A DCL2-dependent DCL genetic pathway is crucial for systemic PTGS in plants.
Abstract
Non-cell autonomous RNA silencing can spread from cell to cell and over long distances in animals and plants. However, the genetic requirements and signals involved in plant mobile gene silencing are poorly understood. Here, we identified a DICER-LIKE2 (DCL2)-dependent mechanism for systemic spread of posttranscriptional RNA silencing, also known as posttranscriptional gene silencing (PTGS), in Nicotiana benthamiana. Using a suite of transgenic DCL RNAi lines coupled with a GFP reporter, we demonstrated that N. benthamiana DCL1, DCL2, DCL3, and DCL4 are required to produce microRNAs and 22, 24, and 21nt small interfering RNAs (siRNAs), respectively. All investigated siRNAs produced in local incipient cells were present at low levels in distal tissues. Inhibition of DCL2 expression reduced the spread of gene silencing, while suppression of DCL3 or DCL4 expression enhanced systemic PTGS. In contrast to DCL4 RNAi lines, DCL2-DCL4 double-RNAi lines developed systemic PTGS similar to that observed in DCL2 RNAi. We further showed that the 21 or 24 nt local siRNAs produced by DCL4 or DCL3 were not involved in long-distance gene silencing. Grafting experiments demonstrated that DCL2 was required in the scion to respond to the signal, but not in the rootstock to produce/send the signal. These results suggest a coordinated DCL genetic pathway in which DCL2 plays an essential role in systemic PTGS in N. benthamiana, while both DCL4 and DCL3 attenuate systemic PTGS. We discuss the potential role of 21, 22, and 24 nt siRNAs in systemic PTGS.
RNA silencing is a cellular gene regulatory mechanism that is conserved across fungal, plant, and animal kingdoms (Baulcombe, 2004; Sarkies and Miska, 2014). Through sequence-specific targeting, RNA silencing can degrade mRNA for posttranscriptional gene silencing (PTGS) or modify related DNA for transcriptional gene silencing (TGS). In plants, primary silencing is triggered by double-stranded RNA (dsRNA) molecules, which are processed into 21 to 24 nucleotide (nt) small-interfering (si) RNA duplexes, known as primary siRNAs, by DICER-LIKE (DCL) RNase III-like enzymes (Baulcombe, 2004; Sarkies and Miska, 2014). For instance, dsRNA can be directly produced from a hairpin transgene in primary silencing (Mlotshwa et al., 2008). Primary silencing differs from transitive silencing, in which the trigger is initially not itself dsRNA. In contrast to primary silencing, the dsRNA in transitive silencing is produced from a single-stranded (ss) RNA template, and requires the coordinated activity of a set of genes. These genes include DCL2, RNA dependent RNA polymerase 6 (RDR6), and SGS3, the latter of which encodes a coiled-coiled domain protein (Mlotshwa et al., 2008; Parent et al., 2015). Transitive silencing cascades and amplifies cell and non-cell autonomous silencing to other parts of the target RNA, leading to generation of secondary siRNAs. It is well known that DCL2 plays an essential role in cell-autonomous silencing transitivity and accumulation of secondary siRNAs in Arabidopsis (Arabidopsis thaliana; Mlotshwa et al., 2008; Parent et al., 2015).
In animals and plants, TGS and PTGS can spread from cell to cell, systemically, or even between different organisms (Weiberg et al., 2013). Intercellular and long-distance movement of non-cell autonomous RNA silencing involves mobile signals and various genetic components (Melnyk et al., 2011b; Molnar et al., 2010). For example, SNF2, a JmjC domain protein JMJ14, the THO/TREX mRNA export complex, and RDR6 are all associated with intercellular RNA silencing (Qin et al., 2012; Searle et al., 2010; Smith et al., 2007; Yelina et al., 2010). Moreover, RDR6 also contributes to initial signal perception for systemic PTGS (Melnyk et al., 2011b). Intriguingly, NRPD1a, encoding RNA polymerase IVa, RDR2, and DCL3, which all function in a chromatin silencing pathway, are required for the reception of long-distance mRNA PTGS, and Argonaute4 is also partially involved in the reception of such long-distance silencing. DCL4 and DCL2 then act hierarchically, as they do in antiviral resistance, to produce 21 and 22 nt siRNAs, respectively, which guide mRNA degradation in systemic tissues (Brosan et al., 2007).
Arabidopsis, and most likely other plant species, possess four DCL RNase III family enzymes for small RNA (sRNA) biogenesis. DCL1 is involved in microRNA (miRNA) production and DCL2, DCL3, and DCL4 are involved in 22, 24, and 21 nt siRNA production, respectively (Henderson et al., 2006; Mukherjee et al., 2013; Xie et al., 2004). DCL1-generated miRNAs can move and act as local and distal signals that play essential roles in plant growth and development, although direct evidence for mobile miRNA signaling is lacking (Carlsbecker et al., 2010; Pant et al., 2008; Skopelitis et al., 2017). The DCL3-processed 24 nt siRNA has been shown to direct systemic TGS that controls genome-wide DNA methylation in recipient cells through RNA-directed DNA methylation of homologous genomic DNA sequences (Henderson et al., 2006; Lewsey et al., 2016; Melnyk et al., 2011a; Molnar et al., 2010; Sarkies and Miska, 2014). However, systemic silencing has also been reported to occur in the absence of sRNAs (Mallory et al., 2001), and no specific siRNAs produced by any of the four DCLs were found to be associated with systemic silencing (Brosnan et al., 2007). Moreover, even in Arabidopsis, different genetic factors required for mobile PTGS have been discovered from mutant screens in very similar experimental systems (Melnyk et al., 2011b). Thus, the precise nature of long-distance mobile signal remains to be elucidated, as this signal is not exclusively produced by any of the four DCLs. Crucial molecular events during systemic RNA silencing reception were elegantly investigated in Arabidopsis (Brosnan et al., 2007). Nevertheless, these findings contrast with systemic silencing, known as RNA interference (RNAi) in animals, where the mobile signal is dsRNA (Jose et al., 2011). In Caenorhabditis elegans, systemic RNAi involves intertissue transfer and entry of gene-specific dsRNAs into the cytosol by the dsRNA-selective importer SID-1 (Winston et al., 2002; Feinberg and Hunter, 2003; Shih and Hunter, 2011). Consequently, mobile dsRNAs expressed in a variety of somatic tissues can lead to SID-1-dependent homologous RNAi of target mRNA in other somatic tissues, or even transgenerational silencing (Jose et al., 2009; Devanapally et al., 2015).
The intricate functions of DCL2 and DCL4 are partially redundant in trans-acting siRNA biogenesis and in plant antiviral defense. However, DCL2 is responsible for 22 nt siRNA biosynthesis and has different roles to DCL4 in the production of primary and secondary siRNAs (Chen et al., 2010; Henderson et al., 2006; Qu et al., 2008; Wang et al., 2011; Xie et al., 2005). DCL4-processed 21nt siRNA is mainly involved in intracellular PTGS as well as represents the signaling molecule that moves from leaf companion cells to adjacent cells to induce limited intercellular PTGS in Arabidopsis (Dunoyer et al., 2005). Interestingly, DCL2 expression in leaf vascular tissues enhances PTGS in surrounding cells in Arabidopsis. DCL2 is thought to promote the production of 22 nt siRNAs that then stimulate 21 nt siRNA biogenesis via RDR6 and DCL4, resulting in increased cell-to-cell spread of PTGS (Parent et al., 2015). By contrast, DCL4 inhibits the cell-to-cell spread of virus-induced gene silencing (VIGS), a form of PTGS, while DCL2, likely along with a DCL2-dependent RNA signal, is required for efficient trafficking of VIGS from epidermal to adjacent cells (Qin et al., 2017). However, the roles of 21 and 24 nt siRNAs and other types of RNA molecules in mobile PTGS and TGS are under debate (Mallory et al., 2001; Brosnan et al., 2007; Melnyk et al., 2011b; Sarkies and Miska, 2014) and remain highly controversial (Berg, 2016).
On the other hand, impairment of DCL2 or DCL4 can reduce or promote cell-autonomous PTGS, and DCL2 promotes intracellular transitive silencing in Arabidopsis (Mlotshwa et al., 2008; Parent et al., 2015). Moreover, two different strong viral suppressors of silencing (Turnip crinkle virus P38 and Turnip mosaic virus HC-Pro) block the accumulation of secondary siRNAs produced by transitive silencing (Mlotshwa et al., 2008). Thus, DCL2 may have a role in viral defense that can be coupled to intercellular and systemic PTGS. This view is indeed supported by the requirement of DCL2 and DCL2-dependent mobile signals for intercellular antiviral silencing in N. benthamiana (Qin et al., 2017). A genetic screen for impaired systemic RNAi revealed that DCL2 is crucial for RDR6-dependent systemic PTGS (Taochy et al., 2017). Nevertheless, it is not known whether, and to what extent, DCL2 and other DCLs regulate systemic silencing. However, DCL2 and its cognate 22 nt siRNA are clearly able to affect secondary siRNA biogenesis, antiviral defense, and plant development (Bouché et al., 2006; Chen et al. 2010; Garcia-Ruiz et al., 2010; Qin et al., 2017; Wang et al., 2011).
In this article, we report the genetic requirement of a DCL network and the involvement of mobile siRNAs in the systemic spread of PTGS in N. benthamiana.
RESULTS
Genetic Resources for Examining Mobile RNA Silencing in N. benthamiana
To investigate systemic RNA silencing in N. benthamiana, we used gene-specific RNAi to inhibit the four DCLs orthologous to Arabidopsis DCL1, DCL2, DCL3, and DCL4 (Supplemental Data Set S1). Specific genes or gene fragments were PCR-amplified and cloned into the overexpression vector pCAMBIA1300 to produce p35S-GFP714 (the GFP coding sequence of this variant is 714 nt long; Ryabov et al., 2004) or the hairpin RNAi vector pRNAi-LIC (Xu et al., 2010) to generate pRNAi-GFP714 and pRNAi-DCLs (Supplemental Fig. S1). These cloned DCL gene fragments shared no detectable nucleotide sequence similarity (Supplemental Table S1). Plants were then transformed with each of the pRNAi-DCL constructs. We selected two single-copy homozygous RNAi lines, DCL2Ai and DCL2Bi for DCL2, DCL3Ai and DCL3Bi for DCL3, and DCL4Ai and DCL4Bi for DCL4, and produced a double RNAi line DCL24i through crossing DCL2Ai with DCL4Bi (Table I). We crossed all of the individual DCL RNAi lines with the 16cGFP plants bearing a single copy of a GFP transgene (referred to as GFP792, as the coding region of this gene variant is 792 nt long) under the control of the 35S promoter (Haseloff et al., 1997; Ruiz et al., 1998) to create the hybrid lines Gfp, GfpDCL2Ai, GfpDCL2Bi, GfpDCL3Ai, GfpDCL3Bi, GfpDCL4Ai, GfpDCL4Bi, GfpDCL24Ai, and GfpDCL24Bi (Table I). Through selfing, we generated homozygous lines homGfpDCL2Ai, homGfpDCL3Bi, and homGfpDCL4Ai. An additional double DCL2-DCL4 RNAi line GfpDCL24Ci was obtained through crossing homGfpDCL2Ai and homGfpDCL4Ai (Table I). However, we only obtained one hemizygous DCL1i or GfpDCL1i line by transforming N. benthamiana or 16cGFP with pRNAi-DCL1, as homozygous DCL1 RNAi was lethal (Table I). Lines RDR6i and GfpRDR6i were included in this work (Schwach et al., 2005). To confirm the inhibition caused by RNAi, we performed RT-qPCR and analyzed DCL mRNA levels in the RNAi lines. Gene-specific RNAi repression of DCL2, DCL3, and DCL4 was between 70% and 90%, and approximately 45% suppression was achieved for DCL1 (Supplemental Fig. S2), consistent with our previous analyses (Qin et al., 2017).
Table I. Summary of transgenic lines generated and used in this study.
| Wild-Type and Transgenic Lines | Genetic Background and Cross | Transgene Information |
|---|---|---|
| Nb | N. benthamiana | Wild-type, no transgene |
| DCL1i | RNAi of DCL1 in Nb | pRNAi-DCL1, single copy, hemizygous |
| DCL2Ai | RNAi of DCL2 in Nb | pRNAi-DCL2, single copy, homozygous |
| DCL2Bi | ||
| DCL3Ai | RNAi of DCL3 in Nb | pRNAi-DCL3, single copy, homozygous |
| DCL3Bi | ||
| DCL4Ai | RNAi of DCL4 in Nb | pRNAi-DCL4, single copy, homozygous |
| DCL4Bi | ||
| RDR6i (Gift from David Baulcombe) | RNAi of NbRDR6 in Nb | RDR6 hairpin, single copy, homozygous |
| DCL24i | Cross between DCL2Ai and DCL4Bi | pRNAi-DCL2, single copy, hemizygous |
| pRNAi-DCL4, single copy, hemizygous | ||
| 16cGFP | GFP transgenic line in Nb | 35S-GFP, single copy, homozygous |
| GfpRDR6i (Gift from David Baulcombe) | RNAi of RDR6 in 16cGFP | 35S-GFP, single copy, homozygous |
| RDR6 hairpin, single copy, homozygous | ||
| GfpDCL1i | RNAi of DCL1 in 16cGFP | 35S-GFP, single copy, homozygous |
| pRNAi-DCL1, single copy, hemizygous | ||
| GfpDCL2Ai | Cross between 16cGFP and DCL2Ai | 35S-GFP, single copy, hemizygous |
| GfpDCL2Bi | Cross between 16cGFP and DCL2Bi | pRNAi-DCL2, single copy, hemizygous |
| GfpDCL3Ai | Cross between 16cGFP and DCL3Ai | 35S-GFP, single copy, hemizygous |
| GfpDCL3Bi | Cross between 16cGFP and DCL3Bi | pRNAi-DCL3, single copy, hemizygous |
| GfpDCL4Ai | Cross between 16cGFP and DCL4Ai | 35S-GFP, single copy, hemizygous |
| GfpDCL4Bi | Cross between 16cGFP and DCL4Bi | pRNAi-DCL4, single copy, hemizygous |
| GfpDCL24Ai | Triple cross among 16c, DCL2Ai and DCL4Ai | 35S-GFP, single copy, hemizygous |
| GfpDCL24Bi | Triple cross among 16c, DCL2Ai and DCL4Bi | pRNAi-DCL2, single copy, hemizygous |
| pRNAi-DCL4, single copy, hemizygous | ||
| homGfpDCL2Ai | Self from GfpDCL2Ai | 35S-GFP, single copy, homozygous |
| pRNAi-DCL2, single copy, homozygous | ||
| homGfpDCL3Bi | Self from GfpDCL3Bi | 35S-GFP, single copy, homozygous |
| pRNAi-DCL3, single copy, homozygous | ||
| homGfpDCL4Ai | Self from GfpDCL4Ai | 35S-GFP, single copy, homozygous |
| pRNAi-DCL4, single copy, homozygous | ||
| GfpDCL24Ci | Cross between homGfpDCL2Ai and homGfpDCL4Ai | 35S-GFP, single copy, homozygous |
| pRNAi-DCL2, single copy, hemizygous | ||
| pRNAi-DCL4, single copy, hemizygous | ||
| Gfp | Cross between Nb and 16cGFP | 35S-GFP, single copy, hemizygous |
DCLs Function Differentially to Generate Small RNAs in N. benthamiana
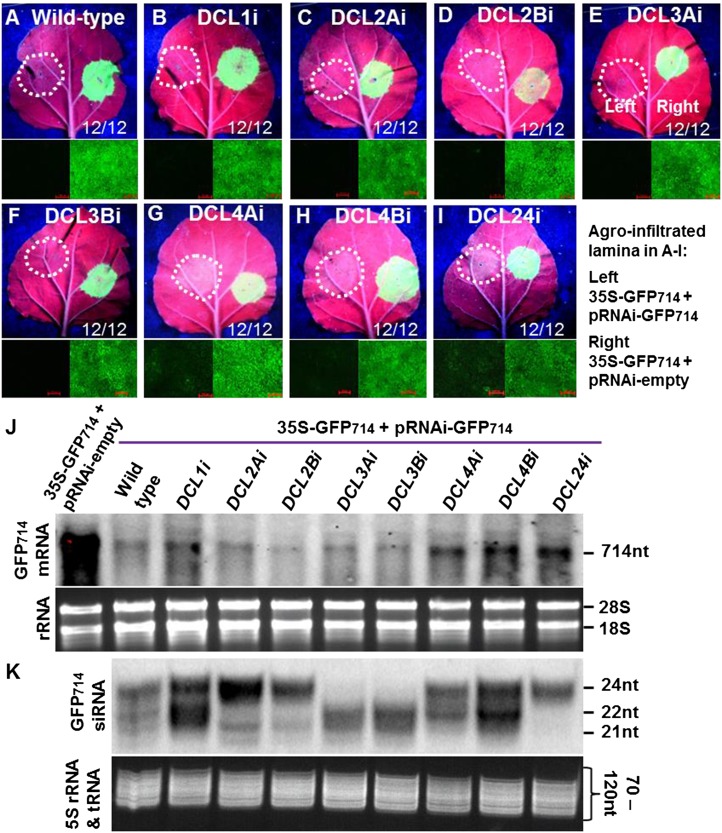
To determine whether individual DCLs function differently to generate 21, 22, and 24 nt siRNAs in N. benthamiana (Fig. 1), we used a GFP reporter system and performed a local PTGS assay (Ruiz et al., 1998; Ryabov et al., 2004). Leaves of the DCL RNAi plants at the six-leaf stage were co-infiltrated with Agrobacterium harboring p35-GFP714 and Agrobacterium harboring pRNAi-GFP714 or an empty pRNAi-LIC vector (Supplemental Figs. S1 and S3A). Compared to the non-RNAi wild-type plant (Fig. 1A), RNAi of DCL1 (DCL1i; Fig. 1B), DCL2 (DCL2Ai and DCL2Bi; Fig. 1, C and D), and DCL3 (DCL3Ai and DCL3Bi; Fig. 1, E and F) did not affect local RNA silencing, but RNAi of DCL4 (DCL4Ai, DCL4Bi, and DCL24i; Fig. 1, G–I) weakened local PTGS. This is evident by almost complete (Fig. 1, A–F, left) or partial reduction (Fig. 1, G–I, left) in GFP fluorescence intensity in the coagroinfiltrated lamina (Supplemental Fig. S4). Consistent with the level of RNAi achieved, only a low level of GFP714 mRNA was detected in wild-type, DCL2Ai, DCL2Bi, DCL3Ai, and DCL3Bi plants, whereas a relatively high level of GFP714 mRNA was found in DCL4Ai, DCL4Bi, and DCL24i plants (Fig. 1J). We also noticed that the GFP714 mRNA levels were slightly higher in DCL1i plants (Fig. 1J).
Figure 1.
Impact of DCL RNAi on local hairpin dsRNA-mediated PTGS. A to I, Local PTGS assay. The left half of leaves of N. benthamiana (A), DCL1i (B), DCL2Ai and DCL2Bi (C and D), DCL3Ai and DCL3Bi (E and F), DCL4Ai and DCL4Bi (G and H), and DCL24i (I) plants were co-infiltrated with agrobacterium harboring p35S-GFP714 and pRNAi-GFP714 and the right half with p35S-GFP714 and the pRNAi-empty vector. Without PTGS, GFP714 expression from p35S-GFP714 shows strong green fluorescence. Induction of PTGS by the hairpin GFP714 dsRNA generated from pRNAi-GFP eliminates GFP714 expression and the infiltrated lamina show red auto-fluorescence. DCL4 RNAi attenuated intracellular silencing and the level of PTGS decreased in DCL4Ai, DCL4Bi, and DCL24i plants. Some green fluorescence is still visible in the infiltrated lamina of these plants (G–I). Photographs were taken under long-wavelength UV light (top) or through a stereo-fluorescent microscope (bottom) at 4 dpa. Bar = 100 μm. The ratio in the bottom right corner of each panel indicates the number of plants out of the number of agro-infiltrated plants that developed local PTGS in two experiments. The green fluorescence intensity was measured using ImageJ software (Supplemental Fig. S4). J, Analysis of GFP714 mRNA in DCL RNAi lines. Northern detection was performed using GFP714-specific probes (top). Equal loading of total RNAs extracted from leaf tissues at 4 dpa is indicated by the equal amount of 18S and 28S rRNAs shown on the gel (bottom). The position and size of GFP714 mRNA and 18S and 28S rRNA are indicated. K, Detection of local L-siRNAs. L-siRNAs were analyzed by small RNA northern blots (top). Equal loading of sRNAs extracted from leaf tissues at 4 dpa is indicated by the equal amount of 5S rRNA/tRNAs shown on gel (bottom). The position of 21, 22, and 24 nt siRNA as well as 5S rRNA/tRNAs are indicated.
We then used northern hybridizations to characterize siRNAs present in the infiltrated leaf tissues of different DCL RNAi lines (Fig. 1K). These local siRNAs are hereafter designated L-siRNAs. L-siRNAs include primary siRNAs that are directly generated from the hairpin GFP714 dsRNA and secondary siRNAs that are generated from the p35S-GFP714 expressed GFP714 mRNA. There is some variation in the levels of siRNAs detected in the two independent RNAi lines with respect to a given DCL gene. Nevertheless, 21, 22, and 24 nt L-siRNAs were readily detectable in the control and DCL1i plants. This contrasted with the specific loss of 22 nt L-siRNAs in DCL2Ai and DCL2Bi, 24 nt L-siRNAs in DCL3Ai and DCL3Bi, and 21 nt L-siRNAs in DCL4Ai and DCL4Bi (Fig. 1K). We also noticed a large amount of 22 and 24 nt L-siRNAs in DCL4Ai and DCL4Ai, and a high level of 24 nt L-siRNAs in DCL2Ai and DCL2Bi. However, 21 and 22 nt L-siRNAs were undetectable in the DCL24i plants (Fig. 1K). These data reveal that in N. benthamiana, as in Arabidopsis, DCL4, DCL2, and DCL3 are required to produce 21, 22, and 24 nt siRNAs, respectively.
RNAseq Analysis Confirms DCLs for Biosynthesis of sRNAs in N. benthamiana
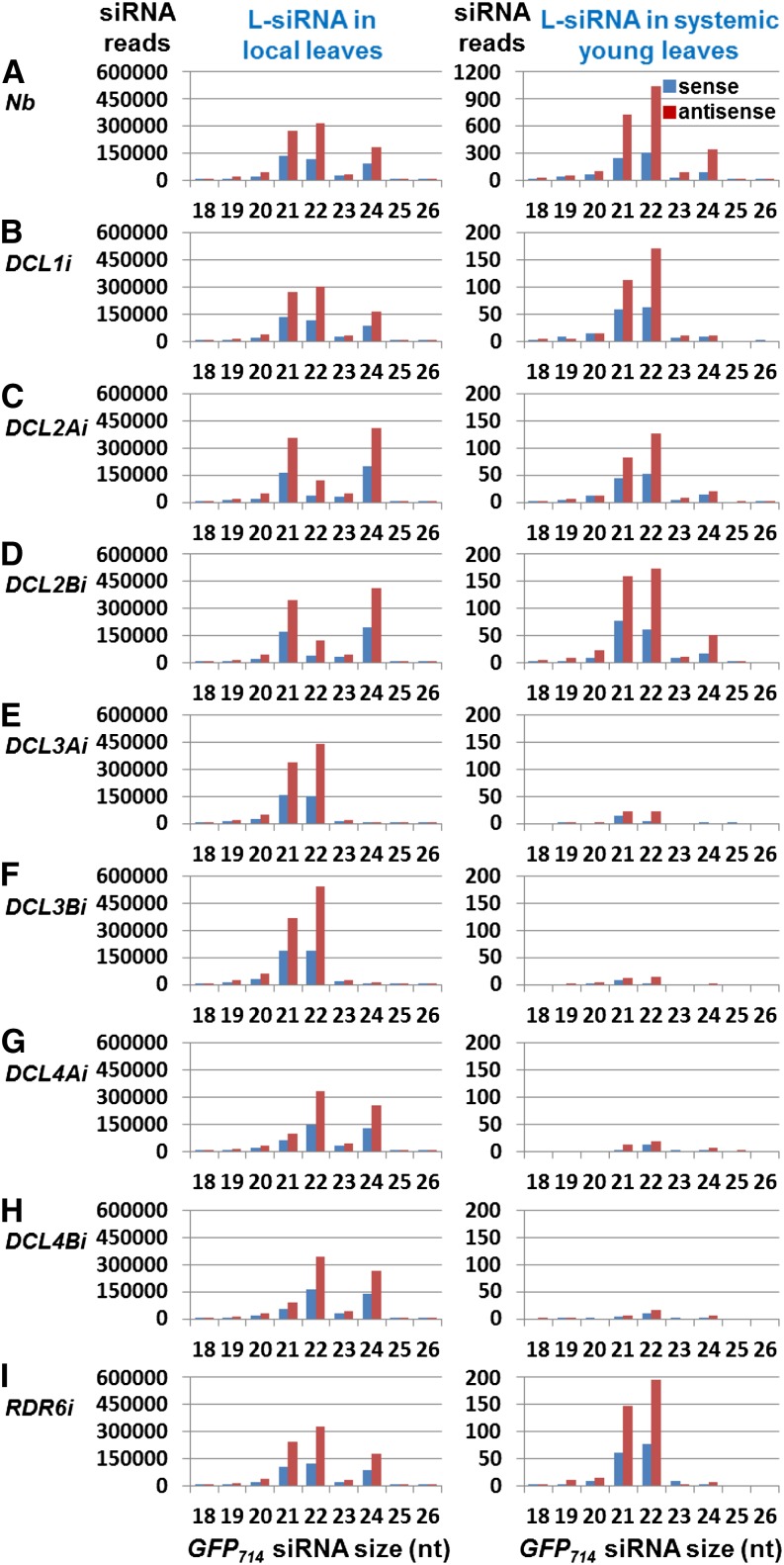
To further investigate how DCL RNAi affects sRNA synthesis, we collected agro-infiltrated leaf tissues from N. benthamiana (Nb), DCL1i, DCL2Ai, DCL2Bi, DCL3Ai, DCL3Bi, DCL4Ai, DCL4Bi, and RDR6i plants at 7 d post agro-infiltration (dpa). The leaves were co-infiltrated with agrobacterium harboring p35S-GFP714 and agrobacterium harboring pRNAi-GFP714 (Fig. 2; Supplemental Fig. S3A). We then constructed 9 individual local sRNA libraries for next generation sequencing (NGS) and generated approximately 11 million sRNA reads for each library (Supplemental Table S2). We found that (1) gene-specific RNAi of DCLs or RDR6 had no off-target effect on other silencing-related genes (Supplemental Fig. S5); (2) correlation analyses of total miRNAs among these sRNA libraries validated the authenticity and direct comparability of the detected sRNA profiles (Supplemental Tables S3 and S4); and (3) DCL1 RNAi resulted in a clear reduction of miRNAs (Supplemental Fig. S6; Supplemental Table S2). Compared to Nb, DCL1i, and RDR6i (Fig. 2, A, B, and I, left), decreased levels in 21, 22, and 24 nt GFP714 L-siRNAs and total sRNAs correlated with RNAi of DCL4, DCL2, and DCL3 (Fig. 2, C–H, left; Supplemental Fig. S3, B–J). These data are consistent with the sRNA northern detection (Fig. 1K) and further show that as in Arabidopsis, DCL1 is also required for miRNA biosynthesis in N. benthamiana.
Figure 2.
Detection of L-siRNAs in local and systemic leaves. A to I, GFP714 L-siRNAs generated from hairpin dsRNA-mediated local PTGS can be detected in distal systemic young leaves. Sense (blue) and antisense (red) 21, 22, and 24 nt L-siRNAs were present in local (left) and systemic (right) tissues of N. benthamiana (Nb, A) and RNAi lines DCL1i (B), DCL2Ai (C), DCL2Bi (D), DCL3Ai (E), DCL3Bi (F), DCL4Ai (G), DCL4Bi (H), and RDR6i (I). Outline of mobile siRNA assay is indicated in Supplemental Figure S3A.
Detection of L-siRNAs in Distal Systemic Leaves
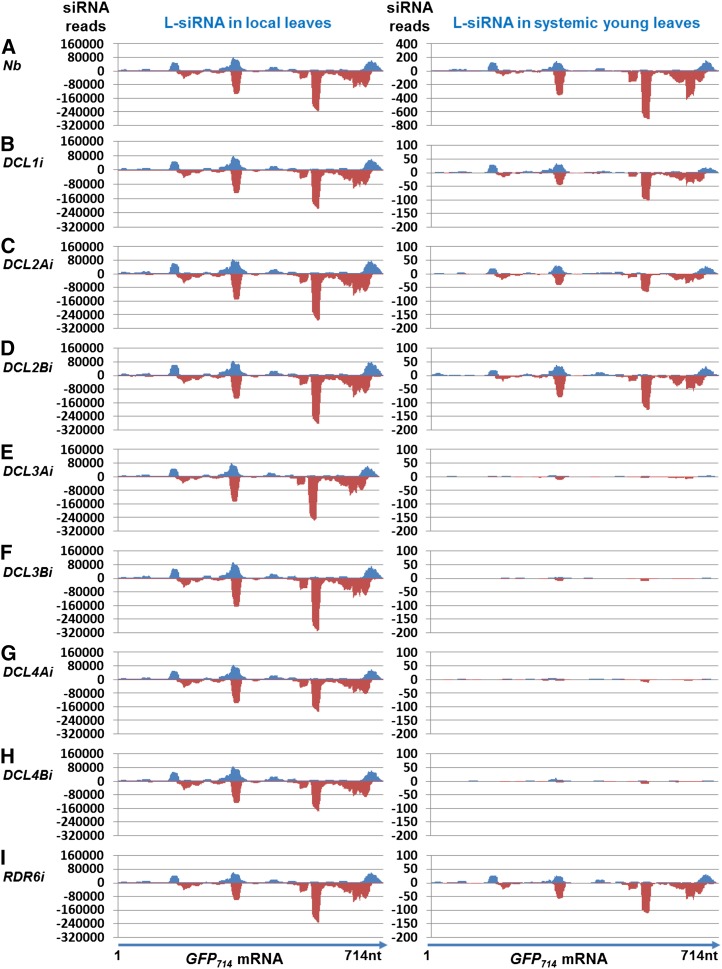
To investigate whether GFP714 L-siRNAs generated in local leaves are able to move systemically to distant tissues (Fig. 3), we collected a pool of young systemic leaves from the agro-infiltrated DCL RNAi or control plants at 14 dpa and constructed 9 systemic leaf sRNA libraries for NGS (Supplemental Fig. S3A). Similar to the local sRNA libraries made from the infiltrated leaf tissues, we generated approximately 11 million sRNA reads for each of the systemic libraries, and DCL-specific RNAi was again found to have little off-target effect on other silencing-related genes in the systemic leaves (Supplemental Table S2; Supplemental Fig. S5). We then scrutinized the systemic leaf sRNA datasets for reads matching the GFP714 mRNA. Different levels of both sense and antisense 21, 22, and 24 nt GFP714 L-siRNAs were detected in systemic leaves of Nb, RDR6i, and DCL RNAi plants (Fig. 2, A–I, right), although the total number of GFP714 L-siRNA (and total sRNA) reads in the infiltrated leaves was similar among all samples (Fig. 2, A–I, left; Supplemental Table S2). We detected high levels of 22 nt L-siRNAs, some 21 nt L-siRNAs, and a low level of 24 nt L-siRNAs in systemic Nb leaves (Fig. 2A, right). However, in the young leaves of DCL1i, DCL2Ai, DCL2Bi, and RDR6i plants (Fig. 2, B–D and I, right), the number of 22 or 21 nt L-siRNA reads was 5 to 8 times lower than that in Nb. In systemic leaves of DCL3Ai and DCL3Bi as well as those of DCL4Ai and DCL4Bi, only very low levels of 22 or 21 nt L-siRNAs were identified (Fig. 2, E–H, right), 45- to 68-fold lower than in Nb. The distribution across the GFP714 mRNA of both sense and antisense 21 to 24 nt L-siRNAs detected in local or systemic leaf tissues was almost identical among all samples (Fig. 3, left vs right panels A–I). However, we failed to detect any full-length or partial GFP714 RNA in systemic young leaves in any of the agro-infiltrated plants by RT-PCR using GFP714-specific primers (Supplemental Fig. S7; Supplemental Table S5).
Figure 3.
Distribution of 20 to 25nt L-siRNAs across the GFP714 mRNA. A to I, Distribution of 20 to 25nt GFP714 L-siRNAs for N. benthamiana (Nb; A); DCL1i (B); DCL2Ai and DCL2Bi (C and D); DCL3Ai and DCL3Bi (E and F); DCL4Ai and DCL4Bi (G and H); and RDR6i (I). Total GFP714 L-siRNA reads are from sRNA libraries of agro-infiltrated leaves (7 dpa; left) and systemic young leaves (14 dpa; right). Outline of mobile siRNA assay is indicated in Supplemental Figure S3A.
DCL2 Facilitates, While DCL4 and DCL3 Attenuate, Systemic PTGS
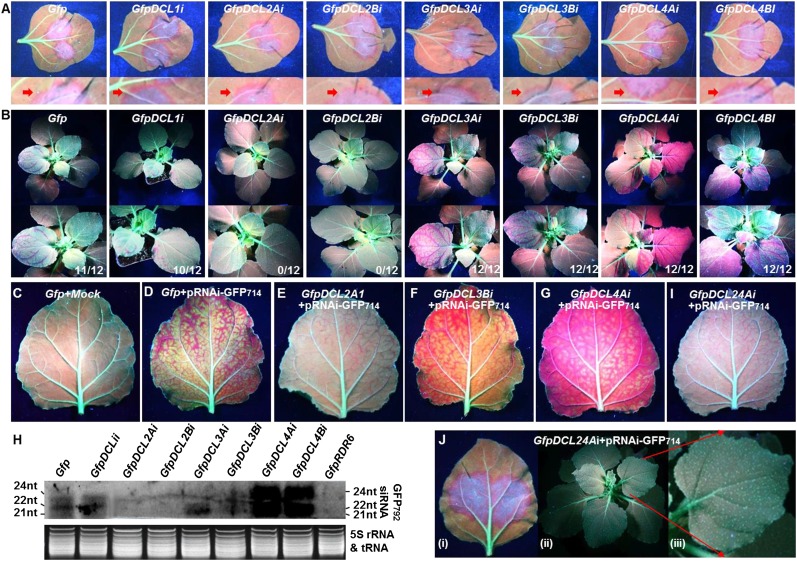
To analyze the impact of DCL RNAi on systemic PTGS (Fig. 4), we infiltrated leaves of Gfp (resulting from a cross between Nb and 16cGFP), GfpDCL1i, GfpDCL2Ai, GfpDCL2Bi, GfpDCL3Ai, GfpDCL3Bi, GfpDCL4Ai, or GfpDCL4Bi plants at the six-leaf stage with Agrobacterium harboring pRNAi-GFP714. It is important to note that local PTGS is induced by the hairpin GFP714 that directly produces long dsRNA in the agro-infiltrated leaves. However, sense transgene GFP792 silencing in local and systemic young leaves requires the activity of multiple genes, including RDR6, to produce dsRNA from GFP792 ssRNA (Mlotshwa et al., 2008). Under long-wavelength UV light, we observed red fluorescence in halos around the infiltrated areas on Gfp and RNAi line leaves at 7 dpa (Fig. 4A; Supplemental Table S6), indicating local silencing.
Figure 4.
Antagonistic roles of DCL2 against DCL3 and DCL4 in systemic PTGS. A and B, Impact of DCL RNAi on local and systemic PTGS. Local GFP silencing was induced by the hairpin GFP714 dsRNA in all plants by infiltration of leaves with agrobacterium harboring pRNAi-GFP714, evident by the red halos around the infiltrated lamina. A close-up of the red halo (red arrow) below each image shows clearer silencing effects (A). Sense transgene GFP792-mediated systemic PTGS was assessed on young leaves (B). At 14 dpa, all infiltrated plants of GfpDCL4Ai and GfpDCL4Bi lines developed very strong systemic GFP792 PTGS in young leaves. GfpDCL3Ai and GfpDCL3Ai plants also developed strong systemic PTGS in young leaves, although some relatively weak GFP792 silencing was observed in some Gfp and GfpDCL1i plants, and no systemic silencing appeared on any GfpDCL2Ai and GfpDCL2Bi plants. An enlarged snapshot (lower) of each photograph in its panel shows a clearer image of the silencing phenotypes (B). The ratio in the bottom right corner of each photo in B indicates the number of plants out of the number of agro-infiltrated plants that developed systemic PTGS in two separate experiments. Photographs were taken at 7 (A) and 14 dpa (B). C to G, Systemic transgene GFP792 silencing in young leaves. Transgene GFP792 expression was not silenced in the mock-infiltrated Gfp plant, and uniform green fluorescence occurred in the whole lamina of the noninfiltrated young leaf (C). Systemic GFP792 silencing in young leaf of Gfp plants infiltrated with agrobacterium harboring pRNAi-GFP714 was evident by the red fluorescence of chlorophyll across the leaf lamina of a newly grown young leaf (D). Only very weak GFP792 silencing was observed in the minor veins of a leaf immediately above the infiltrated leaves of GfpDCL2Ai (E), but extremely strong systemic GFP792 silencing appeared in GfpDCL3Bi (F) and GfpDCL4Ai (G). I, Weak minor vein-restricted systemic silencing in GfpDCL24Ai. Photographs were taken at 18 dpa. H, Northern-blot detection of Sy-siRNAs in the systemic young leaves at 14 dpa. Total sRNA extracted from systemic leaves of GfpRDR6i plants that were agro-infiltrated with pRNAi-GFP714 was included as a negative control. Top: siRNA blot, bottom: 5S rRNA/tRNA control showing equal loading of sRNA samples. The sizes and positions of siRNAs and 5S rRNA/tRNA are indicated. J, Local (i) and systemic (ii, iii) GFP792 silencing in GfpDCL24Ai. A section of Jii is enlarged to show a clearer young leaf image (Jiii). Photographs were taken at 7 dpa (Ji) or 14 dpa (Jii and iii). Influence of DCL RNAi on the development of local and systemic transgene GFP792 silencing is also summarized in Supplemental Table S6.
DCL1 RNAi had no obvious effect on systemic silencing in GfpDCL1i; however, DCL2 RNAi resulted in no PTGS in systemic young leaves of GfpDCL2Ai and GfpDCL2Bi (Fig. 4B; Supplemental Fig. S8; Supplemental Table S6). This contrasts with enhancement of systemic PTGS in GfpDCL4Ai and GfpDCL4Bi or GfpDCL3Ai and GfpDCL3Bi (Fig. 4B; Supplemental Fig. S8; Supplemental Table S6). Systemic silencing was further illustrated in single young leaves (Fig. 4, C–G). In contrast to the positive control (Fig. 4D), systemic PTGS was completely absent in the young leaves of GfpDCL2Ai and GfpDCL2Bi, as in Gfp (Fig. 4C). Only limited silencing was observed in one leaf located immediately above the infiltrated leaves at 18 dpa or later and such limited systemic PTGS was restricted to minor veins of DCL2 RNAi plants (Fig. 4E). However, strong systemic silencing appeared in young leaves of DCL3 and DCL4 RNAi lines (Fig. 4, F and G). Similar results of DCL RNAi on local or systemic silencing were observed in 16cGFP, homGfpDCL2Ai, homGfpDCL3Bi, and homGfpDCL4Ai plants as in Gfp and GfpDCL RNAi lines (Supplemental Table S6).
We then used northern hybridizations to detect systemic siRNA (dubbed Sy-siRNAs; Fig. 4H). Sy-siRNAs are siRNAs resulting from transitive silencing of the sense transgene GFP792 mRNA in nonagroinfiltrated leaves. Such systemic transitive silencing was triggered by mobile signals that were generated in agro-infiltrated local leaves. Sy-siRNAs also include two types of secondary siRNAs generated either from the GFP792 mRNA sequences that were directly targeted by the mobile signals or from the other parts of the GFP792 mRNA sequences that were not directly targeted by the mobile signals. No 21, 22, and 24 nt Sy-siRNAs were detected in the young leaves of GfpDCL2Ai and GfpDCL2Bi by northern blot (Fig. 4H). We were also unable to detect 24 nt Sy-siRNA in GfpDCL3Ai and GfpDCL3Bi, in which systemic silencing was apparently enhanced (Fig. 4, B, F, and H; Supplemental Fig. S8). However, 21, 22, and 24 nt Sy-siRNAs accumulated to a higher level in systemic leaves of GfpDCL4Ai and GfpDCL4Bi plants compared to Gfp (Fig. 4H). Considering the differential functions of DCL4 and DCL2 in primary and secondary siRNA biosynthesis (Chen et al., 2010; Cuperus et al., 2010), and the up-regulated DCL2 expression in the DCL4 or DCL3 RNAi lines (Qin et al., 2017), we suspect that these Sy-siRNAs are secondary siRNAs generated from the sense-transgene GFP792 mRNA. Taken together, our data demonstrate that DCL2 facilitates, while DCL4 and DCL3 attenuate, systemic PTGS, at least for the transgene-induced PTGS in N. benthamiana.
Genetic Interplay between DCL4 and DCL2 to Influence Systemic PTGS
To investigate a potential genetic link between DCL2 and DCL4 in systemic PTGS (Fig. 4, I and L), we infiltrated leaves of the triple-cross GfpDCL24Ai and GfpDCL24Bi (Table I) with Agrobacterium harboring pRNAi-GFP714 at the six-leaf stage. Local RNA silencing was sufficiently induced, as evidenced by the appearance of red halos around the agro-infiltrated lamina (Fig. 4Ji). No long-distance spread of PTGS occurred to reach systemic leaves, which showed uniform green fluorescence under long-wavelength UV light (Fig. 4J, ii and iii; Supplemental Table S6), although some minor vein-restricted silencing was observed in one leaf immediately above the agro-infiltrated leaves (Fig. 4I). Similar systemic PTGS patterns were also found in an additional DCL2-DCL4 double RNAi line GfpDCL24Ci (Table I; Supplemental Table S6). These findings show that 16cGFP plants with simultaneous RNAi against DCL2 and DCL4 develop systemic PTGS in a manner similar to GfpDCL2Ai and GfpDCL2Bi lines, but in contrast to GfpDCL4Ai and GfpDCL4Bi lines.
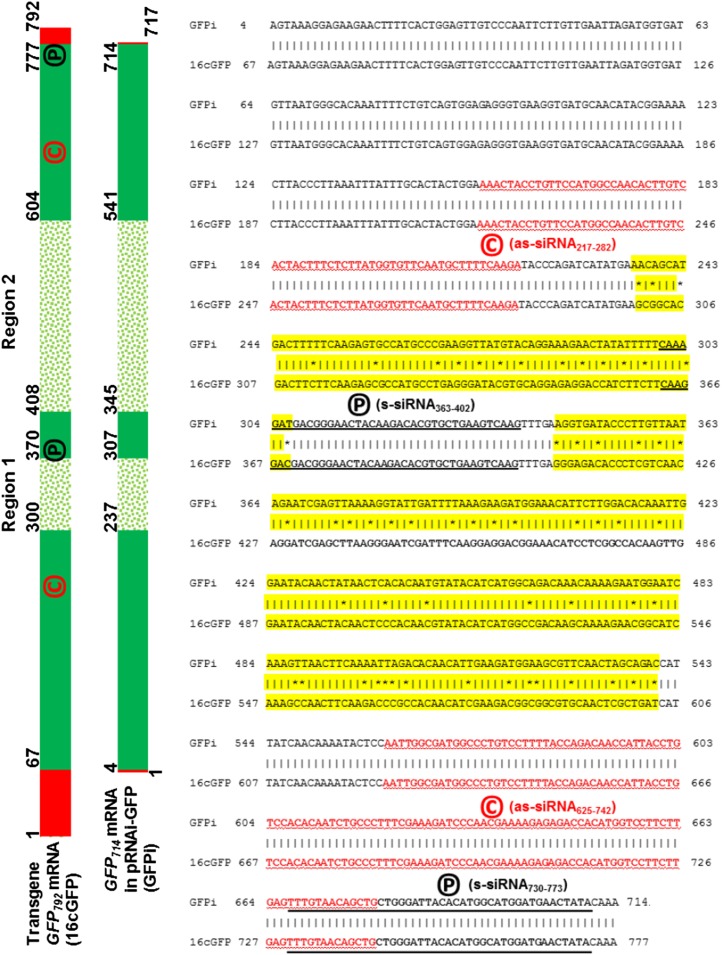
Comparison of Hairpin GFP714 and Transgene GFP792 Sequences
As indicated in Figure 5, sequences of transgene 792nt-GFP792 (Haseloff et al., 1997; Ruiz et al., 1998) in all transgenic Gfp and GfpDCL RNAi lines and hairpin 714nt-GFP714 (Ryabov et al., 2004) in the pRNAi-GFP714 vector share three identical and two less-similar regions, the latter named Region 1 and Region 2, respectively. GFP792 also possesses nucleotides 1 to 66 and 778 to 792 at both 5′-and 3′-ends that are completely absent from the GFP714 sequence. Within Region 1 or Region 2, the maximum interval between any two consecutive mismatched bases is 14 nt. This information is essential and relevant to our subsequent siRNA profiling and bioinformatic analyses of siRNAs that may or may not be associated with transitive silencing. We only counted 20 to 25 nt siRNAs that were perfectly (100%) matched to target gene mRNAs. This means that the 20 to 25 nt siRNAs mapped to Region 1/Region 2 of the GFP792 mRNA will not be mapped to the equivalent Region 1/Region 2 of the GFP714 mRNA, or vice versa. Thus, these siRNAs are exclusive to either GFP792 or GFP714, and most importantly, any Sy-siRNA unique to the transgene GFP792 Region 1/Region 2 mRNA should be generated by transitive silencing in systemic tissues.
Figure 5.
Comparisons of the transgene GFP792 and the hairpin GFP714 sequences. Completely distinct sequences between the GFP792 transgene (16cGFP) present in the Gfp and GfpDCL RNAi lines and the hairpin GFP714 (GFPi) in the pRNAi-GFP vector are outlined in red boxes, identical sequences are in green boxes, and similar sequences are in dotted boxes (highlighted yellow for the actual sequences). Sequence coordinates are indicated. The four clusters of the sense (s) and antisense (as) siRNAs corresponding to the positive (p) and complementary (c) stands (underlined) of the GFP714 and GFP792 mRNA are also indicated.
Mapping L-siRNAs and Sy-siRNAs to Transgene GFP792 mRNAs and to Hairpin GFP714 dsRNA
To investigate whether L-siRNAs are associated with Sy-siRNA biosynthesis and systemic PTGS in distal recipient tissues, we performed systemic transgene GFP792 silencing assays (Supplemental Fig. S9A) and sequenced sRNA libraries generated from the infiltrated or systemic leaves of Gfp, GfpDCL1i, GfpDCL2Ai, GfpDCL3Bi, GfpDCL4Ai, and GfpDCL24Ai plants. Systemic transgene GFP792 silencing was triggered by mobile signals originating from local PTGS that was initiated by the hairpin GFP714 dsRNA (Fig. 4; Supplemental Fig. S9B). Similar to the sRNA results obtained from the nonGFP-transformed RNAi lines, we generated approximately 12 million sRNA reads for each library (Supplemental Table S7). Interrogation of these sRNA datasets further confirmed DCL-specific RNAi and their roles in siRNA biogenesis (Supplemental Figs. S9, B–P and S10). Moreover, RNAi of DCL1 resulted in a clear reduction of miRNAs, although the total number of miRNAs was generally higher in systemic leaves than in local infiltrated leaves (Supplemental Fig. S11).
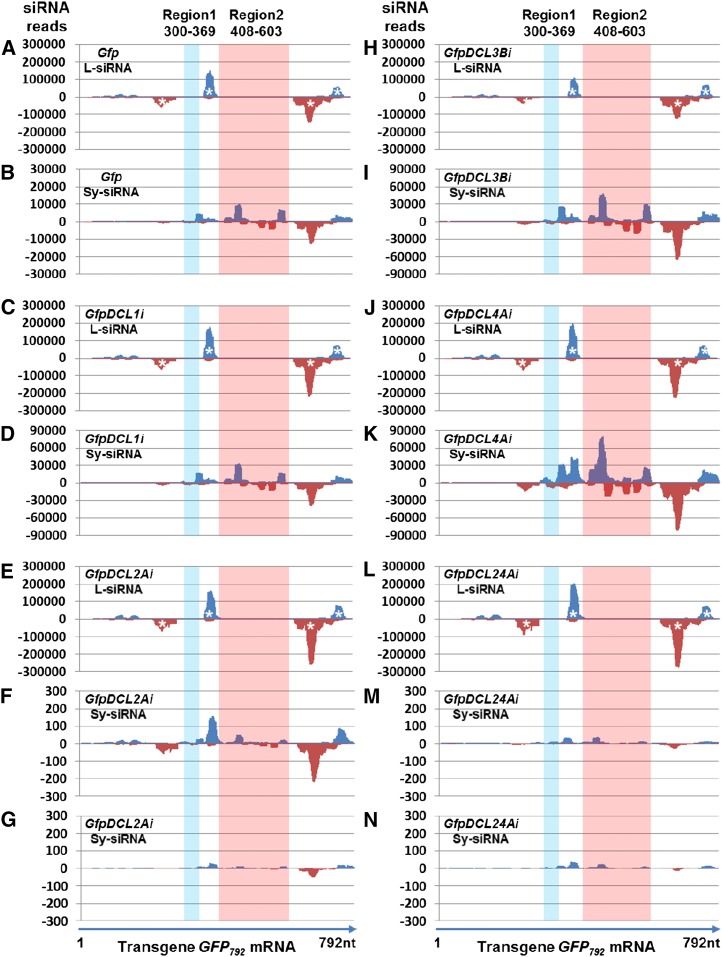
We mapped l- and Sy-siRNAs onto the transgene GFP792 mRNA. The distribution of antisense l- and Sy-siRNAs mapped to nucleotides 217-282 and 625-742 (Fig. 5, (c)-labeled) were essentially the same and sense l- and Sy-siRNAs mapped to nucleotides 363 to 402 and 730 to 773 of the transgene GFP792 mRNA (Fig. 5, (p)-labeled) in both local and systemic leaves of Gfp (Fig. 6, A and B), GfpDCL1Ai (Fig. 6, C and D), GfpDCL2Ai (Fig. 6, E–G), GfpDCL3Bi (Fig. 6, H and I), GfpDCL4Ai (Fig. 6, J and K), and GfpDCL24Ai (Fig. 6, L–N). The L-siRNAs in these four clusters (Fig. 6, A, C, E, H, J, and L; asterisked) corresponded to the occurrence of systemic GFP792 PTGS (Fig. 4; Supplemental Fig. S9B). We also found relatively high numbers of Sy-siRNA reads in young leaves where strong systemic GFP792 PTGS occurred in Gfp, GfpDCL1i, GfpDCL3Bi, and GfpDCL4Ai (Fig. 6, B, D, I and K). However, between the Gfp control and each of GfpDCL RNAi lines, we observed distinct profiles of l- and Sy-siRNAs across Region 1/Region 2 of the transgene GFP792 mRNA. Sy-siRNAs in young leaves specifically mapped to Region 1/Region 2 (Fig. 6, B, D, F, G, I, K, M, and N) were absent in the agro-infiltrated leaves where local silencing occurred efficiently in Gfp, GfpDCL1i, GfpDCL2Ai, GfpDCL3Bi, GfpDCL4Ai, and GfpDCL24Ai (Fig. 6, A, C, E, H, J, and L).
Figure 6.
Distribution of l- and Sy-siRNA across the transgene GFP792 mRNA. A and B, Gfp. C and D, GfpDCL1i. E to G, GfpDCL2Ai. H and I, GfpDCL3Bi. J and K, GfpDCL4Ai. L to N, GfpDCL24Ai. Total l- and Sy-siRNAs are from local agro-infiltrated leaves (A, C, E, H, J, and L) and systemic young leaves (B, D, F, G, I, K, M, and N). Sense- (blue) and antisense (red) siRNAs were mapped to the transgene GFP792 mRNA sequence. The profiles of L-siRNAs generated largely from the hairpin GFP714 dsRNA differ from that of Sy-siRNAs that were derived from the GFP792 mRNA, particularly in the two less similar Region 1/Region 2 (Fig. 5). These differences indicate that elevated levels of L-siRNAs (*) together with their long RNA precursor RNAs, despite the latter being unlikely (Supplemental Fig. S7), moved from local to systemic leaves and contributed to silencing signal. Such mobile signals might act as the primary trigger for production of Sy-siRNAs and for systemic transgene GFP792 silencing in distal young leaves. Sense and antisense Sy-siRNAs associated with Region 2 and Region 1 (highlighted) in systemic recipient cells are likely secondary and resulted from transitive silencing. Experimental design for systemic silencing assays is indicated in Supplemental Figure S9A. Systemic transgene GFP792 PTGS are shown in Figure 4, Supplemental Figure S8, and Supplemental Figure S9B. Compared to Gfp (B) and GfpDCL1i (D), strong systemic PTGS occurred in GfpDCL3Bi (I) and GfpDCL4i (K); no systemic PTGS was observed in young leaves of GfpDCL2Ai (F) and GfpDCL24Ai (M), and only very weak vein-restricted silencing in the leaf located immediately about the agro-infiltrated leaves of GfpDCL2Ai (G) and GfpDCL24Ai (N).
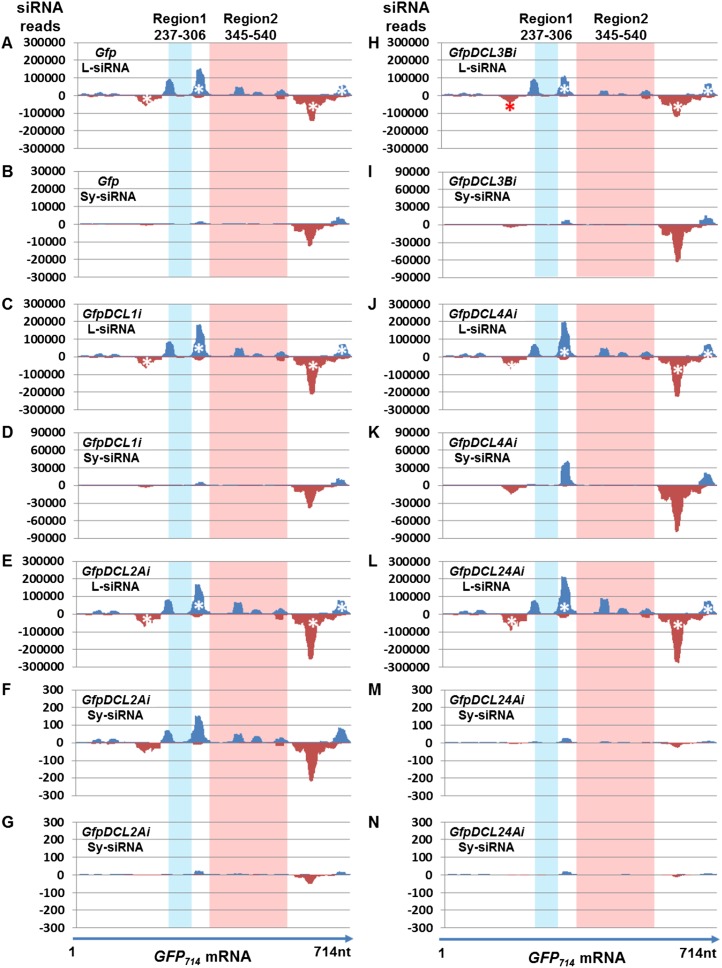
We also mapped the l- and Sy-siRNAs across the GFP714 mRNA, which was only expressed from pRNAi-GFP714 and formed hairpin dsRNA in the agro-infiltrated leaf lamina of Gfp (Fig. 7, A and B), GfpDCL1i (Fig. 7, C and D), GfpDCL2Ai (Fig. 7, E–G), GfpDCL3Bi (Fig. 7, H and I), GfpDCL4Ai (Fig. 7, J and K), and GfpDCL24Ai (Fig. 7, L–N). All four clusters of siRNAs mapped to the transgene GFP792 mRNA nucleotides 217 to 282, 363 to 402, 625 to 742, and 730 to 773, which also mapped to the equivalent identical sequences of the GFP714 RNA. However, L-siRNAs originating from the hairpin GFP714 dsRNAs were more abundant than the Sy-siRNAs derived from the transgene GFP792 mRNA. Moreover, we were able to map some L-siRNAs to Region 1 (nucleotide 237–306) and Region 2 (nucleotide 354–540) of the hairpin GFP714 RNA (Fig. 7, A, C, E, H, J, and L). These two regions differ from equivalent nucleotides 300 to 369 and 409 to 603 of the transgene GFP792 mRNA (Fig. 5). Intriguingly, only extremely low reads of such L-siRNAs were found in systemic leaves (Fig. 7, B, D, F, G, I, K, M, and N).
Figure 7.
Distribution of l- and Sy-siRNAs across the hairpin GFP714 RNA. A and B, Gfp. C and D, GfpDCL1i. E to G, GfpDCL2Ai. H and I, GfpDCL3Bi. J and K, GfpDCL4Ai. L to N, GfpDCL24Ai. Total l- or Sy-siRNAs are from local agro-infiltrated leaves (A, C, E, H, J, and L) or systemic young leaves (B, D, F, G, I, K, M, and N). Sense- (blue) and antisense (red) siRNAs were mapped to the hairpin GFP714 RNA. The L-siRNA profiles differ from Sy-siRNAs in the two less similar Region 1/Region 2 (Fig. 5). Experimental design for systemic silencing assays is indicated in Supplemental Figure S9A. Systemic transgene GFP792 PTGS are shown in Figure 4, Supplemental Figure S8, and Supplemental Figure S9B.
Taken together, these data (Figs. 5–7) suggest that (1) the hairpin GFP714 dsRNA is the main source for biogenesis of L-siRNA in local PTGS; (2) GFP714-specific L-siRNAs mapped to Regions 1/2 are present in systemic leaf tissues; (3) L-siRNAs are associated with the Sy-siRNA biogenesis and the induction of systemic GFP792 silencing; and (4) Sy-siRNAs unique to the two specific regions, that is Regions 1/2 of the transgene GFP792 mRNA, are generated by transitive silencing in systemic young leaves.
Influence of DCLs on Biogenesis of 21, 22, and 24 nt L-siRNAs for Systemic Transgene GFP792 Silencing
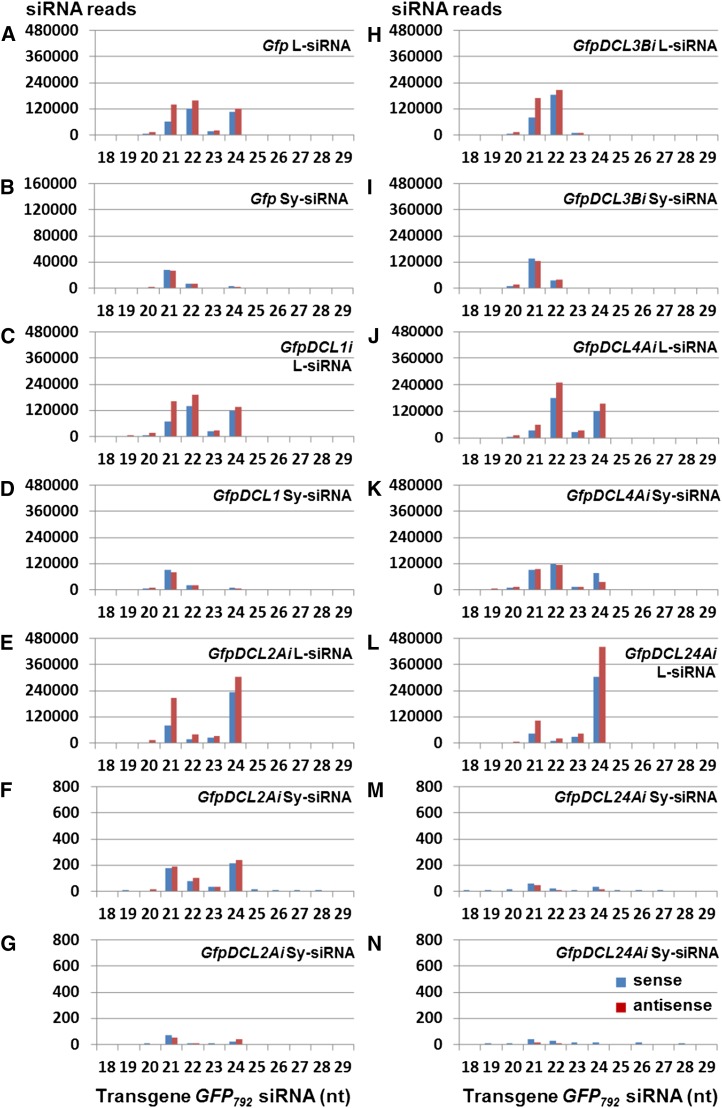
To further examine the association of L-siRNAs with systemic GFP792 PTGS in Gfp (Fig. 8, A and B), GfpDCL1i (Fig. 8, C and D), GfpDCL2Ai (Fig. 8, E–G), GfpDCL3Bi (Fig. 8, H and I), GfpDCL4Ai (Fig. 8, J and K), and GfpDCL24Ai (Fig. 8, L–N), we analyzed the size distribution of sense and antisense l- and Sy-siRNAs (Fig. 8). In local silencing induced by the hairpin GFP714 dsRNA, there were generally more antisense than sense L-siRNAs (Fig. 8, A, C, E, H, J, and L). However, in systemic GFP792 PTGS, sense- and antisense Sy-siRNAs were approximately equal (Fig. 8, B, D, F, G, I, K, M, and N). Furthermore, the distribution of 21, 22, and 24 nt L-siRNAs differed from that of Sy-siRNAs in infiltrated and systemic leaves for Gfp (Fig. 8, A and B), GfpDCL1i (Fig. 8, C and D), GfpDCL2Ai (Fig. 8, E–G), GfpDCL3Bi (Fig. 8, H and I), GfpDCL4Ai (Fig. 8, J and K), and GfpDCL24Ai (Fig. 8, L–N).
Figure 8.
Size profiles for l- and Sy-siRNAs. A to N, Fourteen sRNA libraries were generated from sRNA samples extracted from agroinfiltrated leaves at 7 dpa (A, C, E, H, J, and L) and systemic young leaves at 14 dpa (B, D, F, G, I, K, M, and N) of Gfp (A and B), GfpDCL1i (C and D), GfpDCL2Ai (E–G), GfpDCL3Bi (H and I), GfpDCL4Ai (J and K), and GfpDCL24Ai (L–N). Plants were infiltrated with agrobacterium harboring pRNAi-GFP714. Blue and red bars represent l- and Sy-siRNAs aligned to the sense and antisense strand of GFP792 mRNA, respectively. The abundance of the 22-nt L-siRNA is closely correlated with the induction and reduction of systemic transgene GFP792 silencing in systemic young leaves. Experimental design for systemic silencing assays is indicated in Supplemental Figure S9A. Systemic transgene GFP792 PTGS are shown in Figure 4, Supplemental Figure S8, and Supplemental Figure S9B.
In Gfp and GfpDCL1Ai, the siRNA profile was almost identical, but shifted from 22, 21, and 24 nt L-siRNAs (approximately 2.5:2:1 ratio for sense L-siRNA and about 1.4:1.2:1 for antisense L-siRNA) for local silencing to predominantly 21 nt along with some 22 nt (approximately 6 fold less than 21 nt) and low level of 24 nt (around 300 times less than 21 nt) Sy-siRNAs for systemic PTGS (Fig. 8, A–D). However, RNAi of DCL2, DCL3, and DCL4 had different impacts on the accumulation of 21, 22, and 24 nt l- and Sy-siRNAs. In local silencing, high levels of 24 and/or 21nt L-siRNAs were generated in GfpDCL2Ai and GfpDCL24Ai (Fig. 8, E and L). Only a very low level of Sy-siRNAs (103–105 fold less than L-siRNAs) was found in young leaves, which showed no systemic silencing (Fig. 8, G and N), although 100 to 200 or less of 21, 22, and 24 nt Sy-siRNAs were detected in leaves with minor vein-restricted systemic PTGS (Fig. 8, F and M). The decrease in the 22 nt L-siRNAs was closely correlated with the decrease in the levels of Sy-siRNAs, as well as with the abolition and/or reduction of systemic GFP792 silencing in young leaves of GfpDCL2Ai and GfpDCL24Ai (Fig. 4; Supplemental Figs. S8 and S9B).
By contrast, the enhanced systemic transgene GFP792 PTGS and the increased Sy-siRNA reads in young leaves were positively linked with the elevated levels of 22 nt L-siRNAs in local tissues of GfpDCL3Ai (Fig. 8H) and GfpDCL4Ai (Fig. 8J). However, we observed a reduction in the level of 24 nt L-siRNAs (nearly none) in GfpDCL3Bi (Fig. 8H) as well as a decrease in 21 nt L-siRNAs (3- to 4-fold less than 22 nt L-siRNAs) in GfpDCL4Ai (Fig. 8J), consistent with sRNA northern detection (Fig. 4H). Further siRNA size profiling confirmed that the levels of 22 nt L-siRNA were linked with efficient systemic transgene GFP792 PTGS, whereas variations in 21 and 24 nt L-siRNAs did not affect Sy-siRNA production and the occurrence of systemic GFP792 silencing (Supplemental Fig. S12).
Requirement of DCL2 to Respond to Mobile Systemic PTGS
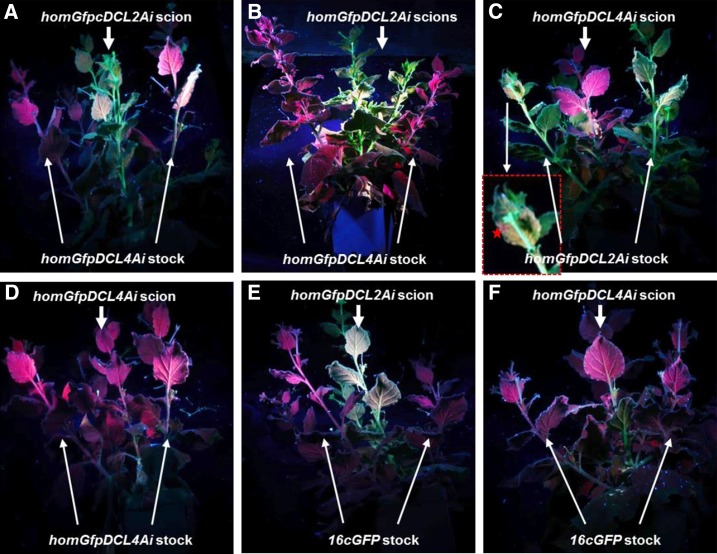
To further investigate the genetic mechanism involved in the control of systemic PTGS, we used 16cGFP, homGfpDCL4Ai, and homGfpDCL2Ai (Table I) as either rootstocks or scions in reciprocal grafting experiments (Fig. 9). When grafted onto the sense transgene GFP792-silenced homGfpDCL4Ai stocks, homGfpDCL2Ai scions developed no PTGS and showed strong GFP green fluorescence (Fig. 9, A and B). However, in the reciprocal grafting, strong sense transgene-mediated PTGS occurred in homGfpDCL4Ai scions 3 weeks after grafting (wag), even if homGfpDCL2Ai stocks had very limited transgene GFP792 silencing (Fig. 9C, inlet). In controls, efficient PTGS took place at 3 wag in homGfpDCL4Ai scions that were grafted onto GFP792-silenced homGfpDCL4Ai stocks (Fig. 9D). In a different experimental setting, homGfpDCL2Ai scions grafted onto presilenced 16cGFP stocks remained unsilenced throughout the experiments, while homGfpDCL4Ai scions developed systemic PTGS at 3 wag (Fig. 9, E and F). These results demonstrate that (1) DCL4 is unlikely required to produce and respond to mobile silencing signals for systemic PTGS, and (2) DCL2 is required in the scion to respond to the signal, but not in the rootstock to produce/send the signal.
Figure 9.
Systemic silencing in grafted homGfpDCL2Ai and homGfpDCL4Ai plants. A and B, 16cGFP, homGfpDCL2Ai, and homGfpDCL4Ai plants were used as silenced stocks to produce or as scions to receive systemic silencing signals in three experiments. No systemic transgene GFP792 silencing was observed in homGfpDCL2Ai scions grafted onto GFP792-silenced homGfpDCL4Ai stocks 3 wag (A) and 6-wag (B). C, Systemic GFP792 silencing at 3 wag in a homGfpDCL4Ai scion grafted onto a homGfpDCL2Ai stock. An enlarged section (boxed) of the stock plant showed very limited GFP792 silencing with red fluorescence marked with an asterisk in the inset leaf. D, Systemic GFP792 silencing in a homGfpDCL4Ai scion grafted onto a GFP792-silenced homGfpDCL4Ai stock at 3 wag as a control. E, A homGfpDCL2Ai scion grafted onto GFP792-silenced 16c stocks remained unsilenced at 3 wag and afterward. F, Systemic transgene GFP792 silencing in a homGfpDCL4Ai scion grafted onto a GFP792-silenced 16cGFP stock at 3 wag. All plants are photographed under long-wavelength UV illumination. Red fluorescence shows transgene GFP792 silencing while green fluorescence indicates no silencing. Scions and stocks are indicated by arrows in each panel.
DISCUSSION
Genetic insights into mobile RNA silencing and involvement of mobile signal(s) in systemic silencing remain two of the least understood, but yet most debated and divisive topics in the field of plant RNA silencing. Using a suite of DCL RNAi lines together with a GFP reporter, grafting, sRNA hybridization, and NGS approaches, we have found that gene-specific suppression of DCL2 expression reduces systemic PTGS. DCLs play central roles in the biogenesis of 21, 22, and 24 nt antisense and sense L-siRNAs in incipient cells. DCL2 is required to respond to mobile signals in recipient cells, but to produce such signals in incipient cells. DCL4 or DCL3 inhibit non-cell autonomous PTGS in systemic tissues. We also extensively profiled l- and Sy-siRNAs associated with both local hairpin dsRNA-mediated silencing and systemic transitive sense-transgene PTGS and examined how L-siRNAs generated by DCLs affect long-distance spread of PTGS. We propose that DCL2 is the key player along with DCL4 and DCL3, as well as mobile siRNAs in systemic PTGS (Fig. 10). Thus, we report several novel findings that shed light on how systemic PTGS is genetically and molecularly regulated in N. benthamiana.
Figure 10.
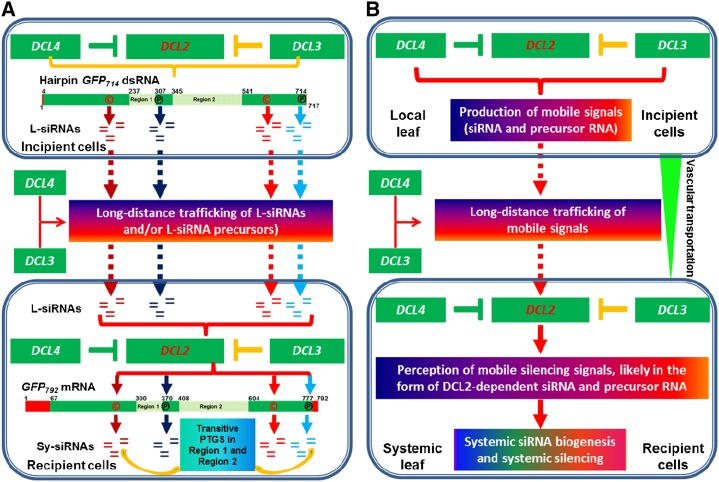
Systemic PTGS in N. benthamiana. A, A regulatory complex among DCL2, DCL3, and DCL4 contributes to biogenesis of mobile silencing signals for systemic PTGS. Regulation of DCL2 expression by DCL4 or DCL3 is demonstrated in our recent work (Qin et al., 2017). DCL1 is not included, because it seems to have no obvious effect on systemic PTGS. In this model, mobile 21, 22, and 24 nt L-siRNAs are generated from the hairpin GFP714 dsRNA by DCL2, DCL3, or DCL4 in the incipient cells of local leaf tissues. However, only the levels of the DCL2-processed 22 nt L-siRNAs, particularly those L-siRNAs matching to the four identical regions indicated by (p) (positive strand) or (c) (complementary strand) of the hairpin GFP714 and the transgene GFP792 mRNA (Fig. 5), are somewhat correlated with the DCL2-dependent induction of systemic transgene GFP792 PTGS and Sy-siRNA biogenesis in the recipient cells of systemic leaf tissues. Sy-siRNAs unique to Region 1/Region 2 of the transgene GFP792 mRNA are generated by transitive PTGS in recipient cells. Different color “=” signs represent siRNAs matching to each of the four identical sequences between GFP714 and GFP792 (Fig. 5). B, DCL2-dependent DCL genetic network in systemic silencing. Reduction of DCL2 reduces systemic PTGS, suggesting that DCL2 acts an important activator of the long-distance spread of PTGS. DCL2 is also required to perceive incoming signals, likely in the form of sense and antisense siRNAs and their long RNA precursor (although the latter is unlikely to contribute to systemic silencing), for induction of systemic siRNA biogenesis and execution of systemic PTGS in distal recipient cells. DCL4 or DCL3 may contribute to systemic silencing through their positive influences on long-distance movement of siRNAs; however, both are negative regulators (T sign) that down-regulate DCL2 expression (Qin et al., 2017). Thus, DCL2 is a key player in the systemic silencing network. Suppression of either DCL4 or DCL3 alleviates the negative control of DCL2 expression and the elevated DCL2 could then respond more effectively to mobile signals for more efficient systemic PTGS in N. benthamiana.
-
1)
We have generated transgenic DCL RNAi lines for dissecting intra/intercellular and systemic PTGS (Table I). DCL1, DCL2, DCL3, and DCL4 are responsible for the biogenesis of miRNAs and 22, 24, and 21 nt siRNAs, respectively, in N. benthamiana. DCL4 and the DCL4-processed 21 nt siRNAs are essential for local hairpin dsRNA-mediated PTGS, while DCL2 or DCL3 and their cognate 22 or 24 nt L-siRNAs are not critical for such intracellular silencing (Fig. 1).
-
2)
Local PTGS induced by the hairpin GFP714 dsRNA can lead to efficient biogenesis of mobile L-siRNAs in incipient cells and local leaf tissues (Figs. 1–3). Our experimental system (Supplemental Fig. S3A) avoids the amplification and cascading production of siRNAs in remote recipient cells and enables unambiguous detection of different sized L-siRNAs in systemic leaves. However, neither long (full-length or partial) GFP714 RNA (Supplemental Fig. S7) nor agrobacterium-originated siRNA (Supplemental Table S2) was detectable in young leaves, although these results do not rule out the possibility that long RNAs are present and serve as the mobile signal. Moreover, the total number of GFP714 L-siRNA (and total sRNA) reads in local leaf tissues and their distributions across the GFP714 RNA are almost identical among Nb, RDR6i, and all DCL RNAi lines (Figs. 2 and 3). The L-siRNAs detected in systemic leaves are mainly 22 and 21 nt, but some are 24 nt (Fig. 2). These mobile L-siRNAs were not particularly abundant in systemic young leaf tissues, but consistent with the levels of mobile siRNAs reported in Arabidopsis (Molnar et al., 2010). These data indicate that all sized sense and antisense L-siRNAs are capable of trafficking over long distances (Fig. 10A). Compared to Nb, the reduced levels of L-siRNAs in distal tissues of all RNAi lines suggest that RDR6 and DCLs, particularly DCL3 and DCL4, contribute to the systemic spread of L-siRNAs (Fig. 2). Indeed, genetic analyses have demonstrated that RDR6 is required for perception of long-distance silencing signals (Schwach et al., 2005; Melnyk et al., 2011b) and DCL2 responds to mobile silencing for efficient systemic PTGS in plants (Fig. 4; Taochy et al., 2017). However, even if locally produced siRNAs are mobile, this does not prove that siRNAs are the signal.
-
3)
Genetic analysis indicates that DCL1 is unlikely to be involved in local hairpin dsRNA-mediated PTGS, although a slightly elevated level of target mRNA was detected in DCL1i plants (Fig. 1) and sense-transgene induced systemic PTGS (Figs. 4). This conclusion is supported by the identical profiles of L-siRNAs or Sy-siRNAs in both Gfp and GfpDCL1i plants (Figs. 6–8). Further, the altered size profiles between l- and Sy-siRNAs in Gfp and GfpDCL1i (Fig. 8) implies that the local and systemic PTGS are likely two linked processes with distinct molecular and genetic requirements. Indeed, local PTGS is induced by a hairpin GFP714 RNA that directly produces long dsRNA, which differs from the sense transgene GFP792-mediated PTGS in systemic leaf tissues. In the latter case, dsRNA is not produced directly, but its production from ssRNA requires the activity of several genes, including RDR6 and DCL2 (Mlotshwa et al., 2008).
-
4)
DCL2 RNAi reduces systemic PTGS and blocks the exit of the mobile silencing signal from vascular tissues to the surrounding mesophyll cells, indicating that DCL2 is required for long-distance spread of PTGS (Fig. 4), consistent with DCL2 promoting, while DCL4 inhibits, cell-to-cell spread of VIGS (Qin et al., 2017). Moreover, DCL2 is required to respond to mobile signals for systemic PTGS (Fig. 9), supported by a recent report of a critical role of DCL2 in systemic PTGS in Arabidopsis (Taochy et al., 2017). DCL2 is thus involved in promoting the long-distance (leaf-to-leaf) and short-distance movement (vascular cells to neighboring cells) of PTGS in addition to its role in generating the 22 nt siRNAs. On the other hand, our grafting experiments showed that DCL2 is not required in the rootstock for producing/sending the signal (Fig. 9). However, in contrast to complete loss-of-function mutants, RNAi lines are partial loss-of-function. It thus remains possible that in the DCL2i rootstock, residual DCL2 could still produce some transportable molecules, and such signals could then be perceived by DCL2 in recipient cells where DCL4 is knocked-down by RNAi to trigger systemic transitive PTGS (Fig. 9). Taochy et al. (2017) have elegantly shown that Arabidopsis DCL2 is required in both the source rootstock and recipient shoot tissue for RDR6-dependent systemic PTGS, although this latest discovery is in contrast to their previous finding (Brosnan et al., 2007).
-
5)
Contrary to DCL2, DCL4 and DCL3 inhibit sense transgene-mediated systemic PTGS, as RNAi of DCL4 or DCL3 enhances systemic PTGS in distant young leaves (Fig. 4). Our data also reveal that DCL4 has an epistatic effect on DCL2, thereby influencing systemic PTGS in N. benthamiana (Fig. 4, I and J). Only DCL2 is required to respond to mobile signals for systemic PTGS in the reciprocal DCL2i-DCL4i grafting experiments (Fig. 9), and RNAi of DCL4 or DCL3 has been found to up-regulate DCL2 expression (Qin et al., 2017). Taken together, our findings suggest that a DCL2-dependent DCL genetic network regulates non-cell autonomous systemic silencing (Fig. 10B). A hierarchical interaction between DCL4 and DCL2 to affect cell autonomous PTGS was also reported in Arabidopsis (Bouché et al., 2006; Henderson et al., 2006; Xie et al., 2005).
-
6)
Our work implicates or excludes the involvement of certain types of RNAs in mobile systemic PTGS (Fig. 10A). First, combined with earlier elegant work showing that DCL2 is required for transitive silencing (Mlotshwa et al., 2008; Parent et al., 2015), our results suggest that the plant’s response to the signal requires a set of genes that are unique to transitive silencing (i.e. genes that play a role in the production of dsRNA). Thus, these results suggest that the signal for systemic silencing in plants is not itself dsRNA, in contrast to dsRNA being required for the systemic spread of RNAi in animals (Jose et al., 2011).
Second, detection of L-siRNAs in systemic tissues implies that mobile L-siRNAs might contribute to systemic PTGS. This notion is supported by the l- and Sy-siRNA profiles associated with the hairpin GFP714 dsRNA-mediated local silencing and systemic transgene GFP792 silencing. The identical distributions of the four clusters of l- and Sy-siRNAs across nucleotides 625 to 742 and 217 to 282, 730 to 773 and 363 to 402 of the transgene GFP792 mRNA and the equivalent sequences of the hairpin GFP714 dsRNA (Fig. 5, labeled (c) or (p)) suggest that L-siRNAs originated from these parts of the hairpin GFP714 dsRNA might represent a component of mobile signals for the biosynthesis of Sy-siRNA and for systemic induction of GFP792 silencing in distal recipient cells (Figs. 6 and 7, asterisk). Consistent with this, the abundance of the four clusters of Sy-siRNAs associated with systemic GFP792 PTGS are much less than the corresponding L-siRNAs associated with local silencing induced by the hairpin GFP714 dsRNA in control and all RNAi lines (Figs. 6 and 7, asterisk). Furthermore, L-siRNAs specific to Regions 1/2 of the hairpin GFP714 dsRNA were found in systemic leaf tissues (Fig. 7). By contrast, Sy-siRNAs unique to Regions 1/2 of the transgene GFP792 mRNA (these two regions have different sequences between GFP714 and GFP792; Fig. 5) were not found in local leaves where primary silencing efficiently occurred (Fig. 6). These results suggest that Sy-siRNAs distinct to Regions 1/2 of the GFP792 mRNA must have been generated from transitive sense-transgene silencing in systemic leaf cells and tissues. Such transitive PTGS was likely induced by secondary siRNAs that were produced from primary silencing of the GFP792 mRNA sequences, directly targeted by the four clusters of mobile L-siRNAs that were generated from the identical GFP714 sequences in local PTGS (Figs. 6–8; Fig. 10A). However, these results do not exclude the possible involvement of other types of RNAs in systemic RNA silencing in plants.
Third, the 21 or 24 nt L-siRNAs processed by DCL4 or DCL3 and their precursor RNAs are unlikely to be a major component of the mobile signal for systemic PTGS (Fig. 8), in line with our genetic and functional analysis of DCLs in systemic silencing (Fig. 4). That suppression of DCL4 or DCL3 enhanced systemic silencing and the levels of 21 or 24 nt L-siRNAs were not correlated with the biogenesis of GFP792 Sy-siRNAs (Fig. 8) and the intensity of systemic PTGS in all GfpDCL RNAi lines (Fig. 4) provides evidence to support such a conclusion. This is also consistent with the fact that DCL4 and DCL3 proteins are not required for the production of the sRNA signals in Arabidopsis (Brosnan et al., 2007; Taochy et al., 2017).
Fourth, the levels of DCL2-processed 22 nt L-siRNAs were weakly correlated with the induction of systemic PTGS. Mapping l- and Sy-siRNAs onto the transgene GFP792 mRNA and hairpin GFP714 dsRNA identified sense and antisense 22 nt L-siRNAs that might be required for systemic GFP792 PTGS in distal recipient cells. It is evident that the varied levels of the 22 nt L- and Sy-siRNAs mapped to the transgene GFP792 antisense sequences 625 to 742 and 217 to 282, and sense strand 730 to 773 and 363 to 402 (Supplemental Fig. S12) were linked to the induction of systemic GFP792 silencing in each RNAi lines (Fig. 4). This is also consistent with the high levels of 21, 22, and 24 nt Sy-siRNAs in GfpDCL4Ai, likely resulting from DCL2 and DCL2-processed 22 nt siRNAs that may trigger efficient biosynthesis of secondary Sy-siRNAs in the systemic leaf tissues (Chen et al., 2010; Cuperus et al., 2010; Mlotshwa et al., 2008). However, these findings per se are not direct evidence that DCL2-processed 22 nt L-siRNAs and/or their precursors are the bona fide signals for induction of Sy-siRNA biogenesis and systemic transitive PTGS.
Nonetheless, the molecular nature of systemic PTGS signals needs further characterization. DCL4-processed 21 nt siRNA or DCL3-processed 24 nt siRNA and their precursors may not be involved in long-distance PTGS signaling, even though 21 and 24 nt siRNAs act primarily as intra- or intercellular triggers for RNA-directed degradation of target mRNA or RNA-directed DNA methylation (Lewsey et al., 2016; Melnyk et al., 2011a, 2011b; Sarkies and Miska, 2014). In Arabidopsis, the DCL3-processed 24 nt siRNAs are thought to be the main signals for systemic TGS (Lewsey et al., 2016; Melnyk et al., 2011a; Molnar et al., 2010). However, we have not examined the roles of DCLs and mobile siRNAs in systemic TGS in N. benthamiana. Furthermore, whether the DCL2-processed/dependent 22 nt siRNAs and/or their precursors could serve as signals for systemic PTGS requires further investigation, even though 22 nt sRNAs can effectively trigger biogenesis of secondary siRNAs of various sizes in plants (Chen et al., 2010; Cuperus et al., 2010; Mlotshwa et al., 2008) and DCL2 and DCL2-processed 22 nt siRNAs are involved in the cell-to-cell spread of VIGS in N. benthamiana (Qin et al., 2017).
Our findings also differ from early works showing that long-distance movement of transitive silencing occurs in the absence of sRNAs in the rootstock (Mallory et al., 2001), and that no specific sRNAs are associated with systemic silencing in Arabidopsis (Brosnan et al., 2007; Taochy et al., 2017). Considering the transitivity and sequence-specificity of RNA silencing along with the fact that various types of RNA molecules can trigger cell-autonomous silencing, we concede that various forms of transportable RNAs, if they can be perceived and processed by DCL2 or a DCL2-dependent pathway in recipient cells, could function as systemic silencing signals (Fig. 10A). On the other hand, DCL4 and DCL3 could affect DCL2-mediated systemic PTGS through their negative regulation of DCL2 expression (Qin et al., 2017), which would indirectly influence DCL2 activity in systemic PTGS. Additionally, DCLs may compete with each other for dsRNA substrates. The loss of DCL4 or DCL3 could enhance the DCL2 output, because the level of dsRNA substrate for DCL2 is increased in both incipient and recipient cells. Cross-regulation among DCLs could also occur at transcriptional, posttranscriptional, and/or translational levels. These mechanisms may act synergistically for DCLs to maximize production of the mobile signals for systemic PTGS (Fig. 10B), in contrast to what has been described in Arabidopsis (Melnyk et al., 2011b; Sarkies and Miska, 2014). Nevertheless, findings from different non-cell autonomous silencing systems may not necessarily be exclusive of each other. There may be various routes to mobile silencing in different plants. N. benthamiana is a natural rdr1 mutant and might behave differently compared to Arabidopsis (Nakasugi et al., 2013). The occurrence of cell-type or tissue dependent functions and specificities of different DCL proteins or different domain arrangements of the DCL proteins has also been reported in N. benthamiana (Andika et al., 2015; Nakasugi et al., 2013).
In conclusion, along with our recent work (Qin et al., 2017), we demonstrate that a complex genetic and regulatory network involving DCL2, DCL4, and DCL3 may govern the rate and characteristics of the mobile PTGS (Fig. 10). DCL2 plays a center role in response to mobile signals for systemic PTGS in distal recipient cells. We have also highlighted the potential (non)-involvement of RNAs, including different sized siRNAs, in systemic PTGS. Arabidopsis DCLs have distinct functions in mobile silencing, but in N. benthamiana their counterparts appear to behave similarly in intracellular silencing to generate miRNAs and 21, 22, or 24 nt siRNAs (this study; Katsarou et al., 2016; Qin et al., 2017), but act coordinately to fulfill different roles in systemic PTGS. Thus, our work provides a new framework to test whether the DCL2-centered DCL genetic pathway for systemic PTGS is the representative model for the majority of plant species and to unravel mobile signals for systemic PTGS in plants, as well as in (and across) organisms of different kingdoms.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Non-transformed Nicotiana benthamiana and transgenic N. benthamiana plants (Table I) were grown and maintained in insect-free glasshouses and growth-rooms at 25°C with supplementary light (13,000 Lux intensity/Visible 40W LED Light Bulbs) to give a 16-h photoperiod.
Plasmid Constructs and Plant Transformation
The GFP714 coding sequence (Ryabov et al., 2004) was PCR-amplified and cloned between the duplicated 35S CaMV promoter and the NOS terminator of pJG045, a pCAMBIA1300-based vector, to produce p35S-GFP714 (Supplemental Fig. S1A). To generate gene-specific hairpin DCL- and GFP714-RNAi constructs (Supplemental Fig. S1B), cDNA fragments corresponding to nucleotides 4835 to 5075 of DCL1 (GenBank no. FM986780), nucleotides 3382 to 3582 of DCL2 (GenBank no. FM986781), nucleotides 4167 to 4347 of DCL3 (GenBank no. FM986782), nucleotides 3283 to 3482 of DCL4 (GenBank no. FM986783), or the full-length GFP714 (nucleotides 1–714) sequence were cloned into the RNAi vector pRNAi-LIC as described (Xu et al., 2010). There is no sequence similarity among the DCL fragments (Supplemental Table S1; Supplemental Data Set S1). The full-length GFP714 sequence (Ryabov et al., 2004) was also cloned into pMD18-T (Takara) to produce pT7.GFP from which GFP714 RNA transcripts were produced by in vitro transcription using the T7 RNA polymerase. Primers used for these constructs are listed in Supplemental Table S5. All constructs were confirmed by DNA sequencing.
pRNAi-DCL1, pRNAi-DCL2, pRNAi-DCL3, and pRNAi-DCL4 were transformed into the Agrobacterium tumefaciens LBA4404, and pRNAi-GFP714 and p35S-GFP714 were introduced into A. tumefaciens EHA105. Transgenic N. benthamiana plants were generated by Agrobacterium-mediated leaf disc transformation as described (Hong et al., 1996). Two independent homozygous lines (DCL2Ai and DCL2Bi, DCL3Ai and DCL3Bi, DCL4Ai and DCL4Bi) with a single copy of each RNAi construct were obtained. Line DCL4Bi was crossed with DCL2Ai to produce DCL24i. DCLi lines were crossed with the GFP transgenic line 16cGFP that constitutively expresses GFP792 (Haseloff et al., 1997; Ruiz et al., 1998) to produce GfpDCL2Ai, GfpDCL2Bi, GfpDCL3Ai, GfpDCL3B, GfpDCL4Ai, GfpDCL4Bi, GfpDCL24Ai, and GfpDCL24Bi, respectively. At the later stage of this project, we also generated homozygous homGfpDCL2Ai, homGfpDCL3Bi, homGfpDCL4Ai, and GfpDCL24Ci through selfing. Only one hemizygous DCL1i or GfpDCL1i line was generated through direct transformation of N. benthamiana or 16cGFP with pRNAi-DCL1 (Supplemental Fig. S1B). All transgenic lines used in this study are summarized in Table I. It should be noted that DCL RNAi is gene-specific and has no obvious off-target effect on other DCL and RNA silencing-associated genes (Supplemental Data Set S1; Supplemental Fig. S5 and S10).
Local RNA Silencing and Small RNA Mobility Assay
The experimental design for local RNA silencing and small RNA mobility assay to detect long-distance movement of siRNAs from local to systemic young leaves is outlined in Supplemental Figure S3A. Two young leaves per N. benthamiana or DCLi plant (six plants at the six-leaf stage in each experiment) were co-infiltrated with 1 OD600 agrobacterium harboring p35S-GFP714 and agrobacterium harboring pRNAi-GFP714 in repeated experiments. In co-infiltrated leaf cells, pRNAi-GFP714 is expected to generate hairpin GFP714 dsRNA that are diced into siRNAs. These siRNAs targeted and degraded GFP714 mRNA transcribed from the p35S-GFP714 expression cassette, leading to disappearance of GFP fluorescence. Green fluorescence intensity was measured using ImageJ software following the software provider’s guidance (https://imagej.nih.gov/ij/). Infiltrated leaf tissues from three to four different plants at 7 d post agro-infiltration (dpa), and 4 newly developed young leaves of each of 3 to 4 different plants at 14 dpa were collected and pooled for sRNA extraction and construction of sRNA libraries.
Systemic RNA Silencing and Grafting Experiments
Systemic RNA silencing was performed for each line as outlined in Supplemental Figure S9A. Briefly, leaves from six seedlings at the six-leaf stage of transgenic GFP792 line Gfp and GfpDCL RNAi lines were agroinfiltrated with freshly cultured agrobacterium (1 OD600) carrying pRNAi-GFP714 as previously described (Hong et al., 2003). Plant grafting was performed as described by Schwach et al. (2005). Induction and spread of GFP silencing was routinely examined under long-wavelength UV light and recorded photographically using a Nikon Digital Camera D7000. Red halos on the infiltrated leaves and regions of leaf lamina where GFP silencing occurred show red chlorophyll fluorescence, while tissues expressing GFP show green fluorescence under long-wavelength UV light. Local and systemic RNA silencing assays were performed for each line in at least two separate experiments. N. benthamiana is an allotetraploid (Nakasugi et al., 2014); however, the specific RNAi constructs described above only knock down specific DCL expression in spite of the homeologous and duplicated gene copies in the genome. This should not interfere with the analysis of the impact of DCL RNAi on cell and noncell autonomous RNA silencing. Agroinfiltrated leaf tissues were collected from 3 to 4 plants at 7 dpa and pooled for sRNA extraction and construction of local sRNA libraries. Young leaves with systemic GFP792 PTGS from 3 to 4 plants of each of the Gfp, GfpDCL1i, GfpDCL3Bi, and GfpDCL4Ai lines, leaves with weak vein-restricted systemic PTGS from 3 to 4 plants of each of the GfpDCL2Ai and GfpDCL24Ai lines, or young leaves without systemic PTGS from 3 to 4 plants of each of the GfpDCL2Ai and GfpDCL24Ai lines were collected at 14 dpa and pooled for sRNA extraction and construction of systemic sRNA libraries.
RNA Extraction and Northern Detection of mRNA and siRNA
For RT-PCR and quantitative real-time PCR (RT-qPCR), total RNAs were extracted from leaf tissues using the RNAprep Pure Plant Kit as recommended by the manufacturer (Tiangen). For northern blot, total RNAs were extracted from leaf tissues with TRIzol reagent as recommended by the manufacturer (Invitrogen). RNAs (5 μg) extracted from infiltrated tissues were separated on a 1% formaldehyde agarose gel, transferred to Hybond-N+ membranes (Amersham Biosciences) by upward capillary transfer in 20× SSC buffer, then cross-linked to the membrane with an UVP CX 2000 UV crosslinker four times (upside, downside, upside, downside) at 120 mJ/cm2, 1 min each. Membranes were hybridized with digoxigenin-labeled GFP DNA probes prepared using the DIG High Prime DNA Labeling Kit (Roche). RNAs were detected using a DIG Nucleic Acid Detection Kit (Roche) as recommended by the manufacturer. Chemilluminescent signals were detected with a ChemiDoc XRS+ imaging System (Bio-Rad).
To analyze siRNAs, low-molecular-mass sRNAs were enriched from total RNA as described (Hamilton and Baulcombe, 1999). The enriched sRNAs (2.5 μg) were fractionated on an 18% denaturing polyacrylamide/7 m urea gel in 1 x Tris-borate-EDTA buffer. Small RNAs were transferred to Hybond-N+ membranes (Amersham Biosciences) by upward capillary transfer in 20× SSC buffer, then cross-linked to the membranes with an UVP CX 2000 UV Crosslinker four times (upside, underside, upside, underside) at 120 mJ/cm2, for 1 min each time. The membranes were hybridized with digoxigenin (Dig)-labeled GFP RNA probes using a DIG RNA Labeling Kit (Roche) as recommended by the manufacturer. The hybridization chemiluminescence signals were detected with a ChemiDoc XRS+ imaging System (Bio-Rad).
RT-PCR and RT-qPCR
First-strand cDNA was synthesized using 1 μg total RNAs that were pretreated with 1 unit RNase-free DNase I as templates by the M-MLV Reverse Transcriptase (Promega). RT-PCR analyses of GFP714 RNA were performed using various sets of primers listed in Supplemental Table S5. The RT-qPCR analyses of DCL mRNA levels were performed using DCL-specific primers (Supplemental Table S5) and the SYBR Green Mix. The amplification program for SYBR Green I was performed at 95°C for 10 s, 58°C for 30 s, and 72°C for 20 s on a CFX96 real-time PCR detection system (Bio-Rad) following the manufacturer’s instructions. Quadruplicate quantitative assays were performed on cDNA of each of the four biological duplicates (leaf tissues from four different treated plants). The relative quantification of DCL mRNA was calculated using the Equation 2–ΔΔCt and normalized to the amount of GAPDH transcripts (GenBank accession no. TC17509) as described (Qin et al., 2012). RT-qPCR data between control and various treatments were analyzed by Student’s t test (http://www.physics.csbsju.edu/stats/t-test.html). The statistical significance threshold was P ≤ 0.05.
Construction of sRNA Library and sRNA NGS
Fragments of 18- to 30-bases-long RNA were isolated from total RNA extracted from pooled samples of three to four different plants for each of the biological and technical duplicates after being separated through 15% denaturing PAGE. The pooled samples contained leaf tissues from three to six leaves per plant. Then sRNAs were excised from the gel and sequentially ligated to a 3′ adapter and a 5′ adapter. After each ligation step, sRNAs were purified after 15% denaturing PAGE. The final purified ligation products were reverse-transcribed into cDNA using reverse transcriptase (Finnzymes Oy). The first-strand cDNA was PCR amplified using Phusion* DNA Polymerase (Finnzymes Oy). The purified DNA fragments were used for clustering and sequencing by Illumina HisEquation 2000 (Illumina) at the Beijing Genomics Institute.
Bioinformatics Analysis of sRNA Sequences
Illumina HighSEquation 2000 sequencing produced a similar number of 11 to 12 million reads per sRNA library. The reads were cropped to remove adapter sequences and were aligned to the reference sequences using Bowtie2 (Langmead and Salzberg, 2012). Reference sequences include hairpin GFP714 (Ryabov et al., 2004), GFP792 transgene (Haseloff et al., 1997), DCL1, DCL2, DCL3, and DCL4 gene sequences (Nakasugi et al., 2013) and a set of 50 tobacco miRNAs identified in Nicotiana plants (Pandey et al., 2008; Figure 5; Supplemental Data Set S1). SAMtools pileup was used to produce the siRNA and miRNA coverage profiles (Li et al., 2009). For correlation analyses for sRNA libraries, the number of miRNA hits corresponding to the previously identified set of 50 Nicotiana miRNAs was determined. All analyzed sRNA libraries for local and systemic leaf samples contained similar proportions of host-encoded miRNA reads (Nakasugi et al., 2013; Ryabov et al., 2014), indicating equivalence and direct comparability of the datasets and no variation due to the efficiency of library preparation and sequencing (Supplemental Tables S2 and S7; Supplemental Figures S5, S6, S10, and S11). Outcomes of comparisons between normalized siRNAs against the total sRNA reads (per 10 million sRNA reads) are consistent with the numbers of siRNA reads directly compared.
Accession Numbers
DCL1: GenBank number FM986780; DCL2: GenBank number FM986781; DCL3: GenBank number FM986782; DCL4: GenBank number FM986783; and GAPDH: GenBank accession number TC17509.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Nucleotide sequence similarity (%) between DCL fragments in the pRNAi-DCLs constructs.
Supplemental Table S2. Summary of sRNAseq datasets of 18 GFP714 sRNA libraries for long-distance spread of siRNAs.
Supplemental Table S3. Correlation analyses of miRNA profiles among agro-infiltrated leaf sRNA libraries (Nb background).
Supplemental Table S4. Correlation analyses of miRNA profiles among systemic leaf sRNA libraries (Nb background).
Supplemental Table S5. Primers used in this study.
Supplemental Table S6. Impact of DCL RNAi on systemic PTGS.
Supplemental Table S7. Summary of sRNAseq datasets of 14 sRNA libraries for local and systemic PTGS.
Supplemental Table S8. Correlation analyses of miRNA profiles among agro-infiltrated leaf sRNA libraries (16cGFP transgenic background).
Supplemental Table S9. Correlation analyses of miRNA profiles among systemic leaf sRNA libraries (16cGFP transgenic background).
Supplemental Figure S1. Construction of gene expression and RNAi vectors.
Supplemental Figure S2. RNAi effects on DCL gene expression.
Supplemental Figure S3. NGS detection of total small RNAs in local and systemic leaves.
Supplemental Figure S4. Effect of DCL RNAi on local GFP714 RNA silencing.
Supplemental Figure S5. DCL RNAi does not cause off-target silencing of genes associated with RNAi pathways in Nb background plants: sRNA reads in 18 sRNA libraries.
Supplemental Figure S6. miRNA reads in 18 sRNA libraries (Nb background).
Supplemental Figure S7. No long-distance movement of large GFP714 RNA.
Supplemental Figure S8. Impact of DCL RNAi on systemic PTGS.
Supplemental Figure S9. Size profiles for total sRNAs in local and systemic leaves.
Supplemental Figure S10. DCL RNAi does not cause off-target silencing of genes associated with RNAi pathways in 16cGFP transgenic background plants: sRNA reads in 14 sRNA libraries.
Supplemental Figure S11. miRNA reads in 14 sRNA libraries (16cGFP transgenic background).
Supplemental Figure S12. Specific 21-24nt siRNA distributions across the transgene GFP792 mRNA.
Supplemental Data Set S1. Reference sequences used in this study.
Supplemental Data Set S2. DCL RNAi does not cause off-target silencing of genes associated with RNAi pathways in Nb backgound plants: sRNA reads in 18 sRNA libraries.
Supplemental Data Set S3. miRNA reads in 18 sRNA libraries (Nb background).
Supplemental Data Set S4. DCL RNAi does not cause off-target silencing of genes associated with RNAi pathways in 16cGFP transgenic background plants: sRNA reads in 14 sRNA libraries.
Supplemental Data Set S5. miRNA reads in 14 sRNA libraries (16cGFP transgenic background).
Acknowledgments
We are indebted to David Baulcombe for his kind gift of 16cGFP, RDR6i, and GfpRDR6i seeds and for his constructive and critical comments on the manuscript. The corresponding author thanks Mahmut Tör for reading the manuscript. No conflict of interest is declared.
Footnotes
This work was supported by grants from the National Natural Science Foundation of China (NSFC 31370180 to Y.H.); Ministry of Agriculture of the People’s Republic of China (the National Transgenic Program of China 2016ZX08009001-004 to Y.H.); Ministry of Science and Technology of the People's Republic of China (2017YFE0110900 to Y.H.); Hangzhou Normal University (Pandeng Program 201108 to Y.H.); the Hangzhou City Government (Innovative Program for Science Excellence 20131028 to Y.H.); NSFC (31601765 to W.C., 31201490 to X.Z., 31500251 to C.Q.); Zhejiang Provincial Natural Science Foundation (LY15C140006 to X.Z., LY14C010005 to N.S.); and the UK Biotechnology and Biological Sciences Research Council (UK-China Partnering Award BB/K021079/1 to S.J. and Y.H.).
Articles can be viewed without a subscription.
References
- Andika IB, Maruyama K, Sun L, Kondo H, Tamada T, Suzuki N (2015) Differential contributions of plant Dicer-like proteins to antiviral defences against potato virus X in leaves and roots. Plant J 81: 781–793 [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Berg JM. (2016) Retraction. Science 354: 190. [DOI] [PubMed] [Google Scholar]
- Bouché N, Lauressergues D, Gasciolli V, Vaucheret H (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25: 3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan CA, Mitter N, Christie M, Smith NA, Waterhouse PM, Carroll BJ (2007) Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc Natl Acad Sci USA 104: 14741–14746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH (2010) 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC (2010) Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanapally S, Ravikumar S, Jose AM (2015) Double-stranded RNA made in C. elegans neurons can enter the germline and cause transgenerational gene silencing. Proc Natl Acad Sci USA 112: 2133–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O (2005) DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet 37: 1356–1360 [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Hunter CP (2003) Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301: 1545–1547 [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC (2010) Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22: 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38: 721–725 [DOI] [PubMed] [Google Scholar]
- Hong Y, Saunders K, Hartley MR, Stanley J (1996) Resistance to geminivirus infection by virus-induced expression of dianthin in transgenic plants. Virology 220: 119–127 [DOI] [PubMed] [Google Scholar]
- Hong Y, Stanley J, van Wezel R (2003) Novel system for the simultaneous analysis of geminivirus DNA replication and plant interactions in Nicotiana benthamiana. J Virol 77: 13315–13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose AM, Garcia GA, Hunter CP (2011) Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Nat Struct Mol Biol 18: 1184–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose AM, Smith JJ, Hunter CP (2009) Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc Natl Acad Sci USA 106: 2283–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsarou K, Mavrothalassiti E, Dermauw W, Van Leeuwen T, Kalantidis K (2016) Combined activity of DCL2 and DCL3 is crucial in the defense against potato spindle tuber viroid. PLoS Pathog 12: e1005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewsey MG, Hardcastle TJ, Melnyk CW, Molnar A, Valli A, Urich MA, Nery JR, Baulcombe DC, Ecker JR (2016) Mobile small RNAs regulate genome-wide DNA methylation. Proc Natl Acad Sci USA 113: E801–E810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Ely L, Smith TH, Marathe R, Anandalakshmi R, Fagard M, Vaucheret H, Pruss G, Bowman L, Vance VB (2001) HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk CW, Molnar A, Bassett A, Baulcombe DC (2011a) Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr Biol 21: 1678–1683 [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Molnar A, Baulcombe DC (2011b) Intercellular and systemic movement of RNA silencing signals. EMBO J 30: 3553–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S, Pruss GJ, Peragine A, Endres MW, Li J, Chen X, Poethig RS, Bowman LH, Vance V (2008) DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS One 3: e1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC (2010) Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875 [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Campos H, Kolaczkowski B (2013) Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol 30: 627–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasugi K, Crowhurst R, Bally J, Waterhouse P (2014) Combining transcriptome assemblies from multiple de novo assemblers in the allo-tetraploid plant Nicotiana benthamiana. PLoS One 9: e91776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasugi K, Crowhurst RN, Bally J, Wood CC, Hellens RP, Waterhouse PM (2013) De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS One 8: e59534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Shahi P, Gase K, Baldwin IT (2008) Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata. Proc Natl Acad Sci USA 105: 4559–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible W-R (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent J-S, Bouteiller N, Elmayan T, Vaucheret H (2015) Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J 81: 223–232 [DOI] [PubMed] [Google Scholar]
- Qin C, Li B, Fan Y, Zhang X, Yu Z, Ryabov E, Zhao M, Wang H, Shi N, Zhang P, et al. (2017) Roles of DCL2 and DCL4 in Intra- and Intercellular Antiviral Silencing in Nicotiana benthmiana. Plant Physiol 174: 1067–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Shi N, Gu M, Zhang H, Li B, Shen J, Mohammed A, Ryabov E, Li C, Wang H, et al. (2012) Involvement of RDR6 in short-range intercellular RNA silencing in Nicotiana benthamiana. Sci Rep 2: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci USA 105: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabov EV, van Wezel R, Walsh J, Hong Y (2004) Cell-to-cell, but not long-distance, spread of RNA silencing that is induced in individual epidermal cells. J Virol 78: 3149–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, Mead A, Burroughs N, Evans DJ (2014) A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog 10: e1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Miska EA (2014) Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat Rev Mol Cell Biol 15: 525–535 [DOI] [PubMed] [Google Scholar]
- Searle IR, Pontes O, Melnyk CW, Smith LM, Baulcombe DC (2010) JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 24: 986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC (2005) An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol 138: 1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih JD, Hunter CP (2011) SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 17: 1057–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitis DS, Benkovics AH, Husbands AY, Timmermans MCP (2017) Boundary formation through a direct threshold-based readout of mobile small RNA gradients. Dev Cell 43: 265–273.e6 [DOI] [PubMed] [Google Scholar]
- Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC (2007) An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19: 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taochy C, Gursanscky NR, Cao J, Fletcher SJ, Dressel U, Mitter N, Tucker MR, Koltunow AMG, Bowman JL, Vaucheret H, et al. (2017) A genetic screen for impaired systemic RNAi highlights the crucial role of DICER-LIKE 2. Plant Physiol 175: 1424–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW (2011) The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23: 1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H (2013) Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342: 118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP (2002) Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295: 2456–2459 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Sui N, Tang Y, Xie K, Lai Y, Liu Y (2010) One-step, zero-background ligation-independent cloning intron-containing hairpin RNA constructs for RNAi in plants. New Phytol 187: 240–250 [DOI] [PubMed] [Google Scholar]
- Yelina NE, Smith LM, Jones AM, Patel K, Kelly KA, Baulcombe DC (2010) Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc Natl Acad Sci USA 107: 13948–13953 [DOI] [PMC free article] [PubMed] [Google Scholar]