ABSTRACT
Histone ubiquitination regulates sperm formation and is important for nucleosome removal during spermatogenesis. RNF8 is an E3 ubiquitin ligase, and RAD6B is an E2 ubiquitin-conjugating enzyme. Both proteins participate in DNA damage repair processes via histone ubiquitination. Loss of RNF8 or RAD6B can lead to sterility in male mice. However, the specific mechanisms regulating these ubiquitin-mediated processes are unclear. In this study, we found that RNF8 knockout mice were either subfertile or sterile based on the numbers of offspring they produced. We explored the mechanism by which RAD6B and RNF8 knockouts cause infertility in male mice and compared the effects of their loss on spermatogenesis. Our results demonstrate that RAD6B can polyubiquitinate histones H2 A and H2B. In addition, RNF8 was shown to monoubiquitinate histones H2 A and H2B. Furthermore, we observed that absence of histone ubiquitination was not the only reason for infertility. Senescence played a role in intensifying male sterility by affecting the number of germ cells during spermatogenesis. In summary, both histone ubiquitination and senescence play important roles in spermatogenesis.
KEYWORDS: RAD6B, RNF8, ubiquitin, histone, senescence, spermatogenesis
Introduction
Ubiquitin (ub) is a small protein containing only 76 amino acids and is present in all cells. The ubiquitin system plays key roles in regulating protein activity and in several essential cellular processes, such as cell cycle regulation, DNA repair, apoptosis and protein degradation in eukaryotic cells.1-6 Three types of enzymes play essential roles in the ubiquitination process: the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and the ubiquitin ligase enzyme (E3).7,8 Substrate ubiquitination can occur by the addition of a single ubiquitin molecule (monoubiquitination) or by the addition of different types of ubiquitin chains (polyubiquitination).9 Polyubiquitination often results in substrate targeting for proteasomal degradation.10 In contrast, monoubiquitination is involved in processes such as DNA repair and regulation of gene expression.11
Histones are highly basic proteins that bind tightly to acidic DNA to form chromatin. Nucleosomes are the fundamental packing unit of chromatin and are formed by 2 molcules each of H2 A, H2B, H3, and H4, as well as one linker H1 that associates with 147 bp of DNA.12,13 Histone modifications are linked to various developmental defects and human diseases.14,15 Histone ubiquitination is unique among histone modifications because of the relatively bulky size of ubiquitin, and histone ubiquitination has previously been shown to stimulate or repress various cellular processes.16-24
In addition to playing important roles in DNA damage, histone ubiquitination is important for spermatogenesis.25 Spermatogenesis is a continuous and precisely controlled biologic process that transforms diploid spermatogonial stem cells into haploid spermatozoa in the seminiferous tubules of testes. Spermatogenesis consists of mitosis, meiosis and spermatogenesis processes.26 Spermatogenesis is divided into 12 stages (stages I–XII) in mouse, and spermiogenesis is further divided into 16 steps (steps 1–16) based on changes in acrosome structure and the nuclear morphology of the maturing spermatids27-29 During spermatogenesis, most nucleosomal histones are replaced by transition proteins and subsequently by protamines.30,31 The biologic functions of chromatin remodeling events remain unclear, but it is hypothesized that protamines facilitate the packaging of DNA into sperm heads by promoting DNA condensation.31 Failure to accomplish this global chromatin restructuring causes male sterility.32-34
RAD6B functions as an E2 ubiquitin-conjugating enzyme that plays a role in DNA damage and in protein degradation via the N-end rule pathway, and along with UBR2, it promotes the ubiquitination of H2 A and H2B but not H3 and H4.35-41 RNF8 encodes an E3 ubiquitin-ligase enzyme, and either RAD6B deficiency or RNF8 inactivation can cause male infertility.42-44 Although the molecular mechanism remains elusive, it has been shown that RAD6B promotes H2 A and H2B ubiquitination and that RNF8 regulates both H2 A and H2B ubiquitination at DNA damage sites.19,20,24,45-48 These previous findings indicate that RAD6B and RNF8 may play important roles in chromatin remodeling.
In this study, we demonstrate that RNF8 knockout results in subfertility or sterility in mice. Additionally, RAD6B knockout mice were shown to be sterile. H2 A and H2B in testes are monoubiquitinated by RNF8 and polyubiquitinated by RAD6B. Interestingly, we found that senescence may play an important role during spermatogenesis.
Results
Infertility of RAD6B-deficient males and reduced fertility of RNF8-deficient males
RNF8−/− and RAD6B−/− males appeared abnormal during reproduction (Table 1). In mating tests, each male was paired with 2 females for all genotypes. Matings between 3-month-old WT male and WT female mice produced an average of 18 pups per cage (Fig. 1), and matings between 6-month-old WT male and WT female mice produced an average of 21 pups per cage (Fig. 1). In contrast, 3-month-old RNF8 homozygous knockout males mated with WT female mice produced an average of 6 pups per cage, and 6-month-old RNF8 homozygous knockout males mated with WT female mice produced only 2 pups per cage. As we expected, matings between WT female mice and either 3-month-old or 6-month-old RAD6B homozygous knockout males did not produce pups after the mating test. Interestingly, WT female mice when mated with either RNF8+/− or RAD6B+/− male mice had normal litter sizes. Hence, heterozygous knockout of RNF8 and RAD6B expression permitted normal spermatogenesis to occur. Furthermore, loss of RNF8 or RAD6B in females did not affect their fertility, as evidenced by the observation of normal litter sizes when RNF8 KO and RAD6B KO females were mated with WT male mice.
Table 1.
The number of pups per litter with genotypes for various paternal combinations.
| Genotype (n = 6) | NO. of pups (3 months) | NO. of pups (6 months) |
|---|---|---|
| WT(♂) × WT(♀) | 12,13,18,21,21,24 | 15,17,18,30,23,25 |
| RNF8−/−(♂) × WT(♀) | 2,3,8,10,7,6 | 0,1,3,4,2,2, |
| RAD6B−/−(♂) × WT(♀) | 0,0,0,0,0,0 | 0,0,0,0,0,0, |
Figure 1.
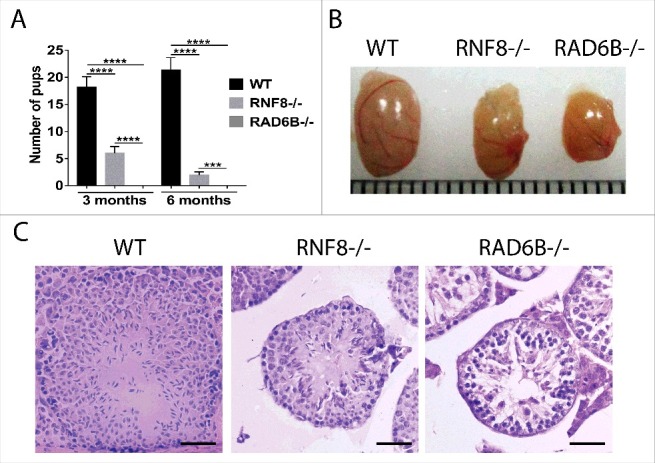
Testicular phenotypes of WT, RNF8−/− and RAD6B−/− mice. (A) Three-month-old (3 m) and 6-month-old (6 m) WT, RNF8−/− and RAD6B−/− males were mated with WT females of the same age. The numbers of pups are shown. All values are presented as the mean ± SEM (WT n = 6; RNF8−/− n = 6; RAD6B−/− n = 6). Student's t-test; ***P < 0.001, ****P < 0.0001. (B) Pictures of normal testes from 6-month-old wild-type (WT) male mice and of testes from 6-month-old RNF8 knockout (KO) male mice and 6-month-old RAD6B knockout male mice are shown. (C) Images of H&E-stained sections of seminiferous tubules from 6-month-old WT, RNF8−/− and RAD6B−/− mice. H&E staining of testis sections from RAD6B−/− males showed a dramatic reduction in germ cells compared with RNF8−/− males. Bar, 20 μm.
Testicular phenotypes of RAD6B- and RNF8-deficient mice
To investigate potential spermatogenesis defects, we analyzed the testes of RAD6B−/− and RNF8−/− mice. Testes from 6-month-old RAD6B−/− mice and RNF8−/− mice were smaller than testes from WT mice (Fig. 1B). H&E staining showed clear differences among testicular sections from WT, RAD6B−/− and RNF8−/− mice. The number of post-meiotic cells and the presence of vacuoles decreased dramatically in RAD6B−/− and RNF8−/− mice, with the largest decrease observed in RAD6B−/− testes (Fig. 1 C). Furthermore, seminiferous tubule cross sections were smaller in RAD6B−/− mice and RNF8−/− mice than in WT mice.
Spermiogenesis is defective in RAD6B- and RNF8-deficient mice
Significant morphological differences were observed among DAPI-stained testicular sections from WT, RAD6B−/− and RNF8−/− males. The knockout mice had testicular degeneration within the seminiferous tubules, and RAD6B−/− tubules were significantly devoid of mature sperm compared with WT and RNF8−/− tubules, suggesting that RAD6B mice were hypocellular for germ cells (Fig. 2 A). We divided spermatogenic stages into 4 groups, I–IV, V–VIII, IX–X, and XI–XII,49 based on the presence and arrangement of specific cell types. Compared with WT mice, fewer spermatids were present in the seminiferous tubules of RAD6B−/− and RNF8−/− mice throughout all 4 stages. RAD6B−/− and RNF8−/− mice showed a failure in spermiation. Light microscopic analysis of RNF8−/− mice revealed that spermatogenesis was normal until stages V–VIII of spermiogenesis and that RAD6B−/− mice showed abnormalities earlier than RNF8−/− mice (Fig. 2B, 2C and 2D). Meiotic division was normal, and in RNF8−/− mice, we observed haploid, round spermatids that had just completed meiosis before beginning acrosome development. However, there was severe sloughing of round and elongated spermatids from RAD6B−/− males, and the number of germ cells in the seminiferous epithelium decreased dramatically in RAD6B−/− male mice. Moreover, RAD6B−/− mice showed a 19.25% decrease in spermatogonia (Spg) and a 49.55% decrease in spermatocytes (Spc) compared with WT mice. The number of post-meiotic spermatids at each stage of spermiogenesis was examined in microscopic sections. In stage I–IV tubule sections, RAD6B−/− mice showed a 56.57% decrease in round spermatids (Rs) compared with WT. In contrast, RNF8−/− mice did not show a significant (ns) difference in Rs compared with WT. In stage V–VIII tubule sections, RAD6B−/− mice showed a 90.73% decrease in Rs compared with WT, and RNF8−/− tubules showed a 16.73% decrease in Rs compared with WT tubules. Therefore, loss of sperm appears to occur post-meiotically in RAD6B−/− and RNF8−/− tubules. These results suggest that developing sperm are progressively removed from RAD6B−/− tubules beginning at the transition occurring between stages I–IV and V–VIII.
Figure 2.
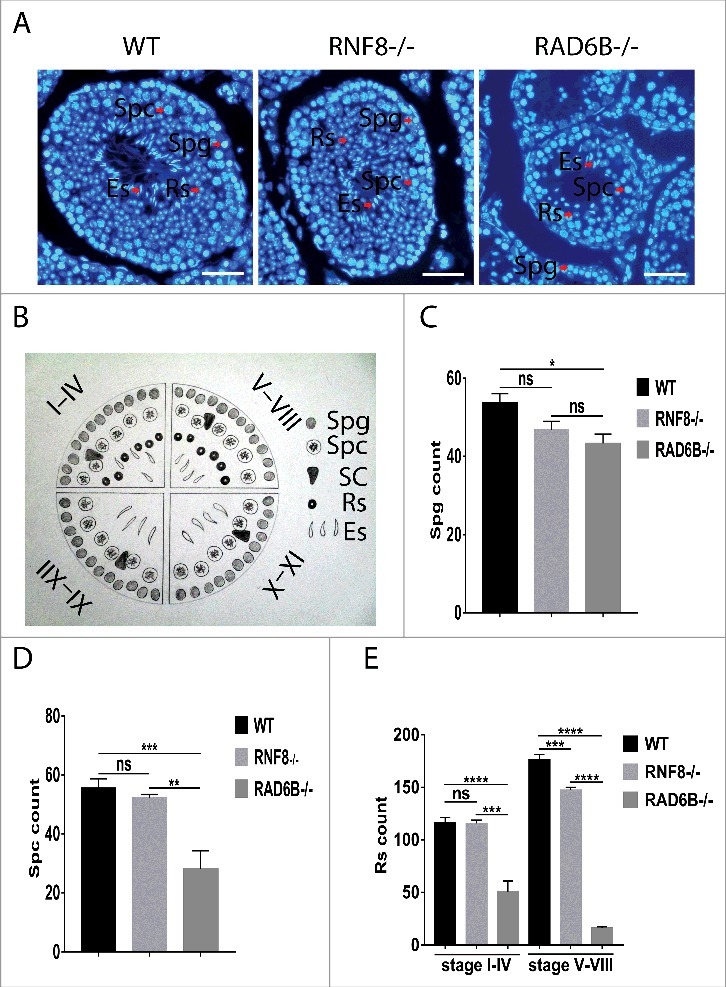
RAD6B and RNF8 KO mice showed abnormalities in spermatogenesis. (A) DAPI staining of seminiferous tubules sections from WT, RNF8−/− and RAD6B−/− males. Bar, 20 μm. (B) The spermatogenic stages were divided into 4 groups based on the presence of specific cell types and their arrangements: I–IV, V–VIII, IX–X and XI–XII. (C and D) The numbers of developing germ cells present during the first wave of spermatogenesis. Spermatogonia (Spg) and spermatocytes (Spc) were counted from DAPI-stained testicular sections at the postnatal days indicated. A significant reduction in Spc counts was observed in RAD6B−/− mice. (E) The numbers of stage I–IV and V–VIII round spermatids (Rs) counted from DAPI-stained testicular sections from WT, RNF8−/− and RAD6B−/− mice. The decrease in the total number of Rs in RNF8−/− and RAD6B−/− mice is attributed to a loss of stage V–VIII spermatids. All values are presented as the mean ± SEM (WT n = 6; RNF8−/− n = 6; RAD6B−/− n = 6). Student's t-test; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
RAD6B- and RNF8-deficient male mice have reduced sperm production and generate defective sperm
To evaluate sperm cells, the epididymis was isolated from WT, RAD6B−/− and RNF8−/− mice, and Giemsa staining was performed. Various hookless heads, which include club-shaped heads, amorphous heads, and those with disintegrated nuclei or no nuclei, were observed from RAD6B−/− and RNF8−/− mice (Fig. 3A). Normal, crescent-shaped sperm heads were not observed in the epididymis of RAD6B KO mice. As expected, the epididymal tubules of WT animals were full of sperm cells, and RNF8 KO mice had some sperm cells (Fig. 3B). In contrast, the epididymal tubules of RAD6B KO mice were almost completely devoid of sperm cells. Thus, a major loss of sperm cells in RAD6B−/− mice and RNF8−/− mice occurs before their release from the testes. We examined the sperm number per epididymis in cross sections obtained from animals (Fig. 3C). The RAD6B−/− males clearly showed the smallest number of sperm. Therefore, variable sperm counts and the uniformity of morphological abnormalities account for the infertility of RAD6B KO mice and the subfertility of RNF8 KO mice.
Figure 3.
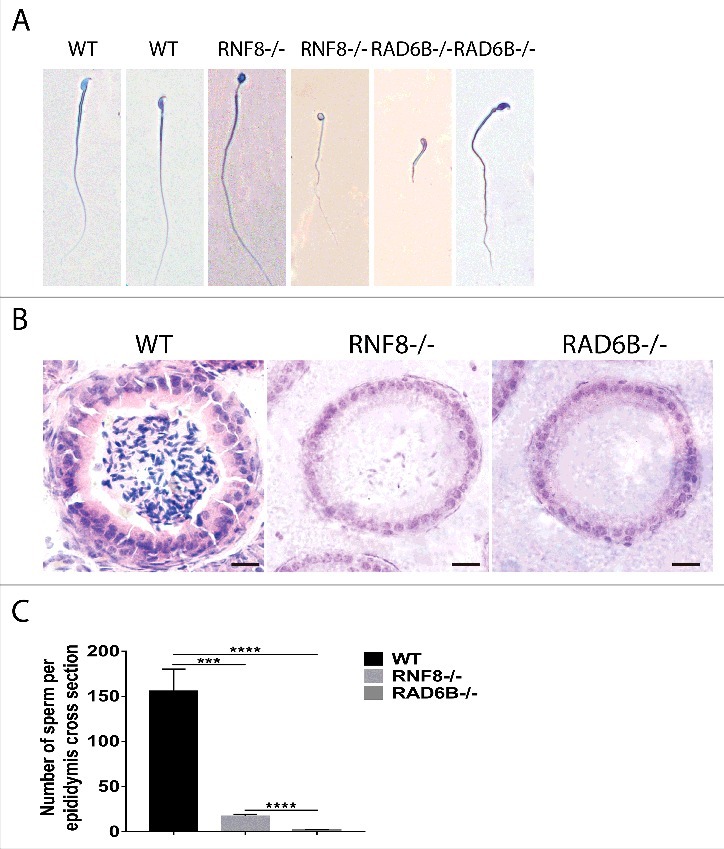
RAD6B and RNF8 knockout mice have developmental sperm disorders. (A) Normal sperm was observed in wild-type (WT) males, whereas abnormal round sperm was observed in RNF8−/− and RAD6B−/− mice. Typical morphologies of sperm from WT, RNF8−/− and RAD6B−/− mice as observed with Giemsa staining. Bar, 20 μm. (B) H&E staining of paraffin sections of cauda epididymis from wild-type (WT) and RNF8−/− and RAD6B−/− mice. Bar, 20 μm. (C) The number of sperm was counted from H&E staining of the cauda epididymis. All values are presented as the mean ± SEM (WT n = 6; RNF8−/− n = 6; RAD6B−/− n = 6). Student's t-test; ***P < 0.001; ****P < 0.0001.
RAD6B and RNF8 deletions both affect the histone-to-protamine replacement and spermatid nucleus head formation
During mammalian spermiogenesis, most nucleosomal histones are removed and replaced by histone variants, transition proteins and subsequently protamines.50-53 The process of histone-to-protamine replacement repackages the sperm genome to be at least 6-fold more compact than its somatic counterpart.52 To evaluate whether histone replacement was affected in RNF8−/− males and RAD6B−/− males, we used immunofluorescence to assess the expression of testes-specific histone H2B (tH2B) during spermatogenesis. In WT testes, histone H2B was quickly depleted after the Rs steps, with minimal retention in condensing and condensed spermatids (Fig. 4A and 4B). In contrast, in RAD6B−/− differentiated spermatids, more histone H2B was retained, indicating ineffective histone removal. Furthermore, immunofluorescent analyses of sperm showed greater expression of H2B in RAD6B−/− and RNF8−/− spermatids than in WT spermatids. Sperm from WT mice showed no staining for tH2B (Fig. 4C), whereas sperm from RNF8 KO mice and RAD6B KO mice contained histones in their rounded heads (Fig. 4C). These results suggest that histones are not properly replaced with protamine in sperm from RNF8 KO mice and RAD6B KO mice.
Figure 4.
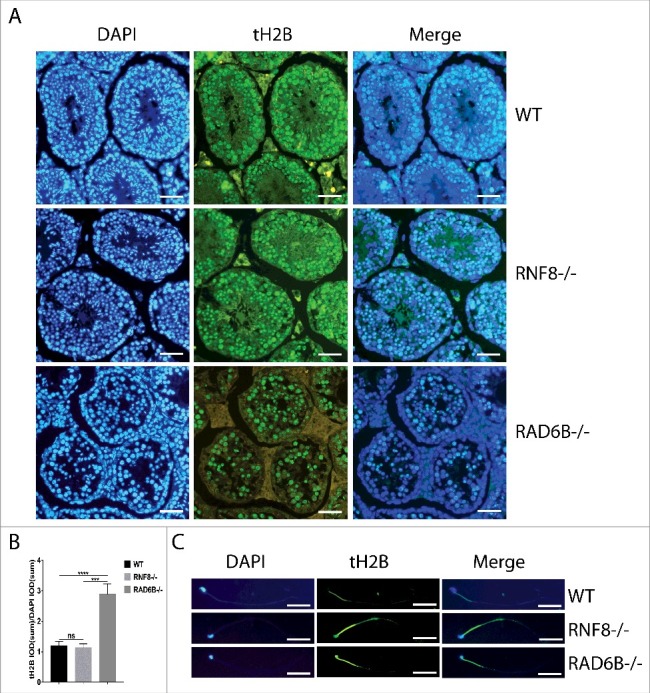
RAD6B-knockout mice and RNF8-knockout mice present a histone replacement disorder during spermatogenesis. (A) Sections of testes from WT, RNF8−/− and RAD6B−/− mice stained with anti-tH2B (testis-specific histone H2B) are shown. Bar, 20 μm. (B) We used the ratio of tH2B IOD (sum) to DAPI IOD (sum) to analyze the difference in expression levels of tH2B among WT, RNF8−/− and RAD6B−/− mice. All values are presented as the mean ± SEM (WT n = 6; RNF8−/− n = 6; RAD6B−/− n = 6). Student's t-test; ***P < 0.001; ****P < 0.0001; ns, not significant. (C) Histone tH2B was not replaced in RNF8−/− and RAD6B−/− mice sperm. Images of immunostained sperm from WT, RNF8−/− and RAD6B−/− mice are shown. Bar, 20 μm.
Immunoblot analysis of ubiquitinated histones in mouse testis
Basic nuclear proteins were isolated from total testis of WT, RNF8−/− and RAD6B−/− mice and analyzed using SDS–PAGE. To determine whether RAD6B and RNF8 are important for quantitative maintenance of overall protein ubiquitination, we studied protein ubiquitination by using one-dimensional SDS-PAGE gels. Coomassie staining revealed global changes in basic protein levels. The levels of histone H1 T and of TP2 (transition protein 2) were higher in the testes of WT mice than in those of RAD6B KO mice (Fig. 5A). However, there were few differences in basic protein levels between RNF8−/− mice and WT mice. The proteins present in Fig. 5 were identified by mass spectrometry (MS). These results indicate that RAD6B and RNF8 may not be required for the maintenance of bulk protein ubiquitination but are involved in the ubiquitination of some specific substrates whose functions are essential for spermatogenesis. To quantitatively assess histone modifications, we performed Western blot analysis and found that levels of H1 T were decreased in testes from RAD6B−/− and RNF8−/− males (Fig. 5B and 5C). During spermatogenesis, histone H1 T is detected in mid-pachytene Spc,54-56 and it then rapidly becomes a part of the chromatin and replaces most of the other H1 subtypes. In pachytene Spc and round spermatids, H1 T represents a major part of the total H1 histone complement,57,58 and persists until the stage of elongating spermatids. Moreover, these findings suggest that loss of germ cells in the testes from RAD6B−/− and RNF8−/− males leads to decreased H1 T. Furthermore, we found that levels of H2 A and H2B were increased in testes from RAD6B−/− males (Fig. 5B and 5C). In contrast, no significant changes were observed in RNF8−/− males (Fig. 5B and 5C). These findings are consistent with the results in Figure 4 and indicates histones cannot be successfully replaced and degraded. Thus, the expression of histones residue is increased. We found that in RNF8−/− males, ub-H2 A and ub-H2B were decreased in the testes, consistent with previous findings made by Linyu Lu43 (Fig. 5D). Interestingly, the levels of mono-ub-H2 A and mono-ub-H2B were increased in testes from RAD6B KO mice, whereas poly-ub-H2 A was decreased (Fig. 5D). We performed the ub blot with WT, RNF8- and RAD6B-deficient MEFs (Fig S1) and the level of ub in RAD6B-deficient showed a significant reduction compared with WT and RNF8-deficient MEFs. We hypothesized that RNF8 may play a role in monoubiquitination and RAD6B may play a role in polyubiquitination. To investigate this hypothesis, we detected the levels of ub-H2 A (Fig. 5E). The H2 A-ub2 in testis cells had almost completely disappeared in RAD6B−/− mice and was significantly decreased in RNF8−/− mice.
Figure 5.
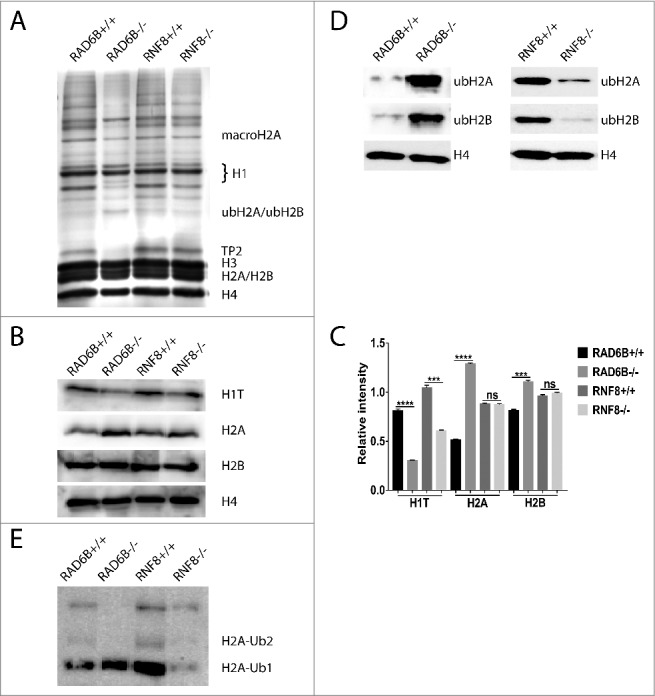
H2 (A)and H2B ubiquitination were increased in testes from RAD6B−/− males but decreased in testes from RNF8−/− males. (A) Basic nuclear proteins were isolated from total testis from WT, RNF8−/− and RAD6B−/− mice, separated using 15% SDS-PAGE, and stained with Coomassie blue. Proteins were excised from the gel and identified by MS. (B) H1 T was decreased in testes from RNF8−/− and RAD6B−/− mice and H2 A and H2B were increased in testes from RAD6B−/− males. In contrast, no significant changes were observed in RNF8−/− males. Western blots of proteins using testes from WT, RNF8−/− and RAD6B−/− mice are shown. Antibodies used are indicated, and H4 was used as a loading control. (C) The relative expression levels of H1 T, H2 A and H2B in panel B are summarized in the histogram (mean ± SEM). (D) ub-H2 A and ub-H2B were decreased in testes from RNF8−/− mice but increased in testes from RAD6B−/− mice. Western blots of proteins from testes of WT, RNF8−/− and RAD6B−/− mice are shown. Antibodies used are indicated, and H4 was used as a loading control. (E) H2 A-ub2 had almost completely disappeared from the testes of RAD6B−/− mice and was decreased significantly in the testes of RNF8−/− mice. Western blots of proteins using testes from WT, RNF8−/− and RAD6B−/− mice are shown.
Analysis of senescence in seminiferous tubule cross sections from 6-month-old mice
β-Gal is a direct evidence of the presence and aggregation of senescent cells in the process of aging. The p53/p21 and p16/pRB pathways are the key regulators of the senescence, activated along with the cellular senescent state. Examination of testis sections via β-Galactosidase (β-Gal) staining was detected in aging cells of WT, RNF8−/− and RAD6B−/− mice. Positive β-Gal staining was detected in Leydig cells that were located near the basal lamina of the seminiferous tubules in 6-month-old RAD6B−/− mice (Fig. 6A and 6B). In contrast, there were few β-Gal-positive cells in WT or RNF8−/− mice. To confirm that aging plays an important role during spermatogenesis, we performed immunohistochemistry. Consistent with β-Gal staining, P16 and P21 were detected in testes cross sections from RAD6B KO mice (Fig. 6C and 6D). This result suggests that aging was essential for spermatogenesis.
Figure 6.
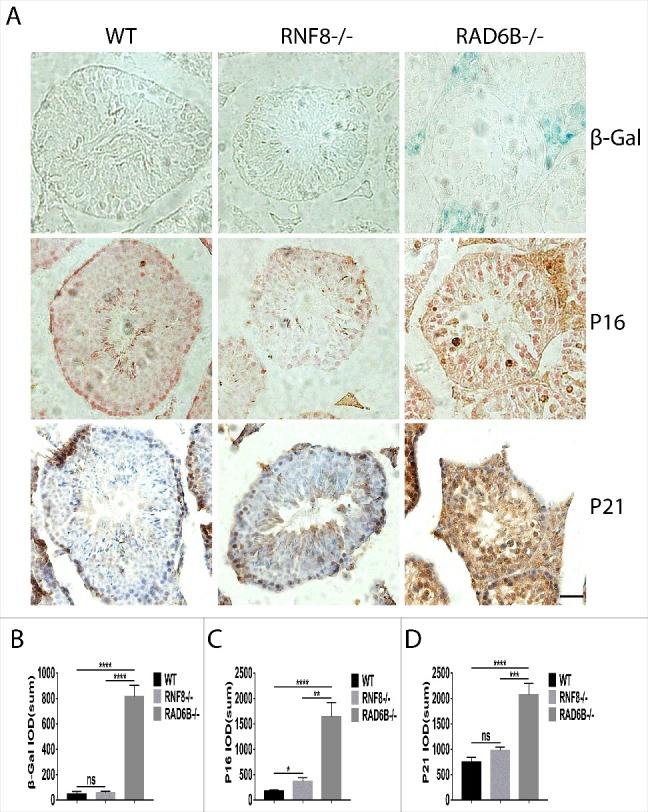
Aging is essential for spermatogenesis. (A) Sections of testes from WT, RNF8−/− and RAD6B−/− mice stained with β-Gal, P16 and P21. Bar, 20 μm. (B–D) Quantification of aging in WT, RNF8−/− and RAD6B−/− mice. We used IOD (sum) of β-Gal, P16 and P21 in A to analyze the aging expression. All values are presented as the mean ± SEM (WT n = 6; RNF8−/− n = 6; RAD6B−/− n = 6). Student's t-test; *P < 0.05, **P < 0.01, ***P < 0.001; ****P < 0.0001; ns, not significant.
Discussion
Global histone removal from the DNA of spermatids followed by replacement with transition proteins and protamines is a critical process for proper spermiogenesis. Previous studies have highlighted the importance of histone ubiquitination and its crucial role in global histone replacement in spermatids.42,43 Transition proteins and protamines are basic proteins that promote the condensation of chromatin, which is packaged into the sperm head. This process is proposed to protect the paternal genome and make it inaccessible to nucleases and mutagens.59 Hence, it is not surprising that several studies have shown that reduced levels of transition proteins or protamines in mice are linked to defects in spermiogenesis.60-62 In our study, we demonstrate that RAD6B-dependent and RNF8-dependent histone ubiquitination affects global histone removal (Fig. 4C). Spermatids with global histone ubiquitination defects display abnormal morphologies and reduced sperm production and generate defective sperm resulting in male sterility (Fig. 3). In summary, histone ubiquitination appears to control histone removal and transition protein incorporation into chromatin. We observed abundant germ cells in the testes of RAD6B−/− and RNF8−/− males, but few sperm were present in their epididymides, and those sperm displayed abnormal morphology. The mechanism by which sperm is screened and eliminated will be the focus of our next study.
H2 A ubiquitination has been shown to play an important role during spermatogenesis. Ubiquitinated H2 A is accumulated in the XY body during the pachytene stage of meiosis I and is enriched again in elongating spermatids after meiosis.25,63 The function of ubiquitinated H2 A in the XY body is unclear, but it is thought that these modifications may mediate meiotic sex chromosome inactivation.64 Interestingly, the absence of ubiquitinated H2 A in the XY body does not affect XY body formation, sex chromosome inactivation, or meiotic progression, indicating that H2 A ubiquitination is not essential for these processes.43 H2B ubiquitination has also been shown to modulate the formation of double-strand breaks during meiosis.65 Protein structure analysis of recombinant nucleosomes indicates that H2B ubiquitination may be involved in the decondensation of chromatin.66 In addition, ubiquitinated H2B is associated with active genes,67,68 which may promote local nucleosome removal during transcription. Genetic deletion of HR6B causes male sterility, due to defects at postmeiotic stages of spermiogenesis.42 The ubiquitination of H2 A was significantly enhanced in the testes of RAD6B knockout mice but was suppressed in those of RNF8 knockout mice (Fig. 5D). Furthermore, we provide evidence that H2B ubiquitination shows the same change in the testes of RAD6B knockout and RNF8 knockout mice. These results suggest that RAD6B and RNF8 have different effects on H2 A and H2B ubiquitination. It is possible that RNF8 and RAD6B work in concert to regulate H2 A/H2B ubiquitination during spermatogenesis, which could prove essential for nucleosome removal.
Polyubiquitination modifications typically target a substrate for proteasomal degradation,10 whereas monoubiquitination is involved in many processes, including DNA repair and regulation of gene expression.11 Interestingly, we found that RAD6B−/− and UBR2−/− males showed very similar phonotypes,69 such as male sterility, reduced Spc and germ cell numbers and hollow lumens.69 It has been reported that mouse UBR2 can mediate the ubiquitination of N-end rule substrates in concert with Ub-conjugating enzymes such as RAD6B via the N-end rule pathway.69 This finding indicates that the infertility phenotype of RAD6B−/− males may be mediated by the interaction of RAD6B with UBR2 and involves the dysregulation of protein degradation via the N-end rule. UBR1, UBR1 and UBR3 form an E2-E3 complex with the E2 enzyme RAD6, which leads to polyubiquitylation of the substrates and their subsequent degradation.36 UBR2 functions as a scaffold E3 that promotes HR6B polyubiquitinates H2 A and H2B but not H3 and H4.69 The levels of both ubiquitinated H2 A and H2B are decreased in testes from RNF8-deficirnt mice.44 We provided evidence that H2 A polyubiquitination was undetectable and that H2 A monoubiquitination was highly expressed in the testes of RAD6B knockout mice (Fig. 5D). In contrast, H2 A polyubiquitination and monoubiquitination were suppressed in the testes of RNF8 knockout mice. Combined with H2 A/H2B monoubiquitination in testes of RAD6B knockout and RNF8 knockout mice, these results suggest that RAD6B may play a role in H2 A/H2B polyubiquitination and that RNF8 may play a role in H2 A/H2B monoubiquitination during spermatogenesis. Since H2 A/H2B monoubiquitination was decreased in the testes of RNF8 knockout mice, we hypothesized that RNF8 may participate in histone ubiquitination during spermatogenesis.23 We believe that polyubiquitination is more essential than monoubiquitination during spermatogenesis because histone degradation is critical for the process of histone-protamine replacement. Furthermore, RAD6B is critical for spermatogenesis. In summary, we hypothesize that H2 A/H2B ubiquitination may be a signal to initiate the replacement process and that histone degradation recruits transition proteins that promote histone-protamine processing, and we plan to investigate this further in future studies.
In this study, we provide evidence that senescence may be critical for the spermatogenesis dysfunction (Fig. 6). We observed small testis size and decreased seminiferous tubule cross section width in RAD6B knockout and RNF8 knockout mice, phenotypes that may be attributable to the senescent cells found to be concentrated in the testis interstitial regions (Fig. 1A and 1B). These results proved that RAD6B- and RNF8-deficient males displayed testicular degeneration and atrophy, which indicates that DNA damage leads to an increase in senescent cells in the testes from RAD6B- and RNF8-deficient males, although the expression of β-Gal, P16 and P21 in RNF8-deficient mice was not significant in the experiment. In addition, we observed significantly lower numbers of Spc from 6-month-old RAD6B−/− mice (Fig. 2C). An intact tubular structure with normal development of Sertoli cells and Spc has been observed in immature mHR6B −/− mice.42 Hence, RAD6B knockout does not affect mitosis and the formation of Spc; instead, senescence affects the number of germ cells formed during spermatogenesis. A similar reduction in the number of Spc was observed in UBR2−/− males.69 These findings suggest that mutations in DNA repair proteins in RAD6B−/− and UBR2−/− mice can lead to DSB repair defects, which intensify the aging process. Thus, the number of Spc is decreased significantly.
Taken together, our results reveal that RAD6B and RNF8 contribute to nucleosome removal during spermiogenesis. RAD6B knockout mice are infertile, and RNF8 knockout mice are either sub-fertile or sterile. RNF8 monoubiquitinates histones H2 A and H2B, and RAD6B polyubiquitinates histones H2 A and H2B. In addition, RAD6B interacts with UBR2 in the N-end rule pathway to promote histone degradation and facilitate the process of histone-protamine replacement during spermatogenesis. Aging also plays an important role during spermatogenesis.
Materials and methods
Animals
We used 6-month-old homozygous male mice to collect all data presented here. RAD6B-deficient mice have been reported previously,42 and the RNF8-deficient mice also have been described.43 RNF8 knockout mice were obtained from Dr. Xiaochun Yu. RAD6B knockout mice were obtained from COMM (MGI: 102944). All animals were maintained at the Lanzhou University Animal Center. The animals were housed under standard housing conditions and were given free access to food and water. The experimental protocols were approved by the Committee on the Ethics of Animal Experiments of Lanzhou University. All procedures were performed under anesthesia with pentobarbital sodium, and efforts were made to reduce the suffering of mice.
Paraffin-embedded sections
Testes and epididymides were isolated and excised, fixed in 4% paraformaldehyde overnight and then embedded in paraffin according to standard procedures and sectioned (5 μm). Sections were used for hematoxylin-eosin (H&E) staining, immunofluorescence, Immunohistochemistry and β-Galactosidase staining.
Histology and immunostaining
Paraffin sections were baked overnight at 55°C, dewaxed and hydrated according to standard methods. Tissue sections were H&E stained following standard procedures. Testicular sections were examined systematically to detail the different germ cell types, and cross sections of the cauda epididymis were observed to determine the numbers of spermatozoa within the lumen. For immunostaining of Spc, spermatids and seminiferous tubules, sections from 6-month-old mice were stained using anti-Histone H2B [testis specific] (ab23913, Abcam, 1:400 dilution). For secondary detection, goat anti-rabbit IgG-FITC (BA1105, BOSTER, 1:400 dilution) was used according to standard immunostaining procedures. Then, slides were mounted with DNA-specific fluorochrome 4, 6-diamidino-2-phenylindole (DAPI; D9542, Sigma) for counterstaining. An Olympus fluorescence microscope (BX53) was used to capture images.
Sperm morphology
The sperm were harvested by dissection and removal of the epididymis. Next, air-dried smears were prepared from spermatozoa suspended in PBS. The smears were used for Giemsa staining and immunofluorescence and were stained with Giemsa for observing normal and abnormal morphology of sperm. For immunostaining, the smears were stained using anti-Histone H2B [testis specific] (ab23913, Abcam, 1:400 dilution) according to standard immunostaining procedures.
SDS-PAGE and immunoblotting
Histone protein extracts were prepared from 6-month-old mouse testes, as described.36 Protein concentrations were determined by using the Bradford assay. Proteins were separated on SDS-PAGE gels (15% SDS-PAGE, 120 V, 0–4°C) and subjected to either staining with Coomassie brilliant blue (CBB) or Western blot using the following antibodies: anti-ubiquityl-histone H2B [clone 56] (05–1312, Millipore, 1:500 dilution), anti-ubiquityl-histone H2 A [clone E6C5] (05–678, Millipore, 1:500 dilution), anti-histone H2B (15857–1-AP, Proteintech, 1:500 dilution), anti-HIST1H1 T (18188–1-AP, Proteintech, 1:500 dilution), anti-histone H4 (16047–1-AP, Proteintech, 1:500 dilution) and anti-ubiquitin (ab7780, Abcam, 1:1000 dilution) .The CBB staining gel was analyzed using liquid chromatography-mass spectrometry (LC-MS).
Cell culture
MEFs were obtained from WT, RNF8- and RAD6B-deficient mice. MEFs were cultured in DMEM (Gibco Invitrogen Corporation) supplemented with 10% FCS.
β-Galactosidase staining
β-Galactosidase reagent was obtained from Beyotime Biotechnology. Paraffin sections were subjected to dewaxing and hydration treatment using standard procedures and added to an appropriate volume of β-galactosidase staining fixative at room temperature for 15 minutes. Tissues were washed 3 times with PBS for 5 minutes. PBS was removed, and the appropriate amount of staining solution added before overnight incubation at 37°C.
Immunohistochemistry
Immunohistochemistry was performed as described previously.42 The following antibodies were used for immunohistochemistry: anti-P16 (ab51243, Abcam, 1:400 dilution), anti-P21 (ab109520, Abcam, 1:400 dilution) and goat anti-rabbit IgG (H&L) biotin-conjugated, affinity-purified antibody (AP132 B, Millipore, 1:500 dilution).
Statistical analysis
Prism software (GraphPad) was used to generate graphs and analyze data. Data are presented as the mean ± SEM for each genotype. Photoshop CC was used to count different germ cells. The data of IOD was obtained using ImageJ software. One-way analysis of variance (ANOVA) with student's t-test was used to compare the difference in means between the variants and WT. P < 0.05 was considered statistically significant.
Supplementary Material
Funding Statement
This work was funded by the National Natural Science Foundation of China [Grant No. 81171954 and 81472541 to D.W. and No. 81576312 to Y.S.]
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422-9. doi: 10.1038/nature07958. PMID:19325621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703-17. doi: 10.1016/j.cell.2006.04.029. PMID:16713563 [DOI] [PubMed] [Google Scholar]
- [3].Fang S, Weissman AM. A field guide to ubiquitylation. Cell Mol Life Sci. 2004;61:1546-61. PMID:15224180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ciechanover A. The ubiquitin- proteasome proteolytic pathway. Cell. 1994;79:13-21. doi: 10.1016/0092-8674(94)90396-4. PMID:7923371 [DOI] [PubMed] [Google Scholar]
- [5].Ubiquitin Hochstrasser M., proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215-23;doi: 10.1016/0955-0674(95)80031-X. PMID:7612274 [DOI] [PubMed] [Google Scholar]
- [6].Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461-7. doi: 10.1038/nature07963. PMID:19325626 [DOI] [PubMed] [Google Scholar]
- [7].Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203-29. doi: 10.1146/annurev-biochem-060310-170328. PMID:22524316 [DOI] [PubMed] [Google Scholar]
- [8].Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 2011;12:35-48. PMID:22158412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430-7. doi: 10.1038/nature07959. PMID:19325622 [DOI] [PubMed] [Google Scholar]
- [10].Hershko A, Ciechanover A. The ubiquitin system. Annual review of biochemistry. 1998;67:425-79. doi: 10.1146/annurev.biochem.67.1.425. PMID:9759494 [DOI] [PubMed] [Google Scholar]
- [11].Varshavsky A. Regulated protein degradation. Trends Biochem Sci 2005;30:283-6. doi: 10.1016/j.tibs.2005.04.005. PMID:15950869 [DOI] [PubMed] [Google Scholar]
- [12].Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610-6. doi: 10.1016/j.cbpa.2004.09.009. PMID:15556404 [DOI] [PubMed] [Google Scholar]
- [13].Carrell DT. Epigenetics of the male gamete. Fertility and sterility 2012;97:267-74. doi: 10.1016/j.fertnstert.2011.12.036. PMID:22289286 [DOI] [PubMed] [Google Scholar]
- [14].Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176-8. doi: 10.1126/science.1070963 [DOI] [PubMed] [Google Scholar]
- [15].Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879-89. doi: 10.1242/dev.020966. PMID:19234061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008-16. doi: 10.1038/nsmb1337 [DOI] [PubMed] [Google Scholar]
- [17].Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008; 29:653-63. doi: 10.1016/j.molcel.2008.02.014. PMID:18374642 [DOI] [PubMed] [Google Scholar]
- [18].Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, et al.. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2 A. Genes & development. 2006;20:1343-52. doi: 10.1101/gad.373706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al.. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435-46. doi: 10.1016/j.cell.2008.12.041. PMID:19203579 [DOI] [PubMed] [Google Scholar]
- [20].Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901-14. doi: 10.1016/j.cell.2007.09.041. PMID:18001825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887-900. doi: 10.1016/j.cell.2007.09.040. PMID:18001824 [DOI] [PubMed] [Google Scholar]
- [22].Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al.. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009; 136:420-34. doi: 10.1016/j.cell.2008.12.042. PMID:19203578 [DOI] [PubMed] [Google Scholar]
- [23].Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, et al.. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25:663-75. doi: 10.1016/j.molcel.2007.01.029. PMID:17349954 [DOI] [PubMed] [Google Scholar]
- [24].Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13-15 on H2 A/H2AX to drive DNA damage signaling. Cell. 2012; 150:1182-95. doi: 10.1016/j.cell.2012.08.005. PMID:22980979 [DOI] [PubMed] [Google Scholar]
- [25].Wu J, Huen MS, Lu LY, Ye L, Dou Y, Ljungman M, Chen J, Yu X. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol Cell Biol. 2009;29:849-60. doi: 10.1128/MCB.01302-08. PMID:19015238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Baarends WM, Hoogerbrugge JW, Roest HP, Ooms M, Vreeburg J, Hoeijmakers JH, Grootegoed JA. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev Biol. 1999;207:322-33. doi: 10.1006/dbio.1998.9155. PMID:10068466 [DOI] [PubMed] [Google Scholar]
- [27].Hess RA, Renato de Franca L. Spermatogenesis and cycle of the seminiferous epithelium. Adv Exp Med Biol. 2008;636:1-15; PMID:19856159 [DOI] [PubMed] [Google Scholar]
- [28].Oakberg EF. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am J Anat. 1956;99:507-16. doi: 10.1002/aja.1000990307. PMID:13402729 [DOI] [PubMed] [Google Scholar]
- [29].Oakberg EF. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am J Anat. 1956; 99:391-413. doi: 10.1002/aja.1000990303. PMID:13402725 [DOI] [PubMed] [Google Scholar]
- [30].Ahmed EA, de Rooij DG. Staging of mouse seminiferous tubule cross-sections. Methods Mol Biol. 2009;558:263-77. doi: 10.1007/978-1-60761-103-5_16 [DOI] [PubMed] [Google Scholar]
- [31].Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111:483-8. doi: 10.1007/s00412-002-0227-z. PMID:12743712 [DOI] [PubMed] [Google Scholar]
- [32].Oliva R. Protamines and male infertility. Hum Reprod Update. 2006;12:417-35. doi: 10.1093/humupd/dml009. PMID:16581810 [DOI] [PubMed] [Google Scholar]
- [33].Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, Eddy EM. Haploinsufficiency of protamine-1 or – 2 causes infertility in mice. Nat Genet. 2001;28:82-6. doi: 10.1038/ng0501-82. PMID:11326282 [DOI] [PubMed] [Google Scholar]
- [34].Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biology of reproduction 2004;71:1220-9. doi: 10.1095/biolreprod.104.029363. PMID:15189834 [DOI] [PubMed] [Google Scholar]
- [35].Zhao M, Shirley CR, Hayashi S, Marcon L, Mohapatra B, Suganuma R, Behringer RR, Boissonneault G, Yanagimachi R, Meistrich ML. Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis. 2004; 38:200-13. doi: 10.1002/gene.20019. PMID:15083521 [DOI] [PubMed] [Google Scholar]
- [36].Sriram SM, Kim BY, Kwon YT. The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nat Rev Mol Cell Biol. 2011;12:735-47. doi: 10.1038/nrm3217. PMID:22016057 [DOI] [PubMed] [Google Scholar]
- [37].Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261-89. doi: 10.1146/annurev-biochem-051710-093308. PMID:22524314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sriram SM, Kwon YT. The molecular principles of N-end rule recognition. Nat Struct Mol Biol. 2010;17:1164-5. doi: 10.1038/nsmb1010-1164 [DOI] [PubMed] [Google Scholar]
- [39].Kwon YT, Xia Z, Davydov IV, Lecker SH, Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol Cell Biol. 2001;21:8007-21. doi: 10.1128/MCB.21.23.8007-8021.2001. PMID:11689692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci U S A. 1996;93:12142-9. doi: 10.1073/pnas.93.22.12142. PMID:8901547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein science: a publication of the Protein Society 2011;20:1298-345. doi: 10.1002/pro.666. PMID:21633985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].An JY, Kim E, Zakrzewska A, Yoo YD, Jang JM, Han DH, Lee MJ, Seo JW, Lee YJ, Kim TY, et al.. UBR2 of the N-end rule pathway is required for chromosome stability via histone ubiquitylation in spermatocytes and somatic cells. PloS one. 2012;7:e37414. doi: 10.1371/journal.pone.0037414. PMID:22616001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Roest HP, van Klaveren J, de Wit J, van Gurp CG, Koken MH, Vermey M, van Roijen JH, Hoogerbrugge JW, Vreeburg JT, Baarends WM, et al.. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell. 1996;86:799-810. doi: 10.1016/S0092-8674(00)80154-3. PMID:8797826 [DOI] [PubMed] [Google Scholar]
- [44].Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell. 2010;18:371-84. doi: 10.1016/j.devcel.2010.01.010. PMID:20153262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Li L, Halaby MJ, Hakem A, Cardoso R, El Ghamrasni S, Harding S, Chan N, Bristow R, Sanchez O, Durocher D, et al.. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med. 2010;207:983-97. doi: 10.1084/jem.20092437. PMID:20385750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501-4. doi: 10.1126/science.287.5452.501. PMID:10642555 [DOI] [PubMed] [Google Scholar]
- [47].Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601-11. doi: 10.1016/j.molcel.2005.09.025. PMID:16307923 [DOI] [PubMed] [Google Scholar]
- [48].Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459-71. doi: 10.1016/j.cell.2009.02.027. PMID:19410543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].An JY, Kim EA, Jiang Y, Zakrzewska A, Kim DE, Lee MJ, Mook-Jung I, Zhang Y, Kwon YT. UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc Natl Acad Sci U S A. 2010;107:1912-7. doi: 10.1073/pnas.0910267107. PMID:20080676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lin CY, Chen CY, Yu CH, Yu IS, Lin SR, Wu JT, Lin YH, Kuo PL, Wu JC, Lin SW. Human X-linked Intellectual Disability Factor CUL4B Is Required for Post-meiotic Sperm Development and Male Fertility. Sci Rep. 2016;6:20227. doi: 10.1038/srep20227. PMID:26832838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cooke HJ, Saunders PT. Mouse models of male infertility. Nat Rev Genet. 2002;3:790-801. doi: 10.1038/nrg911. PMID:12360237 [DOI] [PubMed] [Google Scholar]
- [52].Pang AL, Johnson W, Ravindranath N, Dym M, Rennert OM, Chan WY. Expression profiling of purified male germ cells: stage-specific expression patterns related to meiosis and postmeiotic development. Physiol Genomics. 2006;24:75-85. doi: 10.1152/physiolgenomics.00215.2004. PMID:16291737 [DOI] [PubMed] [Google Scholar]
- [53].Margolin G, Khil PP, Kim J, Bellani MA, Camerini-Otero RD. Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics. 2014;15:39. doi: 10.1186/1471-2164-15-39. PMID:24438502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jackson S, Xiong Y. CRL4 s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562-70. doi: 10.1016/j.tibs.2009.07.002. PMID:19818632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Drabent B, Bode C, Bramlage B, Doenecke D. Expression of the mouse testicular histone gene H1 t during spermatogenesis. Histochem Cell Biol. 1996;106:247-51. doi: 10.1007/BF02484408. PMID:8877387 [DOI] [PubMed] [Google Scholar]
- [56].Moens PB. Histones H1 and H4 of surface-spread meiotic chromosomes. Chromosoma. 1995;104:169-74. doi: 10.1007/BF00352181. PMID:8529456 [DOI] [PubMed] [Google Scholar]
- [57].Oko RJ, Jando V, Wagner CL, Kistler WS, Hermo LS. Chromatin reorganization in rat spermatids during the disappearance of testis-specific histone, H1 t, and the appearance of transition proteins TP1 and TP2. Biol Reprod. 1996;54:1141-57. doi: 10.1095/biolreprod54.5.1141. PMID:8722637 [DOI] [PubMed] [Google Scholar]
- [58].Bucci LR, Brock WA, Meistrich ML. Distribution and synthesis of histone 1 subfractions during spermatogenesis in the rat. Exp Cell Res. 1982;140:111-8. doi: 10.1016/0014-4827(82)90162-8. PMID:7106196 [DOI] [PubMed] [Google Scholar]
- [59].Meistrich ML, Bucci LR, Trostle-Weige PK, Brock WA. Histone variants in rat spermatogonia and primary spermatocytes. Dev Biol. 1985;112:230-40. doi: 10.1016/0012-1606(85)90137-X. PMID:3932111 [DOI] [PubMed] [Google Scholar]
- [60].Oliva R, Dixon GH. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog Nucleic Acid Res Mol Biol. 1991;40:25-94. doi: 10.1016/S0079-6603(08)60839-9. PMID:2031084 [DOI] [PubMed] [Google Scholar]
- [61].Nair M, Nagamori I, Sun P, Mishra DP, Rheaume C, Li B, Sassone-Corsi P, Dai X. Nuclear regulator Pygo2 controls spermiogenesis and histone H3 acetylation. Dev Biol. 2008;320:446-55. doi: 10.1016/j.ydbio.2008.05.553. PMID:18614164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2 A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119-23. doi: 10.1038/nature06236. PMID:17943087 [DOI] [PubMed] [Google Scholar]
- [63].Wu JY, Ribar TJ, Cummings DE, Burton KA, McKnight GS, Means AR. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat Genet. 2000;25:448-52. doi: 10.1038/78153. PMID:10932193 [DOI] [PubMed] [Google Scholar]
- [64].Santos MA, Huen MS, Jankovic M, Chen HT, Lopez-Contreras AJ, Klein IA, Wong N, Barbancho JL, Fernandez-Capetillo O, Nussenzweig MC, et al.. Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J Exp Med. 2010;207:973-81. doi: 10.1084/jem.20092308. PMID:20385748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E, Hoeijmakers JH, de Boer P, Grootegoed JA. Silencing of unpaired chromatin and histone H2 A ubiquitination in mammalian meiosis. Mol Cell Biol. 2005;25:1041-53. doi: 10.1128/MCB.25.3.1041-1053.2005. PMID:15657431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yamashita K, Shinohara M, Shinohara A. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc Natl Acad Sci U S A. 2004;101:11380-5. doi: 10.1073/pnas.0400078101. PMID:15280549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251-60. [DOI] [PubMed] [Google Scholar]
- [68].Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483-8. doi: 10.1038/ncb1712. PMID:18344985 [DOI] [PubMed] [Google Scholar]
- [69].Kwon YT, Xia Z, An JY, Tasaki T, Davydov IV, Seo JW, Sheng J, Xie Y, Varshavsky A. Female Lethality and Apoptosis of Spermatocytes in Mice Lacking the UBR2 Ubiquitin Ligase of the N-End Rule Pathway. Mol Cell Biol. 2003;23:8255-71. doi: 10.1128/MCB.23.22.8255-8271.2003. PMID:14585983 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


