Abstract
The role of adaptive immunity in early cancer development is controversial. Here we show that chronic inflammation and fibrosis in humans and mice with non-alcoholic fatty liver disease is accompanied by accumulation of liver-resident immunoglobulin-A-producing (IgA+) cells. These cells also express programmed death ligand 1 (PD-L1) and interleukin-10, and directly suppress liver cytotoxic CD8+ T lymphocytes, which prevent emergence of hepatocellular carcinoma and express a limited repertoire of T-cell receptors against tumour-associated antigens. Whereas CD8+ T-cell ablation accelerates hepatocellular carcinoma, genetic or pharmacological interference with IgA+ cell generation attenuates liver carcinogenesis and induces cytotoxic T-lymphocyte-mediated regression of established hepatocellular carcinoma. These findings establish the importance of inflammation-induced suppression of cytotoxic CD8+ T-lymphocyte activation as a tumour-promoting mechanism.
The successes of immune checkpoint inhibitors1 and adoptive T-cell transfer2 in cancer therapy demonstrate how activated immune cells eradicate established malignancies. However, the role of adaptive immunity in tumorigenesis and the existence of immunosurveillance remain controversial3,4. We investigated how adaptive immunity affects hepatocellular carcinoma (HCC), a leading cause of cancer deaths. Other than surgical resection or ablation of localized tumours, no effective HCC treatments exist. HCC is initiated by chronic liver inflammation driven by hepatitis virus B or C infections, alcohol consumption, or non-alcoholic fatty liver disease (NAFLD)5. Liver fibrosis is another outcome of chronic hepatitis, but its exact pro-tumorigenic function remains obscure6. The contribution of adaptive immunity to HCC development has also been elusive. T cells activated by viral antigens or during non-alcoholic steatohepatitis (NASH) were suggested to promote HCC by inducing liver damage7,8, whereas alcoholic steatohepatitis (ASH) causes immune dysfunction9, and T-cell-produced interferon γ (IFNγ) attenuates liver fibrosis10. Moreover, a key mediator of liver fibrosis which usually precedes HCC development, transforming growth factor β (TGFβ), is a potent immunosuppressive cytokine that inhibits anti-cancer immunity11. TGFβ stimulates class switch recombination, which converts IgM-expressing B cells to IgA-expressing cells with regulatory activity12. Patients with ASH or NASH who have liver fibrosis exhibit more circulating IgA than patients without fibrosis13, and IgA+ cells interfere with activation of cytotoxic CD8+ T lymphocytes (CTL)14. We now show that IgA+ cells, found in human and mouse NASH-afflicted livers, mediate immunosuppression that fosters HCC development by inhibiting a strongly protective, tumour-directed CTL response. Manipulations that unleash CTL activity cause regression of established HCC in mice and should be applicable to humans.
IgA+ cells accumulate in fibrotic liver
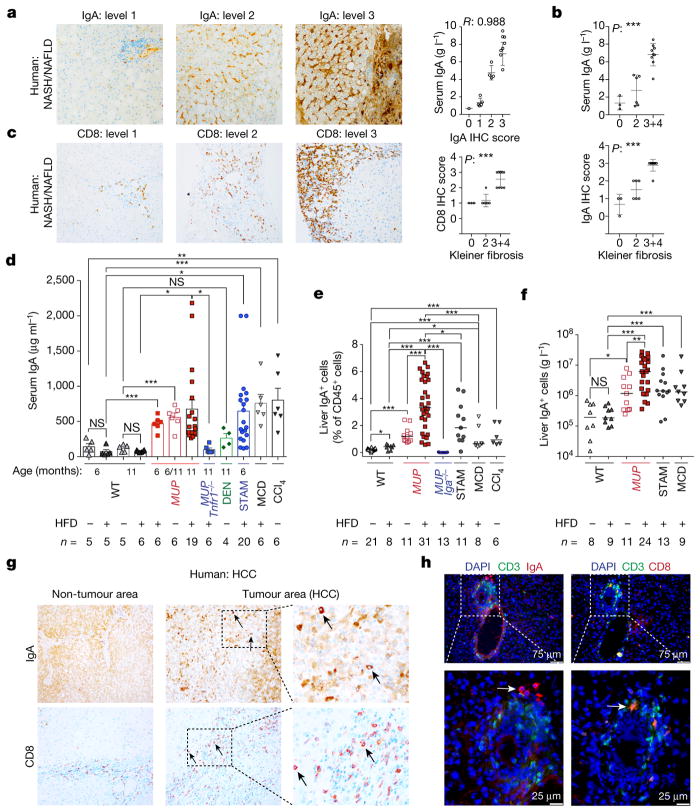
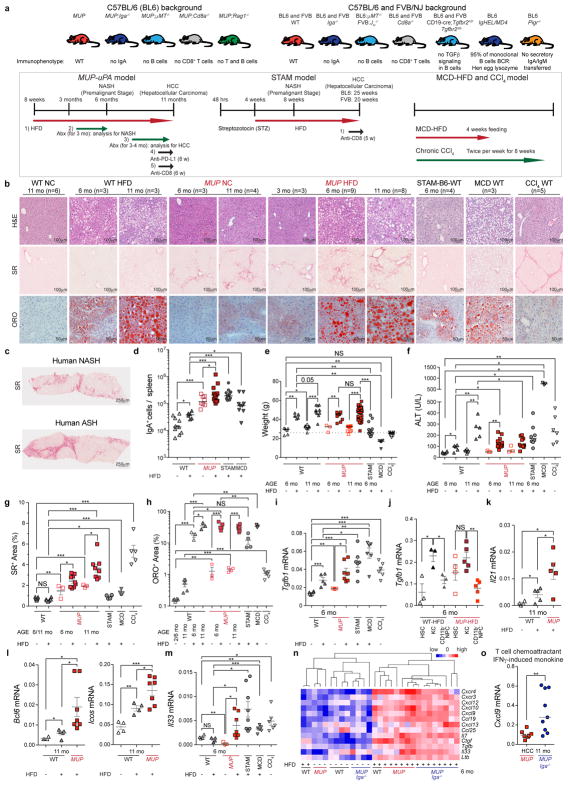
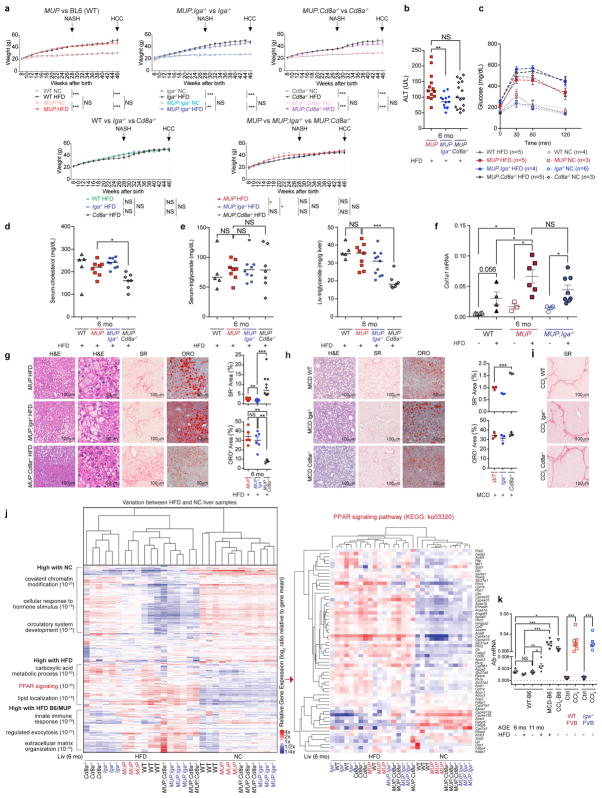
Serum IgA was elevated in two cohorts totalling 598 patients with NASH (Extended Data Fig. 1a, b), paralleling liver-intrinsic cell-bound and interstitial IgA, which correlated with fibrosis scores (Fig. 1a, b and Extended Data Fig. 1c). CD8+ T cells were also elevated in fibrotic livers (Fig. 1c and Extended Data Fig. 1d). Mouse models of NASH-driven HCC (Extended Data Fig. 2a), including high-fat diet (HFD)-fed MUP-uPA mice, which show classical NASH signs including fibrosis15, and mildly fibrogenic HFD-fed streptozotocin-treated mice (STAM)16 (Extended Data Fig. 2b, c), exhibited elevated serum IgA before and after HCC development (Fig. 1d). MUP-uPA mice, which show some fibrosis on normal chow, exhibited elevated serum IgA before HFD feeding that was abrogated by NASH-preventive ablation of tumour-necrosis factor receptor 1 (TNFR1)15. However, diethylnitrosamine-induced HCC (dih) or HFD feeding of wild-type (WT) mice, neither of which are fibrogenic17, did not elevate IgA (Fig. 1d). IgA in mouse serum correlated with liver IgA+ lymphocyte content, which was particularly high in HFD-fed MUP-uPA mice (Fig. 1e, f). Mouse and human NASH-induced HCC also contained IgA+ and CD8+ T cells (Fig. 1g, h). IgA staining was diffuse early and became more defined and perisinusoidal at higher NASH grades, whereas human and mouse HCCs contained IgA+ cells with plasma cell morphology (Fig. 1a, g, h). Spleen IgA+ cells were also elevated (Extended Data Fig. 2d). HFD feeding of WT and MUP-uPA mice caused weight gain and mild liver damage, but methionine and choline deficient diet (MCD)-fed mice exhibited severe weight loss and extensive liver damage, and STAM mice did not gain weight (Extended Data Fig. 2e–h).
Figure 1. NASH-induced IgA+ plasmocyte accumulation in human and mouse liver.
a, c, Liver biopsies from patients with NASH/NAFLD (n = 18) stained for IgA and CD8. Staining intensity was graded from 0 to 3 and plotted against serum IgA (a) or Kleiner fibrosis scores (c). The data were validated in 2 different experiments. b, Serum and liver IgA concentrations (immunohistochemistry scores) plotted against Kleiner fibrosis scores, and were further validated in two different human cohorts (n = 502 and 96; Extended Data Fig. 1a, b). d, Mice of indicated genotypes, ages, and treatments were analysed for serum IgA (n = 4–20). e, f, Liver IgA+ cell (percentage and absolute cell numbers) in different mouse strains and treatment groups (n = 6–31). g, Representative images of paired human non-tumour liver and tumour sections (n = 4) stained for IgA and CD8. h, STAM-B6 liver sections were stained for CD3, CD8, and IgA, and counterstained with DAPI (4′,6-diamidino-2-phenylindole; n = 3). Scale bars, 25–75 μm. Arrows indicate IgA+ and CD3+CD8+ cells in g, h. The data were validated at least in two or three experiments. Linear regression analysis was used to determine the correlation between serum IgA and IgA immunohistochemistry score (a). One-way analysis of variance (ANOVA; means ± s.e.m.; b, c), two-sided t-test (means ± s.e.m.; d), and Mann– Whitney test (median; e, f) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001; NS, not significant. Specific n values are shown in d–f.
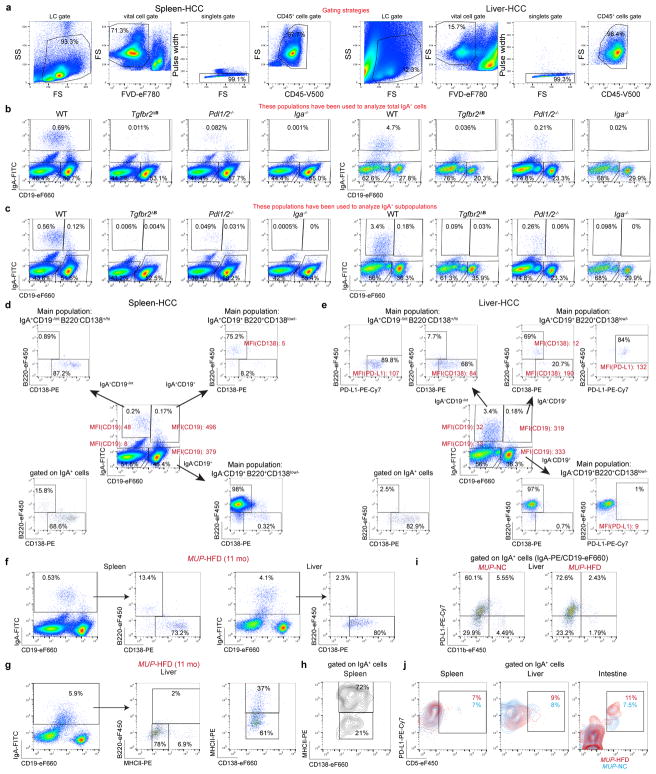
Class switch recombination to IgA is stimulated by TGFβ 1, lipopoly-saccharide, interleukin-33 (IL-33), IL-21, CD40, and lymphotoxin18,19. NASH and fibrosis correlated with elevated liver TGFβ 1, although it was also elevated in non-fibrotic HFD-fed WT mice (Extended Data Fig. 2i). In MUP-uPA mice, TGFβ 1 was mainly expressed by CD11b+ Kupffer cells (Extended Data Fig. 2j). HFD induced IL-21 in liver CD4+ T cells with follicular helper (Tfh) markers only in MUP-uPA but not BL6 mice, as well as IL-33, chemokines, and receptors that control B- and T-cell recruitment and IgA class switch recombination (Extended Data Fig. 2k–o). Thus, NASH-bearing MUP-uPA livers contain factors and cell types needed for generation of IgA+ cells. Flow cytometry revealed that MUP-uPA spleens contained two main IgA+ cell populations: IgA+B220+CD138low/−, which are probably germinal centre or memory B cells but not antibody-secreting cells, and IgA+B220−CD138+, corresponding to plasmablasts or plasma cells (Extended Data Fig. 3a–f). MHCII staining revealed two further subpopulations: MHCIIhi cells that are probably plasmablasts and MHCIIlow plasma cells (Extended Data Fig. 3g, h). The liver harboured both populations, with 5% of cells being IgA+B220+CD138low/− B cells and around 95% being plasmablasts/plasma cells (Extended Data Fig. 3a–f). Most IgA+ cells were CD11b and CD5 negative (Extended Data Fig. 3i, j) and expressed high levels of PD-L1 and IL-10 (Fig. 2a–c). CD19+IgA− cells, however, hardly expressed PD-L1 or IL-10, and PD-L1 in CD138+ plasmablasts/plasma cells was higher in IgA+ cells than in IgA− cells (Fig. 2b and Extended Data Figs 3e and 4a–e). HFD feeding increased liver IgA+ plasmocytes as early as 3 months of age, with a minor effect on intestinal IgA+ cells, which barely expressed PD-L1 (Extended Data Fig. 4f, g). Most liver IgA+ cells in human NASH expressed PD-L1, whereas 10–15% of CD8+ cells were PD-1+ (Extended Data Fig. 1c, d).
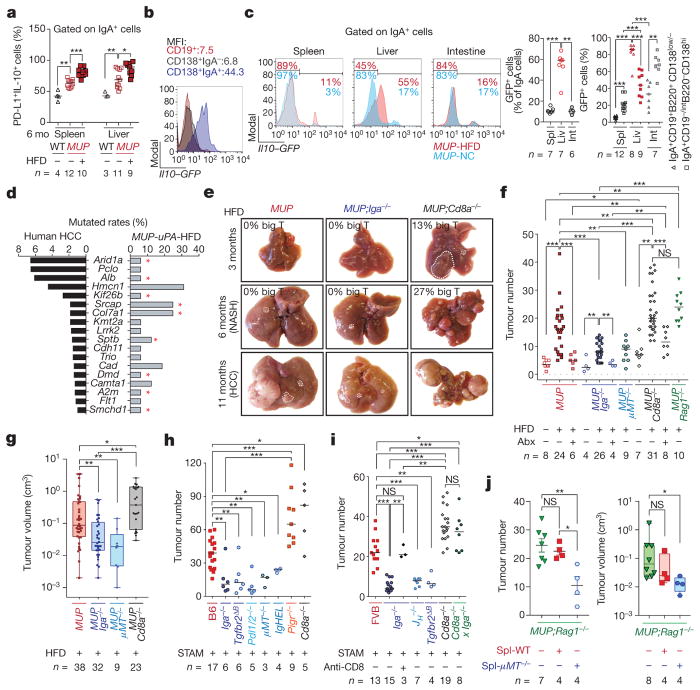
Figure 2. IgA ablation inhibits while CD8a ablation accelerates HCC appearance.
a, Splenic and liver cells from the indicated mice (n = 3–12) were stained after ex vivo stimulation and analysed by flow cytometry. % of IgA+ cells expressing IL-10 and PD-L1. b, c, Splenic, liver, and intestinal cells from Il10-GFP/MUP-uPA mice (n = 6–12) were stained and analysed by flow cytometry. b, Representative flow cytometry analysis of Il10-GFP expression in liver B cells and plasma cell subpopulations. c, Representative flow cytometry plots and percentage of GFP+ cells in IgA+ subpopulations. d, Mutated genes in human HCC on the basis of the COSMIC Database and MUP-uPA-HFD mice are compared. Driver genes20 are marked with red asterisks. e, Representative gross liver morphology and large tumour (T) incidence (> 0.4 cm) at different time points. f, g, Tumour numbers (n = 4–31) (f) and volumes (n = 9–38) (g) in 11-month-old HFD-fed MUP-uPA mice. When indicated, mice were treated with broad spectrum antibiotics (Abx) from 6 to 11 months. h, i, Tumour multiplicity in indicated BL6 (h) or FVB (i) STAM mice was determined at 25 or 20 weeks of age, respectively (n = 3–19). Experiments were repeated at least three times. STAM-Iga−/− mice were injected weekly with anti-CD8 (n = 3) for 5 weeks. j, Splenocytes from WT-BL6 (T+ B cells) or μMT−/− (T cells) mice were transferred into MUP-uPA/Rag1−/− mice (n = 4 per transfer). After 1 month, tumour numbers and volumes were determined. Experiments were performed twice. Tumour volumes (g, j, right) are plotted logarithmically, and minima to maxima with all points are shown. Two-sided t-test (means ± s.e.m.; j, left) and Mann–Whitney test (median; a, c, f–i, j, right) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001. Specific n values are shown in a, c, f–j, and Supplementary Tables 1 and 2.
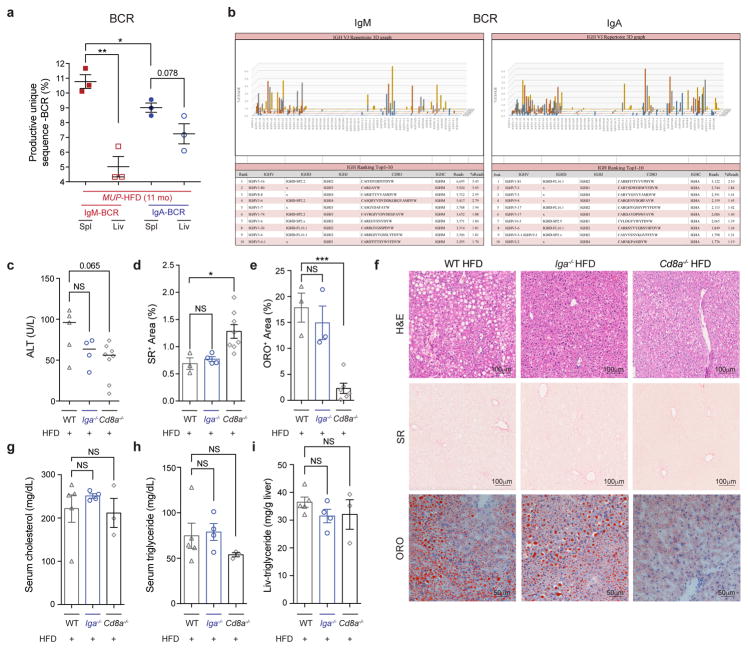
To investigate how IgA+ plasmocytes affect NASH or HCC, we generated MUP-uPA/Iga−/− (Iga is also known as Igha) and STAM/Iga−/− mice (Extended Data Fig. 2a). IgA deficiency abolished IgA+PD-L1+IL-10+ cells and had no effect on other PD-L1- and/or IL-10-expressing CD45+ cells and chemokine expression (Extended Data Figs 2n and 4b, h). IgA ablation barely affected weight gain, liver damage, or glucose tolerance, and did not alter serum and liver lipids, lipogenic and PPAR target genes (elevated in NASH), liver histology, fibrosis, or steatosis in either MUP-uPA or MCD-fed mice (Extended Data Fig. 5a–j). MUP-uPA/Cd8a−/− and STAM/Cd8a−/− mice were generated and showed no alterations in weight gain, glucose tolerance, alanine aminotransferase (ALT) release, serum triglycerides, lipogenic and PPAR target genes, or hepatocyte ballooning, although steatosis, liver triglycerides, and serum cholesterol were reduced (Extended Data Figs 5a–e, g, j and 6i), probably because of early HCC development. Consistent with the anti-fibrogenic effect of IFNγ 10, CD8+ cell ablation enhanced liver fibrosis in MUP-uPA and MCD-fed mice but not in CCl4-treated animals, whose collagen deposition pattern was different (Extended Data Fig. 5g–i). Induction of alpha-fetoprotein and other regeneration antigens in MCD-HFD and CCl4-treated livers can explain the origin of HCC-independent, damage-related CD8+ T-cell activation (Extended Data Fig. 5k). Unlike CDD-HFD mice in which CD8+ T cells promote non-fibrotic liver damage7, CD8+ T cells do not contribute to NASH in MUP-uPA mice, which are highly fibrotic and depend on macrophage TNF production.
IgA+ cell and CTL interplay in HCC development
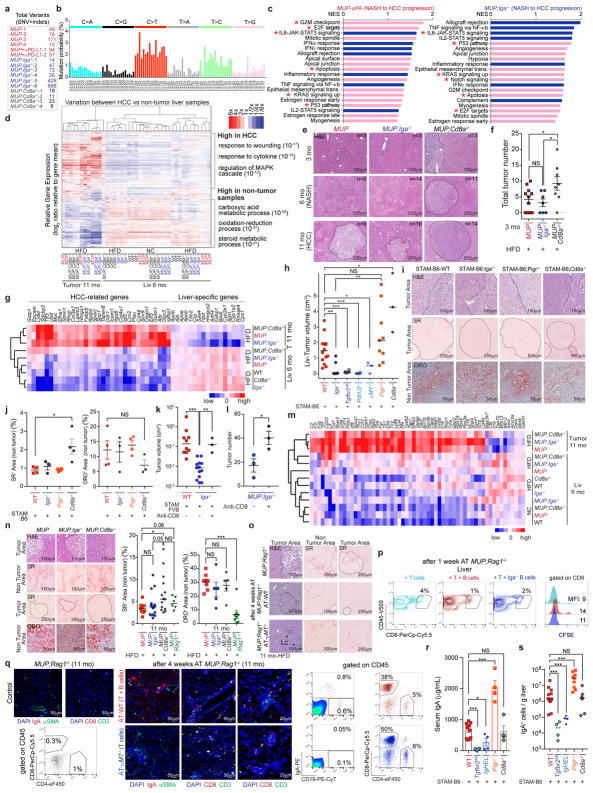
After 7 months of HFD, MUP-uPA mice developed HCC15, which contained numerous (usually 50–100 per tumour) non-recurrent coding mutations in pathways that are mutated in human HCC (Fig. 2d and Extended Data Fig. 6a). Mutational signatures were almost identical between mouse and human HCC (Extended Data Fig. 6b)20. Pathways and responses that were upregulated during MUP-uPA NASH to HCC progression were similar to those altered in human HCC (Extended Data Fig. 6c, d).
Cd8a ablation greatly accelerated HCC development: 12.5% of MUP-uPA/Cd8a−/− mice showed a single large tumour (> 0.4 cm) at 3 months and 27% contained such tumours at 6 months, at which point MUP-uPA or MUP-uPA/Iga−/− mice had no large tumours (Fig. 2e–g and Extended Data Fig. 6e, f). Similar observations were made in STAM mice (Fig. 2h, i). MUP-uPA/Cd8a−/− tumours had typical HCC histology and their early appearance was confirmed by whole-liver RNA sequencing (RNA-seq), showing elevated expression of HCC markers (Extended Data Fig. 6e–g), which are also expressed by HCC progenitor cells21. HCC development and burden were greatly reduced in IgA-deficient mice, some of which contained no large tumours at 11 months (Fig. 2e–g). Complete B-cell deficiency, replacement of the entire immunoglobulin repertoire with hen egg lysozyme (HEL)-specific IgM, B-cell-specific TGFβ receptor 2 ablation, and PD-L1/2 deficiency also inhibited HCC development (Fig. 2f–i and Extended Data Fig. 6h–j). By contrast, ablation or depletion of CD8+ T cells in IgA-deficient mice restored HCC development, whereas absence of T and B cells (Rag1−/−) enhanced HCC development as effectively as Cd8a ablation (Fig. 2f, i, j and Extended Data Fig. 6k, l), indicating that without T cells, B or IgA+ cells do not promote tumorigenesis. IgA ablation enhanced immune activation and graft rejection markers (Extended Data Fig. 6c, m). At the time of HCC analysis, Cd8a−/− livers were more fibrotic than MUP-uPA or STAM livers, but steatosis did not differ between WT, Cd8a−/− and Iga−/− mice (Extended Data Fig. 6i, j, n). These results strongly suggest that reduced tumorigenesis after Iga ablation is due to CTL reactivation. Indeed, adoptive transfer of T, but not T + B, cells into MUP-uPA/Rag1−/− mice suppressed tumorigenesis (Fig. 2j and Extended Data Fig. 6o–q).
Gut microbes promote HCC development
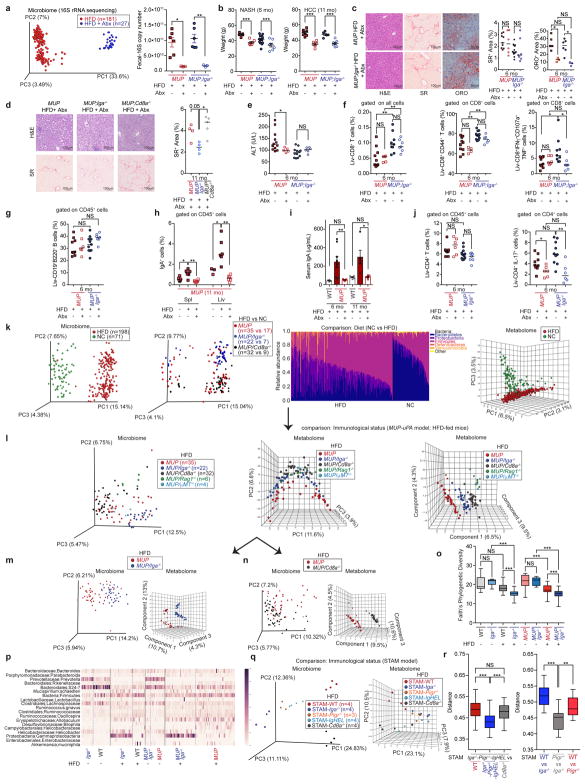
IgA dimers secreted by intestinal IgA+ plasma cells that maintain microbial homeostasis enter the intestinal lumen via the polyimmunoglobulin receptor (PIgR)22. Gut microbial components reach the liver via the portal circulation and enhance inflammation, fibrosis, and carcinogenesis23,24. To identify gut-specific secretory IgA functions, we examined NASH-induced HCC in Pigr−/− mice. Unlike IgA ablation, polyimmunoglobulin receptor deficiency potentiated tumorigenesis and increased circulating and liver IgA (Fig. 2h and Extended Data Fig. 6h, r, s). Congruently, broad spectrum antibiotics, which reduced gut bacteria by more than 90% (Extended Data Fig. 7a), attenuated HCC development in all mouse strains (Fig. 2f). Antibiotics reduced weight gain, liver steatosis, and inflammation, but had a marginal effect on liver fibrosis and damage, barely affected liver CD8+ effector cells and CD19+B220+ cells, and, as expected, reduced liver and circulating IgA and Th17 cells (Extended Data Fig. 7b–j). HFD feeding induced microbial dysbiosis; but even in HFD-fed mice, immune alterations dominantly modulated microbiota composition and faecal metabolites (Extended Data Fig. 7k–r). Despite opposing effects on tumorigenesis, the microbiota of Iga−/−, Pigr−/−, and IgHEL/MD4 mice showed similar deviations from the WT or Cd8a−/− microbiota on the basis of unweighted UniFrac distances (Extended Data Fig. 7r).
IgA+ plasmocytes suppress CTL activation
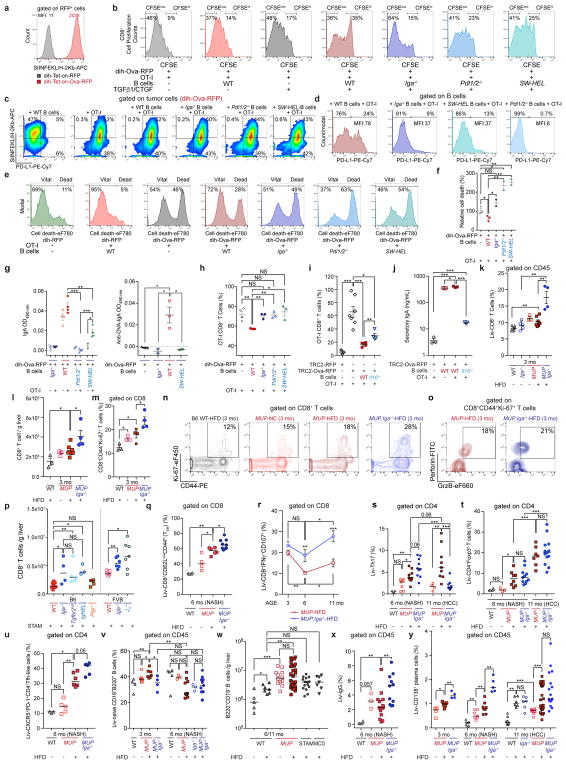
Liver IgA+ cells directly suppressed CTL activation. Ovalbumin (Ova)- expressing HCC-derived dih cells presented the SIINFEKL peptide on H-2Kb and activated ovalbumin-specific, CD8+ T cells (OT-I cells) (Extended Data Fig. 8a, b). Red fluorescent protein (RFP)- (controls) and Ova-expressing cells were starved overnight to induce some (5%) cell death and antigen release, and incubated with or without B cells from WT, Pdl1/2−/−, or SW-HEL25 mice. TGFβ 1 and connective tissue growth factor (CTGF) were added to induce IgA class switch recombination and, after their removal, OT-I cells were added. Whereas WT B cells modestly inhibited basal OT-I cell proliferation, this was strongly potentiated by TGFβ 1 + CTGF (Extended Data Fig. 8b). By contrast, Iga−/−, Pdl1/2−/−, and SW-HEL B cells had compromised inhibitory activity, indicating a requirement for PD-L1 and antigen-specific B-cell receptor (BCR) for CD8+ T-cell suppression. Iga−/− and SW-HEL B cells produced low PD-L1 levels, whereas dih cells incubated with IFNγ-producing OT-I cells expressed more PD-L1 (Extended Data Fig. 8c, d). Cancer cell killing by OT-I cells required Ova expression and was attenuated by WT, but not Iga−/−, Pdl1/2−/−, or SW-HEL B cells (Extended Data Fig. 8e, f). Incubation with dih-Ova cells, TGFβ 1, and CTGF, with or without OT-I cells, induced IgA production by WT B cells and, to a lesser extent, in SW-HEL B cells, but not in Pdl1/2−/− B cells; some of the secretory IgA was directed against Ova (Extended Data Fig. 8g). WT, but not Iga−/−, Pdl1/2−/−, or SW-HEL B cells, suppressed CD8+ T-cell expansion (Extended Data Fig. 8h). Similar results were obtained using Ova-expressing TRAMP-C2 cells14, indicating that suppression of OT-I CD8+ T cells by IgA+ cells was not unique to HCC (Extended Data Fig. 8i, j). These results suggest that effective T-cell suppression required IL-10, which was also needed for maximal IgA production.
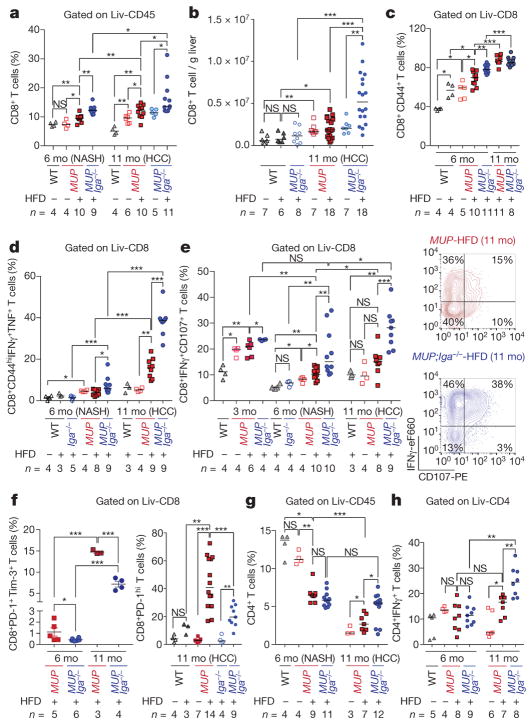
IgA+ cells also inhibited CTL activation in vivo. At 3 months, HFD-fed MUP-uPA/Iga−/− mice had significantly more liver CD8+ T cells than MUP-uPA mice, and this was maintained at later time points (Fig. 3a, b and Extended Data Fig. 8k, l). Importantly, MUP-uPA mice on HFD had more effector and degranulating CD8+ T cells at 3 months than at 6 months; however, such a difference was not found in MUP-uPA/Iga−/− mice, which showed even more activated CTL at 11 months (Fig. 3c–e and Extended Data Fig. 8m–r). Correspondingly, IgA ablation decreased the number of PD-1+Tim3+CD8+ exhausted T cells (Fig. 3f). These results suggest that, before IgA+ plasmocyte accumulation, CD8+ T cells are engaged in immunosurveillance, thereby explaining the absence of HCC in young MUP-uPA mice and its appearance after IgA plasmocyte accumulation and CD8+ T-cell exhaustion. As reported26, HFD feeding decreased liver CD4+ T cells, but increased relative Th17 and regulatory T-cell amounts, as well as Tfh-like (PD1+CXCR5+CD4+) cells (Fig. 3g and Extended Data Fig. 8s–u). IgA ablation had a minimal effect on these changes, although it increased HCC-infiltrating Th1 cells (Fig. 3h), while having little effect on liver B cells and increasing liver IgG+ and CD138+ cells during NASH (6 months) but not HCC (11 months) (Extended Data Fig. 8v–y).
Figure 3. Liver IgA+ cells induce CD8+ T-cell exhaustion.
a–f, Liver cells from indicated 3-, 6-, and 11-month (mo)-old mice (n = 3–18 per group) were stained and analysed by flow cytometry. The percentage of CD8+ T cells (a), absolute CD8+ T-cell number per gram of liver (b), and the percentage of CD44+CD8+ effector T cells (c). d, e, Liver CD8+ T cells from the indicated mice (n = 3–10) were stimulated with phorbol 12-myristate 13-acetate (PMA)/ionomycin medium and stained as indicated. The percentage of CD8+ T cells positive for CD44, IFNγ, and TNF (d), and IFNγ and CD107a (e). f, The percentage of CD8+Tim- 3+PD-1+ T cells at 6 and 11 months (left), and PD-1+ T cells at 11 months (right). g, h, Liver cell suspensions were stained for CD4 (g) and CD4 and IFNγ (h). The gating scheme is indicated above each panel. The data were validated in at least three independent experiments. Two-sided t-test (means ± s.e.m.; f, left) and Mann–Whitney test (median; a–h, f, right) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001. Specific n values are shown in panels.
PD-L1 blockade induces HCC regression
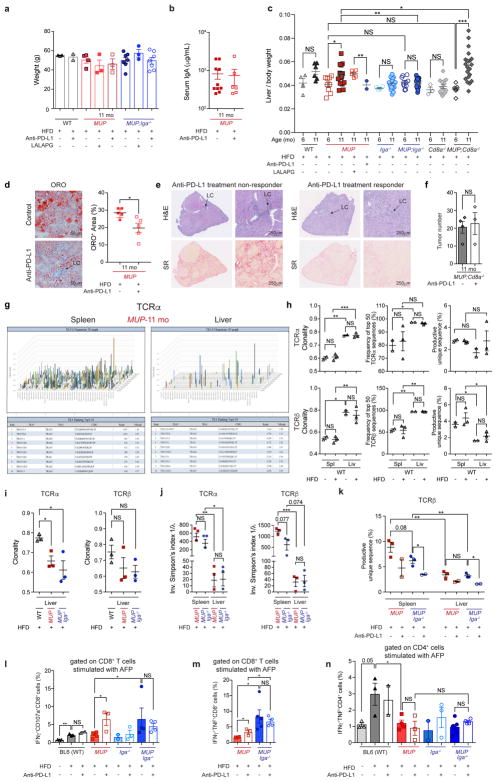
We placed MUP-uPA and MUP-uPA/Iga−/− mice on HFD for 7 months to establish HCC growth and injected them weekly with a PD-L1 blocking antibody (anti-PD-L1) or a control (LALAPG) that did not block PD-1 engagement (Extended Data Fig. 2a). PD-L1 blockade reduced tumour multiplicity and load, and most large tumours disappeared (Fig. 4a, b). The treatment reduced liver-to-body ratio without affecting body weight or reducing serum IgA levels (Extended Data Fig. 9a–c), further demonstrating HCC eradication. This beneficial effect was associated with tumoural lymphocyte accumulation (Fig. 4c, d) and decreased steatosis (Extended Data Fig. 9d). Anti-PD-L1 non-responsive tumours lacked immune cells and were encapsulated by fibrotic tissue (Extended Data Fig. 9e). PD-L1 blockade substantially decreased liver IgA+IL-10+ cell abundance, probably because of disrupted PD-L1:PD-1 B-cell–Tfh interactions, needed for plasma cell maturation27, and increased CD8+ T cells, including proliferating cells expressing IFNγ and GrzB (Fig. 4e, f). PD-L1 neutralization did not decrease tumour burden in MUP-uPA/Iga−/− or MUP-uPA/Cd8a−/− mice (Fig. 4a and Extended Data Fig. 9f), indicating that it acts via IgA+ cells and targets CD8+ T cells.
Figure 4. PD-L1 blockade reactivates and expands antigen-specific CD8+ T cells to induce HCC regression.
Tumour-bearing MUP-uPA mice (n = 7–10) were given weekly injections for 8 consecutive weeks of blocking (anti-PD-L1) or control (LALAPG) PD-L1 antibodies or left untreated. a–c, Tumour number (a), representative gross liver morphology (b), and tumour histology (c). H&E, haematoxylin and eosin. Arrow in c shows a typical inflammatory infiltrate (scale bars, 100 μm). d, Liver sections were stained for CD3 and CD8 and examined by fluorescence microscopy (scale bars, 50 μm). Experiments were repeated at least two or three times. e, f, Liver CD45+ cells were stained as indicated and analysed by flow cytometry. g, h, CD8+ T cells were sorted from spleens and livers of indicated mice (n = 9) and sequenced for TCR α - (g) and β - (h) chain CDR3 region (18 samples). Panels show clonality and frequency of top 50 TCR α - and β-sequences. i, Similar analysis was performed on CD8+ T cells from spleens and livers of anti-PD-L1-treated (eight samples) or control (−) HCC-bearing mice. Panel shows the percentage of productive unique TCRα sequences. Two-sided t-test (means ± s.e.m.; g–i) and Mann–Whitney test (median; a, e, f) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001; Specific n values are shown in a, e, f.
Clonal expansion of liver CD8+ T cells
We sequenced CD8+ TCR α - and β-chains from livers and spleens of HCC-bearing MUP-uPA and MUP-uPA/Iga−/− mice and age-matched controls. In HCC-bearing mice the TCR repertoire was much less diverse in liver than in spleen (Fig. 4g, h, left, and Extended Data Fig. 9g). The liver TCRα β repertoire of control BL6 mice was less diverse than that of HCC-bearing mice and was unaffected by HFD feeding (Fig. 4g, h, right, and Extended Data Fig. 9h, i). Although IgA ablation enhanced CD8+ T-cell activation, it barely effected liver/HCC CD8+ cell TCR diversity (Fig. 4g, h and Extended Data Fig. 9i, j). PD-L1 neutralization clonally expanded liver/HCC CD8+ T cells, some of which were alpha-fetoprotein directed (Fig. 4i and Extended Data Fig. 9k–m). Alpha-fetoprotein-specific CD8+ T cells were elevated after anti-PD-L1 treatment or IgA ablation. By contrast, HFD-fed BL6 mice contained more alpha-fetoprotein-specific CD4+ T cells than HCC-bearing mice, in which CD4+ cell abundance was unaffected by anti-PD-L1 or IgA ablation (Extended Data Fig. 9n). BCR sequencing showed clonal expansion of a small repertoire of liver-infiltrating IgA- and IgM-producing cells in HCC-bearing mice (Extended Data Fig. 10a, b). Both T- and B-cell clonal expansion could be driven by tumour antigens.
Discussion
CD8+ T cells can eradicate established tumours, but evidence that adaptive immunity curtails early cancer development is scant4, or has been derived from chemically induced mouse sarcomas that evade destructive T-cell responses by downregulating rejection antigens3. Our results provide one of the strongest and most direct demonstrations that adaptive immunity actively prevents cancer emergence. In fact, liver immunosurveillance is so effective that, without immunosuppressive IgA+ cells that directly inhibit HCC-directed CTL activity, NASH to HCC progression is severely hampered. Immunosuppressive IgA+ cells are by-products of chronic liver inflammation, which causes Tfh accumulation and elevated TGFβ, IL-21, and IL-33, and they are distinct from intestinal IgA+ B cells that maintain microbial homeostasis and suppress liver inflammation and tumorigenesis. PD-L1, first shown to prevent excessive CD8+ cell activation during viral infections28, is also needed for generation of immunosuppressive IgA+ cells29. Given that intestinal IgA suppresses inflammation and tumorigenesis, the reduction in NASH-induced tumorigenesis in Iga−/− mice is partly compromised by the increase in liver inflammation, and could therefore be greater than actually apparent.
Our ability to unravel the cancer-preventive effect of adaptive immunity is probably due to a mouse model that closely mimics NASH-driven human HCC and does not rely on engineered mutations and tumour suppressor ablation that trigger rapid malignant progression without additional neo-antigen-generating genetic alterations30. NASH-driven liver carcinogenesis depends on ongoing stress and inflammation15,23, which underlie human HCC, whose mutational landscape and signature are almost synonymous with MUP-uPA HCC20. The immunopathogenesis of NASH to HCC progression in MUP-uPA and STAM mice is also very similar to human NASH-driven HCC, with the pre-cancerous liver exhibiting accumulation of PD-L1- and IL-10-expressing IgA+ cells, paralleling the appearance of inflammation-induced HCC progenitor cells. IgA+ cell depletion, via Iga, Tgfbr2, or PD-L1 ablation or blockade, enhances HCC progenitor cell eradication, and regression of established tumours through CTL activation, despite the potential increase in intestinal dysbiosis. Liverinfiltrating IgA+ cells do not affect liver injury and are not unique to NASH, as they are also found in ASH and other chronic hepatides. Another feature of human NASH and our mouse models is accumulation of CD8+ T cells, whose ablation resulted in a sevenfold to eightfold reduction in tumour latency, suggesting they eradicate HCC cells as soon as they appear. Although hepatic CD4+ T cells decline in HFD-fed MUP-uPA mice, as found previously26,31, the frequency of Th17 and regulatory T cells that are cancer supportive increases. Nevertheless, the key immune-related HCC-promoting mechanism triggered during NASH entails IgA+ cell generation. Further evidence for the immunosurveillance function of CD8+ T cells is provided by their oligoclonality and reduced diversity in HCC, and their reinvigoration and expansion after IgA+ cell depletion. Our results appear applicable to human HCC, as suggested by a clinical trial that revealed the utility of PD-1 blockade with only one patient developing treatment-related hepatitis32, showing that activated CD8+ T cells rarely damage the liver.
Methods
Animal models
C57BL/6 and FVB/NJ control mice were purchased from Charles River Laboratories, and Cd8a−/−, Il10-GFP reporter mice33 and μMT−/− mice34 were purchased from the Jackson Laboratory. IgA gene-deficient (Iga−/−) mice35 were obtained from Baylor College of Medicine. MUP-uPA mice were previously described15 and were crossed with B-cell-deficient (μMT−/−) mice34, CTL-deficient (Cd8a−/−) mice36, Iga−/− or Rag1−/− (ref. 37) mice. B-cell-deficient (JH −/−)38, IgHEL/MD4 (ref. 39) (monoclonal for BCR specific for HEL), Cd8a−/−;Iga−/− double knockout, Pdl1/2−/− (Genentech, San Francisco, California, USA), and Pigr−/− (ref. 40) mice were subjected to STAM, MCD, and CCl4-protocol. Cd19-Cre mice were crossed to Tgfbr2F/F (ref. 41) with both BL6 and FVB/NJ background, and were also used as indicated. Cd8a−/−, Iga−/−, and JH −/− were crossed to FVB/NJ for at least ten generations and were used for experiments in both BL6 and FVB/NJ backgrounds. Iga−/−, Pdl1/2−/−, SW-HEL (expressing HEL-specific monoclonal BCR that could switch to IgG and IgA)42, and OT-I mice43 (Taconic) were used for cell culture experiments. All mice were maintained in filter-topped cages on auto-claved food and water at the University of California San Diego (UCSD) animal facility and were used in accordance with UCSD and NIH guidelines and regulations, particularly for experimental neoplasia, which includes tumour size limits (not exceeding 2 cm for a single tumour) and monitoring parameters. Tumour volumes were calculated as (width2 × length)/2, and for multiple spontaneous liver tumours the volumes of single tumours were added for a total tumour volume. When indicated, mice were fed HFD starting at 8 weeks of age for the MUP-uPA HFD-fed model15 or at 4 weeks for the STAM model44, respectively (Extended Data Fig. 2a). In the MCD-HFD model, indicated mice were kept on an MCD diet with 60 kcal% fat, for 4 weeks (Extended Data Fig. 2a). In the CCl4 model, indicated mice were injected intraperitoneally with CCl4 twice a week for 8 weeks (Extended Data Fig. 2a).
Mouse treatment studies were ‘matched design control trials’. Accordingly, mice were randomly chosen and paired on the basis of gender (male), weight, and age. An identification code was assigned to each mouse and the investigators were blinded to treatment allocation at the time of analysis. Mice that did not gain weight or showed sign of dermatitis, a rarely observed phenotype in MUP-uPA HFD-fed mice, were excluded from the study. The numbers of mice used in experiments are shown in Supplementary Table 1 (MUP-uPA model) and Supplementary Table 2 (STAM, MCD, and CCl4 models). MUP-uPA (n = 543 mice), STAM (n = 123 mice), MCD (n = 65 mice), and CCl4 (n = 82 mice) models were analysed in 50, 15, 3 and 6 experiments, respectively. Most of the basic characterization of control BL6, Iga−/−, and Cd8a−/− mice is shown in Extended Data Fig. 10. These groups were compared to exclude the general effect of IgA and CD8 deficiencies on weight gain and immune status. Non-treated control mice that were used are not shown in Supplementary Tables 1 and 2, but are mentioned in each of the figure legends. Flow cytometry. Cell suspensions were prepared from livers, spleens, intestine, and visceral fat as described14. Lymphocytes were isolated from human blood using Ficoll-Paque PLUS (GE Healthcare Life Science) according to the manufacturer’s recommendations. For lymphocyte isolation, tissues (liver, intestine, and HCC tumours) were cut into small pieces and incubated in dissociation solution (DMEM medium supplemented with 5% FBS, collagenase type I (200 U ml−1), collagenase type IV (100 U ml−1) and DNase I (100 μg ml−1)) for 30 min at 37 °C. After incubation, cell suspensions were passed through a 40 μm cell strainer and washed twice. To block Fc-mediated interactions, mouse cells were pre-incubated with 0.5–1 μg of purified anti-mouse CD16/CD32 per 100 μl, and human cells were incubated with FcR blocking reagent (Miltenyi Biotec). Isolated cells were stained with labelled antibodies in PBS with 2% FBS and 2 mM EDTA or cell staining buffer (Biolegend). Dead cells were excluded on the basis of staining with Live/Dead Fixable Viability Dye (FVD-eFluor780, eBioscience). For intracellular cytokine staining, cells were re-stimulated with alpha-fetoprotein (5 μg ml−1) or cell stimulation cocktail (eBioscience; containing PMA and ionomycin), in the presence of a protein transport inhibitor cocktail containing Brefeldin A and Monensin (eBioscience). For measurement of IL-10 production by B cells, lipopolysaccharide was added to the above-described PMA/ionomycin medium. For CD107a measurement, a staining antibody was added to the culture during the stimulation. After 4–5 h, cells were fixed and permeabilized either with BD Cytofix/Cytoperm reagent for cytokine staining, or eBioscience Foxp3/Transcription Factor staining buffer for combined staining of cytokines with transcription factors. After fixation/permeabilization, cells were stained with labelled antibodies of interest. Cells were analysed on a Beckman Coulter Cyan ADP flow cytometer. Data were analysed using FlowJo 10.2 software (Treestar). Absolute numbers of particular immune cells in spleen were calculated by multiplying the total cell numbers from one spleen by the percentages of the particular cell type among total cells. Absolute numbers of particular immune cells (for example, IgA+ and CD8+ cells) in livers were calculated by multiplying the cell number in one liver portion by the percentages of the corresponding cell type in total liver cells divided by the weight of the analysed liver fragment (cell number per gram of liver).
Human specimens and analysis
It has previously been demonstrated that serum IgA levels correlate with hepatic fibrosis stage in the Newcastle NAFLD Clinic cohort13. A larger NAFLD cohort from Newcastle (n = 502) was analysed and is shown in Extended Data Fig. 1a. Furthermore, the study included 18 patients with biopsy-proven NAFLD (11 in initial phase, 7 in second phase) and 5 patients with HCC who attended the Newcastle Hospitals fatty liver clinic between 1999 and 2016. NAFLD cases were selected for this study on the basis of high serum IgA/advanced fibrosis (n = 9) versus low serum IgA/mild fibrosis (n = 9). Liver biopsies were performed as part of the investigation of abnormal liver function tests or to stage disease severity in patients with ultrasound evidence of NAFLD. Clinical and laboratory data were collected from the time of liver biopsy. Alternative diagnoses were excluded, including increased alcohol intake (males and females consuming more than 21 and 14 units of alcohol per week (> 30/20 g d−1 ethanol), respectively, were excluded, as were any individuals with chronic viral hepatitis (hepatitis B and hepatitis C), autoimmune liver diseases, hereditary hemochromatosis, α 1-antitrypsin deficiency, Wilson disease, and drug-induced liver disease. In addition, patients in whom immunoglobulins were not available within 6 months of liver biopsy were excluded. Blood test results were taken from the time of liver biopsy or within 6 months. The normal ranges for serum IgA, IgG, and IgM were 0.64–2.97 g l−1, 5.8–15.4 g l−1, and 0.71–2.3 g l−1, respectively. Percutaneous liver biopsies were performed using a Menghini needle or an 18-gauge BioPince liver biopsy system (Medical Devices Technologies, Gainesville, Florida, USA). Histological scoring was performed according to the NIH NAFLD Clinical Research Network criteria. Liver biopsy specimens were assessed by experienced hepatopathologists (A.D.B., Q.M.A., and J.B.). In the current study, formalin-fixed, paraffin-embedded liver tissue was stained for IgA, CD8, and CD20 and assessed by a single histopathologist (J.B.) using a semi-quantitative score. All statistical analyses were performed using SPSS software version 22.0 (SPSS, Chicago, Illinois, USA). All the Newcastle patients gave informed consent. Ethical approval to establish the Newcastle HPB Biobank from which these samples were used was received from the Newcastle & North Tyneside 1 Research Ethics Committee, REC reference 10/H0906/41.
In a second cohort from UCSD (n = 96), serum IgA, IgM, and IgG levels from corresponding patients were analysed and are shown in Supplementary Table 3.
Anonymized samples from consenting human participants were provided with UCSD Institutional Review Board Ethical Committee approval. Liver biopsy assessment was performed by a trained and experienced hepatopathologist (R.L.). All patients had a clinical indication for a liver biopsy and underwent a core biopsy of the liver using a 16-gauge BioPince needle. All biopsies were read using the NASH CRN Histologic Scoring System45.
Immunostaining
Tissues were embedded in Tissue-Tek OCT compound (Sakura Finetek, Torrance, California, USA) and snap-frozen. Tissue sections were fixed in cold acetone/methanol or 3% PFA for 3–10 min and washed with PBS. Slides were blocked with 1× PBS/1% normal donkey or goat serum for surface staining or 0.2% gelatin (from cold water fish skin; Sigma-Aldrich)/PBS/1% normal donkey or goat serum for intracellular staining for 30 min. Sections were incubated with primary antibodies for 1 or 12 h at room temperature or 4 °C, respectively. After washing with PBS, secondary antibodies were added for 1 h at room temperature. As negative controls, samples were incubated with isotype-matched control antibodies or secondary antibodies only. After staining with DAPI, sections were covered with Vectashield Mounting Medium (Vector Laboratories, Burlingame, California, USA). Paraffin-embedded tissue sections were subjected to de-paraffinization and rehydration, and then were immersed in a pre-heated antigen retrieval water bath with a pH 6.1 citrate buffer, or Dako Target Retrieval Solution for 20 min at 95–96 °C. All stainings were done according to the manufacturer’s protocols (Vector Laboratories). Nuclei were lightly counterstained with a freshly made haematoxylin solution then further washed in water. Sections were examined using an Axioplan 200 microscope with AxioVision Release 4.5 software (Zeiss, Jena, Germany) or a TCS SPE Leica confocal microscope (Leica, Germany).
Histology
Livers were fixed in 10% neutral-buffered formalin or 4% paraformal-dehyde, embedded in paraffin, sectioned, stained with haematoxylin and eosin, Sirius Red, and processed for immunohistochemistry. For frozen-block preparation, tissue was embedded in Tissue-Tek OCT compound, and were stained with Oil Red O and haematoxylin and eosin. Whole-slide images were analysed using a Hamamatsu 2.0-HT Digital slide scanner and processed using the NDP View Hamamatsu software (Hamamatsu Photonics, EU, Japan, and USA). For Sirius Red analysis, areas of at least 1 mm3 were quantitated using Image J 1.50i software (NIH, Maryland, USA) and normalized for vascularization and lipid accumulation using corresponding haematoxylin and eosin areas. For Oil Red O analysis, multiple images (two or three) were examined for each tissue section using an Axioplan 200 microscope with a 20× objective with AxioVision Release 4.5 software and quantitated as averages using the Image J software.
Antibodies
Antibodies specific for the following antigens were used (anti-mouse antigens were abbreviated as ‘m’, and anti-human as ‘h’): Tim-3 (mB8.2C12- AlexaF647; hF38-2E2-PerCP-eF710); CD138 (m281-2-PE/APC; hDL-101- APC); IgA (mA-6E1-PE/FITC; m11-44-2-Biotin; mRMA-1; hIS11-8E10- FITC; DAKO hA0262); CD8α (m53-6.7-PerCp-Cy5.5/PE-Cy7; hRPA-T8-PE; hC8/144B); CD45 (m30-F11-V500; hHI30-eF450); CD20 (mAISB12-PE); CD44 (mIM7-PE/FITC/eF450); CD4 (mRM4-5-eF450; mGk1.5-FITC/PE/PE-Cy7; hRPA-T4-APC); B220 (mRA3-6B2-eF450/FITC); CD19 (meBio1D3-eF660/PE-Cy7; hHIB19-PE); IgM (mII/41-PE-Cy7; hSA-DA4-eF450); IgD (m11- 26c.2a-Pacific-Blue; m11-26c-PerCP-eF710); TNF (mMP6-XT22-FITC/PE); IFNγ (mXMG1.2-eF660); GrzB (mNGZB-eF660); CD107a (meBio1D4B-PE); PD-1 (mJ43-eF450/FITC; mRMP1-30-APC; hMIH4-FITC); PD-L1 (mMIH5- PE-Cy7/PE; hMIH1-PE-Cy7); FAS-L1 (mMFL3-FITC); Ki-67 (mSolA15-eF450); IgG2a (m2a-15F8-FITC/APC; mRMG2a-62-PE); IgG1 (mM1-14D12-eF450; mRMG1-1-FITC); IL-10 (mJES5-16E3-PE); CD11c (mN418-eF450/PE); CD11b (mM1/70-eF450/eF660); MHCII (mM5/114.15.2-FITC/PE); Gr-1 (m1A8-Ly6G-PerCP- eF710); IL-17 (meBio17B7-PE-Cy7); Foxp3 (mFJK-16s-eF450); Perforin (meBioOMAK-D-FITC); CXCR5 (mSPRCL5-PE/APC/Biotin); CD62L (mMEL- 14-PE/APC); SIINFEKL/H-2Kb (meBio25-D1.16-APC); F4/80 (mBM8-PE/FITC); anti-Biotin (BK-1/39-PE); CD5 (m53-7.3-FITC/eF450) and NK1.1 (mPK136- APC); CD31 (m390-eF450) (most from eBioscience and Biolegend); pAb rabbit to CD3 (DAKO, IS503); and alpha-SMA (m-ab5694; DAKO, h1A4). The following Alexa 594-, Alexa 647-, and Alexa 488-conjugated secondary antibodies were used: donkey anti-rat IgG, donkey anti-rabbit IgG, donkey anti-goat IgG, and goat anti-rat IgG (Molecular Probes, Invitrogen).
Cell culture experiments for analysing the ex vivo immunosuppression and IgA class switch recombination
Two inducible Ova–P2A–RFP-expressing dih10 and dihXY HCC cell lines and prostate cancer cell line TRAMP-C2 (ATCC) were developed (dih10–Ova–RFP, dihXY–Ova–RFP, TRC2–Ova–RFP), and ovalbumin processing confirmed, which resulted in presentation of SIINFEKL peptide on MHCI (H-2Kb). This peptide can be recognized by OT-I CD8+ T cells, which express a high-affinity TCR specific for ovalbumin. Both dih cell lines were established previously in our laboratory using diethylnitrosamine-induced HCC and were regularly tested for mycoplasma (MycoAlert Mycoplasma Detection kit) and genomic PCR to confirm their characterization46.
Dih–RFP, TRC2–RFP or ova-expressing dih, TRC2 cells were starved over or treated with low-dose chemotherapy for 24 h (approximately 5–8% cell death). After that, the medium was changed and magnetic-bead-sorted B cells (7 × 105 per well) from either WT, Pdl1/2−/−, Il10−/−, or SW-HEL (monoclonal BCR specific for hen egg lyzosyme-HEL, however, which could still switch to other immunoglobulin classes upon BCR and/or cytokine signalling) mice were added in the presence of TGFβ 1 (5 ng ml−1) and CTGF (3 ng ml−1), for an additional 24 h. After TGFβ 1/CTGF removal, CFSE-labelled OT-I T cells (specific for ovalbumin) were added to the cultures (3 × 105 per well) that either contained or did not contain the B cells described. T cells were isolated from Rag1−/−/OT-I mice using CD8− selection beads and their proliferation was analysed on days 3, 4, 5, 6, and 7. The proliferation and the amount of CFSE-labelled OT-I CD8+ cells were analysed by flow cytometry and quantitated. IgA concentrations in the culture supernatants were determined using ELISA. Induction of cancer cell death was analysed using LIVE/DEAD cell staining dyes (FVD-eF780, eBioscience). The percentage of CD19+ cells, antigen presentation (SIINFEKL/H-2Kb), and PD-L1 expression were also analysed using flow cytometry.
ELISA and colorimetric protein detection
Mouse IgA and IgG ELISA kits from eBioscience were used according to the manufacturer’s protocols. Human serum IgA and IgG were measured in the first cohort from Newcastle, as described previously13, or in cohort two at the UCSD Medical Center Laboratory. ELISA for the determination of OVA-specific IgA (anti-OVA-IgA) in the co-culture experiment was performed by adding culture supernatant to the ovalbumin-coated wells, incubating at 4 °C overnight, and washing with phosphate buffered saline with Tween 20. Rat anti-mouse IgA antibody (RMA-1, Biolegend) was added and incubated at room temperature for 1 h; thereafter, anti-rat HRP-conjugated antibody was added for a further 30 min. After washing with phosphate buffered saline with Tween 20, 1× TMB was added and the reaction was stopped with 2 N H2SO4, followed by detection in a microplate reader at 450 nm. Mouse serum ALT was measured using ALT (GPT) reagent (Thermo Fisher Scientific). Triglyceride and total cholesterol contents were measured using a Triglyceride Colorimetric Assay Kit (Cayman Chemical) and a Cholesterol Fluorometric Assay kit (Cayman Chemical) according to each manufacturer’s instructions.
Glucose tolerance test
Mice were transferred to clean cages with no food or faeces in the hopper or bottom of the cage and fasted for approximately 6 h with access to drinking water. Small drops of blood were obtained from tail cuts and assessed for baseline glucose levels using a One-touch Ultra 2 (Lifescan, Johnson & Johnson) glucometer. A 20% glucose solution was intraperitoneally injected (2 g/kg mouse body mass). Blood was sampled at 30, 60, and 120 min. Dried blood was quickly removed from the tail wounds.
Liver perfusion and isolation of hepatic stellate cells and Kupffer cells
The hepatic stellate cells and Kupffer cells were isolated as previously described47. In particular, the suprahepatic inferior vena cava was tightened and the inferior vena cava was cannulated and perfused with 50 ml Hanks’ balanced salt solutions and then the portal vein was cut. The perfusion was continued with 50 ml Dulbecco’s modified Eagle medium F/12 (DMEM F/12; GibcoBRL) containing 12.5 mg pronase, followed by 50 ml DMEM F/12 containing 33.3 mg collagenase D (Roche). The liver was removed and agitated in 50 ml DMEM (GibcoBRL) containing 0.2 mg DNase I, 17 mg pronase, and 17 mg collagenase D at 42 °C for 10 min. The cell suspension was filtered through a 70 μm cell strainer (Sigma). The hepatic stellate cells were purified by 8.2% Nycodenz (Accurate Chemical and Scientific Corporation), and Kupffer cells were isolated from the non-parenchymal cell fraction by CD11b-conjugated magnetic beads (Miltenyi). The purity was confirmed by flow cytometry (Kupffer cells, ~ 83%; hepatic stellate cells, ~ 85%).
Antibody treatments
Anti-PD-L1 and LALAPG (which binds to PD-L1, but cannot inhibit the cell signalling) antibodies were intraperitoneally injected at 10 mg/kg (body mass) weekly. Mice were treated for 8 weeks in total. Anti-CD8a antibodies were intraperitoneally injected weekly (150 μg per mouse). Mice were treated for 6–8 weeks in total. PD-L1 and control antibodies were provided by I. Mellman (Genentech).
Antibiotic treatment
Ampicillin (1 g l−1; Sigma), vancomycin (500 mg l−1; MIP Pharma, Blieskastel, Germany), neomycin sulfate (1 g l−1; Sigma), and metronidazole (1 g l−1; OTC Pharma GmbH, Kamen, Germany) were applied through the drinking water as described in Extended Data Fig. 2a.
Adoptive lymphocyte transfer
For adoptive splenocyte transfer, single-cell suspensions prepared from spleens were transferred intraperitoneally with one total spleen injected per mouse. Labelling with 5- (and 6-) carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Eugene, Oregon, USA) was done according to the manufacturer’s protocol. Four million CD8+ T cells alone or combined with 10 × 106 B cells were transferred (equivalent to one spleen). For adoptive lymphocyte transfer, T cells were isolated from single-cell suspensions of spleens using a Pan T Cell Isolation Kit (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany), and B cells were isolated using a Pan B Cell Isolation Kit. T cells were transferred intraperitoneally alone or with B cells (3 × 106 to 5 × 106 each). The purity of sorted cells was confirmed by flow cytometry (B cells, ~ 90%; T cells, ~ 95%).
Mouse TCR and BCR repertoire analysis
Total RNA was extracted from sorted CD8+ T or B cells with an RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Next-generation sequencing was performed with an unbiased TCR repertoire analysis technology developed by Repertoire Genesis (Osaka, Japan). An unbiased adaptor-ligation PCR was performed according to the previous report48. In brief, total RNA was converted to complementary DNA (cDNA) with Superscript III reverse transcriptase (Invitrogen, Carlsbad, California, USA)48. Then, double strand (ds)-cDNA was synthesized and an adaptor was ligated to the 5′ end of the ds-cDNA and cut with SphI restriction enzyme. For TCRα, PCR was performed with a P20EA adaptor primer and a TCR α-chain constant region-specific primer (mCA1). The second PCR was performed with mCA2 and P20EA primers using the same PCR conditions. For TCRβ, mCB1 and mCB2 primers were used for the first PCR and the second PCR, respectively. For BCR, the following primers were used: mCM1: GTGTTGGGAAGGTTCTGATA; mCM2: TTCAGTGTTGTTCTGGTAG (IgM); mBCA1: TAGAGTCATGTTGC ACGGAA; mBCA2: GTTATATCCTTCCCACTCT (IgA). After Tag PCR amplification, index (barcode) sequences were added by amplification with a Nextera XT Index Kit v2 setA (Illumina, San Diego, California, USA). Sequence was done with the Illumina MiSeq paired-end platform (2 × 300 base pairs (bp)). Data processing, assignment, and data aggregation were automatically performed using repertoire analysis software originally developed by Repertoire Genesis. TCR and BCR (IgM and IgA) sequences were assigned with a data set of reference sequences from the international ImMunoGeneTics information system (IMGT) database (http://www.imgt.org). Nucleotide sequences of CDR3 regions ranged from conserved cysteine at position 104 (Cys104) of IMGT nomenclature to conserved phenylalanine at position 118 (Phe118) and the following glycine (Gly119) was translated to deduced amino-acid sequences. A unique sequence read was defined as a sequence read having no identity in V, J, and deduced amino-acid sequence of CDR3 with the other sequence reads. The copy numbers of identical unique sequence reads were automatically counted by RG software. TCR and BCR analyses were provided by Repertoire Genesis.
Real-time PCR analysis
Total RNA and DNA was extracted using an AllPrep DNA/RNA Mini Kit (Qiagen). RNA was reverse transcribed using a Superscript VILO cDNA synthesis kit (Invitrogen). Real-time PCR (RT-PCR) was performed as described14. The relative expression levels of target genes were measured in triplicates and normalized against the level of Rpl32 expression. Fold-difference (as relative mRNA expression) was calculated by the comparative CT method (2Ct(Rpl32–gene of interest)). Primers obtained from the NIH qPrimerDepot (http://mouseprimerdepot.nci.nih.gov) were as follows: Rpl32, forward 5′-GGGAGCAACAAGAAAACCAA-3′, reverse 5′-TTGTGAGCAATCTC AGCACA-3′; Tgfb1, forward 5′-GGAGAGCCCTGGATACCAAC-3′, reverse 5′-AAGTTGGCATGGTAGCCCTT-3′; Il21, forward 5′-CCCTTGTCTG TCTGGTAGTCATCTT-3′, reverse 5′-GGAGGCGATCTGGCCC-3′; Bcl6, forward 5′-AGTTTCTAGGAAAGGCCGGA-3′, reverse 5′-ACTAGCG TGCCGGGTAAACT-3′; Icos, forward 5′-ACAGGGGAGGAGAAGACTGC-3′, reverse 5′-CGAGCCATTGATTTCTCCTG-3′; Il33, forward 5′-GAGA AATCACGGCAGAATCA-3′, reverse 5′-CTTCTTATTTTGCAAGGCGG-3′; Cxcr4, forward 5′-AGTGACCCTCTGAGGCGTTT-3′, reverse 5′-TCCAGACCC CACTTCTTCAG-3′; Cxcr3, forward 5′-AAGTGCCAAAGGCAGAGAAG-3′, reverse 5′-CAAAGTCCGAGGCATCTAGC-3′; Cxcl12, forward 5′-TCCTC TTGCTGTCCAGCTCT-3′, reverse 5′-GGTAGCTCAGGCTGACTGGT-3′; Cxcl10, forward 5′-CGTCATTTTCTGCCTCATCC-3′, reverse 5′-CCTATGGC CCTCATTCTCAC-3′; Cxcl9, forward 5′-GAGGCACGATCCACTACAAA-3′, reverse 5′-AGTCCGGATCTAGGCAGGTT-3′; Ccl19, forward 5′-CTTCAGCCT GCTGGTTCTCT-3′, reverse 5′-GGAAGGCTTTCACGATGTTC-3′; Cxcl13, forward 5′-AGGTTGAACTCCACCTCCAG-3′, reverse 5′-TTGTGTAA TGGGCTTCCAGA-3′; Ccl25, forward 5′-GAAACTGTGGCTTTTTGCCT-3′, reverse 5′-TTCCATTTGATCCTGTGCTG-3′; Il7, forward 5′-GCCACATTA AAGACAAAGAAGGT-3′, reverse 5′-TGGTTCATTATTCGGGCAAT-3′; Ctgf, forward 5′-CCACCCGAGTTACCAATGAC-3′, reverse 5′-GCTT GGCGATTTTAGGTGTC-3′; Ltb, forward 5′-CTTTTCTGAGCCTG TGCTCC-3′, reverse 5′-ATCACTGTCCTGGCTGTGC-3′; Col1a1, forward 5′-ACATGTTCAGCTTTGTGGACC-3′, reverse 5′-TAGGCCATTGTG TATGCAGC-3′; Afp, forward 5′-GGCGATGGGTGTTTAGAAAG-3′, reverse 5′-TCTTTCCACTCCACTTTGGC-3′.
RNA-seq library preparation and sequencing
Total RNA was isolated as described above from 30 livers and 11 HCC nodules as indicated. In particular, some of the livers from 6-month-old mice were homogenized with TRIzol reagent and extracted with chloroform/isopropanol, followed by purification using an RNeasy Mini Kit (Qiagen). RNA purity was assessed by an Agilent 2100 Bioanalyzer. Sequencing libraries were prepared from 100 ng of total RNA using a TruSeq Stranded mRNA Library Prep Kit (Illumina) according to the manufacturer’s protocol. Samples were ligated to unique adaptors and subjected to PCR amplification. Libraries were then validated using a 2100 BioAnalyzer (Agilent), normalized, and pooled for sequencing. RNA-seq libraries prepared from at least three biological replicates for each group as indicated were sequenced on an Illumina HiSeq 2500 using barcoded multiplexing and a 100-bp read length. Image analysis and base calling were done with Illumina HiSeq2500 control software v2.17. The RNA-seq data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE90497.
RNA-seq processing and analysis
RNA-seq reads were aligned to the mouse genome (GRCm38/mm10) using STAR (v2.5.2b)49. Only reads that mapped to a single, unique location in the mouse genome were used for downstream analysis. Gene expression counts were counted for reads overlapping exons for all RefSeq transcripts using HOMER50. Differentially expressed genes were determined using DESeq2 (ref. 51) using thresholds of 5% false discovery rate and a fold change of 1.5 to determine the final lists. Gene expression profile clustering was performed by first using the DESeq2 rlog variance stabilization transform to normalize the gene expression values. Heirarchical clustering was performed using Cluster3.0 (ref. 52) and visualized using Java TreeView53. Pathway/functional enrichment calculations were performed using Metascape (http://metascape.org)54. Gene set enrichment analysis was also performed to identify hallmark genesets55,56. The obtained results from the mouse geneset were compared with previous published human data20,57.
Exome sequencing, processing, and analysis
DNA was isolated as described above from 9 spleens and 16 HCC nodules as indicated. Exome sequencing was performed at the University of California San Diego, IGM Genomic Centre using standard protocols. The exome sequencing data have been deposited in the NCBI Short Read Archive under accession numbers SRA556071 and SRP104724.
Exome sequencing data were processed into genetic variants using the Genome Analysis Tool Kit (GATK). First, exome capture reads were aligned with BWA (version 0.7.15)58 to the mouse genome (GRCm38/mm10). GATK (version 3.5) was used to process all exome sequencing data following the recommended best practices workflow59,60. Base recalibration was performed using the Mouse Genome Project SNP database as a reference for known SNPs61. Variant calls were made using the GATK HaplotypeCaller while considering the entire data set together to maximize sensitivity. Somatic mutations were called by comparing HCC samples with a pool of splenic controls. Only sites with at least four variant allele reads and total coverage of at least ten reads in both target or control samples were considered. In addition, a minimum of 10% of target sample reads had to contain the variant allele, and no more than 2% of control sample reads could contain the variant allele. Since not all samples had a matching control, each sample was compared with all background spleen samples to remove any variants that might have been present in any of the splenic control samples. Variants were annotated using snpEff (v4.3)62. Coding mutations were defined as variants that directly impacted amino-acid changes or splice sites. Variant visualization was performed using IGV63.
Comparison of gene expression profile and mutation signature between human and mouse
The direct overlap between genes regulated in human NASH versus control lean patients64 and the mouse RNA-seq were found by converting the genes found on the human Affymetrix HuGene 1.1 ST microarray using NCBI Homologene, which maps one-to-one orthologues between species. Only genes that could be mapped to a unique mouse Entrez Gene ID were considered (retained ~ 75% of the total). Homologene was also used to convert human HCC gene variants found in the COSMIC database to mouse gene identifiers to compare with mouse mutation profiles.
Metabolomics analysis
The metabolomic analysis of samples was conducted using reverse-phase ultra-high-pressure liquid chromatography–mass spectrometry (UHPLC MS). The samples were thawed at room temperature for approximately 10 min. Each Eppendorf tube with sample was weighed and the sample weight logged. An aliquot of 0.5 ml of HPLC-grade 80% v:v ethanol:water was added into each sample tube, and the material was homogenized at 25 Hz for 5 min in Qiagen TissueLyser II (Qiagen, Hilden, Germany). The samples were allowed to extract for 4 h at room temperature. After extraction, the tubes were centrifuged for 10 min at 2,000 r.p.m. and 400 μl of supernatant from each tube was transferred onto a 2 ml deep-well 96-well plate (Thermo Fisher Scientific, Waltham, MA) dried down, then re-suspended in 300 μl of 50% v-v ethanol: water. The plates were sealed, sonicated for 10 min, centrifuged at 2,000 r.p.m. for 10 min, and a 50 μl aliquot of each sample was transferred onto Falcon 96-well MS plate, which was then immediately sealed with sealing film. The MS plates were centrifuged at 2,000 r.p.m. for 10 min and stored at 2 °C until analysis. Blanks of resuspension solvent as well as the solvent that went through the extraction procedure steps (sample extraction blank) were included with each batch. Also, quality control was included at the beginning and end of each sample batch to control instrument performance.
The HPLC MS analysis was done on a Dionex UltiMate 3000 UHPLC system (Thermo Fisher Scientific, Waltham, Massachusetts, USA) coupled to a Bruker impact HD quadrupole time-of-flight mass spectrometer (Bruker, Billerica, Massachusetts, USA). The chromatographic separation was performed on a Kinetex C18 1.7 μm, 100Å UHPLC column (50 mm × 2.1 mm) (Phenomenex, Torrance, California, USA), held at 40 °C during analysis. Five microlitres of sample were injected. Mobile phase A was water, mobile phase B was acetonitrile, both with added 0.1% v:v formic acid. The solvent gradient table was set as follows: 2% B, increased to 10% B over 0.2 min, then to 100% B at 12 min, held at 100% B for 1.5 min, decreased back to 2% B in 0.5 min, followed by washout cycle and equilibration for a total analysis time of 15 min. The scanned m/z range was 80–2000 Th; capillary voltage 4,500 V; nebulizer gas pressure 2 bar, drying gas flow rate 9 l min−1 and temperature 200 °C. The full MS scan was followed by tandem mass spectrometry fragmentation of the seven most abundant ions in the spectrum. For tandem mass spectrometry, the collision cell collision energy was set at 3 eV and the collision energy was stepped 50%, 75%, 150%, and 200%. The scan rate was 3 Hz. A HP-921 lock mass compound was infused during the analysis for post-processing mass correction.
The collected LC/MS raw data files were converted to mzXML format and feature detection was performed with OpenMS 2.0 software65. The alignment window was set at 0.5 min, noise threshold at 1,000 counts, chromatographic peak FWHM at 20, and mass error at 30 p.p.m. (https://github.com/rsilvabio-info/openms_snakemake). All of the peaks that were present in any of the blanks with signal:noise ratio below 3:1 were removed from the final feature table. The extracted feature table was parsed into tables corresponding to different experimental categories, which then were used as input for the MetaboAnalyst software66. The feature tables were filtered with interquantile ranges to remove outliers, the data normalized by the sum total of peak intensities, and autoscaled (mean centring and dividing by the standard deviation for each feature). Principal component analysis and partial least squares discriminant analysis (PLS-DA) were used to explore and visualize variance within data and differences among experimental categories. The HFD-fed MUP-uPA strains were found to be concordant between two different age groups, 6 and 11 months either in the 16S rRNA sequencing or in the metabolomics. For 16S sequencing, the effect of animal age on microbiota was minimal (P = 0.021, pseudo-F statistic = 1.66) compared with immune status (P = 0.001, F statistic = 5.19) by PERMANOVA (999 permutations) on the unweighted UniFrac distance matrix, and age was not significant by PERMANOVA on the weighted UniFrac distance matrix (P = 0.095, pseudo-F statistic = 1.766). Similarly, in metabolomics analysis the effect of age (PLS-DA tenfold cross-validation Q2 value of 0.477) was minor compared with the immune status (PLS-DA tenfold cross-validation Q2 value of 0.926 for WT versus Iga−/− and 0.934 for WT and Cd8a−/− correspondingly). Therefore, for some analyses, data from both time points were pooled to increase the sample numbers. All of the raw data are publicly available at the UCSD Center for Computational Mass Spectrometry (the data set ID MassIVE MSV000080918).
Bacterial genomic DNA extraction and 16S rRNA gene amplification and sequencing
For the 16S rRNA analyses, faeces were taken from the indicated mice strains. The analysed genotypes (Cd8a−/−, Iga−/−, μMT−/−, Rag1−/−, Pigr−/−, IgHEL/MD4) were housed separately. Out of 32 MUP-uPA/Cd8a−/− mice, five were co-housed with Cd8a+/−. For the MUP-uPA group, some MUP-uPA-negative control mice were co-housed with MUP-uPA-positive groups. DNA was extracted using a MagAttract PowerSoil DNA KF Kit (Qiagen, Germany) following protocols benchmarked through the Earth Microbiome Project (http://www.earthmicrobiome.org/protocols-and-standards/dna-extractionprotocol/). The V4 of the 16S rRNA gene was then amplified with PCR again following protocols benchmarked through the Earth Microbiome Project (http://www.earthmicrobiome.org/protocols-and-standards/16s/)67. This approach facilitates the capture of most of the bacteria present in a sample because of the ubiquitous nature of the 16S rRNA gene and the ‘universal’ V4 primer. The V4 region recapitulates the full-length 16S rRNA gene at approximately genus-level resolution. Barcoded amplicons were then sequenced on an Illumina MiSeq using the 2 × 150 bp protocol at the Institute for Genomic Medicine at UCSD. The data are publicly available at the European Bioinformatics Institute under accession number ERP022642.
16S rRNA sequencing data analysis
The data were deposited and prepared in Qiita (http://qiita.microbio.me; study id: 10856). Raw sequences were de-multiplexed using QIIME 1.9.1 (ref. 68) following the default parameters available in Qiita. De-multiplexed sequences were then trimmed to 150 bp and de-noised using Deblur69. Since some downstream analyses require a phylogenetic tree, we used SEPP70 to generate this tree (see https://github.com/smirarab/sepp/tree/master/sepp-package) by inserting the V4 fragment sequences resulting from Deblur into the Greengenes tree71. Taxonomy was assigned to the sequences using the taxonomy present on the internal nodes of the already annotated Greengenes tree, and this taxonomy information was added to the biom-format observation matrix72 containing unique sequences and their abundances in each sample. The samples included in downstream analysis had at least 4,500 sequences per sample. Alpha diversity statistical analyses were performed using a non-parametric t-test with Monte Carlo permutations to calculate the P value73. The unweighted UniFrac beta diversity distances between samples were computed on a rarefied table of 4,500 sequences per sample74. Principal component analysis was applied to the resulting distance matrix and plots were generated using Emperor software75. To analyse the significance of sample groupings, PERMANOVA (non-parametric, 999 permutations) was performed on the UniFrac distance matrices using QIIME 1.9.1. Stacked bar plots of relative bacterial abundance were created using Matplotlib and heat maps were created with Seaborn. Significant differences in Deblur sequences between groups were assessed using the scikit-bio implementation of ANCOM76 with a Bonferroni correction for multiple comparisons (http://scikit-bio.org/docs/0.4.2/generated/generated/skbio.stats.composition.ancom.html).
For the microbiome principal coordinate analysis (PCoA) plots, the samples displayed in each figure are indicated in the publicly available sample information file in Qiita study ID 10856 (https://qiita.ucsd.edu/). Each PCoA plot figure has a column in the sample information file (labelled ‘Figure#_Extended_Data’), which indicates whether that sample was included in the plot with a ‘true’ or ‘false’ value and a column describing the subsequent grouping. This file can be downloaded and used in conjunction with the sequencing data to recreate the plots.
Analysis of bacteria by RT-PCR quantification of 16S rRNA-encoding DNA
For analyses of gut-bacteria content, faecal total DNAs extracts were prepared using a Powersoil DNA isolation kit. The abundance of total faeces bacteria was measured by RT-PCR based on a SYBR Green Assay using universal bacterial 16S rRNA-encoding DNA primers (16S-V6-784F: AGGATTAGATACCCTGGTA; 16S-V6-1061R: CRRCACGAGCTGACGAC). The primers were first used to amplify a 16S rRNA gene fragment from pooled DNA extracts. The DNA fragment was purified and quantified. As reference for quantification, standard curves with serial dilutions of purified 16S rRNA gene fragment were generated for each run.
Statistical analysis and reproducibility
Data are presented either as means ± s.e.m. or medians of continuous values and were analysed by a two-sided Students’ t-test or Mann–Whitney test for comparison of two groups, respectively. Two-way ANOVA was used to compare three or more groups in mouse data analyses. One-way ANOVA or a Kruskal–Wallis test was used to compare three or more groups in human data analyses. Bonferroni’s or Dunn’s multiple comparison test was applied to compare all pairs of groups. A D’Agostino and Pearson test and/or Shapiro–Wilk test was used to test the normality of sample distribution. Fisher’s exact χ2 t-test was used to calculate statistical significance of categorical values between groups. Two-tailed P values of no more than 0.05 were considered significant. Linear regression was used to determine the correlation between two different variables. GraphPad PRISM software was used for statistical analyses. An unpaired t-test was used to determine the power with GraphPad StatMate software.
Representative examples of power calculations (α = 0.05, two-tailed) for important groups are shown as follows. 1. MUP-uPA model: MUP-uPA + HFD (24) versus MUP-uPA + normal chow (8) ≈ 99%; MUP-uPA + HFD (24) versus MUP-uPA + HFD + antibiotics (6) ≈ 85%; MUP-uPA + HFD (24) versus MUP-uPA/Iga−/− + HFD (26) ≈ 95%; MUP-uPA + HFD (24) versus MUP-uPA/μMT −/− + HFD (9) ≈ 75%; MUP-uPA + HFD (24) versus MUP-uPA/Cd8a−/− + HFD (31) ≈ 85%. 2. STAM model: WT-FVB (13) versus FVB/Iga−/− (15) > 99%; WT-FVB (13) versus FVB/JH −/− (7) > 99%; WT-FVB (13) versus FVB/Tgfbr2ΔB (4) ≈ 95%; WT-FVB (13) versus FVB/Cd8a−/− (19) > 99%; WT-BL6 (17) versus BL6/μMT−/− (3) ≈ 75%; WT-BL6 (17) versus BL6/Pigr−/− (9) ≈ 99%; WT-BL6 (17) versus BL6/Cd8a−/− (5) ≈ 99%. 3. CD8 depletion: STAM-FVB: Iga−/− (15) versus Iga−/− + anti-CD8 (3) > 99%; MUP-uPA + HFD: Iga−/− (3) versus Iga−/− + anti-CD8 (3) ≈ 70%. 4. Adoptive lymphocyte transfer in MUP-uPA + HFD model: Rag1−/− (7) versus Rag1−/− (T cells) (4) ≈ 90%; Rag1−/− (T + B cells) (4) versus Rag1−/− (T cells) (4) ≈ 85%. 5. anti-PD-L1 treatment in MUP-uPA + HFD model: control (8) versus anti-PD-L1 (7) ≈ 80%; LALAPG (7) versus anti-PD-L1 (7) ≈ 99%.
The number of analysed and treated mice are shown in Supplementary Tables 1 and 2, with a total mouse number of 813 from 74 single experiments. Experiments were repeated independently at least two or three times with similar results.
Data and code availability
The RNA-seq (GSE90497; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90497), exome sequencing (SRA556071, SRP104724; https://www.ncbi.nlm.nih.gov/sra/?term=SRA556071), and 16S rRNA sequencing (ERP022642; http://www.ebi.ac.uk/ena/data/view/PRJEB20486) data are available in the NCBI Gene Expression Omnibus, NCBI Short Read Archive, and European Bioinformatics Institute, respectively. Samples displayed in the microbiome PCoA plots are indicated in the publicly available sample information in Qiita study ID 10856 (https://qiita.ucsd.edu/). The metabolomics raw data are publicly available at the UCSD Center for Computational Mass Spectrometry (the data set ID MassIVE MSV000080918): ftp://massive.ucsd.edu/MSV000080918.
The raw and processed feature tables used in the preparation of the ‘Metabolome’ panels in Extended Data Fig 7k–n, q as well as the data processing reports generated by the MetaboAnalyst are included along with the raw data (the data set ID MassIVE MSV000080558) at ftp://massive.ucsd.edu/MSV000080558.
Source data are available in the online version of the paper. All other data are available from the corresponding authors upon reasonable request.
The microbiome data analysis was primarily done using Qiita and QIIME, and the custom code used for taxonomic assignment from the phylogenetic tree generated with SEPP (https://github.com/smirarab/sepp/blob/master/sepp-package/README.md) is available online at Github (https://github.com/knightlabanalyses/shalapour-nature-iga-liver-cancer/blob/master/assignTaxa.ipynb). Scripts used to parse, filter, and organize data and results for RNA-seq and DNA exome sequencing are available upon request.
Extended Data
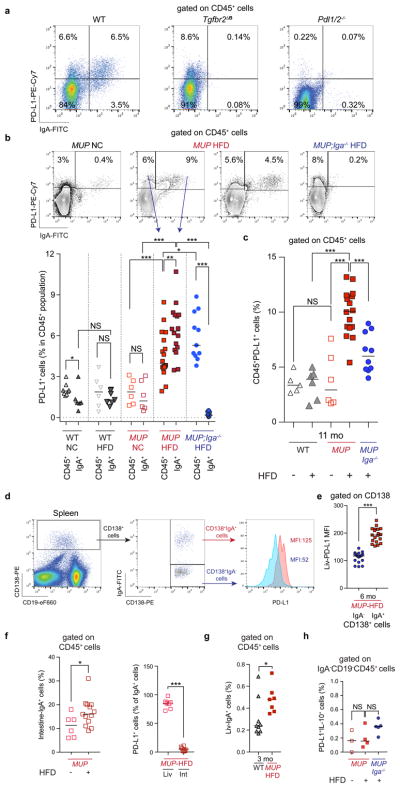
Extended Data Figure 1. NASH-induced accumulation of IgA+ plasmocytes in human liver.
a, Serum IgA levels of patients with NAFLD/NASH (Newcastle cohort; n = 502) were analysed and compared with clinical data. b, Serum was collected from a second cohort of patients with NAFLD/NASH (San Diego cohort; n = 96), whose degree of liver fibrosis was measured by MRI and classified as grades 0 (n = 51), 1–2 (n = 33), and 3–4 (n = 12). Serum concentrations of IgA, IgG, and IgM were determined by ELISA and plotted against the fibrosis grade. c, Single-cell suspensions were prepared from liver biopsies taken from patients with NASH (n = 4) with fibrosis, stained with antibodies to IgA, CD19, and PD-L1, and analysed by flow cytometry. Blood collected from three healthy donors (used as a surrogate for liver biopsies that could not be obtained from such individuals; HD), stained with antibodies, as indicated. Shown are representative scatter plots from two patients and two healthy donors, and bar graphs compiling data from all samples and depicting PD-L1 expression by IgA+ cells. d, Single-cell suspensions were prepared from liver biopsies as above (n = 3 per group) and stained with antibodies to CD4, CD8, Tim-3, and PD-1. Shown are representative data from one patient and one healthy donor with bar graphs compiling results obtained from three patients and depicting frequencies of PD-1+CD8+ T cells. χ2 test (a) and two-sided t-test (means ± s.e.m.; a–d) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001; NS, not significant.
Extended Data Figure 2. Experimental scheme and comparison of different NASH, HCC, and liver damage mouse models.
a, In the HFD-fed MUP-uPA model, MUP-uPA, MUP-uPA/Iga−/−, MUP-uPA/μMT−/−, MUP-uPA/Rag1−/−, and MUP-uPA/Cd8a−/− male mice were kept on normal chow (NC) or placed on high-fat diet (HFD) starting at 8 weeks of age. Mice were analysed for NASH-related immune parameters and pathology at 6 months of age and for HCC-related parameters at 11 months of age (n = 543 mice analysed in 50 experiments). Mice were treated with a cocktail of broad spectrum antibiotics (Abx) from 3 to 6 months or from 6 to 11 months to determine microbe impact on NASH and HCC, respectively. Mice were also subjected to weekly anti-PD-L1 or anti-CD8 antibody injections starting at 9 months of age for a total of 8 or 6 weeks before being evaluated for HCC. In the STAM model, male mice of the indicated genotype were subcutaneously injected with 200 μg streptozotocin (STZ) 2 days after birth and fed with HFD after 4 weeks of age. Tumour multiplicity, immune parameters, and pathology were determined when the mice were 25 or 20 weeks of age in either the BL6 or FVB backgrounds, respectively (n = 123 mice analysed in 15 experiments). STAM-Iga−/− mice were also subjected to weekly anti-CD8 antibody injections for a total of 5 weeks before being evaluated for HCC. In the MCD-HFD-fed model, mice were kept on a methionine and choline deficient (MCD) diet with 60 kcal% fat, for 4 weeks (n = 65 analysed in three experiments). In the CCl4 model, mice were injected intraperitoneally with CCl4 twice a week for 8 weeks (n = 82 analysed in six experiments). b, Paraffin-embedded and frozen mouse liver sections (n = 3–9 per group as indicated) were stained with haematoxylin and eosin (H&E) to determine liver histology (scale bar, 100 μm), Sirius Red (SR) to determine collagen fibre deposition (scale bar, 100 μm), or Oil Red O (ORO) to determine lipid droplet accumulation (scale bar, 50 μm). c, Paraffin-embedded human liver sections from patients with NASH and ASH were stained with Sirius Red to determine collagen fibre deposition (n = 5 per group with one representative shown; scale bar, 250 μm). The data were validated in at least two or three experiments. d, Spleen content of IgA+ cells (absolute cell number per spleen) in different mouse strains and treatment groups (n = 11, 6, 8, 13, 11, 9). e, f, Indicated mice were placed on the different models described in a. At the endpoints, mouse body weights were measured (e) (n = 6, 5, 8, 10, 3, 10, 11, 31, 15, 6, 9) and sera were analysed for ALT (f) (n = 6, 5, 6, 5, 3, 13, 3, 10, 8, 3, 6), an indicator of liver damage. g, h, Collagen deposition and lipid droplets were quantified using image analysis software, and data points for individual mice are shown (g: n = 11, 6, 3, 9, 3, 8, 4, 6, 5; h: n = 6, 5, 3, 3, 3, 5, 4, 8, 4, 3, 6). i, Total RNA was extracted from livers of indicated mice and analysed by RT-PCR using primers for Tgfb1 (n = 6, 5, 3, 7, 7, 6, 6). j, Hepatic stellate cells (HSC), CD11b+ Kupffer cells (KC), and CD11b− non-parenchymal cells (NPC) from the indicated mice were isolated from livers. Total RNA was extracted from these cells and analysed for Tgfb1 mRNA expression (n = 3–5). k, l, CD4+ T cells were sorted from livers of normal chow- and HFD-fed WT or HFD-fed MUP-uPA mice using CD4, CD8, CD45, and CD3 antibodies. RNA was extracted and analysed by RT-PCR using primers for Il21, Bcl6, and Icos (n = 4–7 as indicated). m, Total RNA was extracted from livers of indicated mice and analysed by RT-PCR for Il33 (n = 4, 5, 3, 7, 8, 6, 6). n, o, Total RNA was extracted from livers of indicated mice at 6 months (n = 29 mice in total, with 3–8 mice per group as indicated) (n) or tumours of indicated mice at 11 months of age (n = 7, 9) (o), and analysed by RT-PCR for chemokine and cytokine mRNA. The heat map in n depicts the differential expression of immune-regulatory genes. Two-sided t-test (means ± s.e.m.; g–m) and Mann–Whitney test (median; d–f, o) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001. N values for each group are shown either in individual panels or in legends for each group from left to right accordingly.
Extended Data Figure 3. IgA characterization in the indicated tissue of STAM and MUP-uPA mice.
a–e, Single splenocyte or liver cell suspensions from tumour (HCC)-bearing mice (STAM-B6) were stained with CD45, CD19, IgA, CD138, B220, and PD-L1, and FVD-eF780 was used to exclude dead cells. a, The gating strategies for splenocytes and liver lymphocytes: lymphocyte gate, dead cell exclusion, doublets exclusion, and CD45+ population gate. b, Flow cytometry analysis of IgA and CD19 expression of indicated strains, gated on the CD45+ population. Spleen or liver from Iga−/− mice was used to set up the gating for the populations. Tgfbr2ΔB and Pdl1/2−/− mice clearly showed less IgA+ cells than WT mice. c–e, IgA subpopulations were gated as indicated and analysed for CD138, B220, and PD-L1 expression. IgA subpopulations were gated on the basis of CD19 expression, in the indicated strains, and the IgA+CD19+, IgA+CD19−/low/int, and IgA−CD19+ subpopulations were further analysed for their B220 and CD138 expression. The decisions for CD19 and CD138 levels were also based on their mean fluorescence intensity (MFI; red). The analyses showed two main populations: (1) IgA+CD19+B220+CD138low/− and (2) IgA+CD19−/intB220−CD138+/hi. These two populations and the IgA−CD19+ populations were further analysed for their ability to express PD-L1 (percentage and mean fluorescence intensity). f–j, Single-cell suspensions were prepared from the spleen, liver, or intestine of MUP-uPA mice kept on normal chow or HFD, and were stained with CD45, CD19, IgA, B220, CD138, CD11b, MHCII, PD-L1, and CD5, and analysed by flow cytometry. f, The B220 and CD138 expression in total IgA+ cells (CD19+ and CD19− populations) confirmed that most of the IgA+ cells in spleen and liver of HFD-fed MUP-uPA mice are CD138+ cells. g, h, IgA subpopulations in liver (g) and spleen (h) were gated as indicated and analysed for CD138, B220, and MHCII expression. i, Representative dot-plots gated on IgA+ cells, showing that most of the IgA+ cells are not CD11b+. j, PD-L1 and CD5 expression of IgA+ cells in spleen, liver, and intestine.
Extended Data Figure 4. IgA+ cells in NASH livers express PD-L1.
a, Single-cell suspensions were prepared from the livers of tumour-bearing mice (STAM-B6) as indicated. Shown are the PD-L1 and IgA staining in the indicated strains. Spleen or liver from Iga−/− and Pdl1/2−/− mice were used as control for IgA and PD-L1 staining. b, Liver single-cell suspensions of MUP-uPA mice and indicated strains were stained for CD45, IgA, and PD-L1. Percentages of liver PD-L1+ cells in CD45+IgA− (labelled: CD45+) or IgA+ cell populations from the indicated strains are shown (n = 8, 6, 6, 7, 6, 6, 18, 16, 11, 12). c, Percentages of liver PD-L1+CD45+ cells are shown in the indicated strains (n = 5, 6, 6, 16, 10). The data were validated in at least three different experiments. d, e, CD138+ cells were divided into IgA+ and IgA− populations and analysed for PD-L1 expression. d, Shown is the mean fluorescence intensity of PD-L1 expression for CD138+ cells from spleen (gated on either CD138+IgA− or CD138+IgA+ cells). e, Mean fluorescence intensity of PD-L1 expression is shown for CD138+ plasmocytes from livers of 19 mice (gated on either CD138+IgA− or CD138+IgA+ plasma cells). f, Percentages of intestinal IgA+ cells in MUP-uPA mice kept on normal chow or HFD (n = 6, 13; left). Percentages of PD-L1+IgA+ cells in intestine and liver are included for comparison (n = 7, 8; right). g, Percentages of liver IgA+ cells are shown in the indicated strains kept on HFD at the age of 3 months (n = 9, 7). h, Percentages of PD-L1+ cells are shown gated on CD45+CD19−IgA− cells in the indicated strains (n = 3, 4, 5). The data were validated at least in two or three experiments. Two-sided Mann– Whitney tests (median; b, c, e–h) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001. N values for each group in each panel are provided from left to right accordingly.
Extended Data Figure 5. CD8+ T cells and IgA+ plasmocytes are only marginal players in the MUP-uPA NASH model.
a, Comparison of weight gain by different strains of mice kept either on normal chow or HFD. Mice were continuously monitored from 8 to 46 weeks, and 20 data points were used to analyse weight gain, with at least 11–52 mice per time point (see Supplementary Source Data), and 5 BL6 mice on normal chow were used as controls. b–e, Sera, blood, and liver samples were collected from mice of the indicated strains and ages. Serum and blood samples were analysed for ALT (n = 13, 13, 16) (b), glucose tolerance test (n = 3–6 as indicated) (c), cholesterol (n = 5, 8, 9, 7) (d), or triglycerides (n = 5, 8, 9, 7) (e, left). Liver extracts were analysed for triglycerides (e, right) (n = 5, 9, 11, 7). f, Total RNA was extracted from livers of indicated mice and analysed by RT-PCR for Col1a1 mRNA (n = 4, 4, 3, 6, 4, 8). g–i, Paraffin-embedded and frozen liver sections were stained with haematoxylin and eosin to determine liver histology (scale bar, 100 μm except 50 μm in the second panel of g), Sirius Red to determine collagen fibre deposition (scale bar, 100 μm), or Oil Red O to determine lipid droplet accumulation (scale bar, 50 μm) in the indicated strains and models (Extended Data Fig. 2a). Representative images are shown and were validated at least two or three times. Collagen deposition and lipid droplets were quantified using image analysis software and data points for individual mice are shown on the right for the MUP-uPA (g) (Sirius Red: n = 9, 14, 10; Oil Red O: 5, 6, 3) and MCD (h) (n = 3 per group) models. i, Representative Sirius Red staining for the CCl4 model (n = 4 or 5 mice per group). j, Total RNA from livers of 6-month-old mice of the indicated genotypes was subjected to RNA-seq analysis. Shown is the hierarchical clustering of gene expression profiles comparing HFD- and normal-chow-fed mouse liver samples. The top three enriched pathways/functional categories from Metascape were reported for major clusters of genes. k, Total RNA from livers of indicated mice was analysed by RT-PCR for alpha-fetoprotein (Afp) mRNA (n = 5, 5, 6, 5, 6, 6, 5, 8, 6, 6). The data were validated at least in two or three experiments (see Supplementary Tables 1 and 2). Two-way ANOVA (means ± s.e.m.; a, c), two-sided t-test (means ± s.e.m.; f–h, k), and Mann–Whitney test (median; b, d, e, g, h) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001. N values for each group are shown either in individual panels or in legends for each group from left to right accordingly.
Extended Data Figure 6. IgA ablation inhibits, while CD8 deficiency accelerates HCC development.
a, b, Total DNA was extracted from HCC nodules of 11-month-old mice (n = 16) of the indicated genotypes with or without anti-PD-L1 treatment, and subjected to exome sequencing. Shown are the number of point mutations identified per sample (a) and mutational signatures (b). Horizontal axis shows the 96-substitution patterns with substitution subtypes on top, and vertical axis indicates the probability of each pattern in b. c, Top 20 hallmark gene sets sorted by normalized enrichment score (NES) are shown to depict HCC progression in MUP-uPA and MUP-uPA/Iga−/− mice by gene set enrichment analysis. Immune-related gene sets are coloured blue. The gene sets previously described20,57 for human HCC are marked with a red asterisk. d, Total RNA was extracted from livers of 6-month-old mice and from HCC nodules of 11-month-old mice of the indicated genotypes and subjected to RNA-seq analysis. Hierarchical clustering of gene expression profiles comparing HCC and non-HCC mouse samples according to the RNA-seq. The top three enriched pathways/functional categories from Metascape are reported for major clusters of genes. e, Representative images of liver histology at different time points and indicated strains are shown with detailed n values. Scale bars, 100 μm (3 months), 250 μm (6, 11 months). f, Total tumour numbers in 3-month-old MUP-uPA mice (n = 11, 7, 8). g, Heat map depicting differential expression of 17 liver-specific genes and 33 HCC-related genes in the indicated strains, illustrating the upregulation of some HCC-related genes in MUP-uPA/Cd8a−/− livers at 6 months of age (total mice number 29; n = 3 or 4 per group). h–j, BL6 mice of the indicated phenotype were subjected to the STAM protocol and their tumour volumes (n = 14, 6, 6, 4, 3, 9, 3) (h) and histopathology (i, j) were evaluated at 25 weeks of age. The data were validated at least in two or three experiments. Paraffin-embedded and frozen liver sections from these mice were stained with haematoxylin and eosin, Sirius Red, or Oil Red O, as indicated. Shown are typical images of tumour-containing and tumour-free areas, the borders between which are marked by the black lines. Scale bars: haematoxylin and eosin, 100 μm; Sirius Red, 100, 250, or 500 μm; Oil Red O, 50 μm. Oil Red O-positive areas were quantitated and are shown on the right. The Sirius Red-stained areas for each mouse were calculated by image analysis of the whole-tissue scan and normalized to the haematoxylin and eosin stain (n = 3 or 4 per group). k, Tumour volumes are shown for STAM-WT (n = 10), STAM-Iga−/− (n = 13), and STAM-Iga−/− after CD8 depletion (n = 3). l, MUP-uPA/Iga−/−-HFD mice were injected weekly with anti-CD8 for 6 weeks and tumour multiplicity was determined (n = 3). k, l, CD8 depletion experiments were repeated using two different HCC models (MUP and STAM). m, Heat map depicting the differential expression of 59 genes involved in allograft rejection, IFNγ response, and inflammation (total mouse number 41; 6 months: n = 3 and 11 months: 3 or 4 per group). n, o, Paraffin-embedded and frozen liver sections from 11-month-old mice (n) and adoptively transferred mice (o) were stained with haematoxylin and eosin, Sirius Red, or Oil Red O as indicated and analysed (Sirius Red: n = 9, 16, 14, 6; Oil Red O: n = 8, 5, 5, 8 for n). Shown are typical images of tumourcontaining and tumour-free areas, the borders between which are marked by the black lines. p, Liver cells from MUP-uPA/Rag1−/− mice 1 week after being adoptively transferred with CFSE-labelled T cells with or without B cells as indicated (n = 3 in each group) were stained and analysed by flow cytometry. Shown are the percentage of CD8+ T cells among CD45+ cells (left), and histogram of proliferating CFSE-labelled T cells with the corresponding mean fluorescence intensity (right). q, Liver sections for MUP-uPA/Rag1−/− and the corresponding adoptive lymphocyte transfer mice (4 weeks after adoptive transfer (AT)) were stained with alpha-SMA, IgA, CD3, and CD8 antibodies, counterstained with DAPI and examined by fluorescent microscopy (scale bars, 50 μm). For CD3/CD8 staining, images with higher magnification are shown (scale bars, 20 μm). Single-cell suspensions were prepared from the corresponding liver, stained with antibodies to CD45, IgA, CD19, B220, CD8, and CD4, and analysed by flow cytometry. Shown are representative scatter plots. MUP;Rag1−/− mouse livers have been used for validation of CD4, CD8, IgA, and CD19, both for flow cytometry and for immunofluorescence analyses. The data were validated in at least two experiments. r, STAM-BL6 mice of the indicated phenotypes were analysed for IgA serum amounts by ELISA (n = 11, 4, 4, 4, 4). s, Absolute IgA+ cell number in livers of indicated STAM-BL6 mice (n = 13, 4, 4, 9, 8). Each dot represents a mouse. Two-sided t-test (means ± s.e.m.; f, j, l, n, r) and Mann–Whitney test (median; h, k, n, s) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001. N values for each group are shown either in individual panels or in legends for each group from left to right accordingly.
Extended Data Figure 7. Gut microbes promote HCC development and microbial translocation does not account for the anti-tumorigenic effect of IgA ablation.
a–j, MUP-uPA and MUP-uPA/Iga−/− mice were placed on HFD and treated with broad spectrum antibiotics (Abx) as described in Extended Data Fig. 2a, from 3 to 6 months (all panels except right part of b, and d, h, i) or 6 to 11 months (right part of b, and d, h, i) of age. At the end of the treatments, the stool contents of the corresponding mice were subjected to eubacterial 16S rRNA encoding DNA sequencing. a, Principal coordinate analysis plot of microbiome data using unweighted UniFrac distances; antibiotic treatment was significant by PERMANOVA (pseudo-F statistic = 105.5, P = 0.001) (left: n = 181, 27 as indicated; right: n = 6, 3, 6, 3). Mouse weight (6 months: n = 5, 6, 13, 7; 11 months: n = 8, 6, 5, 7) (b) and circulating ALT (n = 12, 5, 12, 6) (e) were measured. c, d, Paraffin-embedded and frozen liver sections from the above mice were stained with haematoxylin and eosin, Sirius Red, or Oil Red O, and were analysed for collagen deposition and lipid droplets as indicated. Scale bars, 50 μm for Oil Red O; 100 μm for haematoxylin and eosin, and Sirius Red (Sirius Red: n = 9, 6, 14, 7 for c and 4, 4, 5 for d; Oil Red O: n = 5, 3, 6, 3 mice). The data were validated at least in two or three experiments. f–h, j, Liver cell suspensions were stained with antibodies as indicated, and analysed by flow cytometry. Each dot represents one mouse. Shown are percentages of CD8+ cells in total cells (n = 11, 5, 9, 7), CD8+CD44+ (n = 10, 4, 11, 6) or CD8+IFNγ +CD107a+TNF+ (n = 8, 6, 9, 7) cells in CD8+ T cells (f), CD19+B220+ cells in CD45+ cells (n = 9, 6, 11, 7) (g), IgA+ cells in CD45+ cells (n = 6, 6, 6, 3, 6, 6) (h), and CD4+ cells in CD45+ cells (n = 9, 6, 11, 7) or IL-17+ cells in CD4+ T cells (n = 9, 6, 10, 6) (j). i, MUP-uPA mice placed on HFD and treated with antibiotics were analysed for serum IgA by ELISA (n = 4, 8, 5, 6, 5, 5). Note that flow cytometry data of MUP-uPA and MUP-uPA/Iga−/− mice, which were not treated with antibiotics (control mice), are also shown in Fig. 3 and Extended Data Fig. 8q–y. The data were validated at least in two or three experiments. k–r, Effects of HFD and immunological background on mouse intestinal microbiomes and metabolomes (total mouse number n = 288). Each dot represents one mouse. k, The most pronounced differences are engendered by HFD compared with normal chow. Left to right: principal coordinate analysis (PCoA) plot of microbiome data using unweighted UniFrac distances (PERMANOVA, pseudo-F statistic = 46, P = 0.001 comparing diet); barchart of relative abundances of bacterial phyla; principal component analysis plot of metabolome. k, l, Subsequent effect of immune status for the MUP-uPA HFD-fed mice: WT, Iga−/−, Cd8a−/−, μMT−/−, and Rag1−/− groups. l, Left to right: PCoA plot of microbiome data using unweighted UniFrac distances (PERMANOVA, pseudo-F statistic = 4.37, P = 0.001 comparing immune status); principal component analysis plot of metabolome; partial least squares discriminant analysis (PLS-DA) plot (the tenfold cross validation Q2 value was 0.817) of metabolome. Large differences between categories are evident. Subsequent juxtaposition of the (m) MUP and MUP;Iga−/−: PCoA of plot of microbiome using unweighted UniFrac distances (PERMANOVA, pseudo-F statistic = 7.31, P = 0.001 comparing immune status) (left); PLS-DA (the tenfold cross validation Q2 value of 0.926) plot of metabolome (right) and (n) MUP and MUP;Cd8a−/−: PCoA plot of microbiome data using unweighted UniFrac distances (PERMANOVA, pseudo-F statistic = 4.61, P = 0.001 comparing immune status) (left); PLS-DA plot (the tenfold cross validation Q2 value of 0.934) plot of metabolome (right) illustrates the discordance stemming from these specific immune status differences. o, Bacterial Faith’s phylogenetic diversity metric (alpha diversity, box plot with minimum to maximum) calculated with rarefaction at 4,500 sequences per sample using Faith’s phylogenetic diversity metric (n = 14, 8, 14, 25, 16, 6, 44, 34). p, Heat map of abundant bacterial taxa by immune status, genetic background, and diet. Trends in significantly differing taxa (ANCOM) by immune status include increased Gammaproteobacteria in Iga−/− with HFD and increased Ruminococcaceae in WT versus Iga−/− with HFD. Mucispirillum schaedleri was elevated in Iga−/− for all groups except normal-chow-fed MUP-uPA. q, Discordance according to the immune status for the STAM model mice. Left to right: PCoA plot of microbiome data using unweighted UniFrac distances; principal component analysis plot of metabolome (the tenfold cross validation Q2 value of the corresponding partial least squares discriminant analysis is 0.814). r, Box plot (minimum to maximum) of unweighted UniFrac distances comparing distances within Iga−/−, Pigr−/−, and IgHEL/MD4 strains with distances between these strains (n = 44, 40, 56) and WT or Cd8a−/− STAM mice (n = 16, 12, 12) for microbiome data. Two-sided t-test (means ± s.e.m.; a, b, i, r) and Mann– Whitney test (median; c–h, j, o) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001. N values for each group are shown either in individual panels or in legends for each group from left to right accordingly.
Extended Data Figure 8. IgA+ plasmocytes regulate tumour killing by CD8+ T cells.
a, Dih10 and dihXY HCC cells were transfected with an inducible ovalbumin (Ova) expression vector, and Ova expression and presentation were confirmed by flow cytometry, using an antibody that recognized the SIINFEKL peptide on the MHCI molecule H-2Kb. b–h, Ova-expressing dih cells or controls (dih–RFP) were starved overnight (5% cell death), after which their medium was changed and B cells from WT, Iga−/−, Pdl1/2−/−, or SW-HEL mice were added in the presence of TGFβ (5 ng ml−1) and CTGF (3 ng ml−1), for an additional 24 h. Thereafter, the medium was replaced and CFSE-labelled OT-I T cells were added to the cultures that either contained or did not contain the B cells described above. After 4–6 days, the co-cultured cells were analysed by flow cytometry, while the secretory IgA was analysed by ELISA (n = 2–4 wells per group per day). a–j, Experiments were repeated with two different Ova-expressing HCC and one prostate cancer cell lines. Shown are the representative flow cytometry histograms or plots depicting (b) OT-I CD8+ T-cell proliferation, (c) PD-L1 and SIINFEKL/H-2Kb expression on cancer cells, (d) PD-L1 expression on B cells, (e) cancer cell death. f, Relative dih-Ova–RFP killing by OT-I CD8+ T cells in the presence or absence of the indicated B cells. g, Total secretory IgA and anti-OVA-IgA antibody amounts in culture supernatants. h, Percentages of OT-I CD8+ cells in each culture, as indicated. i, j, TRC2-Ova–RFP cells or its control cell line (TRC2–RFP) were co-cultured with OT-I cells and splenic B cells (WT and Il10−/−), as described for a–h. i, Proliferation of OT-I cells was analysed using CFSE (n = 3, 7, 5, 4). j, The amounts of secretory IgA were analysed using ELISA as indicated (n = 3 per group). k–o, Liver cells from indicated 3-month-old mice were stained and analysed by flow cytometry. Experiments were repeated at least two or three times. Each dot represents one mouse. Shown are the percentage of CD8+ T cells among CD45+ cells (n = 6, 5, 3, 7, 4) (k), absolute CD8+ T-cell number per gram of liver (n = 3, 3, 7, 4) (l), the percentage of CD8+CD44+Ki-67+ T cells with representative scatter plots (n = 3 or 4 per group) (m, n), and the representative scatter plots of perforin and GrzB among CD8+CD44+Ki-67+ T cells (o). p, q, Liver cell suspensions from the indicated mice were stained as shown and analysed by flow cytometry to determine the absolute CD8+ T-cell number in both STAM-BL6 and STAM-FVB mice (n = 8, 4, 5, 3, 5, 5, 6, 7) (p), and the percentage of TEM cells using CD8, CD44, and CD62L (n = 4, 4, 6, 9) (q). r, Liver cells from indicated 3-, 6-, and 11-month-old mice kept on HFD (n = 4–10) were stained and analysed by flow cytometry. Shown are the percentage of CD8+IFNγ +CD107a+ T cells. Detailed n values are shown in Fig. 3e. s–y, Liver cell suspensions from the indicated mice were stained as shown and analysed by flow cytometry to determine the percentage of Th17 cells using CD4 and IL-17a (n = 4, 7, 9, 10, 4, 9, 11) (s), the percentage of regulatory T cells using CD4 and Foxp3 (n = 4, 3, 8, 11, 7, 13) (t), the percentage of Tfh-like cells using CXCR5, PD1 and CD4 (3, 5, 6, 5) (u), the percentage of B220+CD19+ B cells (n = 4, 7, 7, 4, 5, 3, 9, 3, 11) (v), absolute B220+CD19+ B-cell number per gram of liver (n = 5, 7, 12, 27, 15, 7) (w), the percentage of IgG+ cells (n = 4, 3, 14, 11) (x), and CD138+ plasma cells (n = 4, 7, 4, 7, 7, 8, 5, 8, 6, 6, 13, 27, 17) (y). Two-sided t-test (means ± s.e.m.; f–m) and Mann–Whitney test (median; p–y) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001. Mouse ages are indicated in the graphs. N values for each group in each panel are provided from left to right accordingly.
Extended Data Figure 9. The response to PD-L1 blockade is dependent on CD8+ T cells and clonal expansion of HCC-directed CD8+ T cells.
MUP-uPA and MUP-uPA/Iga−/− mice were placed on HFD and treated with anti-PD-L1, as described in Extended Data Fig. 2a. a, At the end of the treatments, mouse weights were measured (n = 3, 2, 4, 3, 3, 7, 3, 7). b, MUP-uPA mice treated with anti-PD-L1 were analysed for serum IgA by ELISA (n = 10, 6). c, Liver/body weight ratio of indicated strains kept on HFD that received the indicated treatments and were of the indicated ages (6 months, 11 months) (n = 4, 6, 9, 19, 6, 2, 4, 15, 11, 17, 3, 14, 7, 20). d, e, Paraffin-embedded and frozen liver sections from HCC-bearing MUP-uPA mice were stained with Oil Red O, haematoxylin and eosin, or Sirius Red and analysed (n = 4 or 5). The experiments were repeated at least two times. Low and high magnifications are shown in e to demonstrate the absence of tumour-invading immune cells in a mouse that failed to respond to anti-PD-L1 and their presence within a tumour of a treatment responsive mouse. Non-responsiveness to anti-PD-L1 treatment correlates with a fibrotic tumour stroma. Scale bars: haematoxylin and eosin, 250 μm; Sirius Red, 250 μm; Oil Red O, 50 μm. f, Response to PD-L1 blockade is dependent on CD8+ T cells. HCC-bearing MUP-uPA/Cd8a−/− mice (n = 3) were treated with anti-PD-L1 for 8 weeks and tumour multiplicity was determined. g, Two-dimensional plot showing the frequency of the top ten TCRα CDR3 sequences expressed by CD8+ T cells from spleens and livers of HCC-bearing MUP-uPA mice. h, CD8+ T cells were sorted from spleens and livers of normal-chow- and HFD-fed WT mice (n = 5 mice), their RNA was extracted, and TCR α-chain (top) and β-chain (bottom) CDR3 sequences were amplified and analysed by deep sequencing (ten samples). The panels show the clonality, the frequency of the top 50 TCR α- and β-chain sequences, and the percentage of productive unique TCR α - and β-chain sequences. i–k, CD8+ T cells were sorted from spleens and livers of HCC-bearing mice of the indicated strains and treatments (n = 13 mice, 26 samples as indicated) and their TCR α - and β-chain CDR3 sequence diversity was analysed. Shown are the clonality (i), the diversity (inverse Simpson’s index) (j), and percentage of productive unique TCR β-chain sequences in the indicated strains (k). Note that TCR sequencing data of WT, MUP-uPA, and MUP-uPA/Iga−/− mice are also shown in Fig. 4g, h and Extended Data Fig. 9h. l–n, Splenocytes from the indicated mice treated with or without anti- PD-L1 were stimulated overnight with alpha-fetoprotein, stained as indicated, and analysed by flow cytometry. Shown are the percentage of IFNγ +CD107a+ cells (n = 3, 3, 2, 5, 3, 2, 3, 4, 4) (l) and IFNγ +TNF+ cells (n = 4, 3, 5, 4) (m) gated on CD8+ T cells. n, The percentage of IFNγ +TNF+ cells gated on CD4+ T cells (n = 3, 3, 2, 4, 3, 2, 3, 6, 4). Two-sided t-test (a–d, f, h–n) and Mann–Whitney test (l) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001. N values for each group in each panel are provided from left to right accordingly.
Extended Data Figure 10. Liver-IgA+ cells are oligoclonal, and effects of HFD on control WT, Iga−/−, and Cd8a−/− mice.
a, b, B cells and plasmocytes (CD19+, CD138+, IgA+ cells) were sorted from spleens (n = 3) and livers (n = 3) of HCC-bearing MUP-uPA mice and their μ (IgM) and α (IgA) locus genetic diversities were determined by BCR sequencing. c, Circulating ALT in HFD-fed WT, Iga−/−, and Cd8a−/− mice at 6 months of age (n = 5, 4, 8). d–f, Paraffin-embedded and frozen liver sections from the above mice were stained with haematoxylin and eosin, Oil Red O, or Sirius Red, as indicated. The data were validated at least twice. Scale bars: haematoxylin and eosin, 100 μm; Sirius Red, 100 μm; Oil Red O, 50 μm. The Sirius Red (n = 3, 4, 8) (d) and Oil Red O (n = 3, 3, 6) (e) stained areas were quantitated. g–i, Serum cholesterol (g), serum triglycerides (h), and liver triglycerides (i) were measured (n = 5, 4, 3). Two-sided t-test (a, d–i) and Mann–Whitney test (c) were used to determine significance. * P < 0.05; * * P < 0.01; * * * P < 0.001. N values for each group in each panel are provided from left to right accordingly.
Supplementary Material
Acknowledgments
We thank S. Choi, B. H. Dang, K. Nguyen, I. Mellman, G. Ackermann, H. Reeves, R. Quinn, G. Di Caro, G. Karin, and J. Haybaeck for help and advice. The research was supported by the National Institutes of Health (NIH), National Institute of Environmental Health Sciences Superfund Research Program, Horizon-2020-Framework Program of the European Union (Q.M.A.), Newcastle NIHR Biomedical Research Centre, CMI seed grant, Irvington Cancer Research Institute (S.S., Z.Z.), Prostate Cancer Foundation Young Investigator Award (S.S.), Southern California Research Center for ALPD and Cirrhosis grant (S.S., I.N.B.), Prevent Cancer Foundation Board of Directors Research Award (Z.Z.), and American Liver Foundation liver scholar award (D.D.). M.K. holds the Ben and Wanda Hildyard Chair. NIH funding was as follows: CA127923, CA118165, AI043477, and U01AA022614 to M.K.; P41GM103484-07 and GMS10RR029121 to P.C.D.; P42ES010337 to M.K., R.M.E., and R.L.; DK106419 to R.L.; and EPoS-634413 to Q.M.A.
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions M.K. and S.S. conceived and designed the project. S.S. designed and performed experiments and analysed data with M.K. X.L., I.N.B., W.L., A.P., Z.Z., D.D., and J.X. assisted with experiments and data analysis. T.M. analysed TCR/BCR repertoires. J.B., A.D.B., R.L., and Q.M.A. collected and analysed human specimens. C.B., M.D., R.T.Y., R.M.E., X.L., and S.S. conducted and analysed sequencing data. A.F.V., A.A.A., J.A.N., P.C.D., and R.K. performed microbiome and metabolomic analyses. M.K. and S.S. wrote the manuscript, with all authors contributing and providing feedback and advice.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer Information Nature thanks A. Biragyn, M. Bodogai, G. Gores and the other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Sharma P, Wagner K, Wolchok JD, Allison JP. Novel cancer immunotherapy agents with survival benefit: recent successes and next steps. Nat Rev Cancer. 2011;11:805–812. doi: 10.1038/nrc3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases — elimination, equilibrium and escape. Curr Opin Immunol. 2014;27:16–25. doi: 10.1016/j.coi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corthay A. Does the immune system naturally protect against cancer? Front Immunol. 2014;5:197. doi: 10.3389/fimmu.2014.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singal AG, El-Serag HB. Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clin Gastroenterol Hepatol. 2015;13:2140–2151. doi: 10.1016/j.cgh.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luedde T, Schwabe RF. NF-κ B in the liver—linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:108–118. doi: 10.1038/nrgastro.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolf MJ, et al. Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell. 2014;26:549–564. doi: 10.1016/j.ccell.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Nakamoto Y, Guidotti LG, Kuhlen CV, Fowler P, Chisari FV. Immune pathogenesis of hepatocellular carcinoma. J Exp Med. 1998;188:341–350. doi: 10.1084/jem.188.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhanda AD, Collins PL. Immune dysfunction in acute alcoholic hepatitis. World J Gastroenterol. 2015;21:11904–11913. doi: 10.3748/wjg.v21.i42.11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baroni GS, et al. Interferon gamma decreases hepatic stellate cell activation and extracellular matrix deposition in rat liver fibrosis. Hepatology. 1996;23:1189–1199. doi: 10.1002/hep.510230538. [DOI] [PubMed] [Google Scholar]
- 11.Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limón P. The polarization of immune cells in the tumour environment by TGFβ. Nat Rev Immunol. 2010;10:554–567. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mauri C, Menon M. The expanding family of regulatory B cells. Int Immunol. 2015;27:479–486. doi: 10.1093/intimm/dxv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McPherson S, Henderson E, Burt AD, Day CP, Anstee QM. Serum immunoglobulin levels predict fibrosis in patients with non-alcoholic fatty liver disease. J Hepatol. 2014;60:1055–1062. doi: 10.1016/j.jhep.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Shalapour S, et al. Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature. 2015;521:94–98. doi: 10.1038/nature14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakagawa H, et al. ER stress cooperates with hypernutrition to trigger TNF-dependent spontaneous HCC development. Cancer Cell. 2014;26:331–343. doi: 10.1016/j.ccr.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Font-Burgada J, et al. Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell. 2015;162:766–779. doi: 10.1016/j.cell.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park EJ, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik A, et al. IL-33 regulates the IgA-microbiota axis to restrain IL-1α-dependent colitis and tumorigenesis. J Clin Invest. 2016;126:4469–4481. doi: 10.1172/JCI88625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulze K, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He G, et al. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155:384–396. doi: 10.1016/j.cell.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brandtzaeg P. Secretory IgA: designed for anti-microbial defense. Front Immunol. 2013;4:222. doi: 10.3389/fimmu.2013.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dapito DH, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seki E, et al. TLR4 enhances TGF-β signaling and hepatic fibrosis. Nat Med. 2007;13:1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 25.Phan TG, et al. B cell receptor-independent stimuli trigger immunoglobulin (Ig) class switch recombination and production of IgG autoantibodies by anergic self-reactive B cells. J Exp Med. 2003;197:845–860. doi: 10.1084/jem.20022144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma C, et al. NAFLD causes selective CD4+ T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 2016;531:253–257. doi: 10.1038/nature16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Good-Jacobson KL, et al. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat Immunol. 2010;11:535–542. doi: 10.1038/ni.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim PS, Ahmed R. Features of responding T cells in cancer and chronic infection. Curr Opin Immunol. 2010;22:223–230. doi: 10.1016/j.coi.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125:3347–3355. doi: 10.1172/JCI80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellman I, et al. De-risking immunotherapy: report of a consensus workshop of the Cancer Immunotherapy Consortium of the Cancer Research Institute. Cancer Immunol Res. 2016;4:279–288. doi: 10.1158/2326-6066.CIR-16-0045. [DOI] [PubMed] [Google Scholar]
- 31.Kang TW, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 32.El-Khoueiry AB, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madan R, et al. Nonredundant roles for B cell-derived IL-10 in immune counter-regulation. J Immunol. 2009;183:2312–2320. doi: 10.4049/jimmunol.0900185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitamura D, Roes J, Kühn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin μ chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 35.Harriman GR, et al. Targeted deletion of the IgA constant region in mice leads to IgA deficiency with alterations in expression of other Ig isotypes. J Immunol. 1999;162:2521–2529. [PubMed] [Google Scholar]
- 36.Koh DR, et al. Less mortality but more relapses in experimental allergic encephalomyelitis in CD8−/− mice. Science. 1992;256:1210–1213. doi: 10.1126/science.256.5060.1210. [DOI] [PubMed] [Google Scholar]
- 37.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, et al. Immunoglobulin gene rearrangement in B cell deficient mice generated by targeted deletion of the JH locus. Int Immunol. 1993;5:647–656. doi: 10.1093/intimm/5.6.647. [DOI] [PubMed] [Google Scholar]
- 39.Silveira PA, et al. The preferential ability of B lymphocytes to act as diabetogenic APC in NOD mice depends on expression of self-antigen-specific immunoglobulin receptors. Eur J Immunol. 2002;32:3657–3666. doi: 10.1002/1521-4141(200212)32:12<3657::AID-IMMU3657>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 40.Shimada S, et al. Generation of polymeric immunoglobulin receptor-deficient mouse with marked reduction of secretory IgA. J Immunol. 1999;163:5367–5373. [PubMed] [Google Scholar]
- 41.Forrester E, et al. Effect of conditional knockout of the type II TGF-beta receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer Res. 2005;65:2296–2302. doi: 10.1158/0008-5472.CAN-04-3272. [DOI] [PubMed] [Google Scholar]
- 42.Phan TG, Gardam S, Basten A, Brink R. Altered migration, recruitment, and somatic hypermutation in the early response of marginal zone B cells to T cell-dependent antigen. J Immunol. 2005;174:4567–4578. doi: 10.4049/jimmunol.174.8.4567. [DOI] [PubMed] [Google Scholar]
- 43.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 44.Fujii M, et al. A murine model for non-alcoholic steatohepatitis showing evidence of association between diabetes and hepatocellular carcinoma. Med Mol Morphol. 2013;46:141–152. doi: 10.1007/s00795-013-0016-1. [DOI] [PubMed] [Google Scholar]
- 45.Kleiner DE, Makhlouf HR. Histology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis in adults and children. Clin Liver Dis. 2016;20:293–312. doi: 10.1016/j.cld.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He G, et al. Hepatocyte IKKβ/NFκ B inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17:286–297. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng F, et al. Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology. 2012;143:765–776. e761–763. doi: 10.1053/j.gastro.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida R, et al. A new method for quantitative analysis of the mouse T-cell receptor V region repertoires: comparison of repertoires among strains. Immunogenetics. 2000;52:35–45. doi: 10.1007/s002510000248. [DOI] [PubMed] [Google Scholar]
- 49.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 53.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 54.Tripathi S, et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for ubr4 in virus budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mootha VK, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 56.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Totoki Y, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 58.Li H, Durbin R. Fast and accurate short read alignment with Burrows- Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinform. 2013;43:11.10.1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teufel A, et al. Comparison of gene expression patterns between mouse models of nonalcoholic fatty liver disease and liver tissues from patients. Gastroenterology. 2016;151:513–525. doi: 10.1053/j.gastro.2016.05.051. [DOI] [PubMed] [Google Scholar]
- 65.Röst HL, et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat Methods. 2016;13:741–748. doi: 10.1038/nmeth.3959. [DOI] [PubMed] [Google Scholar]
- 66.Xia J, Wishart DS. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinform. 2016;55:141011–141091. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- 67.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ammon A, et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2:e00191–16. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mirarab S, Nguyen N, Warnow T. SEPP: SATé-enabled phylogenetic placement. Pac Symp Biocomput. 2012;17:247–258. doi: 10.1142/9789814366496_0024. [DOI] [PubMed] [Google Scholar]
- 71.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McDonald D, et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience. 2012;1:7. doi: 10.1186/2047-217X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 74.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mandal S, et al. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. 2015;26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq (GSE90497; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90497), exome sequencing (SRA556071, SRP104724; https://www.ncbi.nlm.nih.gov/sra/?term=SRA556071), and 16S rRNA sequencing (ERP022642; http://www.ebi.ac.uk/ena/data/view/PRJEB20486) data are available in the NCBI Gene Expression Omnibus, NCBI Short Read Archive, and European Bioinformatics Institute, respectively. Samples displayed in the microbiome PCoA plots are indicated in the publicly available sample information in Qiita study ID 10856 (https://qiita.ucsd.edu/). The metabolomics raw data are publicly available at the UCSD Center for Computational Mass Spectrometry (the data set ID MassIVE MSV000080918): ftp://massive.ucsd.edu/MSV000080918.
The raw and processed feature tables used in the preparation of the ‘Metabolome’ panels in Extended Data Fig 7k–n, q as well as the data processing reports generated by the MetaboAnalyst are included along with the raw data (the data set ID MassIVE MSV000080558) at ftp://massive.ucsd.edu/MSV000080558.
Source data are available in the online version of the paper. All other data are available from the corresponding authors upon reasonable request.
The microbiome data analysis was primarily done using Qiita and QIIME, and the custom code used for taxonomic assignment from the phylogenetic tree generated with SEPP (https://github.com/smirarab/sepp/blob/master/sepp-package/README.md) is available online at Github (https://github.com/knightlabanalyses/shalapour-nature-iga-liver-cancer/blob/master/assignTaxa.ipynb). Scripts used to parse, filter, and organize data and results for RNA-seq and DNA exome sequencing are available upon request.