Abstract
Resistance to chemotherapy and pathway-targeted therapies poses a major problem in cancer research. While the fields of tumor biology and experimental therapeutics have already benefited from ex vivo preclinical tissue models, these models have yet to address the reasons for malignant transformations and the emergence of chemoresistance. With the increasing number of ex vivo models poised to incorporate physiological biophysical properties, along with the advent of genomic sequencing information, there are now unprecedented opportunities to better understand tumorigenesis and to design therapeutic approaches to overcome resistance. Here we discuss that new preclinical ex vivo models should consider—in addition to common biophysical parameters such as matrix stiffness and bioadhesivity—a more comprehensive milieu of tissue signaling, nuclear mechanics, immune response, and the gut microbiome.
Chemoresistance and changes in biophysical factors
Therapeutic resistance in cancer often arises through genetic mutations that enhance drug metabolism, inactivate apoptotic pathways, and activate pro-survival signals [1–3]. The underlying genetic mutations are accompanied and sometimes preceded by changes in the biochemical and biophysical properties of the surrounding tissue. The biophysical factors, such as bioadhesivity, porosity, confinement, and stiffness, have been extensively studied as the response of individual cells to these factors, and are vital for cellular functioning and tissue development. Cells cope up with biophysical stimuli through an integrated mechanosignaling of physically interconnected proteins, starting from the extracellular matrix adhesion molecules (integrin), focal adhesion plaques, actin fibers, and structural components of the cells’ nucleus, among others. The mechanotransduction response includes both the activation of mechanosensitive transcription factors and downstream genes, as well as the rearrangement of cellular structure and organization to adjust to the physical environment [4, 5].factors. Mutations in cellular proteins and alterations in cellular microenvironment aberrantly engage mechanosignaling networks in cancer cells, either by perturbing the mechanical input or by altering the signaling network itself, which can promote cell growth, invasion, migration, and likely chemoresistance. For example, integrin signaling has been shown to increase epidermal growth factor secretion and receptor tyrosine-protein kinase erbB-2 (ERBB2) clustering in breast cancer cells, resulting in resistance to the ERBB2 inhibitor trastuzumab [6]. Increased tumor and stroma stiffness has also become a hallmark of cancer, as evident from the use of palpation for detection of breast tumors and cancerous lymph nodes in case of lymphomas. Increased tissue stiffness in the liver, pancreas, prostate, and lung have also been shown to be positive indicators of disease progression in the corresponding cancers [7–10]. Nevertheless, how chemoresistance and changes in biophysical and biochemical factors relate to one another is poorly understood.
While genomic studies have benefitted from direct patient sample analysis, exploring the role the stiffness of the microenvironment plays in cellular function has only become possible through the use of atomic force microscopy (AFM), microindenters, and engineered tissues. Ex vivo preclinical models that recapitulate tumor microenvironment have been critical for improving our understanding of tumorigenic growth and resistance. In the case of mammary tumors, changes in tissue stiffness are associated with increased deposition and cross-linking of collagen type I, and the stiffness can increases from 100–400 Pa up to 1–5 kPa when comparing normal and cancerous mammary tissue [9, 11]. It is now well accepted that matrix stiffness perturbs epithelial morphogenesis by clustering integrins to enhance extracellular-signal-regulated kinase (ERK) activation and increase Rho-associated protein kinase (ROCK)-generated contractility and focal adhesions. Integrin signaling and stiffness are not only involved in chemoresistance in solid tumors, but also in palpable lymphoid malignancies as shown by us [12], as well as in liquid tumors [13]. Recent work from Shin and Mooney demonstrated that matrix softening leads to resistance against standard chemotherapy in myeloid leukemias [13]. More recently, matrix softness was shown to influence histone methylation and epigenetics of tumor repopulating cells [10], which exhibit high chemoresistance to conventional chemotherapeutic drug treatment. To better understand the role of tissue stiffness in cancer, we refer the readers to excellent recent reviews [11][14]. Nonetheless, these ex vivo models have yet to successfully address the reasons for the emergence of tumor resistance. This is because most ex vivo tissues focus on bioadhesive signaling and stiffness. Although extensively investigated, cell adhesion and stiffness mediated drug resistance are not the only factors that contribute to chemoresistance in vivo. Here we discuss that in addition to matrix stiffness, cellular, biochemical, and biophysical parameters such as stress relaxation, adhesion, spatio-temporal protein signaling, and porosity/confinement need to be considered. For ex vivo models, it will thus be important to incorporate these various biophysical parameters, ideally in a modular fashion to maximize control of cell fate and drivers of oncogenic transformations. We propose several new areas of technological advancement needed for building better ex vivo cancer models to understand tumor resistance. These topics cover integration of biomaterials-based engineering with emerging forefronts of tissue mechanics, nuclear mechanics, immune response, and the gut microbiome.
Integrating independent control of biomechanical and spatio-temporal signaling of tumors
The plasticity of cancer cells to evolve different drug resistant phenotypes is encoded by the organization and spatiotemporal dynamics of signal transduction networks. This plasticity allows them to adapt to challenging microenvironments, remodel them in their own favor, and to withstand highly toxic therapeutic assaults. A recent review discusses the rich molecular signaling dynamics and their impact on cancer cell proliferation, survival, invasiveness and drug resistance [15]. Most prior studies in cancer 3D modeling used hydrogels or scaffolds as hydrated networks of motif-containing bulk proteins or their peptidomeric forms. These peptides are either short peptides that represent adhesive binding motifs (e.g., fibronectin or vitronectin derived RGD), or hydrogel crosslinking peptides that are matrix metalloproteinase (MMP)-degradable and allow for matrix remodeling. While numerous studies have shown the utility of these hydrogels in spread, mesenchymal-like cells, we have engineered modular hydrogels to show that integrin ligands and matrix degradability can serve as pro-survival signals in Band T-cell lymphomas, modulating their 3D aggregation and response to therapeutics [12]. Using RGD-presenting hydrogels and complementary studies in patient-derived xenografts mouse models, we have further shown that integrin αvβ3 acts as a membrane receptor for thyroid hormones to mediate angiogenesis in malignant T cells [16]. This particular study led us to discover novel mechanism for the endocrine modulation of T cell lymphoma pathophysiology. However, the flow of information between tumor cells and surrounding ECM is bidirectional and functions in a spatio-temporal manner. The adhesion process involves dynamic interactions taking place over multiple time- and length-scales, from seconds for nanoscale integrin receptor-ECM ligand binding, to days and weeks for meso/macroscale ECM remodeling and cancerous tissue organization. Similar to the dynamic nature of cell-ECM interactions, engineering of materials to elicit desired tumor cell responses will require precise and independent, multi-dimensional control over matrix spatio-temporal bio-ligand presentation, structural porosity, and the mechanical properties of the materials (Figure 1A, Key Figure). Recent biomaterials designs now allow spatio-temporal control, as reported by several research groups across various cell-tissue models [17–21]. Nonetheless, current hydrogels have yet to demonstrate that the same biomaterials can provide independent control over all of the bioadhesivity, spatio-temporal signaling, stiffness, and porosity of the material. In a major advancement, chemical strategies that allow external manipulation of the ligand presentation in real-time were recently developed. A new class of hydrogels was reported, that used an addition-fragmentation-chain transfer chemistry and permitted repeated exchange of biochemical ligands in a non-destructive manner [22]. Such advances afford powerful designer tools in material engineering to study cancer cell processes.
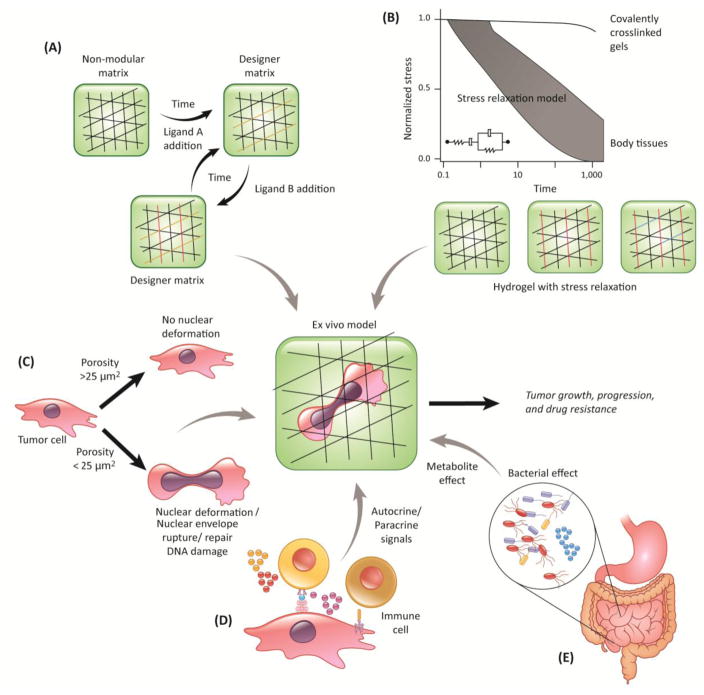
Figure 1. Key Figure. Ex vivo cancer tissues with multi-dimensional control of biophysical and biochemical properties.
The schematic depicts integration of new strategies into existing tumor tissue models: (A) time dependent, controlled, reversible exchange of biochemical ligands; (B) stress relaxation that models viscoelastic behavior of tissues in comparison to current, covalently crosslinked matrices. Simple covalently cross-linked hydrogels (black lined network) with stress relaxation can be designed by including ionic bridges (red lines) and chemical spacers (e.g. polyethylene glycol, blue lines); (C) 3D niche porosity to model nuclear deformability of cancer cells; (D)-(E) integrating cell-cell and autocrine/paracrine effect of immune system and the gut microbiome
Another emerging trend is the understanding of stress relaxation in vivo and ex vivo, and is increasingly becoming a crucial parameter for biomaterials design. Using bead displacement methods, Legant and colleagues showed deformations of 20–30% peak principal strain in the hydrogel surrounding the cell [78]. In 2D culture of cells on acrylamide gels, strains of 3–4% (ratio of traction to elastic modulus) are typically observed [23]. The ability of ex vivo scaffolds to either store (purely elastic) or dissipate (viscoelastic) forces generated by cells in contact with these surfaces can influence cell’s interaction with its surrounding [24], and we believe they may in turn regulate spreading, growth, migration, and possibly chemoresistance. In fact, stem cell growth and differentiation is enhanced in hydrogels with fast stress relaxation characteristics, as reported recently by Chaudhuri and colleagues [24]. Most non-degradable, synthetic hydrogels are purely elastic, whereas many naturally derived matrices and tissues are viscoelastic (Figure 1B, Key Figure), can be degraded/remodeled by cells, and often exhibit partial stress relaxation when a constant strain of 15% is applied [24]. For example, collagen and fibronectin matrices exhibit a decrease in the storage or elastic modulus over time when a constant strain is applied. This likely occurs from the unbinding of weak hydrophobic and electrostatic interactions, which hold the fibers in a network [25, 26]. The elasticity of these materials is also nonlinear. On reconstituted natural matrices, the resistance to cellular traction forces is expected to relax over time due to flow and remodeling of the matrix, dissipating the energy that cell-generated forces impart into the material. Substrates with stress relaxation enhance cell spreading at a low initial elasticity, which is mediated through β1 integrin, actin polymerization, and actomyosin contractility, and is associated with increased YAP (Yes-associated protein) nuclear localization and proliferation [26, 27]. This suggests that increased stress relaxation can compensate for matrices with a lower stiffness. Since several mechanosignaling network components are involved, we suspect that changes in stress relaxation and the resulting cellular response could be key to chemoresistance. Therefore, it is imperative that advanced tissue models for cancer (and also for regenerative medicine) research should consider stress relaxation beyond and independent of stiffness and bioadhesivity. Some of the questions to be considered are: can stress relaxation potentially program tumor cells into a more resistant phenotype? Can stress relaxation cross-talk with genetic mutations? What is the role of time-dependent viscoelastic properties in tumorigenesis, infiltration of immune cells? And what categories of tumors depend on stress relaxation? These questions can be answered using engineered biomaterials that incorporate stress relaxation behaviors and comparing them with purely elastic materials. The challenge will be in developing such materials where stress relaxation, bio-adhesivity, porosity, stiffness, and topography are independently tunable.
Designing 3D niche porosityto accommodate nuclear deformability of cells
In vivo, cells often have to transit through narrow constrictions smaller than the nuclear diameter during migration, for example, when passing through interstitial spaces or endothelial layers during intra- and extravasation. This concept applies to all migrating cells, including immune cells, fibroblasts, invasive cancer cells, and possibly even tissue stem cells. Recent work suggests that the biophysical properties of the nucleus can play a crucial rate-limiting role during cell migration in 3D environments: For pores substantially smaller than the nuclear cross-section, migration efficiency decreases, and cells eventually stall completely when the pore size reaches the ‘nuclear migration limit’ [28]. The ability of cells to pass through such confined spaces is mostly determined by nuclear size, deformability, and cytoskeletal contractility, which are affected by nuclear and cytoskeletal composition and organization [28–31]. As these parameters can vary wildly between cell types, porosity and confinement should be key considerations of engineered microenvironments. Although most engineered niches have considered porosity of the scaffold from the crosslinking density perspective, better material designs are needed to account for nuclear deformability, as some applications may favor designs that prevent (specific) cells from entering, while other designs may benefit from large enough pores to let cells enter the material. In addition to limiting the motility of cells, moving the large cell nucleus through small pores can also have other biological consequences. Migrating through such tight spaces places substantial physical stress on the nucleus, which can result in transient loss of nuclear envelope integrity, herniation of chromatin across the nuclear envelope, DNA damage, and redistribution of mobile nuclear proteins in vitro and in vivo [32–34]. Recent in vitro work by the Discher group further supports the concept that migration through tight spaces results in increased genomic stability [34, 35], but direct observations of migration-induced DNA damage in vivo and resulting genomic instability is still outstanding. Nuclear deformation may also alter chromatin organization and gene expression, which could further modulate tumor progression and resistance to therapy. Nuclear envelope rupture and migration-induced DNA damage may emerge as a potential novel therapeutic approach to specifically target metastatic cells. Supporting this idea, combined inhibition of nuclear envelope repair and DNA damage repair resulted in significant death of cancer cells during migration through confined environments [32]. Finally, our recent work further links nuclear envelope rupture in micronuclei and an inflammatory response, explaining how chromosomal instability drives metastasis through a cytosolic DNA response [36].
Given the importance of cell migration in many biomedical applications, ranging from tissue-engineering, prosthetic device coating, and cancer therapy, these recent findings highlight the importance of considering pore sizes and nuclear deformability of the relevant cells in the design process of biomaterials-based scaffolds. In general, while designing new biomaterials-based ex vivo tissue engineered models of development, wound healing, tumors, and even coating of prosthetic devices, one should consider tissue scaffold pore sizes <25 μm2 in cross section (Figure 1C, Key Figure). These considerations will vary with regard to tumor type and patient-specific attributes. Infiltrating immune cells, such as the leukocytes, neutrophils and dendritic cells, for example, can squeeze through much smaller pores, down to around 1–2 μm2. We suggest that the scaffold pore size is an important design parameter, especially since small pores could not only slow down migrating cells, but also induce nuclear rupture and deformation. We suspect that nuclear rupture and deformation can contribute to DNA damage and genomic instability, promoting drug resistance, through genomic rearrangements or mutations that increase cell proliferationand/or abnormal signaling. Nevertheless, a generalized approach in determining pore size is not feasible because of wide differences in the biophysical characteristics of tumor cell. Application for different cells will require different consideration for the pore sizes, and that degradability of material must also be considered, as many cells can remodel ECM, for example, by secretion of matrix-metalloproteases (MMPs).
Integrating immune system and effect of gut microbiome
In addition to changes in cell and tissue stiffness, other hallmarks of cancer include chronic inflammation and altered immune responses [37]. While modeling tumor-immune interaction in engineered tissues is important, another point of view is that tumor-infiltrating immune cells differentiate into phenotypes that support each step of the metastatic cascade, and thus, are novel targets for therapy. In vivo, tumor-infiltrating T-cells (including regulatory T-cell), B-cells, immunosuppressive myeloid-derived suppressor cells (MDSCs) and macrophages continuously interact with tumor cells through direct cell contact or by the secretion of a milieu of pro-inflammatory cytokines and chemokines. This immune-privileged microenvironment could selectively impair the recognition of tumor antigens by cytotoxic T-cells and also protect residual tumor cells against cytotoxic destruction [38–41]. Immune cell populations, like T-cells and natural killer cells, can control metastases of cancer cells by either restricting them to the primary tumor niche or promoting migration away from the primary tumor site [42, 43]. Even macrophages, the cells of innate immune system, adapt to tumor microenvironment and polarize to the M1 extreme and secrete large levels of interleukins (e.g. interleukin 8 and 10) and granulocyte colony-stimulating factor, which suppress immunity [44]. The secretion of cytokines is dependent on tumor subtype.
Introducing these immune components and recapitulating immune-cancer interaction complex in an ex vivo engineered systems is ideal but non-trivial (Figure 1D, Key Figure). One of the main challenges is that immune cells, when cultured ex vivo, undergo rapid apoptosis over time, unless rescued by anti-apoptotic signals and replenished with a fresh supply of immune cells [45]. Newer approaches that recapitulate continuous replenishment of immune cells in vivo can be achieved by integrating microfluidic platforms [46] with engineered tissues, where immune cells could be added to the feeding lymphatic or vascular networks. Simple encapsulation of immune cells in collagen or Matrigel is suboptimal, as these biomaterials may not provide necessary survival signals and functional immune cells. Alternatively, advanced biomaterials-based scaffolds that support immune cell growth [47–51] could be integrated with the existing tumor niches and other on-chip approaches [52]. Developing such tissues can further be applied to study the efficacy of immunotherapy and effect of immunomodulatory drugs on immune cells.
In addition to the immune cells, tumor progression and the efficacy of anti-tumor therapies are now known to be affected by the microbiome. For example, Fusobacterium nucleatem, amongst other bacteria, is enriched in colorectoral cancer patients’ microbiomes [53, 54], and Helicobacter pylori has also been shown to be abundant in patients with non-cardia gastric cancers [55]. These interactions can be local, as Clostridia, Bifidobacteria and Salmonella (reviewed in [56]) have been shown to grow within the tumor microenvironment and elicit immune reactions that can assist in tumor suppression. Similarly, within the colon, microbial biofilms have also been implicated in the progression of colon cancer [57]. Direct impacts of the microbiome may also be systemic. Recently, the production of a microbe-derived carcinogen, deoxycholic acid, was shown to directly contribute to liver cancer [58]. Although the mechanisms that causally link microbes and tumorigenicity have not been established [59], several mechanisms have been proposed, including metabolic signaling to promote proliferation or angiogenesis pathways [60]. Microbes may also produce reactive oxygen species, which may in turn contribute to DNA damage and ultimately, tumor resistance. Manipulating these systems in vivo can be technically challenging due to the complexity of the communities and the difficulty of working with germ-free or monocolonized mice. An outstanding question is how to best integrate the effects of the microbiome into current in vitro engineered model systems of cancer and tumor resistance to create a more efficient cancer ecosystem (Figure 1E, Key Figure).
The longest standing evidence for a link between the microbiome and cancer is in relation to colon cancer. In vitro models of colon cancer utilize organoid models, which are extremely useful for testing drug delivery, genetic manipulation, and incorporation of biopsied tissue. Bacteria can be administered in these models by injecting them directly into the organoid cavity [61], but integrating biofilms into the organoid model of colon cancer has yet to be done. An alternative to organoids for intestinal cancer models are 3-D scaffold models of intestinal tissue. Several studies have been completed with co-culture of organisms on these scaffolds [62, 63], yet these experiments are limited in scope to a handful of organisms and are constrained by the difficulties of co-culturing intestinal cells with organisms that are normally found in an anaerobic environment. There are additional engineering challenges in introducing the range of biologically relevant cell types, which would both enhance mucous production and maintain barrier integrity. More attention is warranted in areas of understanding microscale interactions of these organisms and cancer cells, and in shear stress caused by the laminar flow of intestinal contents, peristalsis and large-scale mechanical reflexes and cellular apoptosis.
The mechanistic effects of the microbiome in colon cancer may be direct (e.g., signaling and small molecule delivery directly to intestinal stem cells, affecting their proliferation) or indirect, via the immune system function. The most striking example of this relates the microbiome and the efficacy of cancer immunotherapies in rodent models of colon cancer and melanoma [64, 65]. It is currently thought that that the microbiome alters the function of the immune system, specifically through changes in immune cell composition, maturation, and the inflammatory milieu. Several questions remain: do in vitro models of cancer lacking microbes adequately recapitulate in vivo malignancies, or must we include microbes? Are there effects on tumors or the immune system that are specific to certain microbes or microbiome compositions? Are microbiome-mediate effects on cancer progression direct or predominantly mediated through the immune system? Can microbiome-mediated effects on cancer progression be modeled by simply introducing metabolites, cellular lysate or supernatant to tumor cell culture, or do we need to co-culturing bacterial cells via ex vivo gut reactors integrated with immune and cancer tissues to elicit real-time feedback? Additionally, this feedback likely impacts the composition of the microbiome as well and its function. The promise of integrating complex microbial communities is that it will permit bidirectional analysis into microbiome effects on cancerous cells and the effects of tumorigenic cells on those bacteria. Single-cell analysis of individual bacteria [66] will provide information on selective pressures exerted on bacteria within the tumor micro-environment; metagenomic and strain-level analyses of microbial communities [67] co-cultured with tumor cells can be used to examine the function of these cells, including information on the metabolites they produce that might in turn influence tumorigenic progression and the formation of resistance. In vitro systems may prove fruitful in testing the effects of these different communities on immune or cancer tissue.
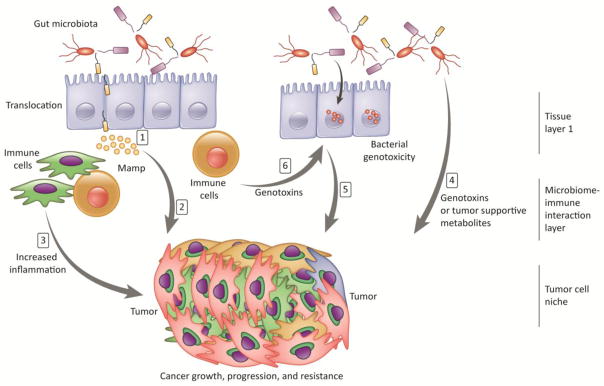
Lastly, because we know that the tumor microenvironment is permissive to bacterial colonization, can we study the effects of tumor-targeting synthetic bacteria in vitro? Din and colleagues engineered a side trap array microfluidic platform to co-culture human cervical cancer HeLa cells with S. Typhimurium [68]. Ingber and colleagues have also reported a simple biomimetic ‘human gut-on-a-chip’ microdevice that recapitulates epithelium polarization and villi-like folds, and can be used for co-culture of a normal intestinal microbe (Lactobacillus rhamnosus GG) for extended periods of time (>1 week) without compromising epithelial cell viability [69]. Yissachar et. al. [70] have taken an alternative approach, whereby segments of intestinal tissue are excised and cultured in vitro, linking intestinal inputs and outputs directly to cultured immune and neuronal cells. This system may prove valuable in linking microbiome outputs to other in vitro models of tumor progression. The next challenge will be in determining how to integrate the diversity of the microbiome, so that we can study its effect on overall tumor progression, tumor resistance, and drug targeting while modulating other aspects of the tumor microenvironment cross-talk (Figure 2).
Figure 2. Exvivo cancer models with possible microbiome-tumor interactions.
The figure shows an example of a three-layer ex vivo model.1) Alterations in gut microbiota may result in increased bacterial translocation, 2)Increased abundance of microorganism-associated molecular patterns (MAMPs) directly influences tumor cells through local or distant mediators, 3)MAMPs stimulate Toll-Like-Receptors on immune and other niche cells, leading to increased tumor supportive milieu of cytokines (e.g., interleukins) and growth factors, 4) The microbiota mediates tumor suppression through generation of short-chain fatty acids and biological activation of cancer-preventing phytochemicals, 5) Bacterial genotoxins, after being delivered to the nucleus of host cells, actively induce DNA damage in organs that are in direct contact with the microbiome, such as the gastrointestinal tract. Other genotoxic components include reactive oxygen species, reactive nitrogen species released from inflammatory cells, and hydrogen sulphide from the microbiota, 6)Gut-mediated metabolites may result in the a) activation of genotoxins such as acetaldehyde; b) activation of the metabolism of hormones; and c) alterations in the in the metabolism of bile acids.
Concluding Remarks
Engineered microenvironments have already emerged as important and useful tools to study tumor cells in vitro. A key advantage of ex vivo models is that the number of features (and correspondingly, the number of variables in the system) is constrained, so that the effects of specific factors can be more clearly delineated. Although the most comprehensive models may not necessarily be the best model for a particular application, there is still a need to address unresolved questions of cellular, molecular, and microenvironment complexity (see outstanding questions). In addition to conventional bioadhesive matrices, decellularized matrices and glycosaminoglycans (GAGs) offer alternative choices for fine tuning the ‘right’ characteristics in the model in terms of both pore size and matrix relaxation. Most previous ex vivo models have focused on stiffness and biochemical ligand presentation as the predominant design parameters and tumor invasion as the primary read-outs. We propose that additional parameters should be considered in the design of such models, including viscoelastic properties, physical pore sizes, and the microbiome. Additional aspects of tumor microenvironments could include cancer associated fibroblasts, stromal cells, immune cells, the role of fluid flow, matrix heterogeneity, and interactions with the vascular and lymphatic circulations [71–73]. In addition, genomic and epigenetic evolution of cancer cells should be considered as key drivers of therapy resistance, and the effect of the physical microenvironment of these processes should be investigated in more detail. We believe that careful modeling of ex vivo niches will result in improved understanding of epigenetic, metabolic, and signaling patterns in cancer, therefore leading to new therapeutics development [16, 74–77]. Modular approaches that enable independently tuning individual biophysical and biochemical parameters will facilitate more systematic studies. Technical advances that permit co-culture of bacteria with cancer cells can not only lead to better understanding of the tumor-microbiome interaction but also drive new directions to systemically deliver an anti-tumor toxin using synthetically engineered bacteria. Finally, newer ex vivo models that recapitulate complete selective aspects of the tumor-immune-microenvironment interactome are needed to maximize our effort towards tumor microenvironment driven precision medicine strategy and evaluate synergies between combination therapies to overcome resistance. Such innovative approaches will increase “predictive power” of pre-clinical inhibitors, provide potential biomarkers for correlative studies in new inhibitor clinical trials, and provide clues towards mechanisms that induce resistance to therapeutic inhibitors by more faithfully representing patient biological features and creating clinically relevant treatment regimens.
OUTSTANDING QUESTIONS.
How do chemoresistance and changes in biophysical and biochemical factors relate to one another?
Can stress relaxation potentially program tumor cells into a more resistant phenotype? Can stress relaxation cross-talk with genetic mutations? And what categories of tumors depend on stress relaxation?
What is the role of time-dependent viscoelastic properties in tumorigenesis, infiltration of immune cells?
How can we engineer ex vivo models with cell-specific consideration for the pore sizes?
Is it sufficient to leave out the microbes from in vitro models of cancer? Are there effects of specific microbes or is it mainly the triggers they provide to the immune system?
Can microbiome-mediated effects on cancer progression be modeled by simply introducing metabolites, cellular lysate or supernatant to tumor cell culture, or do we need to co-culturing bacterial cells via ex vivo gut reactors integrated with immune and cancer tissues to elicit real-time feedback?
How best to integrate the effects of the microbiome into current in vitro engineered model systems of cancer and tumor resistance to create a more efficient cancer ecosystem?
TRENDS BOX.
Matrix stiffness influences phenotype and epigenetics of tumor cells and influences chemoresistance across solid, palpable, and liquid tumors
Biomaterials that independently modulate matrix stiffness from composition and architecture reveal that, in normal mammary epithelial cells, increasing matrix stiffness alone induces malignant phenotypes.
Healthy and malignant cells migrating through narrow confinements undergo nuclear deformation, which can result in transient loss of nuclear envelope integrity, herniation of chromatin across the nuclear envelope, DNA damage, and redistribution of mobile nuclear proteins
Immune-privileged microenvironmentcould selectively impair the recognition of tumor antigens by cytotoxic T cells
Commensal microbiota promotes the efficacy of cancer therapies
Acknowledgments
The authors acknowledge financial support from the National Institutes of Health (1 R01AI132738-01A1, Singh; 1 R33 CA212968-01, Singh; U54 CA210184, Lammerding; R01 HL082792, Lammerding), the U.S. Department of Defense Career Development Award (W81XWH-17-1-0215, Singh), the U.S. Department of Defense Breast Cancer Research Program (Breakthrough Award BC150580, Lammerding), and the National Science Foundation CAREER Awards (DMR-1554275, Singh; CBET-1254846, Lammerding). The authors also acknowledge the financial support from Sloan Foundation Research Fellowship (Brito) and the National Sciences Foundation (MCB-1650122, Brito). This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Cancer Research Program, under Award No. W81XWH-17-1-0215 (Singh). Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense, the NIH, the NSF, or other funding agency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Holohan C, et al. Cancer drug resistance: an evolving paradigm. Nature reviews Cancer. 2013;13(10):714–26. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 2.Correia AL, Bissell MJ. The tumor microenvironment is a dominant force in multidrug resistance. Drug Resist Updat. 2012;15(1–2):39–49. doi: 10.1016/j.drup.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holzel M, Bovier A, Tuting T. Plasticity of tumour and immune cells: a source of heterogeneity and a cause for therapy resistance? Nature reviews Cancer. 2013;13(5):365–76. doi: 10.1038/nrc3498. [DOI] [PubMed] [Google Scholar]
- 4.Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol. 2015;17(8):955–63. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468(7323):580–4. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang SE, et al. Transforming growth factor beta induces clustering of HER2 and integrins by activating Src-focal adhesion kinase and receptor association to the cytoskeleton. Cancer Res. 2009;69(2):475–82. doi: 10.1158/0008-5472.CAN-08-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acerbi I, et al. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integrative biology : quantitative biosciences from nano to macro. 2015;7(10):1120–34. doi: 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apoorva FNU, et al. Lymph node stiffness-mimicking hydrogels regulate human B-cell lymphoma growth and cell surface receptor expression in a molecular subtype-specific manner. J Biomed Mater Res A. 2017;105(7):1833–1844. doi: 10.1002/jbm.a.36031. [DOI] [PubMed] [Google Scholar]
- 9.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer cell. 2005;8(3):241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Tan Y, et al. Matrix softness regulates plasticity of tumour-repopulating cells via H3K9 demethylation and Sox2 expression. Nature communications. 2014;5:4619. doi: 10.1038/ncomms5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nature reviews Cancer. 2009;9(2):108–22. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian YF, et al. Integrin-specific hydrogels as adaptable tumor organoids for malignant B and T cells. Biomaterials. 2015;73:110–119. doi: 10.1016/j.biomaterials.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin JW, Mooney DJ. Extracellular matrix stiffness causes systematic variations in proliferation and chemosensitivity in myeloid leukemias. Proc Natl Acad Sci U S A. 2016;113(43):12126–12131. doi: 10.1073/pnas.1611338113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15(12):1243–53. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolch W, et al. The dynamic control of signal transduction networks in cancer cells. Nature reviews Cancer. 2015;15(9):515–27. doi: 10.1038/nrc3983. [DOI] [PubMed] [Google Scholar]
- 16.Cayrol F, et al. Integrin alphavbeta3 acting as membrane receptor for thyroid hormones mediates angiogenesis in malignant T cells. Blood. 2015;125(5):841–51. doi: 10.1182/blood-2014-07-587337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKinnon DD, et al. Biophysically Defined and Cytocompatible Covalently Adaptable Networks as Viscoelastic 3D Cell Culture Systems. Advanced materials. 2014;26(6):865–72. doi: 10.1002/adma.201303680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alge DL, et al. Synthetically tractable click hydrogels for three-dimensional cell culture formed using tetrazine-norbornene chemistry. Biomacromolecules. 2013;14(4):949–53. doi: 10.1021/bm4000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeForest CA, Anseth KS. Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions. Nature chemistry. 2011;3(12):925–31. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhuri O, et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater. 2014;13(10):970–8. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 21.Lee TT, et al. Light-triggered in vivo activation of adhesive peptides regulates cell adhesion, inflammation and vascularization of biomaterials. Nat Mater. 2015;14(3):352–60. doi: 10.1038/nmat4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gandavarapu NR, Azagarsamy MA, Anseth KS. Photo-click living strategy for controlled, reversible exchange of biochemical ligands. Adv Mater. 2014;26(16):2521–6. doi: 10.1002/adma.201304847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 24.Chaudhuri O, et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater. 2016;15(3):326–34. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munster S, et al. Strain history dependence of the nonlinear stress response of fibrin and collagen networks. Proc Natl Acad Sci U S A. 2013;110(30):12197–202. doi: 10.1073/pnas.1222787110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nam S, et al. Strain-enhanced stress relaxation impacts nonlinear elasticity in collagen gels. Proc Natl Acad Sci U S A. 2016;113(20):5492–7. doi: 10.1073/pnas.1523906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhuri O, et al. Substrate stress relaxation regulates cell spreading. Nature communications. 2015;6:6364. doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf K, et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201(7):1069–84. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harada T, et al. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol. 2014;204(5):669–82. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidson PM, et al. Nuclear Deformability Constitutes a Rate-Limiting Step During Cell Migration in 3-D Environments. Cellular and molecular bioengineering. 2014;7(3):293–306. doi: 10.1007/s12195-014-0342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lautscham LA, et al. Migration in Confined 3D Environments Is Determined by a Combination of Adhesiveness, Nuclear Volume, Contractility, and Cell Stiffness. Biophysical journal. 2015;109(5):900–13. doi: 10.1016/j.bpj.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denais CM, et al. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352(6283):353–8. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raab M, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352(6283):359–62. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- 34.Irianto J, et al. DNA Damage Follows Repair Factor Depletion and Portends Genome Variation in Cancer Cells after Pore Migration. Curr Biol. 2017;27(2):210–223. doi: 10.1016/j.cub.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfeifer CR, et al. Genome variation across cancers scales with tissue stiffness - an invasion-mutation mechanism and implications for immune cell infiltration. Curr Opin Syst Biol. 2017;2:103–114. doi: 10.1016/j.coisb.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakhoum SF, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553(7689):467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol. 2015;15(2):73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143(3):355–66. doi: 10.1016/j.cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert LA, Hemann MT. Chemotherapeutic resistance: surviving stressful situations. Cancer Res. 2011;71(15):5062–6. doi: 10.1158/0008-5472.CAN-11-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shree T, et al. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011;25(23):2465–79. doi: 10.1101/gad.180331.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakasone ES, et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer cell. 2012;21(4):488–503. doi: 10.1016/j.ccr.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bidwell BN, et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat Med. 2012;18(8):1224–31. doi: 10.1038/nm.2830. [DOI] [PubMed] [Google Scholar]
- 43.Fridlender ZG, Albelda SM, Granot Z. Promoting metastasis: neutrophils and T cells join forces. Cell research. 2015;25(7):765–6. doi: 10.1038/cr.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang R, et al. Increased IL-10 mRNA expression in tumor-associated macrophage correlated with late stage of lung cancer. J Exp Clin Cancer Res. 2011;30:62. doi: 10.1186/1756-9966-30-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh A. Biomaterials innovation for next generation ex vivo immune tissue engineering. Biomaterials. 2017;130:104–110. doi: 10.1016/j.biomaterials.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 46.Shin Y, et al. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat Protoc. 2012;7(7):1247–59. doi: 10.1038/nprot.2012.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purwada A, et al. Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials. 2015;63:24–34. doi: 10.1016/j.biomaterials.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purwada A, et al. Modular Immune Organoids with Integrin Ligand Specificity Differentially Regulate Ex vivo B Cell Activation. ACS Biomater Sci Eng. 2017;3(2):214–225. doi: 10.1021/acsbiomaterials.6b00474. [DOI] [PubMed] [Google Scholar]
- 49.Purwada A, Singh A. Immuno-engineered Organoids for Regulating the Kinetics of B cell Development and Antibody Production. Nature Protocols. 2017;12:168–182. doi: 10.1038/nprot.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan Y, et al. Bioengineering Thymus Organoids to Restore Thymic Function and Induce Donor-Specific Immune Tolerance to Allografts. Mol Ther. 2015;23(7):1262–77. doi: 10.1038/mt.2015.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hun M, et al. Native thymic extracellular matrix improves in vivo thymic organoid T cell output, and drives in vitro thymic epithelial cell differentiation. Biomaterials. 2017;118:1–15. doi: 10.1016/j.biomaterials.2016.11.054. [DOI] [PubMed] [Google Scholar]
- 52.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. 2014;32(8):760–72. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 53.Kostic AD, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–15. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castellarin M, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuccio L, Eusebi LH, Bazzoli F. Gastric cancer, Helicobacter pylori infection and other risk factors. World J Gastrointest Oncol. 2010;2(9):342–7. doi: 10.4251/wjgo.v2.i9.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pawelek JM, Low KB, Bermudes D. Bacteria as tumour-targeting vectors. The Lancet Oncology. 2003;4(9):548–56. doi: 10.1016/s1470-2045(03)01194-x. [DOI] [PubMed] [Google Scholar]
- 57.Johnson CH, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21(6):891–7. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshimoto S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499(7456):97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 59.Thomas RM, Jobin C. The Microbiome and Cancer: Is the ‘Oncobiome’ Mirage Real? Trends Cancer. 2015;1(1):24–35. doi: 10.1016/j.trecan.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lax AJ, Thomas W. How bacteria could cause cancer: one step at a time. Trends in microbiology. 2002;10(6):293–9. doi: 10.1016/s0966-842x(02)02360-0. [DOI] [PubMed] [Google Scholar]
- 61.Bartfeld S, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148(1):126–136 e6. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shah P, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nature communications. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim HJ, et al. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A. 2016;113(1):E7–15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–9. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vetizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–84. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu L, et al. Virtual microfluidics for digital quantification and single-cell sequencing. Nature methods. 2016;13(9):759–62. doi: 10.1038/nmeth.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brito IL, Alm EJ. Tracking Strains in the Microbiome: Insights from Metagenomics and Models. Front Microbiol. 2016;7:712. doi: 10.3389/fmicb.2016.00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Din MO, et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature. 2016;536(7614):81–5. doi: 10.1038/nature18930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim HJ, et al. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab on a chip. 2012;12(12):2165–74. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 70.Yissachar N, et al. An Intestinal Organ Culture System Uncovers a Role for the Nervous System in Microbe-Immune Crosstalk. Cell. 2017;168(6):1135–1148 e12. doi: 10.1016/j.cell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Calvo F, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol. 2013;15(6):637–46. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seo BR, et al. Obesity-dependent changes in interstitial ECM mechanics promote breast tumorigenesis. Science translational medicine. 2015;7(301):301ra130. doi: 10.1126/scitranslmed.3010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Polacheck WJ, et al. Mechanotransduction of fluid stresses governs 3D cell migration. Proc Natl Acad Sci U S A. 2014;111(7):2447–52. doi: 10.1073/pnas.1316848111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang L, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med. 2015;21(11):1364–71. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beguelin W, et al. EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb-E2F1 feedback loop. Nature communications. 2017;8(1):877. doi: 10.1038/s41467-017-01029-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stephenson R, Singh A. Drug discovery and therapeutic delivery for the treatment of B and T cell tumors. Adv Drug Deliv Rev. 2017 doi: 10.1016/j.addr.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jarde T, et al. Wnt and Neuregulin1/ErbB signalling extends 3D culture of hormone responsive mammary organoids. Nature communications. 2016;7:13207. doi: 10.1038/ncomms13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Legant WR, et al. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]