Abstract
Background
Epidemiological studies have found that triple-negative breast cancer (TNBC) and TN inflammatory breast cancer (IBC) are associated with lower frequency and duration of breast-feeding compared to non-TNBC and non-TN IBC, respectively. Limited breast-feeding could reflect abrupt or premature involution and contribute to a “primed” stroma that is permissive to the migration of cancer cells typical of IBC. We hypothesized that gene expression related to abrupt mammary gland involution after forced weaning may be enriched in the tissues of IBC patients and, if so, provide a potential correlation between limited breast-feeding and the development of aggressive breast cancer.
Methods
We utilized the Short Time-series Expression Miner (STEM) program to cluster significant signatures from two independent studies that analyzed gene expression at multiple time-points of mouse mammary gland involution. Using 10 significant signatures, we performed gene ontology analysis and gene set enrichment analysis (GSEA) on training and validation sets from human breast cancer gene expression data to identify specific genes that are enriched in IBC compared to non-IBC and in TN compared to non-TN in IBC and non-IBC groups.
Results
Examining the combined data, we identified 10 involution gene clusters (Inv1-10) that share time-dependent regulation after forced weaning. Inv5 was the only cluster significantly enriched in IBC in the training and validation set (nominal p-values <0.05) and only by unadjusted p-values (FDR q-values 0.26 and 0.46 respectively). Eight genes in Inv5 are upregulated in both the training and validation sets in IBC. Combining the training and validation sets, both Inv5 and Inv6 have nominal p-values <0.05 and q-values 0.39 and 0.20, respectively. The time course for both clusters includes genes that change within 12 hours after forced weaning.
Conclusions
Results from this in silico study suggest correlation between molecular events during abrupt involution and aggressive breast cancer. Specifically, candidate genes from Inv5 merit functional investigation regarding the role of limited breast-feeding in IBC development.
Introduction
The mammary gland undergoes various stages of development during the embryonic, pubertal, reproductive, and post-reproductive stages of life. Involution is a term that has been described as the reverse of development [1]. Post-lactational involution is a complex multistage process characterized by regression of the mammary gland epithelium to its non-lactating state through apoptosis and tissue remodeling [2]. Clarkson et al. [3] and Stein et al. [4] conducted gene expression profiling studies of these changes in the mouse mammary gland with the induction of forced weaning at the peak of lactation. Results of these two studies highlight distinct molecular characteristics between the virgin, pregnant, lactating, and involuting states of the mammary gland. It has been shown that the mammary gland microenvironment undergoing tissue remodeling during post-lactational involution mimics that of pathological conditions like wound healing and tumorigenesis [5]. Inflammation and would-healing responses have been found to be associated with tumor growth and progression [6]. Findings from several animal and in vitro studies indicate that involution may create a microenvironment that promotes breast cancer growth and progression [7–12].
Breast cancer is the most common cancer and the second leading cause of cancer mortality among women in the United States [13]. In the absence of estrogen receptor (ER) expression, progesterone receptor (PR) expression, and HER2-neu amplification, breast cancer is termed triple-negative breast cancer (TNBC). TNBC accounts for approximately 15% of all breast cancer incidents and has the lowest survival rates among all subtypes of breast cancer [14]. Inflammatory breast cancer (IBC) is a distinct subtype of breast cancer characterized pathologically by the presence of tumor emboli in the dermal lymphatics and clinically by its rapid and diffuse onset with erythematous and edematous presentation of the breast [15]. IBC accounts for approximately 1–5% of all breast cancers but at all stages has significantly lower 5-year survival rates than that for non-IBC [15–17]. Little-to-no breast-feeding has been found to correlate with an increased risk of developing aggressive breast cancer subtypes [18, 19]. In Gaudet et al, TNBC was associated with a shorter duration of breast-feeding in a cohort of 890 young (≤56 years) breast cancer patients [18]. Atkinson et al found also that in a cohort of 224 women with IBC, those that did not breast-feed were more likely to develop TN IBC and luminal IBC [19]. Furthermore, Lyons et al. showed that TN ductal carcinoma in-situ (DCIS) cells exposed to the involuting mammary microenvironment formed large, invasive tumors characterized by abundant fibrillar collagen and high COX-2 expression, which both correlate with a poor prognosis [10]. Additionally, both luminal and myoepithelial lineages in the mammary gland contain long-lived stem cells and stem-like cells, and pregnancy leads to a transient 11-fold increase in their habitation there throughout lactation [11, 12, 20]. In particular, parity-induced mammary epithelial cells (PI-MECs) are an epithelial subpopulation that arise from differentiating cells during the first pregnancy and persist after postlactational remodeling. They later serve as committed alveolar progenitors along the luminal epithelium during subsequent pregnancies and exhibit two important features of multipotent stem cells: self-renewal and contribution to diverse epithelial populations in the ducts and alveoli [20, 21]. PI-MECs have been identified as primary targets of malignant transformation [22]. Thus, it is possible that abrupt involution leaves persistent, likely receptor-negative stem cells or PI-MECs within the mammary gland microenvironment, increasing the chance of an initiating TNBC and TN IBC event.
Given the correlation between minimal breast-feeding and aggressive breast cancers, we hypothesized that gene expression signatures of the abrupt post-lactational involution stage of mammary gland development persist, are complicit in the development and progression of these cancers, and are therefore enriched in TNBC and IBC bulk tumor samples. To test our hypothesis, we identified gene expression signatures for the abrupt post-lactational involution stages of mammary gland development using gene expression data from post-natal mouse mammary gland development and evaluated whether or not these gene expression signatures were enriched in IBC versus non-IBC as well as TNBC versus non-TNBC. We found significant enrichment of one post-lactational involution gene signature in IBC compared to non-IBC. This enriched signature represents genes showing initial up-regulation and later down-regulation during the involution process and significant overlap with genes upregulated in vascular smooth muscle cells (VSMC) by c-Jun N-terminal protein kinase (JNK1). Specifically, we identified 3 genes–Involucrin (IVL), Cluster of Differentiation 79B (CD79B), and leptin (LEP)–that were significantly enriched in IBC compared to non-IBC. Thus, it is possible that these genes play a role in IBC development and progression.
Materials and methods
Development of involution-specific gene signatures
Gene expression dataset on mouse mammary gland involution
In the study by Clarkson et al. [3], genome-wide expression profiles were measured with Affymetrix GeneChip MGU74ver2a arrays at the 12 stages of adult mouse mammary gland development (virgin, 8 week; pregnancy days 5, 10 and 15; lactation days 0, 5 and 10; and involution hours 12, 24, 48, 72 and 96 after forced weaning at 10 days of lactation). In the study by Stein et al. [4], gene expression profiles were measured at the 17 stages of adult mouse mammary gland development (virgin, 10 and 12 weeks; pregnancy days 1, 2, 3, 8.5, 12.5, 14.5 and 17.5; lactation days 1, 3 and 7; and involution days 1, 2, 3, 4 and 20 after forced weaning at 7 days of lactation). The gene expression data for both studies can be downloaded from the webpage of the Mammary Apoptosis and Development Group at the University of Cambridge and from the NCBI Gene Expression Omnibus (GSE12247).
Preprocessing of gene expression data
Raw gene expression profiles were preprocessed using Guanine Cytosine Robust Multi-Array (GCRMA) analysis [23] with quantile normalization, and probeset-level signals were summarized in log base 2 scale. We selected a custom Chip Definition File (CDF) MGU74Av2_Mm_ENTREZG version 18 for more accurate probe mapping to the genome [24]. There are 7952 probe sets with a CDF MGU74Av2_Mm_ENTREZG version 18 representing 7882 genes as per the annotation database available for a CDF MGU74Av2_Mm_ENTREZG version 18 at the BrainArray. After preprocessing gene expression data, further analyses were conducted using information available for 7882 probe sets representing 7882 genes with a one probe set–one gene relationship.
Identification of differentially expressed genes across time points
Tests of differences in expression were performed with the limma package (version 3.22.1) [25] from the Bioconductor project. The limma package uses the moderated t-statistic. A total of 1,055 genes were identified as the most significantly differentially expressed genes across time points (q-value <0.05) with greater than two-fold changes in at least one pair comparing time points.
Clustering analysis of time-series expression data
Ernst et al. presented an algorithm specifically designed for clustering time-series expression data [26] and developed the Short Time-series Expression Miner (STEM) program for analysis of time-series gene expression data [27]. The STEM program was obtained from the website of the Systems Biology Group of the School of Computer Science of Carnegie Mellon University. The STEM program first defines a set of representative model profiles that correspond to possible patterns of gene expression across the conditions examined in the experiment. Each gene is, then, assigned to the closest profile on the basis of correlation coefficients. The expected number of genes for each profile is also computed using random permutation, renormalization, and assignment of original values for each gene to profiles with over 500 repeated permutations. This serves as a basis for the calculation of the statistical significance of each profile. Statistically significant profiles represent the dominant expression profiles in the data set. The parameters used for STEM clustering were set at a maximum of 50 model profiles, a maximum unit change between time points of 3 and a minimum correlation for clustering similar profiles >0.7. Significant expression profiles were identified with a false discovery rate (FDR) <0.05.
Ontology analysis of significant clusters
An ontology-based analysis was performed on genes of significant clusters identified through the STEM program. We used gene ontology (GO) annotations for Mus musculus gene products available from Mouse Genome Informatics. Enrichment analysis for GO annotations was performed using a hypergeometric distribution in the STEM program, and multiple hypothesis correction was done using a randomization test. For gene-ontology enrichment with this program, p-values were corrected with 500 randomizations and were considered significant with an FDR of <0.05.
Gene set analysis of involution-specific gene signatures
Gene expression dataset on IBC and non-IBC cases
Gene expression data for IBC and non-IBC cases were obtained from the NCBI Gene Expression Omnibus (GSE22597) and the EBI ArrayExpress (E-MTAB-1006 and E-MTAB-1547) and collected through the World IBC Consortium [28]. These databases include the largest series of IBC samples ever reported, and tumor samples were obtained from patients treated in three institutions: the Institut Paoli-Calmettes (IPC, Marseille, France: 71 IBC and 139 non-IBC cases), the MD Anderson Cancer Center (MDA, Houston, TX, USA: 25 IBC and 58 non-IBC cases), and the General Hospital Sint-Augustinus (TCRU, Antwerp, Belgium: 41 IBC and 55 non-IBC cases) [28].
We also examined benign-appearing breast tissues from both IBC and non-IBC patients, 44 in total (19 with IBC and 25 with non-IBC) for further analysis. All were treated with neoadjuvant chemotherapy and mastectomy from March 2004 –May 2012. Clinical details for all patients was recorded as part of an institutional database or prospective tumor registry. All patients gave written informed consent to banking surplus tissue for future research prior to study enrollment. This specific study was separately approved by the appropriate institutional review board of The University of Texas MD Anderson Cancer Center to examine these banked tissues and correlate findings to clinical demographics.
Preprocessing of gene expression data
Raw gene expression profiles were preprocessed using GCRMA analysis [23] with quantile normalization, and probeset-level signals were summarized in log base 2 scale. We selected custom Chip Definition Files (CDFs) HGU133A_Hs_ENTREZG version 18 for preprocessing GSE22597 data and HGU133Plus2_Hs_ENTREZG version 18 for preprocessing E-MTAB-1006 and E-MTAB-1547 data [24]. There are 12,135 probe sets with a CDF HGU133A_Hs_ENTREZG version 18 with 12,064 probe sets representing 12,064 genes as per the annotation database available at the BrainArray. There are 19,674 probe sets with a CDF HGU133Plus2_Hs_ENTREZG version 18 with 19,544 probe sets representing 19,544 genes as per the annotation database available at the BrainArray. After preprocessing gene expression data, all 3 data sets were merged using common informative probe sets (n = 12,129). To remove the batch effect, we used the removeBatchEffect function from the limma package from the Bioconductor [25]. This function fits a linear model to the data and removes the components due to the batch effects. The principal component analysis plots were generated prior and after removing the batch effect to of the removeBatchEffect function (data not shown). The final merged dataset consisted of 388 samples (137 IBC cases and 251 non-IBC cases) with 12,129 probe sets with 12,063 probe sets representing 12,063 genes with a one probe set–one gene relationship.
Non-tumor breast gene expression
We employed the previously described methods for gene expression studies from normal adjacent breast tissue [29, 30]. Briefly, RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA, USA) and the RNeasy kit (Qiagen, Valencia, CA, USA). A flouorescently labeled T7 RNA polymerase promotor was used to synthesize cDNA. Reverse transcription was performed and followed by RNA labeling. The labeled RNA samples were hybridized onto U133 Plus2 GeneChips (Affymetrix, Santa Clara, CA). dChip analyzer software was used to estimate expression values, as previously described [29, 30].
Involution-specific gene signatures
Involution-specific gene signatures were identified from the results of STEM cluster analysis conducted on post-natal mouse mammary gland development as previously described here. We identified orthologous genes for genes that were found to form significant clusters in STEM cluster analysis by using ENSEMBL gene id on the orthologous data downloaded from the ENSEMBL website. Involution-specific gene signatures have also been reported in the study by Stein et al. [9]. We downloaded those signatures and identified orthologous genes for each signature (S1 Table) and used them to evaluate their enrichment in IBC versus non-IBC and TN versus non-TN subtypes.
Gene set enrichment analysis (GSEA) of involution-specific signatures
We used GSEA algorithm as mentioned in [31] to evaluate enrichment of involution-specific gene signatures in IBC cases compared to non-IBC cases and TN BC cases compared to non-TN BC cases. We ranked genes in the GSEA using the student’s t-test, and all other options in the GSEA were kept as default.
Training and validation data
A training set is a set of data used to discover potentially predictive relationships, and a validation set is used to assess the strength and utility of said predictive relationships. We divided the merged dataset into the training set to run the GSEA and into the validation set to validate the GSEA results for reproducibility. We used the stratified random sampling method with inclusion of information on IBC status, TN status, and age at diagnosis (<50 years or > = 50 years) to divide the merged dataset into the training and validation sets (Table 1).
Table 1. Final merged dataset and training and validation sets.
| Total cases |
IBC* | Non-IBC* | |||||
|---|---|---|---|---|---|---|---|
| Total | TN | Non-TN | Total | TN | Non-TN | ||
| Merged Dataset | 388 | 137 | 20 | 101 | 251 | 34 | 197 |
| Training Set | 195 | 68 | 10 | 50 | 127 | 18 | 99 |
| Validation Set | 193 | 69 | 10 | 51 | 124 | 16 | 98 |
*16 cases in IBC and 20 cases in non-IBC groups did not have information available on TN status.
Results
Development of involution-specific gene signatures
Differentially expressed genes across time points
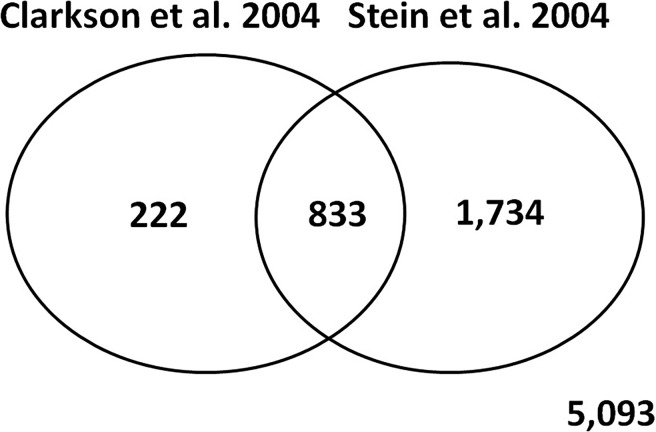
We used the limma package [25] from the Bioconductor project to identify differentially expressed genes across time points from lactation day 10 to involution day 4. We identified 1,055 differentially expressed genes across time points from lactation day 10 to involution day 4 in the data from Clarkson et al. [3]. To verify our results, we conducted an analysis for differentially expressed genes using data from Stein et al. [4] and identified 2,567 genes differentially expressed across time points from lactation day 7 to involution day 20. 79% of the genes identified as differentially expressed in the data from both studies. Fig 1 shows the Venn diagram of the overlap and discrepancies between genes differentially expressed in [3] and [4].
Fig 1. Venn diagram showing the overlap and discrepancies between genes differentially expressed in Clarkson et al. [3] and Stein et al. [4].
Clusters of differentially expressed genes
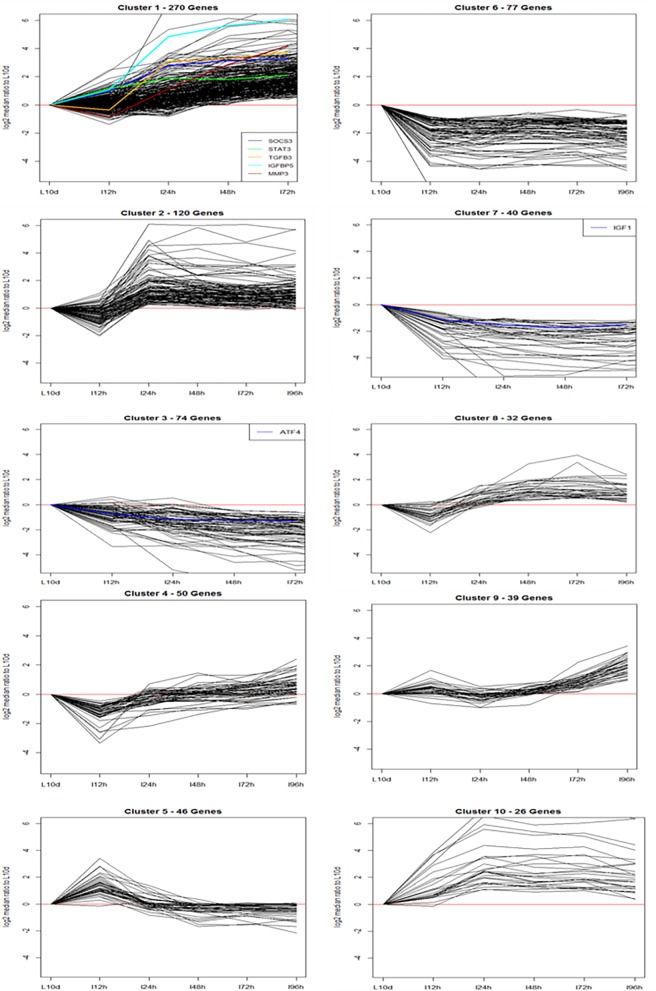
Given the limited overlap between the two datasets, we sought to identify relevant gene clusters over time from the two published studies together. We used the STEM algorithm developed by Ernst et al. [26, 27] to cluster genes identified as differentially expressed between the last day of lactation and the last time point of involution. We used c = 3 and m = 50 for input parameters, where c indicates units of change and m the number of candidate profiles. This run significantly clustered 774 genes out of 1,055 differentially expressed genes (73.4%). Table 2 lists the patterns, size, and p-value of significant clusters out of 50 possible cluster profiles. Patterns indicate the log2 fold change in expression of genes in clusters compared to the lactation day 10 levels. Fig 2 shows the log2 fold change in gene expression profiles for the ten significant clusters.
Table 2. Patterns, size and p-values of significant clusters identified through the STEM algorithm using data from Clarkson et al. [3].
The far right column indicates Involution Specific Gene Signatures through the STEM clustering using data from Clarkson et al. [3].
| Cluster | Pattern* | Size | p-Value | Number of human orthologous genes identified | Signature name |
|---|---|---|---|---|---|
| #1 | 0,1,2,3,4,5 | 270 | 1.30E-234 | 256 | Inv1 |
| #2 | 0,-1,2,1,1,1 | 120 | 1.10E-54 | 118 | Inv2 |
| #3 | 0,-1,-2.-3.-4.-5 | 74 | 3.00E-26 | 69 | Inv3 |
| #4 | 0,-3,-1,-1,0,1 | 50 | 5.10E-11 | 47 | Inv4 |
| #5 | 0,3,0,-3,-3,-2 | 46 | 7.30E-08 | 43 | Inv5 |
| #6 | 0,-3,-4,-2,-3,-3 | 77 | 1.20E-07 | 76 | Inv6 |
| #7 | 0,-3,-4,-5,-6,-3 | 40 | 2.30E-05 | 39 | Inv7 |
| #8 | 0,-2,1,2,4,1 | 32 | 1.10E-04 | 31 | Inv8 |
| #9 | 0,2,1,0,2,5 | 39 | 5.70E-04 | 37 | Inv9 |
| #10 | 0,2,5,3,6,3 | 26 | 7.30E-04 | 26 | Inv10 |
*Pattern indicates the log2 fold change in gene expression levels of lactation day 10 and involution days 0.5, 1, 2, 3 and 4 days compared lactation day 10.
Fig 2. The log2 fold change in gene expression profiles for the ten significant clusters identified through the STEM Clustering.
Y-axis represents the relative gene expression levels of involution days 0.5, 1, 2, 3 and 4 days compared lactation day 10 in log base 2 scale. X-axis represents the time points (L10, lactation day 10; I12h, involution day 0.5; I24h, involution day 1; I48h, involution day 2; I72h, involution day 3; I96h, involution day 4). SOCS3, suppressor of cytokine signaling 3; IGF1, insulin-like growth factor 1 (somatomedin C); STAT3, signal transducer and activator of transcription 3 (acute-phase response factor); TGFB3, transforming growth factor, beta 3; ATF4, activating transcription factor 4; IGFBP5, insulin-like growth factor binding protein 5; MMP3, matrix metallopeptidase 3.
Involution-specific gene signatures
To use the involution-specific gene signatures to conduct gene set analysis on human IBC and non-IBC gene expression profiles, we identified human orthologous genes for genes of significant clusters identified through STEM (S1 Table). Ortholog data were downloaded from the ENSEMBL website and orthologous genes were identified using ENSEMBL gene id. See Table 2 for a list of the number of human orthologous genes identified for each of 10 significant clusters.
Gene set analysis of involution-specific gene signatures
Results of GSEA of involution-specific signatures in IBC versus non-IBC
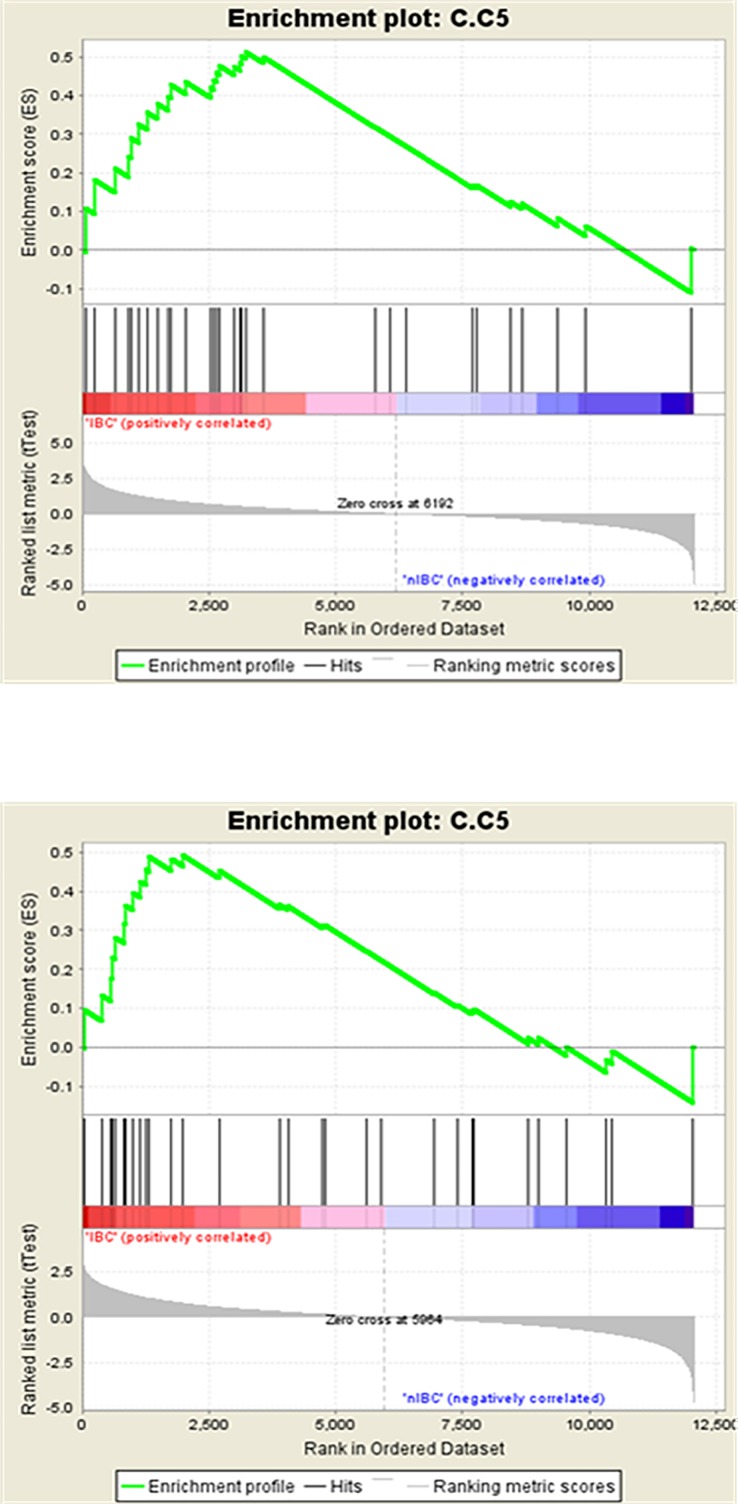
Out of 10 gene signatures developed through the STEM clustering using data from Clarkson et al. [3], 7 signatures have a normalized enrichment score (NES) >1 in the training set of IBC phenotype with 2 signatures, Inv5 and Inv6, having nominal p-values <0.05. In the validation set, we found 9 gene signatures with NES >1 in IBC cases with Inv5 having a nominal p-value of <0.05. Three signatures had NES >1 in non-IBC phenotype with nominal p-values >0.05 and no gene signatures significant at FDR <25% in the training set (Table 3). In the validation set, 1 gene signature had NES >1 in non-IBC phenotype with no gene signature significant at FDR <25% (Table 3). When comparing the results in the training and validation sets, we found that 6 out of 10 gene signatures were enriched in IBC in both training and validation sets. In the merged analysis repeated using the entire data set, we found that 2 out of 10 gene signatures (Inv5 and Inv6) were significantly upregulated in IBC versus non-IBC phenotype at nominal p-value of 0.05 (Table 4). Fig 3 represents the enrichment plot from the GSEA for Inv5 signature in IBC versus non-IBC and Table 5 shows the list of genes in the Inv5 signature as well as genes enriched in IBC. For the involution specific signatures reported by Stein et al. [4], we found that no gene signature was significantly enriched in IBC or non-IBC at FDR <25% or nominal p-value of 0.05 in both the training and validation sets (Table 3) or in the total data set (Table 4).
Table 3. GSEA results of involution-specific signatures in IBC versus non-IBC in the training and validation sets.
| Gene Signature | Results on Training Set | Results on Validation Set | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enriched in IBC vs non-IBC | Size | ES | NES | Nominal p-value | FDR q-value | Enriched in IBC vs non-IBC | Size | ES | NES | Nominal p-value | FDR q-value | |
| Involution specific signatures developed through STEM clustering using data from Clarkson et al. 2004 | ||||||||||||
| Inv1 | IBC | 205 | 0.360 | 1.064 | 0.415 | 0.443 | IBC | 205 | 0.212 | 0.643 | 0.912 | 0.927 |
| Inv2 | Non-IBC | 100 | -0.139 | -0.552 | 0.933 | 0.976 | IBC | 100 | 0.169 | 0.674 | 0.849 | 1.000 |
| Inv3 | Non-IBC | 57 | -0.299 | -0.917 | 0.625 | 1.000 | IBC | 57 | 0.322 | 1.030 | 0.403 | 0.836 |
| Inv4 | IBC | 33 | 0.392 | 1.108 | 0.360 | 0.469 | IBC | 33 | 0.307 | 0.907 | 0.581 | 0.835 |
| Inv5 | IBC | 30 | 0.514 | 1.492 | 0.014 | 0.263 | IBC | 30 | 0.492 | 1.436 | 0.028 | 0.433 |
| Inv6 | IBC | 62 | 0.402 | 1.399 | 0.043 | 0.222 | IBC | 62 | 0.326 | 1.122 | 0.258 | 1.000 |
| Inv7 | IBC | 36 | 0.295 | 0.843 | 0.719 | 0.650 | Non-IBC | 36 | -0.284 | -0.810 | 0.790 | 0.709 |
| Inv8 | Non-IBC | 24 | -0.294 | -0.883 | 0.647 | 0.897 | IBC | 24 | 0.294 | 0.878 | 0.621 | 0.774 |
| Inv9 | IBC | 33 | 0.611 | 1.363 | 0.131 | 0.184 | IBC | 33 | 0.409 | 0.915 | 0.592 | 0.975 |
| Inv10 | IBC | 21 | 0.296 | 0.897 | 0.583 | 0.654 | IBC | 21 | 0.339 | 1.030 | 0.414 | 1.000 |
| Involution specific signatures reported in Stein et al. 2009 | ||||||||||||
| S.C1 | IBC | 182 | 0.42 | 1.16 | 0.274 | 0.818 | IBC | 182 | 0.35 | 0.94 | 0.522 | 1 |
| S.C2 | Non-IBC | 205 | -0.27 | -0.78 | 0.737 | 0.746 | Non-IBC | 205 | -0.2 | -0.63 | 0.945 | 0.962 |
| S.C3 | IBC | 252 | 0.2 | 0.74 | 0.766 | 0.922 | Non-IBC | 252 | -0.22 | -0.81 | 0.695 | 1 |
| S.C4 | IBC | 258 | 0.18 | 0.74 | 0.753 | 0.832 | Non-IBC | 258 | -0.29 | -1.25 | 0.215 | 0.718 |
| S.C5.I3VL7 | IBC | 117 | 0.21 | 0.8 | 0.743 | 0.906 | IBC | 117 | 0.23 | 0.83 | 0.672 | 1 |
| S.C6 | Non-IBC | 100 | -0.25 | -0.94 | 0.534 | 0.905 | Non-IBC | 100 | -0.26 | -1 | 0.438 | 0.715 |
| S.C7 | IBC | 225 | 0.29 | 1.17 | 0.19 | 1 | IBC | 225 | 0.24 | 0.97 | 0.504 | 1 |
| S.C8 | Non-IBC | 153 | -0.25 | -1.01 | 0.413 | 1 | IBC | 153 | 0.29 | 1.17 | 0.171 | 1 |
| S.C9 | Non-IBC | 66 | -0.25 | -0.8 | 0.818 | 0.952 | Non-IBC | 66 | -0.39 | -1.24 | 0.149 | 0.365 |
| S.I1VL7 | IBC | 495 | 0.19 | 0.85 | 0.693 | 1 | Non-IBC | 495 | -0.16 | -0.7 | 0.962 | 1 |
| S.I2VL7 | IBC | 612 | 0.21 | 0.87 | 0.647 | 1 | IBC | 612 | 0.2 | 0.79 | 0.784 | 0.977 |
| S.I3VL7 | IBC | 648 | 0.2 | 0.83 | 0.708 | 0.968 | IBC | 648 | 0.19 | 0.75 | 0.836 | 0.779 |
| S.I4VL7 | IBC | 894 | 0.23 | 0.91 | 0.579 | 1 | IBC | 894 | 0.2 | 0.78 | 0.777 | 0.835 |
ES, enrichment score; NES, normalized enrichment score.
Table 4. GSEA results of involution-specific signatures in IBC versus non-IBC in the merged 3 breast cancer data sets.
| Gene Signature | Merged 3 breast cancer data sets IBC = 137 and non-IBC = 251 # Genes = 12064 |
|||||
|---|---|---|---|---|---|---|
| Enriched in IBC versus non-IBC | Size | ES | NES | Nominal p-value | FDR q-value | |
| Involution specific signatures developed through STEM clustering using data from Clarkson et al. 2004 | ||||||
| Inv1 | IBC | 205 | 0.29 | 0.88 | 0.584 | 0.869 |
| Inv2 | IBC | 100 | 0.13 | 0.52 | 0.97 | 0.988 |
| Inv3 | IBC | 57 | 0.25 | 0.81 | 0.807 | 0.902 |
| Inv4 | IBC | 33 | 0.32 | 0.94 | 0.541 | 0.873 |
| Inv5 | IBC | 30 | 0.51 | 1.47 | 0.03 | 0.392 |
| Inv6 | IBC | 62 | 0.42 | 1.47 | 0.021 | 0.204 |
| Inv7 | IBC | 36 | 0.26 | 0.75 | 0.867 | 0.896 |
| Inv8 | Non-IBC | 24 | -0.3 | -0.92 | 0.573 | 0.541 |
| Inv9 | IBC | 33 | 0.54 | 1.22 | 0.279 | 0.407 |
| Inv10 | IBC | 21 | 0.43 | 1.34 | 0.122 | 0.298 |
| Involution specific signatures reported in Stein et al. 2009 | ||||||
| S.C1 | IBC | 182 | 0.4 | 1.08 | 0.379 | 0.614 |
| S.C2 | Non-IBC | 205 | -0.25 | -0.77 | 0.735 | 0.902 |
| S.C3 | Non-IBC | 252 | -0.19 | -0.72 | 0.783 | 0.849 |
| S.C4 | Non-IBC | 258 | -0.21 | -0.89 | 0.568 | 1 |
| S.C5.I3VL7 | IBC | 117 | 0.29 | 1.1 | 0.315 | 0.818 |
| S.C6 | Non-IBC | 100 | -0.26 | -0.99 | 0.48 | 1 |
| S.C7 | IBC | 225 | 0.28 | 1.13 | 0.26 | 1 |
| S.C8 | Non-IBC | 153 | -0.22 | -0.88 | 0.717 | 0.835 |
| S.C9 | Non-IBC | 66 | -0.37 | -1.21 | 0.189 | 0.847 |
| S.I1VL7 | IBC | 495 | 0.19 | 0.83 | 0.717 | 0.639 |
| S.I2VL7 | IBC | 612 | 0.21 | 0.87 | 0.671 | 0.798 |
| S.I3VL7 | IBC | 648 | 0.21 | 0.84 | 0.694 | 0.729 |
| S.I4VL7 | IBC | 894 | 0.23 | 0.9 | 0.591 | 0.882 |
Fig 3. Enrichment plots form GSEA for Inv5 signature for IBC versus non-IBC in the training and validation sets.
nIBC, non-Inflammatory breast cancer.
Table 5. List of genes in Inv5 signature and genes enriched in IBC versus non-IBC.
| Gene Symbol | Gene Name | Enrichment in IBC (Training Set) | Enrichment in IBC (Validation Set) |
|---|---|---|---|
| CD79B | CD79b molecule, immunoglobulin-associated beta | Yes | Yes |
| IVL | involucrin | Yes | Yes |
| KIF2C | kinesin family member 2C | Yes | Yes |
| NOP2 | nucleolar protein 2 | Yes | Yes |
| LDHB | lactate dehydrogenase B | Yes | Yes |
| LEP | leptin (obesity homolog, mouse) | Yes | Yes |
| AVIL | advillin | Yes | Yes |
| DKK2 | dickkopf homolog 2 (Xenopus laevis) | Yes | Yes |
| ARAP3 | ankyrin repeat and PH domain 3 | Yes | Yes |
| YBX2 | Y box binding protein 2 | Yes | No |
| CEP250 | centrosomal protein 250kDa | Yes | No |
| RPS6KB2 | ribosomal protein S6 kinase, 70kDa, polypeptide 2 | Yes | No |
| STX3 | syntaxin 3 | Yes | No |
| NFKBIE | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | Yes | No |
| GSTO1 | glutathione S-transferase omega 1 | Yes | No |
| WT1 | Wilms tumor 1 | Yes | No |
| DYNC1I1 | dynein, cytoplasmic 1, intermediate chain 1 | Yes | No |
| RET | ret proto-oncogene (multiple endocrine neoplasia and medullary thyroid carcinoma 1, Hirschsprung disease) | Yes | No |
| TIPIN | TIMELESS interacting protein | Yes | No |
| LLGL2 | lethal giant larvae homolog 2 (Drosophila) | No | Yes |
| TG | thyroglobulin | No | Yes |
| DDIT4 | DNA-damage-inducible transcript 4 | No | Yes |
| HPCA | hippocalcin | No | Yes |
| GRAMD3 | GRAM domain containing 3 | No | No |
| PAX4 | paired box gene 4 | No | No |
| KCNJ4 | potassium inwardly-rectifying channel, subfamily J, member 4 | No | No |
| NPY | neuropeptide Y | No | No |
| CHRNA6 | cholinergic receptor, nicotinic, alpha 6 | No | No |
| FRAT2 | frequently rearranged in advanced T-cell lymphomas 2 | No | No |
| ERBB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | No | No |
Results of GSEA of involution-specific signatures in TN subtype versus non-TN subtype in IBC and non-IBC
Out of 10 gene signatures developed through STEM clustering using data from Clarkson et al. [3], we found that no gene signature was significantly enriched in TN subtype versus non-TN subtype in IBC cases at FDR <25% or nominal p-value of 0.05 in both the training and validation sets. For involution specific signatures reported by Stein et al. [4], we also did not find any gene signature that was significantly enriched in TN subtype versus non-TN subtype in IBC cases at FDR <25% or nominal p-value of 0.05 in both the training and validation sets. We found similar results when comparing gene signatures in TN subtype versus non-TN subtype in non-IBC cases.
GSEA results of involution-specific signatures in IBC versus non-IBC normals
Unfortunately there are not additional samples from which to validate the findings beyond the report above. Nevertheless, we speculated that the relevant signatures might be expressed in gene expression data from histopathologically normal breast isolated from IBC (N = 19) and non-IBC (N = 25) mastectomy patients given our hypothesis that the unique normal tissue changes influence the symptoms of IBC.
Out of 10 gene signatures developed through STEM clustering using data from Clarkson et al. [3], we found that one gene signature (Inv1) was significantly enriched in IBC versus non-IBC Normal samples and one gene signature (Inv5) was significantly enriched in non-IBC normal versus IBC samples at FDR <25% (0.202 and 0.249, respectively). For involution-specific signatures reported by Stein et al. [4], no gene signature that was significantly enriched in IBC versus non-IBC normal samples and in non-IBC normal versus IBC samples at FDR <25%.
This again demonstrates enrichment of involution signatures in human breast tissues from women with breast cancer, but the significant enrichment pattern is different in normal versus tumor. Again, this is limited by a small sample size (19 IBC and 25 non-IBC normals). Of the three genes mentioned from Inv5 tumor data as upregulated, only Leptin is upregulated in IBC in normal breast tissue. Of note, Inv1 genes with 2-fold enrichment are the following: CYP4B1, ACADL, PCK1, RASA3, CDO1, ABCA1 (see S4 Table for details). Interestingly, ABCA1, the primary cholesterol transporter in HDL trafficking previously described by our group as important in mediating the beneficial effects of HDL in IBC patients and pre-clinical studies is upregulated in IBC [32].
Ontology analysis for significant clusters
We also conducted ontology analysis on genes of significant clusters identified through the STEM using GO annotations for mus musculus gene products available from the Mouse Genome Informatics to understand biologically relevant processes. We did not find any significant biologic processes by the method for the IBC-enriched involution signature (S2 Table).
Discussion
We reanalyzed the previously published expression profiling data set obtained from mammary glands derived from mice at various stages of post-lactational mammary gland involution (12 gene expression profiles with two hybridizations for each of 10 day lactation time points, and 12, 24, 48, 72 and 96 hour involution time points). We focused on genes that were differentially expressed during time periods spanning from the last day of lactation (day 10) to the fourth day of involution by greater than 2-fold (p <0.05) and performed STEM cluster analysis to discern time-varied expression patterns. We identified 10 time-based gene clusters that represent this time period, Inv1-10. Broadly, many are enriched versus depleted in IBC samples, but only Inv5 is significantly enriched in both the training and validation set based on nominal p-values, and note that FDR adjusted p-values are not significant for this cluster. Nevertheless, given the limitations of the data and the lack of larger datasets or similar extensive gene array studies of the involuting human breast, these hypothesis-generating findings may merit further study.
Up to now, the two most comprehensive studies examining global gene expression on the post-lactational mammary gland have been conducted by Clarkson et al. [3, 5] and Stein et al. [4]. Clarkson et al. [3] used the K means clustering method and Stein et al. used the self-organizing map in [4] and the hierarchical ordered partitioning and collapsing hybrid (HOPACH) method in [9] to find the patterns among differentially expressed genes in the post-lactational involution period. They discovered that apoptotic pathways and immunomodulatory signals are induced during the process of post-lactational involution. During our reanalysis, we used the STEM clustering method on the dataset by Clarkson et al. [3] to find the time-varied patterns among differentially expressed genes in the post-lactational involution period. We identified 10 separate and significant time-varied expression patterns from 774 genes out of 1,055 significantly differentially expressed genes. Gene ontology analyses of these clusters showed that cluster 1, which represented genes showing gradual up-regulation during the first 4 days of involution, had over-representation of numerous biological processes whereas relatively few are noted in other clusters. Representation of fatty acid oxidation and ER and membrane biology in Inv6 may be noteworthy given the nominal significance of Inv6 in analysis of the full data set and relevance of these systems in published IBC studies [28]. The over-representation of biological processes that we found are in general agreement with findings by Clarkson et al. [3], who used a different analytical approach, and with findings by Stein et al. [4, 9], who used a different mouse system and a different analytical approach.
We examined the enrichment of post-lactational mammary gland involution gene expression patterns in TNBC and IBC using the GSEA method. First, we used 10 significant time varied gene expression patterns that we found in the dataset from Clarkson et al. (3). We found that only one gene expression pattern was enriched in IBC compared to non-IBC in both training and validation sets at the nominal p-value. None of these gene expression patterns was enriched in TNBC compared to non-TN BC for both IBC and non-IBC groups in both training and validation sets. Second, we used 13 gene expression patterns on post-lactational involution as reported by Stein et al (9) and found that none of these gene expression patterns was significantly enriched in IBC compared to non-IBC and TN BC compared to non-TN BC in both training and validation sets. To investigate further, we examined the overlap between the involution-specific signatures and the IBC-like signature (79 genes) [28]. We found that there was minimal overlap between the involution-specific signatures and the IBC-like signature, and no gene overlapped between the Inv5 signature and the IBC-like signature (S3 Table).
One gene signature that showed nominal enrichment in IBC compared to non-IBC, Inv5, contained genes that showed initial up-regulation and later down-regulation during the involution process. This might suggest that genes that upregulate during an initial phase of involution after abrupt weaning might not be turning off and, therefore, could be responsible for facilitating an IBC-like phenotype after a tumor-initiating event. We examined the overlap of this gene expression pattern with the existing gene signatures using the Molecular Signatures Database (MSigDB) v4.0 [28]. We found that the genes up-regulated in VSMC by JNK1 [33] showed the most significant overlap (FDR q value = 6.07E-5). Among these overlapped genes, 3 genes–Involucrin (IVL), Cluster of Differentiation 79B (CD79B), and leptin (LEP)–were significantly enriched in IBC compared to non-IBC in both training and validation data sets in our analysis. Involucrin is a transglutaminase substrate protein present in keratinocytes of epidermis and other stratified squamous epithelia [34]. Tsuda et al [35] investigated the expression of Involucrin in breast cancer and found that Involucrin expression was detected in 27% of breast cancer cases and was associated with high-grade atypia, a solid-nest pattern, cancer cell necrosis on histology, and negative ER status. Leptin is a product of the obese (OB) gene, an important regulator of energy balance and necessary for normal mammary gland development [36]. In ER-positive breast cancer cell lines, leptin has been shown to stimulate cell growth through activation of multiple signaling pathways including the Janus Kinase/Signal Transducer and Activator of Transcription (JAK/STAT) pathway [37]. Thus, our results along with the published functions of the above-mentioned genes indicate that they play a role within the tumor microenvironment and may merit functional study of their role in promoting IBC development and progression.
A major strength of our study is the use of the largest series of IBC samples ever reported by the World IBC Consortium. Furthermore, this work is novel in part due to the use of updated Chip Definition Files from the BrainArray [25] during preprocessing of gene expression data for accurate probe mapping to the genome. Also, we used the GSEA [31] to examine enrichment of involution signatures in IBC and TNBC phenotype. The GSEA gives more statistical power to detect smaller changes in genes of a gene set compared to other methods of enrichment analysis.
Major limitations of our study include the cross-sectional analysis of enrichment of involution specific signatures in breast cancer and array-based measurement of gene expression profiles, which limit the detection of differentially expressed genes with lower levels of expression. Also, although having the parity status of the patients within the World IBC consortium would be valuable to our study, that information is not recorded or available to us and, as such, we cannot determine or comment on whether parity-related effects persist or were present prior to the time of analysis. Further, there are surprisingly few TNBC in the cohort given the established over-representation of these subtypes in IBC which may influence the findings overall and regarding TNBC versus non-TNBC. Additionally, the studies by Clarkson et al (3) and Stein et al (4) did not include a corresponding group that underwent non-abrupt involution or that was not force weaned, as the authors did not distinguish between abrupt involution versus the normal involution process. Thus, the correlation between limited nursing and abrupt involution signatures remains unstudied here. Although inclusion of such a control group in these studies would be useful to our analysis, we can speculate that Inv5 and IVL, CD79B, and LEP in particular may play in role in IBC development after abrupt or involution. Given the limitations of this study, however, additional research is warranted before a concrete conclusion can be made.
In conclusion, our results provide some evidence that molecular events after abrupt involution are identifiable in IBC patient tissues from the uninvolved breast and tumor; however, they are hypothesis-generating given the potential for false discovery after multiple comparisons as well as the other noted limitations of our study. Whether or not Inv5 or Inv6 related genes or signaling are upregulated in the normal tissues around IBC tumors, and if breast-feeding or abrupt cessation of breast-feeding contributes to the persistence of related genes in the normal breast will be investigated in future studies.
Supporting information
(DOCX)
BP, biological process; CC; cellular components.
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Supported by the National Institutes of Health Grants RO1CA180061 and RO1CA138239 to Wendy Ann Woodward, the State of Texas Grant for Rare and Aggressive Breast Cancer Research Program to Wendy Ann Woodward, and the National Center for Clinical and Translational Science Grant TL1-TR000369 to Wendy Ann Woodward. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Weber A. Fundamentals of the Histology of Domestic Animals. Alfred Trautmann and Josef Fiebiger; trans. and rev. from the 8th and 9th German eds. of 1949 by Robert E. Havel and Ernst L. Biberstein. Ithaca, NY.: Comstock Pub., Cornell Univ. Press, 1952. 426 pp. Ill. Science (80-). 1952;116(3024):671–671. [Google Scholar]
- 2.Macias H, Hinck L. Mammary gland development Wiley Interdisciplinary Reviews: Developmental Biology; 2012. p. 533–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarkson RWE, Wayland MT, Lee J, Freeman T, Watson CJ. Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression. Breast Cancer Res [Internet]. 2004;6(2):R92–109. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=400653&tool=pmcentrez&rendertype=abstract doi: 10.1186/bcr754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein T, Morris JS, Davies CR, Weber-Hall SJ, Duffy M-A, Heath VJ, et al. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res. 2004;6:R75–91. doi: 10.1186/bcr753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarkson RWE, Watson CJ. Microarray analysis of the involution switch. J Mammary Gland Biol Neoplasia. 2003;8(3):309–19. [DOI] [PubMed] [Google Scholar]
- 6.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–45. doi: 10.1016/S0140-6736(00)04046-0 [DOI] [PubMed] [Google Scholar]
- 7.McDaniel SM, Rumer KK, Biroc SL, Metz RP, Singh M, Porter W, et al. Remodeling of the mammary microenvironment after lactation promotes breast tumor cell metastasis. Am J Pathol [Internet]. 2006;168(2):608–20. Available from: http://www.scopus.com/inward/record.url?eid=2-s2.0-33144465789&partnerID=tZOtx3y1 doi: 10.2353/ajpath.2006.050677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard A, Shiu R, Soresnsen G, DeCorby N, Nistor A, Wong P, et al. Gene expression profiling of early involuting mammary gland reveals novel genes potentially relevant to human breast cancer. Front Biosci. 2007;12(1):2221–32. [DOI] [PubMed] [Google Scholar]
- 9.Stein T, Salomonis N, Nuyten DSA, Vijver MJ, Gusterson BA. A mouse mammary gland involution mRNA signature identifies biological pathways potentially associated with breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2009;14(2):99–116. doi: 10.1007/s10911-009-9120-1 [DOI] [PubMed] [Google Scholar]
- 10.Lyons TRTR, O’Brien J, Borges VFVF, Conklin MWMW, Keely PJPJ, Eliceiri KWKW, et al. Postpartum mammary gland involution drives progression of ductal carcinoma in situ through collagen and COX-2. Nat Med [Internet]. 2011;17(9):1109–15. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3888478&tool=pmcentrez&rendertype=abstract doi: 10.1038/nm.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479(7372):189–93. doi: 10.1038/nature10573 [DOI] [PubMed] [Google Scholar]
- 12.Asselin-Labat M-L, Vaillant F, Sheridan JM, Pal B, Wu D, Simpson ER, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465(7299):798–802. doi: 10.1038/nature09027 [DOI] [PubMed] [Google Scholar]
- 13.American Cancer Society. Cancer Facts & Figures 2017. 2017; Available from: https://old.cancer.org/acs/groups/content/@editorial/documents/document/acspc-048738.pdf
- 14.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15):4429–34. [DOI] [PubMed] [Google Scholar]
- 15.Woodward WA, Cristofanilli M. Inflammatory Breast Cancer. Seminars in Radiation Oncology. 2009. p. 256–65. doi: 10.1016/j.semradonc.2009.05.008 [DOI] [PubMed] [Google Scholar]
- 16.Anderson WF, Schairer C, Chen BE, Hance KW, Levine PH. Epidemiology of inflammatory breast cancer (IBC). Breast Dis [Internet]. 2010;22:9–23. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2852616/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hance KW, Anderson WF, Devesa SS, Young HA, Levine PH. Trends in inflammatory breast carcinoma incidence and survival: the surveillance, epidemiology, and end results program at the National Cancer Institute. J Natl Cancer Inst. 2005;97(13):966–75. doi: 10.1093/jnci/dji172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudet MM, Press MF, Haile RW, Lynch CF, Glaser SL, Schildkraut J, et al. Risk factors by molecular subtypes of breast cancer across a population-based study of women 56 years or younger. Breast Cancer Res Treat. 2011;130(2):587–97. doi: 10.1007/s10549-011-1616-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkinson RL, El-zein R, Valero V, Lucci A, Bevers TB, Fouad T, et al. Epidemiological risk factors associated with inflammatory breast cancer subtypes. Cancer Causes Control. 2016;27(3):359–66. doi: 10.1007/s10552-015-0712-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner K-U, Smith GH. Pregnancy and stem cell behavior. J Mammary Gland Biol Neoplasia. 2005;10(1):25–36. doi: 10.1007/s10911-005-2538-1 [DOI] [PubMed] [Google Scholar]
- 21.Wagner K-U, Boulanger C a, Henry MD, Sgagias M, Hennighausen L, Smith GH. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development [Internet]. 2002;129(6):1377–86. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11880347 [DOI] [PubMed] [Google Scholar]
- 22.Henry MD, Triplett A a, Oh KB, Smith GH, Wagner K-U. Parity-induced mammary epithelial cells facilitate tumorigenesis in MMTV-neu transgenic mice. Oncogene. 2004;23(41):6980–5. doi: 10.1038/sj.onc.1207827 [DOI] [PubMed] [Google Scholar]
- 23.Wu C, Irizarry R. gcrma: Background Adjustment Using Sequence Information. 2005. p. 2.2.0.
- 24.Sandberg R, Larsson O. Improved precision and accuracy for microarrays using updated probe set definitions. BMC Bioinformatics [Internet]. 2007;8(1):48 Available from: http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-8-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smyth G. Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions using R and Bioconductor. 2005. p. 397–420. [Google Scholar]
- 26.Ernst J, Nau G, Bar-Joseph Z. Clustering short time series gene expression data. Bioinformatics. 2005; [DOI] [PubMed] [Google Scholar]
- 27.Ernst J, Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics [Internet]. 2006;7(1):0 Available from: citeulike-article-id%5Cn984928%5Cnhttp%5Cn//dx.doi.org/10.1186/1471-2105-7-191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Laere SJ, Ueno NT, Finetti P, Vermeulen P, Lucci A, Robertson FM, et al. Uncovering the molecular secrets of inflammatory breast cancer biology: An integrated analysis of three distinct affymetrix gene expression datasets. Clin Cancer Res. 2013;19(17):4685–96. doi: 10.1158/1078-0432.CCR-12-2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atkinson RL, Yang WT, Rosen DG, Landis MD, Wong H, Lewis MT, et al. Cancer stem cell markers are enriched in normal tissue adjacent to triple negative breast cancer and inversely correlated with DNA repair deficiency. Breast Cancer Res [Internet]. 2013;15(5):R77 Available from: http://www.ncbi.nlm.nih.gov/pubmed/24008095%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4053576 doi: 10.1186/bcr3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci [Internet]. 2001;98(1):31–6. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.98.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette M a, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A [Internet]. 2005;102(43):15545–50. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16199517 doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfe AR, Bambhroliya A, Reddy JP, Debeb BG, Huo L, Larson R, et al. MiR-33a decreases high-density lipoprotein-induced radiation sensitivity in breast cancer. Int J Radiat Oncol Biol Phys. 2016;95(2):791–9. doi: 10.1016/j.ijrobp.2016.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshimura K, Aoki H, Ikeda Y, Fujii K, Akiyama N, Furutani A, et al. Regression of abdominal aortic aneurysm by inhibition of c-Jun N-terminal kinase. Nat Med. 2005;11(12):1330–8. doi: 10.1038/nm1335 [DOI] [PubMed] [Google Scholar]
- 34.Djian P, Phillips M, Easley K, Huang E, Simon M, Rice RH, et al. The involucrin genes of the mouse and the rat: study of their shared repeats. Mol Biol Evol [Internet]. 1993;10(6):1136–49. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8277848 doi: 10.1093/oxfordjournals.molbev.a040069 [DOI] [PubMed] [Google Scholar]
- 35.Tsuda H, Sakamuri C, Fukutomi T, Hirohashi S. Squamoid features and expression of involucrin in primary breast carcinoma associated with high histological grade, tumour cell necrosis and recurrence sites. Br J Cancer. 1997;75(10):1519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garofalo C, Surmacz E. Leptin and cancer. Journal of Cellular Physiology. 2006. p. 12–22. [DOI] [PubMed] [Google Scholar]
- 37.Yin N, Wang D, Zhang H, Yi X, Sun X, Shi B. Molecular mechanisms involved in the growth stimulation of breast cancer cells by leptin. Cancer Res [Internet]. 2004;64(16):5870–5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15313931%5Cnhttp://cancerres.aacrjournals.org/content/64/16/5870.short doi: 10.1158/0008-5472.CAN-04-0655 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
BP, biological process; CC; cellular components.
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.