ABSTRACT
Chromosome morphology in Saccharomyces cerevisiae is only visible at the microscopic level in the ribosomal DNA array (rDNA). The rDNA has been thus used as a model to characterize condensation and segregation of sister chromatids in mitosis. It has been established that the metaphase structure (“loop”) depends, among others, on the condensin complex; whereas its segregation also depends on that complex, the Polo-like kinase Cdc5 and the cell cycle master phosphatase Cdc14. In addition, Cdc14 also drives rDNA hypercondensation in telophase. Remarkably, since all these components are essential for cell survival, their role on rDNA condensation and segregation was established by temperature-sensitive (ts) alleles. Here, we show that the heat stress (HS) used to inactivate ts alleles (25 ºC to 37 ºC shift) causes rDNA loop condensation in metaphase-arrested wild type cells, a result that can also be mimicked by other stresses that inhibit the TORC1 pathway. Because this condensation might challenge previous findings with ts alleles, we have repeated classical experiments of rDNA condensation and segregation, yet using instead auxin-driven degradation alleles (aid alleles). We have undertaken the protein degradation at lower temperatures (25 ºC) and concluded that the classical roles for condensin, Cdc5, Cdc14 and Cdc15 still prevailed. Thus, condensin degradation disrupts rDNA higher organization, Cdc14 and Cdc5 degradation precludes rDNA segregation and Cdc15 degradation still allows rDNA hypercompaction in telophase. Finally, we provide direct genetic evidence that this HS-mediated rDNA condensation is dependent on TORC1 but, unlike the one observed in anaphase, is independent of Cdc14.
KEYWORDS: Heat stress, rapamycin, TORC1, ribosomal DNA, condensin, Cdc5, Cdc14, Cdc15, Cdc60, aid degron, temperature-sensitive allele
Introduction
The generation of conditional alleles has been of the upmost importance to address the biological function of essential genes. These alleles let the carrying organism to live and reproduce without major problems in what is called the permissive condition, whereas viability is severely compromised in the restrictive condition. Conditional alleles carry mutations that make the encoded protein hypersensitive to a restrictive condition that is not deleterious for the organism per se. Thermosensitive (ts) alleles are the most widely used conditional alleles in all organisms, from virus to cell lines [1,2]. These alleles have the advantage of allowing the experimenter to shift between permissive and restrictive conditions in a very straightforward and inexpensive manner. In the yeast Saccharomyces cerevisiae, ts alleles have been critical to understand the logic behind the cell cycle, DNA replication, transcription, etc. [3–6] When performing experiments with ts alleles, yeast cells are grown at 21°-25°C (permissive condition) and then shifted to 34°–37°C (restrictive condition) to investigate the function of the essential protein. A prerequisite for the use of ts alleles is that the biological process the protein participates in is affected by neither the permissive nor the restrictive temperature. In the past, temperature has been often neglected as a variable in the experiment because wild type yeast cells seem to grow well at 37 °C.
Chromosome reshaping during the cell cycle is among the many biological processes where multiple essential proteins have been identified, such as histones, topoisomerase II (Top2) and those that comprise the cohesin and condensin complexes [7,8]. Other essential proteins linked to the different chromosome morphologies observed within a round of cell division are cell cycle regulators that indirectly drive chromosome reorganization through the posttranslational modification of the aforementioned players [7]. In eukaryotes, the most striking chromosome reorganizations take place during the last stage of the cell division (M phase). Five are the major signatures: chromosome axial compaction and lateral condensation; sister chromatid resolution of the chromosome arms, chromosome alignment at the metaphase plate; sister chromatid segregation; and chromosome decondensation. Whereas in higher eukaryotes these signatures proceed more or less in that order from prophase to telophase, in yeast and other fungi both the signatures and their order appear to be slightly different. Thus, in S. cerevisiae, it is not simple to divide M phase into substages: G2, prophase and metaphase are indistinguishable; they are collectively referred to as G2/M. On the one hand, there is a very modest general condensation of chromosomes, which make impossible to visualize them as single entities [9]. On the other hand, there is no chromosome alignment prior to segregation and, as opposed to higher eukaryotes, sister chromatid resolution goes from centromeres to telomeres right at the time of segregation [10,11]. In spite of these differences, cohesin, condensin, Top2 and many cell cycle regulators of chromosome morphology are found in yeast. Not only that, their functions seem to be conserved with respect to higher eukaryotes; indeed, S. cerevisiae has importantly contributed to our knowledge on the function of these proteins, often with the aid of the corresponding ts alleles [9,12–22].
Since individual chromosomes cannot be visualized within the yeast nucleus, at least in mitosis, yeast researchers have relied on the only chromosome portion whose sequence is repetitive enough to infer structural patterns, the ribosomal DNA array (rDNA). This array can be studied either by Fluorescence In Situ Hybridization (FISH) with probes against the rDNA sequence or by coating the repetitive sequence with specific binding proteins fused to fluorescent reporters (e.g., Net1-GFP) [9,10]. These studies first established the rDNA structural reorganization along the cell cycle and then the roles of cohesin, condensin and cell cycle regulators such as Cdc5, Cdc14 and Ipl1 [9,18,19,22]. The most impacting visual structure that came up from these studies was the rDNA metaphase loop. Initially thought to be a condensed structure, its axial length is actually the longest during the cell cycle. Nevertheless, its thin linear pattern points out that it is a highly organized structure with probable lateral condensation [9,22,23]. Previous studies with ts alleles showed that both the condensin and cohesin complexes were needed for the rDNA loop establishment and maintenance in metaphase [18]. Condensin is also a critical player in sister chromatid resolution, a needed step for faithful chromosome segregation in anaphase. The rDNA is particularly troublesome for resolution and segregation because the very high level of transcription blocks condensin from loading onto. Thus, transcription must be temporarily shut down in anaphase. This is accomplished through the activation of the master cell cycle phosphatase Cdc14, which is preceded by that of the Polo-like kinase Cdc5 [24–28]. Noteworthy, the rDNA itself is responsible for keeping Cdc14 inactive for most of the cell cycle. Thus, Cdc14 binds tightly to the rDNA all throughout the cell cycle but anaphase through its inhibitor Net1 [25,28–30]. Upon anaphase onset, Cdc5 gets active and aids in diminishing the strong interaction between Cdc14 and Net1 [31–34]. This allows Cdc14 to temporarily abandon the rDNA in two waves. The first (minor) wave is controlled by the Cdc Fourteen Early Anaphase Release (FEAR) network and the second (major) wave is triggered by the Mitotic Exit Network (MEN). These two activation waves allow Cdc14 to reach its multiple targets and co-ordinately execute anaphase first (Cdc14/FEAR) and then telophase-to-G1 transition (Cdc14/MEN) [32,35–37]. The role of Cdc14 is not only to allow rDNA segregation but also to carry out rDNA axial compaction in order to avoid severing by cytokinesis. This latter role was inferred because a telophase block by a ts allele for the MEN component CDC15 showed a hypercondensed rDNA [9,22].
Herein, we present data that the rDNA loop is dramatically changed upon incubation at 37°C in wild type yeast strains. Hence, we have checked again the roles of condensin, Cdc5, Cdc14 and Cdc15, but making use of the novel auxin-mediated degradation of aid alleles as an alternative to ts alleles. We show that the classical findings of ts alleles for these four players still prevail. Finally, we demonstrate that this novel mechanism of rDNA condensation in metaphase is dependent on TORC1 but independent of Cdc14.
Results
Heat stress causes condensation of the metaphase rDNA loop
We began this work because of a casual observation while filming Cdc14-GFP in yeast cells blocked in metaphase. We were initially interested in recording any possible Cdc14 relocalization upon different kinds of environmental stresses. Cytologically, Cdc14-GFP relocalization is seen as an overall drop of the rDNA/nucleolus signal. Most clearly of all, Cdc14 binds to the Spindle Pole Body (SPB) upon its release, which is then easily visible as a fluorescent focus [31,36].
Incidentally, we blocked cells in metaphase at 25 °C with the microtubule depolymerizing drug Nocodazole (Nz), which also elicits the most spectacular rDNA loops in vivo [10,22]. As expected from its binding to the rDNA, Cdc14-GFP appeared as a loop in 80% of cells blocked in Nz (Figure 1A). To our great surprise, we observed that the Cdc14-GFP loop was largely lost when the cell culture was shifted to 37 °C for 1 h while keeping the metaphase block with Nz. A temperature shift from 25 °C to 37 °C is considered to be a physiologically meaningful heat stress [38]. Instead of keeping the loop appearance, an oval- or line-like Cdc14-GFP signal was seen in 80% of cells at 37 °C (Figure 1A). Even in the remaining 20% of cells where a loop-like structure could still be distinguished at 37 °C, the loop was shorter than the one at 25 °C (see below). We performed this experiment in an YPH499/S288C background, but similar results were obtained in other backgrounds (data not shown).
Figure 1.
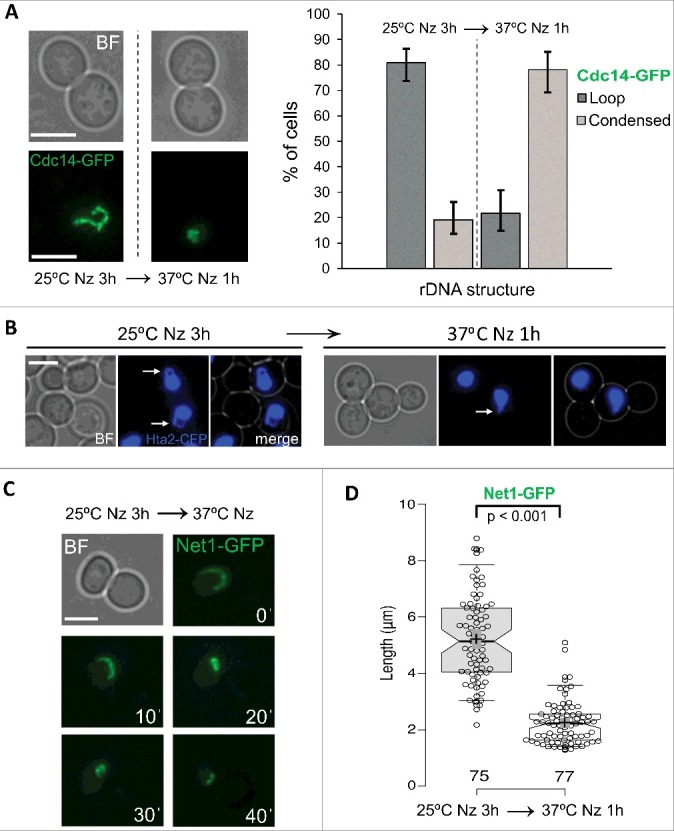
The rDNA metaphase loop gets condensed upon heat stress (HS). (A) Strain FM230 (CDC14:GFP, YPH499 background) was blocked in metaphase with Nocodazole (Nz) for 3h at 25 ºC and then shifted to 37 ºC for 1 h. Samples were taken and visualized under the fluorescence microscope before and after the temperature shift. Representative photos are shown on the left and a quantification chart on the right. (B) Strain yED233 (HTA2:yEmCFP) was Nz-blocked at 25 ºC and then shifted to 37 ºC for 1 h. Representative photos are shown before and after the temperature shift. White arrows point to the metaphase loop seen with the HTA2-CFP histone signal. (C) Strain FM225 (NET1:GFP, YPH499 background) was Nz-blocked for 3h at 25 ºC before being sawed onto an agarose patch with Nz and filmed while cells were being heated up to 37 ºC. A representative cell filmed in 10 min intervals during this HS time-lapse is shown. (D) Strain FM931 (OsTIR1 NET1:GFP, W303 background) was Nz-blocked at 25 ºC and then shifted to 37 ºC for 1 h. Box plot of the quantification of the length of the loop, the line or the major axis of the oval shape (when neither a loop nor a line could be inferred) before and after the HS. White scale bars represent 5 μm. BF, bright field; Nz, nocodazole.
The pattern of the Cdc14-GFP change pointed towards a reorganization of the rDNA rather than a Cdc14 relocalization; i.e., there was no fainting and diffusion of Cdc14-GFP, nor was there appearance of the extra SPB foci (see other chapters below for further support). We assumed that these oval- or line-like signals were similar to the “condensed” rDNA phenotype described earlier on [9,19]. In order to get further support for this temperature-dependent loss of the rDNA loop, we checked a strain with a histone protein (Hta2) labelled with CFP. Histones are unlikely to detach from the DNA upon this moderate heat stress (HS). Indeed, we were able to observe a histone-labelled loop upon Nz arrest at 25 °C and how this loop disappeared after the HS (Figure 1B), strongly supporting that the observed phenotype is a morphological change of the rDNA rather than a relocalization of Cdc14. Although Hta2-CFP can be used a marker for presence/absence of the loop it fails to address whether the loop always corresponds to the rDNA and also what happens to the loop after the HS. Thus, we next chose a constitutive marker for the rDNA: Net1-GFP. Unlike Cdc14, Net1 is known to be an exclusive rDNA-binding protein throughout the entire cell cycle and has been broadly used as a first-class rDNA reporter for in vivo studies [10,22,29,32,39]. We again confirmed this change from looped to condensed rDNA upon the 25 °C to 37 °C shift. We further filmed this change and observed that the transition from the loop to the condensed form seems to occur through axial compaction of the rDNA until it loses its loop-like shape (Figure 1C).
Anticipating the consequences that this finding might have for previous works on rDNA structure and segregation that were carried out with ts alleles, we further included for analysis the reference strain created for degradation of essential proteins by C-terminal tagging with the auxin-based degron sequence (aid) [40]. At present, auxin-degron tagging of essential genes seems to be the best alternative to ts alleles. Again, we observed the Net1-GFP condensation upon the temperature shift in Nz. Quantification of the rDNA length before (Nz, 25 °C) and after (Nz, 37 °C x 1h) the HS showed that the transition goes from a loop with a median length of ∼5 µm into an oval/line condensed form which is ∼2 µm long in its major axis (Figure 1D).
Finally we wondered if the temperature might have an effect on the rDNA morphologies observed with the Net1-GFP marker in cycling yeast cells, as opposed to cells blocked in metaphase with Nz. Strikingly, rDNA loops were rare or small in cycling cells, even in G2/M cells at 25 °C (Figure S1). In addition, no clear differences could be inferred between 25 °C and 37 °C, or throughout the cell cycle (e.g., small loops were also observed in G1 and early S phase). Noteworthy, most cycling G2/M cells has a condensed morphology for the rDNA rather similar to what we observe after the HS in Nz-arrested cells.
From this set of experiments we concluded that moderate HS (25 °C to 37 °C incubations) reshapes the canonical rDNA loop morphology observed in Nz-blocked cells into a more condensed organization. Because this might have enormous consequences for previous works where rDNA structure and segregation were studied with ts alleles, we decided to revisit some of the most remarkable previous findings, yet making use of the auxin-degron system as an alternative to ts alleles.
Auxin-mediated degradation of Cdc14 still forms an rDNA anaphase bridge
Amongst the previous findings obtained with ts alleles that more closely relate to the rDNA we find the involvement of Cdc14 in the segregation of this locus in anaphase. We and others have previously reported that cells bearing cdc14-ts alleles get blocked in anaphase with the rDNA forming an anaphase bridge at the restrictive temperature [10,21–23,41]. Since the HS-mediated rDNA condensation takes place before cdc14-ts cells notice the absence of Cdc14 by anaphase, it is conceivable that the rDNA condensation itself is linked to the segregation problems seen in cdc14-ts; i.e., Cdc14 might not be required for rDNA segregation if the locus maintained the loop structure at anaphase onset. In order to address this doubt, we tagged CDC14 with several versions of the auxin-degron sequence; including the one developed originally (simply referred to as aid) [40], and newer and shorter versions of it (aid*) further tagged with sequences for detection by Western blotting (9xmyc) and fluorescence microscopy (e.g., GFP) [42]. All these tags conferred strong sensitivity to the auxin 3-indolacetic acid (IAA) in the micromolar range (Figure 2A). As expected, this sensitivity fully depended on the co-expression of a plant-based auxin-degron targeting ubiquitin ligase (OsTIR1 in this case) (Figure S2A). Thus, Cdc14 tagged with different versions of aid appeared to be functional, and growth was only abrogated when both the auxin and OsTIR1 were present. Furthermore, all these strains got arrested as dumbbells within one cell cycle (>95% cells after 3 h with 1 mM IAA, data not shown). We determined the fate of Cdc14-aid* after IAA treatment and could confirm that the protein levels quickly dropped >85% by western blotting (aid*-9myc, Figure 2B) and up to becoming undetectable by fluorescence microscopy (aid*-GFP, Figure S2B).
Figure 2.
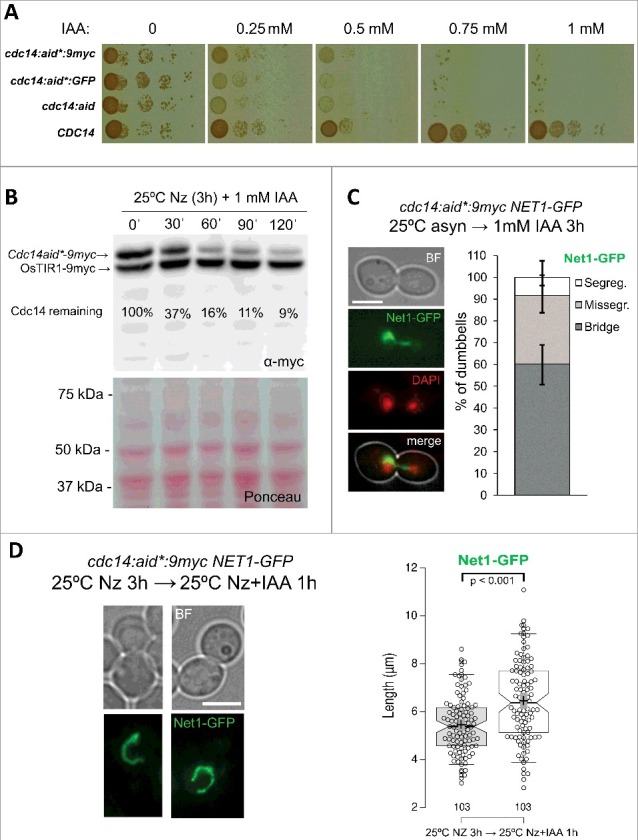
Auxin-driven depletion of Cdc14 at 25 ºC still resembles the terminal phenotype of cdc14-ts alleles at 37 ºC. (A) Strains FM931 (OsTIR1 NET1:GFP), FM1391 (OsTIR1 NET1:GFP cdc14:aid), FM1784 (OsTIR1 cdc14:aid*:GFP) and FM1785 (OsTIR1 NET1:GFP cdc14:aid*:9myc) were spotted on YPD plates containing increasing concentrations of the auxin IAA and incubated for 3 days at 25 ºC. (B) Strain FM1785 was first blocked 3h with Nz and then incubated with 1 mM IAA. At the indicated time points, a sample was taken and processed for western blotting against the myc epitope (upper picture depicts the actual western blotting and lower picture the Ponceau staining of the membrane). The amount of Cdc14 was normalized to OsTIR1 and the relative Cdc14 abundance compared to the time of IAA addition. (C) A log-phase culture of FM1785 was treated with 1 mM IAA for 3h at 25ºC. A representative picture of the major phenotype is shown on the right: a cell arrested in anaphase with DAPI-stained segregated nuclear masses and a Net1-GFP anaphase bridge. On the left, a quantification of the segregation pattern of Net1-GFP. (D) The same strain was blocked 3h with Nz before adding 1 mM IAA for an extra hour at 25 ºC. Representative cells before and after IAA addition are shown on the left. A box plot of the loop lengths is included on the right. Note how neither auxin addition nor Cdc14 depletion caused the condensation of the rDNA loop. White scale bars represent 5 μm. BF, bright field; Nz, nocodazole; IAA, 3-indolacetic acid.
Importantly, when we looked at the nuclear masses and the rDNA (Net1-GFP) upon Cdc14-aid*-9myc depletion (2-3 h after 1 mM IAA addition), we still found that the rDNA formed a bridge connecting the DAPI-stained daughter nuclei (Figure 2C). Likewise, the nucleolar marker Nop1-CFP was abnormally segregated when depleting Cdc14-aid*-GFP (Figure S2C). We confirmed that these segregation problems still happened even though the metaphase rDNA loop remained as such in the new Cdc14 depleting conditions (Figure 2D, S2D). In fact, the loop got slightly extended after the extra hour in Nz+IAA (Figure 2D, box-plot). Additionally, two important corollaries can be driven from this latter experiment. Firstly, auxin treatment, unlike HS, does not stress the cells to condense the rDNA loop (also confirmed in cells lacking either aid tagging or OsTIR1, data not shown). Secondly, Cdc14 depletion does not disarray the rDNA loop in Nz-blocked cells.
Altogether, we conclude in this chapter that Cdc14 depletion through the auxin-based degron resembles the rDNA segregation problems previously reported with cdc14-ts alleles.
Disruption of condensin by auxin-mediated degradation wholly disorganizes and spreads the rDNA structure
Probably the most important works where the rDNA has been used as a model for chromosome structure in yeast relate to condensin function [18,19]. Condensin is a multimeric complex thought to either staple distant regions within the same chromosome or topologically modify the chromosome to allow such stapling [7,8,43]. It is a highly conserved across all life kingdoms. In humans, there are two complexes and their disruption causes clearly visible condensation abnormalities during prophase and metaphase, as well as chromosome bridges during anaphase. In yeast, the most obvious problem of knocking down its only condensin complex is the presence of anaphase bridges [17]. The rDNA has been used to address if condensin disruption also caused signs of condensation problems [18,19]. The main conclusion from these works was that condensin was essential to establish and maintain the highly organized metaphase rDNA loop structure. Thus, when condensin function was abrogated by means of ts alleles for its subunits (e.g., smc2-8, ycs4-2, etc.), the loop got disorganized into the G1-like structure termed “puff”; i.e., the rDNA appears spread and punctuated by FISH.
We have now revisited condensin results on the rDNA, yet making use of aid-tagged alleles for condensin subunits (SMC2:aid* and YCS4:aid) and the rDNA marker Net1-GFP. As shown for Cdc14-aid, cells carrying either Smc2-aid* or Ycs4-aid were fully viable in the absence of auxins, yet become highly hypersensitive to 1 mM IAA, and this sensitivity also depended on OsTIR1 (Figure 3A). In integral wild type cells (i.e., non-treated for FISH analysis), there is barely a sign of a puff-like structure for Net1-GFP, even in G1 cells (Figure S1) [44,45]. This was also true for either condensin subunit tagged with aid, provided that auxin was absent. In addition, Nz brought about the Net1-coated rDNA loop in all strains (Figure 3B). Nevertheless, auxin caused a complete disruption of Net1-GFP signal in both SMC2:aid* and YCS4:aid strains when Nz was added together with the auxin to block the cells in metaphase (Figure 3B, C). In all cases, Net1-GFP appeared randomly dotted and, very often, a diffuse GFP pool was also discernible. Co-staining with DAPI showed that Net1-GFP could still partly localize within a region next to the main nuclear DNA signal, although it seems that Net1-GFP has also invaded the nuclear DNA (Figure 3C). In many ways, this clearly aberrant phenotype resembles the puff structure described years ago by FISH in ts alleles for condensin subunits [18,19]. Therefore, we again concluded that the classical role of condensin as a key organizer of the rDNA loop prevails.
Figure 3.
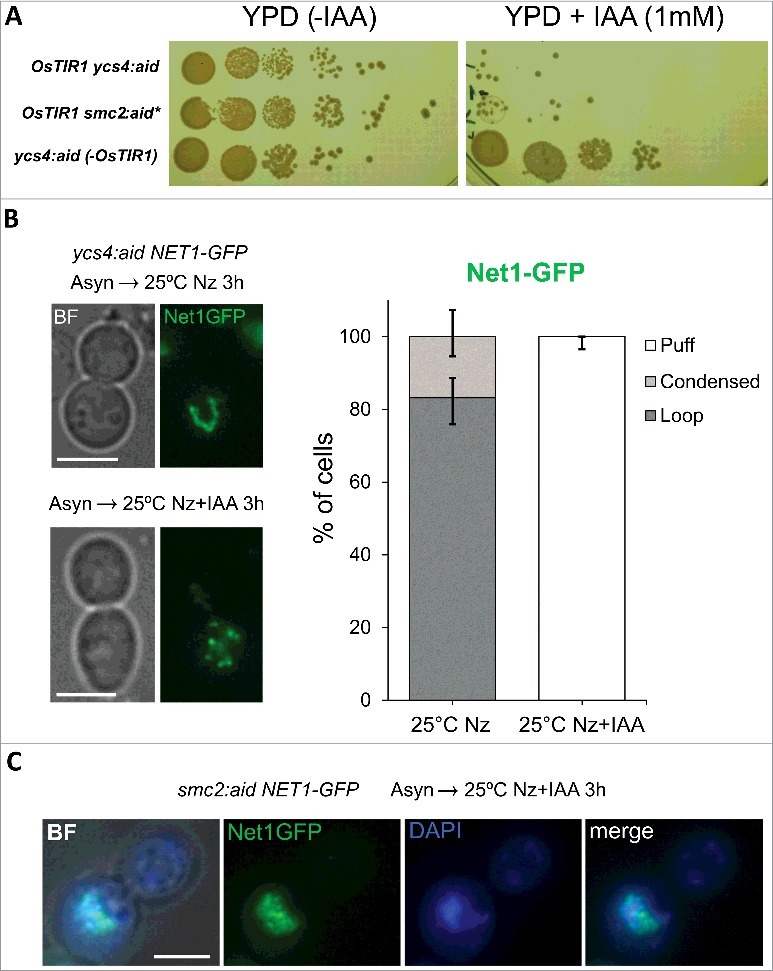
Condensin depletion disorganizes the rDNA loop at 25 ºC. (A) Strains FM1009 (OsTIR1 NET1:GFP ycs4:aid), FM2055 (NET1:GFP ycs4:aid) and FM2150 (OsTIR1 NET1:GFP smc2:aid*) were spotted on YPD plates with and without 1 mM IAA and incubated for 3 days at 25 ºC. (B) Strain FM1009 was blocked in metaphase (Nz x 3h) at 25 ºC with or without the simultaneous addition of 1 mM IAA. Samples were taken for each condition and immediately visualized under the microscope without further processing. (C) Strain FM2150 was treated like in B. Samples were frozen 24 h at -20 ºC before staining with DAPI and visualized under the microscope. White scale bars represent 5 μm. BF, bright field; Nz, nocodazole; IAA, 3-indolacetic acid.
Auxin-mediated degradation of Cdc15 also blocks telophase with a hypercondensed rDNA
Another important rDNA phenotype described previously with ts alleles was that the locus appears fully segregated and hypercondensed (line-like) in a cdc15-2 telophase block [9,21,22,44]. Cdc15 is a key kinase in the MEN-driven Cdc14 activation in late anaphase [35]. Thus, the terminal hypercondensed phenotype in cdc15-2 has been attributed to the action of Cdc14 during its first wave of activation by FEAR; i.e., Cdc14 allows condensin binding to the rDNA, which condenses the locus, resolves sister chromatid entanglement and allows segregation [17,21–23,27,46]. Again, it might happen that this hypercondensed state is actually a consequence of the temperature (37 °C) at which the cells are incubated to bring about the cdc15-2 block. Thus, we studied the terminal rDNA appearance in an auxin-driven cdc15-aid* telophase block at 25 °C. First, we checked that Cdc15-aid worked as expected from the essential functions attributed to this kinase; i.e., (i) cdc15:aid* strains grow normally without auxin and were highly hypersensitive to it (Figure S3A), (ii) incubation of the strain with the auxin led to a telophase block in the first cell cycle (Figure 4A, chart). After showing that both requirements were fulfilled, we checked the rDNA with Net1-GFP and found that it appears evenly segregated and condensed (Figure 4A, photos). We therefore concluded that Cdc14 does trigger an rDNA condensation in anaphase, ruling out any confounding effect due to the HS-mediated rDNA condensation just described above.
Figure 4.
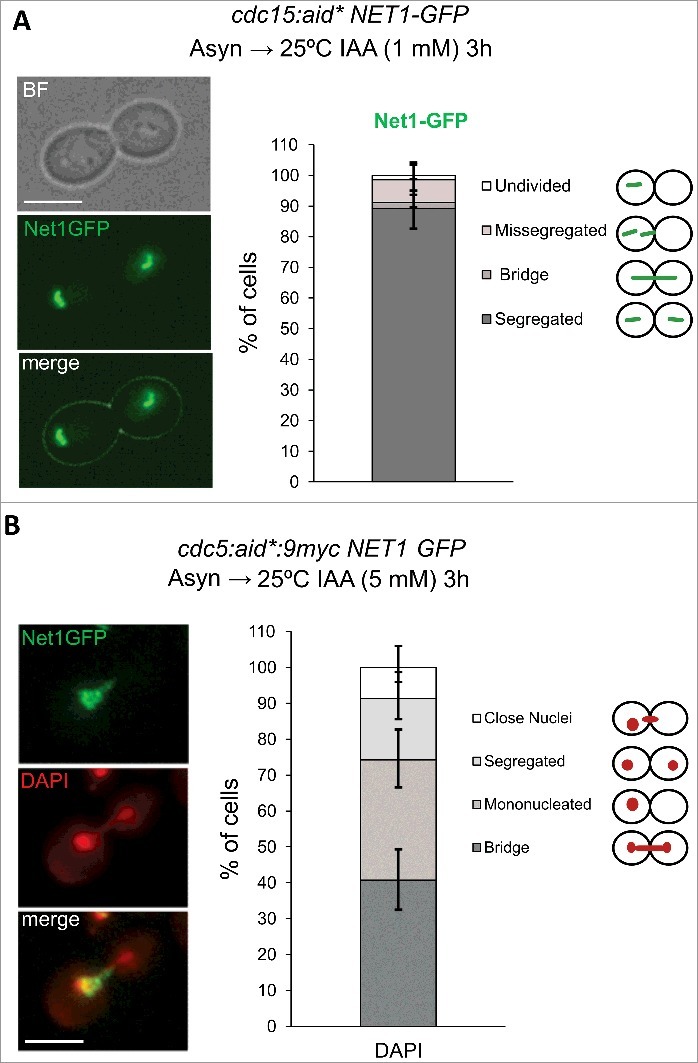
Cdc15 depletion at 25 ºC blocks cells with the rDNA hypercondensed and segregated, whereas Cdc5/Polo depletion at 25 ºC leads to an anaphase block with missegregated rDNA. (A) A log-phase 25 ºC culture of the strain FM2177 (OsTIR1 NET1:GFP cdc15:aid*:9myc) was treated with 1 mM IAA for 3h at 25ºC. A representative picture of the major phenotype is shown. A quantification chart of the Net1-GFP segregation pattern is included on the right. (B) A log-phase 25 ºC culture of the strain FM2833 (OsTIR1 NET1:GFP cdc5:aid*:9myc) was treated with 5 mM IAA for 3h at 25 ºC. A representative cell with the major phenotype is shown: Missegregated rDNA with a DAPI bridge. Whereas missegregated rDNA was seen in almost all cells, the nuclear morphology was more variable. A chart quantifying the different nuclear DAPI signals in dumbbell cells is included on the right. White scale bars represent 5 μm. BF, bright field; IAA, 3-indolacetic acid.
Auxin-mediated degradation of Cdc5 prevents segregation of the rDNA in anaphase
Another important factor that regulates Cdc14 throughout anaphase, both by FEAR and MEN, is the Polo kinase Cdc5 [32]. An anaphase block with an rDNA anaphase bridge is the terminal phenotype of cdc5-ts alleles [22]. In addition, cdc5-ts fails to hyperactivate condensin, which results in non-cannonical rDNA structures in anaphase; i.e., rDNA bunches into a cluster rather the classical line seen in cdc15-ts [47]. More recently, it has been shown that cdc5-ts also abolishes a protruding nucleolus in metaphase, which might also influence the rDNA loop shape and its later segregation [48]. We thus checked the main terminal cdc5-aid* phenotype at 25 °C in order to see if it also mimicked that of the cdc5-ts alleles (Figure 4B). We found that 1 mM of IAA caused severe but not complete growth impairments (Figure S3B). Increasing the IAA concentration up to 5 mM caused complete growth inhibition, but still some remaining Cdc5-aid*9myc could be detected by Western blotting (Figure S3C). Despite this caveats, 3h incubations with 5 mM IAA led to a complete cell cycle block reminiscent of that shown above for cdc14-aid; i.e., dumbbell morphology and Net1-GFP forming an anaphase bridge (Figure 4B). In most cells, the segregation of the main nuclear mass was also seriously compromised, as it has been reported for cdc5-ts alleles. We concluded here that auxin-driven Cdc5 depletion at 25°C leads to a terminal phenotype which is similar to cdc5-ts alleles at 37°C.
The mechanism of condensation of the rDNA loop by heat stress does not involve Cdc14
Having addressed the consequences of the HS-mediated rDNA condensation over our previous knowledge with ts alleles, we went back to the primary phenotype in order to shed more light on the mechanism.
At present, the best known mechanism involved in rDNA condensation is the one that takes place at anaphase onset [21–23,28]. The FEAR-driven Cdc14 release from the nucleolus transiently shuts down overall transcription which, in turn, allows condensin to fully interact with chromatin [26,27,46,49]. Condensin-rDNA overloading is particularly critical due to the special requirements of this locus. It is also likely that the role of Cdc14 on condensation goes beyond the control of transcription; Cdc14 also posttranslationally regulate condensin [21]. So far, Cdc14 has been shown to get activated exclusively by FEAR and, later, by MEN. No other mechanism out of anaphase is known to activate Cdc14. This, in principle, would discard Cdc14 as a player in the HS-mediated rDNA condensation in metaphase and, as noted above, there were no signs of Cdc14 release when Cdc14-GFP was used as the reporter for the HS-mediated condensation (Figure 1A). However, previous reports have suggested that Cdc14 might somehow get activated out of anaphase as a response, for example, to environmental conditions (i.e., DNA damage) [50]. Therefore, we decided that more investigation on the relationship between Cdc14 and the HS-mediated metaphase rDNA condensation was needed.
Firstly, we carried out HS experiments with a Cdc14-GFP strain that also bears the nucleolar marker Nop1-CFP and the SPB reported Spc42-RedStar. We further stained the nuclear DNA with DAPI. We reasoned that if Cdc14 gets released upon the HS in metaphase, though transiently, it will spread across the nucleus and further colocalize with the SPB. We never observed this happening, even in 15’ intervals after the temperature shift (data not shown). Indeed, we confirmed that upon the HS the Cdc14-GFP loop shifted to an arc next to the main DAPI signal (Figure 5A). Cdc14 was mostly embedded into the Nop1-CFP signal both before and after the HS, and never colocalized with the SPB.
Figure 5.
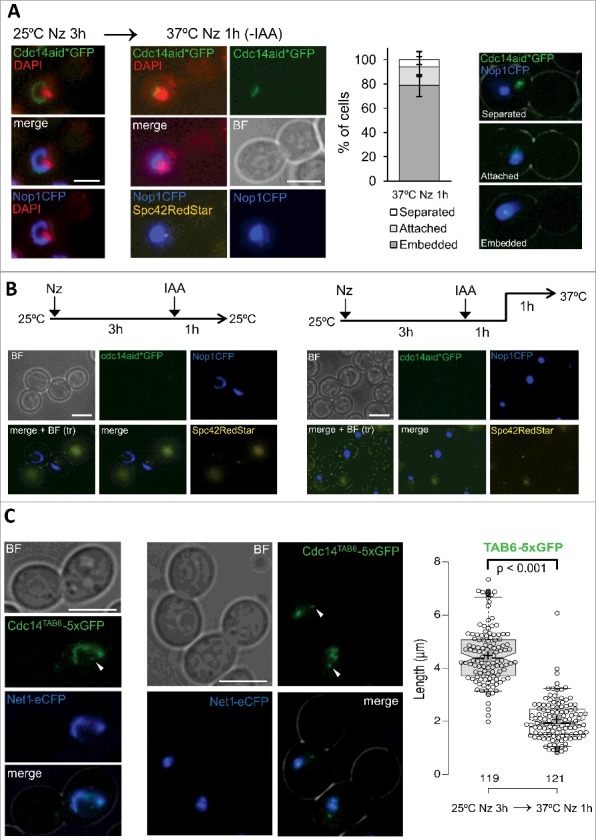
The condensation of the rDNA mediated by heat stress in metaphase is independent of Cdc14. (A) The strain FM2113 (OsTIR1 cdc14:aid*:GFP NOP1:CFP SPC42:RedStar) was first blocked with Nz for 3h at 25 ºC and then incubated for 1h at 37 ºC. At the indicated time points, a sample was taken and processed for DAPI staining. Representative cells are depicted for each incubation condition. A chart quantifying the relative co-localization of Cdc14-GFP and Nop1-CFP at the end of the experiment is also shown (with representative pictures on its right). “Separated”, both signals did not colocalized; “attached”, Cdc14 mostly colocalized at the edge of the Nop1 signal; “embedded”, Cdc14 was all within Nop1. (B) The same strain was blocked for 3h at 25 ºC with Nz, then incubated with 1 mM IAA for an extra hour and finally shifted to 37 ºC for another 1h incubation. Samples were collected just before and after the HS step. (C) The strain FM2333 (CDC14TAB6:5xGFP NET1:eCFP) was first Nz-blocked for 3h at 25 ºC and then incubated for 1h at 37 ºC. Representative cells before and after the HS are shown on the left. On the right, a box plot of the loop length before and after the HS. The white arrowheads point to the probable small pool of Cdc14TAB6 that binds to the Spindle Pole Bodies in a cell cycle independent manner (see text for more details). White scale bars represent 5 μm. BF, bright field; Nz, nocodazole; IAA, 3-indolacetic acid.
Secondly, we carried out a HS experiment in cells blocked in metaphase where we had previously knocked down Cdc14-aid*-GFP with IAA. We reasoned that rDNA condensation (in this case the nucleolar marker Nop1-CFP) would not take place upon HS if Cdc14 was needed. However, Nop1-CFP became an oval/round signal upon the HS treatment (Figure 5B). Importantly, this experiment also strongly argues against Cdc14 getting activated without being released from the nucleolus, a hypothesis we could not rule out with the first experiment and totally feasible taking into account that Cdc14 and its target (i.e., rDNA) already colocalize in metaphase.
Finally, we also used a CDC14 allele which is constitutively active, i.e., CDC14TAB6. Cdc14Tab6-GFP appears coating the rDNA in interphase, yet weaker than Cdc14-GFP, and can also be distinguished as a diffuse signal within the nucleus plus a distinctive focus, which likely corresponds to one SPB [24,51]. Our hypothesis was that, if Cdc14 was needed for the loop-to-condensed transition, the loop would not arise at all upon Nz treatment in this strain. Nevertheless, Cdc14Tab6-GFP was indeed seen as a loop in the Nz block, fully colocalizing with Net1-CFP, and HS made this loop to be remodelled into a condensed form (Figure 5C).
Putting all these data together, we conclude that the rDNA condensation observed after the HS is independent of Cdc14.
TORC1 inhibition condensates the rDNA loop
Given the Cdc14-independent mechanism for the HS-mediated rDNA condensation, we next wondered if this dramatic rDNA reshaping was exclusive of the HS treatment or other stresses could lead to the same outcome. HS has been shown to inhibit TORC1, the master complex that regulates cell growth and proliferation [52]. Thus, we tested if the TORC1 inhibitor rapamycin could lead to a similar phenotype. Notably, rapamycin has been shown to condensate the yeast interphase nucleolus [39,53–55]. Thus, we added rapamycin to the metaphase rDNA loop at 25 °C and also observed the rDNA condensation (Figure 6A, B). Measurement of the looped and condensed signals showed that the transition was equivalent to that of the HS. Similar results were obtained with other stresses known to inhibit TORC1 (Fig 6C) [56]. Thus, oxidative stress, calorie restriction, glucose depletion and nitrogen starvation led to rDNA condensation in metaphase-arrested cells. Nevertheless, this was not the case for another stress that shares with the HS its ability to misfold proteins but does not inhibit TORC1; i.e., treatment with the proline analogue Azetidine-2-carboxylic acid (AZC) [57,58].
Figure 6.
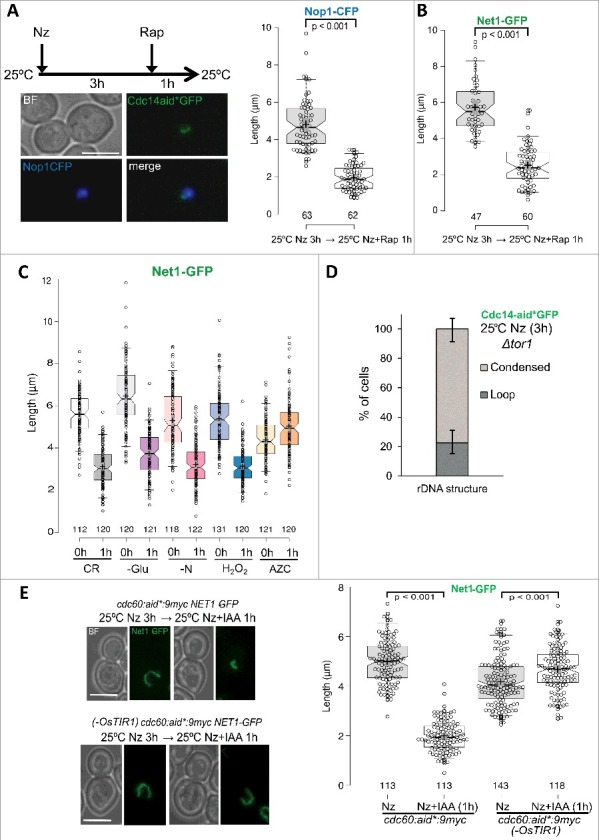
Condensation of the the rDNA loop depends on inhibition of TORC1. (A) The strain FM2113 (OsTIR1 cdc14:aid*:GFP NOP1:CFP SPC42:RedStar) was first metaphase-blocked with Nz for 3h at 25 ºC and then incubated for 1h with 200 nM rapamycin. A cell, representative of the condensed rDNA/nucleolus seen after rapamycin addition, is shown on the left. Box plots of loop lengths before and after rapamycin treatment is depicted on the right. (B) Box plots of loop lengths before and after rapamycin addition for the strain FM931 (OsTIR1 NET1:GFP) under the same experimental conditions. (C) Box plots of loop lengths before and after other stresses. The FM931 strain was blocked like in A and then shifted to different media while keeping the Nz: CR (YPD with just 0.005% glucose; i.e., calorie restriction); -Glu (YP without any cabon source); -N (Yeast Nitrogen Base media without amino acids and ammonium sulphate); H2O2 (YPD plus 1 mM H2O2); AZC (YPD plus 10 mM AZC). (D) A chart quantifying the presence of the rDNA loop after blocking the strain FM2179 (OsTIR1 cdc14:aid*-GFP Δtor1) with Nz for 3h at 25 ºC in YPD. Note how the rDNA loop is mostly absent in this strain. (E) Auxin-mediated depletion of TORC1 activator Cdc60 causes rDNA loop condensation at 25 ºC (see also Fig. S4C). Strains FM2292 (OsTIR1 cdc60:aid*9myc NET1:GFP) and FM2332 (cdc60:aid*9myc NET1:GFP without OsTIR1) were treated with Nz for 3h at 25 ºC and then incubated with 1 mM IAA for an extra hour at the same temperature. Representative cells before and after IAA addition on the left and box plots of the rDNA loop length on the right. White scale bar represents 5 μm. BF, bright field; Nz, nocodazole; Rap, rapamycin; IAA, 3-indolacetic acid; AZC, L-Azetidine-2-carboxylic acid.
Next, we chose a genetic approach for TORC1 inhibition. Firstly, we deleted TORC1-specific TOR1 gene and were able to preclude the formation of the rDNA loop after Nz treatment at 25 °C (Fig 6D, S4A). Secondly, we degraded through the auxin system one of the most important nutrient sensors for TORC1 activation, Cdc60 (Figure S4B, S4C) [59]. We did so once the rDNA loop had been mounted in Nz at 25°C, and this was enough to trigger loop condensation in the absence of any external stress (Figure 6E).
Altogether, we concluded that stresses that inhibit TORC1, including HS, elicit rDNA condensation in metaphase-blocked cells.
Discussion
In this report we described novel stressful conditions able to condense the yeast mitotic ribosomal DNA (rDNA). The most widely studied in this work relates to an acute heat stress (HS) that affect cells growing at 25 °C when they are shifted to 37 °C for just one hour. Such HS leads to an axial shortening of the rDNA until it goes from the structure known as “loop”, which is maximized in metaphase-blocked cells, to a line or oval shape we refer to as “condensed”. We have studied this HS-mediated rDNA condensation in vivo by means of proteins that specifically target the rDNA (Net1, Cdc14 and Nop1). While writing of this manuscript was in progress, another report described the same phenotype by DAPI staining of the rDNA loop in fixed cells treated for spreading the nuclear DNA onto two dimensions (i.e., a protocol for FISH but without all probe-related steps) [60]. Based on our and their data, we propose that the large rDNA loop visible after a protracted metaphase arrest by nocodazole (Nz) is axially shortened upon HS. We observed this kind of compaction in movies (Figure 1B), and they found that the rDNA could still be spread as a loop out of the main nuclear mass, yet having a much shorter length. In vivo, this shorter loop could be still visible in around 20% of cell, but in most cells it actually appeared as an amorphous oval shape or just a line. This probably reflects the three dimensional condition of our study whereby shorter loops could be less discernible. This is also the likely cause of why the loop was mostly absent in rapidly cycling cells (Fig S1). Interestingly, in other previous works in which the rDNA loop was assessed by FISH, authors reported that the loop was still the major category present in wild type cells shifted to 37 °C [9,17–19,23,48,61]. Unlike the recent report by Shen and Skibbens, there were no comparisons of the loop length between 23–25 °C and 34–37 °C. Thus, we assume that the rDNA metaphase loop conformation is not dismantled into something else but it is shortened until getting a condensed appearance in vivo.
Why and how does the loop become shorter after the HS? Several hypotheses and models are plausible (Figure 7A, B). It might happen that cells sense the HS and shut down transcription of rRNAs to concentrate available resources in dealing with this stress. Less transcription would allow overloading of condensin on the rDNA and condensation of the locus, as it has been shown in anaphase in a Cdc14-dependent manner [26,27,46,49]. We call this kind of model “active rDNA condensation” (Figure 7A). We here provide direct genetic evidence that the nutrient sensor TORC1 complex is at the core of the molecular mechanism behind this phenotype (Fig 6D, E). In addition, rapamycin and nutrient starvation, which are well-established triggers of TORC1 inhibition [56], condensed the rDNA loop as well (Figure 6A, B, C). In cycling cells, TORC1 inhibition has been shown to downregulate rRNA production and concomitantly recruit condensin and Sir2 to the rDNA in order to stabilize the repetitive locus [53–55,62]. An alternative mechanism for the HS-mediated condensation, the stress response due to protein misfolding at higher temperatures, is unlikely; a toxic proline analogue that causes this stress could not condense the loop at 25 °C (Fig 6C). Other previous findings also support this TORC1-dependent misfolding-independent mechanism for HS-mediated rDNA condensation. Thus, HS mostly triggers TORC1 inhibition, being the protein misfolding response secondary to this stress in yeast [63,64]. Therefore, we propose that solely TORC1 inhibition by HS is needed to exert the observed condensation. Remarkably, we also show here that this condensation is independent of Cdc14 (Fig 5, S2D). On support of this finding, a very recently published paper has shown that nutrient starvation and rapamycin do not cause Cdc14 release in metaphase [65].
Figure 7.
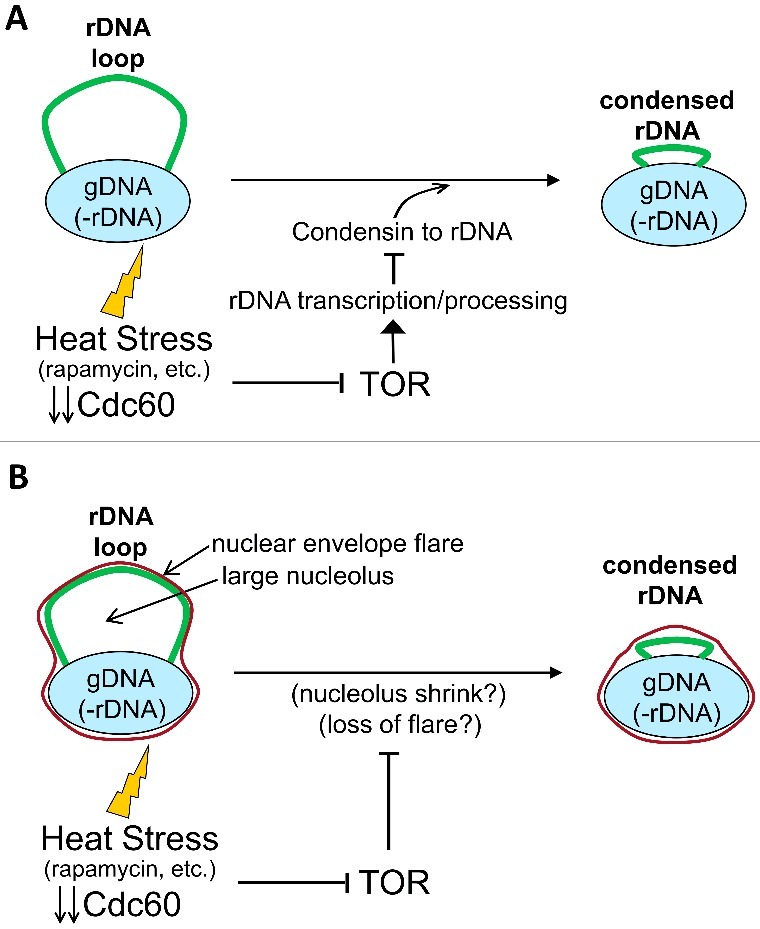
“Active” vs “passive” models of rDNA condensation by HS and other TORC1-dependent stresses. (A) An active model for rDNA condensation in metaphase upon stress. Heat stress, rapamycin, nutrient starvation, etc. would temporarily stop rRNA transcription through TORC1 inhibition. This, in turn, would recruit condensin to the locus to drive condensation of the loop. (B) Passive models for rDNA condensation in metaphase upon the same stresses. The rDNA loop in metaphase would be a result of the rDNA being accommodated to either nucleolus flattening or nuclear envelope flaring. Either of these protrusions would act as rDNA scaffolds and depend on an active TORC1. Environment stress would shrink these scaffolds (see text for more details) and the rDNA would accommodate to the reduced space. Note that both models are not necessarily mutually exclusive. Light blue oval, the bulk of nuclear DNA; Thick green line, the rDNA loop; Thin red line, the nuclear envelope.
Although the above-mentioned regulatory networks favour the “active rDNA condensation” model, we contemplate that other “passive rDNA condensation” models cannot be ruled out at present (Figure 7B). For example, the protracted metaphase block may cause an accumulation of an intermediate product of the rRNA/ribosome processing into the nucleolus, which would fatten as a result, pushing the rDNA away from the main nuclear mass to become the loop. This intermediate product would be a rate-limiting step at low temperatures but not at 37 °C, so the temperature shift would reverse the situation. In this scenario, HS and TORC1-inhibiting stresses would lead to the same phenotype but through different mechanisms: HS would speed up the rate-limiting step and TORC1 inhibition would let flux forward the intermediate because its substrate (i.e., transcription) has been removed. Another “passive model” is based on the recent finding that the nuclear envelope associated to the rDNA forms a flare during an extended metaphase [66]. This flare results from the accumulation of phospholipids, which are diverted towards the nuclear envelope that surrounds the nucleolus. Since the rDNA is tethered to the nuclear envelope [67], it might well happen that the rDNA loop is just a consequence of the adaptation of this locus to the physical space that occupies the flare. We thus speculate that the HS somehow changes this flare/rDNA relationship, either the flare changes with temperature (e.g., phospholipids are recycled from the nuclear envelope to other membranes upon HS) or the rDNA gets detached by the stress. Finally, other alternative explanations include either that TORC1 itself is a structural component of the loop, as it gets detached from rDNA promoter upon inhibition [68], or that the change in the way nascent rRNAs are processed after TORC1 inhibition modifies the rDNA morphology [69]. Current work in our lab has in mind these passive models of rDNA condensation.
A major orientation of the work undertaken in this report relates to revisiting previous results obtained with temperature-sensitive (ts) alleles for essential genes; more specifically, several genes in which a role was given for the rDNA structure or segregation. We chose five genes CDC14, CDC15, CDC5, SMC2 and YCS4 (the latter two encoding proteins which belong to the condensin complex), although there are many others that should be studied as well. The main reason for revisiting results obtained through ts alleles with new conditional alleles whose restrictive condition can be induced at 25 °C (auxin-driven aid alleles here) is that the HS-driven rDNA condensation in metaphase could have misled us about the real role of these genes. Thus, Cdc14 downregulates transcription and targets condensin to the rDNA in anaphase [26,27,49]. However, if HS did the same in metaphase this could mask the real contribution of Cdc14 at a later cell cycle stage: rDNA segregation and subsequent hypercompaction in anaphase (as previously addressed with cdc14-ts and cdc15-ts alleles) [9,21,22,44]. Remarkably, we still observed gross rDNA missegregation in cdc14-aid and rDNA hypercompaction in cdc15-aid (i.e., it blocks cells in telophase but allows Cdc14 transient activation to segregate the rDNA). Likewise, cdc5-aid still missegregates the rDNA, which also fails to get condensed in anaphase, in accordance to expectation for the Cdc5 role in Cdc14 and condensin activation as previously reported with ts alleles [22,32,47]. It is important to note that Cdc5 and Cdc15 are kinases, so conditional alleles sensitive to ATP analogues are available (i.e., cdc5-as1 and cdc15-as1). This has allowed to inhibit the enzymes at temperatures other than 37 °C, with results similar to what we find with aid alleles [33,70]. Condensin was a remarkable case to study since it has been shown to disorganize the rDNA structure into a randomly spread punctuated pattern by FISH, the so-called “puff”. Puffed rDNA has also been observed in G1 by FISH, but in live cells coated with Net1-GFP the rDNA appears as a condensed line/oval shape in cycling G1 cells (Figure S1) [44,45], rather similar to what we observed upon HS in metaphase. Strikingly, we observed puffed rDNA in virtually all Net1-GFP cells upon condensin depletion. Because of this great dependency on condensin for any coherent in vivo rDNA structure, regardless the cell cycle stage and degree of condensation, we believe condensin is always loaded onto the rDNA and we prefer the use of the above-mentioned term “overloading” when Cdc14 and TORC1 inhibition (and perhaps HS) target further condensin to the locus. In summary, the four players we have re-checked here by aid alleles mimicked previous findings with ts alleles, so the novel HS-mediated rDNA condensation have a minor effect on previous results obtained with these ts alleles. Nevertheless, it is premature to say that this is going to be the case for other players as well. Cohesin, Aurora kinase Ipl1, Top2 and essential factors implicated in rRNA maturation are among these other players which are worth revisiting in the future.
A final consideration we would like to highlight is the possible implications of the HS-mediated rDNA condensation in thermotherapy. Hyperthermia is a well known sensitizer in antitumor therapy, although its mechanism of action is poorly understood at present [71], It has been speculated that there is a link between hyperthermia and ribosome biogenesis in humans since HS inhibits mTOR, and the activity of this complex is needed for RNA pol I transcription of the rDNA [72,73]. Therefore, our new findings on the rDNA compaction upon heat stress could shed more light on why cancer cells are hypersensitive to hyperthermia and help to envision new therapeutical approaches against this devastating disease.
Material and methods
General experimental conditions
Yeast strains used in this study are listed on Table 1. Genetic backgrounds of these strains are W303 and YPH499 (congenic to S288C). Genetic engineering was carried out through standard PCR-based procedures [74]. We used two aid-based degron systems, the originally reported and an improved version formed by a chimera between a shorter aid (aid*) and epitopes such as 9xmyc and GFP [40,42].
Table 1.
Strains used in this work.
| Strain | Genotype | Origin |
|---|---|---|
| AS499 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 bar1-Δ | Strunnikov lab |
| FM225 | AS499 NET1:GFP(LEU2) | Aragon lab |
| FM230 | AS499 CDC14:GFP:KanMX | Aragon lab |
| YNK54 | Mata ura3-1::ADH1-OsTIR1:9Myc(URA3) ade2-1 his3-11,15 leu2-3,112 trp1-1 can1-100 | Kanemaki lab |
| FM931 | YNK54 NET1:GFP(LEU2) | This work |
| FM1009 | FM931 ycs4:aid:KanMX | This work |
| FM1391 | FM931 cdc14:aid:KanMX | This work |
| FM1784 | YNK54 cdc14:aid*-GFP:HphNT | This work |
| FM1785 | FM931 cdc14:aid*-9myc:HphNT | This work |
| FM1934 | FM1784 ura3-1 (-OsTIR1)1 | This work |
| FM2055 | FM1009 ura3-1 (-OsTIR1)1 | This work |
| FM2113 | FM1784 SPC42:RedStar:KanMX [NOP1-CFP(LEU2)] | This work |
| FM2150 | FM931 smc2:aid*:hphNT | This work |
| FM2177 | FM931 cdc15:aid*:hphNT | This work |
| FM2179 | FM1784 Δtor1::kanMX4 | This work |
| FM2283 | FM931 cdc5:aid*-9myc:HphNT | This work |
| FM2292 | FM931 cdc60:aid*-9myc:HphNT | This work |
| FM2332 | FM2292 ura3-1 (-OsTIR1)1 | This work |
| yED233 | Mata ura3-1::Hta2:CFP(URA3) ade2-1 his3-11,15 leu2-3,112 trp1-1 can1-100 | Pelet lab |
| BY23836 | MATα TAB6-5GFP ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | Toh-e lab2 |
| FM2333 | BY23836 NET1:eCFP:KanMX | This work |
1OsTIR1was removed by reverting the ura+ phenotype on 5-FOA plates.
2Obtained from the NBRP repository (http://yeast.lab.nig.ac.jp/yeast/)
For experiments, strains were routinely grown overnight in YPD broth (http://www.bd.com/ds/productCenter/242820.asp) at 25°C with moderate shaking (200 rpm). Synthetic complete media (SC) supplemented with the corresponding drop-outs was also used in specific experiments. This media was made from Yeast Nitrogen Base without Amino Acids (YNB). Two versions of YNB were used: with and without ammonium sulphate as the only nitrogen source. Both YNB media were also purchased from BD™. Chemical agents used in experiments were: Nocodazole, Nz (http://www.sigmaaldrich.com/catalog/product/sigma/m1404); 3-indolacetic acid, IAA (http://www.sigmaaldrich.com/catalog/product/sigma/i2886); rapamycin, Rap (http://www.sigmaaldrich.com/catalog/product/sigma/r8781); hydrogen peroxide, H2O2 (http://www.sigmaaldrich.com/catalog/product/sigma/h1009); and L-Azetidine-2-carboxylic acid, AZC (http://www.sigmaaldrich.com/catalog/product/sigma/a0760).
For HS treatment in Nz-blocked cells, cultures were first grown at 25°C until an OD660 of 0.4 was reached. Cells were then blocked in metaphase by adding Nz (15 μg/ml in 1% v/v DMSO, final concentration) and waiting for 3h at 25°C. Usually, Nz re-addition (50% of the working concentration) was employed after 2h of the first addition to avoid release from the arrest. Then, the temperature was set for 1h at 37°C to induce the HS. In the case of experiments with IAA, a stock of 200 mM in Ethanol 100% v/v was used for a final concentration of 1 mM or less, whereas a stock of 500 mM in DMSO was used for 5 mM final concentration. For the rapamycin experiment, a final concentration of 200 nM was used (from a 2.74 mM stock in DMSO). For the H2O2 and AZC experiments, final concentrations of 1 and 10 mM were used, respectively.
For spot dilution assays on plates, cultures were grown exponentially and adjusted to an OD660 of 0.4 and then serially diluted in 10-fold steps in distilled water. Volumes of 5 μl where spotted onto the corresponding plates and incubated at the indicated temperatures for 3–4 days before taking the photo.
Western blotting
For western blotting, 5 ml of the yeast liquid culture was used to extract total protein using the alkaline method [75]. During IAA treatment, cells were incubated with the appropriate concentration range and at the indicated time points. Briefly, cell pellets were re-suspended in 100 μl of distilled water and then 100 μl of 0.2 M of NaOH was added. After 5 min incubation at room temperature, the sample was pelleted again and re-suspended in 50 μl of PAGE sample buffer (http://www.bio-rad.com/es-es/sku/1610737-2x-laemmli-sample-buffer), then boiled for 3 min and pelleted again. Next, 6–10 μl per lane of the supernatant (containing proteins) was resolved on a 10% SDS-PAGE gel electrophoresis. After transferring the proteins to PVDF membranes, the myc epitope was detected with successive incubations with a primary monoclonal mouse anti-myc antibody (http://www.sigmaaldrich.com/catalog/product/sigma/m4439; 1:20,000) and a secondary rabbit anti-mouse conjugated to alkaline phosphatase (S3721; https://www.promega.com/; 1:60,000). Light emission was detected with CDP-Star detection reagent (RPN3682; http://www.gelifesciences.com) on a Vilber-Lourmat Fusion Solo S chemiluminescence detector. Finally, a staining of the membrane with Ponceau S-solution (https://www.applichem.com/en/shop/product-detail/as/ponceau-s-loesung/) was also carried out for a loading reference. The myc specific signal was quantified in non-saturated conditions with the Bio1D software.
The relative amount of aid*-9myc tagged proteins after IAA addition was estimated using OsTIR1-9myc as an internal housekeeping control. We validated that OsTIR1-9myc amounts did not change over the course of an IAA experiment. For that purpose, we compared OsTIR1-9myc signal with overall lane loading as reported by the Ponceau staining.
Fluorescence microscopy
A Leica DMI6000B epifluorescence microscope with an ultrasensitive DFC350 digital camera was employed for single cell visualization as we have reported before [76,77]. Briefly, 250 μl of cell culture was collected at each time point, centrifuged at 3,000 r.p.m. for 2 min at room temperature, the supernatant carefully retired, and ∼1 μl of the pellet was added to ∼1 μl of water on the microscope slide. Samples were visualized directly using a 63X/1.30 immersion objective, immersion oil with a refractive index of 1.515-1.517, and the appropriate filter cube for each tag/stain. In the case of DAPI staining, the cell pellet was frozen 24h at −20°C and then ∼1 μl of the pellet was added to ∼1 μl of 4 μg/ml of DAPI (http://www.sigmaaldrich.com/catalog/product/sigma/32670) on the microscope slide. For each field, we first captured a series of 20 z-focal plane images (0.3 μm depth between each consecutive image), and then we processed images with the Leica AF6000 software and ImageJ (https://imagej.nih.gov/ij/). For time-lapse movies of living cells, a small number of Nz-blocked cells were spread onto small patches of agarose medium as we have reported before [76]. The patches had 15 μg/ml Nz as well.
Loop length quantification was performed on a z-stack image with the AF6000 software.
Statistics
Nz-blocked cells were dichotomically categorized when possible (e.g., rDNA loop vs condensed shape, loop/condensed vs puff, etc.), and the corresponding proportions calculated and represented in bar charts. Error bars in these charts depict 95% confidence intervals under the assumption of a binomial distribution.
Continuous data (i.e., loop length in μm) were represented in box-plots (http://shiny.chemgrid.org/boxplotr/). In these plots, centre lines depict the medians; box limits indicate the 25th and 75th percentile and notches roughly give the 95% confidence intervals for each median. Sample means are represented by a cross and confidence interval of means is represented as grey boxes inside. Altman whiskers were depicted, which extend to 5th and 95th percentiles (outliers are represented by dots outside whiskers). Numbers at the bottom of the box plots indicate the rDNA entities measured in the analysis. Statistical significance of the difference between medians was calculated applying a Mann-Whitney U test.
R software (https://www.r-project.org/) was used for calculation of confidence intervals and statistical tests.
Supplementary Material
Funding Statement
This work was supported by the Instituto de Salud Carlos III under Grant PI12/00280 and Spanish Ministry of Economy and Competitiveness (MINECO) under Grant BFU2015-63902-R. Both grants were co-financed with the European Commission's ERDF structural funds.
Disclosure of Potential Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
We would like to thank Damien D'Amours for fruitful ideas on how to develop this story when we first presented the compaction of the rDNA upon heat stress in the EMBO Workshop on SMC proteins (Vienna, May 2015), as well as David Pincus for later insights on the putative role of TORC1 in this phenomenon. We also thank Jessel Ayra-Plasencia for technical help.
References
- [1].Edgar RS, Lielausis I. Temperature-sensitive mutants of bacteriophage T4D: Their isolation and genetic characterization. Genetics. 1964;49:649–662. PMID:14156925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Basilico C. Temperature-sensitive mutations in animal cells. Adv Cancer Res. 1977;24:223–266. doi: 10.1016/S0065-230X(08)61016-7. PMID:322459 [DOI] [PubMed] [Google Scholar]
- [3].Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93:1662–1670. PMID:5337848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hartwell LH. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J Bacteriol. 1973;115:966–974. PMID:4580573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hartwell LH, Culotti J, Pringle JR, et al. . Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. PMID:4587263 [DOI] [PubMed] [Google Scholar]
- [6].Warner JR, Udem SA. Temperature sensitive mutations affecting ribosome synthesis in Saccharomyces cerevisiae. J Mol Biol. 1972;65:243–257. doi: 10.1016/0022-2836(72)90280-X. PMID:4557193 [DOI] [PubMed] [Google Scholar]
- [7].Kschonsak M, Haering CH. Shaping mitotic chromosomes: From classical concepts to molecular mechanisms. Bioessays. 2015;37:755–766. doi: 10.1002/bies.201500020. PMID:25988527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hirano T. Condensin-Based Chromosome Organization from Bacteria to Vertebrates. Cell. 2016;164:847–857. doi: 10.1016/j.cell.2016.01.033. PMID:26919425 [DOI] [PubMed] [Google Scholar]
- [9].Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. PMID:8175878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Torres-Rosell J, Machín F, Jarmuz A, et al. . Nucleolar segregation lags behind the rest of the genome and requires Cdc14p activation by the FEAR network. Cell Cycle. 2004;3:496–502. doi: 10.4161/cc.3.4.802. PMID:15004526 [DOI] [PubMed] [Google Scholar]
- [11].Renshaw MJ, Ward JJ, Kanemaki M, et al. . Condensins promote chromosome recoiling during early anaphase to complete sister chromatid separation. Dev Cell. 2010;19:232–244. doi: 10.1016/j.devcel.2010.07.013. PMID:20708586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Strunnikov AV, Larionov VL, Koshland D. SMC1: An essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. PMID:8276886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. PMID:7698648 [DOI] [PubMed] [Google Scholar]
- [14].Holm C, Goto T, Wang JC, et al. . DNA topoisomerase II is required at the time of mitosis in yeast. Cell 1985;41:553–563. doi: 10.1016/S0092-8674(85)80028-3. PMID:2985283 [DOI] [PubMed] [Google Scholar]
- [15].Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/S0092-8674(01)80007-6. PMID:9335333 [DOI] [PubMed] [Google Scholar]
- [16].Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. PMID:10403247 [DOI] [PubMed] [Google Scholar]
- [17].Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. PMID:10811823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lavoie BD, Hogan E, Koshland D. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J Cell Biol. 2002;156:805–815. doi: 10.1083/jcb.200109056. PMID:11864994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lavoie BD, Hogan E, Koshland D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 2004;18:76–87. doi: 10.1101/gad.1150404. PMID:14701879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Alexandru G, Uhlmann F, Mechtler K, et al. . Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/S0092-8674(01)00362-2. PMID:11371343 [DOI] [PubMed] [Google Scholar]
- [21].D'Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/S0092-8674(04)00413-1. PMID:15137939 [DOI] [PubMed] [Google Scholar]
- [22].Machín F, Torres-Rosell J, Jarmuz A, et al. . Spindle-independent condensation-mediated segregation of yeast ribosomal DNA in late anaphase. J Cell Biol. 2005;168:209–219. doi: 10.1083/jcb.200408087. PMID:15657393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sullivan M, Higuchi T, Katis VL, et al. . Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 2004;117:471–482. doi: 10.1016/S0092-8674(04)00415-5. PMID:15137940 [DOI] [PubMed] [Google Scholar]
- [24].Yoshida S, Toh-e A. Budding yeast Cdc5 phosphorylates Net1 and assists Cdc14 release from the nucleolus. Biochem Biophys Res Commun. 2002;294:687–691. doi: 10.1016/S0006-291X(02)00544-2. PMID:12056824 [DOI] [PubMed] [Google Scholar]
- [25].Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. PMID:15568976 [DOI] [PubMed] [Google Scholar]
- [26].Machín F, Torres-Rosell J, De Piccoli G, et al. . Transcription of ribosomal genes can cause nondisjunction. J Cell Biol. 2006;173:893–903. doi: 10.1083/jcb.200511129. PMID:16769819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Clemente-Blanco A, Mayán-Santos M, Schneider DA, et al. . Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature. 2009;458:219–222. doi: 10.1038/nature07652. PMID:19158678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Machín F, Quevedo O, Ramos-Pérez C, et al. . Cdc14 phosphatase: warning, no delay allowed for chromosome segregation! Curr Genet. 2016;62:7–13. doi: 10.1007/s00294-015-0502-1. PMID:26116076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Straight AF, Shou W, Dowd GJ, et al. . Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/S0092-8674(00)80734-5. PMID:10219245 [DOI] [PubMed] [Google Scholar]
- [30].Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. PMID:10235265 [DOI] [PubMed] [Google Scholar]
- [31].Yoshida S, Asakawa K, Toh-e A. Mitotic exit network controls the localization of Cdc14 to the spindle pole body in Saccharomyces cerevisiae. Curr Biol. 2002;12:944–950. doi: 10.1016/S0960-9822(02)00870-9. PMID:12062061 [DOI] [PubMed] [Google Scholar]
- [32].Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/S0092-8674(02)00618-9. PMID:11832211 [DOI] [PubMed] [Google Scholar]
- [33].Rahal R, Amon A. The Polo-like kinase Cdc5 interacts with FEAR network components and Cdc14. Cell Cycle. 2008;7:3262–3272. doi: 10.4161/cc.7.20.6852. PMID:18927509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liang F, Jin F, Liu H, et al. . The molecular function of the yeast polo-like kinase Cdc5 in Cdc14 release during early anaphase. Mol Biol Cell. 2009;20:3671–3679. doi: 10.1091/mbc.E08-10-1049. PMID:19570916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shou W, Seol JH, Shevchenko A, et al. . Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/S0092-8674(00)80733-3. PMID:10219244 [DOI] [PubMed] [Google Scholar]
- [36].Pereira G, Manson C, Grindlay J, et al. . Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J Cell Biol. 2002;157:367–379. doi: 10.1083/jcb.200112085. PMID:11970961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Weiss EL. Mitotic exit and separation of mother and daughter cells. Genetics. 2012;192:1165–1202. doi: 10.1534/genetics.112.145516. PMID:23212898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Morano Ka, Grant CM, Moye-Rowley WS. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190:1157–1195. doi: 10.1534/genetics.111.128033. PMID:22209905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ha CW, Huh WK. Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2011;39:1336–1350. doi: 10.1093/nar/gkq895. PMID:20947565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nishimura K, Fukagawa T, Takisawa H, et al. . An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. PMID:19915560 [DOI] [PubMed] [Google Scholar]
- [41].Granot D, Snyder M. Segregation of the nucleolus during mitosis in budding and fission yeast. Cell Motil Cytoskeleton. 1991;20:47–54. doi: 10.1002/cm.970200106. PMID:1661641 [DOI] [PubMed] [Google Scholar]
- [42].Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 2013;30:341–351. doi: 10.1002/yea.2967. PMID:23836714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kalitsis P, Zhang T, Marshall KM, et al. . Condensin, master organizer of the genome. Chromosom Res. 2017;25:61–76. doi: 10.1007/s10577-017-9553-0. [DOI] [PubMed] [Google Scholar]
- [44].Varela E, Shimada K, Laroche T, et al. . Lte1, Cdc14 and MEN-controlled Cdk inactivation in yeast coordinate rDNA decompaction with late telophase progression. EMBO J. 2009;28:1562–1575. doi: 10.1038/emboj.2009.111. PMID:19387493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tong K, Skibbens RV. Pds5 regulators segregate cohesion and condensation pathways in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2015;112:7021–7026. doi: 10.1073/pnas.1501369112. PMID:25986377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang B-D, Yong-Gonzalez V, Strunnikov AV. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 2004;3:960–967. doi: 10.4161/cc.3.7.1003. PMID:15190202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].St-Pierre J, Douziech M, Bazile F, et al. . Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol Cell. 2009;34:416–426. doi: 10.1016/j.molcel.2009.04.013. PMID:19481522 [DOI] [PubMed] [Google Scholar]
- [48].Walters AD, May CK, Dauster ES, et al. . The yeast polo kinase Cdc5 regulates the shape of the mitotic nucleus. Curr Biol. 2014;24:2861–2867. doi: 10.1016/j.cub.2014.10.029. PMID:25454593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tomson BN, D'Amours D, Adamson BS, et al. . Ribosomal DNA transcription-dependent processes interfere with chromosome segregation. Mol Cell Biol. 2006;26:6239–6247. doi: 10.1128/MCB.00693-06. PMID:16880532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Villoria MT, Ramos F, Dueñas E, et al. . Stabilization of the metaphase spindle by Cdc14 is required for recombinational DNA repair. EMBO J. 2017;36:79–101. doi: 10.15252/embj.201593540. PMID:27852625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Shou W, Sakamoto KM, Keener J, et al. . Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol Cell. 2001;8:45–55. doi: 10.1016/S1097-2765(01)00291-X. PMID:11511359 [DOI] [PubMed] [Google Scholar]
- [52].Takahara T, Maeda T. Transient Sequestration of TORC1 into Stress Granules during Heat Stress. Mol Cell. 2012;47:242–252. doi: 10.1016/j.molcel.2012.05.019. PMID:22727621 [DOI] [PubMed] [Google Scholar]
- [53].Tsang CK, Bertram PG, Ai W, et al. . Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. PMID:14609951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tsang CK, Li H, Zheng XS. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J. 2007;26:448–458. doi: 10.1038/sj.emboj.7601488. PMID:17203076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tsang CK, Zheng XFS. Opposing role of condensin and radiation-sensitive gene RAD52 in ribosomal DNA stability regulation. J Biol Chem. 2009;284:21908–21919. doi: 10.1074/jbc.M109.031302. PMID:19520859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. PMID:22174183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Trotter EW, Berenfeld L, Krause SA, et al. . Protein misfolding and temperature up-shift cause G1 arrest via a common mechanism dependent on heat shock factor in Saccharomycescerevisiae. Proc Natl Acad Sci U S A. 2001;98:7313–7318. doi: 10.1073/pnas.121172998. PMID:11416208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Trotter EW, Kao CM-F, Berenfeld L, et al. . Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J Biol Chem. 2002;277:44817–44825. doi: 10.1074/jbc.M204686200. PMID:12239211 [DOI] [PubMed] [Google Scholar]
- [59].Bonfils G, Jaquenoud M, Bontron S, et al. . Leucyl-tRNA Synthetase Controls TORC1 via the EGO Complex. Mol Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. PMID:22424774 [DOI] [PubMed] [Google Scholar]
- [60].Shen D, Skibbens RV. Temperature-dependent regulation of rDNA condensation in Saccharomyces cerevisiae. Cell Cycle. 2017;4101:00–00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Robellet X, Thattikota Y, Wang F, et al. . A high-sensitivity phospho-switch triggered by Cdk1 governs chromosome morphogenesis during cell division. Genes Dev. 2015;29:426–439. doi: 10.1101/gad.253294.114. PMID:25691469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Medvedik O, Lamming DW, Kim KD, et al. . MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. PMID:17914901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Solís EJ, Pandey JP, Zheng X, et al. . Defining the Essential Function of Yeast Hsf1 Reveals a Compact Transcriptional Program for Maintaining Eukaryotic Proteostasis. Mol Cell. 2016;63:60–71. doi: 10.1016/j.molcel.2016.05.014. PMID:27320198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Urban J, Soulard A, Huber A, et al. . Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. PMID:17560372 [DOI] [PubMed] [Google Scholar]
- [65].de los Santos-Velázquez AI, de Oya IG, Manzano-López J, et al. . Late rDNA Condensation Ensures Timely Cdc14 Release and Coordination of Mitotic Exit Signaling with Nucleolar Segregation. Curr Biol. 2017;:1–16. PMID:27916526 [DOI] [PubMed] [Google Scholar]
- [66].Witkin KL, Chong Y, Shao S, et al. . The budding yeast nuclear envelope adjacent to the nucleolus serves as a membrane sink during mitotic delay. Curr Biol. 2012;22:1128–1133. doi: 10.1016/j.cub.2012.04.022. PMID:22658600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Grunstein M, Gasser SM. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb Perspect Biol. 2013;5. doi: 10.1101/cshperspect.a017491. PMID:23818500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Li H, Tsang CK, Watkins M, et al. . Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. PMID:16900101 [DOI] [PubMed] [Google Scholar]
- [69].Kos-Braun IC, Jung I, Koš M. Tor1 and CK2 kinases control a switch between alternative ribosome biogenesis pathways in a growth-dependent manner. PLOS Biol. 2017;15:e2000245. doi: 10.1371/journal.pbio.2000245. PMID:28282370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Rodriguez-Rodriguez JA, Moyano Y, Játiva S, et al. . Mitotic Exit Function of Polo-like Kinase Cdc5 Is Dependent on Sequential Activation by Cdk1. Cell Rep. 2016;15:2050–2062. doi: 10.1016/j.celrep.2016.04.079. PMID:27210759 [DOI] [PubMed] [Google Scholar]
- [71].Mallory M, Gogineni E, Jones GC, et al. . Therapeutic hyperthermia: The old, the new, and the upcoming. Crit Rev Oncol Hematol. 2016;97:56–64. doi: 10.1016/j.critrevonc.2015.08.003. PMID:26315383 [DOI] [PubMed] [Google Scholar]
- [72].Hein N, Hannan KM, George AJ, et al. . The nucleolus: An emerging target for cancer therapy. Trends Mol Med. 2013;19:643–654. doi: 10.1016/j.molmed.2013.07.005. PMID:23953479 [DOI] [PubMed] [Google Scholar]
- [73].Dokladny K, Myers OB, Moseley PL. Heat shock response and autophagy—Cooperation and control. Autophagy. 2015;11:200–213. doi: 10.1080/15548627.2015.1009776. PMID:25714619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Smith JS, Burke DJ. Yeast Genetics: Methods and Protocols. New York, NY: Springer New York; 2014. [Google Scholar]
- [75].Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9%3c857::AID-YEA561%3e3.0.CO;2-B. PMID:10861908 [DOI] [PubMed] [Google Scholar]
- [76].Quevedo O, García-Luis J, Matos-Perdomo E, et al. . Nondisjunction of a single chromosome leads to breakage and activation of DNA damage checkpoint in g2. PLoS Genet. 2012;8:e1002509. doi: 10.1371/journal.pgen.1002509. PMID:22363215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].García-Luis J, Machín F. Mus81-Mms4 and Yen1 resolve a novel anaphase bridge formed by noncanonical Holliday junctions. Nat Commun. 2014;5:5652. doi: 10.1038/ncomms6652. PMID:25466415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


