ABSTRAT
Alcoholic liver disease (ALD) and its complication continued to be a major health problem throughout the world. Increasing evidence suggests that microRNA (miRNA) that regulate apoptosis, inflammation and lipid metabolism are affected by alcohol in ALD. MiR-200a has emerged as a major regulator in several liver diseases, but its role in ALD has not been elucidated. The aim of this study is to figure out the biological function of miR-200a in ALD and to explore its underlying mechanism. The expression pattern of miR-200a were analyzed in vitro and in vivo, we showed that miR-200a was up-regulated in ALD in AML-12 and primary hepatocyte. We then examined it's effect on cell apoptosis and identified zinc finger E-box binding homeobox 2 (ZEB2; also known as SIP1) as a direct target gene of miR-200a. Furthermore, reintroduction of ZEB2 could reverse the pro-apoptosis of miR-200a on AML-12. Taken together, our study demonstrated that miR-200a regulates the apoptosis of hepatocyte in ALD by directly target ZEB2, both of which could serve as new therapeutic targets for ALD.
KEYWORDS: Alcoholic liver disease, ALD, microRNA, miR-200a, ZEB2, Apoptosis
Introduction
In recent years, the global number of drinking has increased significantly, especially in the developed countries [1]. For example, in the United States about 67.3% of people drinking of alcohol with about 7.4% meet criteria for alcohol consumption. According to the World Health Organization (WHO) [2], Alcohol abuse either directly or indirectly to 3.3 million people die annually. More notably, chronic excessive alcohol intake confers detrimental effects on almost all human organs and associated with 60 types of diseases and injuries [3,4]. Liver is the main organ of alcohol metabolism and the most critical target of alcohol-induced injury. ALD includes a broad spectrum of clinical features, including simple alcoholic steatosis, acute alcoholic hepatitis(AH) cirrhosis and increased risk of hepatocellular carcinoma [5,6]. Because alcohol and its metabolites targets numerous signaling pathways and the precise mechanisms of alcohol-induced liver damage are yet to be elucidated. Thus a master regulatory which can modulate multiple signal processes is very attractive.
Recently, increasing studies put attention into the contribution of miRNAs to ALD due to the fine-tuning roles in all important cellular processes, including metabolism, apoptosis regulation, cell-cycle control, and oncogenesis [5]. MiRNAs are highly conserved small noncoding RNAs, about 20–22 nucleotides long, were first described in 1993 [7]. MiRNAs are transcribed from DNA by RNA polymerase II or III in the nucleus as pri-miRNAs and transported to the cytoplasm as pre-miRNAs, where they are processed by Dicer into mature miRNAs [8]. The small single-strand of the mature miRNA is incorporated into the RNA-induced silencing complex (RISC), where miRNAs can target genes by binding specifically to the 3' or 5' untranslated (UTR) region of mRNA [9], miRNAs regulate diverse pathophysiological processes by reduced or increased target gene expression [10]. The roles of miRNAs in initiation and progression of various cancers have been well documented. We have seen that some specific miRNAs, such as miR-451 in Gastric Cancer, microRNA-21 in hepatocellular carcinoma and microRNA-133a in osteosarcoma manipulate disease progression via regulate cell survival, migration and invasion [11–13]. However, the biological significance of miRNA in alcoholic liver disease is unclear but is getting attention lately.
Recently, miR-200s as one of the miRNA family, has been well demonstrated to participate in virtually all stages of tumor progression. The miR-200 family is comprised of 5 members (miR-200a, -200b, -200c, -141, -429), which are organized in two gene clusters. The tricistronic miR-200b/a/429 cluster on chromosome 1p36, and the bicistronic miR-200c/141 cluster on chromosome [14]. Meanwhile, miRNA has been found abundantly in the liver [9]. It is notable that miR-200s not only mediate the initiation and maintenance of Hepatocellular carcinoma [15], but also involved in Hepatic fibrosis [15]. So far, only several miRNAs have been implicated in alcoholic liver disease [16–18]. We therefore aimed to ascertain the function of miR-200a in alcoholic liver disease and dig involved molecular mechanisms.
In this study, we confirmed that miR-200a was up-regulated and clarify the function of miR-200a is inducing hepatocyte apoptosis. By use Luciferase reporter assays and gain-of-function and loss-of-function studies, we also identified ZEB2 as the direct downstream targets of miR-200a, and involved in the regulation of apoptosis in hepatocyte.
Results
MiR-200a is up-regulated in mice after chronic and binge ethanol feeding
Both group mice body weight was increased after a short-time adaptation period, but Ethanol-fed mice body weight was still lower than the beginning (Figure 1a). Interestingly, the liver to body weight ratio of ethanol group was higher than the control group (Figure 1a). TG, TCH, ALT, AST levels were dramatic increased in serum of ethanol-fed mice versus to the CD-fed mice (Figure 1b). Compared to the CD-fed group, there exhibited abundant fat vacuoles of different sizes, and scrambled cell cord in Ethanol-fed group. The degree of liver injury was evaluated and histological features was assessed by HE staining and Oil Red O staining (Figure 1c).Which suggesting that Ethanol consumption can induce hepatic steatosis, aggravate the liver injury in this mouse model. The expression of miR-200 were analyzed by qPCR analysis and as shown in Figure 1d, miR-200a expression was increased compared with normal primary hepatocyte.
Figure 1.
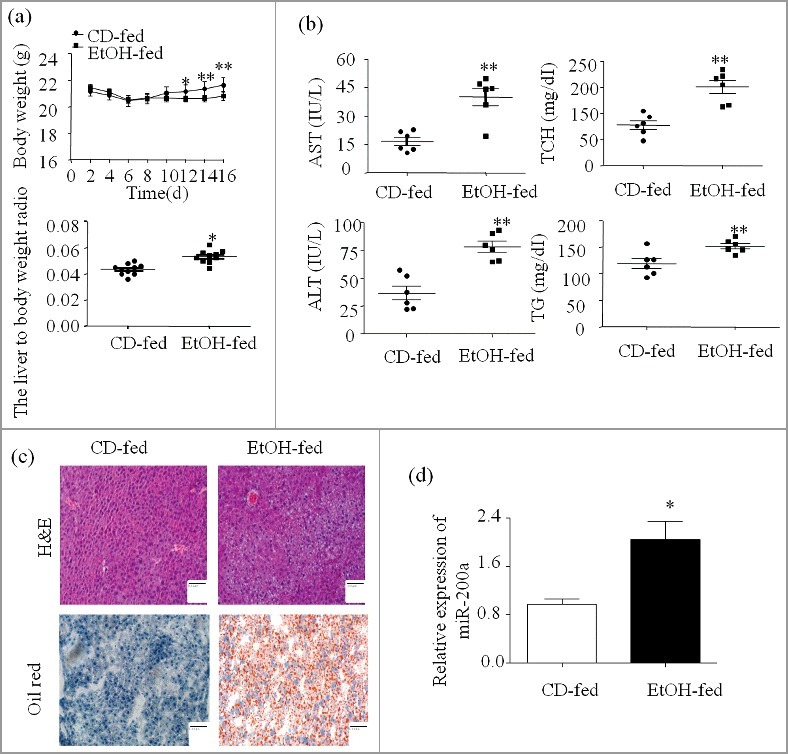
MiR-200a is up-regulated in mice after chronic and binge ethanol feeding. (a) The integrated liver tissue was separate from sacrificed mice. Body weights and the liver to body weight ratio after ethanol feeding. (b) Serum AST, ALT, Hepatic TG, TCH levels. (c) Representative views of H&E staining and Oil Red O staining (original magnification, ×20). (d) Total miR-200a were isolated from the primary hepatocyte and were analyzed by qRCR. The results are shown as relative expression against control expression without treatment. The values represent means ± SD (n = 6 in CD-fed group, n = 6 in EtOH-fed group). * P < 0.05, ** P < 0.01 versus control.
MiR-200a is up-regulated in AML-12 stimulated by ethanol
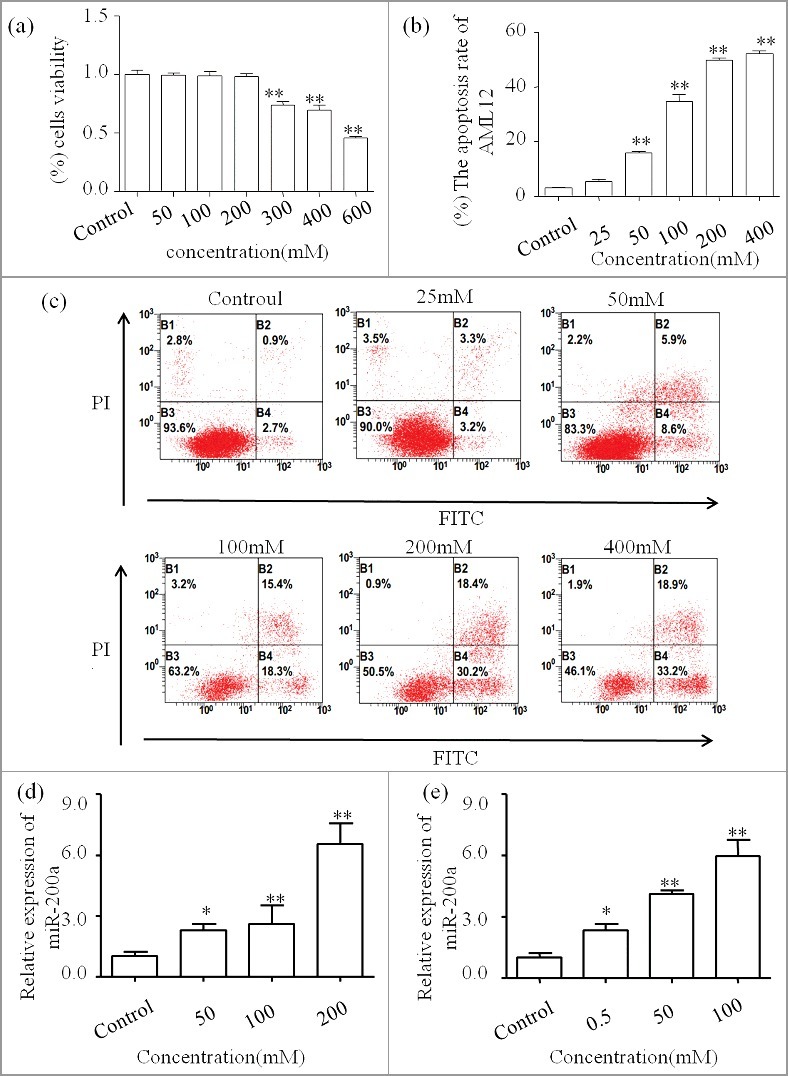
MTT assay combined with Flow cytometric analysis were used to choose the perfect concentration of ethanol in AML-12 cells (Figure 2a, 2b and 2c). Firstly, we exclude concentration more than 538.55±5.12 mM, which is the IC50 calculated form the MTT assay. Next, we find that the apoptosis rate (include right two quadrants) increased graded within 200mM. The lower or upper right represent the early or late apoptotic cells respectively. Finally, we pick 200mM as the best stimulation concentration. Then we used qPCR to analyze the expression of miR-200a. As shown in Figure 2d, we find the expression pattern of miR-200a is up-regulated in vitro. In addition, we also explore the miR-200a expression pattern treated by acetaldehyde in AML-12 cells by qPCR (Figure 2e).
Figure 2.
MiR-200a is up-regulated in AML-12 stimulated by ethanol.
a MTT assay was performed to assess the cytotoxicity of ethanol on AML-12 cells. The IC50 is equal to 538.55±5.12 mM. b The apoptosis rate calculated by three independent Annexin V-FITC staining experent. c Annexin V-FITC staining represent the apoptosis rate caused by ethanol treatment. One representative experiment of the three independent experiments is demonstrated. d MiR-200a expression in ethanol-stimulated AML-12 was analyzed by qPCR. The result are shown as relative expression against control expression without treatment. e MiR-200a expression in acetaldehyde-stimulated AML-12 was analyzed by qPCR. The result are shown as relative expression against control expression without treatment. * P <0.05,** P <0.01 versus control.
MiR-200a induce hepatocyte apoptosis in vivo
Firstly, ALT, AST, TCH, TG were used to evaluate the effect of miR-200a on liver injury in ALD mice (Figure 3a). Next, we want to gain the role of miR-200a played in the regulation of hepatocyte apoptosis in vivo, ALD mouse models were obtained and utilized lentivirus to knockdown miR-200a. qPCR was used to examine the effect of lentivirus injection (Figure 3b), then TUNEL staining and western blot were performed to find the effect of miR-200a on the cell apoptosis. Done-regulated expression of miR-200a displayed a decreased of apoptotic rates, as shown in Figure 3c. The apoptosis related proteins levels was altered corresponding to the inhibition of miR-200a (Figure 3d).
Figure 3.
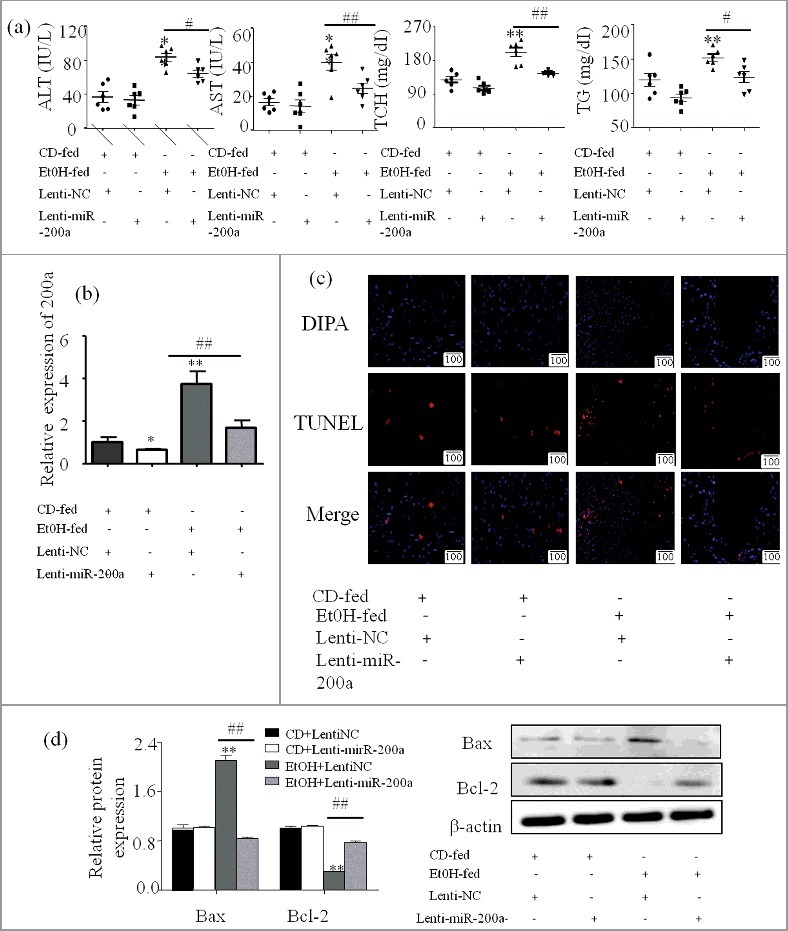
miR-200a induced hepatocyte apoptosis in vivo (a) Serum AST, ALT, Hepatic TG and TCH levels were calculated to evaluate the degree of liver injury. (b) Effect of injected letivirus in mice was evaluated by alteration of miR-200a expression. MiR-200a expression in different groups was analyzed by qPCR. The results are shown as relative expression against control expression without treatment. The values represent means± SD (n = 3 in CD-fed group, n = 3 in EtOH-fed group). (c) The apoptosis cells staining with TUNEL were visualized with a fluorescence microscope (original magnification, ×20). (d) The expression of Bax and Bcl-2 upon treatment with ethanol was determined by western blot in primary hepatocyte derive from experiment mice. The results are shown as relative expression against control expression without treatment. Data shown are the mean± SD from 3 independent experiments. ** P < 0.01 versus control. # P< 0.05, ## P< 0.01 versus ethanol treatment group.
MiR-200a induce hepatocyte apoptosis in vitro
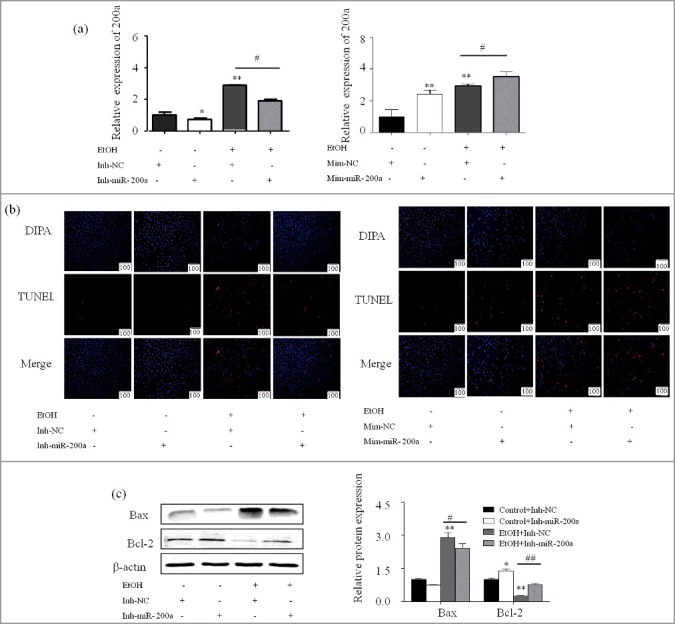
We further investigate the responsibility of miR-200a in AML-12 cells apoptosis. Cells were transfected with 60nM miRNA-200a mimics or 80nM miR-200a inhibitor without or with the treatment of Ethanol. qPCR showed that the expression of miR-200a was successfully increased or decrease after transfected with mimics or inhibitor (Figure 4a). The protein levels of iconic apoptosis protein including Bax, Bcl-2, were altered due to the over-expression of miR-200a, while miR-200a inhibitor led to the opposite effect (Figure 4c and4d). Additionally, Forced expression of miR-200a induced an increased apoptosis rate in AML-12 cells, conversely, decrease was found in AML-12 cells transfected with miR-200a inhibitor (Figure 4b and 4e).
Figure 4.
miR-200a induced hepatocyte apoptosis in vitro.
(a) qPCR showed that transfect miR-200a inhibitor and mimics decrease or increase the expression of miR-200a in AML-12 cells. The results are shown as relative expression against control expression without treatment. Data were presented as mean ± SD from three independent experiments. (b) Representative photomicrographs of group data for TUNEL positive cells (original magnification, × 20). (c) The protein expression of Bax and Bcl-2 were analyzed by western blot in Inh-miR-200a or Mim-miR-200a transfected ethanol-induced AML-12 cells. The results are shown as relative expression against control expression without treatment. Data shown treatment are the mean ±SD from 3 independent experiments. (d) The number of apoptosis cells were presented by flowcytometry using AnnexinV- PI staining. Values are mean ± SD of data from three experiments. * P < 0.05, ** P < 0.01 versus control. # P< 0.05, ## P<0.01 versus ethanol treatment group, ns, not significant versus ethanol treatment group.
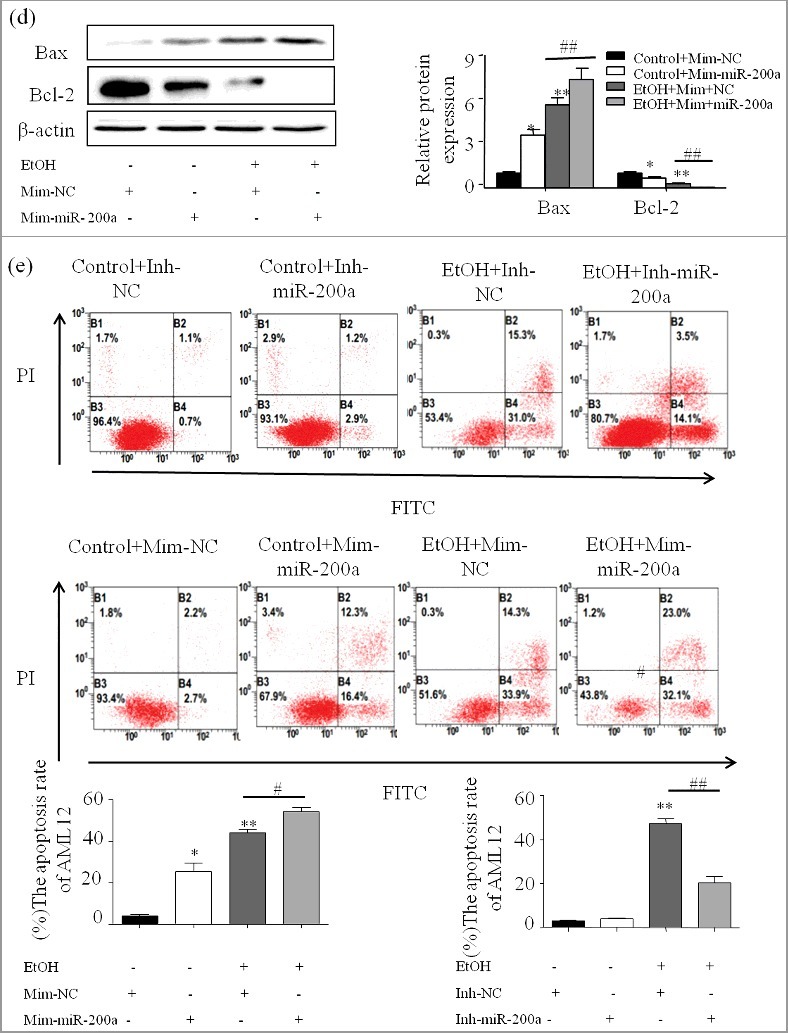
ZEB2 expression pattern and MiR-200a-3p directly targets and inhibits ZEB2 expression
Firstly, we analyzed the expression pattern of ZEB2 by Immunofluorescent (Figure 5a), Western blot and qPCR. The results show that ZEB2 expression is significantly decreased in vivo and in vitro (Figure 5b;5c). To further ascertain the underlying mechanism of the pro-apoptosis of miR-200a in ALD, ZEB2 was identified as a putative target of miR-200a by bioinformatics analysis using TargetScan and miRanda (Figure 5d). Next, the luciferase reporter assay was used to detect the direct interaction between miR-200a and 3′UTR of ZEB2 (Figure 5e). Our results shows that miR-200a mimics repressed wild-type ZEB2 3′UTR, while mutant ZEB2 reporter was unaffected in miR-200a over-expressing cells compared to the miR-NC control. Additionally, we used the lentivirus and mimics, inhibitor to up or down regulate the expression of miR-200a. As shown in Figure 4a, miR-200a was successful regulated in vivo and in vitro. RNA and protein of ZEB2 were analyzed by qPCR and Western blots. We found that the expression of ZEB2 was attenuated compared with control groups due to the over-expression of miR-200a but miR-200a inhibitor have the opposite effect in vivo and in vitro (Figure 5f and 5g).
Figure 5.
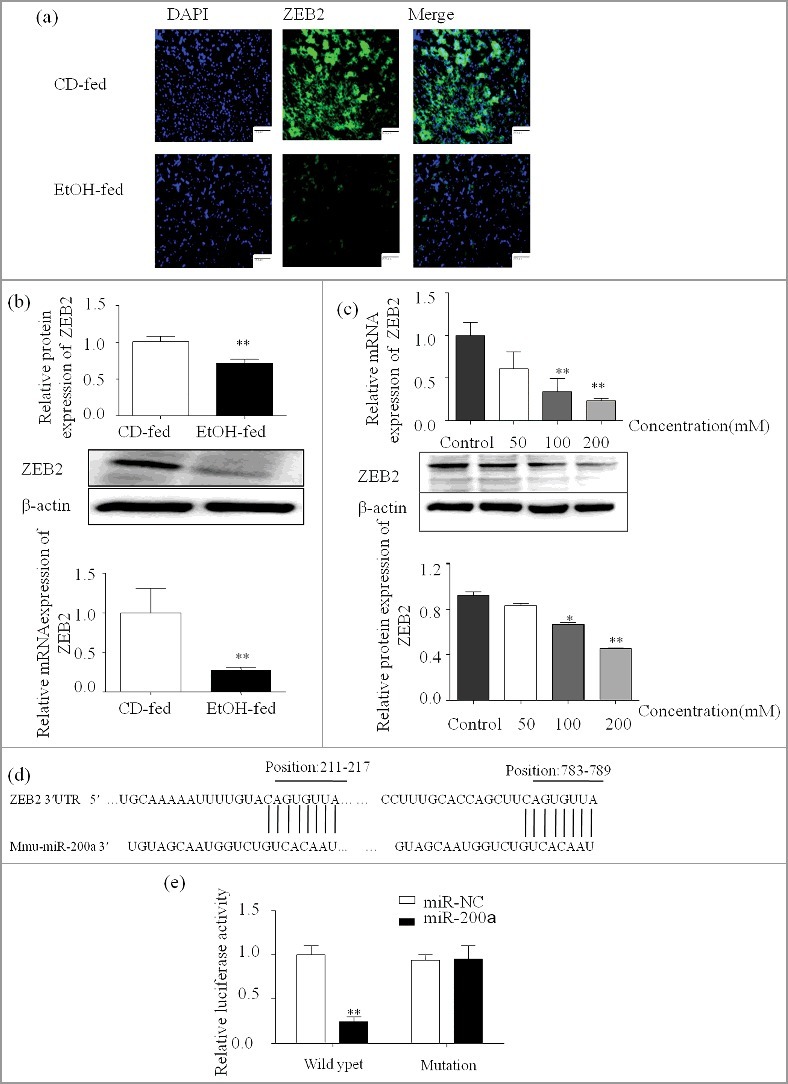
ZEB2 expression pattern and MiR-200a directly targets and inhibits ZEB2 424 expression
(a) The expression of ZEB2 in ethanol treated mice was analyzed by immunofluorescence assay (original magnification, × 20). (b) Total mRNA and protein of ZEB2 extracted from the primary hepatocyte and were analyzed by qPCR and western blot. The results are shown as relative expression against control expression without treatment. (c) The expression of ZEB2 was analyzed by qPCR and western blot in ethanol stimulated AML-12 cells. The results are shown as relative expression against control expression without treatment. (d) Computational analysis of the ZEB2 3′UTR revealed putative conserved miR-200-binding sites. (e) Luciferase reporter assay was performed on AML-12 cells to detect the relative luciferase activities of Wild type and mutation ZEB2 reporters. Data shown are the mean ± SD from 3 independent experiments. (f) The successful over-expression of ZEB2 was verify by western blot and qPCR. The results are shown as relative expression against control expression without treatment. (g) ZEB2 protein and mRNA expression were analyzed by western blot and qPCR in AML-12 cells transfected with miR-200a inhibitor or mimics. Data were presented as mean ± SD from three independent experiments. * P < 0.05, ** P < 0.01 versus control. # P< 0.05, ## P< 0.01 versus ethanol treatment group, ns, not significant versus ethanol treatment group.

Up-regulated ZEB2 expression contributes to inhibition of hepatocyte apoptosis
To explore the role of ZEB2 played in hepatocyte apoptosis, ZEB2 plasmid was transiently transfected into AML-12 cells with or without the treatment of ethanol. The result of transfect was shown by qPCR and Western blot (Figure 6a). As shown in Figure 6b, amount of apoptosis hepatocyte was significant decreased due to the up-regulated ZEB2 in AML-12 cells treated with ethanol. The protein levels of Bcl-2, Bax, were in line with the above result. (Figure 6c).
Figure 6.
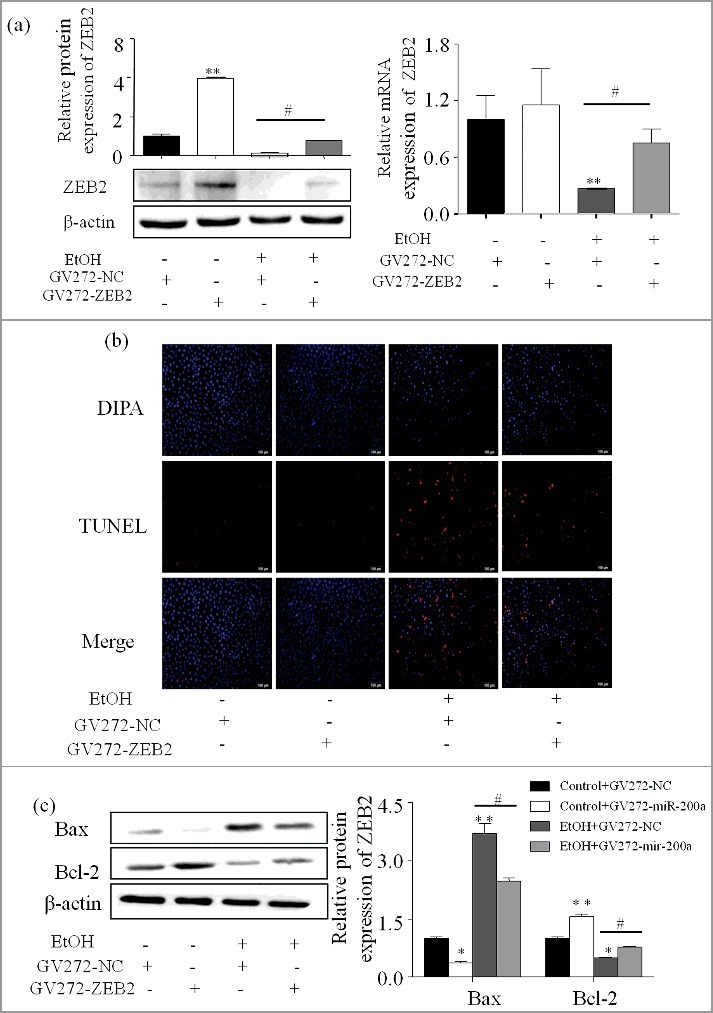
Up-regulated ZEB2 expression contributes to the inhibition of hepatocyte apoptosis (a) ZEB2 successful over-expression was affirmed by western blot and qPCR. The result are shown as relative expression against control expression without treatment. Data were presented as mean ±SD from three independent experiments. (b) AML-12 cells were transfected with ZEB2 or vector for 24 h alone or in combination with ethanol. The stained apoptosis cells with TUNEL were visualized with a fluorescence microscope (original magnification, ×20). (c) The expression of Bax and Bcl-2 were determined by western blot in AML-12 cells. The results are shown as relative expression against control expression without treatment. Data were presented as mean ± SD from three independent experiments. * P < 0.05, ** P < 0.01 versus control. # P< 0.05 versus ethanol treatment group, ns, not significant versus ethanol treatment group.
Restoration of ZEB2 attenuates the effects of miR-200a
Our results have indicated that miR-200a and ZEB2 play opposite roles in the apoptosis in ALD, we speculated that ZEB2 might be involved in the pro-apoptosis effect of miR-200a in ALD. Annexin V-FITC was then conducted and the cell apoptosis rate was significantly decreased in the Mim-miR-200a + GV272-NC group compared to Mim-miR-200a + GV272-ZEB2 group (Figure 7b). Accordingly, over-expressed ZEB2 also reduce the effects of miR-200a on apoptosis-related proteins (Figure 7a).
Figure 7.
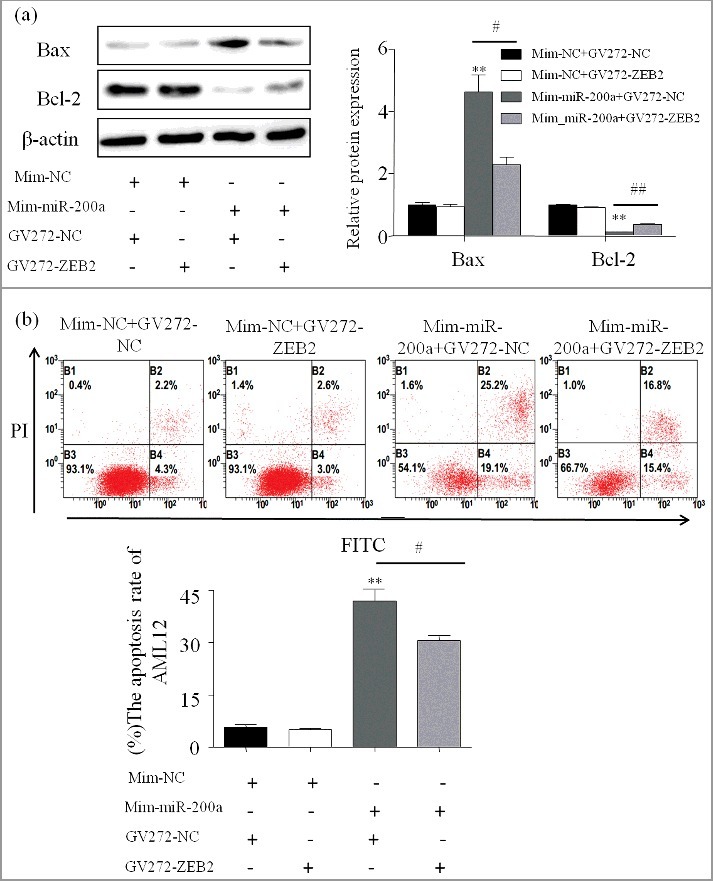
Restoration of ZEB2 attenuates the effects of miR-200a (a) The protein expression of Bax and Bcl-2 were analyzed by western blot in AML-12 transfected with miR-200a mimics and ZEB2. The results are shown as relative expression against control expression without treatment. Data were presented as mean ±SD from three independent experiments. (b) Effect of over-expressed ZEB2 on apoptosis in AML-12 cells transfected with miR-200a mimics. One representative experiment of the three independent experiments is demonstrated. ** P < 0.01 versus control. # P< 0.05 versus ethanol treatment group.
Discussion
Increased apoptosis during ethanol exposure is an indispensable contributor to liver injury [19]. It is well documented that ethanol administration increases the percentage of apoptotic cells significantly in experimental animals. The mechanisms of apoptosis in ALD are being increasingly implicated [20]. Apoptosis is an organized and ordered form of cell death, which is characterized by chromatin shrinkage and condensation, nuclear fragmentation [21]. Cell apoptosis is an active process in which cell strive to death to adapt to the environment [22]. Consequently, the occurrence of apoptosis involve a series of gens activation and expression. Among the numerous regulators of apoptosis, Bcl-2 family proteins are key players which include three clusters, the pro-survival subfamily includes Bcl-2, Bcl-w, Bcl-XL, cl-1and A1, the pro-apoptotic subfamily divide into multidomain group (Bax, Bok, and Bak) and BH3-only group (Bad, Bim, others) [23,24]. Among them, multidomain group act as xecutioners of apoptosis, whereas pro-survival and BH3-only group act as inhibitors and initiators of apoptosis, respectively. Bcl-2 family are frequently up- or down- regulated in some specific pathology and physiology situation. Therefore we choose Bcl-2 and Bax protein level as index of apoptosis [25].
Dysregulation of microRNA expression is highly correlated with several liver disease [26]. There is a considerable body of literature suggest that abundant expression of miR-200a was closely correlated with decreased cell survival rate. It has been reported that miR-200a suppresses cell growth in hepatocellular carcinoma [27–29], and over-expressed miRNA-200a inhibits the HSC proliferation [30]. In this study, microRNA mimics and inhibitor was employed to explore the role of altered expression of miR-200a. We have shown that over-expressed miR-200a facilitated hepatocyte apoptosis. Usually, if a cell undergoes apoptosis hinges on the balance between pro-survival and pro-apoptotic proteins. By transfected miRNA-200a inhibitor into AML-12, we find positive regulator Bax was significantly decreased and negative regulator Bcl-2 was increased, which are in accordance with the pro-apoptosis role of miR-200a. However, the mechanism at epigenetic scale of this function is still remain unclear.
ZEB1 (also shown as dEF1, TCF8) and ZEB2 (also shown as SIP1) encoded by the ZFHX1a and ZFHX1b gens are belong to ZEB family, which is crucial EMT (Epithelial-to-mesenchymal transition activator) activator [31]. EMT means convert epithelial cells into mesenchymal cells, is a crucial part of human development and disease. It is already known that EMT confers cellular motility, now dates show that EMT also control other cellular functions, such as senescence, differentiation, proliferation, especially apoptosis [32]. miR-200 family is highly implicated in the regulation of ZEB2 expression, and it is reported that miR-200 are inducers of epithelial differentiation and can revert directly an EMT [33]. Notably, not only miR-200 have the inhibitory effect on ZEB2, we also found a reverse control of each other [34,35]. The consequence of such double-negative feedback loop is that the apoptosis result not only because increased miR-200a due to ethanol treatment decrease ZEB2 expression, but also due to the down-regulated ZEB2 induce the elevated miR-200a expression.
To further investigate the mechanism, we identified ZEB2 as a direct target of miR-200a by luciferase reporter system. The low expression of ZEB2 in ALD mouse liver and AML-12 cells was associated with high miR-200a expression. The posttranscriptional level expression of ZEB2 was inverse altered in response to treatment with inhibitor or mimics. Having established the directly targeted relationship between ZEB2 and miR-200a, we next examined weather ZEB is a essential mediator for apoptosis of miR-200a. ZEB2 is an usual tumor suppressor, capable of facilitating proliferation and suppressing apoptosis. We established miR-200a over-expressing meanwhile up-regulated ZEB2 cell lines, and our dates indicated that up-regulated expression of ZEB2could decreased apoptosis rate of hepatocyte by over-expressed miR-200a. Taken together, our findings suggest that miR-200a contributes to alcoholic liver injury by directly target and down-regulate ZEB2 which modulate apoptosis course (Figure 8).
Figure 8.
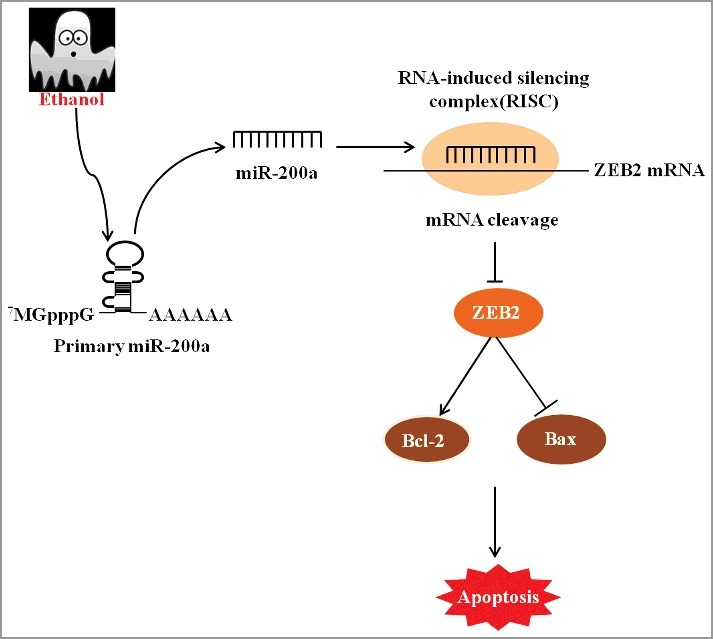
The major pro-apoptosis mechanism contributed by miR-200a in ALD. Ethanol treatment elevate the level of miR-200a expression, which could directly binding to ZEB2 and subsequently suppress ZEB2 expression. ZEB2 depletion resulted in the inhibition of Bcl-2 and facilitation of Bax, which are landmark protein of apoptosis.
In recent decades, the incident of ALD has increased significantly, but the diagnosis of ALD is still difficult and gentle form of liver damage often occurs long before clinically symptomatic. In this study, for the first time, we characterized the pro-apoptosis function of miR-200a in ALD. Additionally, we demonstrated that ZEB2 was a direct target and essential mediator for apoptosis of miR-200a. The data presented here may suggest that miR-200a and ZEB2 has the potential to be the treatment target for ALD.
Materials and methods
Cell culture and treatment
Mice normal hepatocyte cell lines (AML-12) were purchased from American Type Culture Colection (MTCC) (Shanghai, China), they were cultured in F-12 medium (Gibco,USA), with 8% fetal bovine serum USA) and 100 units/ml penicillin, 100 mg/ml streptomycin and incubated at 37 °C with 5% CO2.
Cell viability assay (cytotoxicity test)
MTT (3-(4, 5-dimethylthiazol-2yl)-2, 5-diphenyl tetrazolium bromide) assay was used to quantitatively evaluate the influence of ethanol on cell viability. Cells were seeded at a density of 3 × 103 cells/well into a 96-well plate. And next day, the supernatant was replaced with fresh medium containing ethanol (0, 50, 100, 200, 300, 400, 600 mM) for 24 h. After that 20ul of MTT solution (5 mg/ml) was added to each well and still incubated in 37 °C at least 4h. Then suction abandon all supermatant replace with 150ul DMSO (Dimethyl sulfoxide). The optical density at 490 nm was determined at a microplate reader. Each group was repeated in triplicate.
Reverse transcription and quantitative real-time PCR analysis (qPCR)
The expression pattern of miR-200a and ZEB2 in primary hepatocyte from CD-fed versus Ethanol-fed mouse and in AML-12 was measured by reverse transcription and qPCR. EzOmicsTM One-Step qPCR Kit (Biomics, USA) was used for miR-200a, but ZEB2 need converted to cDNA bytranscription kit (Thermo, USA) first. The sequence were as flows: ZEB2 (Forword: GGCAAGCGCTTCTCACACTC and Reverse: TGCAGGTAAGCCCGGTTCAT).
Plasmid construction and cell transfection
After cell adherence, inhibitor and mimics was added to each well at a final concentration of 60nM and 80nM in Opti-MEM medium (Gibco, USA). This form of transient transfection use Lipofectamine 2000 as vector following the Manufacture's protocol.
Histopathological examination and Immunofluorescent Staining
For histologic analysis, fixed in 10% neutral neutral formalin solution, dehydrated in an ethanol gradient and xylene treatment, after then embedded in paraffin. the paraffin liver tissue cut into 5 μm thick sections, and stained with hematoxylin and eosin. For immunofluorescent staining, firstly tissue sections ware fixed with 4% paraformaldehyde and blocked with 10% BSA for 10min. Then the sectionwere incubated with mouse anti-mouse ZEB2 (1:800, CST, USA) overnight at 4 °C. On the following day, incubated with anti-mouse FITC (1:200) for 2 h at temperature. The images were taken using fluorescence microscopy.
Animal, mouse model of ALD
8-week-old male C57BL/6J mice were obtained from the Experimental Animal Center of Anhui Medical University. The animal experimentation were performed in adherence with the Guideline of Animal Care and Use Committee of Anhui Medical University and approved by the Ethics Committees of Anhui Medical University. All animals were divided randomly into 4 groups, CD-fed + lenti-miR-NC, CD-fed + lenti-miR-200a group and Ethanol-fed + lenti-NC, Ethanol-fed+lenti-miR-200a group after acclimatization for at least 1-2 week. The period of entire molding is 16 days [36], except the CD-fed group, the first 5 day was liquid diet adaptation period, next is 10 days modeling (5% v/v), then is one time gavage (5 g/kg, body weight, 20% ethanol) and a day time specimens. The lentivirus was inject twice in mice caudal vein, respectively at the first day and the eighth day of the building period. All of the groups were sacrificed 9h after the last injection, blood and liver tissue were prepare for further experiments.
Western blot analysis
Primary hepatocyte and AML-12 cells were lysed with RIPA lysis buffer and protease inhibitors (100:1, Beyotime, Shanghai, China). Equal amounts of protein (30-50 mg) were separated by 10% SDS-PAGE (sodium dodecyl sulfate– polyacrylamide gel electrophoresis) and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After blockade in TBST containing 5% skim milk 3h at room temperature, the membranes were incubated in Rabbit polyclonal anti-Bcl2, anti-Bax (Abcam, USA,1:800), mouse anti-ZEB2 (CST, USA.1:800) over night at 4 °C. Subsequently washing three times for 15min with TBS/ Tween-20 (0.075%) containing 3% Marvel, and then incubated with a 1:1000 dilution of the HRP- secondary antibodies at 37 °C for 1h. β-actin was used as an loading control. After washing again, the membrane was exposed in the ECL-chemiluminescent kit (ECL-plus, Thermo Scientific).
Apoptosis assessment
Apoptosis was evaluated by FITC-Annexin V apoptosis detection kit (BestBio, china) or terminal deoxymu-cleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) assay kit (ROCHE, USA). Cells were collected, washed with ice-cold PBS three times and then re-suspended in binding buffer (1 × 106 cells/ml), Next, add 5ul Annexin V-FITC into the cell suspension then incubation 15 min at 4 °C in the dark. after PI staining 10min, stained cells were analyzed with BD LSR flow cytometer (BD Biosciences). For TUNEL staining, cell slides were firstly fixed with 1% paraformaldehyde, then permeabilized with proteinase K and H2O2. Digoxigenin -conjugated dUTP and recombinant terminal deoxynucleotidyl transferase (TdT) were added onto the sections and incubated for 10min at 15–25 °C, subsequently incubation with 50ul reaction mixture for 1h at 37 °C. The labeled cells were visualized with a fluorescence microscope.
Statistical analysis
Statistical analyses were performed by using SPSS 15.0 software and all results are presented as means ± SD. Statistical significances were analyzed by one-way ANOVA and Student's t-test. In all cases, value of P< 0.05 was considered to be statistically significant.
Funding Statement
National Natural Science Foundation of China [grant number 81473268]; Anhui University of Science and Technology [grant number 1704a0802161].
Acknowledgments
This study is supported by the National Science Fundations of China (no. 81473268) and Science and Technology Project of Anhui (no.1704a0802161).
Conflict of interest statement
The authors declared that they have no conflicts of interest to this work. We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
References
- [1].Dunn W, Shah VH. Pathogenesis of alcoholic liver disease. Clin Liver Dis. 2016;20:445–456. doi: 10.1016/j.cld.2016.02.004. PMID:27373608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chacko KR, Reinus J. Spectrum of alcoholic liver disease. Clinics in Liver Disease. 2016;20:419–427. doi: 10.1016/j.cld.2016.02.002. PMID:27373606 [DOI] [PubMed] [Google Scholar]
- [3].McDaniel K, Herrera L, Zhou T, et al. The functional role of microRNAs in alcoholic liver injury. J Cell Mol Med. 2014;18:197–207. doi: 10.1111/jcmm.12223. PMID:24400890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pompili M, Serafini G, Innamorati M, et al. Suicidal behavior and alcohol abuse. Int J Environ Res Public Health. 2010;7:1392–431. doi: 10.3390/ijerph7041392. PMID:20617037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Szabo G, Satishchandran A. MicroRNAs in alcoholic liver disease. Semin Liver Dis. 2015;35:36–42. doi: 10.1055/s-0034-1397347. PMID:25632933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Massey VL, Arteel GE. Acute alcohol-induced liver injury. Front Physiol. 2012;3:193. doi: 10.3389/fphys.2012.00193. PMID:22701432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bala S, Szabo G. MicroRNA Signature in Alcoholic Liver Disease. Int J Hepatol. 2012;2012:498232. doi: 10.1155/2012/498232. PMID:22518321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Stickel F, Dubuquoy L. MicroRNA in alcoholic hepatitis: implications for pathophysiology and treatment. Gut. 2016;65:1400–1401. doi: 10.1136/gutjnl-2016-312101. PMID:27252446 [DOI] [PubMed] [Google Scholar]
- [9].Natarajan SK, Pachunka JM, Mott JL. Role of microRNAs in Alcohol-Induced Multi-Organ Injury. Biomolecules. 2015;5:3309–3338. doi: 10.3390/biom5043309. PMID:26610589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shah N, Nelson JE, Kowdley KV. MicroRNAs in Liver Disease: Bench to Bedside. J Clin Exp Hepatol. 2013;3:231–242. doi: 10.1016/j.jceh.2013.09.001. PMID:25755505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gao X, Han D, Fan W. Down-regulation of RBP-J mediated by microRNA-133a suppresses dendritic cells and functions as a potential tumor suppressor in osteosarcoma. Exp Cell Res. 2016;349:264–272. doi: 10.1016/j.yexcr.2016.10.019. PMID:27794430 [DOI] [PubMed] [Google Scholar]
- [12].You W, Xu L, Zhang X, et al. High-Throughput Screening Identifies miR-451 as a Pleiotropic Modulator That Suppresses Gastric Cancer Metastasis. SLAS Technol. 2017;22:136–143. PMID:27780852 [DOI] [PubMed] [Google Scholar]
- [13].Jiang J, Yang P, Guo Z, et al. Overexpression of micro RNA-21 strengthens stem cell-like characteristics in a hepatocellular carcinoma cell line. World J Surg Oncol. 2016;14:278. doi: 10.1186/s12957-016-1028-9. PMID:27793160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang HF, Xu LY, Li EM. A family of pleiotropically acting microRNAs in cancer progression, miR-200: potential cancer therapeutic targets. Curr Pharm Des. 2014;20:1896–1903. doi: 10.2174/13816128113199990519. PMID:23888967 [DOI] [PubMed] [Google Scholar]
- [15].Feng J, Wang J, Chen M, et al. miR-200a suppresses cell growth and migration by targeting MACC1 and predicts prognosis in hepatocellular carcinoma. Oncol Rep. 2015;33:713–720. PMID:25482402 [DOI] [PubMed] [Google Scholar]
- [16].Francis H, McDaniel K, Han Y, et al. Regulation of the extrinsic apoptotic pathway by microRNA-21 in alcoholic liver injury. J Biol Chem. 2014;289:27526–27539. doi: 10.1074/jbc.M114.602383. PMID:25118289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Blaya D, Coll M, Rodrigo-Torres D, et al. Integrative microRNA profiling in alcoholic hepatitis reveals a role for microRNA-182 in liver injury and inflammation. Gut. 2016;65:1535–1545. doi: 10.1136/gutjnl-2015-311314. PMID:27196584 [DOI] [PubMed] [Google Scholar]
- [18].Li M, He Y, Zhou Z, et al. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6-p47phox-oxidative stress pathway in neutrophils. Gut. 2017;66:705–715. doi: 10.1136/gutjnl-2016-311861. PMID:27679493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Casey CA, Nanji A, Cederbaum AI, et al. Alcoholic liver disease and apoptosis. Alcohol Clin Exp Res. 2001;25:49S–53S. doi: 10.1111/j.1530-0277.2001.tb02373.x. PMID:11391049 [DOI] [PubMed] [Google Scholar]
- [20].Nanji AA. Apoptosis and alcoholic liver disease. Semin Liver Dis. 1998;18:187–190. doi: 10.1055/s-2007-1007154. PMID:9606815 [DOI] [PubMed] [Google Scholar]
- [21].Wang S, Pacher P, De Lisle RC, et al. A Mechanistic Review of Cell Death in Alcohol-Induced Liver Injury. Alcoholism, Clinical and Experimental Research. 2016;40:1215–1223. doi: 10.1111/acer.13078. PMID:27130888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kemp MG. Crosstalk between apoptosis and autophagy: environmental genotoxins, infection, and innate immunity. J Cell Death. 2017;9:1179670716685085. PMID:28469477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vervliet T, Parys JB, Bultynck G. Bcl-2 proteins and calcium signaling: complexity beneath the surface. Oncogene. 2016;35:5079–5092. doi: 10.1038/onc.2016.31. PMID:26973249 [DOI] [PubMed] [Google Scholar]
- [24].Um HD. Bcl-2 family proteins as regulators of cancer cell invasion and metastasis: a review focusing on mitochondrial respiration and reactive oxygen species. Oncotarget. 2016;7:5193–5203. doi: 10.18632/oncotarget.6405. PMID:26621844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Renault TT, Manon S. Bax: Addressed to kill. Biochimie. 2011;93:1379–1391. doi: 10.1016/j.biochi.2011.05.013. PMID:21641962 [DOI] [PubMed] [Google Scholar]
- [26].Wang XW, Heegaard NH, Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431–1443. doi: 10.1053/j.gastro.2012.04.007. PMID:22504185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xu Y, Zhang Y, Wang L, et al. miR-200a targets Gelsolin: A novel mechanism regulating secretion of microvesicles in hepatocellular carcinoma cells. Oncol Rep. 2017;37:2711–2719. doi: 10.3892/or.2017.5506. PMID:28440466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pendlebury A, Hannan NJ, Binder N, et al. The circulating microRNA-200 family in whole blood are potential biomarkers for high-grade serous epithelial ovarian cancer. Biomed Rep. 2017;6:319–322. doi: 10.3892/br.2017.847. PMID:28451393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Huang C, Zeng X, Jiang G, et al. XIAP BIR domain suppresses miR-200a expression and subsequently promotes EGFR protein translation and anchorage-independent growth of bladder cancer cell. J Hematol Oncol. 2017;10:6. doi: 10.1186/s13045-016-0376-9. PMID:28057023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yang JJ, Tao H, Hu W, et al. MicroRNA-200a controls Nrf2 activation by target Keap1 in hepatic stellate cell proliferation and fibrosis. Cell Signal. 2014;26:2381–2389. doi: 10.1016/j.cellsig.2014.07.016. PMID:25049078 [DOI] [PubMed] [Google Scholar]
- [31].Qi S, Song Y, Peng Y, et al. ZEB2 mediates multiple pathways regulating cell proliferation, migration, invasion, and apoptosis in glioma. PLoS One. 2012;7:e38842. doi: 10.1371/journal.pone.0038842. PMID:22761708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop–a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. PMID:20706219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hill L, Browne G, Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int J Cancer. 2013;132:745–754. doi: 10.1002/ijc.27708. PMID:22753312 [DOI] [PubMed] [Google Scholar]
- [34].Bracken CP, Gregory PA, Kolesnikoff N, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. PMID:18829540 [DOI] [PubMed] [Google Scholar]
- [35].Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. PMID:18483486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bertola A, Mathews S, Ki SH, et al. Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat Protoc. 2013;8:627–637. doi: 10.1038/nprot.2013.032. PMID:23449255 [DOI] [PMC free article] [PubMed] [Google Scholar]


