Abstract
Regeneration is a fascinating phenomenon that allows organisms to replace or repair damaged organs or tissues. This ability occurs to varying extents among metazoans. The rebuilding of the damaged structure depends on regenerative proliferation that must be accompanied by proper cell fate respecification and patterning. These cellular processes are regulated by the action of different signaling pathways that are activated in response to the damage. The imaginal discs of Drosophila melanogaster have the ability to regenerate and have been extensively used as a model system to study regeneration. Drosophila provides an opportunity to use powerful genetic tools to address fundamental problems about the genetic mechanisms involved in organ regeneration. Different studies in Drosophila have helped to elucidate the genes and signaling pathways that initiate regeneration, promote regenerative growth, and induce cell fate respecification. Here we review the signaling networks involved in regulating the variety of cellular responses that are required for discs regeneration.
1. Introduction
Regeneration is the ability that presents some organisms and allows them to partially or fully replace missing or damaged organs. This capacity is conserved among different phyla and it involves a wide range of processes, from wound healing to the induction of regenerative growth to reconstruct a whole new organ, as in urodele amphibians [1, 2]. Studying regeneration should shed light on the mechanisms that regulate this process, paving the way for their potential therapeutic applications in regenerative medicine.
Drosophila melanogaster is a powerful model system to perform genetic analysis, and it has provided much of the information of our understanding about the genetic basis of organ morphogenesis. While the adult organs of Drosophila are incapable of regenerating, the primordia of these structures, known as imaginal discs, can undergo regenerative growth. Imaginal discs are epithelial sac-like structures that develop from the embryonic ectoderm and after a period of cell proliferation in the larval stages, they give rise to the adult cuticle.
A series of classic experiments from the mid-1940s to the 1970s laid the groundwork for our current understanding of regeneration in Drosophila imaginal discs [3–5]. The classic approach used to study regeneration in Drosophila was to grow the regenerating discs in vivo culture. The disc was extracted from the larva and after amputating a fragment of it, it was transplanted into the abdomen of an adult host, where the cells of the discs could proliferate and the disc would regenerate [3, 6]. After regeneration the disc was then extracted from the adult host and examined [5, 7].
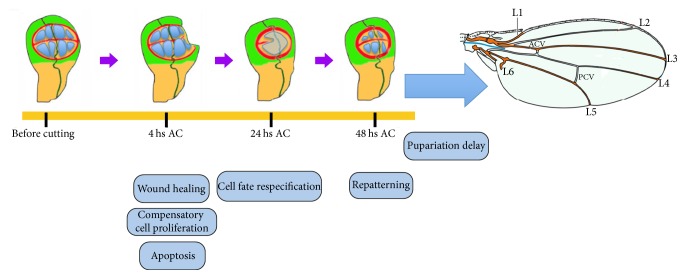
One of the problems of these studies is that regeneration did not occur under physiological conditions, since the disc regenerates in an adult host. Moreover, the process of disc extraction and transplantation will cause some stress to the cells of the discs, provoking apoptosis and halting cell proliferation in the first few hours after transplantation [8]. An alternative approach based on the transient induction of cell death in specific regions of the discs has resolved some of these problems. This method implies the use of the Gal4/UAS binary system in combination with Gal80ts to express a cell-death inducer, which makes it possible to genetically ablate a region of the disc in vivo for a predetermined period of time, after which the disc recovers [9, 10]. Although this technique mimics some aspects of surgical amputation, there are important differences between these two approaches. For instance, unlike amputation the overexpression of a cell-death inducer may not be sufficient to eliminate all the cells in the region targeted, and thus dying cells will coexist with living cells during the period of recovery. A further method has been developed to study disc regeneration in its normal developmental context. This system consists of removing a section of the disc “in situ” inside the larvae without extracting the discs from the larvae [11, 12]. The results obtained using these different approaches revealed that the initial stages of disc regeneration involve different processes. Thus, after wound healing a zone with a high rate of cell proliferation appears at the edges of the wound, similar to the blastema that originates during limb regeneration in amphibians and teleost fish. In addition, there is a temporary loss of markers of cell fate commitment and repatterning (Figure 1) [5, 7, 10, 12, 13]. All these cellular processes are triggered and regulated by the action of different signaling pathways. In this article we will focus on the signaling networks that regulate the various cell responses required to control the early stages of imaginal disc regeneration.
Figure 1.
Drosophila wing imaginal discs regeneration process. After a cut or genetic ablation in the third instar wing imaginal discs various cellular processes occur. First, four hours after cut (AC), the wound heals restoring the epithelial continuity. Around 24 hours AC, some cells lose expression of markers of cell fate commitment. Finally, the pattern is restored and the discs give rise to a normal pattern and sized adult wings. However, there is a delay in pupariation to allow the tissue to regenerate.
2. Reactive Oxygen Species (Ros) Are Induced in Response to Disc Damage
One of the first reactions of the imaginal disc in response to damage is the production of reactive oxygen species (Ros). Ros have been increasingly implicated in the physiological regulation of many developmental processes, including the emergence of stem cells, inflammatory cell recruitment, or the differentiation of embryonic cardiomyocytes. Ros act at many distinct levels in biological processes, affecting gene expression, protein translation, and protein-protein interactions [14–17]. Ros are the by-products of aerobic metabolism and they include superoxide (O2−) and hydrogen peroxide (H2O2). There are numerous potential sources of Ros within the cells and various organelles within the cell can produce Ros. One important generator of oxidant is a family of NADPH-dependent oxidases (Nox/Duox). These transmembrane proteins regulate the generation of Ros and an increase in their activity is produced in response to different stimuli after damage, such as the liberation of Ca2+. Indeed, wounding in Drosophila embryos provokes the binding of Ca2+ to Duox and its activation [18].
The specific effects of Ros are largely modulated by the reversible oxidation and reduction of reactive Cys residues, which in turn provokes the reversible modification of enzymatic activity of redox-sensitive targets, such as Tyrosine phosphatases or kinases [16, 17].
There is increasing evidence indicating that, during regeneration in vertebrates, the response to damage involves oxidative stress and, consequently, the stimulation of stress-activated protein kinases [19–21]. Ros have been also proposed to play a key role in disc regeneration [22–27]. Physical injury or genetic ablation of part of the imaginal disc provokes Ros production in cells at the wound site [22–25]. The initial burst of Ros acts as a chemoattractant for macrophages and it is necessary for the activation of different signaling pathways [22, 25, 27] (reviewed in [26]) (Figure 2). It is not clear how the first burst of Ros is generated, although different mechanisms have been identified that contribute to the increase in Ros levels in response to damage (see below). Shortly after trauma, Ros is also detected in cells adjacent to the injured region, although at a lower concentration [22].
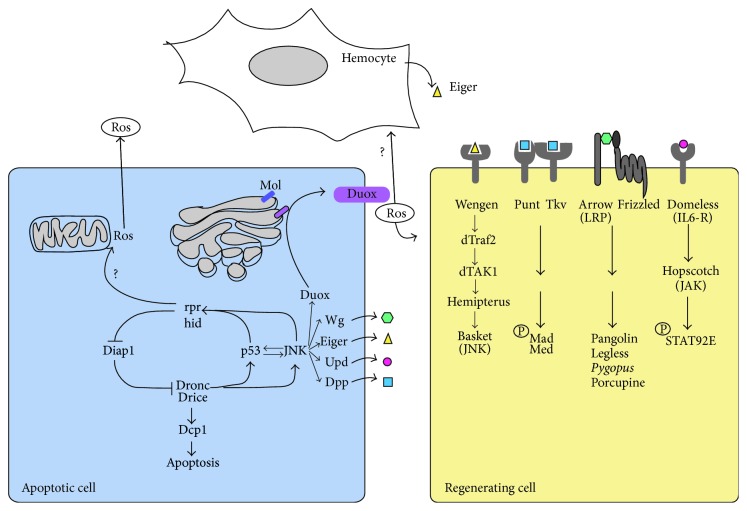
Figure 2.
Signaling network promoting apoptosis and Ros production after discs damage. Damage causes the activation of different signals that lead to apoptosis and high levels of Ros accumulation in dying cells. The function of Duox leads to extracellular Ros production that is responsible for hemocyte recruitment and promotes the activation of different signaling pathways in surrounding cells. Hemocytes secrete Eiger, which activates JNK pathway in the adjacent cells. Apoptotic cells produce also different signals that influence surrounding cells.
3. Activation of JNK Signaling Promotes Ros Production and Triggers Multiple Responses
JNK signaling is initially triggered at the wound site in response to Ros, and probably by other cellular stress signals [28]. Ros has been proposed to regulate the activity of MAP3K (MEKK1) and apoptosis signal-regulating kinase 1 (ASK1 or MAP3K5), kinases that reside upstream of JNK [16, 17, 29–31]. In addition, Ros can also block the function of the MAP kinase phosphatases that inhibit JNK signaling [30, 32]. JNK signaling promotes the activation of proapoptotic genes like reaper (rpr), head involution defective (hid), and grim. The proteins encoded by these genes bind to and inhibit the activity of Drosophila IAP-1 (Diap1), which in turn blocks the initiator caspase-9 orthologue Dronc (Drosophila NEDD2-like caspase). Dronc activates the effector caspases Dcp1 and Drice (Drosophila interleukin converting enzyme), inducing apoptosis. It has been shown that rpr and hid alter cytochrome Cytochrome C in the mitochondria, leading to mitochondrial disruption [33–36]. While the origin of Ros after damage remains unclear, the action of rpr and hid on mitochondria could favor Ros production by apoptotic cells (Figure 2).
The tumor suppressor Dp53 is another factor activated by JNK signaling and that is necessary to trigger apoptosis, playing a fundamental role in the elimination of cells that cannot complete DNA repair [37]. Both dp53 and JNK can activate each other in a Dronc-independent manner, and they establish a feedback loop that amplifies the initial apoptotic signals [38]. This loop is very important to promote cell death in response to the activation of the apoptotic pathway (Figure 2) [38]. Dp53 is required to induce compensatory proliferation and to reestablish the patterning of the damaged discs. Interestingly, it has been proposed that these functions are not dependent on apoptosis [39, 40].
JNK signaling also increases the levels of Ros by transcriptionally activating the gene moladietz (mol) [23]. This gene encodes the Duox-maturation factor NIP that is required for the production of Ros. Therefore, the activation of mol favors the production of Ros and it promotes a positive feedback signal that ensures the prolonged JNK activation necessary for regenerative growth (Figure 2) [23, 25]. Ros are also involved in the activation of Cap-n-collar (cnc). The transcriptional targets of cnc constrain Ros levels within a range in which regeneration is most efficient [24]. The cells with the highest levels of JNK signaling die and produce high levels of Ros. Indeed, at the wound edge there are apoptotic cells that have high levels of Ros in conjunction with high levels of JNK [22]. Ros propagate from dying or dead cells to the nearby surviving cells, and they activate multiple factors and signaling pathways. Indeed, Ros can propagate from cell to cell through aquaporins [41] or gap junctions [42].
Like other signals generated by apoptotic cells, Ros can affect surrounding cells. Various studies have shown that apoptotic cells can influence the proliferation and survival of nearby surviving cells. Indeed, apoptotic cells have been proposed to send signals that induce surrounding cells to divide or to die; events are known as apoptosis-induced proliferation (AiP) or apoptosis-induced apoptosis (AiA) [43]. It has been suggested that apoptotic cells can release mitogenic factors, such as the Drosophila Wnt1 homologue Wingless (Wg), the bone morphogenetic protein (BMP) Decapentaplegic (Dpp), and the leptin-like (IL-6 family) cytokine ligands Unpaired proteins (Upd, Upd2, Upd3) (Figure 2) (see below). Therefore, apoptosis at the wound site might fulfill a fundamental role in regulating regenerative proliferation. However it has been shown that the partial inhibition of apoptosis does not have a major effect on disc regeneration [8, 44]. A possible explanation for this paradox might be that apoptosis was not fully suppressed in any of these analysis. Thus, the signals emitted by the few apoptotic cells that remain might be sufficient to induce proliferation of the surrounding cells. More studies will be necessary to clarify the true role of apoptotic cells during regeneration.
4. P38 and JNK Signaling Promotes JAK/STAT Activation
Ros generated during apoptosis promotes tolerable levels of JNK in nearby surviving cells. Thus, in addition to apoptotic cells at the wound site, nonapoptotic cells also appear that have nondeleterious levels of JNK and low levels of Ros [22]. Hemocytes stimulated by Ros are also involved in activating JNK in surviving cells adjacent to the wound, since hemocytes release the TNF ligand Eiger that can induce JNK signaling [25]. JNK signaling plays a key role in regulating many biological processes involved in regeneration, including wound healing, compensatory proliferation, apoptosis, and cytoskeletal rearrangement. Inhibition of JNK during disc regeneration impairs wound healing and reduces regenerative proliferation [8, 28, 45, 46]. These regenerative responses depend on the activation of several downstream pathways by JNK.
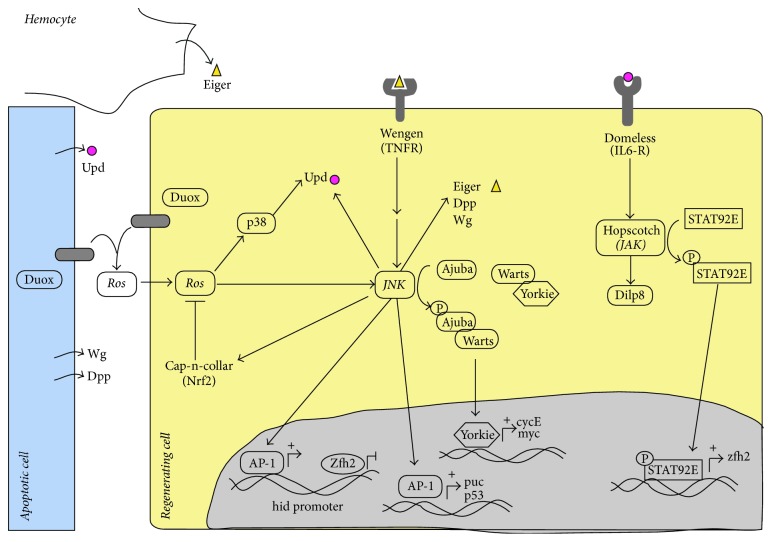
In addition to the activation of JNK signaling, Ros can regulate the P38 stress-activated MAP kinases in surviving cells. The activation of P38 signaling is independent of the JNK pathway [22] and it has been proposed that Ros may promote the P38 pathway through the oxidative modification of intracellular kinases, such as the redox-sensitive activating protein-1 ASK1 [31]. The nondeleterious activation of JNK and P38 MAP kinases by Ros may have multiple effects, among them the induction of cytokine expression [11, 22, 29, 47, 48]. Drosophila has three leptin-like (IL-6 family) cytokine ligands known as the Unpaired proteins (Upd, Upd2, and Upd3). After physical injury or cell death, the three upd genes are upregulated in the wound's edges in a manner dependent on JNK signaling [22, 47, 48]. It is unclear whether these factors are expressed exclusively in dead cells (as we mentioned before), or they are also expressed in surviving cells surrounding the damage region. In this review we have considered that both dead and surviving cells can express these ligands. The Upd ligands bind to the IL-6R type receptor Domeless (dome), which activates the Janus kinase Hopscotch (Hop), and this phosphorylation cascade promotes the translocation of a Stat3-like transcription factor (Stat92E) to the nucleus (Figure 3). All these factors constitute the JAK/STAT signaling pathway, which has important roles in disc development, for instance appendage patterning [49–51] and the control of cell proliferation [49, 52–54]. The elimination of JAK/STAT components during leg disc regeneration impairs local cell proliferation [48]. Similarly, during wing disc regeneration, reduced JAK/STAT activity also partially disrupts adult wing recovery, leading to the generation of much smaller adults wings [22, 47]. As such, it was proposed that JAK/STAT signaling functions downstream of JNK/P38 signaling and that it is necessary to induce compensatory cell proliferation and to form the blastema (Figure 4) [48]. However, instead of promoting compensatory cell proliferation, it has been proposed that JAK/STAT might be necessary to restrain the excessive tissue damage caused by the activation of the JNK pathway, which would facilitate the initiation of compensatory responses [47]. JAK/STAT could act as a suppressor of JNK signaling and this repression could either be mediated by direct transcriptional effects on JNK components or indirectly, by suppressing apoptosis. This mechanism could restrain the nonautonomous activation of JNK and excessive apoptosis [47]. This function of JAK/STAT would be mediated by Zfh1 and Zfh2 (Zinc-finger homeobox) proteins. These ZEB proteins that act as transcriptional repressors [47] have been previously identified to be downstream effectors of JAK/STAT [47, 49, 55]. In the promoter region of hid as well as in the promoter of the gene key, which encodes for the AP-1 component dFos, appears multiple, highly clustered mammalian ZEB1-binding motifs [47]. Therefore, it has been proposed that Zfh1 and Zfh2 might be restraining JNK activation by repressing kay. In addition, ZEB proteins might be also competing with AP-1 for transcriptional repression of hid, thereby limiting the apoptosis induced by JNK signaling through hid (Figure 3) [47].
Figure 3.
Schematic of regulatory interactions between components of signaling pathways involved in promoting Drosophila wing disc regeneration. See text for details.
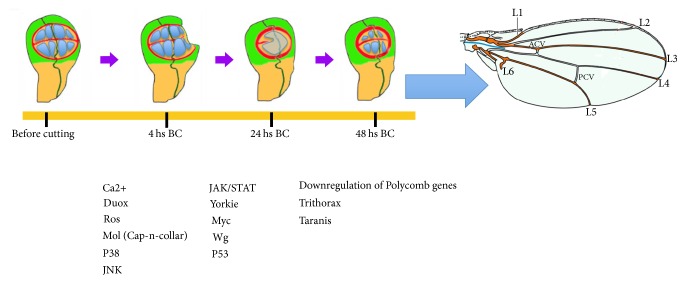
Figure 4.
Schematic representation of the different factors and signaling pathways involved in the regulation of the cellular processes that occur during wing disc regeneration; see text for details.
In addition to the possible role that JAK/STAT might have in controlling cell proliferation, this pathway also induces a physiological response by activating Drosophila insulin-like peptide (dilp8) [47, 48]. This paracrine factor is activated after damage and it regulates the timing of pupariation [56, 57]. It has been reported that Dilp8 regulates both developmental delay and growth coordination between regenerating and undamaged tissue. Dilp8 inhibits the production of the neuropeptide prothoracicotropic hormone (PTTH), causing developmental delay [56, 57]. Moreover Dilp8 activates Nitric oxide synthase (NOS) in the prothoracic gland. NOS limits the growth of undamaged tissues by reducing ecdysone biosynthesis [58, 59]. The function of Dilp8 is mediated by the Orphan leucine-rich G-protein coupled receptor Lgr3. Lgr3 activity is necessary in the Central nervous system (CNS), as well as in the prothoracic gland, for NOS activation following damage [60–62].
As we mentioned the activation of dilp8 after damage depends on the JAK/STAT pathway [47, 48]; therefore, JAK/STAT signaling might favor regeneration by delaying development [47, 48, 56, 57].
5. The Role of Wg and Dpp Signaling in Disc Regeneration
The Wingless family of proteins (Wnt class) are involved in regeneration in different organisms with this capacity [2, 63, 64]. Intriguingly, different responses to wg expression have been observed during disc regeneration depending upon the proapoptotic gene employed or the methods of inducing the wound. Thus, wg (the Drosophila Wnt1 homologue) is ectopically expressed near the lesion edges before blastema formation in amputated leg and eye imaginal discs [48, 65–67]. In addition, wg is upregulated in the wing cells that form the blastema after genetic ablation by expressing Eiger or the proapoptotic gene reaper [9]. During these processes wg is activated by JNK signaling [48]. Using these experimental approaches regenerative proliferation was impaired when wg was reduced [9, 48]. Accordingly, it was proposed that wg is required for regenerative proliferation (Figure 4). This effect is at least in part due to the down regulation of Notch, which leads to Myc upregulation [9]. It has also been proposed that JAK/STAT signaling cooperates with Wg signaling to induce regenerative cell proliferation [48].
Paradoxically, when apoptosis was induced by overexpressing the proapoptotic gene head involution defective (hid) or when a portion of a disc is eliminated in situ [12], wg expression was not altered during disc regeneration [68]. Moreover, knocking down wg did not block discs regeneration after in situ amputation or hid expression [12, 68]. The basis for these differences in wg expression and its requirements are not yet clear. They might in part reflect differences in the efficiency of genetic ablation, or that different methods of inducing a wound elicit different responses in terms of gene expression. Alternatively, wg function might be redundant with the activity of other genes of Wnt family present in Drosophila, such as wnt6. In fact, wnt6 and wg share the same regenerative enhacer (see below). Therefore, more work is needed to define the role of wg signaling in the regenerative response.
The bone morphogenetic protein (BMP) Decapentaplegic (Dpp) activates a signaling pathway that plays an important role in inducing growth and patterning during imaginal disc development [69–72]. Therefore, it was suggested that the Dpp pathway might be redeployed to control regenerative growth. However, the contribution of the Dpp signal to this process remains unclear. Thus, although dpp is transcriptionally activated in response to genetic ablation in wing discs [9], this is not the case in amputated wing discs [46]. Moreover, while Dpp is required for the hyperplastic growth caused by “undead” cells, when apoptotic cells are protected with P35 [73, 74], this factor is dispensable for compensatory cell proliferation when P35 is not ectopically expressed in apoptotic cells, even though Dpp is expressed in apoptotic cells [74, 75]. As yet, the basis for these differences remains unclear.
It has been proposed that both wg and dpp are activated in apoptotic cells and diffuse to surrounding cell to promote proliferation [73–75]. However, as JNK signaling pathway is active, although at low levels, in surviving regenerating cells [22], and wg and Dpp are targets of JNK signal, we do not exclude the possibility that these factors might be also expressed in some regenerating cells (Figure 3).
6. The Hippo Pathway Is Necessary for Regenerative Growth
Hippo signaling is a conserved pathway that regulates growth during development and regeneration, and its deregulation is associated with oncogenesis (reviewed in [76, 77]). This signaling pathway is constituted by a kinase cascade that can be activated by different stimuli. Hippo signaling is mediated by a transcriptional coactivator protein, Yorkie (Yki in Drosophila, YAP in vertebrates: reviewed in [77, 78]) (Figure 2). Yki remains inactive when the signaling pathway is active and it is retained in the cytoplasm due to its phosphorylation by the kinase Warts (Wts) [79]. When Wts is inactive, unphosphorylated Yki accumulates in the nucleus [77, 78] and in conjunction with different DNA-binding proteins, it promotes the transcription of downstream genes necessary to promote cell proliferation, such as Cyclin E and cMyc [77, 78] (Figures 3 and 4).
The Hippo pathway plays a key role in inducing regenerative growth after disc damage [80–82]. This pathway can be activated by multiple upstream inputs, including Fat–Dachsous signaling, sense tissue damage, and JNK signaling [82, 83]. JNK signaling can directly promote the activation of Yki by phosphorylating Ajuba family LIM proteins and enhancing their binding to Wts, thereby preventing their activation by Hippo [84]. Interestingly, the ability of JNK to activate YAP is conserved in mammalian cells [83, 84]. Thus, JNK increases Yki activity after wounding, a process essential to induce compensatory cell proliferation and regeneration.
7. The Control of Cell Plasticity during Imaginal Disc Regeneration by the Polycomb Group (PcG)
One of the processes associated with organ regeneration is the repatterning of the regenerating tissue, which implies genetic reprogramming of cells in order to switch their fates (Figure 4). After damage, newly formed tissue is derived from surviving cells that lie nearby and some of these cells must change their state of determination to contribute to the lost region.
During disc regeneration several observations indicate that cell fates are respecified and that there is a process of cell reprograming. For example, there is a temporary loss of markers of cell fate commitment after genetic ablation or disc amputation [9, 12, 85]. It has also been reported that after genetic ablation in the wing pouch, the cells of the hinge generate cells that become part of the pouch [68, 86]. Moreover, cell fate changes between compartments have been reported after surgical excision [66] or genetic ablation [87]. Indeed, cells near the anterior/posterior or dorsal/ventral boundary can change their identities and contribute to the compartment on the other side of the boundary [87]. Finally, during regeneration the cells of one disc occasionally acquire the identities of different imaginal discs, switching cell fate and generating disc-inappropriate structures, a process known as transdetermination [88].
The preservation of a specific cell fate or determination state depends on a particular genetic program, which is largely maintained through epigenetic modifications that are established during development. The polycomb group (PcG) proteins function as epigenetic modifiers and they are required to maintain cell fates by controlling the expression of developmental regulators [89]. This group of proteins forms two different types of complexes, Polycomb repressive complex 1 (PRC1) and Polycomb repressive complex 2 (PRC2). PcG can silence large numbers of genes by establishing repressive marks like histone H3 lysine 27 trimethylation (H3K27me3). There is evidence that JNK signaling downregulates PcG genes during regeneration, thereby allowing the transcription of otherwise silenced genes [90]. This process is important for cell reprogramming during regeneration. Inappropriate or excessive downregulation of the PcG by JNK during regeneration may activate genes that induce a genetic program corresponding to a different disc, provoking transdetermination. Indeed, the frequency of transdetermination is enhanced in PcG mutant discs [90]. Interestingly, ectopic activation of wg can induce transdetermination, possibly because wg might be a direct target of the PcG [90].
The preservation of the anterior/posterior compartment identity during regeneration is mediated by taranis (tara), that is, the homologue in Drosophila of the vertebrate TRIP-Br (Transcriptional Regulators Interacting with plant homeodomain (PHD) zinc fingers and/or Bromodomains) family of proteins. In mutant conditions for tara, regenerating wing disc undergoes posterior-to-anterior transformations late in regeneration. These changes are consequence of the misregulation of posterior selector gene engrailed (en). The deregulation of en leads to the autoregulatory silencing of the engrailed locus, which requires the PRC1. The misregulation and subsequent silencing of en are induced by JNK signaling. It has been proposed that Tara stabilizes engrailed expression downstream of JNK signaling to maintain the posterior cell fate identity during regeneration [91].
Recently, a defined regulatory element was identified that is responsible for the activation of wg expression after damage [92]. Interestingly, this regenerative enhancer (BRV118) regulates the expression of wg and wnt6. This observation suggests that the function of different members of Wnt family might be involved during regeneration in Drosophila. It has been described that within this enhancer there is a damage-responsive module that remains active throughout the third instar stage and an adjacent silencing element that nucleates increasing levels of epigenetic silencing during development. This latter element can restrict the activity of this enhancer [92]. Therefore, the loss of the regenerative capacity of the discs as development proceeds might be explained by a blockade of the damage-responsive enhancers through the activity of the silencing elements. This mechanism might prevent gene expression in the mature organism without compromising the gene activity regulated by developmental signals [92]. Interestingly, PcG-mediated epigenetic silencing is required to regulate the activity of this enhancer. Hence, the inability of the cells in adult tissue to reactivate programs necessary to promote regenerative growth or cell fate respecification could limit regeneration in adult stages.
8. Perspectives
The urodele amphibians have been used extensively as a model system to study regeneration as they present a remarkable regenerative capacity and they can fully regenerate amputated appendages [93]. While the studies carried out on these organisms allowed multiple cellular processes involved in limb regeneration to be identified [93], much less is known about the genetic mechanisms that control them, as amphibians are not the best model organisms for genetic analyses. Moreover, most studies into regenerative biology aimed at developing biomedical applications have been carried out on stem cells cultivated in vitro. To better understand the processes that occur during regeneration, these phenomena must be studied in vivo, in the context of the complex genetic and cellular interactions that take place. Drosophila is a complex model organism in which the mechanistic details of genetic and cellular processes can be defined. In addition, Drosophila has been extensively used as a model system to carry out unbiased genetic screens to identify genes involved in different cellular processes. These features make Drosophila an excellent model to identify and characterize genes involved in all aspects of regeneration. In fact, different genetic screens and studies of the changes in gene expression during disc regeneration have identified multiple signaling pathways and genes required for different processes associated with regeneration [23, 24, 94].
The conservation between flies and vertebrates of basic signaling pathways and their regulatory elements justifies using Drosophila as a model organism to establish mechanisms and genetic processes that can be translated to vertebrates. Different studies have confirmed that most of the signaling pathways required for disc regeneration are also involved in regeneration in vertebrates; for example, the Hippo pathway appears to play a fundamental role in vertebrate limb regeneration and in skin wound healing [95, 96]. JNK is very important in mammalian liver regeneration and one of its targets, the AP-1 transcription factor subunit c-Jun, is activated during liver regeneration [97–99], the cytokines TNF-α and IL-6 also being required during this process [99]. Finally, and as in Drosophila, PcGs are downregulated during murine skin repair, which provokes the derepression of dmyc [100]. Moreover, it has been suggested that that loss of polycomb-mediated silencing might contribute to the induction of repair genes in mammals.
In summary, basic regenerative research carried out in Drosophila can provide insights into the genetic and cellular responses involved in mammalian regeneration. This knowledge might serve to develop new therapies in regenerative medicine.
Acknowledgments
The authors thank Ali Kiai for helping to improve the manuscript. This work was supported by a grant obtained from the MEC (BFU2014-54153-P) and it has greatly benefited from the financial support of the Fundacion Ramon Areces.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Tanaka E. M., Reddien P. W. The cellular basis for animal regeneration. Developmental Cell. 2011;21(1):172–85. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez Alvarado A., Tsonis P. A. Bridging the regeneration gap: genetic insights from diverse animal models. Nature Reviews Genetics. 2006;7(11):873–84. doi: 10.1038/nrg1923. [DOI] [PubMed] [Google Scholar]
- 3.Hadorn E., Anders G., Ursprung H. Combination derived from partial dissociated imaginal disks of various mutants and types of Drosophila. Journal of Experimental Zoology. 1959;142:159–175. doi: 10.1002/jez.1401420107. [DOI] [PubMed] [Google Scholar]
- 4.Hadorn E., Buck D. Ueber Entwicklungsleistungen transplantierter teilstücke von Flügel-Imaginalschiben von Drosophila melanogaster. Revue suisse de Zoologie. 1962;69:302–310. [Google Scholar]
- 5.Hadorn E., Hürlimann R., Mindek G., Schubiger G., Staub M. Developmental capacity of embryonal blastema in Drosophila following cultivation in an adult host. Revue suisse de Zoologie. 1968;75(3):557–569. [PubMed] [Google Scholar]
- 6.Bodenstein. Hormones and tissue competence in the development of Drosophila. The Biological Bulletin. 1943;84:34–58. [Google Scholar]
- 7.Bryant P. J. Regeneration and duplication following operations in situ on the imaginal discs of Drosophila melanogaster. Developmental Biology. 1971;26(4):637–651. doi: 10.1016/0012-1606(71)90146-1. [DOI] [PubMed] [Google Scholar]
- 8.Bosch M., Baguna J., Serras F. Origin and proliferation of blastema cells during regeneration of Drosophila wing imaginal discs. The International Journal of Developmental Biology. 2008;52(8):1043–1050. doi: 10.1387/ijdb.082608mb. [DOI] [PubMed] [Google Scholar]
- 9.Smith-Bolton R. K., Worley M. I., Kanda H., Hariharan I. K. Regenerative Growth in Drosophila Imaginal Discs Is Regulated by Wingless and Myc. Developmental Cell. 2009;16(6):797–809. doi: 10.1016/j.devcel.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergantinos C., Corominas M., Serras F. Cell death-induced regeneration in wing imaginal discs requires JNK signaling. Development. 2010;137(7):1169–1179. doi: 10.1242/dev.045559. [DOI] [PubMed] [Google Scholar]
- 11.Pastor-Pareja J. C., Wu M., Xu T. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Disease Model Mechanisms. 2008;1(2-3):144–54. doi: 10.1242/dmm.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz-Garcia S., Baonza A. Pattern reorganization occurs independently of cell division during Drosophila wing disc regeneration in situ. Proceedings of the National Academy of Sciences of the United States of America; 2013; p. p. 13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergantinos C., Vilana X., Corominas M., Serras F. Imaginal discs: Renaissance of a model for regenerative biology. Bioessays. 2010;32(3):207–17. doi: 10.1002/bies.200900105. [DOI] [PubMed] [Google Scholar]
- 14.Harris J. M., Esain V., Frechette G. M., et al. Glucose metabolism impacts the spatiotemporal onset and magnitude of HSC induction in vivo. Blood. 2013;121(13):2483–2493. doi: 10.1182/blood-2012-12-471201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez-Garcia D., Wood C. D., Castro-Obregon S., Covarrubias L. Reactive oxygen species: A radical role in development? Free Radical Biology and Medicine. 2010;49(2):130–43. doi: 10.1016/j.freeradbiomed.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Holmstrom K. M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signaling. Nature Reviews Molecular Cell Biology. 2014;15(6):411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 17.Finkel T. Signal transduction by reactive oxygen species. The Journal of Cell Biology. 2011;194(1):7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Razzell W., Evans I. R., Martin P., Wood W. Calcium flashes orchestrate the wound inflammatory response through duox activation and hydrogen peroxide release. Current Biology. 2013;23(5):424–429. doi: 10.1016/j.cub.2013.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauron C., Rampon C., Bouzaffour M., et al. Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed. Scientific Reports. 2013;3, article 2084 doi: 10.1038/srep02084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love N. R., Chen Y., Ishibashi S., et al. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nature Cell Biology. 2013;15(2):222–228. doi: 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y., Nikulenkov F., Zawacka-Pankau J., et al. ROS-dependent activation of JNK converts p53 into an efficient inhibitor of oncogenes leading to robust apoptosis. Cell Death & Differentiation. 2014;21(4):612–623. doi: 10.1038/cdd.2013.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santabarbara-Ruiz P., Lopez-Santillan M., Martinez-Rodriguez I., et al. ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Drosophila Regeneration. PLoS Genet. 2015;11(10) doi: 10.1371/journal.pgen.1005595.e1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan S. J., Abidi S. N. F., Skinner A., Tian Y., Smith-Bolton R. K. The Drosophila Duox maturation factor is a key component of a positive feedback loop that sustains regeneration signaling. PLoS Genetics. 2017;13(7) doi: 10.1371/journal.pgen.1006937.e1006937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brock A. R., Seto M., Smith-Bolton R. K. Cap-n-Collar promotes tissue regeneration by regulating ROS and JNK signaling in the drosophila melanogaster wing imaginal disc. Genetics. 2017;206(3):1505–1520. doi: 10.1534/genetics.116.196832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fogarty C. E., Diwanji N., Lindblad J. L., et al. Extracellular Reactive Oxygen Species Drive Apoptosis-Induced Proliferation via Drosophila Macrophages. Current Biology. 2016;26(5):575–584. doi: 10.1016/j.cub.2015.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serras F. The benefits of oxidative stress for tissue repair and regeneration. Flight Journal. 2016;10(3):128–133. doi: 10.1080/19336934.2016.1188232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diwanji N., Bergmann A. An unexpected friend - ROS in apoptosis-induced compensatory proliferation: Implications for regeneration and cancer. Seminars in Cell & Developmental Biology. 2017 doi: 10.1016/j.semcdb.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosch M., Serras F., Martin-Blanco E., Baguna J. JNK signaling pathway required for wound healing in regenerating Drosophila wing imaginal discs. Developmental Biology. 2005;280(1):73–86. doi: 10.1016/j.ydbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Lo Y. Y., Wong J. M., Cruz T. F. Reactive oxygen species mediate cytokine activation of c-Jun NH2-terminal kinases. Journal of Biological Chemistry. 1996;271(26):15703–15707. doi: 10.1074/jbc.271.26.15703. [DOI] [PubMed] [Google Scholar]
- 30.Son Y., Kim S., Chung H. T., Pae H. O. Reactive oxygen species in the activation of MAP kinases. Methods Enzymol. 2013;528:27–48. doi: 10.1016/B978-0-12-405881-1.00002-1. [DOI] [PubMed] [Google Scholar]
- 31.Tobiume K., Matsuzawa A., Takahashi T., et al. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Reports. 2001;2(3):222–228. doi: 10.1093/embo-reports/kve046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ilmarinen P., Moilanen E., Kankaanranta H. Mitochondria in the center of human eosinophil apoptosis and survival. International Journal of Molecular Sciences. 2014;15(3):3952–3969. doi: 10.3390/ijms15033952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varkey J., Chen P., Jemmerson R., Abrams J. M. Altered cytochrome c display precedes apoptotic cell death in Drosophila. The Journal of Cell Biology. 1999;144(4):701–710. doi: 10.1083/jcb.144.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdelwahid E., Yokokura T., Krieser R. J., Balasundaram S., Fowle W. H., White K. Mitochondrial Disruption in Drosophila Apoptosis. Developmental Cell. 2007;12(5):793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Thomenius M., Freel C. D., Horn S., et al. Mitochondrial fusion is regulated by Reaper to modulate Drosophila programmed cell death. Cell Death & Differentiation. 2011;18(10):1640–1650. doi: 10.1038/cdd.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clavier A., Rincheval-Arnold A., Colin J., Mignotte B., Guénal I. Apoptosis in Drosophila: Which role for mitochondria? Apoptosis. 2016;21(3):239–251. doi: 10.1007/s10495-015-1209-y. [DOI] [PubMed] [Google Scholar]
- 37.Brodsky M. H., Nordstrom W., Tsang G., Kwan E., Rubin G. M., Abrams J. M. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101(1):103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 38.Shlevkov E., Morata G. A dp53/JNK-dependant feedback amplification loop is essential for the apoptotic response to stress in Drosophila. Cell Death & Differentiation. 2012;19(3):451–460. doi: 10.1038/cdd.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells B. S., Yoshida E., Johnston L. A. Compensatory Proliferation in Drosophila Imaginal Discs Requires Dronc-Dependent p53 Activity. Current Biology. 2006;16(16):1606–1615. doi: 10.1016/j.cub.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wells B. S., Johnston L. A. Maintenance of imaginal disc plasticity and regenerative potential in Drosophila by p53. Dev Biol. 2012;361(2):263–76. doi: 10.1016/j.ydbio.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pichiule P., Schmidt A., Vannucci S. Hypoxia-inducible factor-1 mediates neuronal expression of the receptor for advanced glycation end products following hypoxia/ischemia. The Journal of Biological Chemistry. 2007;282:36330–36340. doi: 10.1074/jbc.M706407200. [DOI] [PubMed] [Google Scholar]
- 42.Feine I., Pinkas I., Salomon Y., Scherz A. Local oxidative stress expansion through endothelial cells—a key role for gap junction intercellular communication. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0041633.e41633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Garijo A., Steller H. Spreading the word: Non-autonomous effects of apoptosis during development, regeneration and disease. Development. 2015;142(19):3253–3262. doi: 10.1242/dev.127878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diaz-Garcia S., Ahmed S., Baonza A. Analysis of the function of apoptosis during imaginal wing disc regeneration in Drosophila melanogaster. PLoS One. 2016;11(11) doi: 10.1371/journal.pone.0165554.e0165554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramet M., Lanot R., Zachary D., Manfruelli P. JNK signaling pathway is required for efficient wound healing in Drosophila. Developmental Biology. 2002;241(1):145–56. doi: 10.1006/dbio.2001.0502. [DOI] [PubMed] [Google Scholar]
- 46.Mattila J., Omelyanchuk L., Kyttala S., Turunen H., Nokkala S. Role of Jun N-terminal Kinase (JNK) signaling in the wound healing and regeneration of a Drosophila melanogaster wing imaginal disc. The International Journal of Developmental Biology. 2005;49(4):p. 391. doi: 10.1387/ijdb.052006jm. [DOI] [PubMed] [Google Scholar]
- 47.La Fortezza M., Schenk M., Cosolo A., Kolybaba A., Grass I., Classen A.-K. JAK/STAT signalling mediates cell survival in response to tissue stress. Development. 2016;143(16):2907–2919. doi: 10.1242/dev.132340. [DOI] [PubMed] [Google Scholar]
- 48.Katsuyama T., Comoglio F., Seimiya M., Cabuy E., Paro R. During Drosophila disc regeneration, JAK/STAT coordinates cell proliferation with Dilp8-mediated developmental delay. Proceedings of the National Acadamy of Sciences of the United States of America. 2015;112(18):E2327–E2336. doi: 10.1073/pnas.1423074112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayala-Camargo A., Anderson A. M., Amoyel M., Rodrigues A. B., Flaherty M. S., Bach E. A. JAK/STAT signaling is required for hinge growth and patterning in the Drosophila wing disc. Developmental Biology. 2013;382(2):413–426. doi: 10.1016/j.ydbio.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arbouzova N. I., Zeidler M. P. JAK/STAT signalling in Drosophila: Insights into conserved regulatory and cellular functions. Development. 2006;133(14):2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- 51.Zeidler M. P., Bach E. A., Perrimon N. The roles of the Drosophila JAK/STAT pathway. Oncogene. 2000;19(21):2598–2606. doi: 10.1038/sj.onc.1203482. [DOI] [PubMed] [Google Scholar]
- 52.Tsai Y.-C., Sun Y. H. Long-range effect of Upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004;39(2):141–153. doi: 10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- 53.Mukherjee T., Hombria J. C., Zeidler M. P. Opposing roles for Drosophila JAK/STAT signalling during cellular proliferation. Oncogene. 2005;24(15):2503–2511. doi: 10.1038/sj.onc.1208487. [DOI] [PubMed] [Google Scholar]
- 54.Bach E. A., Vincent S., Zeidler M. P., Perrimon N. A Sensitized Genetic Screen to Identify Novel Regulators and Components of the Drosophila Janus Kinase/Signal Transducer and Activator of Transcription Pathway. Genetics. 2003;165(3):1149–1166. doi: 10.1093/genetics/165.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leatherman J. L., Di Nardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3(1):44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garelli A., Gontijo A. M., Miguela V., Caparros E., Dominguez M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science. 2012;336(6081):579–582. doi: 10.1126/science.1216735. [DOI] [PubMed] [Google Scholar]
- 57.Colombani J., Andersen D. S., Léopol P. Secreted peptide dilp8 coordinates Drosophila tissue growth with developmental timing. Science. 2012;336(6081):582–585. doi: 10.1126/science.1216689. [DOI] [PubMed] [Google Scholar]
- 58.Jaszczak J. S., Wolpe J. B., Dao A. Q., Halme A. Nitric oxide synthase regulates growth coordination during Drosophila melanogaster imaginal disc regeneration. Genetics. 2015;200(4):1219–1228. doi: 10.1534/genetics.115.178053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jaszczak J. S., Halme A. Arrested development: coordinating regeneration with development and growth in Drosophila melanogaster. Current Opinion in Genetics and Development. 2016;40:87–94. doi: 10.1016/j.gde.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaszczak J. S., Wolpe J. B., Bhandari R., Jaszczak R. G., Halme A. Growth coordination during Drosophila melanogaster imaginal disc regeneration is mediated by signaling through the relaxin receptor Lgr3 in the prothoracic gland. Genetics. 2016;204(2):703–709. doi: 10.1534/genetics.116.193706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vallejo D. M., Juarez-Carreño S., Bolivar J., Morante J., Dominguez M. A brain circuit that synchronizes growth and maturation revealed through Dilp8 binding to Lgr3. Science. 2015;350(6262, article no. 6767) doi: 10..1126/science.aac6767. [DOI] [PubMed] [Google Scholar]
- 62.Garelli A., Heredia F., Casimiro A. P., et al. Dilp8 requires the neuronal relaxin receptor Lgr3 to couple growth to developmental timing. Nature Communications. 2015;6, article no. 8732 doi: 10.1038/ncomms9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stoick-Cooper C. L., Moon R. T., Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes & Development. 2007;21(11):1292–1315. doi: 10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- 64.Stoick-Cooper C. L., Weidinger G., Riehle K. J., et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134(3):479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 65.McClure K. D., Sustar A., Schubiger G. Three genes control the timing, the site and the size of blastema formation in Drosophila. Developmental Biology. 2008;319(1):68–77. doi: 10.1016/j.ydbio.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gibson M. C., Schubiger G. Hedgehog is required for activation of engrailed during regeneration of fragmented Drosophila imaginal discs. Development. 1999;126(8):1591–1599. doi: 10.1242/dev.126.8.1591. [DOI] [PubMed] [Google Scholar]
- 67.Sustar A., Bonvin M., Schubiger M., Schubiger G. Drosophila twin spot clones reveal cell division dynamics in regenerating imaginal discs. Developmental Biology. 2011;356(2):576–587. doi: 10.1016/j.ydbio.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herrera S. C., Martin R., Morata G. Tissue homeostasis in the wing disc of Drosophila melanogaster: immediate response to massive damage during development. PLoS Genet. 2013;9(4) doi: 10.1371/journal.pgen.1003446.e1003446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin-Castellanos C., Edgar B. A. A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development. 2002;129(4):1003–1013. doi: 10.1242/dev.129.4.1003. [DOI] [PubMed] [Google Scholar]
- 70.Nellen D., Burke R., Struhl G., Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85(3):357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 71.St Johnston R. D., Hoffmann F. M., Blackman R. K., et al. Molecular organization of the decapentaplegic gene in Drosophila melanogaster. Genes & Development. 1990;4(7):1114–1127. doi: 10.1101/gad.4.7.1114. [DOI] [PubMed] [Google Scholar]
- 72.Burke R., Basler K. Dpp receptors are autonomously required for cell proliferation in the entire developing Drosophila wing. Development. 1996;122(7):2261–2269. doi: 10.1242/dev.122.7.2261. [DOI] [PubMed] [Google Scholar]
- 73.Morata G., Shlevkov E., Perez-Garijo A. Mitogenic signaling from apoptotic cells in Drosophila. Dev Growth Differ. 2011;53(2):168–76. doi: 10.1111/j.1440-169X.2010.01225.x. [DOI] [PubMed] [Google Scholar]
- 74.Perez-Garijo A., Shlevkov E., Morata G. The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development. 2009;136(7):1169–1177. doi: 10.1242/dev.034017. [DOI] [PubMed] [Google Scholar]
- 75.Perez-Garijo A., Martin F. A., Morata G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development. 2004;131(22):5591–5598. doi: 10.1242/dev.01432. [DOI] [PubMed] [Google Scholar]
- 76.Reddy B. V. V. G., Irvine K. D. The fat and warts signaling pathways: New insights into their regulation, mechanism and conservation. Development. 2008;135(17):2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- 77.Zhao B., Tumaneng K., Guan K.-L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nature Cell Biology. 2011;13(8):877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oh H., Irvine K. D. Yorkie: the final destination of Hippo signaling. Trends in Cell Biology. 2010;20(7):410–417. doi: 10.1016/j.tcb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang J., Wu S., Barrera J., Matthews K., Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122(3):421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 80.Sun G., Irvine K. D. Control of growth during regeneration. Curr Top Dev Biol. 2014;108:95–120. doi: 10.1016/B978-0-12-391498-9.00003-6. [DOI] [PubMed] [Google Scholar]
- 81.Su X., Fricke J., Kavanagh D. G., Irvine D. J. In vitro and in vivo mRNA delivery using lipid-enveloped pH-responsive polymer nanoparticles. Molecular Pharmacology. 2011;8(3):774–87. doi: 10.1021/mp100390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grusche F. A., Degoutin J. L., Richardson H. E., Harvey K. F. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Developmental Biology. 2011;350(2):255–266. doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 83.Sun G., Irvine K. D. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Developmental Biology. 2011;350(1):139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun G., Irvine K. D. Ajuba family proteins link JNK to hippo signaling. Science Signaling. 2013;6(292, article no. ra81) doi: 10.1126/scisignal.2004324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Repiso A., Bergantiños C., Serras F. Cell fate respecification and cell division orientation drive intercalary regeneration in Drosophila wing discs. Development. 2013;140(17):3541–3551. doi: 10.1242/dev.095760. [DOI] [PubMed] [Google Scholar]
- 86.Verghese S., Su T. T. Drosophila Wnt and STAT Define Apoptosis-Resistant Epithelial Cells for Tissue Regeneration after Irradiation. PLoS Biol. 2016;14(9) doi: 10.1371/journal.pbio.1002536.e1002536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herrera S. C., Morata G. Transgressions of compartment boundaries and cell reprogramming during regeneration in Drosophila. ELife. 2014;3:p. e01831. doi: 10.7554/eLife.01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hadorn E. Transdetermination in cells. Scientific American. 1968;219(5):110–114. doi: 10.1038/scientificamerican1168-110. [DOI] [PubMed] [Google Scholar]
- 89.Ringrose L., Paro R. Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annual Review of Genetics. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 90.Lee N., Maurange C., Ringrose L., Paro R. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature. 2005;438(7065):234–237. doi: 10.1038/nature04120. [DOI] [PubMed] [Google Scholar]
- 91.Schuster K. J., Smith-Bolton R. K. Taranis Protects Regenerating Tissue from Fate Changes Induced by the Wound Response in Drosophila. Developmental Cell. 2015;34(1):119–128. doi: 10.1016/j.devcel.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 92.Harris R. E., Setiawan L., Saul J., Hariharan I. K. Localized epigenetic silencing of a damage-activated WNT enhancer limits regeneration in mature Drosophila imaginal discs. eLife. 2016;5(2016) doi: 10.7554/eLife.11588.e11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brockes J. P., Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol. 2008;24:525–49. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- 94.Blanco E., Ruiz-Romero M., Beltran S., et al. Gene expression following induction of regeneration in Drosophila wing imaginal discs. Expression profile of regenerating wing discs. BMC Developmental Biology. 2010;10, article no. 94 doi: 10.1186/1471-213X-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hayashi S., Tamura K., Yokoyama H. Yap1, transcription regulator in the Hippo signaling pathway, is required for Xenopus limb bud regeneration. Developmental Biology. 2014;388(1):57–67. doi: 10.1016/j.ydbio.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 96.Lee M. J., Byun M. R., Furutani-Seiki M., Hong J. H., Jung H. S., YAP. AP and TAZ regulate skin wound healing. Journal of Investigative Dermatology. 2014;134(2):518–525. doi: 10.1038/jid.2013.339. [DOI] [PubMed] [Google Scholar]
- 97.Akerman P., Cote P., Yang S. Q., et al. Antibodies to tumor necrosis factor-alpha inhibit liver regeneration after partial hepatectomy. American Journal of Physiology. 1992;263(4):579–585. doi: 10.1152/ajpgi.1992.263.4.G579. [DOI] [PubMed] [Google Scholar]
- 98.Schwabe R. F., Bradham C. A., Uehara T., et al. c-Jun-N-terminal kinase drives cyclin D1 expression and proliferation during liver regeneration. Hepatology. 2003;37(4):824–832. doi: 10.1053/jhep.2003.50135. [DOI] [PubMed] [Google Scholar]
- 99.Yamada Y., Kirillova I., Peschon J. J., Fausto N. Initiation of liver growth by tumor necrosis factor: Deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proceedings of the National Acadamy of Sciences of the United States of America. 1997;94(4):1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shaw T., Martin P. Epigenetic reprogramming during wound healing: Loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Reports. 2009;10(8):881–886. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]