Abstract
Age-related loss of muscle mass and function is associated with increased frailty and loss of independence. The mechanisms underlying the susceptibility of different muscle types to age-related atrophy are not fully understood. Reactive oxygen species (ROS) are recognised as important signalling molecules in healthy muscle and redox sensitive proteins can respond to intracellular changes in ROS concentrations modifying reactive thiol groups on Cysteine (Cys) residues. Conserved Cys residues tend to occur in functionally important locations and can have a direct impact on protein function through modifications at the active site or determining protein conformation. The aim of this work was to determine age-related changes in the redox proteome of two metabolically distinct murine skeletal muscles, the quadriceps a predominantly glycolytic muscle and the soleus which contains a higher proportion of mitochondria. To examine the effects of aging on the global proteome and the oxidation state of individual redox sensitive Cys residues, we employed a label free proteomics approach including a differential labelling of reduced and reversibly oxidised Cys residues. Our results indicate the proteomic response to aging is dependent on muscle type but redox changes that occur primarily in metabolic and cytoskeletal proteins are generally preserved between metabolically distinct tissues.
Biological significance
Skeletal muscle containing fast twitch glycolytic fibres are more susceptible to age related atrophy compared to muscles with higher proportions of oxidative slow twitch fibres. Contracting skeletal muscle generates reactive oxygen species that are required for correct signalling and adaptation to exercise and it is also known that the intracellular redox environment changes with age. To identify potential mechanisms for the distinct response to age, this article combines a global proteomic approach and a differential labelling of reduced and reversibly oxidised Cysteine residues in two metabolically distinct skeletal muscles, quadriceps and soleus, from adult and old mice. Our results indicate that the global proteomic changes with age in skeletal muscles are dependent on fibre type. However, redox specific changes are preserved across muscle types and accompanied with a reduction in the number of redox sensitive Cysteine residues.
Keywords: Skeletal muscle, Aging, Redox proteomics, Cysteine oxidation, Sarcopenia, skNAC
Graphical abstract

Highlights
-
•
Skeletal muscle fast twitch fibres are particularly susceptible to age-related atrophy.
-
•
Proteomic changes with age in skeletal muscles are dependent on fibre type.
-
•
Redox specific changes are preserved across fibres with different susceptibility to age-related atrophy.
-
•
Decreased number of redox sensitive Cysteine residues detected with age in all muscle fibres.
1. Introduction
Skeletal muscle is the largest organ in the body accounting for up to 50% of total body mass. The age-related loss of skeletal muscle mass and function can have a major impact on quality of life and result in loss of independence and an increase in frailty. Skeletal muscle is composed of a mixture of fibre types that can be broadly categorised into fast and slow twitch fibres depending on their myosin heavy chain content. Quadriceps muscle is generally referred to as a fast twitch muscle that primarily produces ATP via glycolysis [1]. The soleus, on the other hand, predominantly uses mitochondrial dependent oxidative phosphorylation via the electron transport chain for ATP generation. In mice the quadriceps is considered a glycolytic muscle and in common with similar muscles such as the tibialis anterior, has a high proportion of type IIB fibres (~60%) while the soleus has a higher proportion of oxidative Type I (~37%) and IIA (~38%) fibres [2]. Human and rodent studies have revealed that fast twitch fibres such as the quadriceps are more susceptible to age-related atrophy than their slow-twitch counterparts such as soleus [[3], [4], [5]]. Nevertheless age-related loss of mitochondrial content and altered mitochondrial morphology has been reported in all skeletal muscle [[6], [7], [8], [9]].
Reactive oxygen species (ROS) are involved in the degeneration of muscle with age but it is unclear how this affects different fibre types [10]. In skeletal muscle there are a number of potential sites for ROS generation including electron leakage from the electron transport chain and endogenous generators of ROS species such as NADPH oxidase (NOX) enzymes that are activated by muscle contractions [[11], [12], [13]]. Although ROS were traditionally thought to be responsible for oxidative damage more recent evidence would suggest that they also play a crucial signalling role within the cell that is dependent on the location, species and concentration of ROS generated [14]. A number of recent studies have identified a beneficial role of exercise induced ROS in redox signalling that is required for the adaptation and repair of injured skeletal muscle [15,16]. ROS are primarily thought to mediate their signalling activities through the selective oxidation/reduction of specific Cysteine (Cys) residues on target proteins. Cys residues have a functionally important thiol group which generally has a low mean pKa and exist as nucleophiles at physiological pH, although it is dependent on the local environment which can impact Cys reactivity [17]. Reversible oxidation of Cys residues into sulfenic acids (-SOH) and subsequent formation of disulfide bonds (-S-S-) between thiol groups can potentially alter the functional role of sensitive proteins by changing their conformation and affecting their activity or metal ion binding [18,19]. Further oxidation of thiols via sustained exposure to ROS can lead to the formation of sulfinic (-SO2H) and sulfonic acids (-SO3H). Reversible modifications of Cys residues enable these residues to participate in local redox signalling events in response to ROS, however a sustained increase in ROS over time can potentially lead to irreversible modifications.
Skeletal muscle from old mice have a blunted response to the adaptations induced by exercise in part due to an altered intracellular redox environment [20]. A chronically elevated intracellular ROS environment would modify redox sensitive proteins in skeletal muscle affecting redox signalling and the adaptive response to exercise. In this study we have utilised a global label free and redox proteomic approach [21] to identify the age related redox proteomic changes in two skeletal muscle types that differ in their primary sources of energy for contraction, quadriceps and soleus from adult and old mice. The redox state of individual Cys residues was calculated using the parent ion intensities of Cys containing peptides labelled with either a light (reduced Cys residues and labelled with d0 N-Ethylmaleimide (NEM)) or heavy (reversibly oxidised labelled Cys residues and labelled with d5 NEM) alkylating agent. Our results demonstrate that the majority of proteins identified between two metabolically distinct muscles was consistent but the effects of age on the protein composition of the two muscle groups was very different. In glycolytic quadriceps tissue there was an increase in cytoskeletal proteins with age and in oxidative soleus a decrease in mitochondrial related proteins. In contrast redox analysis of Cys residues indicated that in both tissues the number of Cys residues labelled as reduced and reversibly oxidised decreased with age, but that the core redox sensitive proteins involved in metabolic processes are preserved between muscle types.
2. Materials and methods
All reagents and chemicals unless otherwise stated were obtained from Sigma Aldrich (Dorset, UK) and were of analytical grade or above.
2.1. Animals
Adult (12 months) and old (25 months) C57BL/6 male mice were purchased from Charles River and housed in the Specific Pathogen-Free (SPF) Facility at the University of Liverpool for at least 2 weeks before use. Experiments were performed in accordance with U.K. Home Office Guidelines under the U.K. Animals (Scientific Procedures) Act 1986 and received ethical approval from the University of Liverpool Animal Welfare and Ethical Review Board. Body and muscle weights were compared using a two tailed Student's t-test. Animals were sacrificed by cervical dislocation, white glycolytic quadriceps tissue and whole soleus muscle were dissected and placed immediately in thiol blocking buffer containing 25 mM d0 N-Ethylmalemide (NEM) and 50 mM ammonium bicarbonate, pH 8, for redox proteomic analysis. The contralateral muscles were dissected, orientated transversely, mounted on a cork block and frozen in optimal cutting temperature compound (OCT) for subsequent histological examination.
2.2. Succinate dehydrogenase staining
Dissected skeletal muscles (quadriceps or soleus at either 12 or 25 months of age), that were transversely orientated in OCT were moved from −80 °C storage to the Cryostat (Leica, Milton Keynes, UK) to acclimatise to −20 °C. Cryo sections were cut at a thickness of 10 μm and stained for succinate dehydrogenase (SDH) activity as described [22]. Histological analysis was performed on a Zeiss Axiovert 200M microscope connected to a PC running Windows XP and Axiovision 4.8.2 (SP3 08-2013). Images were taken using Axiocam ICc5 under bright field conditions at 10×/0.30 magnification for an exposure time of 2 ms giving a 2452 × 2056 pixel image with scale factors of x 0.55 μm/pixel, y 0.55 μm/pixel and z 1.00 pixel/pixel. The % of stained fibres was calculated using a modified 3-point scale: 0 - unstained fibres; 1 – lightly stained fibres, 2 – dark stained fibres, for the semi quantification of SDH staining intensity as described previously [23]. The data shows % unstained/stained fibres of total fibres and analysed by two-way ANOVA followed by Bonferroni multiple comparison test (95% Confidence Interval), a P-value <0.05 was considered as statistically significant * p < 0.05; ** p < 0.01; *** p < 0.01. Statistical analysis was performed using GraphPad Prism version 5.01 for Windows (GraphPad Software, La Jolla California USA). All visible fibres in the section of the muscle were calculated using N = 3 biological replicates. Total SDH staining intensity was measured by mean pixel density using ImageJ software (National Institute of Health (Bethesda, USA)) from grayscale images manually traced at muscle boundaries as described previously [24].
2.3. Redox proteomics sample preparation
Sample preparation was performed as previously described [21], briefly each muscle (N = 5 for soleus tissue from adult and old mice and quadriceps tissue from old mice, N = 6 for quadriceps tissue from adult mice) was dissected and immediately placed in d0 NEM blocking buffer to prevent Cys oxidation. Tissues were homogenised in ice cold buffer (25 mM Ammonium bicarbonate containing 25 mM NEM and 0.1% Rapigest (Waters, Manchester, UK) pH 8) using a hand homogeniser. Samples were passed through a Zeba desalting column 7K MWCO (Thermo Scientific, Hempstead, UK) to remove excess d0 NEM and protein concentrations were calculated using the Bradford (BioRad, Hertfordshire, UK) method using BSA as a standard. 100 μg of protein was aliquoted and reversibly oxidised Cys residues were reduced using tris (2-carboxyethyl) phosphine (TCEP) at a final concentration of 10 mM, newly reduced Cys residues were subsequently labelled with d5 NEM final concentration 20 mM. Protein extracts were digested overnight at 37 °C with trypsin, Rapigest was precipitated by addition of trifluoroacetic acid (TFA) and subsequent centrifugation before analysis by MS.
2.4. Lc-MS/MS and label free quantification
Proteomics was performed using an Ultimate 3000 RSLC Nano system (Thermo Scientific) coupled to a Q-Exactive mass spectrometer (Thermo Scientific) as previously described. [21]. Detection of the peptides was performed by data dependent acquisition (DDA) which takes a select number of peptide peaks from the initial scan according to a rule set and the corresponding ions are then verified against this initial set via tandem mass spectrometry (MS/MS).
Raw spectra were converted to Mascot Generated Files (.mgf) using the Proteome Discoverer software (Thermo Scientific). The .mgf files were searched against the Uniprot mouse sequence database (Mus musculus database from 12th May 2012; 16,376 sequences) via an in-house Mascot database server (Matrix Science, London, UK). The search parameters were: peptide mass tolerances, 10 ppm; fragment mass tolerance, 0.01 Da, 1+, 2+ and 3+ ions; missed cleavages, 1; instrument type, ESI-TRAP. Variable modifications were included as: d0-NEM, d5-NEM, mono-, di-, and tri-oxidation of Cys residues and oxidation of methionine.
Label-free relative quantification software PEAKS7 (Bioinformatics Solutions Inc., Waterloo, Canada) was used to analyse RAW data files against the same mouse protein database for identifications with Mascot [25]. Normalisation was carried out using the total ion current and proteins were considered significantly changed between adult and old mouse protein samples using the following criteria: at least 3 unique peptide, −10 log P score of ≥20 (equivalent to a p value of 0.01), a fold change ≥1.5 and using a quality value of 0.8. PEAKS7 software includes a post-translational modification (PTM) algorithm applying the de novo sequencing module to search for a limited number of PTMs. All identified PTMs using this method adhere to the above search criteria and FDR validation.
Data visualization of significant changes from the label free proteomic data (max fold change ≥ 1.5 and –10logP ≥ 20) was performed using Perseus software and z-scoring of quantitative data for Heatmaps [26]. Label free data of all identified proteins was visualised using Volcano plots using Log2 (Fold Change) plotted against significance in R studio. Pathway analysis of label free quantitative proteomic data was performed using PathVisio [27] together with WikiPathways [28] to visualize and highlight altered pathways from identified proteins.
2.5. Targeted analysis of redox sensitive Cys residues
In these experimental conditions we considered Cys containing peptides that were identified as labelled with both d5-NEM and d0-NEM as redox sensitive Cys residues, reduced Cys were only labelled with d0-NEM and reversibly oxidised Cys were labelled with only d5-NEM. Redox Cys peptides were identified by detecting identical amino acid sequences containing d0 and d5 NEM independently and having a Mascot peptide score of >20. Peptides detected from Proteome Discoverer analysis of RAW files were selected for targeted analysis with the open software Skyline [29]. Targeted analysis applying m/z, retention times and fragmentation spectra for peptide selection allowed the calculation of the reduced/oxidised ratio (or d0/d5 NEM ratio) of the individual Cys residues using their parent ion intensities with Skyline [29]. For redox peptides the individual reduced/oxidised ratio for each redox Cys peptide was used to calculate an average ratio of specific Cys oxidised/reduced peptides. For identification of peptides, the result filtration parameters employed a false detection rate (FDR) of 1%, a mass error tolerance of 10.0 ppm and a retention time shift tolerance of 0.5 min. Search parameters for the MASCOT database defined peptide significance as equal to or greater than a MASCOT Score of 20.
The surrounding amino acid sequences (−6 to +6) of identified redox sensitive Cys residues identified were blasted against the Mus database to find potential common motifs in the surrounding amino acid residues using Motif-X [30]. In order to determine if particular molecular pathways were over-represented or potential protein-protein interactions between proteins identified as containing redox sensitive Cys residues String-DB was used [31].
2.6. Immunoblotting
Protein samples (20 μg) from three biological replicates were loaded on 10% or 12% gels for SDS PAGE and subsequently transferred to nitrocellulose membranes at 100 mA per gel over 1 h. Ponceau S staining of the membrane was used to insure equal protein loading. Membranes were blocked for 1 h with 3% non-fat dry milk in Tris buffered saline-tween (TBS-T) 1 h at room temperature. Membranes were incubated overnight at 4 °C with primary antibodies (HSC70 Stressgen SMC-104A, HSP70 Enzo ADI-SPA-810, PDI Abcam ab137110, SOD1 Enzo ADI-SOD-100, PRDX2 Abcam ab59539 and PRDX5 Abcam ab16944). Following washing with TBS-T membranes were incubated for 1 h with Horseradish peroxidase-conjugated secondary antibodies (anti-mouse and anti-rabbit IgG (Cell Signalling Technologies, Hitchin, UK). Peroxidase activity was detected using the enhanced chemiluminescence (ECL) kit (Amersham International, Cardiff, UK) and band intensities were analysed using ImageLab (v5; BioRad, US). Band intensities were normalised using corresponding Ponceau S stain for differences in protein loading and analysed using a two tailed Student's t-test.
3. Results
3.1. Age-related decrease in quadriceps tissue mass and alterations in SDH activity in both quadriceps and soleus
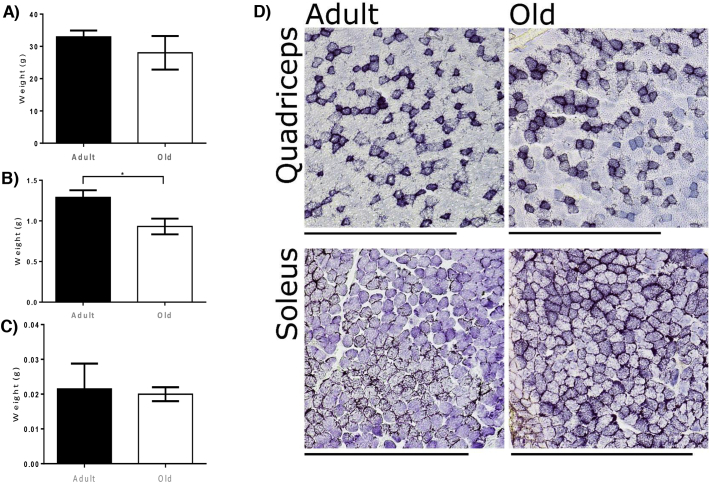
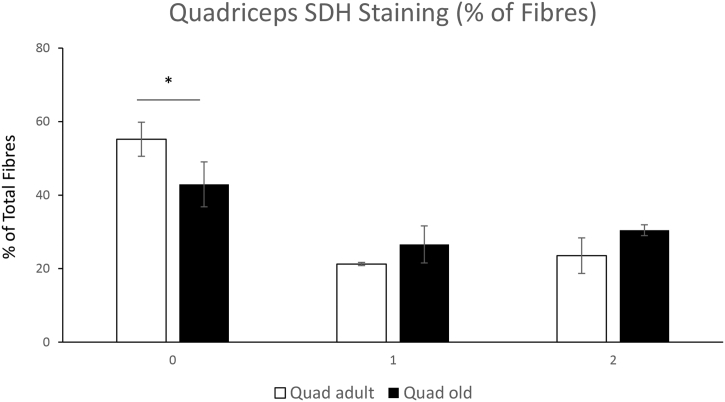
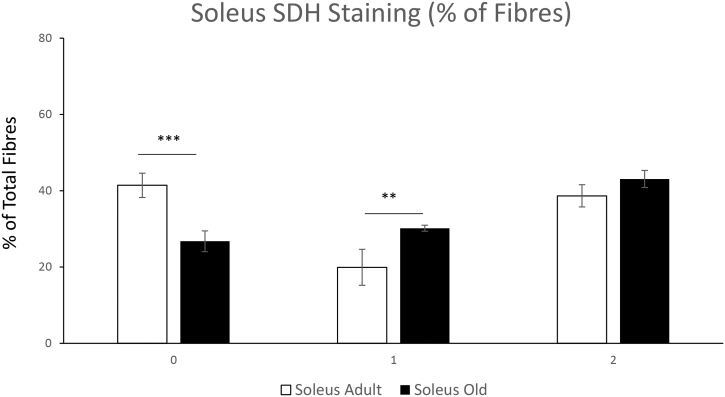
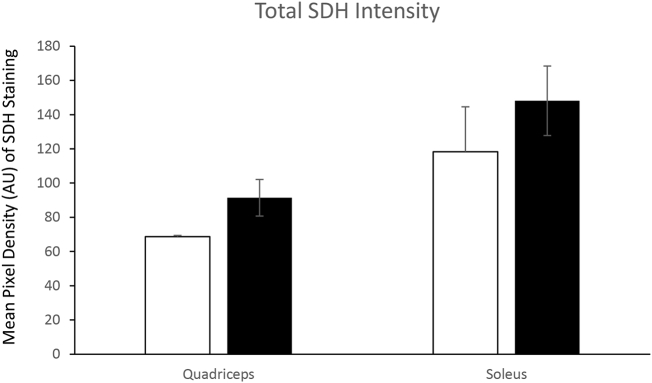
There was no significant decrease in overall body weight between adult and old mice. There was a significant reduction in quadriceps mass in old mice but no change in soleus mass Fig. 1A, B & C. In order to determine whether there was any major shifts in the proportion of oxidative fibres with age we performed SDH staining on transverse sections of the quadriceps and soleus muscles from adult and old mice. The % of stained fibres of quadriceps and soleus are presented in Supplementary Fig. 1, Supplementary Fig. 2, total SDH staining intensity is presented in Supplementary Fig. 3. These images confirm the greater proportion of oxidative fibres in soleus compared with quadriceps muscles and also that aging increased the proportion of oxidative fibres in muscle from old mice compared with those from adult mice.
Fig. 1.
Body A) and Tissue Weights of B) Quadriceps and C) Soleus, D) Representative SDH staining of sections from adult and old quadriceps and soleus muscles (Scale bar represents 500 μm).
Supplementary Fig. 1.
The % of SDH stained fibres from adult and old quadriceps tissue was calculated using a modified 3-point scale: 0 - unstained fibres; 1 – lightly stained fibres, 2 – dark stained fibres for semi quantification. The data shows % unstained/stained fibres of total fibres. All visible fibres in the section of the muscle were calculated. N = 3 biological replicates.
Supplementary Fig. 2.
The % of SDH stained fibres from adult and old quadriceps tissue was calculated using a modified 3-point scale: 0 - unstained fibres; 1 – lightly stained fibres, 2 – dark stained fibres for semi quantification. The data shows % unstained/stained fibres of total fibres. All visible fibres in the section of the muscle were calculated. N = 3 biological replicates.
Supplementary Fig. 3.
The total intensity of SDH staining of adult and old quadriceps and soleus tissue as measured by mean pixel intensity.
3.2. Label free proteomics reveal an increase in cytoskeletal proteins in quadriceps and decrease in mitochondrial proteins in soleus with age
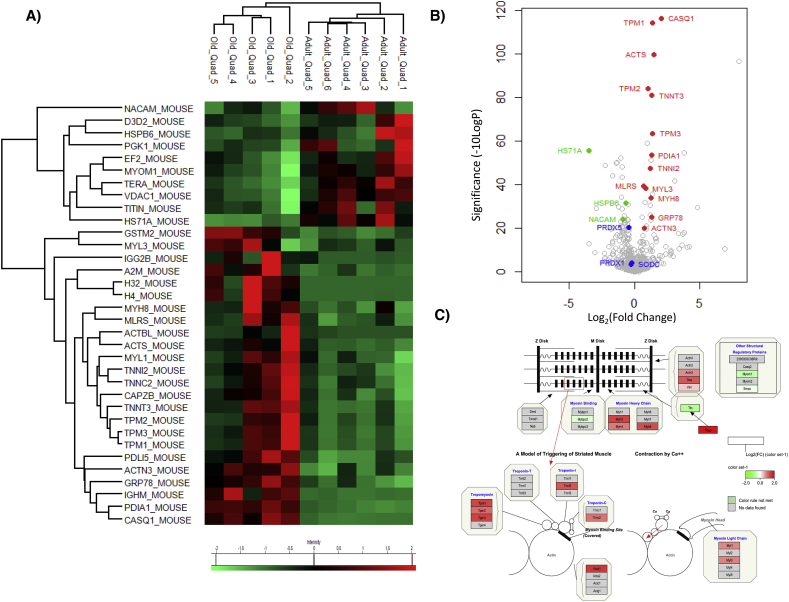
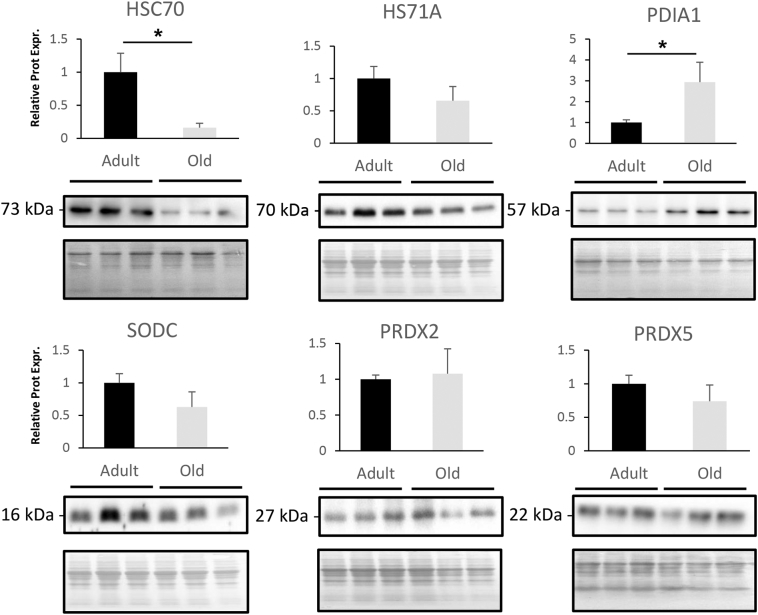
Label free proteomics was performed using Peaks7 quantification software and overall 946 proteins were quantified between the tissues from adult and old mice. Of the total number of identified proteins 434 were common to both quadriceps and soleus tissues, 205 were identified only in quadriceps and 307 only in soleus. Proteins were considered as significantly changed between adult and old mice in each tissue using the parameters P < 0.01 (−10 Log P ≥ 20), fold change ≥1.5 and with at least 3 unique peptides. The label free quantitative proteomic data from this study has been deposited in Mendeley data repository with doi:10.17632/x9wxcnc67r.1. In Quadriceps, 639 proteins were identified (Supplementary File 1 in data repository) of which 34 were considered significantly changed between adult and old mice, 23 up regulated and 11 down regulated with age (Fig. 2A and B). In quadriceps from old mice there was an increased abundance of cytoskeletal proteins including Tropomyosin (TPM1 and TPM2), Troponin (TNNI2 and TNNT3), Myosin (MYL3 and MYH8) and Actin (ACTS). There was a decrease in chaperone proteins including HS71A and HSPB6. Interestingly NACAM (Nascent polypeptide-associated complex subunit alpha, muscle-specific form or skNAC) was decreased and this protein is involved in the expression of genes in development of myotubes and assembly of thick and thin myofibril assembly [32]. Pathway analysis of the quantitative proteomic data confirmed an increase in proteins involved in the muscle contractile apparatus and cytoskeletal proteins (Fig. 2C). We selected a number of proteins that showed a significant differential expression for analysis by immunoblotting including Heat shock cognate 71 kDa (HSC70) and Protein disulfide isomerase (PDI) that indicated similar changes to the label free proteomic data, but no significant change in Heat shock protein 70 kDa (HS71A) was detected (Fig. 3). No significant changes were detected in antioxidant proteins with age using label free proteomics and immunoblotting confirmed these results for PRDX2, PRDX5 and SODC (Fig. 3) and highlighted in blue in the volanco plot of all label free proteomic data (Fig. 2B).
Fig. 2.
Label free proteomics of quadriceps from adult and old mice (A) Heatmap of significantly changed proteins (fold change >1.5 and –10logP > 20 equivalent to P < 0.01) (B) Volcano plot of quantified protein changes with age and significance, proteins highlighted in red are significantly increased with age, proteins highlighted in green are decreased with age (C) Pathway analysis of quantitative proteomic data reveals an upregulation of proteins involved in muscle contraction.
Fig. 3.
Western blot analysis and relative quantification of selected proteins from protein extracts of quadriceps protein extracts from adult and old mice (N = 3). *P < 0.05 using a two tailed unpaired student t-test. Corresponding Ponceau S stain of total protein on membranes were used as loading controls.
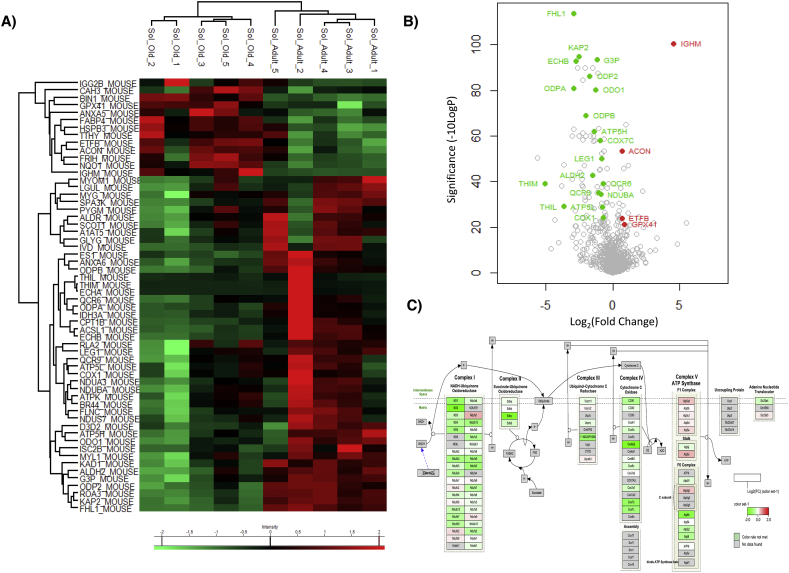
In soleus muscle from adult and old mice 741 proteins were quantified (Supplementary File 2 in data repository). There was a significant change in abundance of 58 proteins between adult and old mice, 45 proteins had decreased content and 13 proteins increased content in muscle from old mice (Fig. 4A and B). Proteins that decreased with age included a large number of mitochondrial proteins and proteins involved in the electron transport chain Fig. 4C. A reduced respiratory capacity with age has previously been demonstrated in soleus muscle [33]. Comparing the significant protein changes between both sets of tissues, 2 proteins had decreased protein content in both muscle types with age (3 2-trans-enoyl-CoA isomerase mitochondrial, Dci and Myomesin-1, Myom1) and 2 proteins had increased protein content (Ig mu chain C, Igh 6 and Ig gamma-2B chain C region, Igh 3). One protein, Myosin light chain 1/3 skeletal muscle isoform, (Myl1) had increased protein content in quadriceps and decreased protein content in soleus with age.
Fig. 4.
Label free proteomics soleus from adult and old mice, (A) Heatmap of significantly changed proteins with fold change >1.5 and –10logP > 20 equivalent to P < 0.01 (B) Volcano plot of quantified protein changes with age and significance, proteins highlighted in red are significantly increased with age and proteins highlighted in green are decreased with age. Proteins highlighted in blue were antioxidant proteins that did not significantly change in abundance and were selected for immunoblotting. (C) Pathway analysis of quantitative proteomic data using PathVisio reveals a downregulation of proteins involved in mitochondrial electron transport chain.
3.3. Age-related changes in redox state of Cys residues in metabolic and cytoskeletal proteins in both quadriceps and soleus
Cys containing peptides that were identified with high confidence (Mascot score > 20) labelled with both d0 and d5 NEM were selected for targeted analysis using the Skyline software. In the Quadriceps 75 redox Cys peptides from 45 proteins were quantified using Skyline. The list of the proteins and changes in the redox state of specific Cys residues with age are shown in Table 1, label free quantification at the protein level is included and proteins that significantly changed in abundance are in bold. In Soleus 44 redox peptides from 30 proteins were quantified using Skyline, Table 2. The changes with age in the ratio of red/rev oxidised redox state of the Cys residues for quadriceps and soleus tissues are presented in Supplementary Fig. 4, Supplementary Fig. 5.
Table 1.
Proteins detected in quadriceps tissue containing redox sensitive Cys residues including the relative quantification of oxidative state of susceptible Cys residues and changes in the redox ratio with age. Relative protein abundance is included and proteins in bold change significantly with age.
| Accession | Protein | Adult:Old | Significance (–10logP) | Peptide | Cysteine | Adult Red/Ox | Old Red/Ox | Change with age |
|---|---|---|---|---|---|---|---|---|
| P62259 | 14-3-3 Protein epsilon (Ywhae) | 1.00:1.16 | 6.06 | LICCDILDVLDK | 97/98* | 7.69 | 22.47 | 2.92 |
| P61982 | 14-3-3 Protein gamma (Ywhag) | 1.00:1.04 | 6.62 | NCSETQYESK | 112 | 14.21 | 36.02 | 2.53 |
| Q60597 | 2-Oxoglutarate dehydrogenase (Ogdh) | 1.00:1.12 | 2.15 | DVVVDLVCYR | 507 | 0.53 | 0.71 | 1.35 |
| P47857 | 6-Phosphofructokinase (Pfkm) | 1.00:0.70 | 8.36 | GITNLCVIGGDGSLTGADTFR | 114 | 5.63 | 9.68 | 1.72 |
| LPLMECVQVTK | 351 | 8.65 | 15.08 | 1.74 | ||||
| CNENYTTDFIFNLYSEEGK | 631 | 5.22 | 21.04 | 4.03 | ||||
| IFANTPDSGCVLGMR | 709 | 3.37 | 14.03 | 4.16 | ||||
| Q99KI0 | Aconitate hydratase (Aco2) | 1.00:1.00 | 1.85 | VAVPSTIHCDHLIEAQVGGEK | 126 | 61.41 | 26.17 | 0.43 |
| VGLIGSCTNSSYEDMGR | 385 | 0.66 | 1.12 | 1.70 | ||||
| DVGGIVLANACGPCIGQWDR | 448/451* | 0.13 | 0.33 | 2.55 | ||||
| CTTDHISAAGPWLK | 592 | 19.33 | 22.92 | 1.19 | ||||
| O88990 | Actin, alpha skeletal muscle (Acta1) | 1.00:2.82 | 99.81 | LCYVALDFENEMATAASSSSLEK | 218 | 4.65 | 6.16 | 1.33 |
| CDIDIR | 286 | 4.36 | 6.32 | 1.45 | ||||
| P45376 | Aldose reductase (Akr1b1) | 1.00:0.74 | 15.33 | LIEYCHSK | 200 | 0.71 | 0.81 | 1.14 |
| P05201 | Aspartate aminotransferase (Got1) | 1.00:1.10 | 4.28 | INMCGLTTK | 391 | 3.54 | 7.45 | 2.10 |
| P05202 | Aspartate aminotransferase (Got2) | 1.00:1.13 | 4.92 | EYLPIGGLAEFCK | 106 | 5.36 | 10.69 | 1.99 |
| TCGFDFSGALEDISK | 187 | 4.31 | 3.99 | 0.93 | ||||
| VGAFTVVCK | 295 | 0.16 | 0.06 | 0.36 | ||||
| Q9DCX2 | ATP synthase dubunit d mitochondrial (Atp5h) | 1.00:1.27 | 8.20 | SCAEFVSGSQLR | 101 | 51.73 | 37.60 | 0.73 |
| P45591 | Cofilin-2 (Cfl2) | 1.00:1.07 | 5.60 | LLPLNDCR | 80 | 1.90 | 13.71 | 7.21 |
| P07310 | Creatine kinase M-type (Ckm) | 1.00:1.14 | 3.01 | GYTLPPHCSR | 146 | 6.40 | 18.06 | 2.82 |
| FCVGLQK | 254 | 11.50 | 28.41 | 2.47 | ||||
| Q6P8J7 | Creatine kinase S-type (Ckmt2) | 1.00:0.95 | 5.14 | SEVELVQIVIDGVNYLVDCEK | 397 | 3.60 | 7.42 | 2.06 |
| Q9CZ13 | Cytochrome b-c1 complex subunit 1 (Uqcrc1) | 1.00:0.92 | 9.70 | LCTSATESEVTR | 380 | 1.96 | 1.60 | 0.82 |
| Q9DB77 | Cytochrome b-c1 complex subunit 2 (Uqcrc2) | 1.00:1.09 | 6.20 | NALANPLYCPDYR | 192 | 0.02 | 0.02 | 0.95 |
| O08749 | Dihydrolipoyl dehydrogenase (Dld) | 1.00:1.99 | 8.53 | NETLGGTCLNVGCIPSK | 80/85* | 71.21 | 307.42 | 4.32 |
| Q99LC5 | Electron transfer flavoprotein subunit alpha (Etfa) | 1.00:0.85 | 12.77 | LGGEVSCLVAGTK | 53 | 15.40 | 5.53 | 0.36 |
| P58252 | Elongation factor 2 (Eef2) | 1.00:0.59 | 25.73 | ETVSEESNVLCLSK | 591 | 2.86 | 6.59 | 2.30 |
| EGALCEENMR | 693 | 14.47 | 17.27 | 1.19 | ||||
| P17182 | Alpha-enolase (Enoa) | 1.00:1.32 | 1.46 | FGANAILGVSLAVCK | 119 | 2.14 | 8.36 | 3.91 |
| TGAPCR | 398 | 4.18 | 8.93 | 2.14 | ||||
| P21550 | Beta-enolase (Enob) | 1.00:0.89 | 8.60 | SGETEDTFIADLVVGLCTGQIK | 389 | 3.60 | 6.42 | 1.79 |
| P05064 | Fructose bisphosphate aldolase A (Aldoa) | 1.00:1.04 | 3.97 | YASICQQNGIVPIVEPEILPDGDHDLK | 178 | 0.53 | 2.08 | 3.91 |
| CQYVTEK | 202 | 2.50 | 6.56 | 2.62 | ||||
| ALANSLACQGK | 339 | 5.77 | 11.37 | 1.97 | ||||
| P06745 | Glucose 6-phosphate isomerase (Gpi) | 1.00:1.25 | 6.57 | MIPCDFLIPVQTQHPIR | 404 | 0.48 | 2.89 | 6.06 |
| P16858 | Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) | 1.00:1.02 | 8.18 | IVSNASCTTNCLAPLAK | 150/154* | 127.86 | 554.21 | 4.33 |
| VPTPNVSVVDLTCR | 245 | 17.90 | 26.74 | 1.49 | ||||
| Q9WUB3 | Glycogen phosphorylase (Pygm) | 1.00:0.98 | 10.01 | ICGGWQMEEADDWLR | 172 | 6.47 | 10.09 | 1.56 |
| TCAYTNHTVLPEALER | 373 | 4.81 | 10.53 | 2.19 | ||||
| WLVLCNPGLAEVIAER | 496 | 14.05 | 18.31 | 1.30 | ||||
| QLLNCLHIITLYNR | 581 | 30.97 | 12.73 | 0.41 | ||||
| P63017 | Heat shock cognate 71 kDa protein (Hspa8) | 1.00:0.79 | 11.10 | VCNPIITK | 603 | 10.88 | 8.79 | 0.81 |
| P02089 | Hemoglobin subunit beta-2 (Hbb-b2) | 1.00:1.54 | 5.99 | GTFASLSELHCDK | 94 | 100.34 | 91.44 | 0.91 |
| P06151 | l-Lactate dehydrogenase A chain (Ldha) | 1.00:0.71 | 23.73 | DYCVTANSK | 84 | 13.62 | 20.01 | 1.47 |
| VIGSGCNLDSAR | 163 | 1.33 | 4.29 | 3.23 | ||||
| P14152 | Malate dehydrogenase (Mdh1) | 1.00:1.06 | 6.17 | VIVVGNPANTNCLTASK | 137 | 15.57 | 35.51 | 2.28 |
| ENFSCLTR | 154 | 0.94 | 6.36 | 6.80 | ||||
| P08249 | Malate dehydrogenase (Mdh2) | 1.00:0.76 | 10.78 | GCDVVVIPAGVPR | 93 | 0.69 | 2.52 | 3.63 |
| TIIPLISQCTPK | 212 | 22.06 | 82.92 | 3.76 | ||||
| EGVVECSFVQSK | 275 | 2.44 | 9.84 | 4.02 | ||||
| ETECTYFSTPLLLGK | 285 | 4.35 | 9.62 | 2.21 | ||||
| Q5XKE0 | Myosin-binding protein C fast-type (Mybpc2) | 1.00:0.81 | 12.24 | IFSENICGLSDSPGVSK | 1007 | 48.64 | 77.79 | 1.60 |
| P97457 | Myosin regulatory light chain 2 (Mylpf) | 1.00:1.63 | 39.47 | QFLEELLTTQCDR | 128 | 16.04 | 22.38 | 1.40 |
| NICYVITHGDAK | 157 | 7.32 | 13.91 | 1.90 | ||||
| P15532 | Nucleoside diphosphate kinase A (Nme1) | 1.00:0.59 | 12.77 | GDFCIQVGR | 109 | 14.89 | 14.54 | 0.98 |
| P17742 | Peptidyl prolyl cis-trans isomerase (PpiA) | 1.00:0.90 | 2.84 | IIPGFMCQGGDFTR | 62 | 2.52 | 5.76 | 2.29 |
| P09411 | Phosphoglycerate kinase 1 (Pgk1) | 1.00:0.59 | 23.68 | GCITIIGGGDTATCCAK | 367/379/380* | 47.50 | 95.40 | 2.01 |
| O70250 | Phosphoglycerate mutase 2 (Pgam2) | 1.00:0.71 | 30.92 | FCGWFDAELSEK | 23 | 7.19 | 9.20 | 1.28 |
| IEFDICYTSVLK | 55 | 1.31 | 1.11 | 0.85 | ||||
| Q9D0F9 | Phosphoglucomutase 1 (Pgm1) | 1.00:0.75 | 14.98 | TIEEYAICPDLK | 160 | 15.52 | 16.76 | 1.08 |
| LSLCGEESFGTGSDHIR | 374 | 3.25 | 8.42 | 2.59 | ||||
| Q99LX0 | Protein DJ-1 (park7) | 1.00:0.81 | 6.52 | DPVQCSR | 46 | 8.50 | 17.53 | 2.06 |
| Q9QYG0 | Protein NDRG2 (Ndrg2) | 1.00:0.83 | 9.86 | CPVMLVVGDQAPHEDAVVECNSK | 255/274* | 14.57 | 32.14 | 2.21 |
| Q9D051 | Pyruvate dehydrogenase E1 component subunit beta (Pdhb) | 1.00:0.82 | 6.78 | EGIECEVINLR | 263 | 6.80 | 9.97 | 1.47 |
| P52480 | Pyruvate kinase isozymes M1/M2 (Pkm2) | 1.00:0.98 | 3.86 | NTGIICTIGPASR | 49 | 12.06 | 17.11 | 1.42 |
| CDENILWLDYK | 152 | 5.75 | 8.62 | 1.50 | ||||
| GIFPVLCK | 474 | 23.16 | 29.64 | 1.28 | ||||
| P07724 | Serum albumin (Alb) | 1.00:1.25 | 9.00 | CSYDEHAK | 58 | 2.95 | 2.78 | 0.94 |
| P17751 | Triosephosphate isomerase (Tpi1) | 1.00:0.84 | 8.68 | IAVAAQNCYK | 117 | 0.63 | 2.40 | 3.80 |
| VSHALAEGLGVIACIGEK | 177 | 9.15 | 11.75 | 1.28 | ||||
| IIYGGSVTGATCK | 268 | 7.29 | 13.17 | 1.81 | ||||
| P58771 | Tropomyosin alpha-1 chain (Tpm1) | 1.00:2.60 | 115.68 | CAELEEELK | 190 | 5.65 | 9.46 | 1.68 |
| P58774 | Tropomyosin beta chain (Tpm2) | 1.00:2.04 | 84.11 | CGDLEEELK | 190 | 6.34 | 7.89 | 1.25 |
| P13412 | Troponin-1 fast skeletal muscle (Tnni2) | 1.00:2.32 | 52.67 | VCMDLR | 134 | 6.79 | 10.27 | 1.51 |
Signifies that 2 or more Cys residues present in tryptic peptide.
Table 2.
Proteins detected in soleus tissue containing redox sensitive Cys residues including the relative quantification of oxidative state of susceptible Cys residues and changes in the redox ratio with age. Relative protein abundance is included and proteins in bold changed significantly with age.
| Accession | Protein | Adult:Old | Significance (–10logP) | Peptide | Cysteine | Adult Red/Ox | Old Red/Ox | Change with age |
|---|---|---|---|---|---|---|---|---|
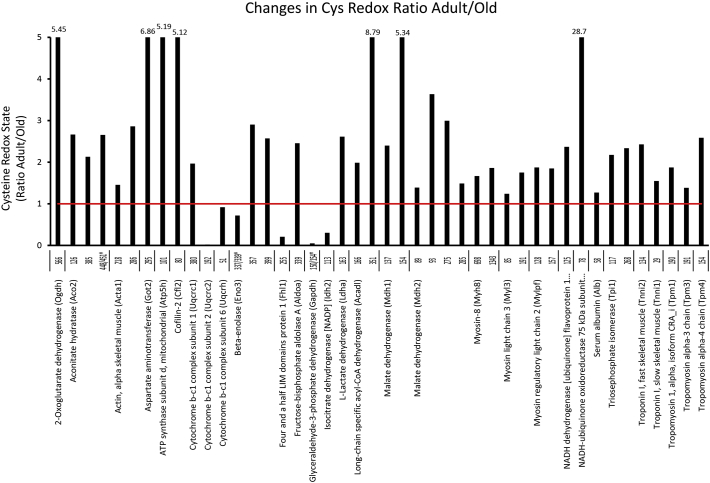
| Q60597 | 2-Oxoglutarate dehydrogenase (Ogdh) | 1.00:0.41 | 80.24 | ICEEAFTR | 566 | 4.02 | 21.91 | 5.45 |
| Q99KI0 | Aconitate hydratase (Aco2) | 1.00:1.60 | 53.50 | VAVPSTIHCDHLIEAQVGGEK | 126 | 4.35 | 11.59 | 2.66 |
| VGLIGSCTNSSYEDMGR | 385 | 1.85 | 3.95 | 2.13 | ||||
| DVGGIVLANACGPCIGQWDR | 448/451* | 3.18 | 8.43 | 2.66 | ||||
| O88990 | Actin, alpha skeletal muscle (Acta1) | 1.00:1.17 | 12.03 | LCYVALDFENEMATAASSSSLEK | 218 | 11.14 | 16.23 | 1.46 |
| CDIDIR | 286 | 13.81 | 39.52 | 2.86 | ||||
| P05202 | Aspartate aminotransferase (Got2) | 1.00:1.02 | 7.39 | VGAFTVVCK | 295 | 1.90 | 13.05 | 6.86 |
| Q9DCX2 | ATP synthase subunit d, mitochondrial (Atp5h) | 1.00:0.38 | 61.87 | SCAEFVSGSQLR | 101 | 24.99 | 129.81 | 5.19 |
| P45591 | Cofilin-2 (Cfl2) | 1.00:1.43 | 18.30 | LLPLNDCR | 80 | 14.54 | 74.39 | 5.12 |
| Q9CZ13 | Cytochrome b-c1 complex subunit 1 (Uqcrc1) | 1.00:0.89 | 8.96 | LCTSATESEVTR | 380 | 23.64 | 46.49 | 1.97 |
| Q9DB77 | Cytochrome b-c1 complex subunit 2 (Uqcrc2) | 1.00:1.04 | 3.16 | NALANPLYCPDYR | 192 | 4833.44 | 90.84 | 0.02 |
| P99028 | Cytochrome b-c1 complex subunit 6 (Uqcrh) | 1.00:0.61 | 39.21 | ERLELCDNR | 51 | 25.97 | 23.86 | 0.92 |
| P21550 | Beta-enolase (Eno3) | 1.00:0.79 | 14.85 | ACNCLLLK | 337/339* | 84.66 | 60.82 | 0.72 |
| VNQIGSVTESLQACK | 357 | 7.14 | 20.73 | 2.91 | ||||
| TGAPCR | 399 | 8.89 | 22.85 | 2.57 | ||||
| P97447 | Four and a half LIM domains protein 1 (Fhl1) | 1.00:0.13 | 113.70 | CSVNLANKR | 255 | 476.66 | 99.38 | 0.21 |
| P05064 | Fructose-bisphosphate aldolase A (Aldoa) | 1.00:1.43 | 23.11 | ALANSLACQGK | 339 | 21.69 | 53.27 | 2.46 |
| P16858 | Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) | 1.00:0.44 | 93.31 | IVSNASCTTNCLAPLAK | 150/154* | 5524.28 | 278.52 | 0.05 |
| P54071 | Isocitrate dehydrogenase [NADP] (Idh2) | 1.00:0.93 | 7.60 | CATITPDEAR | 113 | 0.02 | 0.01 | 0.30 |
| P06151 | l-Lactate dehydrogenase (Ldha) | 1.00:0.68 | 25.24 | VIGSGCNLDSAR | 163 | 22.79 | 59.57 | 2.61 |
| P51174 | Long-chain specific acyl-CoA dehydrogenase (Acadl) | 1.00:1.08 | 10.15 | CIGAIAMTEPGAGSDLQGVR | 166 | 7.54 | 14.97 | 1.99 |
| AFVDSCLQLHETK | 351 | 2.73 | 23.99 | 8.79 | ||||
| P14152 | Malate dehydrogenase (Mdh1) | 1.00:0.97 | 5.01 | VIVVGNPANTNCLTASK | 137 | 9.02 | 21.65 | 2.40 |
| ENFSCLTR | 154 | 8.23 | 43.95 | 5.34 | ||||
| P08249 | Malate dehydrogenase (Mdh2) | 1.00:1.01 | 3.84 | GYLGPEQLPDCLK | 89 | 27.10 | 37.69 | 1.39 |
| GCDVVVIPAGVPR | 93 | 4.48 | 16.30 | 3.63 | ||||
| EGVVECSFVQSK | 275 | 13.51 | 40.48 | 3.00 | ||||
| ETECTYFSTPLLLGK | 285 | 24.59 | 36.59 | 1.49 | ||||
| P13542 | Myosin-8 (Myh8) | 1.00:1.22 | 3.97 | CNGVLEGIR | 698 | 43.95 | 73.27 | 1.67 |
| HDCDLLR | 1343 | 24.04 | 44.75 | 1.86 | ||||
| P09542 | Myosin light chain 3 (Myl3) | 1.00:1.17 | 20.73 | ITYGQCGDVLR | 85 | 54.39 | 67.53 | 1.24 |
| LMAGQEDSNGCINYEAFVK | 191 | 15.56 | 27.26 | 1.75 | ||||
| P97457 | Myosin regulatory light chain 2 (Mylpf) | 1.00:0.73 | 45.56 | QFLEELLTTQCDR | 128 | 27.50 | 51.56 | 1.87 |
| NICYVITHGDAK | 157 | 23.14 | 42.82 | 1.85 | ||||
| Q91YT0 | NADH dehydrogenase [ubiquinone] flavoprotein 1 (Ndufv1) | 1.00:1.01 | 5.73 | YLVVNADEGEPGTCK | 125 | 295.18 | 699.84 | 2.37 |
| Q91VD9 | NADH-ubiquinone oxidoreductase 75 kDa subunit (Ndufs1) | 1.00:0.87 | 9.42 | MCLVEIEK | 78 | 115.96 | 3328.04 | 28.70 |
| P07724 | Serum albumin (Alb) | 1.00:0.90 | 12.32 | CSYDEHAK | 58 | 3.83 | 4.87 | 1.27 |
| P17751 | Triosephosphate isomerase (Tpi1) | 1.00:0.98 | 14.07 | IAVAAQNCYK | 117 | 731.14 | 1591.58 | 2.18 |
| IIYGGSVTGATCK | 268 | 19.41 | 45.36 | 2.34 | ||||
| P13412 | Troponin I, fast skeletal muscle (Tnni2) | 1.00:0.81 | 39.05 | VCMDLR | 134 | 18.38 | 44.64 | 2.43 |
| Q9WUZ5 | Troponin I, slow skeletal muscle (Tnni1) | 1.00:0.91 | 12.86 | ECWEQEHEER | 29 | 85.64 | 132.57 | 1.55 |
| P58771 | Tropomyosin 1, alpha, isoform CRA_i (Tpm1) | 1.00:1.00 | 7.40 | CAELEEELK | 190 | 16.71 | 31.32 | 1.87 |
| P21107 | Tropomyosin alpha-3 chain (Tpm3) | 1.00:1.19 | 13.81 | CSELEEELK | 191 | 55.27 | 76.49 | 1.38 |
| Q6IRU2 | Tropomyosin alpha-4 chain (Tpm4) | 1.00:1.77 | 5.77 | CGDLEEELK | 154 | 14.01 | 36.23 | 2.59 |
Signifies that 2 or more Cys residues present in tryptic peptide.
Supplementary Fig. 4.
Changes in red/reversibly oxidised ratio of individual Cys residues with age in quadriceps muscle, ratio change <1 reflects increased reversible oxidation with age, ratio > 1 reflects decreased cysteine oxidation with age.
Supplementary Fig. 5.
Changes in red/reversibly oxidised ratio of individual Cys residues with age in soleus skeletal muscle, ratio change <1 reflects increased reversible oxidation with age, ratio > 1 reflects decreased cysteine oxidation.
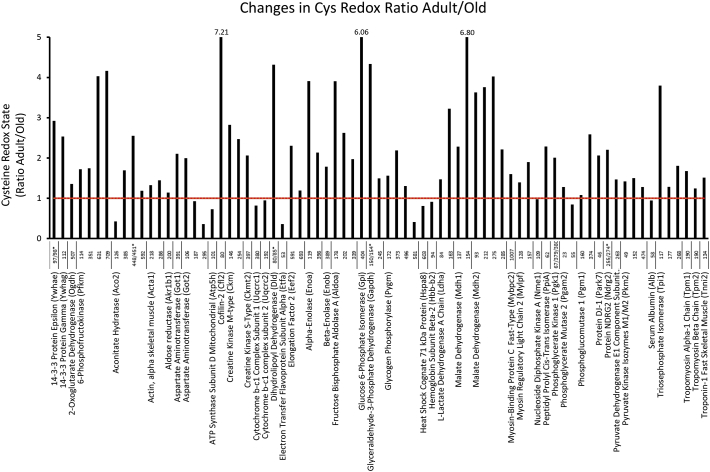
The redox state of peptides has been found to both decrease and increase with age, indicating a more oxidised or reduced state of the Cys residue respectively. The redox state of 25 peptides from 17 proteins were quantified from both quadriceps and soleus, 5 of these Cys residues had an increased oxidation state with age in quadriceps and more reduced redox state in soleus, Aconitase Cys126, mitochondrial Aspartate Aminotransferase Cys295, mitochondrial ATP synthase subunit d Cys101, Cytochrome b-c1 complex subunit 1 Cys 380 and Serum albumin Cys58. The peptide containing the catalytic site of glyceraldehyde dehydrogenase Cys150/154 was more oxidised with age in the soleus and more reduced in quadriceps. There were no significant changes in abundance for these proteins with age and the muscle specific changes in redox state of individual Cys residues could indicate an altered intracellular redox environment. The changes in the redox state of individual Cys residues and the potential effects on protein activity need to be considered on an individual basis. For instance Cys residues involved in the co-ordination of iron in Fe-S proteins have been identified such as aconitase (Cys126, 286, 385, 448/451) and isocitrate dehydrogenase (Cys113). Metabolic enzymes involved in glycolysis and electron transport chain had substantial shifts in the redox state of Cys residues such as pyruvate kinase (Cys49, 152, 474), enolase alpha (Cys119, 399) and beta (Cys389), aldolase (Cys178, 202), malate dehydrogenase 1 (Cys137, 154) and 2 (Cys93, 212, 275, 285), ATP synthase (Cys101) and cytochrome b-c1 complexes 1 (Cys380), 2 (Cys192) and 6 (Cys80/85). There were also changes in the redox state of many of the cytoskeletal proteins that form part of contractile machinery for excitation contraction coupling and force generation; actin (Cys218, 286), myosin light chain 3 (Cys85, 191), myosin-8 (Cys698, 1343), myosin binding protein C (Cys1007), myosin regulatory light chain 2 (Cys128, 157), tropomyosin (Cys190), troponin I fast (Cys134) and slow (Cys29). Many of these sarcomeric proteins have previously been identified as modified by S-nitrosylation including myosin, actin, myosin-binding protein C, troponin C and I [34]. Cys134 from Troponin I in both quadriceps and soleus had a shift towards a more reduced state with age, this Cys residue has been reported to be both S-nitrosylated and S-glutathionylated in a recent study with opposite effects on protein function, S-nitrosylation decreases and S-glutathionylation of Cys134 increases the calcium-sensitivity of contractile machinery [35]. The technique used here does not allow the discrimination of specific reversible redox modifications but it does highlight the sensitivity of the approach for the detection of key Cys residues that affect protein activity and contractility.
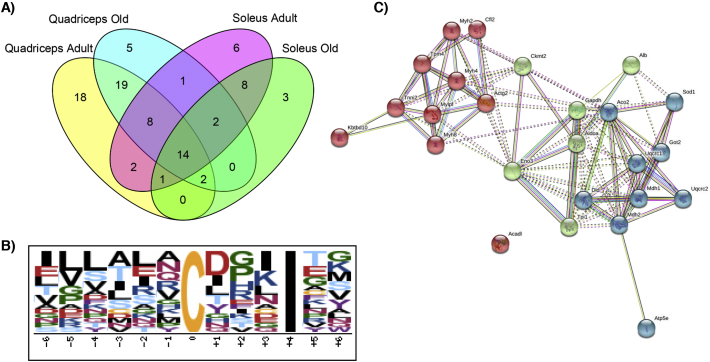
The number of proteins identified as containing redox sensitive Cys residues decreases with age in both quadriceps and soleus (Fig. 5A), which may indicate a reduced redox flexibility in both these tissues with age. It should be pointed out that not all of the redox sensitive peptides initially identified were quantifiable using Skyline and were not included in Table 1, Table 2. Using the identified redox sensitive Cys residues we searched for possible conserved redox motifs or over representation of certain amino acids in the mouse proteome. The highest scoring motif from a Motif-X blast [30] had an Isoleucine residue on the +4 C-terminal side of the redox sensitive Cys (Fig. 5B). A String-DB analysis [31] of identified redox proteins (Fig. 5C) indicate that peptides identified in both tissues are enriched for proteins involved in the generation of precursor metabolites and energy (GO:0006091, FDR 2.06−09).
Fig. 5.
Redox changes between Quadriceps and Soleus a) numbers of redox proteins detected in the different groups, in quadriceps adult muscle there were 64 compared to 53 in old and in soleus there were 28 redox proteins in old compared to 42 in adult. B) To determine if there was a consensus motif surrounding the redox sensitive Cys residues identified in this analysis we used Motif-X. A protein blast against the Mus proteome was performed using the amino acid sequences −6 to +6 of the redox sensitive Cys residues identified to determine potential motifs in surrounding amino acids. The highest scoring motif (6.14 and P < 0.000001) included Isoleucine (I) at a position +4 from the identified redox sensitive peptides using Motif-X. C) To determine if there was an over representation of particular metabolic pathways or protein-protein interactions of identified proteins containing redox sensitive Cys residues we used String-DB. Identified redox sensitive Cys containg proteins revealed a highly interconnected network and an enrichment for proteins involved in the generation of precursor metabolites and energy (GO:0006091, FDR value 2.06e−09).
The data presented here combined a label free proteomic approach with differential Cys labelling to analyse changes in the global proteome and for the first time, identified how the redox state of sensitive Cys residues change with age in two metabolically distinct skeletal muscles. Overall the global proteomic changes were found to be fibre type dependent with significant increases in cytoskeletal proteins in quadriceps and significant decreases in electron transport chain proteins in soleus but the redox proteome was more preserved between tissues with specific changes in redox state of metabolic and cytoskeletal proteins.
4. Discussion
The results of this study describe the effects of age on the protein composition of two distinct muscle groups. There was an increase in cytoskeletal proteins in glycolytic quadriceps muscle and a decrease in mitochondrial proteins involved in electron transport chain in more oxidative soleus muscle. Analysis of the redox state of sensitive Cys residues indicate that age-related changes are identified predominantly in cytoskeletal and metabolic proteins in both muscles. This study provides further information on the effects of age on skeletal muscle groups with distinct susceptibility to age-related atrophy. In humans the age-related loss of skeletal muscle can result in frailty and loss of independence, and is a major factor in contributing to the co-morbidities of many diseases. One of the most effective therapies to delay age-related atrophy of skeletal muscle mass and function is regular exercise while disuse can accelerate the loss of muscle mass [36,37]. Contracting skeletal muscle generates ROS [[11], [12], [13]] that are thought to have a vital role in the beneficial responses to exercise and aid in the repair of injured skeletal muscle [15,16,38,39]. Previous studies using rodent models have shown that the response to redox generated signals for the activation of key transcription factors such as NFκB and Hsf-1 are blunted in muscle from old mice and there is a chronic activation of the inflammatory response [20].
Skeletal muscle that contains higher proportions of glycolytic fibres appear to be more susceptible to age-related atrophy [[3], [4], [5]]. Our results indicate that quadriceps had a reduced muscle mass and increased SDH staining with age, no significant decrease in muscle mass was detected in soleus but there was an increased proportion of SDH stained fibres. Aging results in a reduced respiratory capacity of rat soleus (mainly type I fibres) and glycolytic (type IIB and IIA) from white and red portions of Vastus lateralis [4]. Fast to slow fibre type remodelling with age has been reported, in particular a shift from IIB to IIX fibres in rodent fast twitch muscles such as the tibialis anterior [5] and Type IIA to Type I in plantaris [4,40]. In soleus it has also been reported that there is an increase in single muscle fibres that have increased co-expression of myosin heavy chain isoforms in older rats as a result of the reinnervation of denervated fibres [41]. The increased SDH staining in muscle fibres as representative of Krebs cycle mitochondrial activity could be a compensatory response, therefore to investigate the intracellular changes in the metabolically distinct skeletal muscles a global and redox proteomic approach was undertaken.
Global proteomic approaches that have utilised 2D electrophoresis including differential in gel electrophoresis [[42], [43], [44]] and gel-free approaches [45,46] to analyse the effects of age on skeletal muscles from rodents have reported similar changes in glycolytic, mitochondrial and cytoskeletal proteins with age as described in this study. Alterations in the proteome have been confirmed in more detail in a proteomic study of slow and fast human skeletal muscle single fibres isolated from biopsies of young (22–27) and older (67–75) individuals [47]. In a previous study examining the effects of age on gastrocnemius muscle, an increase in the abundance of cytoskeletal proteins and a decrease in metabolic proteins was also reported [21]. However, in the gastrocnemius, the increase in cytoskeletal proteins in response to age was not as dramatic as what we detected in quadriceps nor the decrease in mitochondrial proteins comparable to what was observed in soleus, which may reflect the more heterogeneous fibre type content. One protein Ig mu chain C increased in abundance across all three muscles with age, while Myl1 increased in abundance in quadriceps and decreased in soleus, also had increased abundance in gastrocnemius muscle in response to age [21]. The greater susceptibility of fast fibres to age-related atrophy could be related to the higher glycolytic flux within fast fibres similar to the metabolic reprogramming in tumour cells [48] affecting ROS generation and downstream pathways including autophagy and the proteasome [49]. It has also been suggested that lower mitochondrial protein turnover in fast fibres could result in reduced mitochondrial quality and increased susceptibility to proteasomal degradation [33]. Higher mitochondrial turnover in slow fibres may offer greater protection against dysfunctional metabolic pathways related to sarcopenia. However, a decrease in the abundance of proteins involved in mitochondrial electron transport proteins with age in soleus yet an increase in mitochondrial SDH activity could be a compensatory mechanism for incomplete metabolism by oxidative phosphorylation.
In comparison to the global changes in proteome abundance, the changes in the redox proteome appeared to be preserved between muscles. In this study, 25 of the Cys containing peptides that had an altered redox state with age were common to both quadriceps and soleus (Table 1, Table 2). In comparison to a previous study of the effects of age on the gastrocnemius muscle 23 of these 25 Cys residues were also reported as having an altered redox state [21]. The transient redox state of key individual Cys residues has an important role in skeletal muscle redox signalling, allowing redox sensitive proteins to rapidly respond to local changes in ROS concentrations. Analysis of individual Cys residues indicate that in a number of instances there was greater oxidation in the redox state with age but this was not always the case and often there were large shifts to a more reduced state in individual Cys residues from both tissues. However, there were 6 Cys residues quantified in both quadriceps and soleus where the redox state shifted in the opposite direction with age. Apart from glyceraldehyde dehydrogenase which requires the catalytic Cys residues in the active site (Cys150/154) to be in a reduced form [50], it was not possible to determine how these redox changes would affect protein activity from the current literature or structural data. Many proteins require their key redox sensitive Cys residues to be in a disulfide bond for functional activity e.g. triose phosphate isomerase and pyruvate kinase while others require the Cys to be reduced [50]. Therefore, it is necessary to consider the age-related changes in the redox state of specific Cys residues on an individual basis for each protein taking into account the location and function of the Cys residue.
A recent top down proteomic approach of rat slow and fast skeletal muscles identified changes with age in the phosphorylation status of a number of the cytoskeletal proteins identified as having an altered abundance and redox state in this study including Tropomyosin alpha and beta, Myosin regulatory light chain and Troponin I [51]. Troponin I was also found to have a reduced level of S-glutathionylation with age but the Cys residue modified was not reported [51]. The S-gluthathionylation and S-nitrosylation of Cys134 of Troponin I has been reported to increase and decrease respectively the Ca2+ sensitivity of this protein [35], highlighting the complexity of Cys specific redox modifications. In this study we identified Cys134 of Troponin I as having an altered redox state with age but could not discriminate the specific redox modifications and hence effects on Ca2+ sensitivity for contractile function, but our approach is sensitive for the identification of regulatory Cys residues. Similarly, oxidation of myosin residues will affect actin binding [52], identification and the relative quantification of the specific residues susceptible to oxidation may provide an indication of the events that lead to defects in mechanical function of skeletal muscle. Cross talk between Cys redox specific and PTMs such as phosphorylation in the context of protein abundance and subsequent effects on contractile function requires further study. It is unlikely that a protein is exclusively reduced or reversibly oxidised but in the approach used here we attempt to estimate the redox state of individual Cys residues. However, the sensitivity of this approach depends on the proportion and concentration of the peptide containing the Cys residue labelled, if it was identified with a high confidence in the initial search of raw data and with a consistent retention time and fragmentation across samples to allow targeted quantification using Skyline.
Overall, there was a reduction in the number of peptides that contain redox Cys residues labelled with both light and heavy isoform of NEM with age in quadriceps and soleus (Fig. 5A). In order to determine if there was particular amino acids over represented or potentially a conserved motif surrounding the redox sensitive Cys residues identified we used Motif-X [30]. Using the amino acid sequences −6 to +6 of the redox sensitive Cys, there was an over representation of Isoleucine at +4 C-terminal side of the redox sensitive Cys residues (Fig. 5B). Isoleucine does not contain reactive side chains but has an important role in protein stability and ligand binding. A String-DB analysis of identified proteins containing redox sensitive Cys residues indicates multiple interactions between proteins identified and an enrichment for proteins involved in precursor metabolites and energy generation (Fig. 5C), consistent with a previous study analyzing the effects of age on the gastrocnemius [21].
Previous studies have detected an increase in irreversible protein oxidation with age in skeletal muscle [53,54], the increase in proportion of specific Cys residues to a more reduced state seems to be counter intuitive in this intracellular environment. Many of the proteins identified as irreversibly modified appear to be highly abundant cytoskeletal proteins and metabolic enzymes that also have an increase in abundance with age. These proteins that could potentially act as a redox buffer inhibiting ROS induced redox signalling. The identification of highly abundant cytoskeletal and sarcomeric proteins also highlights one of the limitations of the data dependent acquisition mass spectrometry approach, as low abundant proteins and their parent ions may not get fragmented [55]. Sarcomeric and cytoskeletal proteins dominate the skeletal muscle proteome and can account for >50% of the proteome [56], thus comparing low to high abundant proteins can span up to seven orders of magnitude [47]. Increasing the sensitivity of detection by sample fractionation or data independent approaches would increase the accuracy of both detection and quantification of redox specific changes and for global label free proteomics.
5. Conclusions
The redox proteome was relatively preserved between metabolically distinct muscles. This is likely due to the nature of the regulatory nature of these residues in proteins involved in processes such as glycolysis, oxidative phosphorylation, translation, transcription and degradation [57]. It has previously been demonstrated that Cys residues have the most extreme conservation patterns across all of the amino acids, they are highly conserved when they play a fundamental role in protein activity in active sites or co-factor binding but poorly conserved otherwise [58]. Cysteine residues in myofilament proteins are generally conserved across different protein species and accessibility can depend on the contractile status of the muscle [59]. Nevertheless our observation of a reduction in available Cys residues for redox signalling during aging is likely to result in a reduced flexibility of the redox response and impair the ability of skeletal muscle to optimally respond and adapt to exercise.
6. Data repository
Quantitative proteomic data from this study has been deposited in Mendeley data repository with doi: 10.17632/x9wxcnc67r.1. Files include all protein detected, no of identified and unique peptides for individual proteins, intensities from each biological replicate, fold change including significance (–10logP).
The following are the supplementary data related to this article.
Transparency document
Transparency document.
Acknowledgements
NTS was funded by a BBSRC DTP PhD studentship and BMcD was funded by Wellcome ISSF fund (097826/Z/11/Z) and with MJJ by MRC grant MR/M012573/1. Proteomics was performed in the Centre for Proteome Research, University of Liverpool with excellent technical help from Dr. Philip Brownridge.
Footnotes
The Transparency document associated with this article can be found, in online version.
References
- 1.Saey D., Lemire B.B., Gagnon P., Bombardier E., Tupling A.R., Debigare R. Quadriceps metabolism during constant workrate cycling exercise in chronic obstructive pulmonary disease. J. Appl. Physiol. 2011;110:116–124. doi: 10.1152/japplphysiol.00153.2010. [DOI] [PubMed] [Google Scholar]
- 2.Augosto V., Padovani C.R.C., Gerson E.R. Skeletal muscle fiber Tyoes in C57BL6J mice. J. Morphological Sci. 2004;21:89–94. [Google Scholar]
- 3.Lexell J. Human aging, muscle mass, and fiber type composition. J. Gerontol. A Biol. Sci. Med. Sci. 1995;50 doi: 10.1093/gerona/50a.special_issue.11. Spec No:11-6. [DOI] [PubMed] [Google Scholar]
- 4.Holloszy J.O., Chen M., Cartee G.D., Young J.C. Skeletal muscle atrophy in old rats: differential changes in the three fiber types. Mech. Ageing Dev. 1991;60:199–213. doi: 10.1016/0047-6374(91)90131-i. [DOI] [PubMed] [Google Scholar]
- 5.Larsson L., Biral D., Campione M., Schiaffino S. An age-related type IIB to IIX myosin heavy chain switching in rat skeletal muscle. Acta Physiol. Scand. 1993;147:227–234. doi: 10.1111/j.1748-1716.1993.tb09493.x. [DOI] [PubMed] [Google Scholar]
- 6.Bratic A., Larsson N.G. The role of mitochondria in aging. J. Clin. Invest. 2013;123:951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chabi B., Ljubicic V., Menzies K.J., Huang J.H., Saleem A., Hood D.A. Mitochondrial function and apoptotic susceptibility in aging skeletal muscle. Aging Cell. 2008;7:2–12. doi: 10.1111/j.1474-9726.2007.00347.x. [DOI] [PubMed] [Google Scholar]
- 8.Hepple R.T. Mitochondrial involvement and impact in aging skeletal muscle. Front. Aging Neurosci. 2014;6:211. doi: 10.3389/fnagi.2014.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rygiel K.A., Picard M., Turnbull D.M. The ageing neuromuscular system and sarcopenia: a mitochondrial perspective. J. Physiol. 2016;594:4499–4512. doi: 10.1113/JP271212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller F.L., Lustgarten M.S., Jang Y., Richardson A., Van Remmen H. Trends in oxidative aging theories. Free Radic. Biol. Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 11.Sakellariou G.K., Vasilaki A., Palomero J., Kayani A., Zibrik L., McArdle A. Studies of mitochondrial and nonmitochondrial sources implicate nicotinamide adenine dinucleotide phosphate oxidase(s) in the increased skeletal muscle superoxide generation that occurs during contractile activity. Antioxid. Redox Signal. 2013;18:603–621. doi: 10.1089/ars.2012.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies K.J., Quintanilha A.T., Brooks G.A., Packer L. Free radicals and tissue damage produced by exercise. Biochem. Biophys. Res. Commun. 1982;107:1198–1205. doi: 10.1016/s0006-291x(82)80124-1. [DOI] [PubMed] [Google Scholar]
- 13.Jackson M.J., Edwards R.H., Symons M.C. Electron spin resonance studies of intact mammalian skeletal muscle. Biochim. Biophys. Acta. 1985;847:185–190. doi: 10.1016/0167-4889(85)90019-9. [DOI] [PubMed] [Google Scholar]
- 14.Winterbourn C.C. Are free radicals involved in thiol-based redox signaling? Free Radic. Biol. Med. 2015;80:164–170. doi: 10.1016/j.freeradbiomed.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Margaritelis N.V., Theodorou A.A., Paschalis V., Veskoukis A.S., Dipla K., Zafeiridis A. Adaptations to endurance training depend on exercise-induced oxidative stress: exploiting redox interindividual variability. Acta Physiol (Oxf.) 2017;222 doi: 10.1111/apha.12898. [DOI] [PubMed] [Google Scholar]
- 16.Horn A., Van der Meulen J.H., Defour A., Hogarth M., Sreetama S.C., Reed A. Mitochondrial redox signaling enables repair of injured skeletal muscle cells. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aaj1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulsen C.E., Carroll K.S. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones D.P. Radical-free Biology of oxidative stress. Am. J. Physiol. Cell Physiol. 2008;295:C849–68. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonagh B. Detection of ROS induced proteomic signatures by mass spectrometry. Front. Physiol. 2017;8:470. doi: 10.3389/fphys.2017.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vasilaki A., McArdle F., Iwanejko L.M., McArdle A. Adaptive responses of mouse skeletal muscle to contractile activity: the effect of age. Mech. Ageing Dev. 2006;127:830–839. doi: 10.1016/j.mad.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 21.McDonagh B., Sakellariou G.K., Smith N.T., Brownridge P., Jackson M.J. Differential cysteine labeling and global label-free proteomics reveals an altered metabolic state in skeletal muscle aging. J. Proteome Res. 2014;13:5008–5021. doi: 10.1021/pr5006394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel T.J., Cuizon D., Mathieu-Costello O., Friden J., Lieber R.L. Increased oxidative capacity does not protect skeletal muscle fibers from eccentric contraction-induced injury. Am. J. Phys. 1998;274:R1300–8. doi: 10.1152/ajpregu.1998.274.5.R1300. [DOI] [PubMed] [Google Scholar]
- 23.Ross J.M. Visualization of mitochondrial respiratory function using cytochrome c oxidase/succinate dehydrogenase (COX/SDH) double-labeling histochemistry. J. Vis. Exp. 2011;57 doi: 10.3791/3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Philp L.K., Heilbronn L.K., Janovska A., Wittert G.A. Dietary enrichment with fish oil prevents high fat-induced metabolic dysfunction in skeletal muscle in mice. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.010587. M111 010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 27.Kutmon M., van Iersel M.P., Bohler A., Kelder T., Nunes N., Pico A.R. PathVisio 3: an extendable pathway analysis toolbox. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kutmon M., Riutta A., Nunes N., Hanspers K., Willighagen E.L., Bohler A. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44:D488–94. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou M.F., Schwartz D. Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics. 2011;Chapter 13(Unit 13):5–24. doi: 10.1002/0471250953.bi1315s35. [DOI] [PubMed] [Google Scholar]
- 31.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkw937. D362-D8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park C.Y., Pierce S.A., von Drehle M., Ivey K.N., Morgan J.A., Blau H.M. skNAC, a Smyd1-interacting transcription factor, is involved in cardiac development and skeletal muscle growth and regeneration. Proc. Natl. Acad. Sci. U. S. A. 2010;107:20750–20755. doi: 10.1073/pnas.1013493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruse S.E., Karunadharma P.P., Basisty N., Johnson R., Beyer R.P., MacCoss M.J. Age modifies respiratory complex I and protein homeostasis in a muscle type-specific manner. Aging Cell. 2016;15:89–99. doi: 10.1111/acel.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Figueiredo-Freitas C., Dulce R.A., Foster M.W., Liang J., Yamashita A.M., Lima-Rosa F.L. S-nitrosylation of sarcomeric proteins depresses myofilament Ca2+ sensitivity in intact cardiomyocytes. Antioxid. Redox Signal. 2015;23:1017–1034. doi: 10.1089/ars.2015.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dutka T.L., Mollica J.P., Lamboley C.R., Weerakkody V.C., Greening D.W., Posterino G.S. S-nitrosylation and S-glutathionylation of Cys134 on troponin I have opposing competitive actions on Ca2+ sensitivity in rat fast-twitch muscle fibers. Am. J. Physiol. Cell Physiol. 2017;312 doi: 10.1152/ajpcell.00334.2016. C316-C27. [DOI] [PubMed] [Google Scholar]
- 36.Ferrando A.A., Lane H.W., Stuart C.A., Davis-Street J., Wolfe R.R. Prolonged bed rest decreases skeletal muscle and whole body protein synthesis. Am. J. Phys. 1996;270:E627–33. doi: 10.1152/ajpendo.1996.270.4.E627. [DOI] [PubMed] [Google Scholar]
- 37.Koltai E., Hart N., Taylor A.W., Goto S., Ngo J.K., Davies K.J. Age-associated declines in mitochondrial biogenesis and protein quality control factors are minimized by exercise training. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012;303:R127–34. doi: 10.1152/ajpregu.00337.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ristow M., Zarse K., Oberbach A., Kloting N., Birringer M., Kiehntopf M. Antioxidants prevent health-promoting effects of physical exercise in humans. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8665–8670. doi: 10.1073/pnas.0903485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakellariou G.K., Lightfoot A.P., Earl K.E., Stofanko M., McDonagh B. Redox homeostasis and age-related deficits in neuromuscular integrity and function. J. Cachexia. Sarcopenia Muscle. 2017;8:881–906. doi: 10.1002/jcsm.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciciliot S., Rossi A.C., Dyar K.A., Blaauw B., Schiaffino S. Muscle type and fiber type specificity in muscle wasting. Int. J. Biochem. Cell Biol. 2013;45:2191–2199. doi: 10.1016/j.biocel.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 41.Snow L.M., McLoon L.K., Thompson L.V. Adult and developmental myosin heavy chain isoforms in soleus muscle of aging Fischer Brown Norway rat. Anat. Rec. A: Discov. Mol. Cell. Evol. Biol. 2005;286:866–873. doi: 10.1002/ar.a.20218. [DOI] [PubMed] [Google Scholar]
- 42.Capitanio D., Vasso M., Fania C., Moriggi M., Vigano A., Procacci P. Comparative proteomic profile of rat sciatic nerve and gastrocnemius muscle tissues in ageing by 2-D DIGE. Proteomics. 2009;9:2004–2020. doi: 10.1002/pmic.200701162. [DOI] [PubMed] [Google Scholar]
- 43.Doran P., O'Connell K., Gannon J., Kavanagh M., Ohlendieck K. Opposite pathobiochemical fate of pyruvate kinase and adenylate kinase in aged rat skeletal muscle as revealed by proteomic DIGE analysis. Proteomics. 2008;8:364–377. doi: 10.1002/pmic.200700475. [DOI] [PubMed] [Google Scholar]
- 44.Piec I., Listrat A., Alliot J., Chambon C., Taylor R.G., Bechet D. Differential proteome analysis of aging in rat skeletal muscle. FASEB J. 2005;19:1143–1145. doi: 10.1096/fj.04-3084fje. [DOI] [PubMed] [Google Scholar]
- 45.Rakus D., Gizak A., Deshmukh A., Wisniewski J.R. Absolute quantitative profiling of the key metabolic pathways in slow and fast skeletal muscle. J. Proteome Res. 2015;14:1400–1411. doi: 10.1021/pr5010357. [DOI] [PubMed] [Google Scholar]
- 46.Chaves D.F., Carvalho P.C., Lima D.B., Nicastro H., Lorenzeti F.M., Siqueira-Filho M. Comparative proteomic analysis of the aging soleus and extensor digitorum longus rat muscles using TMT labeling and mass spectrometry. J. Proteome Res. 2013;12:4532–4546. doi: 10.1021/pr400644x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murgia M., Toniolo L., Nagaraj N., Ciciliot S., Vindigni V., Schiaffino S. Single muscle fiber proteomics reveals fiber-type-specific features of human muscle aging. Cell Rep. 2017;19:2396–2409. doi: 10.1016/j.celrep.2017.05.054. [DOI] [PubMed] [Google Scholar]
- 48.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pajares M., Jimenez-Moreno N., Dias I.H., Debelec B., Vucetic M., Fladmark K.E. Redox control of protein degradation. Redox Biol. 2015;6:409–420. doi: 10.1016/j.redox.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shenton D., Grant C.M. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 2003;374:513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei L., Gregorich Z.R., Lin Z., Cai W., Jin Y., McKiernan S.H. Novel sarcopenia-related alterations in sarcomeric protein post-translational modifications in skeletal muscles identified by top-down proteomics. Mol. Cell. Proteomics. 2018;17:134–145. doi: 10.1074/mcp.RA117.000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prochniewicz E., Lowe D.A., Spakowicz D.J., Higgins L., O'Conor K., Thompson L.V. Functional, structural, and chemical changes in myosin associated with hydrogen peroxide treatment of skeletal muscle fibers. Am. J. Physiol. Cell Physiol. 2008;294:C613–26. doi: 10.1152/ajpcell.00232.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng J., Xie H., Meany D.L., Thompson L.V., Arriaga E.A., Griffin T.J. Quantitative proteomic profiling of muscle type-dependent and age-dependent protein carbonylation in rat skeletal muscle mitochondria. J. Gerontol. A Biol. Sci. Med. Sci. 2008;63:1137–1152. doi: 10.1093/gerona/63.11.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lourenco dos Santos S., Baraibar M.A., Lundberg S., Eeg-Olofsson O., Larsson L., Friguet B. Oxidative proteome alterations during skeletal muscle ageing. Redox Biol. 2015;5:267–274. doi: 10.1016/j.redox.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Picotti P., Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 56.Deshmukh A.S., Murgia M., Nagaraj N., Treebak J.T., Cox J., Mann M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol. Cell. Proteomics. 2015;14:841–853. doi: 10.1074/mcp.M114.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Margaritelis N.V., Cobley J.N., Paschalis V., Veskoukis A.S., Theodorou A.A., Kyparos A. Principles for integrating reactive species into in vivo biological processes: examples from exercise physiology. Cell. Signal. 2016;28:256–271. doi: 10.1016/j.cellsig.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 58.Marino S.M., Gladyshev V.N. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 2010;404:902–916. doi: 10.1016/j.jmb.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gross S.M., Lehman S.L. Accessibility of myofilament cysteines and effects on ATPase depend on the activation state during exposure to oxidants. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.