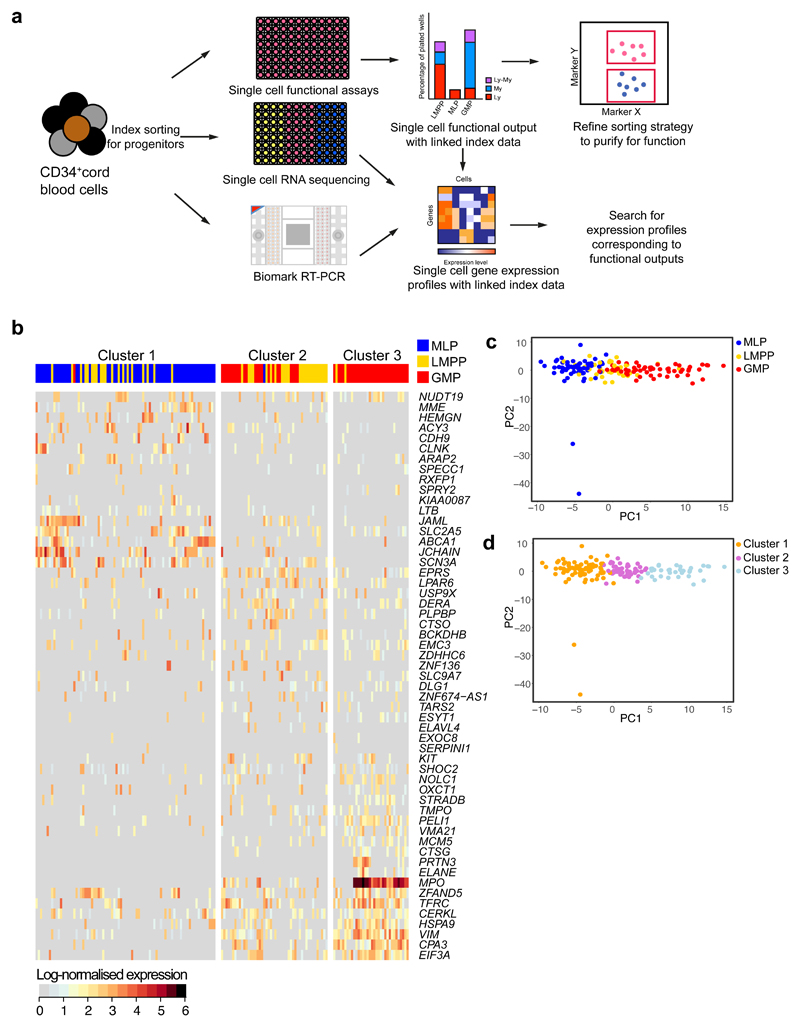
Abstract
The human hemopoietic progenitor hierarchy producing lymphoid and granulocytic-monocytic (myeloid) lineages is unclear. Multiple progenitor populations produce lymphoid and myeloid cells, but remain incompletely characterized. Here, we demonstrated cord blood lympho-myeloid containing progenitor populations - the lymphoid-primed multi-potential progenitor (LMPP), granulocyte-macrophage progenitor (GMP) and multi-lymphoid progenitor (MLP) - were functionally and transcriptionally distinct and heterogeneous at the clonal level, with progenitors of many different functional potentials present. Though most progenitors had uni-lineage myeloid or lymphoid potential, bi- and rarer multi-lineage progenitors occurred in LMPP, GMP and MLP. This, coupled with single cell expression analyses, suggested a continuum of progenitors execute lymphoid and myeloid differentiation rather than only uni-lineage progenitors being present downstream of stem cells.
Human hemopoiesis produces 10 billion new, terminally mature, blood cells daily; a production that is also rapidly responsive to external change. Most of this production generates red cells, short-lived myeloid cells and platelets. It also replenishes long-lived acquired immune cells and innate immune natural killer (NK) cells. Dysregulation of this complex process can lead to hemopoietic and immune deficiencies and blood cancers. Active debate continues about the heterogeneity and plasticity of hemopoietic cell populations, in steady state and in response to stimuli. At the hierarchy apex lie multi-potent hemopoietic stem cell (HSC) populations, heterogeneous with respect to differentiation potential, cell cycle, self-renewal capacity, stability over time and contribution to hemopoiesis in steady state versus transplantation1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11. Downstream of murine long-term HSCs are heterogeneous short-term HSC (HSCST), multipotent (MPP) and early lineage-biased progenitors5, 7, 12, 13, 14. The human HSCST/MPP population has not been fully defined15, 16. In terms of lineage potential restriction, the erythroid and megakaryocyte fates likely diverge early from other myeloid and lymphoid potentials in mouse14, 17, 18, 19, 20 and human21, 22, 23, 24, 25 and may arise directly from either HSC6 or immediate downstream MPP14, 16, 26.
Focusing on the first human lympho-myeloid progenitors downstream of HSC and MPP, two progenitor populations have been identified within the immature Lin-CD34+CD38-CD90-/lo compartment. These include a Lin-CD34+CD38-CD90-/loCD45RA+CD10- lymphoid-primed multi-potential progenitor (LMPP) with granulocytic, monocytic, B and T cell potential, but unable to generate erythrocytes or megakaryocytes22. These data support prior studies showing human CD34+CD10- cells retain lympho-myeloid potential, progressively losing myeloid potential with CD10 expression27, 28. In contrast, the multi-lymphoid progenitor (MLP), which was initially reported as Lin-CD34+CD38-CD90-/loCD45RA+CD10+, has lymphoid (B, T, NK), monocytic and dendritic cell (DC) potential but cannot make granulocytes21. However, recent CD10- MLP populations29 have been reported that may overlap with the LMPP. Within the Lin-CD34+CD38+CD45RA+ compartment, there are at least two lympho-myeloid progenitors: a CD62LhiCD10- lymphoid-primed progenitor with lymphoid, monocytic and DC potential23 and the granulocyte-monocyte progenitor (GMP; Lin-CD34+CD38+CD45RA+CD123+). GMP contains both CD62hi and CD62lo subpopulations and has mainly myeloid potential but retains residual lymphoid potential22, 30 consistent with the murine pre-GM progenitor31. Finally, the human Lin-CD34+CD38+CD45RA+ compartment also contains a CD10+ subpopulation with T, B, NK and DC potential but lacking myeloid potentials32.These prior observations raise questions about whether these progenitor populations are pure or heterogeneous, how distinct they are and the nature of the functional, transcriptional and hierarchical relationships between them.
Taken together, lympho-myeloid progenitors have been described in the Lin-CD34+CD45RA+CD90- compartment that can be either CD38+ or CD38- and CD10+ or CD10-. This led us to directly, and rigorously, compare the in vitro and in vivo functional potential and transcriptional programs of human LMPP, MLP and GMP. We have shown these progenitors are distinct and heterogeneous. Single cell gene expression demonstrated a continuum of progenitors with lymphoid and myeloid potential downstream of stem cells. Using novel flow purification strategies, the bulk of multi-lineage lympho-myeloid progenitors were contained within a sub-compartment of LMPP.
Results
In vitro assays reveal the potential of distinct lympho-myeloid progenitors
We improved prior flow cytometric staining and sorting strategies21, 22 to prospectively purify eight human hemopoietic stem/progenitor cell (HSPC) populations (Supplementary Table 1, Supplementary Fig. 1a) in human cord blood (CB) and bone marrow (BM). These HSPC populations included: haematopoietic stem cells (HSC: Lin-CD34+CD38-CD90+CD45RA-CD10-), multipotent progenitors (MPP: Lin-CD34+CD38-CD90-CD45RA-CD10-), lymphoid-primed multipotent progenitor (LMPP: Lin-CD34+CD38-CD90-/loCD45RA+CD10-), multi-lymphoid progenitor (MLP: Lin-CD34+CD38-CD90-/loCD45RA+CD10+), common myeloid progenitor (CMP: Lin-CD34+CD38+CD123+CD45RA-CD10-), granulocyte macrophage progenitor (GMP: Lin-CD34+CD38+CD123+CD45RA+CD10-), megakaryocyte erythroid progenitor (MEP: Lin-CD34+CD38+CD123-CD45RA-CD10-) and B and NK cell progenitor (B/NK: Lin-CD34+CD38+CD90-CD45RA+CD10+). Within CB Lin-CD34+ cells these eight HSPC populations accounted for 82% of cells. The remaining cells did not constitute separate populations. The Lin-CD34+CD38-CD45RA+ compartment contained a mixture of CD10- LMPP and CD10+ MLP progenitors (Supplementary Fig. 1a). Furthermore, the more mature Lin-CD34+CD38+ compartment was separated into the CD10+ B-NK progenitor population32 and CD10- heterogeneous myeloid progenitors. Immunophenotypic LMPP and MLP were rare (Supplementary Table 1). Using analysis gates they constituted 0.2% of the BM Lin-CD34+ compartment and ~2/105 of BM mononuclear cells (MNCs). Though more frequent in CB, they still only constituted ~1/104 MNCs. GMPs were 20-fold more abundant in CB and 100-fold more abundant in BM than LMPPs and MLPs (~1.5-2/103 MNCs).
As the frequency of adult BM LMPP and MLP was extremely low, we used fresh CB cells as a source of HSPCs. Cells were double-sorted to high purity (>99%, except the CMP 97%). In methylcellulose-based colony forming unit assays the LMPP and MLP had low myeloid clonogenic potential (6% and <1%) compared to GMP (31%) (Fig. 1a-b). GMP and LMPP generated granulocytic (G), monocyte/macrophage (M) and GM colonies with either no, or minimal, erythroid (E) potential (<0.5%) (Fig. 1a). MLP only generated very few monocyte colonies (Fig. 1a), consistent with previous data15, 21, 22, 33.
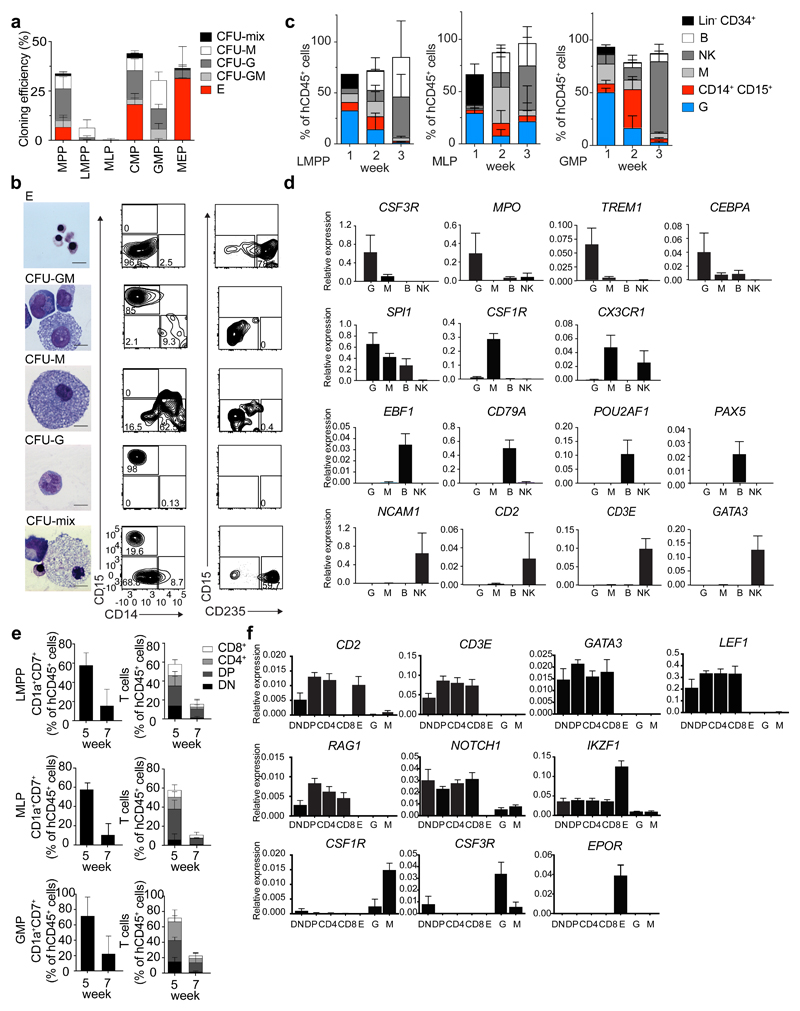
Figure 1. Human CB lympho-myeloid populations have distinct functional potential in vitro.
(a) Cloning efficiency and lineage affiliation of myelo-erythroid colonies in a CFU assay (150 CB HSPCs plated). Error bars are ± SD. n=5. CFU-mix, mixed erythro-myeloid colony; CFU-M, monocyte/macrophage colony; CFU-G, granulocyte colony; CFU-GM, granulocyte and monocyte/macrophage colony; E, erythroid colony (BFU-E and CFU-E). (b) Morphology of May-Grunwald-Giemsa stained cells from CFU assay (left, bar size 10 µm) and flow cytometric plots of cells harvested from indicated colony types (right). (c) Lineage output after culturing 150 LMPP, MLP and GMP cells for 1, 2 or 3 weeks on MS-5 stroma with SCF, G-CSF, FLT3L, IL15, IL2 and DuP-697. Data represent mean from 3 CB donors ± SD. Flow cytometric plots for two week cultures shown in Supplementary Fig. 1b. (d) Gene expression analysis of flow cytometric-purified output cells from (c). (e) T cell output after culturing LMPP, MLP and GMP cells in bulk for 5 or 7 weeks on OP9-hDL1 stroma with SCF, FLT3L and IL7. Data represent percentage from hCD45+ cells from 5 CB donors (mean± 1SD). DN, CD7+CD1a+ CD4-CD8-; DP, CD7+CD1a+CD4+CD8+; CD4, CD7+CD1a+CD4+CD8-; CD8, CD7+CD1a+CD4-CD8+. Flow cytometric plots for 5 week cultures shown in Supplementary Fig. 1c. (f) Gene expression analysis from flow-purified output cells from (e) and control mature non-T cells, obtained from sorting cells from E, G and M colonies.
We analyzed the lymphoid and myeloid differentiation potential of a population of 150 LMPP, MLP and GMPs using an optimized, new in vitro liquid culture on MS-5 stroma supplemented with Stem Cell Factor (SCF), Granulocyte Colony Stimulating Factor (G-CSF), Fms-Related Tyrosine Kinase 3 Ligand (FLT3L), Interleukin 2 (IL2), IL15 and DUP-697 (SGF15/2 condition). By performing a kinetic analysis of lineage outputs, we determined that 2 weeks was the optimal timing to detect hCD45+CD15+ neutrophils (G), hCD45+CD14+ monocytes (M), hCD45+CD19+ B cells and hCD45+CD56+ natural killer (NK) cells from the culture (Fig. 1c and Supplementary Fig. 1b). Flow cytometry purified G, M, B or NK cells from SGF15/2 in vitro culture expressed appropriate lineage-affiliated genes (Fig. 1d, Supplementary Table 2). Therefore, we analyzed all subsequent limiting dilution and single cell cultures at week 2 to capture all four myeloid (G, M) and lymphoid (B, NK) outputs. We tested T cell production of LMPP, MLP and GMP populations at weeks 5 and 7 using an in vitro liquid culture assay on OP9-hDL1 stroma with SCF, FLT3L and IL7 (SF7a condition, Fig 1e, Supplementary Fig. 1c). LMPP, GMP and MLP generated hCD7+CD1a+ immature T cells, more mature hCD7+CD1a+hCD4+CD8+ double positive (DP), and hCD7+CD1a+CD4-CD8+ and hCD7+CD1a+CD4+CD8- single positive T cells. Flow cytometric purified T cell subpopulations expressed appropriate lineage-affiliated genes (Fig 1f, Supplementary Table 2).
In summary, we established conditions to prospectively purify eight HSPC populations and test in vitro potential into myeloid and lymphoid lineages.
Functional heterogeneity of lympho-myeloid progenitors
We next used four in vitro liquid culture assays to test the clonal potential of CB LMPP, MLP and GMP (Supplementary Fig. 2a-b). First, limiting dilution assay (LDA) was performed in the SGF15/2 condition and lineage output assessed by flow cytometry (Supplementary Fig. 2c). 1 in 2 LMPP cells produced B cells, 1 in 3 NK cells, but only 1 in 5 monocytes and 1 in 10 granulocytes (Table 1, Supplementary Fig. 2d). GMPs generated myeloid cells with higher frequency (1 in 2 for M, 1 in 4 for G) and lymphoid cells at lower frequency (1 in 22 for B, 1 in 8 NK) (Table 1, Supplementary Fig. 2d). 1 in 11 MLP cells produced B cells and 1 in 18 generated NK cells, whereas myeloid output was rare (Table 1), indicating that MLPs were lymphoid-biased. Bi-lineage and multi-lineage cells were detected at lower frequencies (1 in 6 to 1 in 789) (Table 1, Supplementary Table 3a).
Table 1. Frequencies of lineage outputs from limiting dilution assay (LDA).
| Frequency | LMPP | MLP | GMP |
|---|---|---|---|
| B | 2 | 11 | 22 |
| NK | 3 | 18 | 8 |
| M | 5 | 194 | 2 |
| G | 10 | 394 | 4 |
| B_NK | 6 | 38 | 31 |
| M_G | 16 | ND | 7 |
| B_M | 10 | 392 | 32 |
| B_G | 13 | 789 | 38 |
| M_NK | 9 | 392 | 11 |
| NK_G | 15 | ND | 17 |
| B_NK_M | 16 | 392 | 44 |
| B_NK_G | 18 | ND | 49 |
| B_M_G | 22 | ND | 43 |
| NK_M_G | 23 | ND | 20 |
| B_NK_M_G | 29 | ND | 56 |
Shown as “1 in X cells can give rise to”. ND – not detected.
As limit dilution analysis does not rigorously define frequency of multi-lineage functional potential at a clonal level, we assessed lympho-myeloid (B, NK, G, M) potential, in the second assay, the optimized liquid culture SGF15/2 condition. We tested potential of 1136 LMPPs, 710 MLP and 1622 GMPs as single cells, isolated from 22 biological CB donors (totaling 6.3x109 MNCs), to provide robust quantitative data, especially for rare functional potentials (Supplementary Table 3b). At a single cell level, LMPP and GMP had higher cloning efficiency (54% and 71% respectively) than MLP (11%) (Fig. 2a). LMPP and MLP were primarily lymphoid progenitors, whereas GMP mainly a myeloid progenitor (Fig. 2a). Focusing on productive wells, 69% of LMPP, 88% of MLP and 63% of GMP gave uni-lineage output (Fig. 2b). When there was uni-lineage output, 92% of LMPP cells had lymphoid output (B or NK) and 8% myeloid output (G or M). The MLP was virtually exclusively a lymphoid progenitor with very low myeloid output (3%). 79% of GMP cells had myeloid and 21% lymphoid output. Bi-lineage output was detected in 24% of LMPP, 12% of MLP and 33% of GMP (Fig. 2c) and output of three or more lineages was rare (6% of LMPP, 0% of MLP and 3% of GMP) (Fig. 2d). Only 8% of all plated LMPPs, 7% of GMPs and hardly any MLPs (0.3%) exhibited combined lympho-myeloid potential (Fig. 2e). Lympho-myeloid output from LMPP was significantly higher compared to GMP (p=0.0125) and MLP (p=0.0019, Supplementary Table 3c).
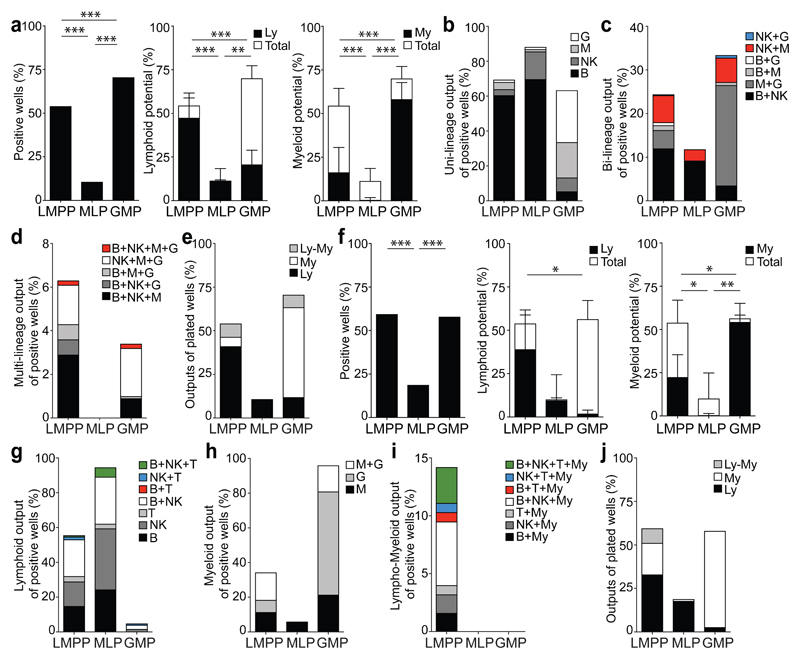
Figure 2. CB LMPP and GMP are lympho-myeloid progenitors, while MLP is mainly a lymphoid progenitor in clonal in vitro assays.
(a) Total cloning efficiency (left) of single LMPP, MLP and GMP in SGF15/2 condition (LMPP: 615/1136 cells, MLP: 76/710, GMP: 1145/1622). Significance defined by Fisher’s exact test. Cloning efficiency of lymphoid (Ly, middle) and myeloid lineages (My, right). Bars indicate total cloning efficiency; filled portion indicates the proportion of lymphoid (lymphoid plus mixed) or myeloid potential (myeloid plus mixed clones). Mean ± SD is shown. Significance is defined using students t-test. (b) Single-, (c) bi- and (d) multi-lineage outputs from single cells, presented as a percentage of positive wells in SGF15/2 condition. (e) Lymphoid (Ly), myeloid (My) and lympho-myeloid (Ly-My) outputs presented as a percentage of all plated cells in SGF15/2 condition. (f) Total cloning efficiency (left) of single cell progenitors in SF7b/Dox condition (LMPP: 128/215 cells, MLP: 37/197, GMP: 127/219). Cloning efficiency of lymphoid (middle) and myeloid lineages (right). Bars indicate total cloning efficiency; filled portion indicates the proportion of lymphoid or myeloid potential. Mean ± SD is shown. Significance is defined as in (a). (g) Single-, (h) bi- and (i) multi-lineage outputs from single cells, presented as a percentage of the positive wells in SF7b/Dox condition. (j) Lymphoid (Ly), myeloid (My) and lympho-myeloid (Ly-My) outputs presented as a percentage of all plated cells in SF7b/Dox condition. For the single cell assay in SGF15/2 condition: 22 CB donors; SF7b/Dox condition: 3 CB donors.
We also tested lympho-myeloid (B, NK, G, M) potential of 96 LMPPs, 52 MLPs and 110 GMPs as single cells, in a third in vitro liquid culture assay, on MS5 stroma with SCF, IL7, thrombopoietin (TPO), IL2, Granulocyte-Macrophage Colony Stimulating Factor (GM-CSF), G-CSF and Macrophage Colony Stimulating Factor (M-CSF) (S7T2GM/G/M condition) that was used to define the MLP21 (Supplementary Fig. 2b). Similar results to SGF15/2 condition were obtained but S7T2GM/G/M condition was less permissive for granulocytic output (Supplementary Fig. 2e-i, Supplementary Table 3d). Most output from LMPP was uni-lineage with rarer bi-lineage and less frequent multi-lineage outputs. MLP exhibited only lymphoid uni-lineage output.
Finally, we assessed the lympho-myeloid potential of 215 LMPP, 197 MLP and 219 GMP single cells in a fourth assay, with an independent culture condition, optimized for detecting combined lymphoid (B, NK, T) and myeloid (M, G) potential. Single LMPP, MLP and GMP were cultured on MS-5/hDL-1IND stroma with SCF, FLT3L, IL7 (condition SF7b/Dox) and the B-NK-M-G output was analyzed at 3 weeks and the T cell output at 6 weeks (Fig. 2f-j, Supplementary Table 3e). Uni-lineage T cell output was detected in LMPP and MLP populations (3% of positive wells) but was virtually absent in GMP (<0.1%) (Fig. 2g). T cell combined with other lymphoid output was detected in 1-5% of LMPP and MLP and rarely in GMP (Fig. 2g). Lympho-myeloid output was only detected in LMPP (14%) (Fig. 2i). Overall, 24 functionally different progenitor types were identified in the three single cell in vitro clonal assays; all 24 progenitor types were observed in the LMPP and only subsets of them were seen in MLP and GMP (Supplementary Fig. 2j).
Ossicle assay defines the in vivo potential of LMPP, MLP and GMP
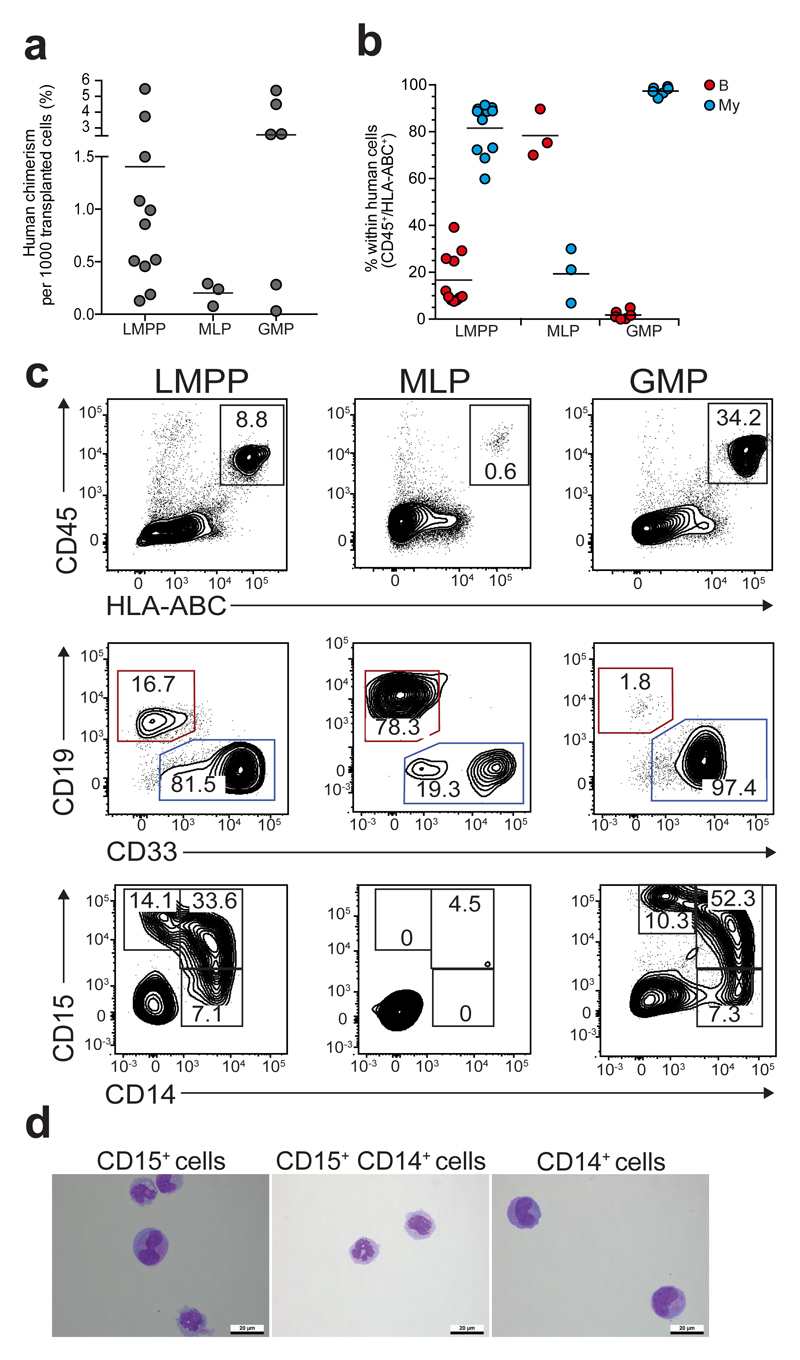
Successful single cell transplantation of human progenitors in xenotransplantation assays is not feasible. Furthermore, direct injection of progenitor cell populations into immunodeficient mice yields low (<0.1%) engraftment15, 21, 22, 25. Therefore, we tested in vivo progenitor function in new humanized ossicle model34. Human BM-derived mesenchymal stromal cells were subcutaneously injected into immunodeficient mice, where over 8 weeks they form a humanized ossicle. LMPP, MLP and GMP progenitors were injected into the ossicle and lineage output was analyzed 1 and 2 weeks post-injection (Supplementary Fig. 3a-b). Engraftment was detected at both time points, with greater hCD45+hCD33+hCD14+ (M), hCD45+hCD33+hCD15+ (G) and hCD45+hCD33-hCD19+ (B) engraftment at week 2 compared to week one (data not shown). All subsequent analyses were done at 2 weeks post-transplantation. As the number of cells injected varied (~300-60,000 cells depending on the progenitor subset, Supplementary Fig. 3c), we report mean cell engraftment per 1000 transplanted cells. GMPs had the highest mean engraftment (2.6%), followed by LMPP (1.4%) and MLP (0.2%). LMPP produced more CD33+ myeloid cells (82%) than CD19+ B cells (17%) (Fig 3b-c). MLP generated more B cells (78±5.9%) than myeloid cells (19±6.7%) (Fig 3b-c). There was no correlation between the number of the transplanted cells and the lympho-myeloid ratio (Supplementary Fig. 3d). GMP generated mainly myeloid cells (97%, Fig. 3b-c). Myeloid cells generated from LMPP and GMP expressed monocytic (CD14) and granulocytic (CD15) markers. No CD14+ and/or CD15+ cells were detected from MLPs (Fig. 3c). Morphology analysis of the engrafted cells confirmed CD15+ cells were granulocytic and CD14+ monocytic (Fig. 3d), Double positive CD14+CD15+ cells, generated by LMPP and GMP (Fig. 3c), were more immature myeloid cells by morphology (Fig. 3d). Thus, LMPP, MLP and GMP have different lymphoid and myeloid potentials in the humanized ossicle assay.
Figure 3. Human CB LMPP, MLP and GMP progenitors have distinct differentiation potential in vivo.
(a) Percentage human engraftment 2 weeks after progenitor transplantation, normalized to 1000 transplanted cells. (b) Percentage B and myeloid cells within human CD45+/HLA-ABC+ cells. (c) Representative flow cytometric plots of percentage human engraftment (CD45+HLA-ABC+), B cells (CD19+) and myeloid cells (CD33+), and percentage CD14+ and CD15+ myeloid cells 2 weeks after transplantation. Frequencies shown are an average from 11 CB donors for LMPP, 3 from MLP, 6 for GMP. (d) Representative images of May-Grunwald-Giemsa stained CD15+, CD15+/CD14+ and CD14+ myeloid cells generated by LMPP 2 weeks after transplantation, n=2.
Transcriptional programs of LMPP, MLP and GMP correlate with their functional potential
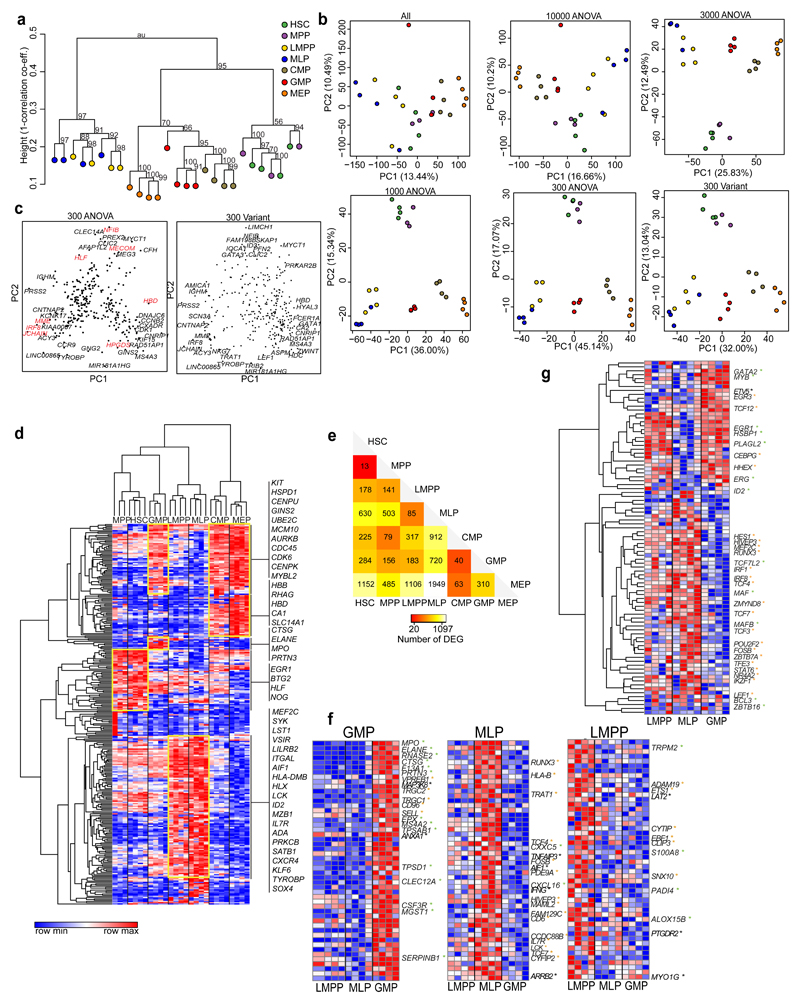
We performed RNA-sequencing of human CB HSPC populations (HSC, MPP, LMPP, MLP, CMP, GMP and MEP). Hierarchical clustering using all expressed genes separated LMPP and MLP from the other HSPCs. HSC and MPP clustered away from mature progenitors (Fig. 4a and Supplementary Fig. 4a). We used ANOVA analysis to obtain differentially expressed genes (DEG) between HSPC populations (Supplementary Table 4). We performed principal component analysis (PCA), using all expressed genes or between 300 to 10000 of the most DEG (Fig. 4b). The best separation of HSPC populations on a PCA plot was achieved using the 300 most DEG (Fig. 4b). Principal component (PC) 1 separated HSPCs by lineage potential and PC2 by maturation. By comparing the eigenvalues of the 300 most DEG with those from a randomized data set, we demonstrated that PCs 1-3 captured most of the variation between populations (Supplementary Fig. 4b-c). We also identified genes with highest variance across all populations without assuming population identity. PCA plot using this gene set gave similar results (Fig. 4b and Supplementary Fig. 4d). The loadings plot for the PCA using the ANOVA define 300 most DEG identified stem- (HLF, MECOM, NFIB), lymphoid- (IGJ, IRF8, MME) and erythroid-megakaryocytic-affiliated genes (HBD, HPGDS) (Fig. 4c). Hierarchical clustering using the 300 ANOVA gene set separated HSPC populations (Fig. 4d). ELANE, MPO and PRTN3 were most strongly expressed in the GMP, whereas the LMPP and MLP shared expression of many lymphoid-affiliated genes (e.g. IL7R, LCK, SYK, ADA, HLX, LST1 and ITGAL).
Figure 4. Distinct transcriptional patterns of human CB HSPC populations.
(a) Hierarchical clustering of HSPC populations using all genes and 1000 bootstrap permutation analyses; “au” = approximate unbiased p-values. Height values expressed as (1- [correlation co-efficient]). (b) PCA plots showing CB HSPC when using varying number of ANOVA genes (ranked by ANOVA p-value) and 300 most variant genes (bottom right). Percentage variance represented by each Principal Component (PC) is shown. (c) Loadings plot, showing the genes with the most extreme loadings scores for the PCA run with top 300 ANOVA (top) or variant (bottom) genes. (d) Heatmap showing hierarchical clustering and the expression of the top 300 ANOVA genes by HSPC populations. Clusters highlighted in yellow show distinct expression patterns across HSPC populations. Expression values are normalized per gene. (e) Summary of all differentially expressed genes between HSPC populations. (f-g) Heatmaps showing the expression of top 50 genes from the LMPP, MLP and GMP gene signatures (f) and transcription factors differentially expressed across HSPC populations (g). Genes affiliated with the lymphoid or myeloid lineages have color-coded asterix (lymphoid: orange, myeloid: green) and genes associated with immune function are labeled with black asterix. Expression values are normalized per gene. RNA seq data come from 4 CB donors (MPP: 3 donors).
Transcriptional relatedness between HSPC populations, without assuming any hierarchical relationships, was further analyzed through pairwise comparisons (Fig. 4e, Supplementary Fig. 4e, Supplementary Table 5-11). The most closely related populations were HSC and MPP (only 13 separating DEG), while LMPP and MLP were closely related (85 DEG). GMP were most closely related to the CMP (40 DEG), but retained a similarity to LMPP (183 DEG). We derived gene expression signatures for LMPP, MLP and GMP from DEG in one versus all population comparisons, filtered for uniquely expressed genes (Fig. 4f, Supplementary Table 12a-c). The GMP signature contained many myeloid genes and the MLP signature many lymphoid genes (Fig. 4d). By contrast, the LMPP signature contained both lymphoid (ETS1, EBF1, CYTIP) and myeloid genes (TRPM2, S100A8, PADI4, ALOX15B).
To validate these findings, we investigated the profiles of LMPP, GMP and MLP using recently published gene sets25. GMP expressed immature myeloid genes whereas LMPP and MLP expressed genes affiliated with B cells, monocytes and DCs, but not neutrophils (with the exception of FOSB) (Supplementary Fig. 4f). Additionally, the GMP was enriched for MetaCore pathways associated with myeloid maturation (e.g. granulocyte development: FDR=0.0136), whereas MLP was enriched for lymphopoiesis pathways (e.g. Notch signaling: FDR<0.001). The LMPP had more balanced enrichment for both lymphoid and myeloid pathways (e.g. M-CSF signaling: FDR<0.001 and BCR signaling: FDR=0.049) (Supplementary Table 13).
We used two approaches to pinpoint transcription factors (TFs) driving these programs. First, we identified TFs differentially expressed between the MLP and GMP (Supplementary Fig. 4g). Second, we examined expression of previously identified hematopoietic TFs35 (Fig. 4g). In both analyses, GMP expressed mainly myeloid TFs (e.g. ERG, GATA2, MYB, EGR1), while lymphoid TFs (e.g. HES1, RUNX3, POU2F2, LEF1, IKZF1, IRF8, TCF4) showed highest expression in MLP. LMPP showed balanced expression of both myeloid and lymphoid TFs. A similar trend was seen with cytokine and chemokine receptor genes (Supplementary Fig. 4h). Therefore, the transcriptional programs of LMPP, MLP and GMP reflect their functional potentials.
Single cell RNA analyses reveals a continuum of differentiation
To begin to separate distinct progenitors within the heterogeneous GMP, LMPP and MLP populations, we index flow sorted single cells for functional analysis, RNA-sequencing and quantitative RT-PCR (qRT-PCR). Index data allowed correlation of function and transcriptional state36 (Fig. 5a). First, we profiled expression of 96 genes, encoding lineage-affiliated transcriptional regulators, cell surface and lineage-affiliated markers (Supplementary Table 14), in a total of 919 single LMPPs, MLPs and GMPs. Genes with low variance and levels of detection were excluded. Expression of 74 genes was taken forward for analysis. Hierarchical clustering assigned GMPs, LMPPs and MLPs to three clusters (Supplementary Fig. 5a-b). Cluster 1 (543 cells) was mainly MLPs and LMPPs, cluster 2 (150 cells) was a mix of GMPs, LMPPs and MLPs, and cluster 3 (226 cells) mainly GMPs. Cluster 1 showed higher expression of lymphoid-affiliated genes, cluster 3 showed increased expression of myeloid genes (Supplementary Fig. 5b). Cluster 2 had a mixed lympho-myeloid expression profile. The cellular composition in each gene expression cluster mirrored the single cell functional output (Supplementary Fig. 5b).
Figure 5. Transcriptional heterogeneity of CB lympho-myeloid progenitor cells from single cell RNA-sequencing.
(a) Experimental scheme used to combine single cell functional analysis, single cell RNA-sequencing and single cell qRT-PCR based on flow cytometric index data. (b) Heatmap showing clustering of single LMPP, GMP and MLP using the 55 most highly and variably expressed genes between clusters. Heatmap shows clustering from one of two donors analyzed. Data from the other donor are in Supplementary Fig. 5d. Log-normalized gene expression (rows) for each single cell (columns) is shown. (c-d) PCA plot colored by cell type (c) or cluster membership (d).
We performed dimensionality reduction on gene expression data using a diffusion map method adapted for single cell data37,38. By indicating progenitor identity on the diffusion map (Supplementary Fig. 5c), MLP, LMPP and GMP cells form a continuum in agreement with the hierarchical clustering (Supplementary Fig. 5a). Next, we colored the diffusion map by cluster assignment (Supplementary Fig. 5c). Cluster 2 was positioned between clusters 1 and 3, in agreement with its mixed lympho-myeloid transcriptional signature (Supplementary Fig. 5b).
To overcome gene selection bias in qRT-PCR data, we performed single cell RNA-sequencing and correlated this with function of 91 LMPP, 110 MLP and 119 GMP from two different donors (157 and 163 from each donor). Clustering using the combined gene set, variable in both donors, identified 3 clusters (Fig. 5b and Supplementary Fig. 5d). Most cluster 1 cells were MLP; most cluster 3 cells were GMP, while cluster 2 was comprised of LMPP and GMP cells (Supplementary Fig. 5e). Cluster 1 showed high expression of lymphoid-affiliated genes (e.g. MME, JCHAIN and ABCA1). Cluster 3 showed increased expression of myeloid genes (e.g. CPA3, MPO and VIM). Cluster 2 showed a mixed transcriptional signature and increased expression of hematopoietic progenitor gene KIT. PCA revealed a transcriptional continuum of LMPP, MLP and GMP populations (Fig. 5c-d). Identical analysis on the second donor provided similar conclusions (Supplementary Fig. 5f). Overall, single cell transcriptional profiles of the LMPP, MLP and GMP suggest a continuum of lympho-myeloid differentiation in the currently defined LMPP, MLP and GMP.
Refined sorting strategies further purify the LMPP and GMP
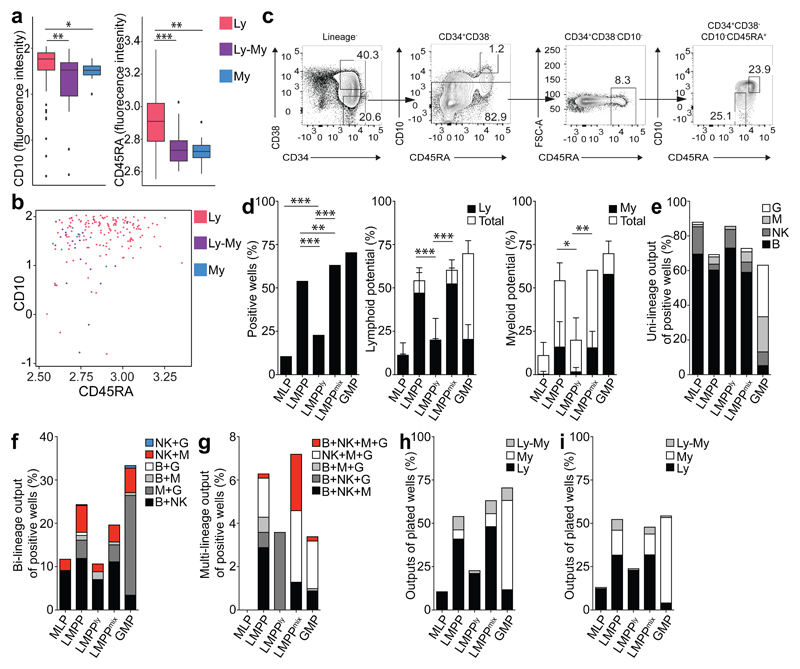
As our data showed that current flow sorting does not purify functionally homogenous populations, we correlated surface marker expression with function in the LMPPs and GMPs as they showed the greatest functional heterogeneity. Flow indexing data showed that single LMPP cells with lymphoid output had significantly higher CD10 and CD45RA expression compared to those with myeloid and lympho-myeloid output (Fig. 6a-b; CD10: Ly vs Ly-My p=0.0052, Ly vs My p=0.027; CD45RA: Ly vs Ly-My p=4.8x10-6, Ly vs My p=0.0027, Wilcoxon rank sum test). This was confirmed by higher CD10 expression in single LMPPs in lymphoid-biased cluster 1, compared to myeloid-biased cluster 3 (Supplementary Fig. 6a). Therefore, we developed a new LMPP flow sorting strategy to purify CD10hi CD45RAhi LMPP, here termed LMPPly, and CD10loCD45RAlo LMPPs, hereafter LMPPmix (Supplementary Fig. 6b), aiming to maximize the lymphoid-only and mixed myeloid and lympho-myeloid potential, respectively. 26% of total LMPP were LMPPly and 27% LMPPmix (Fig. 6c). When cultured in SGF15/2 conditions and analyzed after 2 weeks, LMPPly had significantly lower cloning efficiency compared to LMPP and LMPPmix but significantly higher than MLP (Fig. 6d and Supplementary Table 15; Fisher’s exact test p<0.0001 for all comparisons). LMPPly were lymphoid progenitors with virtually no myeloid potential and significantly lower myeloid potential than LMPP and LMPPmix (Fig. 6d and Supplementary Table 15; Fisher’s exact test p=0.0496 and p=0.0280 respectively). LMPPly had very small residual (1.6%) lymphoid-myeloid potential (Fig. 6h). LMPPmix cells retained virtually all the myeloid potential and most of the lympho-myeloid potential (Fig. 6e-h and Supplementary Table 15). This suggests that functionally LMPPly were intermediate between LMPP and MLP. This was confirmed using a second in vitro culture condition (SF7b) (Fig. 6i, Supplementary Fig. 6c-f and Supplementary Table 15).
Figure 6. New flow sorting strategy to purify functional potential within CB LMPP compartment.
(a) Logicle transformed CD10 and CD45RA surface marker levels in LMPPs, grouped by functional output. Ly - uni-lymphoid (B or NK) or bi-lymphoid output (B+NK), My - uni-myeloid (M or G) or bi-myeloid output (M+G), Ly-My - lympho-myeloid output. n=2 CB donors. (b) CD10 and CD45RA expression levels in LMPPs, measured by flow cytometry, colored by output from functional assays. Logicle-transformed data are from 2 CB donors. (c) Revised sorting strategy based on CD10 and CD45RA expression levels defined by bioinformatic analyses. Representative plots from 6 CB donors. (d) Total cloning efficiency (left) of single MLP, LMPP, LMPPly, LMPPmix and GMP in SGF15/2 condition (LMPPly: 56/244 cells, LMPPmix: 152/240). Significance defined using Fisher’s exact test. Cloning efficiency of lymphoid (Ly, middle) and myeloid lineages (My, right). Bars indicate total cloning efficiency; filled portion indicates the proportion of lymphoid potential (lymphoid plus mixed) or myeloid potential (myeloid plus mixed clones). Mean ± SD is shown. Significance is defined using students t-test. (e) Single-, (f) bi- and (g) multi-lineage outputs from single cells in SGF15/2 condition, presented as percentage of the positive wells. (h) Lymphoid (Ly), myeloid (My) and lympho-myeloid (Ly-My) outputs presented as percentage of all plated cells in SGF15/2 condition. (i) Lymphoid (Ly), myeloid (My) and lympho-myeloid (Ly-My) outputs presented as percentage of all plated MLP, LMPP, LMPPly, LMPPmix and GMP cells in SF7b condition. For SGF15/2 condition (d-h) data are from 6 CB donors (for LMPP, MLP and GMP controls - 22 CB donors (the same shown in Fig. 2a-e)). For SF7b condition (i) - 6 CB donors (for LMPP, MLP and GMP 9 CB donors (including 3 CB donors in Fig. 2f-j)).
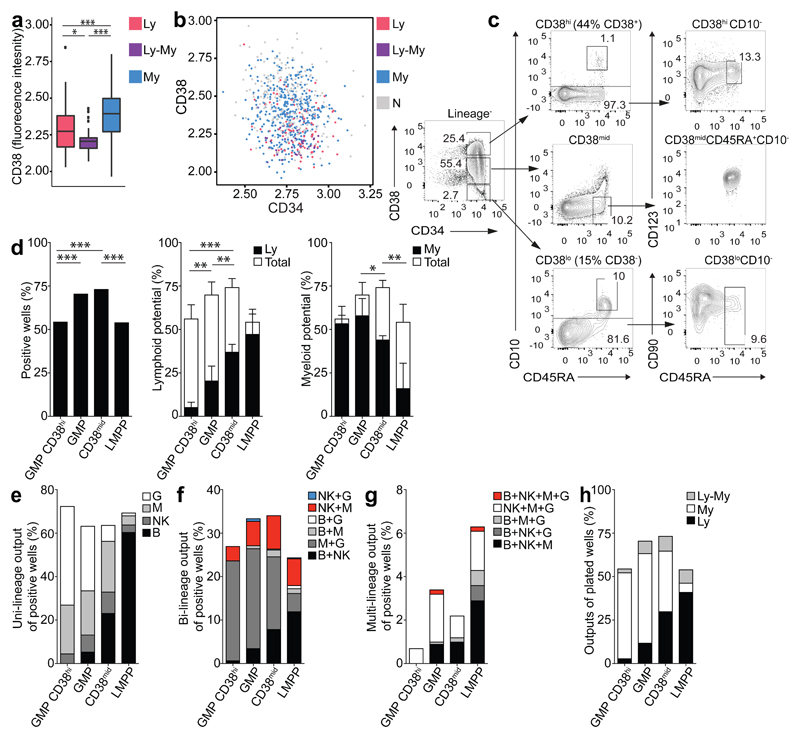
Based on flow indexing data, GMPs with myeloid-only output had significantly higher CD38 expression compared to those with lympho-myeloid or lymphoid output (Fig. 7a-b) (p=1.57x10-11 and p=1.6x10-8 respectively, Wilcoxon rank sum test). Concordantly, CD38 expression in single GMPs in cluster 3 (highest myeloid potential) had significantly higher CD38 expression compared to GMPs in clusters 1 (highest lymphoid potential) and 2 (lymphoid and myeloid potential). (Supplementary Fig. 6g). There was a significant positive correlation between CD38 expression and myeloid gene expression (MPO) and negative correlation between CD38 and lymphoid gene expression (MME and SELL) by single cell qRT-PCR (p=2.2x10-16, ρ=0.53 (MPO), p=7.1x10-5, ρ=-0.22 (MME), p=1.3x10-5, ρ=-0.24 (SELL), Spearman’s rank correlation coefficient, Supplementary Fig. 6h). To purify a GMP sub-population without lymphoid potential based on CD38 expression, we divided the entire Lin-CD34+ population into CD38hi (44% of CD38+), CD38lo (15% of CD38-) and CD38mid (area between the two new gates) (Fig. 7c, Supplementary Fig. 6i). CD38hi, CD38mid and CD38lo cells were further purified to isolate GMP CD38hi, CD38mid (CD38midCD45RA+CD10-) and LMPP CD38lo. LMPP CD38lo cells were rare (1 in 108 MNCs) and no conclusions could be reached about their functional potential. The in vitro lineage potential of single GMP CD38hi (279 cells) and CD38mid (693 cells) was compared to conventionally purified LMPP (1136 cells) and GMP cells (1622) using the SGF15/2 condition. Whereas the GMP CD38hi and LMPP had a similar cloning efficiency of ~55%, the GMP and CD38mid had a slightly higher cloning efficiency of ~70% (Fig. 7d). All four populations produced principally uni-lineage output (63-72%) (Fig. 7e). Compared to conventionally purified GMP, GMP CD38hi had drastically reduced lymphoid (Fisher’s exact test p<0.0001) and lympho-myeloid potential (Fisher’s exact test p=0.0115) (Fig. 7e-h and Supplementary Table 15), indicating a functionally purer population. In summary, the refined sorting strategy enabled purification of functionally homogeneous populations.
Figure 7. New flow cytometric sorting strategy to purify functional potential within CB GMP compartment.
(a) Logicle transformed CD38 surface marker expression levels in GMPs, grouped by functional output. n=5 CB donors. (b) CD38 and CD34 levels in GMPs colored by output from functional assays. Data are from 5 CB donors. (c) Revised sorting strategy, based on CD38 expression levels defined by bioinformatic analysis. Representative plots from 4 CB donors. (d) Total cloning efficiency (left) of the single GMP CD38hi, GMP, CD38mid and LMPP (GMPhi: 152/279 cells, CD38mid: 508/693). Significance defined using Fisher’s exact test. Cloning efficiency of lymphoid (Ly, middle) and myeloid lineages (My, right) of single cell GMP CD38hi, GMP, CD38mid and LMPP. Bars indicate total cloning efficiency; filled portion indicates the proportion of lymphoid (lymphoid plus mixed) or myeloid potential (myeloid plus mixed clones). Mean ± SD is shown. Significance is defined using students t-test. (e) Single-, (f) bi- and (g) multi-lineage outputs from single cells, presented as percentage of the positive wells. (h) Lymphoid (Ly), myeloid (My) and lympho-myeloid (Ly-My) outputs presented as a percentage of all plated GMP CD38hi, GMP, CD38mid and LMPP cells. For the functional assays (d-h), data are from 4 CB donors, for LMPP and GMP controls data are from 22 CB donors (the same shown in Fig. 2a-e).
Taken together, all our single cell observations suggest the progenitor hierarchy downstream of stem cells may be more complex than previous models have suggested (Supplementary Fig. 7).
Discussion
Here we report on the prospective separation and direct comparison of freshly isolated CB LMPP, MLP and GMP. Our results show these lympho-myeloid progenitors were functionally and transcriptionally distinct and heterogeneous at the single cell level. Though uni-lineage progenitors were most abundant, rarer multi-lineage lympho-myeloid progenitors were detected, most frequently in the LMPP. Single cell transcriptional analysis showed that LMPP, MLP and GMP form a transcriptional continuum, with MLP arcing from a lymphoid pole, and GMP from a myeloid pole, to intersect with the LMPP, positioned in the middle. By combining functional and transcriptional analyses with flow cytometric index data, we devised new flow purification strategies to isolate more functionally homogeneous populations within existing LMPP and GMP.
Several issues have prevented a clear understanding of previously identified human lympho-myeloid progenitors. First, these progenitors have been isolated using cell surface markers based on historical precedent rather than marker purifying to functional homogeneity. Second, prospectively isolated lympho-myeloid progenitor populations have never previously been systematically compared. Third, it is unclear if early progenitor populations downstream of HSC contain only uni-lineage cells16, 25 or also bi- and multi-lineage progenitors in the mouse5, 14, 17, 18, 26, 39 or human21, 22, 23, 24. Fourth, functional assays demonstrate potential rather than actual cell fate in vivo in steady state conditions. Finally, failure to register functional potential may reflect the inadequacy of an assay rather than the true potential, or indeed fate, of the cell in vivo. Thus, there is uncertainty about how distinct the differently identified progenitors are and if distinct, what their comparative functional potentials and transcriptional programs are at a clonal level.
Our exhaustive analysis of 4598 single LMPPs, MLPs and GMPs, as well as populations of these progenitors, showed that they were functionally different in vitro and in vivo when transplanted in mice with humanized ossicles. The novel humanized ossicle model allowed ~10-100-fold more human cell output than reported previously21, 23, 25. The GMP was primarily a myeloid progenitor with residual B and NK cell potential. Residual lymphoid potential could be virtually eliminated by purifying the highest 44% of CD38-expressing GMP cells. The MLP was primarily a B, NK and T cell progenitor with residual monocyte output. The LMPP had lymphoid and myeloid potential. Our new flow purification scheme divided the LMPP into two populations based on CD10 and CD45RA: one was almost entirely lymphoid, the other captured most of the myeloid/lympho-myeloid potential. Interestingly, the LMPP produced mainly myeloid cells in vivo. Humanized ossicles may be particularly efficient at promoting human myelopoiesis, unlike naive NSG mice, which better supports lymphopoiesis.
We detected 24 different lineage-affiliated potentials in lympho-myeloid progenitors, a likely underestimate, as we did not test for eosinophil, mast cell, basophil and dendritic cell function. Though the majority of progenitors were uni-lineage, bi- and multi-lineage output was seen (up to 39% and 13%, respectively, of cells in vitro). Lympho-myeloid lineage decisions could occur at multiple levels at the HSC1, 2, 3, MPP5, 14 and presumably more mature LMPP17, 18, 26, MLP and GMP39 populations. Within the LMPP and GMP, true lympho-myeloid progenitors could be rare (up to 10-14% of cells) and concentrated in the LMPP. Importantly, no experiments so far have directly examined the hierarchical relationships between lineage-biased HSC, MPP and lympho-myeloid progenitors. Quantitative differences in multi- versus uni-lineage output have been observed between fetal liver and BM in the broad CD34+CD38± populations16. All our data was in CB and similar experiments to those described here, will be needed to determine the ratio of uni-lineage versus bi- and multi-lineage progenitors in BM.
One separate question is whether diverse lineage-affiliated progenitors identified in vitro have stably different functions or whether there is plasticity such that functional output may be stochastically determined, or variably instructed. Further single cell functional analysis on potentially functionally purer populations will be required with detailed fate mapping in mice.
The rarity of LMPP and MLP (2/105 BM MNCs and 1/104 CB MNCs) and the minor proportion of multi- and bi-lineage progenitors within the LMPP prompted us to study large numbers of single cells to obtain robust information on rare bi- and multi-lineage potentials. The rarity of the LMPP is also noteworthy for two reasons. First, single cell RNA-sequencing programs39 of unfractionated MNCs will have to sequence large numbers of cells to provide adequate representation of these rare progenitors. Second, in acute myeloid leukemia (AML), leukaemic stem cells (LSC) are often arrested at an LMPP-like stage, where they can comprise up to 80% of MNCs22. Given this, we speculate that the small pool size of normal LMPP may be very tightly controlled to minimize oncogenic transformation. Additionally, understanding how normal LMPPs differentiate may provide insight into novel differentiating therapies for AML LMPP LSC.
Online Methods
Normal and patient samples collection
BM or CB samples from normal donors were obtained with informed consent (UK protocol MREC 06/Q1606/ or Administrative Panel on Human Subjects Research Institutional Review Board-approved protocols Stanford IRB no. 18329, no. 6453, and no. 5637). Fresh CB samples were purchased from NHS Cord Blood Bank, UK or from New York Blood Center. They were processed within 16-34h after collection. Mononuclear cells were isolated and CD34+ fraction was separated as described40. Fresh or frozen BM MNCs or CD34+ fractions were used. Human BM stromal cell were obtained from samples according to Medical University of Graz Ethikkommission (Institutional Review Board-approved protocol, MUG Graz IRB no. 19-252). BM mesenchymal stromal cells (MSCs) were isolated and expanded as described34.
Flow cytometric sorting of HSPC populations
Antibodies used for flow cytometric sorting and immunophenotyping are listed in Supplementary Table 16. CB or BM CD34+ enriched fraction was lineage depleted by staining with purified anti-human CD2, CD3, CD4, CD7, CD8a, CD11b, CD14, CD19, CD20, CD56, CD235a followed by Qdot 605 conjugated goat F(ab')2 anti-mouse IgG (H+L). Cells were also stained with anti-human CD38-FITC, CD45RA-PE or -BV650, CD123-PE Cy7, CD90-biotin, CD34-PerCP and CD10-APC. Finally, cells were incubated with streptavidin-conjugated APC-eF780 and Hoechst 33258 (Invitrogen, Loughborough UK; final concentration: 1 μg/ml). For humanized ossicle xenotransplantation assay CD34+ CB was stained with the same panel of anti-human lineage antibodies and anti-CD16. All lineage-antibodies were PE Cy5-conjugated. Cells were then stained with CD38-PE Cy7, CD90-FITC, CD123-PE, CD34-APC, CD10-APC Cy7, CD45RA-BV605 and propidium-iodide (Thermo Fisher, Waltham MA; final concentration: 1 μg/ml). Unstained, single stained and Fluorescence Minus One (FMO) controls were used to determine background staining and compensation in each channel. Single stained controls used anti-mouse compensation particle set (BD, Oxford UK). CB cells were sorted with average purity 99% for in vitro and RNA assays and 96% for humanized ossicle xenotransplantation. Prior to single-cell sorts, single fluorescent beads were deposited directly to a 96-well plate to establish accuracy of single cell deposition (>99%). Sorting was performed on BD Aria III or BD Fusion and flow cytometric analysis was done on LSR Fortessa X20. Data analysis was performed using Diva v8.1 or FlowJo v10.0.06 and v10.0.07r2.
Index sorting for functional and transcriptional analyses
For index sorting we saved information on the following parameters: FSC, SSC, Hoechst and expression of Lineage markers, CD34, CD38, CD45RA, CD10, CD90 and CD123 for each single cell. For 919 index sorted single cells we tested expression of 96 genes qRT-PCR (Supplementary Fig. 5); 74 passed QC. Separately, we performed single cell index sorting and single cell in vitro functional assays on 3458 single cells (from Fig. 2, Supplementary Fig. 2, Fig. 6, Fig. 7, Supplementary Fig. 6). In separate experiments we index sorted 320 single cells for single cell RNA seq (Fig. 5). Using common “position of the cells” in flow cytometric plots we could then map functional potential (i.e. lymphoid, myeloid or lympho-myeloid) to gene expression and cell surface marker expression and forward/side scatter. To purify LMPPly and LMPPmix the thresholds were defined based on maximum CD10 and CD45RA expression of LMPPs with myeloid output. To purify GMP CD38hi thresholds were set using the maximum normalized CD38 level of GMPs with myeloid output and for lympho-myeloid output.
In vitro lympho-myeloid differentiation assays (bulk, single cell, limiting dilution assay)
For population analysis, MS-5 cells were seeded on a 24-well plate coated with 0.1% gelatin at a density of 2x104 cells per well in α-MEM medium (Gibco/Thermo Fisher Scientific Loughborough UK) supplemented with 10% FBS (Hyclone, GE Healthcare, SH30070.03 Amersham Hatfield, UK), 1% Penicillin-Streptomycin, 1% L-Glutamine, 10-7M DuP-697 (Cayman Chemical, Ann Arbor, USA), 20 ng/ml SCF, 10 ng/ml G-CSF, 10 ng/ml FLT3L, 10 ng/ml IL15 and 10 ng/ml IL2 (Peprotech London UK, SGF15/2 condition). 24h after plating of MS-5 cells, 150 highly purified LMPPs, MLPs or GMPs were deposited in each well. Medium was half-changed every week. Harvested cells were flow cytometric analyzed at week 1, 2 and 3.
Limiting dilution assay (LDA) was performed by sorting LMPP, MLP or GMP cells at different cell doses (1, 2, 5, 10 and 20 cells) from 4 different CB samples into 96-well plates pre-plated with 2500 MS-5 cells per well with 100 µl of medium without cytokines. Immediately after sorting 100 µl of 2x SGF15/2 medium was added to each well. Medium was half-changed every week. A total of 833 LMPP, 789 MLP and 1252 GMP cells from 4 different CB samples were analyzed for the LDA at week 2 – 2.5 (Supplementary Table 2a). Frequency calculations were performed using L-Calc software (Stem Cell Technologies) and independently verified by ELDA software (http://bioinf.wehi.edu.au/software/elda/). The LDA plots were generated using R with lines representing the estimates calculated by ELDA software.
For single cell analysis single LMPP, MLP and GMP cells were deposited into 96-well plates pre-plated with 2500 MS-5 cells per well with 100 µl of medium without cytokines. Medium with 2x cytokines was added to each well after sorting. Medium was half-changed every week. After culture for 2-2.5 weeks flow cytometric analysis was performed and wells with more than 15 human CD15+, CD14+, CD56+ or CD19+ cells were scored positive (details in Supplementary Table 2b). To compare with previous published conditions21, single cell LMPP, MLP and GMPs were cultured for 4 weeks on MS-5 stroma in H5100 medium (StemCell Technologies Cambridge UK) supplemented with 100 ng/ml SCF, 20 ng/ml IL-7, 50 ng/ml TPO, 10 ng/ml IL-2, 20 ng/ml GM-CSF, 20 ng/ml G-CSF and 10 ng/ml M-CSF (all from Peprotech, S7T2GM/G/M condition) and analyzed by flow cytometric.
To read lineage readouts for all in vitro lympho-myeloid differentiation assays, harvested cells were stained with anti-human CD15-FITC, CD14-PE, CD19-PE Cy7, CD56-APC or -PE Cy5, CD45-APC Cy7 and in some cases with CD34-BV605.
In vitro T cell differentiation assay
OP9-hDL1 cells41 were seeded on a 24-well plate coated with 0.1% gelatin at a density of 2x104 cells per well in freshly prepared α-MEM medium (Gibco/Thermo Fisher Scientific, 12000-063) with 20% heat-inactivated FBS (Hyclone, GE Healthcare, SH30070.03 Amersham Hatfield, UK), 1% Penicillin-Streptomycin, 1% L-Glutamine, 10 ng/ml SCF, 5 ng/ml FLT3L and 5 ng/ml IL7 (Peprotech, London, UK, SF7a condition). 24h after OP9-hDL1 cell plating, 150 highly purified LMPP, MLP or GMP cells were deposited in each well. Cells were dissociated from wells and transferred to new plates with fresh OP9-hDL1 cells weekly. Harvested cells were flow cytometric analyzed at week 4, 5 and 7. Cells were stained with anti-human CD7-FITC, CD1a-PE, CD8-PE Cy7, CD4-APC and CD45-APC Cy7.
In vitro combined T-lympho-myeloid differentiation assay
MS5-hDL1IND100 cells42 (where hDL1 expression could be induced by adding doxycycline) were seeded on 96-well plates coated with 0.1% gelatin at a density of 2500 cells per well in 100 µl freshly prepared α-MEM medium (Gibco/Thermo Fisher Scientific, Loughborough UK) supplemented with 20% FBS (Hyclone, GE Healthcare, SH30070.03HI, Amersham Hatfield, UK), 1% Penicillin-Streptomycin, 1% L-Glutamine. 24h after plating of MS5-hDL1IND cells, single cell LMPP, MLP or GMP cells were deposited into each well and cultured in the presence of 20nM Insulin (Sigma-Aldrich, St Louis, MO), 50 ng/ml SCF, 20 ng/ml FLT3L and 10ng/ml IL7 (Peprotech London UK, SF7b condition). Fresh medium was added every week.
Cells were harvested at 21 days and split into two, half of them were used for flow cytometric analysis and the remaining half were re-seeded on MS5-hDL1IND100 cells and cultured in SF7b/Dox condition with doxycycline (1 μg/ml). Medium was half-changed twice every week. Fresh doxycycline was added to the cultures 3 times a week. At 42 days cells were harvested and flow cytometric analysis was performed. At 21 days wells with more than 8 human CD15+, CD14+, CD56+ or CD19+ cells were scored positive. At 42 days flow cytometric analysis using CD1a, CD7, CD4 and CD8 antibodies was performed and wells with more than 8 CD7+ cells were scored positive for T cells.
Colony Forming Unit assays
Colony formation was tested as before22. Colony identity was confirmed morphologically after cytospin (medium acceleration, 800 rpm 5 min May-Grunwald Giemsa stain (Sigma, Poole UK) and by flow cytometry with anti-human CD15-FITC, CD14-PE, CD235a-PE Cy5.
Humanized ossicle xenotransplantation assay
Protocol was performed as previously described34. In brief, in vitro expanded human BM-MSCs were harvested, resuspended in 60 μl of pooled human platelet lysate (pHPL) and admixed with 240 μl of matrigel-equivalent matrix. The whole matrix-cell mixtures were injected subcutaneously to generate humanized ossicle niches. 8-10 weeks post BM-MSC application transplants were evaluated for bone and marrow formation. Mice with established humanized ossicle niches were conditioned with 200 rad of irradiation 12-24 hours prior to transplantation. Different numbers of LMPP, MLP and GMP cells from at least 3 different CB donors (Supplementary Fig. 3c) were transplanted in total volume of 20 μl by direct intraossicle injection. Experiments were performed in accordance with a protocol approved by Stanford’s Administrative Panel on Laboratory Animal Care (no. 22264) and in adherence to the US National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. Normal multi-lineage engraftment was assessed 1-2 weeks after transplantation and defined by the presence of myeloid cells (CD33+) and B cells (CD19+) among engrafted human CD45+HLA-ABC+ cells. Engrafted mice were antibody stained with CD14-PE or -APC Cy7, CD15-FITC, HLA-ABC-FITC or -PB, CD19-APC, CD33-PE, CD45-V450.
RNA sequencing of bulk HSPC populations
100 highly purified HSPCs from normal CB samples were sorted directly into lysis buffer in RNAse inhibitor (Clontech St Germain-en-Laye France) and stored at -80°C before further processing. cDNA synthesis was done with Smarter Ultra low input RNA kit v1 (Clontech) as previously described43. Illumina libraries were generated using Nextera XT DNA sample preparation kit and Index Kit (Illumina Chesterford UK). Library size and quality were checked using Agilent High-Sensitivity DNA chip with Agilent Bioanalyser (Agilent Technologies Stockport UK). Concentration of indexed libraries was determined using Qubit High-Sensitivity DNA kit (Invitrogen Loughborough, UK). Libraries were pooled to a final concentration of 5-14 nM and were sequenced on an Illumina HiSeq 2000 single-end 50bp reads.
Bulk and single cell gene expression analysis by Dynamic Arrays
Gene expression analysis was performed as described40. TaqMan assays (Applied Biosystems) are listed in Supplementary Table 2 and 8.
Single cell RNA sequencing
Single cell libraries for RNA sequencing were prepared using the Smart-seq2 protocol44, where 23 cycles were used for the cDNA library preamplification. Illumina Nextera XT DNA sample preparation kit and Index Kit (Illumina Chesterford UK) was used for cDNA tagmentation and indexing. ERCC RNA Spike-In Mix (Ambion) was added to the lysis mix at a final dilution of 1:80,000,000. Library size, quality and concentration were checked as done for the bulk RNA sequencing. Libraries were pooled to a final concentration of 7-28 nM and 78 to 95 single cell libraries were combined per pool. Sequencing was done on HiSeq4000 using 75bp paired-end reads. Each pool contained a library generated from an empty well.
Bioinformatic analysis (bulk RNA seq, single cell Biomark and single cell RNA seq)
For 50 bp single end bulk RNA sequencing, alignment to the hg38 reference genome was carried out using TopHat v2.0.1045. Alignments were processed using Picard tools (http://picard.sourceforge.net/). We used R version 3.1.1 http://www.R-project.org. Sequencing reads were filtered for mapq 4 i.e. uniquely mapping reads. This gave a range of 15.1 x 106 to 56.2 x 106 aligned reads. The total number of genes expressed per sample was calculated as an rpkm>1. The number of expressed genes ranged from 7,707 to 11,350, with an average of 9,800. The count matrix was transformed to log2(cpm) scale and Principal Component Analysis was carried out. An ANOVA-like test was performed, using edgeR package for R, to identify differentially expressed genes between the populations. One CB biological replicate MPP population was excluded because when compared to the 3 remaining MPP biological replicates its global gene expression showed higher number of uniquely expressed genes and low correlation to the other three replicates. The genes were ranked by their significance (p-value adjusted for multiple testing) and different numbers of genes were used for PCA and hierarchical clustering of samples. Eigenvalues from PCA were calculated by using the square of the standard deviation of the principle components. Differential gene expression for one versus one and one versus all comparisons were calculated using edgeR. For gene signature generation a cut-off of logFC>1 was used and genes ranked based on p-value. Heatmaps and associated hierarchical clustering were generated using GENE-E software (Broad Institute) or using the R packages pvclust and heatmap.2 (gplots). MetaCore Pathway Map (Thomson Reuters, London UK) enrichment analysis was carried out on genes differentially expressed by each lympho-myeloid population versus all other populations (one versus all). A p-value cut-off of 0.05 was used to identify positively enriched pathway maps.
Analysis of single cell Biomark data was performed in R version 3.3.1 using data exported from the Fluidigm Data Collection software. For quality control, amplification curves with a Quality Score of <0.65 and any Ct values >27 were treated as undetected expression. Any cells where expression of both B2M and GAPDH housekeeping genes was not detected were removed from further analysis (n=7). An additional cell was removed as it had a high outlying number of genes detected. Housekeeping gene ACTB was also measured in the assay, but unlike B2M and GAPDH did not show robust expression across the majority of cells and therefore was not used in further analysis. Normalized ΔCt values were calculated by subtracting the mean of Ct values for B2M and GAPDH in each cell, as previously described19. Housekeeping genes were excluded from further analysis. Genes detected in <20 cells, with variance <1 across all cells or expressed in none of the MLP, GMP or LMPP 10 cell control samples assayed by qRT-PCR alongside single-cell samples were removed from downstream analysis. Post quality control data measured 74 genes in 919 single cells.
Hierarchical clustering was performed on genes and cells by using the hclust function (stats package) with distance measure 1 – Spearman’s correlation and agglomeration method Ward.D2. The heatmap visualizing the clustering was plotted using the heatmap.2 function (gplots package). Cells were divided into three clusters using the cutree function (stats package) on the hierarchical clustering. A gene was classed as differentially expressed between two clusters if it satisfied two criteria: 1) the magnitude of the log2 fold change of mean ΔCt in each cluster was >1 and 2) the adjusted p-value (Benjamini & Hochberg correction for multiple testing) of 2-sided Wilcox test of ΔCt expression values between the two clusters was < 0.01. Diffusion maps46 were used for dimensionality reduction of the single cell gene expression data. This method was implemented using the DiffusionMap function from the destiny R package with Euclidean distance37, 47.
Single cell RNA sequencing reads were aligned using G-SNAP48 and mapped reads were assigned to Ensembl genes (release 8149) by using HTSeq50. Cells with fewer than 500,000 reads mapping to nuclear genes, greater than 20% of mapped reads mapping to mitochondrial genes, greater than 20% of mapped reads mapping to External RNA Controls Consortium (ERCC) spike-ins or with expression of fewer than 750 different genes with at least 10 counts were removed from further analysis. ERCC spike-in controls identified genes exceeding technical variance51. From donors 1 and 2, 163/166 and 157/249 cells passed quality control, respectively. Single cell profiles were normalized using the scran R package52 and variable genes were identified as having variation exceeding technical levels51. Data showed batch effects between different donors. The Seurat R package (https://github.com/satijalab/seurat) was then used to regress out plate effects from the sequencing data, and set more stringent thresholds for variable genes, leading to 1,605 variable genes in donor 1 and 1,273 variable genes in donor 2. Principal component analysis was performed using Seurat, and clusters found using the Seurat::FindClusters function on the first 10 principal components. Heatmaps display the top genes marking these clusters as identified by the Seurat::FindAllMarkers function and were visualized using the gplots::heatmap.2 function.
Statistical analysis
Frequency of populations in flow cytometric plots gates is the mean of the population across all samples analyzed as indicated. Bar graphs of gene expression analysis represent mean +/-SEM or +/-SD as indicated. Two-tailed students unpaired t-test and Fisher's exact test (Excel, GraphPad sortware) were used to determine statistical significance in gene expression analysis data and single cell functional assays respectively. The statistical significance of the P-value was defined as follows for all P-value comparisons made: P>0.05 - not significant, P =0.01-0.05 - significant (*), P= 0.001-0.01 - very significant (**), P<0.001 - extremely significant (***). Wilcoxon rank sum test was done using R. Kruskal-Wallis test, stratified by group was used to define significant differences between LMPP, MLP and GMP in the single cell functional assay in SGF15/2 condition and gave the following p-values: LMPP - 5x10-6, MLP - 0.1725, GMP - 0.7395. Wilcoxon rank sum test confirmed that there was no outlier among single cell LMPPs coming from different CB donors. Prism software was used to plot the gene expression analysis and single cell in vitro data. LDA plots were generated using R and the lines represent the estimates calculated using ELDA software.
A Life Sciences Reporting Summary for this paper is available.
Data availability
Bulk RNA sequencing data have been deposited in Arrayexpress (https://www.ebi.ac.uk/arrayexpress/) with accession number E-MTAB-5456. Single cell RNA sequencing data accession number: GSE100618. All other source data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgements
We thank J. C. Zuniga-Pflucker for OP9-DL4 cells used for initial experiments. We would like to acknowledge funding from the MRC (to PV: MHU Award G1000729, MRC Disease Team Award 4050189188), CRUK (Program Grant to PV C7893/A12796, CRUK program grant to BG C1163/A21762, CRUK Development Fund to DK and PV CRUKDF0176-DK, CRUK Development Fund to BS and PV C5255/A20758), Bloodwise (Specialist Program 13001 and Project grant 12019), an MRC PhD studentship (ZA & FH), The MRC Single Cell Award (MR/M00919X/1) and the Oxford Partnership Comprehensive Biomedical Research Centre (NIHR BRC Funding scheme). We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z) for generation of sequencing data. RM was supported by National Institutes of Health grants R01CA188055 and U01HL099999, New York Stem Cell Foundation Robertson Investigator and Leukemia and Lymphoma Society Scholar Award. AR was supported by an Erwin-Schroedinger Research fellowship from the Austrian Science Fund (FWF).
Footnotes
Author Contributions
D.K., B.S., Z.A., and P.V. designed the experiments; D.K., B.S., Z.A., A.R., M.S., L.Q., and N.G. performed experiments and analyzed data; F.H., G.O., Z.A., E.R. and S.T. performed bioinformatics and statistical analysis; J.D. and B.U. prepared samples; J.C., E.S., F.P, R.M., C.P. and B.G., provided reagents and materials; D.K., B.S. and P.V wrote the paper; All authors edited the manuscript.
Competing Financial Interests
Nil
References
- 1.Dykstra B, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 2.Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6:265–278. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benz C, et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 2012;10:273–283. doi: 10.1016/j.stem.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Chen JY, et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature. 2016;530:223–227. doi: 10.1038/nature16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanjuan-Pla A, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502:232–236. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto R, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Sun J, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busch K, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015 doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 10.Sawai CM, et al. Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity. 2016;45:597–609. doi: 10.1016/j.immuni.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu VW, et al. Epigenetic Memory Underlies Cell-Autonomous Heterogeneous Behavior of Hematopoietic Stem Cells. Cell. 2016;167:1310–1322 e1317. doi: 10.1016/j.cell.2016.10.045. [DOI] [PubMed] [Google Scholar]
- 12.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 13.Cabezas-Wallscheid N, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15:507–522. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Pietras EM, et al. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell. 2015;17:35–46. doi: 10.1016/j.stem.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Notta F, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016;351:aab2116. doi: 10.1126/science.aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Mansson R, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Guo G, et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell. 2013;13:492–505. doi: 10.1016/j.stem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naik SH, et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496:229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- 21.Doulatov S, et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 22.Goardon N, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Kohn LA, et al. Lymphoid priming in human bone marrow begins before expression of CD10 with upregulation of L-selectin. Nat Immunol. 2012;13:963–971. doi: 10.1038/ni.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorgens A, et al. Multipotent Hematopoietic Progenitors Divide Asymmetrically to Create Progenitors of the Lymphomyeloid and Erythromyeloid Lineages. Stem Cell Reports. 2015;5:154–155. doi: 10.1016/j.stemcr.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velten L, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol. 2017;19:271–281. doi: 10.1038/ncb3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perie L, Duffy KR, Kok L, de Boer RJ, Schumacher TN. The Branching Point in Erythro-Myeloid Differentiation. Cell. 2015;163:1655–1662. doi: 10.1016/j.cell.2015.11.059. [DOI] [PubMed] [Google Scholar]
- 27.Berardi AC, et al. Individual CD34+CD38lowCD19-CD10- progenitor cells from human cord blood generate B lymphocytes and granulocytes. Blood. 1997;89:3554–3564. [PubMed] [Google Scholar]
- 28.Ichii M, et al. The density of CD10 corresponds to commitment and progression in the human B lymphoid lineage. PLoS One. 2010;5:e12954. doi: 10.1371/journal.pone.0012954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farlik M, et al. DNA Methylation Dynamics of Human Hematopoietic Stem Cell Differentiation. Cell Stem Cell. 2016;19:808–822. doi: 10.1016/j.stem.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, et al. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J Exp Med. 2015;212:385–399. doi: 10.1084/jem.20141442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pronk CJ, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Galy A, Travis M, Cen D, Chen B. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- 33.Manz MG, Miyamoto T, Akashi K, Weissman IL. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci U S A. 2002;99:11872–11877. doi: 10.1073/pnas.172384399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinisch A, et al. A humanized bone marrow ossicle xenotransplantation model enables improved engraftment of healthy and leukemic human hematopoietic cells. Nat Med. 2016;22:812–821. doi: 10.1038/nm.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novershtern N, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson NK, et al. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell. 2015;16:712–724. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haghverdi L, Buettner F, Theis FJ. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 2015;31:2989–2998. doi: 10.1093/bioinformatics/btv325. [DOI] [PubMed] [Google Scholar]
- 38.Moignard V, et al. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotechnol. 2015;33:269–276. doi: 10.1038/nbt.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul F, et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 40.Quek L, et al. Genetically distinct leukemic stem cells in human CD34- acute myeloid leukemia are arrested at a hemopoietic precursor-like stage. J Exp Med. 2016;213:1513–1535. doi: 10.1084/jem.20151775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Six EM, et al. Cytokines and culture medium have a major impact on human in vitro T-cell differentiation. Blood Cells Mol Dis. 2011;47:72–78. doi: 10.1016/j.bcmd.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Calvo J, BenYoucef A, Baijer J, Rouyez MC, Pflumio F. Assessment of human multi-potent hematopoietic stem/progenitor cell potential using a single in vitro screening system. PLoS One. 2012;7:e50495. doi: 10.1371/journal.pone.0050495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woll PS, et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell. 2014;25:794–808. doi: 10.1016/j.ccr.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 44.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 45.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coifman RR, et al. Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc Natl Acad Sci U S A. 2005;102:7426–7431. doi: 10.1073/pnas.0500334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Angerer P, et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 48.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yates A, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 52.Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk RNA sequencing data have been deposited in Arrayexpress (https://www.ebi.ac.uk/arrayexpress/) with accession number E-MTAB-5456. Single cell RNA sequencing data accession number: GSE100618. All other source data that support the findings of this study are available from the corresponding author upon request.