Abstract
Neuraminidase-1 (NEU1) is the predominant sialidase expressed in human airway epithelia and lung microvascular endothelia where it mediates multiple biological processes. We tested whether the NEU1-selective sialidase inhibitor, C9-butyl-amide-2-deoxy-2,3-dehydro-N-acetylneuraminic acid (C9-BA-DANA), inhibits one or more established NEU1-mediated bioactivities in human lung cells. We established the IC50 values of C9-BA-DANA for total sialidase activity in human airway epithelia, lung microvascular endothelia and lung fibroblasts to be 3.74 µM, 13.0 µM and 4.82 µM, respectively. In human airway epithelia, C9-BA-DANA dose-dependently inhibited flagellin-induced, NEU1-mediated mucin-1 ectodomain desialylation, adhesiveness for Pseudomonas aeruginosa and shedding. In lung microvascular endothelia, C9-BA-DANA reversed NEU1-driven restraint of cell migration into a wound and disruption of capillary-like tube formation. NEU1 and its chaperone/transport protein, protective protein/cathepsin A (PPCA), were differentially expressed in these same cells. Normalized NEU1 protein expression correlated with total sialidase activity whereas PPCA expression did not. In contrast to eukaryotic sialidases, C9-BA-DANA exerted far less inhibitory activity for three selected bacterial neuraminidases (IC50 > 800 µM). Structural modeling of the four human sialidases and three bacterial neuraminidases revealed a loop between the seventh and eighth strands of the β-propeller fold, that in NEU1, was substantially shorter than that seen in the six other enzymes. Predicted steric hindrance between this loop and C9-BA-DANA could explain its selectivity for NEU1. Finally, pretreatment of mice with C9-BA-DANA completely protected against flagellin-induced increases in lung sialidase activity. Our combined data indicate that C9-BA-DANA inhibits endogenous and ectopically expressed sialidase activity and established NEU1-mediated bioactivities in human airway epithelia, lung microvascular endothelia, and fibroblasts in vitro and murine lungs in vivo.
Keywords: lung, NEU1, neuraminidase, PPCA, sialidase
Introduction
The surface of all eukaryotic cells is sialylated (Varki and Varki 2007; Cohen and Varki 2010). Glycoproteins and glycolipids expressed on the cell surface contain oligosaccharide chains whose outermost positions can terminate with sialic acid. The sialylation state of these surface structures is regulated, at least in part, through sialidase catalytic activity (Monti et al. 2002; Monti et al. 2010; Miyagi and Yamaguchi 2012). At least four human sialidases, also referred to as neuraminidases, have been identified, NEU1, -2, -3 and -4 (Pshezhetsky et al. 1997; Miyagi et al. 1999; Monti et al. 1999; Comelli et al. 2003). Superimposed on this complex biological system, the human host also intimately interacts with numerous microbes that express their own neuraminidases (Kim et al. 2011; Lewis and Lewis 2012). Whether eukaryotic sialidases participate in specific biological processes is only now being explored and remains incompletely understood. Sialidases reportedly influence growth factor and Toll-like receptor (TLR) responsiveness and signaling (Hinek et al. 2008; Amith et al. 2009, 2010; Abdulkhalek et al. 2011, 2012; Feng et al. 2012; Lillehoj et al. 2012; Abdulkhalek and Szewczuk 2013), insulin receptor activation and downstream signaling (Arabkhari et al. 2010; Dridi et al. 2013), elastin fiber assembly (Hinek et al. 2006; Duca et al. 2007; Starcher et al. 2008), neuronal and muscle cell differentiation (Fanzani et al. 2003; Kappagantula et al. 2014), angiogenesis (Lee et al. 2014), tumor cell biology and cancer metastasis (Miyagi et al. 2004, 2008; Uemura et al. 2009), sperm function during fertilization (Ma et al. 2012) and bacterial pathogenesis (Chen et al. 2011; Lillehoj et al. 2012). Our research group has focused on sialidase biology in three key cell types within the human lung, i.e. airway epithelia (Lillehoj et al. 2012, 2015), microvascular endothelia (Cross et al. 2012; Lee et al. 2014) and fibroblasts (Luzina et al. 2016). First, we found that NEU1 was the predominant sialidase expressed in human airway epithelia, lung microvascular endothelia and lung fibroblasts (Cross et al. 2012; Lillehoj et al. 2012; Luzina et al. 2016). In human airway epithelial cells (HAECs), NEU1 impaired the migratory response during wound repair (Luzina et al. 2016). In these same cells, we previously established Pseudomonas aeruginosa (Pa)-derived flagellin as a signal-transducing ligand for the ectodomain of mucin-1 (MUC1-ED) (Lillehoj et al. 2012, 2015), and more recently, found that flagellin stimulation increases NEU1 association with and desialylation of the MUC1-ED (Lillehoj et al. 2015). NEU1-mediated MUC1-ED desialylation increased both its adhesiveness for flagellin-expressing Pa and its shedding from the HAEC surface (Lillehoj et al. 2015). In human pulmonary microvascular endothelial cell (HPMEC)s, we found that NEU1 restrained HPMEC migration into a wound (Cross et al. 2012; Lee et al. 2014) and disrupted HPMEC capillary-like tube formation, i.e. in vitro angiogenesis (Lee et al. 2014). More recently, we found that NEU1 expression is increased in lung tissues of patients with idiopathic pulmonary fibrosis (IPF) (Luzina et al. 2016). In these reports, the impact of NEU1 on any specific cellular response was established through prior siRNA-induced silencing of NEU1 and NEU1 overexpression (Cross et al. 2012; Lillehoj et al. 2012, 2015; Lee et al. 2014; Luzina et al. 2016). However, such interventions would not be easily applied to human disease states in vivo.
NEU1 participates in multiple cellular functions (Monti et al. 2002, 2010; Miyagi and Yamaguchi 2012). In most human cells, NEU1 is co-expressed with NEU2, -3 and -4 (Monti et al. 2002, 2010; Miyagi and Yamaguchi 2012). Selective inhibition of NEU1 without off-target cross-inhibition of any of the other three isoforms could provide insight into NEU1 function and/or therapeutic possibilities for clinical conditions in which NEU1 might be overexpressed and/or activated. Furthermore, NEU1-null mice display lung pathology and premature death, thereby limiting studies of NEU1 function (Starcher et al. 2008). Most previous studies of neuraminidase/sialidase inhibition have utilized recombinant enzymes in cell-free experimental systems (Hata et al. 2008; Magesh et al. 2008, 2009; Zhang et al. 2013). In the current studies, we tested the ability of the only reported NEU1-selective inhibitor, C9-butyl-amide-2-deoxy-2,3-dehydro-N-acetylneuraminic acid (C9-BA-DANA) (Magesh et al. 2008), to inhibit NEU1-mediated biological processes in both in vitro cell culture systems and in an intact murine model.
Although the overall sequence identities between individual members of the neuraminidase/sialidase superfamily are low, each contains several conserved motifs and their catalytic domains share a common six-bladed β-propeller fold architecture (Monti et al. 2002; Miyagi and Yamaguchi 2012). Of the four known human sialidases, the crystal structure of only NEU2 has been solved (Chavas et al. 2005). Several research groups have now designed and synthesized pharmacologic inhibitors selective for both prokaryotic neuraminidases (von Itzstein, 2007) and eukaryotic sialidases (Magesh et al. 2006, 2008, 2009; Zhang et al. 2013). One standard inhibitor, 2-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA), is a broad spectrum, transition state analog inhibitor of viral, bacterial and mammalian neuraminidases/sialidases (Meindi and Tuppy 1969; Burmeister et al. 1993). We designed and synthesized a series of 10 N-amide-linked C9-modified analogs of DANA and tested their inhibitory activities against all four human sialidases (Magesh et al. 2008). We found that the C9 N-amide-linked n-butyl derivative of DANA, C9-BA-DANA, displayed ~15-fold increased inhibitory activity for NEU1 and >100-fold decreased inhibitory activity for NEU2, -3 and -4 compared to that seen with unmodified DANA. When C9-BA-DANA was tested against each human sialidase expressed in HEK-293 cells, it was >200-fold more selective for NEU1 (IC50 = 10 µM) versus NEU2, -3 and -4 (IC50 > 1000 µM). Whether C9-BA-DANA might inhibit bacterial neuraminidases has not been tested. In the current studies, we have tested whether C9-BA-DANA might block endogenous and/or ectopically expressed sialidase activity in human airway epithelia, lung microvascular endothelia and/or lung fibroblasts, and inhibit established NEU1-driven biological processes in two of these same three cell systems. The inhibitory activity for C9-BA-DANA against prokaryotic neuraminidases in vitro, and total murine lung sialidase activity in vivo, was also studied.
Results
C9-BA-DANA inhibits sialidase activity in HAECs
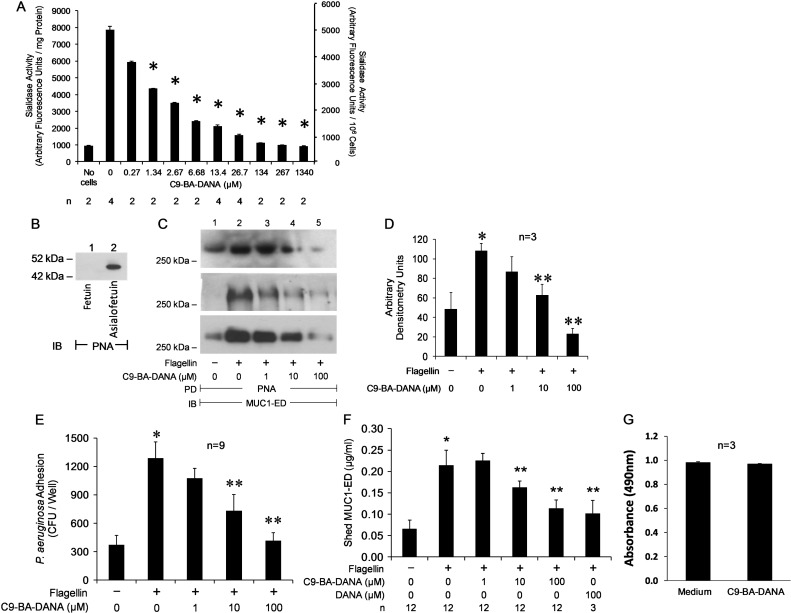
We previously established NEU1 as the predominant sialidase expressed in HAECs (Lillehoj et al. 2012) and defined its participation in multiple bioactivities in these same cells (Lillehoj et al. 2012, 2015; Luzina et al. 2016). To better control NEU1-mediated events in the airway epithelium, we asked whether the NEU1-selective inhibitor, C9-BA-DANA (Magesh et al. 2008), might inhibit sialidase activity in A549 cells. A fixed amount of A549 cellular protein or a fixed number of A549 cells were assayed for sialidase activity for the fluorogenic substrate, 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (4-MU-NANA), in the presence of increasing concentrations of C9-BA-DANA (Figure 1A). C9-BA-DANA, at concentrations ≥1.34 µM, dose-dependently inhibited A549 cell sialidase activity with an IC50 of 3.74 µM.
Fig. 1.
C9-BA-DANA inhibits sialidase activity and flagellin-stimulated, NEU1-mediated MUC1-ED desialylation, increases in adhesiveness for Pa, and shedding in HAECs. (A) A fixed amount of A549 cellular protein (0.63 mg) or a fixed number of A549 cells (106 cells/reaction) were assayed for sialidase activity for the fluorogenic substrate, 4-MU-NANA, in the presence of increasing concentrations of C9-BA-DANA. Vertical bars represent mean (±SE) sialidase activity expressed as arbitrary fluorescence units/mg protein (left Y-axis) or arbitrary fluorescence units/106 cells (right Y-axis) (n = 2/4). *, decreased sialidase activity in the presence versus absence of C9-BA-DANA at P < 0.05. (B) The positive control asialofetuin and the negative control fetuin (1.0 µg of each) were processed for PNA lectin blotting. IB, immunoblot. Molecular weight in kDa indicated on the left. (C-F) A549 cells were incubated for 30 min (C–E) or 24 h (F) with 10 ng/mL of Pa flagellin or medium alone in the presence of increasing concentrations of C9-BA-DANA (C–F) or 100 μM of DANA (F). (C) The cells were lysed and the lysates were incubated with PNA-agarose and the PNA-binding proteins processed for MUC1-ED immunoblotting. PD, pull-down; IB, immunoblot. Molecular weight in kDa indicated on the left. (D) Densitometric analyses of the blots in (C). Vertical bars represent mean (±SE) PNA-bound MUC1-ED signal (n = 3). *, increased MUC1-ED desialylation in flagellin-treated versus untreated cells at P < 0.05. **, decreased MUC1-ED desialylation in C9-BA-DANA treated versus untreated cells at P < 0.05. (E) The cells were fixed, washed and incubated for 30 min with Pa (MOI = 100). Nonadherent Pa was removed by washing, and CFUs of cell-bound Pa quantified. Vertical bars represent mean (±SE) Pa CFUs/well (n = 9). *, increased Pa adhesion to flagellin-treated versus untreated cells at P < 0.05. **, decreased Pa adhesion to C9-BA-DANA treated versus untreated cells at P < 0.05. (F) The cells were cultured for 24 h after which MUC1-ED levels in supernates were quantified by ELISA and normalized to total cellular protein. Vertical bars represent mean (±SE) shed MUC1-ED normalized to total cellular protein (n = 3/12). *, increased shed MUC1-ED in supernates of flagellin-treated versus untreated cells at P < 0.05. **, decreased shed MUC1-ED in supernates in C9-BA-DANA treated versus untreated cells at P < 0.05. (G) A549 cells were cultured for 24 h in the presence of 100 µM of C9-BA-DANA or medium alone after which MTT was added for an additional 4 h. After DMSO solubilization, the A490 was measured. Vertical bars represent the mean (±SE) A490 (n = 3). Each result is representative of ≥3 independent experiments.
C9-BA-DANA inhibits flagellin-stimulated, NEU1-mediated MUC1-ED desialylation, increases in adhesiveness for Pa, and shedding
We previously demonstrated that Pa-derived flagellin is a ligand for the MUC1-ED (Lillehoj et al. 2012, 2015) and that its engagement of this receptor increases NEU1 association with and desialylation of MUC1-ED (Lillehoj et al. 2015). Furthermore, NEU1-mediated desialylation of MUC1-ED renders it hyperadhesive for Pa and increases its shedding from the cell surface (Lillehoj et al. 2015). First, we asked whether C9-BA-DANA could inhibit MUC1-ED desialylation as measured by quantitative peanut agglutinin (PNA) lectin blotting. The PNA lectin probe was validated using asialofetuin and fetuin as nonsialylated positive and sialylated negative controls, respectively (Figure 1B). As anticipated, PNA recognized asialofetuin (lane 2) but not fetuin (lane 1). Flagellin increased PNA recognition of MUC1-ED by 2.2-fold and C9-BA-DANA at concentrations ≥10 µM dose-dependently protected against this desialylation (Figure 1C,D). Although C9-BA-DANA at 1.0 µM diminished MUC1-ED desialylation, this decrease did not achieve statistical significance (P = 0.16) (Figure 1D). C9-BA-DANA at 100 µM completely reversed the flagellin-induced MUC1-ED desialylation. Since C9-BA-DANA clearly inhibited MUC1-ED desialylation, we asked whether it might also diminish flagellin-induced increases in Pa adhesion to MUC1-ED-expressing cells (Figure 1E) and/or MUC1-ED shedding (Figure 1F). As anticipated, flagellin increased Pa adhesion by 3.5-fold, and again, C9-BA-DANA at concentrations ≥10 µM dose-dependently protected against increases in Pa adhesion with almost complete reversal at 100 µM (Figure 1E). Similarly, flagellin increased MUC1-ED shedding 3.2-fold and C9-BA-DANA at concentrations ≥10 µM dose-dependently decreased shedding (Figure 1F). However, C9-BA-DANA at 100 µM only diminished flagellin-provoked shedding by 46.8% (Figure 1F). We asked whether the more broad spectrum, less specific sialidase inhibitor, DANA, might more effectively decrease MUC1-ED shedding through its ability to target sialidase(s) other than NEU1. DANA, at an equivalent concentration, inhibited shedding by 52.7%, comparable to the inhibition seen with the more NEU1-selective inhibitor, C9-BA-DANA (Figure 1F). No difference in viability was detected between A549 cells cultured for 24 h in the presence of 100 µM of C9-BA-DANA or medium alone (Figure 1G), indicating that none of the above results could be ascribed to cytotoxic effects of the compound. These combined data indicate that C9-BA-DANA dose-dependently protected versus flagellin-stimulated MUC1-ED desialylation, which in turn, counter-regulated increases in both Pa adhesion and MUC1-ED shedding.
C9-BA-DANA inhibits sialidase activity in HPMECs
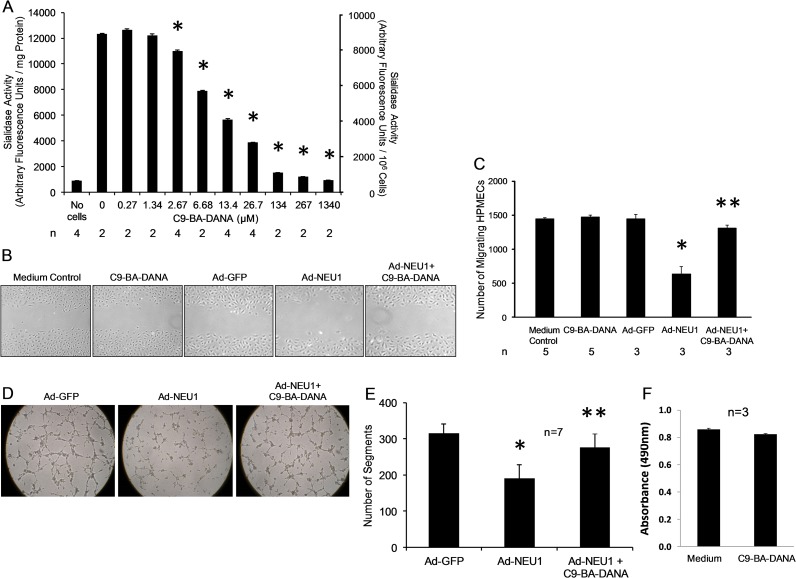
We previously found NEU1 to be the predominant sialidase expressed in HPMECs (Cross et al. 2012). We asked whether C9-BA-DANA might inhibit HPMEC sialidase activity. A fixed amount of HPMEC protein or a fixed number of HPMECs were assayed for sialidase activity in the 4-MU-NANA assay in the presence of increasing concentrations of C9-BA-DANA (Figure 2A). C9-BA-DANA, at concentrations ≥2.67 µM, dose-dependently inhibited HPMEC sialidase activity with an IC50 of 13.0 µM.
Fig. 2.
C9-BA-DANA inhibits sialidase activity and counteracts NEU1-mediated inhibition of HPMEC migration and capillary-like tube formation. (A) A fixed amount of HPMEC protein (0.71 mg) or a fixed number of HPMECs (106 cells/reaction) were assayed for sialidase activity for 4-MU-NANA in the presence of increasing concentrations of C9-BA-DANA. Vertical bars represent mean (±SE) sialidase activity expressed as arbitrary fluorescence units/mg protein (left Y-axis) or arbitrary fluorescence units/106 cells (right Y-axis) (n = 2/4). *, decreased sialidase activity in the presence versus absence of C9-BA-DANA at P < 0.05. (B–E) HPMECs and HPMECs infected with Ad-NEU1 or Ad-GFP (MOI = 100) were cultured for 48 h. (B, C) A single wound was made across the diameter of each confluent monolayer, after which the wounded monolayers were incubated for 24 h in the absence or presence of 134 µM of C9-BA-DANA and photomicrographs taken. (C) The number of migrating HPMECs was calculated using ImageJ software for comparison with that observed in the same wounded monolayer at 0 h, as a measure of migration into the wound. Each vertical bar represents mean (±SE) migration into the wound (n = 3/5). *, decreased migration in Ad-NEU1 infected versus Ad-GFP infected cells at P < 0.05. **, increased migration in C9-BA-DANA treated, Ad-NEU1 infected vs. untreated, Ad-NEU1 infected cells at P < 0.05. (D) HPMECs were seeded on Matrigel-coated wells at 1.5 × 104 cells/well, and after 6 h, tubular structures photographed. (E) Segments of capillary-like tubes per HPF were counted. Vertical bars represent mean (±SE) segments/HPF (n = 7). *, decreased tube formation in Ad-NEU1 infected versus Ad-GFP infected cells at P < 0.05. **, increased tube formation in C9-BA-DANA treated, Ad-NEU1 infected versus untreated, Ad-NEU1 infected cells at P < 0.05. (F) HPMECs were cultured for 24 h in the presence of 100 µM of C9-BA-DANA or medium alone after which MTT was added for an additional 4 h. After DMSO solubilization, the A490 was measured. Vertical bars represent mean (±SE) A490 (n = 3). Each result is representative of ≥3 independent experiments. This figure is available in black and white in print and in color at Glycobiology online.
C9-BA-DANA counteracts NEU1-mediated inhibition of HPMEC migration and capillary-like tube formation
We previously reported that NEU1 overexpression in HPMECs restrains their migration into a wound (Cross et al. 2012; Lee et al. 2014) and disrupts in vitro angiogenesis (Lee et al. 2014). HPMECs and HPMECs infected with adenovirus (Ad) encoding human NEU1 (Ad-NEU1) or green fluorescent protein (Ad-GFP) as a control, were studied in the presence or absence of C9-BA-DANA, for migration in wounding assays (Figure 2B,C) and capillary-like tube formation on Matrigel (Figure 2D,E). In the uninfected HPMECs, C9-BA-DANA did not alter cell migration compared to simultaneous medium controls (Figure 2B,C). NEU1 overexpression diminished HPMEC migration into a wound by 58.6% and the presence of C9-BA-DANA reversed the NEU1-driven inhibition by 84.2% (Figure 2B,C). Similarly, NEU1 overexpression decreased capillary-like tube formation by 39.7% and C9-BA-DANA reversed the NEU1-mediated inhibition by 70.0% (Figure 2D,E). No difference in viability was detected between HPMECs cultured for 24 h in the presence of 100 µM of C9-BA-DANA or medium alone (Figure 2F). These combined data indicate that C9-BA-DANA protects against NEU1-mediated inhibition of both the HPMEC migratory response to injury and capillary-like tube formation or in vitro angiogenesis.
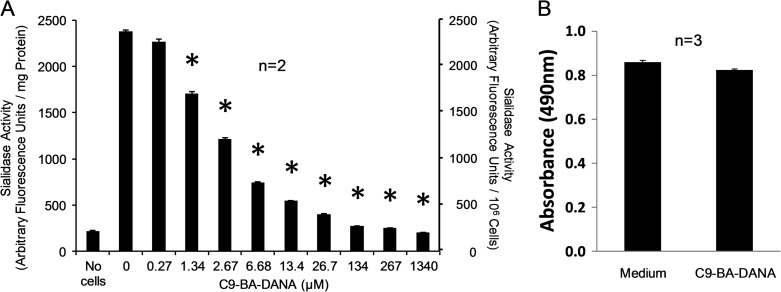
C9-BA-DANA inhibits sialidase activity in HLFs
We recently reported that human lung fibroblasts (HLFs) express NEU1, and NEU1 expression is increased in fibroblasts obtained from the lungs of IPF patients (Luzina et al. 2016). To test whether C9-BA-DANA inhibits sialidase activity in HLFs, a fixed amount of HLF protein or a fixed number of HLFs were assayed for sialidase activity for the fluorogenic substrate, 4-MU-NANA, in the presence of increasing concentrations of C9-BA-DANA (Figure 3A). C9-BA-DANA, at concentrations ≥1.34 µM, dose-dependently inhibited HLF sialidase activity with an IC50 of 4.82 µM. No difference in viability was detected between HLFs cultured for 24 h in the presence of 100 µM of C9-BA-DANA or medium alone (Figure 3B).
Fig. 3.
C9-BA-DANA inhibits sialidase activity in HLFs. (A) A fixed amount of HLF protein (1.00 mg) or a fixed number of HLFs (106 cells/reaction) were assayed for sialidase activity for 4-MU-NANA in the presence of increasing concentrations of C9-BA-DANA. Vertical bars represent mean (±SE) sialidase activity expressed as arbitrary fluorescence units/mg protein (left Y-axis) or arbitrary fluorescence units/106 cells (right Y-axis) (n = 2). *, decreased sialidase activity in the presence versus absence of C9-BA-DANA at P < 0.05. (B) HLFs were cultured for 24 h in the presence of 100 µM of C9-BA-DANA or medium alone after which MTT was added for an additional 4 h. After DMSO solubilization, the A490 was measured. Vertical bars represent mean (±SE) A490 (n = 3). Each result is representative of three independent experiments.
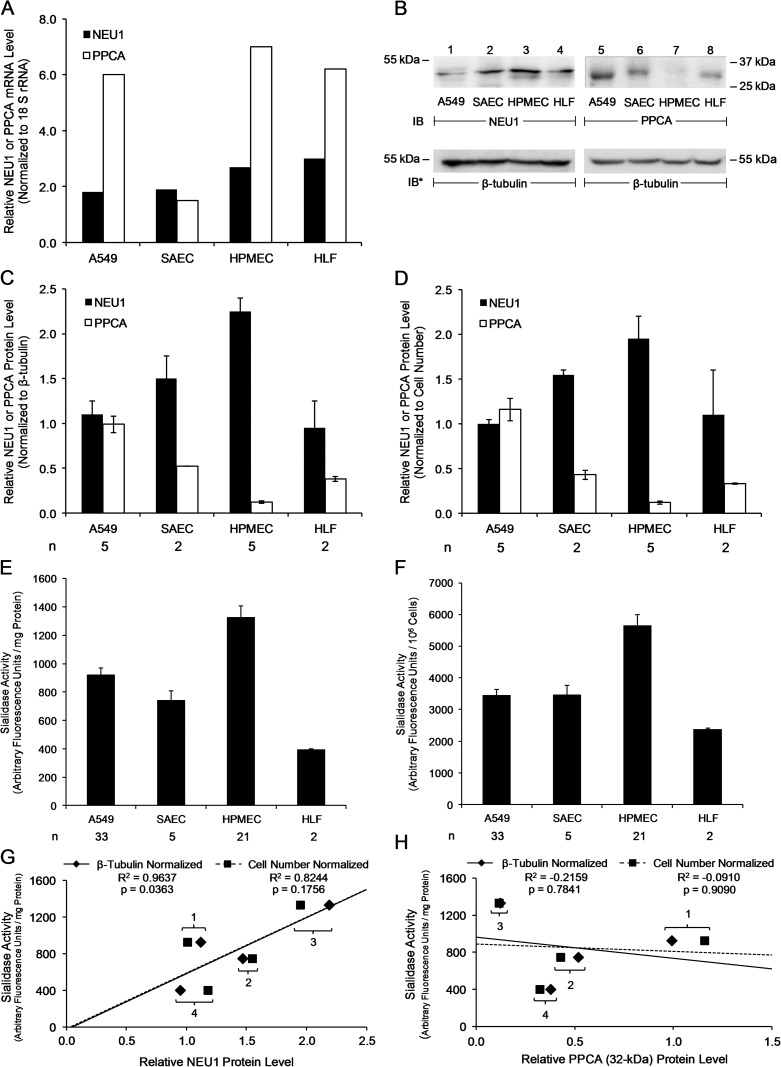
Differential expression of NEU1 and PPCA in HAECs, HPMECs and HLFs
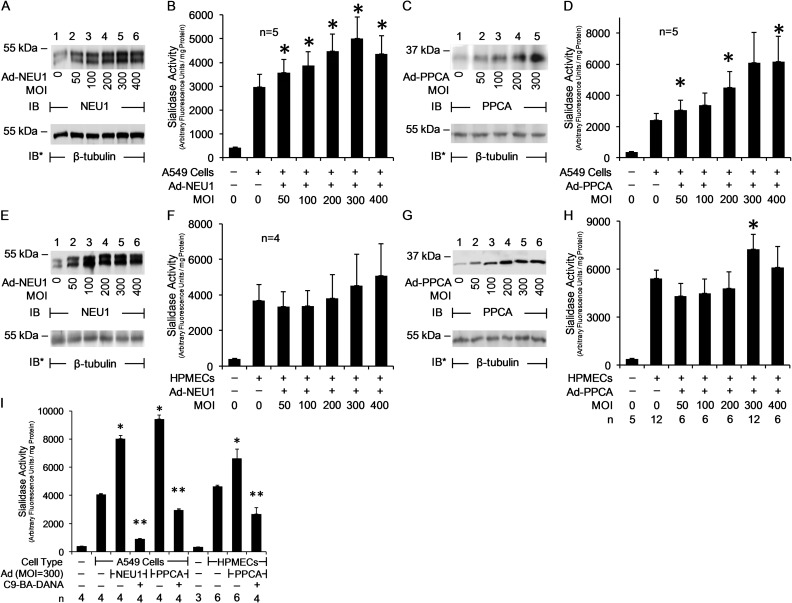
We found that resting baseline total sialidase activities for the 4-MU-NANA substrate, expressed as mean ± SE arbitrary fluorescence units/mg cellular protein, were 7866 ± 206 in A549 cells (Figure 1A), 12,345 ± 51.9 in HPMECs (Figure 2A) and 2384 ± 9.95 in HLFs (Figure 3A). Also resting baseline total sialidase activities, expressed as mean ± SE arbitrary fluorescence units/106 cells, were 5000 ± 128 in A549 cells (Figure 1A), 8705 ± 36.6 in HPMECs (Figure 2A) and 2386 ± 9.97 in HLFs (Figure 3A). The IC50 values for the NEU1 inhibitor, C9-BA-DANA, for sialidase activity in each of these same three cell types were 3.74, 13.0 and 4.82 µM, respectively (Figures 1A, 2A, 3A). The IC50 values for C9-BA-DANA in HPMECs (13.0 µM) were greater than its IC50 value in A549 cells (3.74 µM) or HLFs (4.82 µM), whereas its IC50 values in A549 cells and HLFs were not significantly different. We asked whether differential NEU1 and/or protective protein/cathepsin A (PPCA) expression might explain these differences in total sialidase activity and IC50 values for C9-BA-DANA between these cell types. First, a real-time RT-PCR approach was adopted to detect mRNA for NEU1 and PPCA in A549 cells, small airway epithelial cells (SAECs), HPMECs and HLFs (Figure 4A). Each NEU1 or PPCA mRNA level was normalized to the 18 S rRNA level for the same sample. These data indicate that at the mRNA level NEU1 expression was highest in HLFs, followed by HPMECs, with the lowest in the two airway epithelia. PPCA mRNA expression was highest in HPMECs, followed by HLFs and A549 cells, and lowest in SAECs. To establish relative NEU1 and PPCA protein expression in these cells, quantitative immunoblotting (Figure 4B) was applied where the NEU1 or PPCA signal was normalized to both β-tubulin expression (Figure 4C) and cell number (Figure 4D). When normalized to β-tubulin or cell number, relative NEU1 protein expression in the four cell types was HPMECs > SAECs > HLFs and A549 cells. When normalized to β-tubulin or cell number, relative PPCA protein expression in the four cell types was A549 cells > SAECs and HLFs > HMPECs. Finally, total sialidase activity normalized to mg protein (Figure 4E) or cell number (Figure 4F) was highest in HPMECs, followed by comparable levels in A549 cells and SAECs, and lowest in HLFs.
Fig. 4.
Differential expression of NEU1 and PPCA in HAECs, HPMECs and HLFs. (A) Quantitative RT-PCR analysis of NEU1 and PPCA mRNA levels in A549 cells, SAECs, HPMECs and HLFs. Vertical bars represent mean NEU1 or PPCA mRNA levels normalized to 18 S rRNA mRNA levels. (B) Equivalent numbers of A549 cells (lanes 1, 5), SAECs (lanes 2, 6), HPMECs (lanes 3, 7) and HLFs (lanes 4, 8) were lysed and equivalent volumes were processed for NEU1 (lanes 1–4) and PPCA (lanes 5–8) immunoblotting. To control for protein loading and transfer, blots were stripped and reprobed for β-tubulin. IB, immunoblot; IB*, immunoblot after stripping. Molecular weight in kDa is indicated on left (lanes 1–4) and right (lanes 5–8). (C, D) Densitometric analyses of the blots in (B). Vertical bars represent mean (±SE) NEU1 or PPCA signal normalized to β-tubulin signal in the same lane in the same stripped and reprobed blot (C) or cell number (D) (n = 2/5). (E, F) A fixed amount of cellular protein (A549 cells, 0.63 mg; SAECs, 0.78 mg; HPMECs, 0.71 mg; HLFs, 1.00 mg) (E) or a fixed number of A549 cells, SAECs, HPMECs or HLFs (106 cells/reaction) (F) were assayed for sialidase activity for 4-MU-NANA. Vertical bars represent mean (±SE) sialidase activity corrected for activity in the absence of cells and expressed as arbitrary fluorescence units/mg protein (E) or arbitrary fluorescence units/106 cells (F). In (E, F), the n for each experimental group is indicated beneath each vertical bar. (G, H) Correlation of sialidase activity for 4-MU-NANA in A549 cells (1), SAECs (2), HPMECs (3) and HLFs (4) versus (G) NEU1 protein levels or (H) PPCA protein levels normalized to β-tubulin or cell number. Each result is representative of three independent experiments.
The relative sialidase activity for the 4-MU-NANA substrate directly correlated with NEU1 protein expression levels normalized to β-tubulin (Figure 4G). Although the line of correlation between sialidase activities to NEU1 expression normalized to cell number was superimposable on that calculated for NEU1 expression normalized to β-tubulin, this correlation did not achieve statistical significance (Figure 4G). In contrast to NEU1 levels normalized to β-tubulin, relative sialidase activity did not increase with increasing relative PPCA protein levels normalized to either β-tubulin or cell number (Figure 4H). To clarify this issue, low NEU1-expressing, high PPCA-expressing A549 cells and high NEU1-expressing, low PPCA-expressing HPMECs were infected with increasing multiplicities of infection (MOIs) of Ad-NEU1 or Ad encoding for PPCA (Ad-PPCA), and the cells processed for NEU1 (Figure 5A,E) or PPCA (Figure 5C,G) immunoblotting and sialidase activity for the 4-MU-NANA substrate (Figure 5B,D,F,H). Infection of A549 cells and HPMECs with increasing MOIs of Ad-NEU1 or Ad-PPCA dose-dependently increased NEU1 and PPCA protein expression, respectively (Figure 5A,C,E,G). Ectopic overexpression of either NEU1 or PPCA in A549 cells increased sialidase activity by up to 1.7-fold and >2.6-fold, respectively (Figure 5B,D). However, in high NEU1-expressing, low PPCA-expressing HPMECs, overexpression of NEU1 was not associated with increased sialidase activity (Figure 5F), whereas PPCA overexpression was (Figure 5H). It is likely that NEU1 overexpression in HPMECs fails to increase sialidase activity due to their low expression of PPCA, an established NEU1 chaperone that is required for its catalytic activity. To establish the ability of C9-BA-DANA to inhibit sialidase activity resulting from ectopic expression of either NEU1 or PPCA in A549 cells and HPMECs, these same cells were infected with Ad-NEU1 or Ad-PPCA (MOI = 300), cultured for 48 h and assayed for sialidase activity in the presence or absence of C9-BA-DANA (Figure 5I). In NEU1-overexpressing A549 cells, C9-BA-DANA decreased sialidase activity by 88.8%, and in PPCA-overexpressing A549 cells by 68.6%. In HPMECs, the inhibitor decreased sialidase activity in PPCA-overexpressing cells by 64.1%. These combined data indicate that in either A549 cells or HPMECs with enhanced sialidase activity provoked by either NEU1 or PPCA overexpression, C9-BA-DANA dramatically reduces the activity.
Fig. 5.
Increased sialidase activity in NEU1- or PPCA-overexpressing A549 cells and HPMECs is inhibited by C9-BA-DANA. A549 cells (A–D) and HPMECs (E–H) were infected with increasing MOIs of Ad-NEU1 (A, B, E, F) or Ad-PPCA (C, D, G, H). (A, C, E, G) After 48 h, the cells were lysed and equal protein aliquots of lysates processed for NEU1 (A, E) or PPCA (C, G) immunoblotting. To control for protein loading and transfer, the blots were stripped and reprobed for β-tubulin. IB, immunoblot; IB*, immunoblot after stripping. Molecular weight in kDa is indicated on left. (B, D, F, H) Sialidase activity for the 4-MU-NANA substrate was assayed in A549 cells (B, D) and HPMECs (F, H) infected with increasing MOIs of either Ad-NEU1 (B, F) or Ad-PPCA (D, H). Vertical bars represent mean (±SE) sialidase activity normalized to mg of cellular protein. *, increased sialidase activity in cells infected with Ad-NEU1 or Ad-PPCA versus uninfected cells at P < 0.05. (I) A549 cells and HPMECs infected with Ad-NEU1 or Ad-PPCA (MOI = 300) were cultured for 48 h and assayed for sialidase activity for the 4-MU-NANA substrate in the presence of 134 μM of C9-BA-DANA or medium alone. Vertical bars represent mean (±SE) sialidase activity corrected for activity in the absence of cells and normalized to mg of cellular protein. *, increased sialidase activity in Ad-NEU1-infected or Ad-PPCA-infected cells versus uninfected cells at P < 0.05. **, decreased sialidase activity in C9-BA-DANA-treated cells versus untreated cells at P < 0.05. The n for each experimental group is indicated in each panel. Each result is representative of three independent experiments.
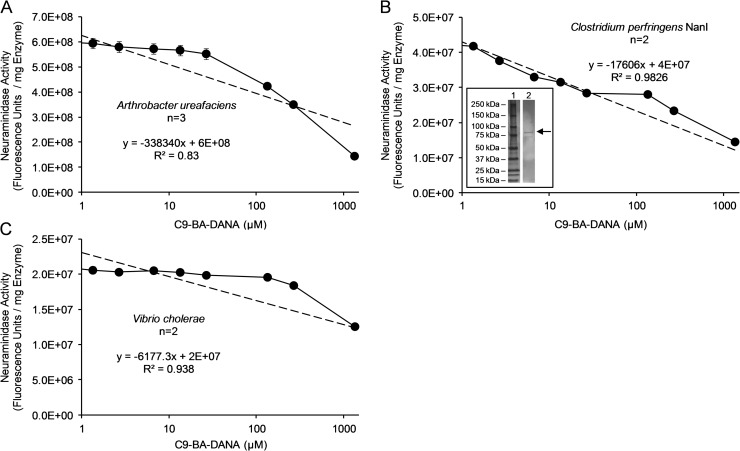
C9-BA-DANA displays less inhibitory activity for neuraminidases expressed by Arthrobacter ureafaciens, Clostridium perfringens and Vibrio cholerae
We reported that C9-BA-DANA selectively inhibited human NEU1 (IC50 = 10 µM) relative to human NEU2, -3 and -4 (IC50 > 1000 µM) (Magesh et al. 2008). Among the four known human NEUs, NEU1 shares the least homology with the other three (Monti et al. 2002, 2010; Miyagi and Yamaguchi 2012). In fact, NEU1 might be more closely related to some prokaryotic neuraminidases than the other human NEUs (Kiyohara et al. 2011). Furthermore, numerous microbes that express their own neuraminidases are known to colonize, or in some instances, infect humans (Kim et al. 2011; Lewis and Lewis 2012). To establish whether C9-BA-DANA might inhibit bacterial neuraminidases, we established the IC50 for C9-BA-DANA for each of three bacterial neuraminidases selected for their phylogenetic relatedness to human NEU1 (Kiyohara et al. 2011) (Figure 6). We found the IC50 for C9-BA-DANA to be 890 µM for Arthrobacter ureafaciens neuraminidase (Figure 6A), and >1000 µM for both Clostridium perfringens neuraminidase NanI (Figure 6B) and Vibrio cholerae neuraminidase (Figure 6C). These combined data indicate that C9-BA-DANA is selective for human NEU1 not only relative to the other human NEUs (Magesh et al. 2008), but also relative to the bacterial neuraminidases studied here.
Fig. 6.
C9-BA-DANA exerts far less inhibitory activity for prokaryotic neuraminidases. Fixed concentrations of the neuraminidases expressed by (A) A. ureafaciens (7.5 ng/mL), (B) C. perfringens (NanI) (40 ng/mL) and (C) V. cholerae (625 ng/mL) were each assayed for sialidase activity for 4-MU-NANA in the presence of increasing concentrations of C9-BA-DANA. The concentration chosen for each bacterial neuraminidase generated catalytic activity for the 4-MU-NANA substrate comparable to that seen with the simultaneous positive control using A549 cells at 106 cells/reaction. Each symbol represents mean (±SE) neuraminidase activity corrected for activity in the absence of bacterial neuramindase and expressed as arbitrary fluorescence units/mg protein (n = 2/3). The broken line in each panel represents the linear trendline with the corresponding equation and correlation coefficient (R2) value determined using Microsoft Excel. Insert in (B) represents the silver-stained gel of resolved C. perfringens neuraminidase preparation. Molecular weight in kDa indicated on left. Arrow on right indicates NanI band of interest. Each result is representative of two or three independent experiments.
Predicted steric hindrances to C9-BA-DANA in bacterial neuraminidases
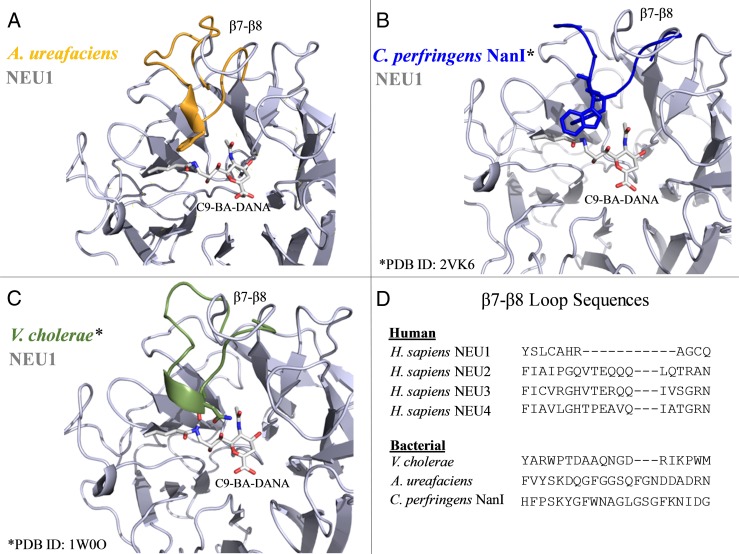
To better understand why C9-BA-DANA is selective for NEU1, not only among the human sialidases (Magesh et al. 2008), but also relative to the bacterial neuraminidases studied here, we compared the X-ray crystal structures of C. perfringens NanI (Protein Database ID 2VK6) and the V. cholerae neuraminidase (Protein Database ID 1W0O) to models of the neuraminidase of A. ureafaciens and NEU1 bound with C9-BA-DANA, for which no high-resolution structures are available. Importantly, DANA, upon which C9-BA-DANA is based, is able to bind and inhibit all of the neuraminidases/sialidases used in this study (Meindi and Tuppy 1969; Burmeister et al. 1993). After superimposition of the X-ray crystal structures and models, we examined the results for any causes of gross steric or electrostatic incompatibility between C9-BA-DANA and the bacterial neuraminidases. We found that universally, the polypeptide loop between the seventh and eighth strands of the β-propeller fold (the β7–β8 loop) is substantially longer in the bacterial neuraminidases compared with the β7–β8 loop of human NEU1 (Figure 7A–C). Modeling studies predict that the longer β7–β8 loops of the bacterial neuraminidases may interfere with C9-BA-DANA accessibility to their catalytic domains. These steric clashes between amino acid side-chains extending from these loops and the n-butyl group linked to the C9 position of the inhibitor are absent in the NEU1-C9-BA-DANA model. Furthermore, amino acid sequence alignments between human NEU1 and human NEU2, -3 and -4 indicate that like the bacterial neuraminidases the β7–β8 loops in NEU2, -3 and -4 are longer than the β7–β8 loop in NEU1 (Figure 7D) (Magesh et al. 2006). The presence of an extended β7–β8 loop in NEU2, -3 and -4 might explain their relative resistance to inhibition by C9-BA-DANA, while absence of the n-butyl group in DANA might account for its ability to inhibit all four human sialidases. However, the ability of DANA molecules with phenyl-containing substituents at C9 to inhibit NEU2 and NEU3 argues that accommodation of larger substituents at C9 is possible for sialidases with longer β7–β8 loops under some circumstances, suggesting that favorable interactions with phenyl rings may drive loop remodeling (Magesh et al. 2008; Zhang et al. 2013). Similarly, a DANA-derivative with a large polar C9-substituent was found to be a highly potent inhibitor of NEU4, suggesting that electrostatic interactions with a methanol moiety at the tip of a triazol ring were similarly stabilizing (Albohy et al. 2013). The ability of NEU1 to accommodate a variety of hydrophobic moieties at C9, unlike NEU2, -3 and -4 (and the bacterial neuraminidases studied here) may indicate that these substituents prevent the β7–β8 loops of NEU2, -3 and -4 from adopting their native structures without stabilizing any alternative conformations. As previously reported (Magesh et al. 2008), other potential sites in NEU2, -3 and -4 for steric hindrance with C9-BA-DANA were identified, including a shared Tyr residue (Y181 in NEU2 and -3, Y179 in NEU4) outside of the β7–β8 loop.
Fig. 7.
Predicted steric hindrance to C9-BA-DANA binding to bacterial neuraminidases. Structural superimposition of the C9-BA-DANA–NEU1 complex (gray) with (A) A. ureafaciens neuraminidase (orange), (B) C. perfringens NanI (blue) and (C) V. cholerae neuraminidase (green). Only the β7–β8 loops of the bacterial neuraminidases are depicted. V. cholerae (National Center for Biotechnology Information, Protein Database ID #1W0O) and C. perfringens (National Center for Biotechnology Information, Protein Database ID #2VK6) neuraminidase structures are X-ray crystal structures. A. ureafaciens neuraminidase and NEU1 structures are based on homology modeling with the NEU2 X-ray crystal structure. (D) Amino acid sequence alignments of the β7–β8 loop from human NEU1, -2, -3 and -4 and the three bacterial neuraminidases are shown. This figure is available in black and white in print and in color at Glycobiology online.
C9-BA-DANA inhibits flagellin-induced increases in mouse lung sialidase activity in vivo
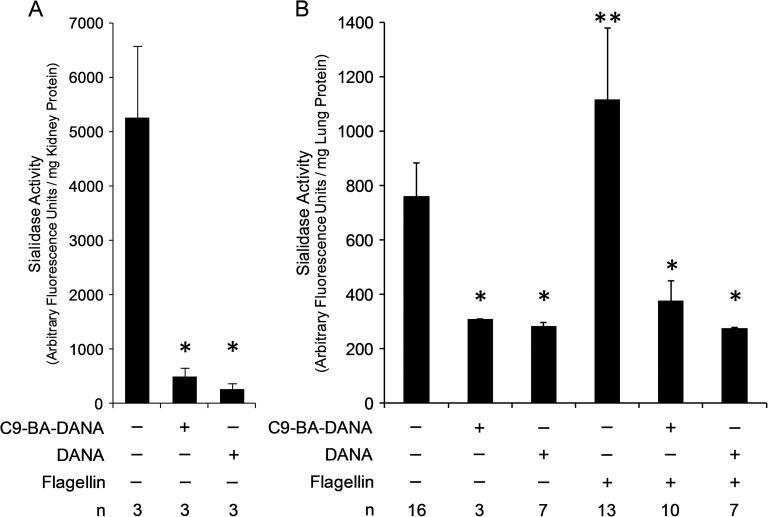
In wild type mice, the highest levels of sialidase activity were detected in kidney tissue, and when compared with the kidney tissues of NEU1-null mice, sialidase activity could be almost exclusively ascribed to NEU1 (de Geest et al. 2002). First, to test whether C9-BA-DANA could inhibit NEU1 catalytic activity in vivo, total sialidase activities for the 4-MU-NANA substrate in kidney tissues obtained from mice preadministered with the inhibitor or PBS vehicle alone were assayed (Figure 8A). Prior administration of C9-BA-DANA inhibited renal sialidase activity by 90.6% compared with the PBS controls. Preadministration of the broad spectrum sialidase inhibitor, DANA, diminished renal sialidase activity to a comparable degree (Figure 8A).
Fig. 8.
C9-BA-DANA prevents flagellin-induced increases in mouse lung sialidase activities in vivo. (A) Mice were injected intraperitoneally with 15 μg/kg of C9-BA-DANA (n = 3) or DANA (n = 3), or PBS vehicle alone (n = 3). After 24 h, mice were euthanized and kidneys were harvested, weighed and homogenized. Kidney homogenates were processed for assays for total protein levels and total sialidase activity for the 4-MU-NANA substrate. Vertical bars represent mean (±SE) total kidney sialidase activity in arbitrary fluorescence units/mg protein. *, decreased sialidase activity in kidneys of C9-BA-DANA- or DANA-treated versus PBS-infused mice at P < 0.05. (B) Mice were injected intraperitoneally with 15 μg/kg of C9-BA-DANA (n = 13) or DANA (n = 14) or PBS vehicle alone (n = 29). After 24 h, mice pretreated intraperitoneally with C9-BA-DANA were challenged intranasally with 100 ng/mouse of Pa-derived flagellin (n = 10) or PBS (n = 3), mice pretreated with DANA were challenged with flagellin (n = 7) or PBS (n = 7), and mice pretreated with PBS were challenged with flagellin (n = 13) or PBS (n = 16). After 4 h, mice were euthanized and lungs were harvested, weighed and homogenized. Lung homogenates were processed for assays for total protein levels and total sialidase activity for the 4-MU-NANA substrate. Vertical bars represent mean (±SE) lung sialidase activity corrected for activity in the absence of a tissue sample and expressed as arbitrary fluorescence units/mg protein. *, decreased sialidase activity in C9-BA-DANA- or DANA-treated lung tissue versus PBS controls at P < 0.05. **, increased sialidase activity in flagellin-stimulated lung tissue versus PBS control at P < 0.05. Each result is representative of three independent experiments.
C9-BA-DANA dose-dependently inhibited sialidase activity in in vitro cultures of HAECs (Figure 1A), HPMECs (Figure 2A) and HLFs (Figure 3A). Furthermore, stimulation of HAECs with Pa flagellin increased NEU1-mediated MUC1-ED desialylation, adhesiveness for Pa and shedding, all of which were inhibited by C9-BA-DANA (Figure 1C–F). We asked whether C9-BA-DANA might inhibit total lung sialidase activity in an in vivo setting. In the lungs of PBS-administered control mice, basal levels of total sialidase activity for the 4-MU-NANA substrate were detected (Figure 8B). Administration of C9-BA-DANA 24 h prior to harvesting lungs reduced baseline sialidase activity by 59.7%. In flagellin-challenged mice, total lung sialidase activity increased by 46.7% compared to the PBS-administered controls in the absence of C9-BA-DANA. Preadministration of C9-BA-DANA completely prevented the flagellin-induced increase in lung sialidase activity. Finally, preadministration of DANA also inhibited lung sialidase activity in the absence or presence of the flagellin stimulus, both of which were equivalent to the inhibition seen by C9-BA-DANA. These findings indicate that the ability of C9-BA-DANA to inhibit sialidase activity in HAECs (Figure 1A), HPMECs (Figure 2A) and HLFs (Figure 3A) can be extended to an intact in vivo model. Sialidases other than NEU1 are known to be expressed in lung tissue (Bonten et al. 1996; Igdoura et al. 1998; Cross et al. 2012; Lillehoj et al. 2012, 2015). However, the decreases in flagellin-provoked lung sialidase activities in response to C9-BA-DANA and the more broad spectrum DANA were comparable (Figure 8B), not supporting the involvement of sialidases other than NEU1. It is still conceivable, however, that one or more of these other sialidases might explain the C9-BA-DANA-unresponsive lung sialidase activity.
Discussion
In the current studies, C9-BA-DANA reduced total sialidase activity in lung epithelia (Figure 1A), endothelia (Figure 2A) and fibroblasts (Figure 3A). In HAECs, at the mRNA level, NEU1 is expressed at levels >7500-fold greater than NEU2, >500-fold greater than NEU3 and >4000-fold greater than NEU4 (Lillehoj et al. 2012). In HPMECs, NEU1 is expressed at levels >8700-fold greater than NEU2, >500-fold greater than NEU3 and >5000-fold greater than NEU4 (Cross et al. 2012). Prior siRNA-induced silencing of NEU1 in both lung epithelia and endothelia reduced total sialidase activity for the 4-MU-NANA substrate by >70% (Cross et al. 2012; Lillehoj et al. 2012). The second most abundant sialidase in these cells, NEU3, may be less active for the 4-MU-NANA substrate (Wada et al. 1999). However, a more recent report indicates otherwise (Smutova et al. 2014). These combined data indicate that C9-BA-DANA can inhibit total sialidase activity in HAECs, HPMECs and HLFs, and that almost all of this sialidase activity can be ascribed to NEU1.
Of the four known mammalian sialidases, NEU1 is the most abundant and widely distributed (Monti et al. 2002, 2010; Miyagi and Yamaguchi 2012). NEU1, like many other gene products, is differentially expressed in various organs and tissues (Bonten et al. 1996; Igdoura et al. 1998). The human NEU1 transcript appears to be most abundant in pancreas, followed in decreasing order by skeletal muscle, kidney, heart, placenta, liver, lung and at very low levels, brain (Bonten et al. 1996). In the mouse, the highest levels of NEU1 mRNA expression were found in kidney and epididymis, with moderate levels in the brain and spinal cord, and lower levels in adrenal, liver, lung, spleen and heart (Igdoura et al. 1998). Although NEU1 gene expression in the lung is relatively modest, it is absolutely essential to normal lung development (Starcher et al. 2008). NEU1-null mice exhibit impaired septation and alveolarization resulting in enlarged air spaces. Just as NEU1 is differentially expressed throughout human tissues (Bonten et al. 1996), we now have demonstrated that its expression within the lung itself is heterogeneous for distinct cell types.
The IC50 in HPMECs (13.0 µM) was 3.5-fold higher than that established for A549 cells (3.74 µM) and 2.7-fold higher than that found for HLFs (4.82 µM). Several properties might explain the requirement for higher concentrations of C9-BA-DANA in HPMECs to provide equivalent inhibition to that seen in the other two cell types. Total baseline sialidase activity in resting, unstimulated HMPECs (Figure 2A) was >1.7-fold higher than that detected in an equivalent number of resting, unstimulated A549 cells (Figure 1A) and >3.6-fold higher than that found in HLFs (Figure 3A). We asked whether differential expression of NEU1 and/or its transport/chaperone protein, PPCA, might explain the higher sialidase activity in HPMECs. We found that NEU1 protein expression normalized to β-tubulin or cell number was >1.9-fold higher in HPMECs compared to its expression in A549 cells, >1.2-fold higher compared to its expression in SAECs and >1.7-fold higher compared to its expression in HLFs (Figure 4C,D). In contrast, normalized PPCA protein expression was lowest in HPMECs and highest in A549 cells (Figure 4C,D). Among the four cell types analyzed, increasing NEU1 protein levels significantly correlated with increasing sialidase activity for the 4-MU-NANA substrate (Figure 4G). Unexpectedly, increasing PPCA protein levels did not correlate with sialidase activity (Figure 4H). To further clarify the contributions of NEU1 and PPCA to cellular sialidase activity, NEU1 and PPCA each were dose-dependently overexpressed in low NEU1-expressing, high PPCA-expressing A549 cells and high NEU1-expressing, low PPCA-expressing HPMECs (Figure 5). In A549 cells, overexpression of either NEU1 or PPCA increased sialidase activity (Figure 5B,D), whereas in HPMECs, PPCA overexpression increased sialidase activity (Figure 5H) while NEU1 overexpression did not (Figure 5F). These combined data indicate that (1) NEU1 and PPCA protein expression and sialidase activity differ in distinct cell types and tissues, (2) NEU1 protein expression correlates with sialidase activity, (3) endogenous PPCA expression can be a limiting factor for NEU1-mediated sialidase activity and (4) differential NEU1 and/or PPCA expression may dictate cellular responsiveness to C9-BA-DANA.
The experimental conditions required for each of the essays described in this study might contribute to the inconsistent effects of C9-BA-DANA. First, the 4-MU-NANA assay for sialidase catalytic activity (Figures 1A, 2A, 3A, 4E,F, 5B,D,F,H,I) involves treatment of cells with the detergent, Triton X-100, which permeabilizes the plasma membrane likely permitting increased entry into and/or exit from cells. The PNA lectin blotting (Figure 1C,D) and adhesion assays (Figure 1E) are performed within 30 min of the flagellin stimulus, whereas the MUC1-ED shedding experiments (Figure 1F) are carried out after 24 h. The stability of C9-BA-DANA in aqueous culture medium at 37°C over time has not been defined. MUC1-ED exhibits a β-turn helix resulting from its high proline content (Fontenot et al. 1995). This extended rod-like conformation allows MUC1 to protrude higher above the airway epithelial surface than most membrane-associated proteins. MUC1-ED desialylation and altered bacterial adhesion likely occur at the outermost position of glycan chains where the terminal sialic acid residues are located (Lillehoj et al. 2013). In the PNA lectin blotting studies which assayed MUC1-ED desialylation, C9-BA-DANA at 10 µM inhibited desialylation by 42.0%, and at 100 µM, by 78.7% (Figure 1C,D). In the Pa adhesion assays, C9-BA-DANA at 10 µM inhibited adhesion by 43.1%, and at 100 µM, by 67.5% (Figure 1E). In contrast, the protease recognition site where proteolytic cleavage and shedding of the MUC1-ED occur is located at a Gly-Ser peptide bond located 58 amino acids upstream of the transmembrane domain within the extracellular, juxtamembranous region (Parry et al. 2001). In the MUC1-ED shedding assays, C9-BA-DANA at 10 µM inhibited shedding only by 23.8%, and at 100 µM, only by 46.7% (Figure 1F). Whether such differences in the site of NEU1 activity and sialic acid removal influences, the ability of C9-BA-DANA to interact with NEU1 to inhibit its catalytic activity is unclear. Other possible explanations might include differences in intracellular and/or cell surface pH, and/or distinct cellular volumes and/or plasma membrane/glycocalyx properties that dictate C9-BA-DANA entry into the intracellular compartment and accessibility to the NEU1 sialidase. Unlike the experiments performed with HAECs (Figure 1C–F), the sialidase activity in HPMECs was comprised of both endogeneous NEU1 and ectopically expressed NEU1 through Ad-NEU1 infection (Figure 2B–E). C9-BA-DANA dramatically reduced sialidase activity in NEU1-overexpressing cells (Figure 5I). Whether endogenous and ectopically expressed NEU1 are equally catalytically active on a molar basis and/or equally susceptible to the inhibitory activity of C9-BA-DANA have not been tested.
The selectivity of C9-BA-DANA for human NEU1, relative to bacterial neuraminidases, may have implications for its potential therapeutic application. A number of commensal bacteria express neuraminidases that can facilitate their acquisition of carbohydrate nutrients and regulate their physical presentation to host tissues and recognition by the immune system (Kim et al. 2011; Lewis and Lewis 2012). Since any intervention that disrupts bacterial neuraminidases might exert an unpredictable impact on host–microbe and microbe–microbe interactions, a more selective approach is theoretically preferable.
NEU1 reportedly influences multiple biological processes. Evidence exists to support a role for NEU1 in aspects of microbial pathogenesis, innate immunity and the sepsis syndrome (Cross et al. 2003; Grewal et al. 2008; Amith et al. 2009, 2010; Seyrantepe et al. 2010; Abdulkhalek et al. 2011, 2012; Feng et al. 2012, 2013; Lillehoj et al. 2012, 2015; Abdulkhalek and Szewczuk 2013). NEU1 participates in TLR activation (Amith et al. 2009, 2010; Abdulkhalek et al. 2011, 2012; Feng et al. 2012; Abdulkhalek and Szewczuk 2013), including TLR4 responsiveness to its ligand, bacterial lipopolysaccharide (LPS) or endotoxin (Amith et al. 2010; Abdulkhalek et al. 2011; Feng et al. 2012), and influences macrophage phagocytosis (Seyrantepe et al. 2010). Pa hijacks human NEU1 to desialylate its receptor, the MUC1-ED, which in turn, increases bacterial adhesion to and invasion of the airway epithelium (Lillehoj et al. 2012, 2015). Sialidase activity regulates complement-mediated pulmonary leukostasis and transendothelial recruitment of granulocytes into the bronchoalveolar compartment (Cross et al. 2003), and finally, the development of endotoxin-induced acute lung injury (Feng et al. 2013). In one study, the coagulopathy associated with Streptococcus pneumoniae sepsis was ascribed to bacterial neuraminidase (Grewal et al. 2008). The role(s) of one or more human sialidase(s) was not considered. Sialic acid and its metabolism also reportedly contribute to the atherogenic process (Tertov et al. 1998; Golovanova et al. 2002; Mochizuki et al. 2002; Cuniberti et al. 2005; Gopaul and Crook 2006; Gayral et al. 2014). Serum sialic acid is a predictive biomarker for coronary artery disease (Gopaul and Crook 2006) and desialylation of low-density lipoproteins (Tertov et al. 1998) and the arterial surface (Cuniberti et al. 2005) facilitates neointimal development and atherogenesis. Sialidase activity is elevated in the atherosclerotic aortic intima (Golovanova et al. 2002) and NEU1 is essential to elastin receptor signaling (Hinek et al. 2006, 2008; Duca et al. 2007; Starcher et al. 2008), which in turn, increases arterial smooth muscle proliferation and enhances atheroma plaque size progression (Mochizuki et al. 2002; Gayral et al. 2014). Finally, NEU1 overexpression impairs the endothelial migratory response to injury (Figure 2C,D) (Cross et al. 2012; Lee et al. 2014). More recently, we reported that NEU1 expression is upregulated in the lungs of IPF patients (Luzina et al. 2016). It is conceivable that a NEU1-selective sialidase inhibitor such as C9-BA-DANA might offer a novel therapeutic intervention for sepsis, atherosclerotic arterial disease, IPF and/or other human disease states.
Materials and methods
Reagents
Precast SDS-polyacrylamide gels were from Novex (San Diego, CA). Polyvinylidene fluoride (PVDF) membrane was from Millipore (Bedford, MA). Enhanced chemiluminescence reagents and prestained protein molecular weight markers were from BioRad (Hercules, CA). Mouse antihuman NEU1 monoclonal antibody was from Origene (Rockville, MD). Rabbit anti-PPCA antibody was from Abcam (Cambridge, MA). Mouse anti-Physarum β-tubulin antibody was from Boehringer-Mannheim (Indianapolis, IN). Horseradish peroxidase-conjugated goat antirabbit and goat-antimouse secondary antibodies were from Cell Signaling Technology (Danvers, MA). PNA and PNA-agarose were from Vector Laboratories (Burlingame, CA). DANA was from Calbiochem (San Diego, CA). C9-BA-DANA was prepared as previously described (Magesh et al. 2008).
Human lung cell cultures
A549 cells are an alveolar type II cell line derived from a lung adenocarcinoma (American Type Culture Collection, Manassas, VA). A549 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (Hyclone Laboratories, Logan, UT), 50 units/mL penicillin and 50 µg/mL streptomycin, as described (Lillehoj et al. 2012). Human primary SAECs isolated from the distal portion of the bronchioles (PromoCell, Heidelberg, Germany) were cultured as described (Lillehoj et al. 2012, 2015). SAECs were studied in passages 2–4. HPMECs (PromoCell) were cultured in endothelial cell growth medium (MV-2, PromoCell) containing growth medium supplement mix (PromoCell) as described (Cross et al. 2012; Lee et al. 2014). HPMECs were studied in passages 4–7. Primary normal adult HLFs (Lonza, Rockland, ME) were cultured in DMEM supplemented with 2.0 mM glutamine, 2.0 mM sodium pyruvate, 50 mg/L gentamicin and 10% fetal bovine serum, as described (Nacu et al. 2008). All HLF cultures were passaged by trypsinization and used for experiments in passages 3−7.
Fluorometric assay for sialidase activity
A549 cells, HPMECs and HLFs (106 cells/reaction) were suspended in 200 µL of 500 mM sodium acetate, pH 4.4 containing 0.1% Triton X-100 and protease inhibitor mixture (Roche Applied Science, Branford, CT) in the presence of increasing concentrations of C9-BA-DANA. The cell-containing mixtures were then incubated for 1 h at 37°C with 25 µL of 2.0 mM 4-MU-NANA, mixing the tubes every 15 min. The sialidase reaction was terminated by addition of 1.0 mL of 133 mM glycine, pH 10.3, 60 mM NaCl and 0.083 M Na2CO3, after which fluorescence intensity was measured with a fluorometer (excitation at 355 nm; emission at 460 nm), as described (Cross et al. 2012; Lillehoj et al. 2012). Half maximal inhibitory concentration (IC50) values of C9-BA-DANA were calculated using the ED50V10 Microsoft Excel add-in (http://www.sciencegateway.org/protocols/cellbio/drug/hcic50.htm) with background corrected fluorescence values and log10-transformed C9-BA-DANA concentrations. The same 4-MU-NANA fluorogenic substrate was used to establish the IC50 values of C9-BA-DANA for neuraminidases expressed by A. ureafaciens (Roche), C. perfringens (Sigma, St. Louis, MO) and V. cholerae (Roche, Indianapolis, IN). C. perfringens is known to express three distinct neuraminidases, NanH (42.8 kDa), NanI (77.4 kDa), and NanJ (~140 kDa) (Newstead et al. 2008). Since the commercial source for the clostridial neuraminidase could not specify, which neuraminidase(s) was contained in their product, the preparation was resolved by SDS-PAGE and the gel processed for silver staining according to the manufacturer's protocol (Life Technologies, Carlsbad, CA). The resolved preparation contained a single protein band with a molecular weight of ~77.0 kDa, compatible with NanI. To control for interassay variability, a simultaneous positive control using A549 cells at 106 cells/reaction was included with each assay. To address the dissimilar purities and specific activities for the bacterial neuraminidases, a concentration range for each enzyme was tested for catalytic activity for the 4-MU-NANA substrate, and a concentration that generated catalytic activity comparable to that seen with the A549 cell control was chosen for IC50 studies for the C9-BA-DANA inhibitor. The concentration chosen was 7.5 ng/mL for A. ureafaciens neuraminidase, 40 ng/mL for C. perfringens NanI and 625 ng/mL for V. cholerae neuraminidase.
Pa culture and flagellin purification
Pa strain PAO1 was cultured overnight in Luria-Bertani broth, washed twice with PBS, resuspended in PBS containing 2.0 mg/mL glucose and quantified spectrophotometrically at A600, as described (Lillehoj et al. 2012, 2015). An overnight culture of Pa was centrifuged at 5000 × g for 30 min, resuspended in Krebs-Ringer buffer, and incubated for 1 h at 37°C, as described (Lillehoj et al. 2012, 2015). The bacteria were removed by centrifugation, and the supernatant was filtered through a 0.22-µm-pore membrane (Millipore) and boiled for 20 min. The supernatant was concentrated by ultrafiltration (Millipore; 30-kDa cutoff), adjusted to pH 6.0, and incubated for 30 min at room temperature with 0.5 mL of Macro-Prep High S support, pH 6.0. The matrix was removed by centrifugation, and the flagellin-containing supernatant was applied to a 2.0-mL Macro-Prep High Q column. The column was washed with 20 mM Tris-HCl, pH 8.0 and flagellin was eluted with 100 mL of a linear 0.0–1.0 M NaCl gradient. Fractions (1.0 mL) were collected and analyzed for the 50-kDa flagellin protein band by SDS-PAGE. Immunoblots were probed with rabbit anti-Pa flagellin antibody (provided by Dr. Dan Wozniak, Wake Forest University, Winston-Salem, NC) and with rabbit antipilin antibody (provided by Dr. Randy Irvin, University of Alberta, Alberta, Canada) to confirm the absence of pilin contamination. Flagellin-containing fractions were preadsorbed with polymyxin B-agarose to remove bacterial LPS, after which less than 0.1 LPS units/µg of protein was detected by the Limulus amebocyte lysate assay.
PNA lectin blotting of desialylated MUC1-ED
A549 cells were incubated for 30 min at 37°C with 10 ng/mL flagellin or medium alone, solubilized with lysis buffer as described (Lillehoj et al. 2012, 2015), and 1.0 mg aliquots were incubated with PNA immobilized on Sepharose beads (Vector Labs, Burlingame, CA) to selectively enrich for PNA-binding proteins. The PNA-bound proteins were resolved by SDS-PAGE, transferred to PVDF, and the blots incubated with mouse antihuman MUC1-ED antibody (clone GP1.4, Biomedia, Foster City, CA), as described (Lillehoj et al. 2012, 2015). Asialofetuin (1.0 µg) was used as a positive control for PNA-binding proteins, whereas fetuin (1.0 µg) was used as a negative control.
Pa adhesion to HAECs
A549 cells were incubated for 30 min at 37°C with 10 ng/mL of Pa-derived flagellin or medium alone, after which they were washed twice with PBS, fixed for 10 min with 2.5% glutaraldehyde in PBS at room temperature, and washed three times with PBS as described (Lillehoj et al. 2012, 2015). Fixed cells (2.0 × 105/well) in 24-well plates were incubated with 2.0 × 107 colony-forming units (CFU)/well of PAO1 in 0.5 mL for 40 min at 37°C and washed three times with PBS. Bound bacteria were released with 0.05% trypsin, and bound CFUs quantified as described (Lillehoj et al. 2012, 2015).
ELISA for MUC1-ED
A549 cell-conditioned culture media were centrifuged at 10,000 × g for 10 min at 4°C, added in triplicate to 96-well ELISA plates (MaxiSorb; Nalge Nunc, Rochester, NY) and incubated overnight at 4°C. Wells were blocked for 1 h at room temperature with PBS, pH 7.0, containing 10 mg/mL bovine serum albumin and 50 mg/mL sucrose, and washed with PBS containing 0.05% Tween-20 (PBS-T). The samples were reacted for 2 h at room temperature with 200 µg/mL of primary MUC1-ED antibody, washed with PBS-T, reacted for 2 h at room temperature with 200 µg/mL of peroxidase-conjugated goat-antimouse IgG secondary antibody (KPL, Gaithersburg, MD) and washed with PBS-T. Bound antibodies were detected with tetramethylbenzidine substrate (SureBlue, KPL). The substrate reaction was stopped with 1 N HCl, and A450 measured using a microplate spectrophotometer (Dexall, Gaithersburg, MD), as described (Lillehoj et al. 2015).
HPMEC migration in a wounding assay
HPMECs were seeded at 2.0 × 105 cells/well in the wells of a 24-well plate (Corning, Corning, NY), and after 3 h, infected with recombinant Ad-NEU1 or Ad-GFP (MOI = 100), as described (Lee et al. 2014). The HPMECs were cultured for 48 h to confluence. Using a sterile 200 µL pipette tip, a single wound was made across the diameter of each monolayer, after which the cell debris was removed by washing with HEPES, as described (Cross et al. 2012; Lee et al. 2014). The wounded monolayers were then incubated for 24 h in the absence or presence of 134 µM of C9-BA-DANA. At 24 h, photomicrographs of each well were taken with a Nikon inverted microscope. HPMEC area was calculated using ImageJ software for comparison with that observed in the same wounded monolayer at 0 h, as a measure of migration into the wound.
In vitro HPMEC capillary-like tube formation
Each well of a 96-well plate was coated with 50 µL of Matrigel (10 mg/mL; BD Biosciences, Sparks, MD), as described with modifications (Lee et al. 2014). Briefly, Matrigel was allowed to polymerize for 1 h at room temperature in each of the wells, followed by 1 h at 37°C in a humidified atmosphere at 5% CO2. HPMECs infected with Ad-GFP or Ad-NEU1 (MOI = 100) were seeded at 7.5 × 103 cells/well onto the Matrigel-coated wells, in the presence 134 µM of C9-BA-DANA or medium alone. At 6 h, tubular structures were photographed through a Nikon inverted microscope, and segments of capillary-like tubes per high power field (HPF) were counted.
Quantitative RT-PCR for human NEU1 and PPCA transcripts
Total cellular RNA was extracted from A549 cells, SAECs, HPMECs and HLFs, as described (Cross et al. 2012; Lillehoj et al. 2012). RNA purity was established with the 260/280 nm absorption ratio (>1.90). Total RNA (1.0 µg) was treated with DNase I (Invitrogen, Carlsbad, CA) for 15 min and reverse transcribed using avian myelobastosis virus reverse transcriptase and poly(T) primer (Promega, Madison, WI). The resulting cDNA was quantified by PCR using SYBR Green PCR Master Mix and an ABI Prism 7900HT cycler. Primers for detection of human NEU1 and PPCA mRNAs were designed using Primer Express 2.0 (Applied Biosystems, Foster City, CA), as described (Cross et al. 2012; Lillehoj et al. 2012). Relative NEU1 and PPCA gene expression was calculated using the 2−ΔΔCt method where NEU1 and PPCA transcripts were normalized to the levels of 18 S rRNA transcripts as the internal control, as described (Cross et al. 2012; Lillehoj et al. 2012).
Quantitative immunoblotting for human NEU1 and PPCA proteins
To establish relative NEU1 and PPCA protein expression in various human lung cells, equivalent numbers of A549 cells, SAECs, HPMECs and HLFs were lysed and equivalent lysate volumes were processed for NEU1 and PPCA immunoblotting, as described (Cross et al. 2012; Lillehoj et al. 2012; Lee et al. 2014). To control for protein loading and transfer for the NEU1 and PPCA immunoblots, the blots were stripped and reprobed for β-tubulin. NEU1 and PPCA signals were normalized to both cell number and β-tubulin signal in the same lane in the same stripped and reprobed blot.
Adenoviral constructs encoding for PPCA
A plasmid encoding for human PPCA (pSCTop-PPCA) was generously provided by Drs E. Bonten and A. d'Azzo (St. Jude Children's Research Hospital, Memphis, TN). A recombinant Ad-PPCA was generated using the AdEasy Ad Vector System (Stratagene, La Jolla, CA) as described for other gene products (Cross et al. 2012; Lillehoj et al. 2012). Briefly, the pSCTop-PPCA was digested with SalI and NotI and subcloned into a shuttle vector (pShuttle CMV, Stratagene) using the same restriction enzymes followed by ligation. The shuttle plasmid was linearized through PmeI digestion and, with the Ad backbone plasmid (pAdEasy-1, Qbiogene, Carlsbad, CA), was used to co-transform electrocompetent Escherichia coli BJ5183 cells to produce the recombinant plasmid, Ad-PPCA. Recombinants were selected for kanamycin resistance and screened for recombination by Pac1 restriction enzyme analysis and agarose gel electrophoresis. The correct recombinant plasmids were used to transform DH5α cells, and bacterial lysates were passed through Maxiprep columns (Qiagen, Valencia, CA) for purification. Ad-PPCA was linearized with PacI digestion and transfected, in the presence of Lipofectamine (Invitrogen), into AD-293 cells. After 7–10 d, cells were scraped off flasks with a rubber policeman and subjected to three freeze-thaw cycles, and virus was harvested in the supernatants for presentation to fresh AD-293 cells and titration in a plaque-forming assay. A549 cells and HPMECs were transiently infected with packaged Ad-PPCA at increasing MOIs and after 48 h were lysed, and the lysates processed for PPCA immunoblotting and the fluorometric assay for sialidase activity.
Cell viability
A549 cells, HPMECs and HLFs were seeded into the wells of 48-well plates and cultured to 80% confluence. The cells were cultured for 20 h in the presence of 100 µM of C9-BA-DANA or medium alone, after which 250 ng/mL of 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) was added for an additional 4 h incubation. The medium was aspirated and 100 µL of DMSO added and mixed by pipette. After 10 min, each well was again mixed and A490 measured.
Homology modeling of the C9-BA-DANA–NEU1 complex with bacterial neruaminidases
Homology models of human NEU1 and A. ureafaciens neuramidase were constructed by multitemplate modeling using Phyre2 protein fold recognition software (Kelley et al. 2015) based on the X-ray diffraction crystal structure of NEU2 (National Center for Biotechnology Information, Bethesda, MD, Protein Database ID #1VCU) (Chavas et al. 2005). C9-BA-DANA was docked onto the NEU1 structure using AutoDock Vina software (Trott and Olson 2010), resulting in a model similar to that previously described by Magesh et al. (2006, 2008). X-ray structures for V. cholerae (Protein Database ID #1W0O) (Moustafo et al. 2004) and C. perfringens NanI (Protein Database ID #2VK6) (Newstead et al. 2008) neuraminidases and the model of A. ureafaciens neuraminidase were superimposed onto the C9-BA-DANA–NEU1 model using the structural alignment algorithm in UCSF Chimera (Petterson et al. 2004) or the cealign command in PyMOL (version 1.3, Schrodinger, LLC, Cambridge, MA). Amino acid sequence alignments of the β7–β8 loop of NEU1, -2, -3 and -4 and the three bacterial neuraminidases were created using PROMALS3D (Pei et al. 2008) and, in the case of C. perfringens NanI, adjusted based on the structural alignment with NEU1 to account for the insertion domain beginning after the β7 strand.
C9-BA-DANA inhibition of flagellin-induced increases in total sialidase activity in mouse lungs
All animal experiments were approved by the University of Maryland Baltimore Institutional Animal Use and Care Committee. Female BALB/c mice (18–20 g) were obtained from Charles River Laboratories (Frederick, MD). The mice were intraperitoneally injected with 15 mg/kg of C9-BA-DANA or DANA dissolved in 25 µL of nonpyrogenic normal saline or saline alone. After 24 h, mice were intranasally administered flagellin (100 ng/mouse) or the PBS vehicle alone. At 4 h, the mice were euthanized by CO2 inhalation and cervical dislocation, and the lungs and kidneys were harvested, weighed and transferred to tubes on ice containing PBS and a protease inhibitor cocktail (Sigma), and homogenized at 4°C. Lung and kidney homogenates were centrifuged at 14,000 × g for 10 min at 4°C and supernates were processed for total protein levels and total sialidase activity for the 4-MU-NANA substrate.
Statistical methods
All values were expressed as mean ± SE. Differences between means were compared using the Student's t-test or ANOVA followed by the Tukey's multiple range test and considered significant at P < 0.05. Correlation coefficients (R2) and P values were calculated using Microsoft Excel.
Funding
This work was supported by U.S. Department of Veterans Affairs grants I01BX002352 (to SEG), I01CX000101 (to IGL), and I01BX002499 (to SPA), and National Institutes of Health grants F32AI110045 (to KHP), R01HL126897 (to SPA), and R01AI090866 (to EJS).
Acknowledgements
We are grateful to Ms. Jennifer Davidson for secretarial support.
Conflict of interest statement
All the authors have no conflict of interest to declare.
Abbreviations
Ad, adenovirus; C9-BA-DANA, carbon 9-butyl-amide-2-deoxy-2,3-dehydro-N-acetylneuraminic acid; CFU, colony-forming unit; DMEM, Dulbecco's modified Eagle medium; GFP, green fluorescent protein; HAEC, human airway epithelial cell; HLF, human lung fibroblast; HPF, high power field; HPMEC, human pulmonary microvascular endothelial cell; IC50, half maximal inhibitory concentration; IPF, idiopathic pulmonary fibrosis; LPS, lipopolysaccharide; MOI, multiplicity of infection; MTT, 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT); MUC1, mucin-1; MUC1-ED; ectodomain of MUC1; 4-MU-NANA, 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid; NEU1, neuraminidase-1; Pa, Pseudomonas aeruginosa; PBS-T, PBS containing 0.05% Tween-20; PNA, peanut agglutinin; PPCA, protective protein/cathepsin A; PVDF, polyvinylidene fluoride; SAEC, small airway epithelial cell; TLR, Toll-like receptor.
References
- Abdulkhalek S, Amith SR, Franchuk SL, Jayanth P, Guo M, Finlay T, Gilmour A, Guzzo C, Gee K, Beyaert R, et al. 2011. Neu1 sialidase and matrix metalloproteinase-9 cross-talk is essential for Toll-like receptor activation and cellular signaling. J Biol Chem. 286:36532–36549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulkhalek S, Guo M, Amit SR, Jayanth P, Szewczuk MR. 2012. G-protein coupled receptor agonists mediate Neu1 sialidase and matrix metalloproteinase-9 cross-talk to induce transactivation of TOLL-like receptors and cellular signaling. Cell Signal. 24:2035–2042. [DOI] [PubMed] [Google Scholar]
- Abdulkhalek S, Szewczuk MR. 2013. Neu1 sialidase and matrix metalloproteinase-9 cross-talk regulates nucleic acid-induced endosomal TOLL-like receptor-7 and -9 activation, cellular signaling and pro-inflammatory responses. Cell Signal. 25:2093–2105. [DOI] [PubMed] [Google Scholar]
- Albohy A, Zhang Y, Smutova V, Pshezhetsky AV, Cairo CW. 2013. Identification of selective nanomolar inhibitors of the human neuraminidase, NEU4. ACS Med Chem Lett. 4:532–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amith SR, Jayanth P, Franchuk S, Finlay T, Seyrantepe V, Beyaert R, Pshezhetsky AV, Szewczuk MR. 2010. Neu1 desialylation of sialyl α-2,3-linked β-galactosyl residues of TOLL-like receptor 4 is essential for receptor activation and cellular signaling. Cell Signal. 22:314–324. [DOI] [PubMed] [Google Scholar]
- Amith SR, Jayanth P, Franchuk S, Siddiqui S, Seyrantepe V, Gee K, Basta S, Beyaert R, Pshezhetsky AV, Szewczuk MR. 2009. Dependence of pathogen molecule-induced Toll-like receptor activation and cell formation on Neu1 sialidase. Glycoconj J. 26:1197–1212. [DOI] [PubMed] [Google Scholar]
- Arabkhari M, Bunda S, Wang Y, Wang A, Pshezhetsky AV, Hinek A. 2010. Desialylation of insulin receptors and IGF-1 receptors by neuraminidase-1 controls the net proliferative response of L6 myoblasts to insulin. Glycobiology. 20:603–616. [DOI] [PubMed] [Google Scholar]
- Bonten E, van der Spoel A, Fornerod M, Grosveld G, d'Azzo A. 1996. Characterization of human lysosomal neuraminidase defines the molecular basis of the metabolic storage disorder sialidosis. Genes Dev. 10:3156–3169. [DOI] [PubMed] [Google Scholar]
- Burmeister WP, Henrissat B, Bosso C, Cusack S, Ruigrok RW. 1993. Influenza B virus neuraminidase can synthesize its own inhibitor. Structure. 1:19–26. [DOI] [PubMed] [Google Scholar]
- Chavas LMG, Tringali C, Fusi P, Venerando B, Tettamanti G, Kato R, Monti E, Wakatsuki S. 2005. Crystal structure of the human cytosolic sialidase Neu2. Evidence for the dynamic nature of substrate recognition. J Biol Chem. 280:469–475. [DOI] [PubMed] [Google Scholar]
- Chen G, Chen X, King S, Cavassani KA, Cheng J, Zheng X, Cao H, Yu H, Qu J, Fang D, et al. 2011. Amelioration of sepsis by inhibiting sialidase-mediated disruption of the CD24-SiglecG interaction. Nat Biotechnol. 29:428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Varki A. 2010. The sialome-Far more than the sum of its parts. OMICS. 14:455–464. [DOI] [PubMed] [Google Scholar]
- Comelli EM, Amado M, Lustig SR, Paulson JC. 2003. Identification and expression of Neu4, a novel murine sialidase. Gene. 321:155–161. [DOI] [PubMed] [Google Scholar]
- Cross AS, Hyun SW, Miranda-Rivera A, Feng C, Liu A, Nguyen C, Zhang L, Luzina IG, Atamas SP, Twaddell WS, et al. 2012. NEU1 and NEU3 sialidase activity expressed in human lung microvascular endothelia. NEU1 restrains endothelial cell migration whereas NEU3 does not. J Biol Chem. 287:15966–15980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross AS, Sakarya S, Rifat S, Held TK, Drysdale B, Grange PA, Cassels FJ, Wang L, Stamatos N, Farese A, et al. 2003. Recruitment of murine neutrophils in vivo through endogenous sialidase activity. J Biol Chem. 278:4112–4120. [DOI] [PubMed] [Google Scholar]
- Cuniberti LA, Martinez V, Schachter J, Magarinos G, Meckert PC, Laguens RP, Levenson J, Werba JP. 2005. Sialic acid as a protective barrier against neointima development. Atherosclerosis. 181:225–231. [DOI] [PubMed] [Google Scholar]
- de Geest N, Bonten E, Mann L, de Sousa-Hitzler J, Hahn C, d'Azzo A. 2002. Systemic and neurologic abnormalities distinguish the lysosomal disorders sialidosis and galactosialidosis in mice. Hum Mol Genet. 11:1455–464. [DOI] [PubMed] [Google Scholar]
- Dridi L, Seyrantepe V, Fougerat A, Pan X, Bonneil E, Thibault P, Moreau A, Mitchell GA, Heveker N, Cairo CW, et al. 2013. Positive regulation of insulin signaling by neuraminidase 1. Diabetes. 62:2338–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca L, Blanchevoye C, Cantarelli B, Ghoneim C, Dedieu S, Delacoux F, Hornebeck W, Hinek A, Martiny L, Debelle L. 2007. The elastin receptor complex transduces signals through the catalytic activity of it's Neu-1 subunit. J Biol Chem. 282:12484–12491. [DOI] [PubMed] [Google Scholar]
- Fanzani A, Guiliani R, Colombo F, Zizioloi D, Presta M, Preti A, Marchesini S. 2003. Overexpression of cytosolic sialidase Neu2 induces myoblast differentiation in C2C12 cells. FEBS Lett. 547:183–188. [DOI] [PubMed] [Google Scholar]
- Feng C, Stamatos NM, Dragan AI, Medvedev A, Whitford M, Zhang L, Song C, Rallabhandi P, Cole L, Nhu QM, et al.. 2012. Sialyl residues modulate LPS-mediated signaling through the Toll-like receptor 4 complex. PLoS One. 7:e32359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Zhang L, Nguyen C, Vogel SN, Goldblum SE, Blackwelder WC, Cross AS. 2013. Neuraminidase reprograms lung tissue and potentiates lipopolysaccharide-induced acute lung injury in mice. J Immunol. 191:4828–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Mariappan SV, Castasti P, Domenech N, Finn OJ, Gupta G. 1995. Structure of a tumor associated antigen containing a tandemly repeated immunodominant epitope. J Biol Struct Dyn. 13:245–260. [DOI] [PubMed] [Google Scholar]
- Gayral S, Garnotel R, Castaing-Berthou A, Blaise S, Fougerat A, Berge E, Montheil A, Malet N, Wymann MP, Maurice P, et al. 2014. Elastin-derived peptides potentiate atherosclerosis through the immune NEU-1 P13K gamma pathway. Cardiovasc Res. 102:118–127. [DOI] [PubMed] [Google Scholar]
- Golovanova NK, Gracheva EV, Il'inskaya OP, Tararak EM, Prokazova NV. 2002. Sialidase activity in normal and atherosclerotic aortic intima. Biochemistry (Moscow). 67:1490–1495. [DOI] [PubMed] [Google Scholar]
- Gopaul KP, Crook MA. 2006. Sialic acid: A novel marker of cardiovascular disease. Clin Biochem. 39:667–681. [DOI] [PubMed] [Google Scholar]
- Grewal PK, Uchiyama S, Ditta D, Varki N, Le DT, Nizet V, Marth JD. 2008. The Ashwell receptor mitigates the lethal coagulopathy of sepsis. Nat Med. 14:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Koseki K, Yamaguchi K, Moriya S, Suzuki Y, Yingsakmongkon S, Hirai G, Sodeoka M, von Itzstein M, Miyagi T. 2008. Limited inhibitory effects of osteltamivir and zanamivir on human sialidases. Antimicrob Agents Chemother. 52:3484–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinek A, Bodaruk TD, Bunda S, Wang Y, Liu K. 2008. Neuraminidase-1, a subunit of the cell surface elastin receptor, desialylates and functionally inactivates adjacent receptors interacting with the mitogenic growth factors PDGF-BB and IGF-2. Am J Pathol. 173:1042–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinek A, Pshezhetsky A, von Itzstein M, Starcher B. 2006. Lysosomal sialidase (neuraminidase-1) is targeted to the cell surface in a multiprotein complex that facilitates elastic fiber assembly. J Biol Chem. 281:3698–3710. [DOI] [PubMed] [Google Scholar]
- Igdoura SA, Gafuik C, Mertineit C, Saberi F, Pschezhetsky AV, Potier M, Trasler JM, Gravel RA. 1998. Cloning of the cDNA and gene encoding mouse lysosomal sialidase and correction of sialidase deficiency in human sialdosis and mouse SM/J fibroblasts. Hum Mol Genet. 7:115–121. [DOI] [PubMed] [Google Scholar]
- Kappagantula S, Andrews MR, Cheah M, Abad-Rodriguez J, Dotti CG. 2014. Neu3 sialidase-mediated ganglioside consersion is necessary for axon regeneration and is blocked in CNS axons. J Neurosci. 34:2477–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Oh D, Kang H, Kwon O. 2011. Features and applications of bacterial sialidases. Appl Microbiol Biotechnol. 91:1–15. [DOI] [PubMed] [Google Scholar]
- Kiyohara M, Tanigawa K, Chaiwangsr T, Katayama T, Ashida H, Yamamoto K. 2011. An exo-a-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology. 21:437–447. [DOI] [PubMed] [Google Scholar]
- Lee C, Liu A, Miranda-Ribera A, Hyun SW, Lillehoj EP, Cross AS, Passaniti A, Grimm PR, Kim B, Welling PA, et al. 2014. NEU1 sialidase regulates the sialylation state of CD31 and disrupts CD31-driven capillary-like tube formation in the human lung microvascular endothelia. J Biol Chem. 289:9121–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AL, Lewis WG. 2012. Host sialoglycans and bacterial sialidases: A mucosal perspective. Cell Microbiol. 14:1174–1182. [DOI] [PubMed] [Google Scholar]
- Lillehoj EP, Hyun SW, Feng C, Zhang L, Liu A, Guang W, Nguyen C, Luzina IG, Atamas SP, Passaniti A, et al. 2012. NEU1 sialidase expressed in human airway epithelia regulates epidermal growth factor receptor and MUC1 signaling. J Biol Chem. 287:8214–8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehoj EP, Hyun SW, Guang W, Liu A, Verceles AC, Luzina IG, Atamas SP, Kim KC, Goldblum SE. 2015. NEU1 sialidase regulates membrane-tethered mucin (MUC1) ectodomain adhesiveness for Pseudomonas aeruginosa and decoy receptor release. J Biol Chem. 290:18316–18331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehoj EP, Kato K, Lu W, Kim KC. 2013. Cellular and molecular biology of airway mucins. Int Rev Cell Mol Biol. 303:139–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzina IG, Lockatell V, Hyun SW, Kopach P, Kang PH, Noor Z, Liu A, Lillehoj EP, Lee C, Miranda-Ribera A, et al. 2016. Elevated expression of NEU1 sialidase in idiopathic pulmonary fibrosis provokes pulmonary collagen deposition, lymphocytosis, and fibrosis. Am J Physiol Lung Cell Mol Physiol. 310:L940–L954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Wu D, Deng L, Secrest P, Zhao J, Varki N, Lindheim S, Gagneax P. 2012. Sialidases on mammalian sperm mediate deciduous sialylation during capacitation. J Biol Chem. 287:38073–38079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magesh S, Moriya S, Suzuki T, Miyagi T, Ishida H, Kiso M. 2008. Design, synthesis, and biological evaluation of human sialidase inhibitors. Part 1: Selective inhibitors of lysosomal sialidase (NEU1). Bioorg Med Chem Lett. 18:532–537. [DOI] [PubMed] [Google Scholar]
- Magesh S, Savita V, Moriya S, Suzuki T, Miyagi T, Ishida H, Kiso M. 2009. Human sialidase inhibitors: Design, synthesis, and biological evaluation of 4-acetamido-5-acylamido-2-fluoro benzoic acids. Bioorg Med Chem. 17:4595–4603. [DOI] [PubMed] [Google Scholar]
- Magesh S, Suzuki T, Miyagi T, Ishida H, Kiso M. 2006. Homology modeling of human sialidase enzymes NEU1, NEU3, and NEU 4 based on the crystal structure of NEU2: Hints for the design of selective NEU3 inhibitors. J Mol Graph Model. 25:196–207. [DOI] [PubMed] [Google Scholar]
- Meindi P, Tuppy H. 1969. 2-Deoxy-2, 3-dehydrosialic acids. II. Competitive inhibition of Vibrio cholerae neuraminidases by 2-deoxy-2, 3-dehydro-N-acylneuraminic acids. Hoppe Seylers Z Physiol Chem. 350:1088–1092. [PubMed] [Google Scholar]
- Miyagi T, Wada T, Iwamatsu A, Hata K, Yoshikawa Y, Tokuyama S, Sawada M. 1999. Molecular cloning and characterization of a plasma membrane-associated sialidase specific for gangliosides. J Biol Chem. 274:5004–5011. [DOI] [PubMed] [Google Scholar]
- Miyagi T, Wada T, Yamaguchi K. 2008. Roles of plasma membrane-associated sialidase NEU3 in human cancers. Biochim Biophys Acta. 1780:532–537. [DOI] [PubMed] [Google Scholar]
- Miyagi T, Wada T, Yamaguchi K, Hata K. 2004. Sialidase and malignancy: A minireview. Glycoconj J. 20:189–198. [DOI] [PubMed] [Google Scholar]
- Miyagi T, Yamaguchi K. 2012. Mammalian sialidases: Physiological and pathological roles in cellular functions. Glycobiology. 22:880–896. [DOI] [PubMed] [Google Scholar]
- Mochizuki S, Brassart B, Hinek A. 2002. Signaling pathways transduced through the elastin receptor facilitate proliferation of arterial smooth muscle cells. J Biol Chem. 277:44854–44863. [DOI] [PubMed] [Google Scholar]
- Monti E, Bonten E, D'Azzo A, Bresciani R, Venerando B, Borsani G, Schauer R, Tettamanti G. 2010. Sialidases in vertebrates: A family of enzymes tailored for several cell functions. Adv Carbohydr Chem Biochem. 64:403–479. [DOI] [PubMed] [Google Scholar]
- Monti E, Preti A, Nesti C, Ballabio A, Borsani G. 1999. Expression of a novel human sialidase encoded by the NEU2 gene. Glycobiology. 9:1313–1321. [DOI] [PubMed] [Google Scholar]
- Monti E, Preti A, Venerando B, Borsani G. 2002. Recent development in mammalian sialidase molecular biology. Neurochem Res. 27:649–663. [DOI] [PubMed] [Google Scholar]
- Moustafo I, Connaris H, Taylor M, Zaitsev V, Wilson JC, Kiefel MJ, von Itzstein M, Taylor GL. 2004. Sialic acid recognition by Vibrio cholerae neuraminidase. J Biol Chem. 279:40819–40826. [DOI] [PubMed] [Google Scholar]
- Nacu N, Luzina IG, Highsmith K, Lockatell V, Pochetuhen K, Cooper ZA, Gillmeister MP, Todd NW, Atamas SP. 2008. Macropages produce TGF-β-induced (β-ig-h3) following ingestion of apoptotic cells and regulate MMP14 levels and collagen turn over in fibroblasts. J Immunol. 180:5036–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newstead SL, Potter JA, Wilson JC, Xu G, Chien CH, Watts AG, Withers SG, Taylor GL. 2008. The structure of Clostridium perfringens NanI sialidase and its catalytic intermediates. J Biol Chem. 283:9090–9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry S, Silverman HS, McDermott K, Willis A, Hollingsworth MA, Harris A. 2001. Identification of MUC1 proteolytic cleavage sites in vivo. Biochem Biophys Res Commun. 283:715–720. [DOI] [PubMed] [Google Scholar]
- Pei J, Kim BH, Grishin NV. 2008. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petterson EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Pshezhetsky AV, Richard C, Michaud L, Igdoura S, Wang S, Elsliger MA, Qu J, Leclerc D, Gravel R, Dallaire L, et al. 1997. Cloning, expression and chromosomal mapping of human lysosomal sialidase and characterization of mutations in sialidosis. Nat Genet. 15:316–320. [DOI] [PubMed] [Google Scholar]
- Seyrantepe V, Iannello A, Liang F, Kanshin E, Jayanth P, Samarani S, Szewczuk MR, Ahmad A, Pshezhetsky AV. 2010. Regulation of phagocytosis in macrophages by neuraminidase I. J Biol Chem. 285:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smutova V, Albohy A, Pan X, Korchagina E, Miyagi T, Bovin N, Cairo CW, Pshezhetsky AV. 2014. Structural basis for substrate specificity of mammalian neuraminidases. PLoS One. 9:e106320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcher B, d'Azzo A, Keller PW, Rao GK, Nadarajah D, Hinek A. 2008. Neuraminidase-1 is required for the normal assembly of elastic fibers. Am J Physiol Lung Cell Mol Physiol. 295:L637–L647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tertov VV, Kaplun VV, Sobenin IA, Orekhov AN. 1998. Low-density lipoprotein modification occurring in human plasma. Possible mechanism of in vivo lipoprotein desialylation as a primary step of atherogenic modification. Atherosclerosis. 138:183–195. [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ. 2010. AutoDock, Vina: Improving the speed and accuracy of docking with a new scoring function, efficient organization, and multithreading. J Comput Chem. 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Shiozaki K, Yamaguchi K, Miyazaki S, Satomi S, Kato K, Sakuraba H, Miyagi T. 2009. Contribution of sialidase NEU1 to suppression of metastasis of human colon cancer cells through desialylation of integrin β4. Oncogene. 28:1218–1229. [DOI] [PubMed] [Google Scholar]
- Varki NM, Varki A. 2007. Diversity in cell surface sialic acid presentations: Implications for biology and disease. Lab Invest. 87:851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Itzstein M. 2007. The war against influenza: Discovery and development of sialidase inhibitors. Nat Rev Drug Discov. 6:967–974. [DOI] [PubMed] [Google Scholar]
- Wada T, Yoshikawa Y, Tokuyama S, Kuwabara M, Akita H, Miyagi T. 1999. Cloning, expression, and chromosomal mapping of a human ganglioside sialidase. Biochem Biophys Res Commun. 261:21–27. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Albohy A, Zou Y, Smutova V, Pshezhetsky AV, Cairo CW. 2013Identification of selective inhibitors of human neuraminidase isoenzymes using C4,C7-modified 2-deoxy-2,3-didehydro-N-acetylneuraminic acid (DANA) analougues. J Med Chem. 56:2948–2958. [DOI] [PubMed] [Google Scholar]