arabidopsis (Fig. 1).
Figure 1.
Schematic representation of the location of HEMA1 (hma1) and HEMA2 (hma2) genes on the Arabidopsis chromosome I. The distance (in centiMorgans [cM]) and the markers nearest to these genes are shown. Numbers in parentheses indicate the marker identification.
Generation and Characterization of Antisense Plants
Infection of Arabidopsis hypocotyls with the A. tumefaciens strain EHA101 harboring the binary pCIT20-HEMA antisense construct resulted in the growth of several hygromycin-resistant shoots. Plantlets from these shoots were generated by transferring onto root-inducing medium followed by growth-inducing medium, as described previously (Akama et al., 1992). Similarly, seeds obtained from Arabidopsis plants vacuum infiltrated with transformed A. tumefaciens were plated on Murashige and Skoog medium containing Suc (3%, w/v) and hygromycin (20 μg/mL) to select hygromycin-resistant growth. The transformation efficiency by this protocol was found to be less than 0.1%. However, all the transgenic plants characterized in this study are derived from hypocotyl infection. From a total of 25 transformants that were established for hygromycin-resistant growth, four lines (2-1B2, 2-10PG1, 2-10PG2, and 17-10) exhibiting varying degrees of chlorophyll deficiency were selected for analysis. The phenotypes of these plants are shown in Figure 2.
Figure 2.
Phenotypes of the selected 3-week-old transgenic Arabidopsis plants. A control plant and four transgenic plants (2-1B2, 2-10PG1, 2-10PG2, and 17-10) representing four different lines exhibiting varying degrees of chlorophyll deficiency are shown. The 2-B12 line represents the severe phenotype and the 17-10 line shows negligible loss of chlorophyll. The other lines (2-10PG1 and 2-10PG2) show intermediary effects of antisense HEMA RNA expression.
Correlation of Chlorophyll and Heme Content
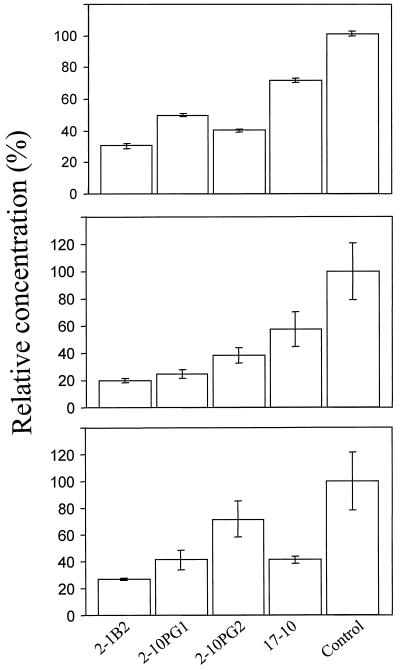
As a first step to demonstrate the effect of the expression of antisense HEMA1 RNA, the levels of chlorophyll and non-covalently bound hemes were measured in the selected four transformants. All of the transgenic plants showed reduced levels of total chlorophyll (23%–82%) compared with the control plants (Fig. 3). Similarly, protoheme content was also lowered (22%–60%) in these plants (Fig. 3). We observed that the levels of chlorophyll and protoheme were proportionally decreased.
Figure 3.
Chlorophyll (top), protoheme (middle), and ALA (bottom) levels in 2-week-old Arabidopsis transgenic lines. The amounts are expressed relative to the amounts present in control plants (taken as 100%). Data are the means of three replicates; ses are shown.
The dithionite-reduced pyridine homochrome spectra showed two absorption peaks (data not shown) around 418 and 557 nm. The authentic protoheme when processed similarly also gave two peaks at 418 and 557 nm. Furthermore, it was previously reported that the protoheme purified from the maize seedlings showed two peaks at 418 and 557 nm (Schneegurt and Beale, 1986). Therefore, it was concluded that the protocol used in the present study measures mostly protoheme. However, heme a could not be measured satisfactorily as we did not detect peaks at 429 nm and 588 nm as shown previously (Schneegurt and Beale, 1986).
Reduced ALA Levels in the Transformants
To determine if the decreased concentrations of chlorophyll and non-covalently bound hemes in the transgenic plants reflect the amount of ALA synthesis, we measured the level of ALA. Plants treated with levulinic acid showed detectable levels of ALA. As in the case of chlorophyll and hemes, plant line 2-1B2 accumulated the least amount of ALA. The levels of ALA in the tested plants ranged from 21% to 56% compared with the control (Fig. 3).
Nucleic Acid Analysis of Transgenic Plants
DNA Analysis
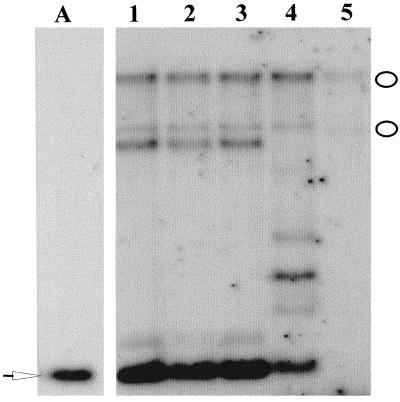
Based on the restriction map of the pCIT20-HEMA antisense construct it was expected that digestion with BamHI would release a 600-bp fragment from the transgenic plant DNA and not from the control (Fig. 4). However, such an analysis with the transgenic plant DNA yielded not only the 600-bp fragment but also multiple fragments, suggesting that one or multiple copies of the antisense HEMA1 gene were integrated into the different locations of the chromosome. The Southern-blot analysis also showed hybridization signals in all the transgenic lines that coincided with the BamHI restriction pattern in the control plant (Fig. 4). This result implied that the native HEMA gene in the transgenic lines is intact and the phenotype in these plants did not arise due to the disruption of the wild-type gene.
Figure 4.
DNA analysis of the transgenic Arabidopsis lines. The BamHI-digested restriction fragments from the pCIT20-HEMA antisense construct (lane A) and from the Arabidopsis plants (lanes 1, 2-1B2; 2, 2-10PG1; 3, 2-10PG2; 4, 17-10; and 5, control plant) were size-fractionated by agarose electrophoresis and probed with the [α-32P]dATP labeled 600-bp NheI HEMA fragment. The white arrow shows the fragment in the plasmid digest and in the transgenic lines, indicating the integration of the fragment from the construct into the genome of the transgenic plants. The white ovals show the positions of the hybridization fragments in the control plant and in the transgenic plants, suggesting that the native HEMA gene in these transgenic plants is not disrupted by the transformation.
RNA Analysis
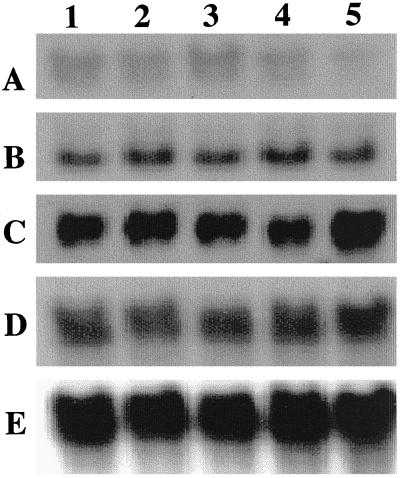
To examine if the observed ALA, chlorophyll, and heme levels of the selected transgenic Arabidopsis plants were due to the inhibition of HEMA mRNA expression by antisense mRNA, the steady-state HEMA mRNA, and the antisense HEMA1 mRNA levels were analyzed. All transgenic lines showed an equal amount of the endogenous HEMA mRNA levels. However, these lines expressed different levels of antisense mRNA (Fig. 5). The transgenic line expressing the severe phenotype (2-1B2) contained an almost 4-fold elevated level of antisense transcript. Interestingly, the transgenic line (17-10) that did not exhibit phenotypic variation showed a 2-fold increase of antisense transcript compared with the control plant. Thus, there is no strict correlation of the antisense transcript level (as measured by northern blot) with the observed phenotype. The GSA gene transcript levels were similar in all the transgenic lines.
Figure 5.
RNA analysis of the transgenic Arabidopsis lines expressing antisense HEMA1 RNA. Total RNA (20 μg) was isolated from 2-week-old Arabidopsis plants and blotted onto a nitrocellulose membrane as described in “Materials and Methods.” Lanes indicate the selected lines: 1, 2-1B2; 2, 2-10PG1; 3, 2-10PG2; 4, 17-10; and 5, control plant. Probes are HEMA1 cDNA sense strand (A), HEMA1 cDNA antisense strand (B), GSA cDNA (C), small subunit of Rubisco (D), and 18S rDNA (E).
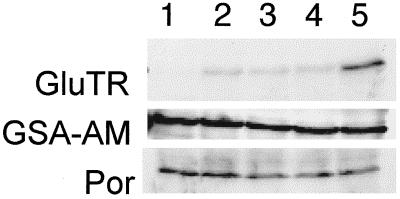
Antisense HEMA1 RNA Expression Lowers GluTR Levels
The GluTR protein level in the extracts was monitored by western-blot analysis using polyclonal anti-GluTR antibodies. These antibodies recognized a 53-kD protein; in the transgenic plants the amount GluTR protein was reduced (Fig. 6). The transgenic line with the most severe phenotype (2-1B2) contained only 1% of the amount found in control plants. To determine if this reduction is specific to the HEMA gene product GluTR, we analyzed the expression of Glu-1-semialdehyde-2,1-aminomutase and protochlorophyllide oxidoreductase by western blots using respective antibodies. We observed that all transgenic lines expressed GSA-AM and Por proteins as much as in they were expressed in control plants. This indicates that the HEMA antisense RNA specifically suppresses the GluTR expression.
Figure 6.
Western-blot analysis of the transgenic Arabidopsis lines expressing antisense HEMA1 RNA. Lane 1, 2-1B2; lane 2, 2-10PG1; lane 3, 2-10PG2; lane 4, 17-10; and lane 5, control plant.
Results obtained by this analysis establish that the expression of antisense HEMA1 RNA suppresses the formation of ALA, resulting in the reduction of chlorophyll and heme levels in Arabidopsis. This is the first direct demonstration of the vital role of GluTR expression in higher plants.
DISCUSSION
The C5-Pathway Is Indispensable for Plant Survival
Transgenic Arabidopsis plants expressing antisense HEMA1 mRNA were generated to monitor the significance of the HEMA gene in the formation of ALA. The coding region of the HEMA1 gene was used to construct the antisense mRNA expression vector. As the HEMA1 and HEMA2 genes have extensive similarity at the nucleotide level in the coding region, an antisense construct made from either HEMA1 cDNA was expected to block the translation of both HEMA1 and HEMA2 mRNAs. Some of the transgenics showed chlorophyll deficiency, ranging from patchy yellow to total yellow. We also observed that plantlets that completely lacked chlorophyll failed to survive under the growth conditions described. Attempts to rescue these plants by growing them in either low light or supplementing the medium with varying concentrations of ALA failed. An earlier study carried out with transgenic tobacco plants expressing antisense GSA mRNA also showed that plants with reduced chlorophyll levels failed to survive (Höfgen et al., 1994). These observations suggest that suppression of the enzymes of the C5 pathway affect the growth of the plant. In addition, an analysis of chlorophyll-deficient Arabidopsis mutants revealed plants with lesions in steps of chlorophyll synthesis beyond the point of ALA formation (Runge et al., 1995); the fact that none were found in the C5 pathway underscores the vital nature of ALA biosynthesis in plants.
The Antisense HEMA1 Phenotype Is Variable
All of the selected transgenic plants expressed different levels of antisense HEMA1 mRNA. The antisense RNA is thought to form a duplex with the endogenous HEMA mRNA and thus to prevent gene expression. As these duplexes are substrates for double-strand-specific RNases, the detection of either sense or antisense mRNAs in transgenic plants is difficult (e.g. Zrenner et al., 1993; Höfgen et al., 1994). However, we detected both antisense and sense mRNAs in the transgenic plants, suggesting that this was either due to sluggish double-stranded RNA-specific RNase activity or a rapid production of the sense and antisense mRNAs that surpasses the RNase activity. The transgenic plant with the weakest phenotype (17-10) was found to contain several copies of the antisense gene (Fig. 4). Nevertheless, the amount of antisense transcript present in this line was not proportional. This phenomenon has been observed in several instances and could be due to silencing phenomena or position effects (Tabler, 1993). On the other hand, although line 2-1B2 showed fewer copies of antisense HEMA1 (as judged by Southern blots), the severe phenotype observed was due to the expression of significant amounts of HEMA antisense RNA reducing the GluTR level (Fig. 6).
While probing the antisense transcripts with the sense RNA probe, we also noticed a hybridization signal in the control plants. This hybridization is either from the minute quantities of antisense probes that are made by aberrant priming process during the probe preparation (Sambrook et al., 1989) or, more interestingly, it may due to the naturally occurring antisense RNA in the wild-type plants. Such complementary RNA species, although not widely reported to be involved in gene regulation in plants, have been described; e.g. there is evidence for naturally occurring antisense RNA of an α-amylase gene in barley (Rogers, 1988).
While there is a slight difference between RNA and protein expression values, it is most satisfying to see that the protein levels correlate with the severity of the antisense plant phenotype.
Does Heme Regulate the ALA Synthesis at the Level of GluTR?
Based on the biochemical analysis it was postulated that plants contain two pools of ALA with distinct regulation, phytochrome or feedback regulated (Huang et al., 1990). In all plants studied so far, expression of at least one of the HEMA and GSA genes is known to respond to light, whereas the other members of these gene families are not responsive to light (Kumar et al., 1996b). It is not clear if expression of this set of HEMA and GSA genes is regulated by a feedback mechanism. The end product, heme, exhibited such a feedback inhibition on ALA biosynthesis (Beale, 1978). Lowering heme levels by iron chelators increased ALA accumulation (Duggan and Gassman, 1974). Of the two enzymes of the C5 pathway, we believe that heme may not have a regulatory role on GSA-AT protein as our results in the present study show that decreased content of non-covalently bound hemes failed elevate the GSA-AT levels (Figs. 5 and 6). While this observation suggests that GluTR probably is the site of heme action, there is no conclusive evidence except an in vitro study in which GluTR activity was shown to be affected by heme (Huang and Wang, 1986). A direct proof would be the examination of the GluTR levels in antisense GSA transgenics or in mutant plants that synthesize suboptimal levels of heme.
Chlorophyll Deficiency in Antisense HEMA1 Transgenics Is Not Due to Lowered Protochlorophyllide Oxidoreductase Levels
NADPH-dependent protochlorophyllide oxidoreductase catalyzes the reduction of protochlorophyllide to chlorophyllide in a light-dependent manner. Similar to HEMA and GSA genes, a set of POR genes (PORA and PORB) is involved in chlorophyll biosynthesis (Armstrong et al., 1995; Holtorf et al., 1995). Transfer of light-grown plants to the dark causes accumulation of Pchlide and decline of ALA synthesis. In light, however, Pchlide is reduced and ALA synthesis increases. The inverse levels of ALA and Pchlide in light and dark suggested the possible existence of a regulatory circuit between ALA and Pchlide synthesis (Beale, 1978). In the present study we observed that the anti-Por antibodies recognized a 36-kD protein in all the Arabidopsis lines including the control plants. While PORA mRNA disappears within a few hours of illumination (Armstrong et al., 1995), the expression of PORB proceeds throughout the greening process; thus, the detected signal was assumed to be the PorB protein. The constant levels of PorB suggested that the chlorophyll deficiency in the transgenic plants was primarily due to insufficient levels of Pchlide that had resulted from decreased ALA synthesis.
Is the C5-Pathway the Sole Source for ALA in Plants?
While the existence of the C5 pathway is widely accepted, other pathways of ALA formation in plants have been reported (Ramaswamy and Nair, 1973; Meller and Gassman, 1982). Efforts to thoroughly characterize these “alternative ALA-forming pathways” have been unsuccessful. More convincing yet indirect evidence for an extraplastidic ALA synthesis was obtained with a barley albina mutant (Hess et al., 1991, 1992). The lack of detectable levels of chloroplastic tRNAGlu, and the presence of minute amounts of Pchlide, chlorophyll, and heme in these mutants indicated an additional pathway for tetrapyrrole biosynthesis.
We show that the levels of protoheme are decreased in the transgenic plants expressing antisense HEMA mRNA. On the other hand, a decrease (more than 50%) in absorption units at 588 nm was noticed in the severe phenotype (compared with the control); however, heme a was not detected as no heme a specific peaks were noticed in the absorption spectra. Nevertheless, radiotracer studies conducted with maize seedlings demonstrated that all of the cellular hemes including protoheme and the heme a (the mitochondrial heme, a marker for alternative pathway for tetrapyrrole biosynthesis) were made from ALA that is derived from C5 pathway (Schneegurt and Beale, 1986). Furthermore, lethal effect produced in some lines by antisense HEMA1 (present study) and antisense GSA (Höfgen et al., 1994) favors the C5 pathway as a sole source for ALA biosynthesis.
ACKNOWLEDGMENTS
We thank Larry Ilag for help, Carl Simmons for suggestions and critical comments regarding the manuscript, Xing-Wang Deng (Yale University) for various plasmids, Gregory Armstrong and Klaus Apel (Eidgenossiche Technische Hochschule, Zurich) for PORA and PORB probes and anti-Por antibodies, and Michael Timko (University of Virginia, Charlottesville) for anti-Por antibodies. We are indebted to Clare Lister and Caroline Dean (John Innes Centre, Norwich, UK) for mapping the HEMA1 and HEMA2 genes.
Footnotes
This work was supported by the Department of Energy (grant no. DE–FG02–87ER13734).
LITERATURE CITED
- Akama K, Shiraishi H, Ohta S, Nakamura K, Okada K, Shimura Y. Efficient transformation of Arabidopsis thaliana: comparison of efficiencies with various organs, plant ecotypes and Agrobacterium strains. Plant Cell Rep. 1992;12:7–11. doi: 10.1007/BF00232413. [DOI] [PubMed] [Google Scholar]
- Armstrong GA, Runge S, Frick G, Sperling U, Apel K. Identification of NADPH:protochlorophyllide oxidoreductases A and B: a branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol. 1995;108:1505–1517. doi: 10.1104/pp.108.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale SI. δ-Aminolevulinic acid in plants: its biosynthesis, regulation, and role in plastid development. Annu Rev Plant Physiol. 1978;29:95–120. [Google Scholar]
- Berthold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Duggan J, Gassman M. Induction of porphyrin synthesis in etiolated bean leaves by chelators of iron. Plant Physiol. 1974;53:206–215. doi: 10.1104/pp.53.2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint DH. Gabaculine inhibits δ-ALA synthesis in chloroplasts (abstract no. S-965) Plant Physiol. 1984;75:170. [Google Scholar]
- Grimm B. Novel insights in the control of tetrapyrrole metabolism of higher plants. Curr Opin Plant Biol. 1998;1:245–250. doi: 10.1016/s1369-5266(98)80112-x. [DOI] [PubMed] [Google Scholar]
- Hess WR, Schendel R, Börner T, Rüdiger W. Reduction of mRNA levels for two nuclear encoded light-regulated genes in barley mutant albostrians is not correlated with phytochrome content and activity. J Plant Physiol. 1991;138:292–298. [Google Scholar]
- Hess WR, Schendel R, Rüdiger W, Fieder B, Börner T. Components of chlorophyll biosynthesis in a barley albina mutant unable to synthesize δ-aminolevulinic acid by utilizing the transfer RNA for glutamic acid. Planta. 1992;188:19–27. doi: 10.1007/BF00198935. [DOI] [PubMed] [Google Scholar]
- Höfgen R, Axelsen KB, Kannangara CG, Schüttke I, Pohlenz HD, Willmitzer L, Grimm B, von Wettstein D. A visible marker for antisense mRNA expression in plants: inhibition of chlorophyll synthesis with glutamate-1-semialdehyde aminotransferase antisense gene. Proc Natl Acad Sci USA. 1994;91:1726–1730. doi: 10.1073/pnas.91.5.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtorf H, Reinbothe S, Reinbothe C, Bereza B, Apel K. Two routes of chlorophyllide synthesis that are differentially regulated by light in barley (Hordeum vulgare L.) Proc Natl Acad Sci USA. 1995;92:3254–3258. doi: 10.1073/pnas.92.8.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DD, Wang WY. Chlorophyll biosynthesis in Chlamydomonas starts with the formation of glutamyl-tRNA. J Biol Chem. 1986;261:13451–13455. [PubMed] [Google Scholar]
- Huang L, Bonner BA, Castelfranco PA. Regulation of 5-aminolevulinic acid (ALA) synthesis in developing chloroplasts. Plant Physiol. 1990;92:172–178. doi: 10.1104/pp.92.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilag LL, Kumar AM, Söll D. Light regulation of chlorophyll biosynthesis at the level of 5-aminolevulinate formation in Arabidopsis. Plant Cell. 1994;6:265–275. doi: 10.1105/tpc.6.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AM, Csankovszki G, Söll D. A second and differentially expressed glutamyl-tRNA reductase gene from Arabidopsis thaliana. Plant Mol Biol. 1996a;30:419–426. doi: 10.1007/BF00049321. [DOI] [PubMed] [Google Scholar]
- Kumar AM, Schaub U, Söll D, Ujwal M. Glutamyl-transfer RNA: at the crossroad of chlorophyll and protein biosynthesis. Trends Plant Sci. 1996b;1:371–376. [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotinoids: pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- May BK, Bhasker CR, Bawden MJ, Cox TC. Molecular regulation of 5-aminolevulinate synthase. Mol Biol Med. 1990;7:405–421. [PubMed] [Google Scholar]
- Meller E, Gassman ML. Biosynthesis of 5-aminolevulinic acid: two pathways in higher plants. Plant Sci Lett. 1982;26:23–29. [Google Scholar]
- Ramaswamy NK, Nair PM. δ-Aminolevulinate synthetase from cold-stored potatoes. Biochim Biophys Acta. 1973;293:269–277. doi: 10.1016/0005-2744(73)90399-9. [DOI] [PubMed] [Google Scholar]
- Rogers JC. RNA complementary to α-amylase mRNA in barley. Plant Mol Biol. 1988;11:125–138. doi: 10.1007/BF00015665. [DOI] [PubMed] [Google Scholar]
- Runge S, van Cleve B, Lebedev N, Armstrong G, Apel K. Isolation and classification of chlorophyll-deficient xantha mutants of Arabidopsis thaliana. Planta. 1995;197:490–500. doi: 10.1007/BF00196671. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sangwan I, O'Brian MR. Expression of a soybean gene encoding the tetrapyrrole-synthesis enzyme glutamyl-tRNA reductase in symbiotic root nodules. Plant Physiol. 1999;119:593–598. doi: 10.1104/pp.119.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneegurt MA, Beale SI. Biosynthesis of protoheme and Hema a from glutamate in maize. Plant Physiol. 1986;81:965–971. doi: 10.1104/pp.81.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön A, Krupp G, Gough S, Berry-Lowe S, Kannangara CG, Söll D. The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature. 1986;322:281–284. doi: 10.1038/322281a0. [DOI] [PubMed] [Google Scholar]
- Stillman LC, Gassman ML. Protoheme extraction from plant tissue. Anal Biochem. 1978;91:166–172. doi: 10.1016/0003-2697(78)90827-8. [DOI] [PubMed] [Google Scholar]
- Tabler M. Antisense RNA in plants: a tool for analysis and suppression of gene function. In: Roubelakis-Angelakis KA, Van KTT, editors. Morphogenesis in Plants. New York: Plenum Press; 1993. pp. 237–258. [Google Scholar]
- Tanaka R, Yoshida K, Nakayashiki T, Masuda T, Tsuji H, Inokuchi H, Tanaka A. Differential expression of two HEMA mRNAs encoding glutamyl-tRNA reductase proteins in greening cucumber seedlings. Plant Physiol. 1996;110:1223–1230. doi: 10.1104/pp.110.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen B, Oelze-Karow H, Schuster C, Mohr H. Stimulation of appearance of extraplastidic tetrapyrroles by photooxidative treatment of plastids. Photochem Photobiol. 1993;58:711–717. [Google Scholar]
- von Wettstein D, Gough S, Kannangara CG. Chlorophyll biosynthesis. Plant Cell. 1995;7:1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JD, Beale SI. Biosynthesis of protoheme and heme a precursors solely from Glu in the unicellular red alga Cyanidium caldarium. Plant Physiol. 1984;74:146–151. doi: 10.1104/pp.74.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JD, Beale SI. Enzymatic conversion of Glu to 5-aminolevulinate in soluble extracts of the unicellular alga Chlorella vulgaris. Arch Biochem Biophys. 1985;237:454–464. doi: 10.1016/0003-9861(85)90299-1. [DOI] [PubMed] [Google Scholar]
- Zrenner R, Willmitzer L, Sonnewald U. Analysis of the expression of potato uridine diphosphate-Glc pyrophosphorylase and its inhibition by antisense RNA. Planta. 1993;190:247–252. doi: 10.1007/BF00196618. [DOI] [PubMed] [Google Scholar]