Abstract
Subtilisin-like proteins represent an ancient family of serine proteases that are extremely widespread in living organisms. We report here the structure and genomic organization of two new transcriptionally active genes encoding proteins that belong to the P69 family of subtilisin-like proteases from tomato (Lycopersicon esculentum) plants. The two new members, P69E and P69F, are organized in a cluster and arranged in a tandem form. mRNA expression analysis and studies of transgenic Arabidopsis plants transformed with promoter-β-glucuronidase fusions for each of these two genes revealed that they are differentially regulated, with each showing a highly specific mRNA expression pattern. P69E mRNA is expressed only in roots, while P69F mRNA is expressed only in hydathodes. A comparison of all the P69 amino acid sequences, gene structure, expression profiles, and clustered organization suggests a working model for P69 gene family evolution.
Ser proteinases are of extremely widespread occurrence. One of the largest families of this type of enzymes is that represented by the subtilisin-like (subtilase) family (EC 3.4.21.14). This family represents an ancient family of proteins with homologs in such diverse organisms as Archae, bacteria, fungi, yeast, and higher eukaryotes including plants. The subtilisin-like Ser proteases are distinguished by the characteristic arrangement of the catalytic His, Asp, and Ser residues that conform the catalytic triad (Siezen and Leunissen, 1997). This active site signature has been used to classify these enzymes into five families: subtilisin, thermitase, kexin, pyrolysin, proteinase K, and lantibiotic peptidases (Siezen and Leunissen, 1997).
Although more than 200 subtilisin-like enzymes are presently known, our information on the existence and role of this type of protease in plants is still scant. So far, subtilisin-like proteases have been identified and the genes cloned in only a few plant species, including Arabidopsis (Ribeiro et al., 1995), cucumber (Cucumis sativus) (Yamagata et al., 1994), Alnus glutinosa (Ribeiro et al., 1995), lily (Lilium longiflorum) (Taylor et al., 1997), and tomato (Lycopersicon esculentum) (Riggs and Horsch, 1995; Tornero et al., 1996, 1997; Meichtry et al., 1999). The plant proteinases can be grouped within the pyrolysin family (Siezen and Leunissen, 1997). In tomato, recent sequence comparison revealed that the subtilase genes fall into five distinct subfamilies (Meichtry et al., 1999), with the P69 subfamily members the best characterized so far. The P69 subtilisin-like proteases are represented by different protein isoforms of approximately 69 kD (P69) that accumulate extracellularly (Tornero et al., 1996, 1997, and refs. therein).
The P69 family members correspond to a multigene family of high complexity (Tornero et al., 1997). Recently, a genomic cluster comprising a tandem array of four closely related P69 subtilin-like proteases (named as P69A, P69B, P69C, and P69D) was identified in tomato plants (Jordá et al., 1999). Detailed analysis of each of these genes revealed that they are tightly regulated by developmental and environmental signals and in a tissue-specific manner (Jordá et al., 1999). The P69A gene was shown to be constitutively expressed in all vegetative organs in the aerial part of the plant except flowers. Conversely, P69D is expressed in flowers and in leaves. However, P69D is under strict transcriptional regulation in young, rapidly expanding leaves. Once the leaf is fully expanded transcription of the P69D gene declines and expression is no longer detected. This suggests that there is a transitory developmental “switch” regulating the coexistence of P69D and P69A activities in the developing leaf. Since the P69-like enzymes are located in the intercellular spaces (Tornero et al., 1996), we suggest that this type of proteinase may play a critical role in the remodeling of the extracellular matrix during rapid cell growth and tissue expansion. Proteinases may be involved in this process which requires the partial separation of cells following cell wall breakdown (Dale, 1988; McQueen-Mason and Cosgrove, 1995).
In contrast to the expression pattern of P69A and P69D, the P69B and P69C genes do not appear to be constitutively expressed at any stage of normal plant development. Instead, they are coordinately and systemically induced de novo by salicylic acid treatment or following infection with the pathogen Pseudomonas syringae (Jordá et al., 1999). This mechanism of gene regulation suggests that both, P69B and P69C, may play roles as active defense weapons against the attacking pathogen. Alternatively, they may take part in the remodeling or reprogramming processes of the extracellular matrix (including the cell wall) that are characteristic of pathogen-afflicted plants (Dixon and Lamb, 1990).
To gain further understanding on the role and complexity of this gene family in tomato, we have characterized two new genes encoding novel members of the P69 family (named as P69E [accession no. Y18931] and P69F [accession no. Y18932]). The two genes are clustered in tandem in the genome and show different expression patterns when analyzed in transgenic Arabidopsis plants containing each of the 5′promoter regions fused to the GUS reporter gene. An evolutionary relationship based on sequence comparison is also presented for these plant proteases.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Treatments
Tomato (Lycopersicon esculentum cv Rutgers) and Arabidopsis (Col-0) plants were grown at 22°C in growth chambers programmed for a 14-h light and 10-h dark cycle. Fully expanded leaves or rosette leaves were either sprayed with salicylic acid (SA) (0.5 mm) or buffer alone (50 mm phosphate buffer, pH 7.2) as described before (Jordá et al., 1999). Leaves were also inoculated with Pseudomonas syringae DC3000 (Avr Rpm1) and samples were analyzed 24 to 48 h post-inoculation as described (Jordá et al., 1999).
Library Screening and DNA Sequence Analysis
A tomato genomic DNA library constructed in λ-EMBL3 was screened as described previously (Jordá et al., 1999) with a radiolabeled p26 cDNA encoding the entire P69A preproprotein (Tornero et al., 1996). The positive clones were isolated and characterized as described (Maniatis et al., 1982). Multiple alignments of the amino acid sequences of the P69-like enzymes and related subtilases were created with the CLUSTAL-X program (Thompson et al., 1997) or alternatively with the GCG9.1 Pileup program (Wisconsin Package, version 9.1, Genetics Computer Group, Madison, WI).
Reverse Transcriptase (RT)-PCR
cDNA synthesis, quantification of the products, and reverse transcriptase-mediated PCR were conducted as described (Jordá et al., 1999). The oligonucleotide primer pairs (50 pmol each), e1 + e2 (TATTTCTTTCTTTAGTAC + ATCCATGGCAGCTAA) and f1 + f2 (ACTCCTCAGACATAC + GTTCGAGTACTTTATGCAC), specific for the amplification of P69E and P69F sequences, respectively, were used to amplify by PCR the in vitro synthesized single-stranded cDNA from the different mRNA sources in a DNA Cycler (Perkin-Elmer/Cetus, Foster City, CA). PCR amplification was programmed as described before (Jordá et al., 1999). The amplified DNA fragments were visualized in agarose gels or, alternatively, they were hybridized with a radiolabeled DNA probe for either P69E or P69F open reading frames (ORFs). The inability of each combination of primers to amplify the closely related P69 sequences of the different family members was confirmed in control PCR reactions that included 10 ng of plasmid DNA containing each of the six P69 ORFs (P69A, B, C, D, E, and F) as template.
Promoter-GUS Fusion, Plant Transformation, and Analysis of Transgenic Plants
Oligonucleotides 5′pEH (5′-AAAAGCTTTGCG ACTATTATCGCCGCTTT-3′)/3′pEB (5′-GGGATCCAGTACTAAAGAAAGAAATATT-3′) and 5′promF (5′-TAGAAAGCTTGTGATGATGACTTCCAG-3′)/3′promF (5′-GCG GATCC- AATTTTACTACTAAAGAAAGAG-3′), served as primers for the incorporation of a synthetic BamHI and HindIII restriction sites in each promoter by site-directed mutagenesis (Kunkel et al., 1987). These primers introduced the BamHI site at positions −1 relative to the translation initiation sites in each gene. BamHI-HindIII fragments encompassing 1.3 and 2 kb of the promoter regions of P69E and p69F, respectively, were cloned upstream of the β-glucuronidase (GUS) gene in pBI101.1 (Jefferson, 1987) to generate plasmids pP69E::GUS and pP69F::GUS. The resulting transcriptional fusions were verified by nucleotide sequence analysis using specific primers. The constructs were introduced into Arabidopsis (Col-0) by Agrobacterium tumefaciens mediated transformation (Bechtold et al., 1993). Transformants were selected on MS agar medium containing kanamycin, transferred to soil, and allowed to self pollinate. The transgenic lines were assayed for GUS activity by a fluorimetric assay or by an in situ assay using the colorigenic substrate X-gluc (Jefferson, 1987).
RESULTS
Characterization of a Genomic Cluster Containing Two New Subtilisin-Like P69 Protease Genes from Tomato Plants
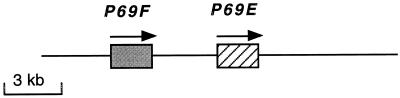
A DNA fragment encoding the complete sequence for a previously identified P69A subtilisin-like protease was obtained from plasmid p26 (Tornero et al., 1996) and used as a radiolabeled probe to screen a tomato genomic library constructed in λ-EMBL3. One clone (named as λ-D), differing from those previously identified in a similar screen (Jordá et al., 1999), was isolated and subjected to restriction analysis and sequencing. These analyses revealed that the genomic DNA insert of λ-D (approximately 25 kb of genomic DNA) contains two intronless transcription units in tandem (Fig. 1) which were highly similar to members of the P69 subtilisin-like protease family (P69A/B/C/D) recently identified (Jordá et al., 1999) (Figs. 2 and 3B). The two new genes present in this cluster were designated as P69E and P69F.
Figure 1.
P69E and P69F genomic cluster. The two P69-like ORF sequences (boxes) are arranged in tandem. Arrows indicate the direction of transcription. The distances are only approximate.
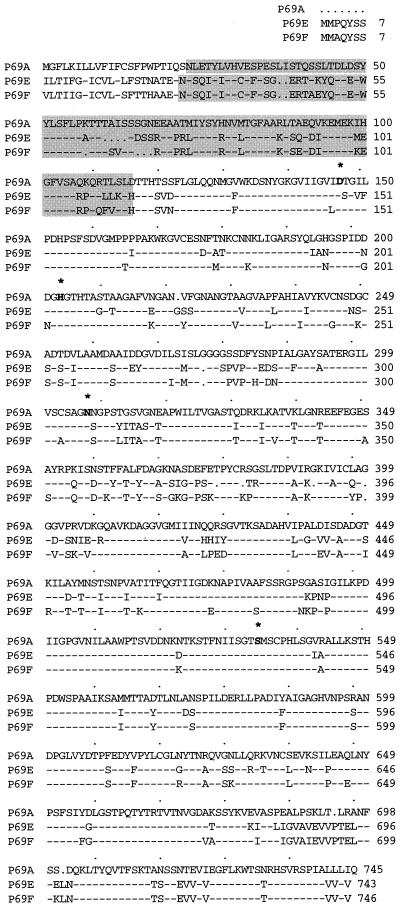
Figure 2.
Amino acid sequence alignment for the predicted P69E and P69F gene products. The sequence of P69A is shown in full and compared with the predicted ORFs of P69E and P69F. Dashes represent sequence identity. The catalytically important Asp, His, Asn, and Ser residues are shown in bold with an asterisk. Dots were introduced to maximize alignment. Amino acid residues of each protease are numbered from the precursor sequence. The propeptide domains are shaded.
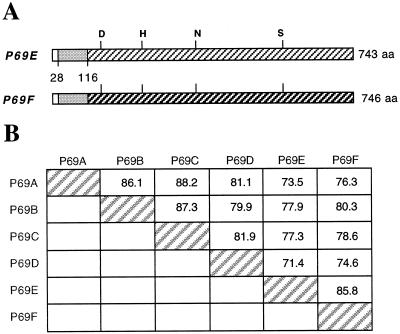
Figure 3.
Schematic representation of P69E and 69F preproenzyme structures and amino acid sequence homology to other P69-like proteases. A, The signal peptide, the propeptide, and the mature peptide region are shown by areas marked in white, gray, and cross-hatching, respectively. Numbers indicate positions of amino acid residues from the N terminus. The amino acids forming the catalytic triad in the active site (D, Asp; H, His; S, Ser) and the conserved N (Asn) residue are marked. B, Identity percentages between the different P69 proteases from tomato plants.
Sequence comparison of P69E and P69F ORFs with that of other P69 members revealed a high degree of similarity (74%–80% identical) (Fig. 3B), however the highest degree of homology was between the P69E and P69F themselves (85.8% identical). Conversely, comparison of the 5′ promoter regions (preceding the ATG initiation codon) or the 3′ region after the polyadenylation signal of each gene revealed no homology between them (not shown). In the two newly identified genes, putative TATA boxes and CAAT boxes located shortly upstream of the ATG initiation codon were observed (data not shown).
Comparison of the P69E and P69F sequences at the amino termini, to the constitutively expressed P69A isoform (Fig. 2), indicated that closely related preprosequences were present in P69E and P69F. However these sequences were clearly distinct from those in the P69A homolog. Both the prosequences of P69E and P69F contain a hydrophobic signal peptide at the extreme N terminus which, accordingly to von Heijne (Von Heijne, 1986), is cleaved C-terminal of the conserved Gln-28 residue. In both cases, the signal peptide is followed by an 87-amino acid prosequence which is a typical feature of zymogens and its cleavage is mandatory for the generation of the active protease from the inactive zymogen (Zhou et al., 1995). The putative N-terminal amino acid of the mature P69E and P69F proteins is the conserved Thr-115, identified also by comparison with other subtilisin-like proteases from plants, including P69A used here as reference (Fig. 2). As deduced from the nucleotide sequences of the ORFs, the P69E and P69F genes encode preproproteins of 743 amino acids (79.1 kD, pI 5.41) and 746 amino acids (79.06 kD; pI 6.73), respectively (Figs. 2 and 3A). The predicted mature enzymes thus contain 628 and 631 amino acids for the P69E and P69D isoforms, respectively. Within the mature proteins the amino acid residues Asp-147, His-204, and Ser-529 (or Ser-532 for the P69F), corresponding to the catalytic site (catalytic triad) essential for the enzymatic activity of subtilisin-like members to function as proteases, were identified (Fig. 2). Also the new P69E and P69F proteases have an Asn residue (Asn-308) that has been found to be highly conserved in this position and that is catalytically important in this type of Ser proteases (Barr, 1991; Steiner et al., 1992). Additionally, none of the P69 isoforms contain the Asp-Asp-Gly conserved motif present at the active site Asp residue of the closely related kexin family members; instead they possess the Asp-Ser/Thr-Gly motif at this position. Thus it is unlikely that any of the P69-like members identified so far have a dibasic cleavage specificity, indicating that they do not belong to the kexin family (Barr, 1991; Steiner et al., 1992) of prohormone-processing proteases.
As was the case for the P69A, B, C, and D isoforms previously identified, sequences close to Asn-308 are also highly variable within the P69E and P69F isoforms (Fig. 2). In these two cases there is also an insertion of a long sequence (222 amino acids [in P69E] or 225 amino acids [P69F]) between the stabilizing Asn-308 and the reactive Ser-529 relative to all other subtilisin-like proteases (Siezen and Leunissen, 1997), in which these two residues are separated by much shorter distances. This displacement has been observed in all the subtilisin-like proteinases recently identified from plants and could represent a characteristic signature of this type of enzyme.
Expression Analysis of P69E and P69F Genes
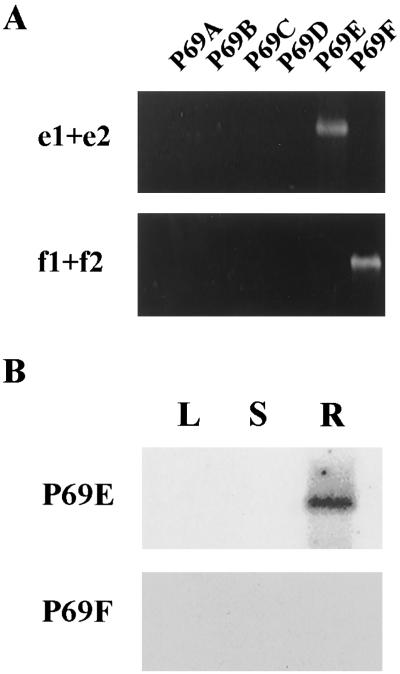
The study of the mRNA expression pattern of the P69E and P69F genes was initially attempted by gene-specific RT-PCR reactions. This technique was used to determine the expression of the P69A/B/C/D family members (Jordá et al., 1999). However, while primer specificity was demonstrated in pilot experiments using each of the individually cloned genes as template in the PCR reaction (Fig. 4A), we could not detect specific RT-PCR products when using the same set of primers specific for the P69E and P69F genes in mRNA preparations obtained from leaf tissues (not shown). Conversely, when these primers were used for RT-PCR reactions with mRNA preparations coming from tissues other than leaf (e.g. stem and roots), a specific amplified DNA product was obtained for P69E in mRNA samples from root tissues (Fig. 4B), thus indicating that this isoform was specifically expressed in root. No amplified product was obtained in these experiments when primers for P69F were used (Fig. 4B), suggesting that this latter gene was very poorly expressed, or not expressed, in the tissues analyzed. Neither P69E norP69F was found to be induced by the pathogen that induces transcription of the P69B and P69C isoforms (Jordá et al., 1999; data not shown).
Figure 4.
RT-PCR detection of P69E and P69F gene expression. A, PCR products derived from amplification of plasmids containing either the P69A, P69B, P69C, P69D, P69E, and P69F ORFs with the set of primers e1 + e2 (specific for the P69E gene) and f1 + f2 (specific for the P69F gene) is shown for comparison. B, Southern blot of DNA products derived from the PCR amplification of reversed transcribed mRNA from leaf (L), stem (S), and root (R) tissue from tomato plants using the same specific sets of primers shown in A. The blots were hybridized with a radiolabeled DNA probe for each gene.
To investigate in more detail the expression pattern of P69E, and ascertain whether or not the P69F gene is expressed in the plant, each of 5′ flanking promoter regions was fused to the GUS reporter gene to generate constructs pP69E::GUS and pP69F::GUS (see “Materials and Methods”). These constructs were introduced separately into Arabidopsis plants by transformation with A. tumefaciens, and five independent transgenic lines were selected for each construct.
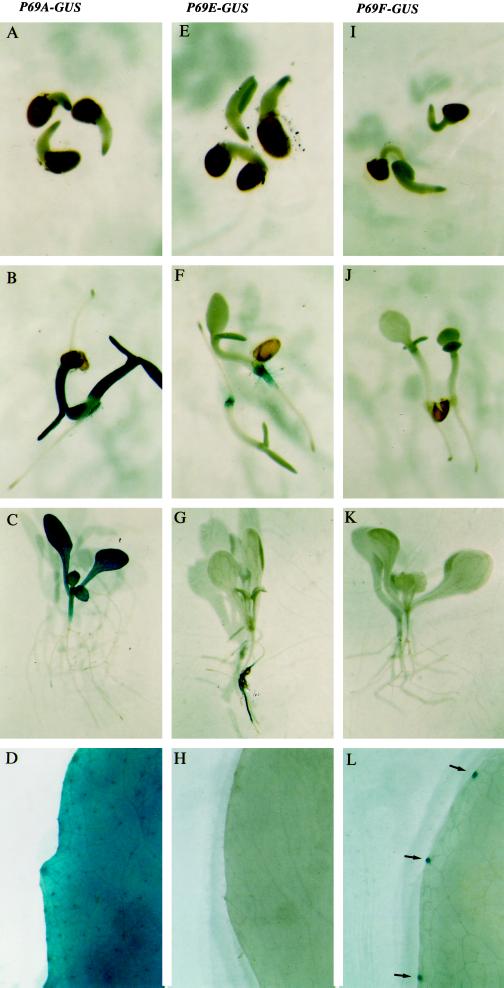
To study the distribution of GUS activity in planta the transgenic lines were analyzed in situ using the chromogenic substrate X-Gluc and compared with the expression pattern of the constitutively expressed P69A::GUS gene (Fig. 5). Expression of GUS activity driven by the P69E promoter was detected in the seedlings as well as in fully grown plants and was limited to root tissues (Fig. 5G), with no expression at all in any aerial tissue of the plant (Fig. 5H). As deduced from the tissue staining pattern in the different P69E::GUS transgenic lines, the P69E gene is transcribed at post-embryonary phases of plant growth, and this root-specific expression pattern maintained during subsequent phases of plant growth and maturation (Fig. 5, E–G). This expression pattern was complementary to that observed for the P69A which is expressed also post-embryonarly but only in the aerial parts of the plant gene (Fig. 5, A–D).
Figure 5.
Comparative GUS staining patterns of transgenic Arabidopsis plants harboring either P69A::GUS, P69E::GUS, or P69E::GUS gene fusions. A, E, and I, Four-day-old GUS-stained intact seedlings. B, F, and J, Ten-day-old GUS-stained intact seedlings. C, G, and K, Fifteen-day-old GUS-stained seedlings. D, H, and L, GUS staining of intact fully expanded leaves from 25-d-old plants. A, B, C, and D correspond to P69A::GUS transgenic plants. E, F, G, and H correspond to P69E::GUS transgenic plants. I, J, K, and L correspond to P69F::GUS transgenic plants. The arrows indicate the positions of hydathodes in a leaf sector of P69F::GUS transgenic plants.
Conversely, transgenic plants in which GUS expression was driven by the P69F promoter revealed that this gene is not transcribed in any part of the plant (Fig. 5, I–K) except in a discrete set of cell clusters located in the margins of full expanded leaves (Fig. 5L) that conform the hydathodes, which are tissue structures organized in the form of water pores located at leaf margins and that exude copious quantities of fluid, which collects around leaf margins as guttation drops (Essau, 1965).
Previously, we demonstrated that both P69B and P69C, but not the P69A and theP69D genes (Jordá et al., 1999), were transcriptionally activated following infection of the plant with the bacteria P. syringae as well as after treatment with salicylic acid (SA), a master regulatory molecule mediating most of the plant defense responses to challenging pathogens (Enyedi et al., 1992). This suggested that P69B and P69C participate in the defense response of the plant. To extend our understanding of P69E and P69F expression pattern, the transgenic plants containing the corresponding promoter-GUS constructs were infected with P. syringae or treated with SA. The extent of gene induction was determined and compared with the induction of the P69C gene. These studies revealed that neither P69E nor P69F is induced by the pathogen or SA (data not shown).
The Relatedness of P69 Amino Acid Sequences
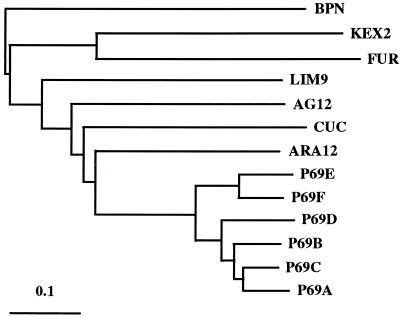
The amino acid sequences of the P69-like proteases and other plant related enzymes were compared using a computer-based “phylogenetic” analysis program, and these were also compared with other canonical subtilases (e.g. kex2 from yeast, human furin, and bacterial BPN) to generate a P69-like family tree (Fig. 6). These studies indicated that the closest relative of the tomato P69s is that of the Arabidopsis Ara12 protease. More interestingly, this amino acid sequence-based analysis grouped the P69s in a manner consistent with our observations regarding the structure and organization of the two P69 gene clusters, and suggests a working model for P69 evolution (see “Discussion”).
Figure 6.
P69 family tree. The amino acid alignment program CLUSTAL-X was used to create a dendogram for subtilisin-like sequences. Available related sequences from plants encoding subtilisin-like proteases (the ARA12 protein from Arabidopsis [Ribeiro et al., 1995], the cucumisin [CUC] protein from C. sativus [Yamagata et al., 1994], the AG12 protein from A. glutinosa [Ribeiro et al., 1995], the LIM9 protein from L. longiflorum [Taylor et al., 1997] as well as the six members of the P69 family from tomato plants [Jordá et al., 1999]) have been included for comparison. The human furin, the yeast Kex2, and the bacteria BPN protease sequences are shown in the same dendogram for evolutionary references.
DISCUSSION
This report describes the characterization of a genomic locus from tomato plants that contains two newly identified members of the subtilisin-like protease family (EC 3.4.21.14) based on amino acid sequence conservation and structural organization (Siezen and Leunissen, 1997). We have named these two new members as P69E and P69F. This finding, together with the previously identified genomic cluster comprising the four other related P69 proteases (P69A, P69B, P69C, and P69D) (Jordá et al., 1999) makes a total of six different transcriptionally active members of this type of protease in the tomato genome.
The predicted primary structure of the two new P69 proteases indicates that they are synthesized as pre-pro-enzymes with three distinct domains: a 28-amino acid signal peptide, an 87-amino acid propolypeptide, and a mature polypeptide of 628 and 631 amino acids for the P69E and P69F, respectively. Within the mature polypeptides, the amino acid sequences surrounding Asp-147, His-204, and Ser-529 (or Ser-532 in the P69F isoform) are the most salient features of these two proteases. These three amino acids constitute the catalytic triad of subtilisins (Siezen and Leunissen, 1997). P69E and P69F possess the conserved Asn (Asn-308) at the position of the oxyanion hole residue (Siezen and Leunissen, 1997). As is the case for other plant subtilisin-like proteases, these two new members also have a long conserved replacement between the conserved Asn-308 residue and the reactive Ser residues of the catalytic triad. The meaning of such a conserved displacement, which is only observed in plant subtilases, remains unknown.
Studies of the mode of gene expression reveal that these two new subtilisin-like protease genes have distinct expression profiles. P69E is transcribed only in root tissues soon after the process of plant germination, and this root-specific expression pattern is maintained at all subsequent stages of plant growth. Conversely, P69F is specifically transcribed in hydathodes. Neither P69E nor P69F gene expression is induced over basal levels during pathogenesis, thus favoring the interpretation that these two new proteases might be involved in basic metabolic functions rather than participating in the response of the plant to challenging pathogens. However, since the plant root is in permanent confrontation with soil-borne pathogens, and the hydathode is a port of entry for pathogens (e.g. Xanthomonas campestry, the agent of black rot) (Cook et al., 1952), we cannot rule out that these two new members are implicated in the defense response of the plant by acting as an early line of defense. It has been observed that other genes whose products may have a role in defense are expressed in hydathodes (Samac and Shah, 1991). The antipathogenic role of the proteinase encoded by P69F could be complemented with the later deployment of the P69B and P69C isoforms which are dramatically induced de novo in the infected plant (Jordá et al., 1999).
The dendogram in Figure 6 suggests that the genes encoding the different P69 isoforms might have arisen by relatively recent gene duplication events in this family of Ser proteases. Indeed, the P69E and P69F genes on the one hand, and the P69A, P69B, P69C, and P69D on the other, are grouped in two different clusters in the nuclear genome of tomato plants. Within the two clusters, each sequence lies adjacent to the other and they share virtually identical structures. From this dendogram, it appears that somewhat earlier in the evolutionary history of this gene family duplication of an ancestral gene occurred that gave rise to the P69E/F and the P69A/B/C/D branches. The two branches appear to evolved independently from each other, and within each branch, subsequent gene duplication events generated the array of P69 isoforms. In the P69A/B/C/D branch, the P69D member appears to evolved independently of a subbranch comprising P69B/C/A. Most recently, this latter subbranch again duplicated and gave rise to the P69B and a new branch that again duplicated and finally gave the P69A and P69C isoforms. The sequence-based relationships depicted in Figure 6 are thus consistent with the structure and positions of the P69 genes in the genome. Additionally, the P69 genes and their plant homologs diverged independently from the other eukaryotic-related sequences as they evolved from the ancestral bacterial subtilisin protease. This may help explaining why plant subtilases have been positioned in a distinct group called the “pyrolysins” according to the classification scheme recently proposed (Siezen and Leunissen, 1997).
From the scenario of P69 evolution, and the very distinct tissue-specific mRNA expression profiles, with constitutive or transitory versus pathogen-inducible expression patterns, how might the distinct roles of the P69 members in plants have evolved? The highly conserved protein sequences and relative positions of the P69 genes in the P69A/B/C/D and P69E/F loci, compared with their highly different promoter sequences and expression profiles, rule out a trivial explanation. However, one can speculate that a constitutively expressed ancestral P69 gene may have been duplicated en bloc to yield two P69 genes with promoters specifying different, but permanent tissue-specific patterns. Such a scenario would explain the distinct expression patterns of the P69A and P69E, with the former expressed only in aerial parts of the plants and the latter only in the roots. This would result in complete constitutive expression throughout the plant. Subsequent gene duplication events accompanied by divergent evolution in the promoter regions, with acquisition of new regulatory elements in cis, may have generated highly specific expression patterns (e.g. transient expression in expanding zones of the leaf or flowers for P69D or specific expression in hydathodes for P69F). A similar mechanism could be explain the pathogen-induced expression of P69B and P69C. However, in this speculative evolutionary model it remains to be demonstrated whether these differential expression patterns also imply different roles for the protein products.
With all of the individual members of the P69 family now identified, we can approach a comparative study of the effect of overexpression of each individual member in transgenic plants and address the identification of the proteolytic substrates.
ACKNOWLEDGMENTS
We thank Dr. S. Ambros and M. De la Peña for help with the construction of dendograms and Dr. C.A. Ryan for constructive critique of the manuscript.
Footnotes
This work was supported by the Spanish Ministry of Science and Education and the Conselleria de Educación y Ciencia de la Generalitat de Valencia (for a fellowship to L.J.).
LITERATURE CITED
- Barr PJ. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991;66:1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Cook AA, Walker JC, Larson RH. Studies on the diseasecycle of black rot of crucifers. Phytopathology. 1952;42:162–167. [Google Scholar]
- Dale JE. The control of leaf expansion. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:267–295. [Google Scholar]
- Dixon RA, Lamb CJ. Molecular communication in interactions between plants and microbial pathogens. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:339–367. [Google Scholar]
- Enyedi AJ, Yalpani N, Silverman P, Raskin I. Localization, conjugation, and function of salicylic acid in tobacco during the hypersensitive reaction to tobacco mosaic virus. Proc Natl Acad Sci USA. 1992;89:2480–2484. doi: 10.1073/pnas.89.6.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essau K. Plant Anatomy. Ed 2. New York: John Wiley & Sons; 1965. [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Jordá L, Coego A, Conejero V, Vera P. A genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J Biol Chem. 1999;274:2360–2365. doi: 10.1074/jbc.274.4.2360. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–381. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ. Expansin mode of action on cell walls. Plant Physiol. 1995;107:87–100. doi: 10.1104/pp.107.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meichtry J, Amrhein N, Schaller A. Characterization of the subtilase gene family in tomato (Lycopersicon esculentum Mill.) Plant Mol Biol. 1999;39:749–760. doi: 10.1023/a:1006193414434. [DOI] [PubMed] [Google Scholar]
- Ribeiro A, Akkermans ADL, van Kammen A, Bisseling T, Pawlowski K. A nodule-specific gene encoding a subtilisin-like protease is expressed in early stages of actinorhizal nodule development. Plant Cell. 1995;7:785–794. doi: 10.1105/tpc.7.6.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs CD, Horsch A. Molecular cloning of an anther specific gene from tomato. Plant Physiol. 1995;108:117. [Google Scholar]
- Samac DA, Shah DM. Developmental and pathogen-induced activation of the Arabidopsis acidic chitinnase promoter. Plant Cell. 1991;3:1063–1072. doi: 10.1105/tpc.3.10.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, Leunissen JAM. Subtilases: the subtilisin-like serine proteases. Protein Sci. 1997;6:501–523. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner DF, Smeekens ST, Ohagi S, Chan SJ. The new enzymology of precursor processing endoproteases. J Biol Chem. 1992;267:23435–23438. [PubMed] [Google Scholar]
- Taylor AA, Horsch A, Rzepczyk A, Hasenkampf CA, Riggs DC. Maturation and secretion of a serine proteinase is associated with events of late microsporogenesis. Plant J. 1997;12:1261–1271. doi: 10.1046/j.1365-313x.1997.12061261.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P. Primary structure and expression of a pathogen-induced protease (PR-P69) in tomato plants: similarity of functional domains to subtilisin-like endoproteases. Proc Natl Acad Sci USA. 1996;93:6332–6337. doi: 10.1073/pnas.93.13.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P. Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J Biol Chem. 1997;272:14412–14419. doi: 10.1074/jbc.272.22.14412. [DOI] [PubMed] [Google Scholar]
- Von Heijne G. A new method for predicting signal sequence cleavage site. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Masuzawa T, Nagaoka Y, Ohnishi T, Iwasaki T. Cucumisin, a serine protease from melon fruits, shares structural homology with subtilisin and is generated from a larger precursor. J Biol Chem. 1994;269:32725–32731. [PubMed] [Google Scholar]
- Zhou A, Paquet L, Mains E. Structural elements that direct specific processing of different mammalian subtilisin-like prohormone convertases. J Biol Chem. 1995;270:21509–21516. doi: 10.1074/jbc.270.37.21509. [DOI] [PubMed] [Google Scholar]