The barley ELIGULUM-A gene regulates lateral branch development and acts to establish the blade-sheath boundary during leaf development.
Abstract
The shoot apical and axillary meristems control shoot development, effectively influencing lateral branch and leaf formation. The barley (Hordeum vulgare) uniculm2 (cul2) mutation blocks axillary meristem development, and mutant plants lack lateral branches (tillers) that normally develop from the crown. A genetic screen for cul2 suppressors recovered two recessive alleles of ELIGULUM-A (ELI-A) that partially rescued the cul2 tillering phenotype. Mutations in ELI-A produce shorter plants with fewer tillers and disrupt the leaf blade-sheath boundary, producing liguleless leaves and reduced secondary cell wall development in stems and leaves. ELI-A is predicted to encode an unannotated protein containing an RNaseH-like domain that is conserved in land plants. ELI-A transcripts accumulate at the preligule boundary, the developing ligule, leaf margins, cells destined to develop secondary cell walls, and cells surrounding leaf vascular bundles. Recent studies have identified regulatory similarities between boundary development in leaves and lateral organs. Interestingly, we observed ELI-A transcripts at the preligule boundary, suggesting that ELI-A contributes to boundary formation between the blade and sheath. However, we did not observe ELI-A transcripts at the axillary meristem boundary in leaf axils, suggesting that ELI-A is not involved in boundary development for axillary meristem development. Our results show that ELI-A contributes to leaf and lateral branch development by acting as a boundary gene during ligule development but not during lateral branch development.
Leaves and tillers, the vegetative branches that form at the base of grass plants, are key determinants of grass shoot architecture. Tillers develop from axillary meristems and undergo three distinct morphological stages: (1) initiation of an axillary meristem in the leaf axil; (2) development of leaf primordia on the axillary meristem to form an axillary bud; and (3) elongation of internodes into a tiller with the potential to form a grain-bearing spike (Schmitz and Theres, 2005). Primary tillers form in leaf axils on the main stem, and secondary and higher order tillers form in axils of leaves on primary tillers and subsequent tillers, respectively. Grass leaves develop from the flanks of the shoot apical meristem and axillary meristems and are composed of a proximal sheath and distal blade divided by the ligular boundary. The ligular region is composed of the ligule, an outgrowth of an epidermal tissue flap, and the auricle. Auricles have two parts, a band of small cells separating the sheath from the blade and a flap of tissue growing out from the leaf margin that wraps around the stem in some species (Becraft et al., 1990; Sylvester et al., 1990). Both tillers and leaves are important agricultural traits for cereal crops and have been studied extensively (for review, see Wang and Li, 2008; Lewis and Hake, 2016; Mathan et al., 2016). However, our understanding of the interrelatedness of their genetic control is early in its fruition.
Positional information is important for morphogenesis, and boundaries between cell types often are the location of new tissue development. Thus, the role of boundary formation in axillary meristem development is an intense area of study (for review, see Žádníková and Simon, 2014; Hepworth and Pautot, 2015; Wang et al., 2016). The Arabidopsis (Arabidopsis thaliana) REGULATORS OF AXILLARY MERISTEMS1 (RAX1) and CUP-SHAPED COTYLEDON2 (CUC2) genes were identified by their expression pattern and reduced-branching mutant phenotypes and were found to establish the boundary for axillary meristem development (Keller et al., 2006; Müller et al., 2006). Other boundary genes show the expected expression pattern but lack a clear axillary meristem phenotype in mutant plants. Plants overexpressing Arabidopsis BLADE-ON-PETIOLE (BOP) show a branching phenotype, producing extra paraclades in leaf nodes (Ha et al., 2007). The role of Arabidopsis LATERAL ORGAN FUSION (LOF1) in axillary meristem development was revealed by double mutants with its homolog, LOF2 (Lee et al., 2009). The Arabidopsis REGULATOR OF AXILLARY MERISTEM FORMATION1 (ROX1) has a subtle phenotype but is involved in axillary meristem development (Yang et al., 2012). However, the role of ROX1 in axillary meristem development is more obvious in other species such as rice (Oryza sativa) and maize (Zea mays), highlighting the importance of comparative work to fully delineate developmental pathways (Komatsu et al., 2003; Gallavotti et al., 2004). These studies, and others, have identified genes acting in axillary meristem boundary formation, and it appears that a number of these genes help establish other developmental boundaries.
Boundary formation also is critical for leaf patterning (for review, see Bar and Ori, 2014; Lewis and Hake, 2016). Tomato (Solanum lycopersicum) plants produce compound leaves with several pairs of lateral leaflets and a terminal leaflet, with each leaflet having multiple lobes. Goblet (Gob) is one gene controlling this process, and Gob encodes a homolog of CUC1/2 (Berger et al., 2009). Gob mutations also repress axillary meristem development (Busch et al., 2011). Potato leaf (C) and blind are recent duplications of the tomato RAX1 homolog, and subfunctionalization of these duplicated genes gave blind a role in axillary meristem development and C a role in leaf development (Busch et al., 2011). In Arabidopsis, CUC2 functions similarly to produce serrated leaves (Nikovics et al., 2006; Bilsborough et al., 2011). It is now evident that many of the same genes act to establish boundaries for meristem and leaf development (Hepworth and Pautot, 2015; Wang et al., 2016)
The identification of genes with dual roles in boundary demarcation and leaf and axillary meristem development prompted Busch and colleagues (2011) to propose a conserved genetic system that establishes axillary meristems and determines leaf shape. A related genetic system for maize leaf and lateral organ initiation was recently proposed as well (Johnston et al., 2014). Transcriptome analysis of laser-dissected tissues from the maize preligule region identified genes expressed at the blade-sheath boundary that are homologs of previously identified genes involved in lateral organ initiation (Johnston et al., 2014). Among the differentially expressed genes were the maize CUC2 and BOP homologs. RNA in situ hybridization experiments showed maize CUC2-like transcripts accumulating in the preligule band, the cleft of developing ligules, and at the location of lateral branch initiation. The maize BOP-like transcripts accumulated in developing ligules, leaf axils, and axillary meristems (Johnston et al., 2014). The barley (Hordeum vulgare) UNICULME4 (CUL4) gene is the barley BOP homolog (Tavakol et al., 2015), and plants carrying mutations in CUL4 are defective in both axillary meristem and ligule development. In addition, CUL4 is expressed in developing ligules, leaf axils, and axillary meristems and defines the boundaries of ligule and axillary bud development like the maize BOP homolog (Tavakol et al., 2015).
In this study, we conducted a genetic suppressor screen using a mutant that does not make tillers, cul2 (Babb and Muehlbauer, 2003), and identified two mutations in the ELIGULUM-A (ELI-A) gene that promoted axillary meristem development and tillering in the cul2 mutant background. Mutations in ELI-A have been described previously as pleiotropic, with altered ligule development, reduced plant height, weak culms, and compact spikes (Lundqvist and Franckowiak, 2002). Additional characterization showed that eli-a mutant plants exhibited reduced tillering and secondary cell wall formation compared with the nonmutant backcross parent line. We isolated the ELI-A gene and determined that it encodes a previously unannotated protein. RNA in situ hybridizations showed that ELI-A transcripts are found in the preligular region, the developing ligule, leaf margins, cells destined to develop secondary cell walls, and cells surrounding leaf vascular bundles. Taken together, these observations show that ELI-A plays a role in ligule and axillary meristem development. We propose that ELI-A functions in establishing a boundary during ligule development but not for axillary meristem development.
RESULTS AND DISCUSSION
Isolation and Genetic Characterization of cul2 Suppressor Mutants
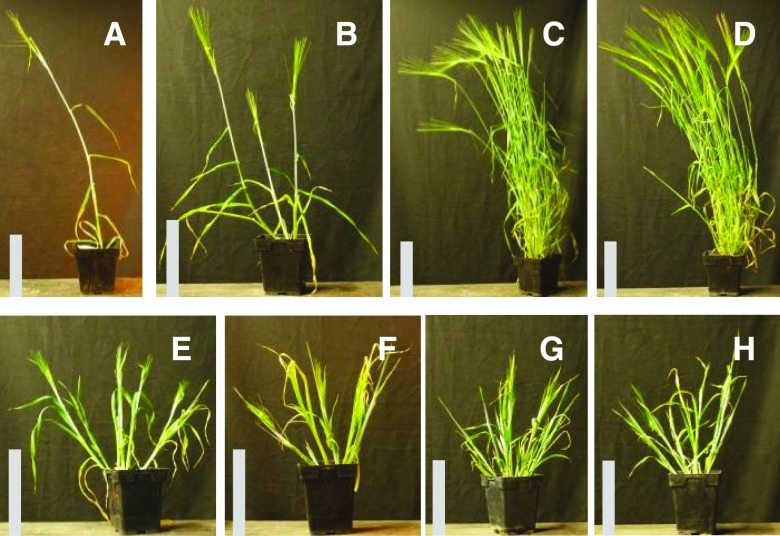
The barley cul2 mutant rarely makes tillers due to its inability to produce axillary buds (Fig. 1; Babb and Muehlbauer, 2003). To identify suppressors of the cul2 mutant phenotype, we mutagenized the Bowman-cul2.b-rob1 stock. Rob1 (orange lemma) is a phenotypic marker tightly linked to cul2 (Franckowiak et al., 1997). Over 15,000 sodium azide-mutagenized, M3 Bowman-cul2.b-rob1 families were screened for plants that produced tillers, and two recessive suppressor mutants were recovered. The two suppressors proved to be alleles of the previously described ELI-A gene (see below) and were named eli-a.17 and eli-a.18. In Bowman-eli-a.17; cul2.b-rob1 and Bowman-eli-a.18; cul2.b-rob1 mutant plants, the uniculm phenotype of cul2 was partially suppressed (Fig. 1). For example, in a greenhouse trial, 28 of 41 Bowman-eli-a.17; cul2.b-rob1 plants produced one or two tillers with the remaining plants having no tillers, and all 21 Bowman-eli-a.18; cul2.b-rob1 plants had one or more tillers (Supplemental Fig. S1). Unexpectedly, homozygous mutant eli-a.18 plants were short with leaves that drooped and lacked ligules (Figs. 1 and 2), whereas these traits were not seen in eli-a.17 plants (Figs. 1 and 2).
Figure 1.
Mutant and nonmutant adult plant characteristics. A, Bowman-cul2.b-rob1. B, Bowman-cul2.b-rob1; eli-a.17. C, Nonmutant cv Bowman. D, Bowman-eli-a.17. E, Bowman-eli-a.18. F, Bowman-cul2.b-rob1; eli-a.18. G, eli-a.14. H, cul2.b-rob1; eli-a.14. The nonmutant cv Bowman and Bowman-eli-a.17 plants in C and D were grown in the field and transferred to pots for photographs. Other plants were grown in a growth chamber. Bars = 20 cm.
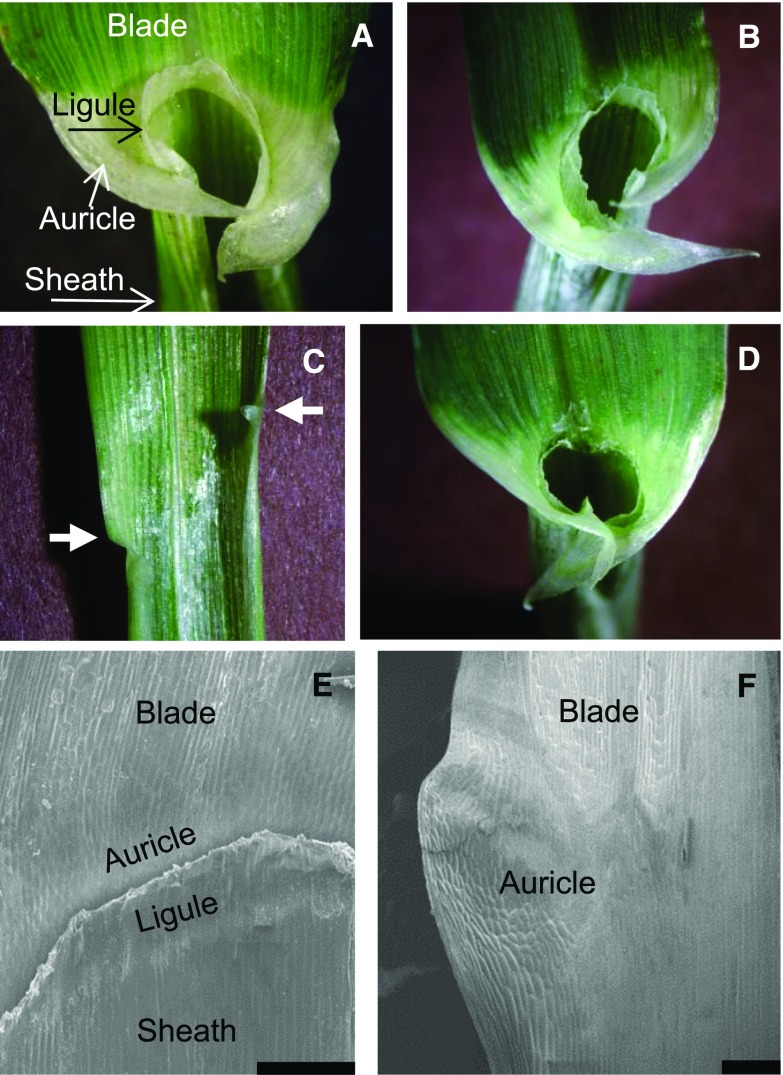
Figure 2.
The ligular region in eli-a alleles. A to D, Ligules and auricles. A, Nonmutant cv Bowman. B, Bowman-eli-a.17. C, Bowman-eli-a.18. Note the reduced auricles at the leaf margin, indicated by the arrows and the absence of the ligule. D, Heterozygous Bowman-eli-a.17/eli-a.18. Note the reduced ligule and auricle development. E and F, Scanning electron micrographs of the ligular regions. E, Nonmutant cv Bowman. The ligule has been trimmed back to uncover the underlying auricle. F, Bowman-eli-a.18 ligular region. Bars = 200 μm.
To determine if eli-a.17 and eli-a.18 were allelic, seven crosses between Bowman-cul2.b-rob1/cul2.b-rob1; eli-a.17/eli-a.17 and Bowman-cul2.b-rob1/cul2.b-rob1; eli-a.18/eli-a.18 were made. Tillers were observed on 18 out of 19 F1 plants, demonstrating that the two suppressors were allelic (Supplemental Fig. S2). In the F2 plants, eli-a.18 mutants exhibited stronger suppression of cul2.b than eli-a.17. Bowman-eli-a.18; cul2.b-rob1 mutant plants were liguleless and developed an average of 2.9 tillers per plant compared with Bowman-eli-a.17; cul2.b-rob1 plants, which developed ligules and had 0.8 tillers per plant (Supplemental Fig. S1). Finally, the heteroallelic combination of eli-a.17/eli.a-18 exhibited an intermediate number of tillers in the cul2.b mutant background, 1.59 tillers per plant, and an intermediate liguleless phenotype (Fig. 2; Supplemental Figs. S1 and S3).
The eli-a.17 and eli-a.18 alleles also mapped to the same region on chromosome 2HS. We mapped eli-a.17 using the cul2 suppressor phenotype. The eli-a.17; cul2.b-rob1 line was crossed with cv Steptoe and a segregating F2 family was developed. The tightly linked rob1 marker and a cleaved-amplified polymorphic sequence (CAPS) marker for single-nucleotide polymorphism (SNP) 1_0964 were used to identify 56 homozygous cul2.b-rob1/cul2.b-rob1 F2 plants (Supplemental Table S1). The eli-a.17 phenotype of these 56 individuals was determined in F3 families, because the suppressor phenotype is not fully penetrant and some F2 eli-a.17/eli-a.17; cul2.b-rob1/cul2.b-rob1 plants were uniculm. eli-a.17 mapped to 2HS 2.2 centimorgan (cM) proximal of SNP 2_0964 at position 17.85 on the SNP map (Supplemental Fig. S4). eli-a.18 was mapped using the liguleless phenotype in 220 F2 individuals from a cross between Bowman-eli-a.18; cul2.b-rob1 and cv Harrington. The liguleless trait was mapped 1.6 cM proximal to SNP 3_1284 at position 19.47 on 2HS (Supplemental Fig. S4).
Barley eli-a mutants were described previously as recessive mutations producing a phenotype of dwarfed liguleless plants with weak culms that break at the nodes (Lundqvist and Franckowiak, 2002). We observed these characteristics in the eli-a.18 mutant. In addition, the attachment of outer tillers to the crown was so poor that tillers leaned outward (Fig. 1). These similarities prompted us to test for allelism between eli-a.18 and the previously described eli-a alleles. Six mutants classified as eligulum that had been backcrossed into the Bowman background were examined (Druka et al., 2011). Genetic stocks carrying three of the mutations, eli.12, eli-b.5, and eli-a.216, had few of the reported eli-a characteristics nor resembled either of our two suppressors and were not pursued. eli-a.3, eli-a.9, and eli-a.14 mutant stocks exhibited the short stature and liguleless characteristics of plants carrying the eli-a.18 allele. An adult eli-a.14 plant is shown in Figure 1, and the liguleless trait from eli-a plants is shown in Supplemental Figure S3. Three crosses of eli-a.18 with eli-a.3, two crosses with eli-a.9, and one cross with eli-a.14 were made. Ten F1 plants were produced, and they all exhibited short and liguleless mutant phenotypes. An example of the heteroallelic combination eli-a.9/eli-a.18 is presented in Supplemental Figure S2. These results confirm that our cul2 suppressors are allelic with eli-a mutants.
To determine if eli-a.3, eli-a.9, and eli-a.14 suppress the cul2 uniculm phenotype, we crossed eli-a.3, eli-a.9, and eli-a.14 with Bowman-cul2.b-rob1. In total, 23 mutant plants were recovered (eli-a.3/eli-a.3; cul2.b-rob1/cul2.b-rob1, eli-a.9/eli-a.9; cul2.b-rob1/cul2.b-rob1, and eli-a.14/eli-a.14; cul2.b-rob1/cul2.b-rob1), and 22 of 23 individuals developed tillers. Examples of the suppression of cul2.b by eli-a.14 and eli-a.9 are shown in Figure 1 and Supplemental Figure S2. All five eli-a alleles tested suppress cul2, thereby establishing a role for ELI-A in axillary meristem development.
Axillary Bud and Tiller Development in eli-a Mutants
To study the impact of eli-a on early axillary bud development, we examined 7-d-old shoot apices from Bowman-eli-a.17; cul2.b-rob1, Bowman-cul2.b-rob1, Bowman-eli-a.17, and the nonmutant cv Bowman. Despite being a weak allele, the eli-a.17 allele was used for this experiment because germination rates were higher and growth more uniform than in other eli-a alleles. Two to three primary axillary buds were typically seen in nonmutant cv Bowman seedlings at 7 d (Fig. 3). In these experiments, no axillary buds were seen in cul2.b seedlings (Fig. 3), but in previous experiments, occasionally an axillary meristem would develop but would be blocked from forming an axillary bud (Babb and Muehlbauer, 2003). One to two primary axillary buds were present in 7-d-old Bowman-eli-a.17 seedlings (Fig. 3). In the Bowman-eli-a.17; cul2.b-rob1 material, zero to two axillary buds were visible at 7 d (Fig. 3). A 7-d-old Bowman-eli-a.18 shoot apex is shown in Supplemental Figure S5 for comparison.
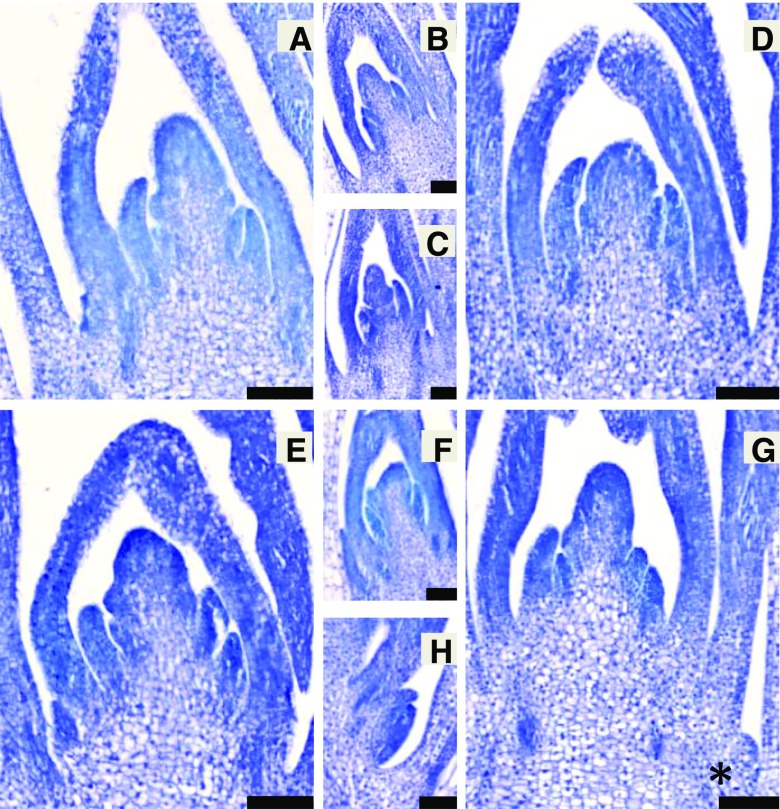
Figure 3.
Longitudinal sections of 7-d-old shoot apices from nonmutant and mutant lines stained with Toluidine Blue. Axillary buds are shown in the smaller images, as median sections of the shoot apex generally do not capture the axillary buds. A, Nonmutant cv Bowman shoot apical meristem. B, Cv Bowman axillary bud 3. C, Cv Bowman axillary bud 2. D, Bowman-cul2.b-rob1 shoot apical meristem. E, Bowman-eli-a.17 shoot apical meristem. F, Bowman-eli-a.17 axillary bud. G, Bowman-eli-a.17; cul2.b-rob1 shoot apical meristem. The edge of a small axillary bud is visible at bottom right (asterisk). H, Section through axillary bud 1 seen in G. Bars = 100 µm.
The rates of axillary bud and tiller development between the eli-a.17 mutant and the nonmutant were compared, and the numbers of tillers on adult plants for eli-a.17 were compared with the nonmutant. Developing axillary buds and tillers were counted weekly in dissected seedlings of eli-a.17 and nonmutant plants at 2 to 6 weeks after planting. Over this period, the rate of axillary bud and tiller emergence was significantly slower in eli-a.17 plants than in nonmutant plants (Supplemental Fig. S6).
Tiller numbers on field-grown plants were determined for both Bowman-eli-a.17 and Bowman-eli-a.18 plants. At maturity, plants carrying the strong mutant allele, eli-a.18, had approximately half as many tillers as nonmutant plants, whereas plants carrying the weak eli-a.17 allele had approximately 20% fewer tillers than nonmutant plants (Table I). This small reduction in tillering in eli-a.17 compared with the nonmutant cv Bowman was consistent with tiller numbers counted from individual families in previous seasons. For example, nonmutant cv Bowman plants had an average of 45.5 (se = 3.4) tillers per plant and an adjacent family of Bowman-eli-a.17 plants had 33.9 (se = 3.41) tillers per plant in the 2013 field. The reduced tiller number in eli-a.17 and eli-a.18 mutants compared with the nonmutant and the increase in tiller number in cul2.b; eli-a double mutants indicates that the mechanism controlling the rate of tillering and adult tiller number is not necessarily the same mechanism that suppresses the cul2 mutant phenotype.
Table I. Tiller development in eli-a.17 and eli-a.18 mutants and in nonmutant cv Bowman.
Asterisks indicate P < 0.05 by two-tailed Student’s t test.
| Genotype | Tiller Number, 4 Weeks | Tiller Number, 6 Weeks | Tiller Number, Maturity |
|---|---|---|---|
| Cv Bowmana | 7.69 | 28.01 | 33.95 |
| Bowman-eli-a.17a | 6.70 | 23.93* | 27.37* |
| Cv Bowmanb | 6.61 | 27.32 | 43.99 |
| Bowman-eli-a.18b | 4.73 | 15.75* | 21.97* |
2015 field.
2016 field.
Ligule and Auricle Development in eli-a Mutants
The grass leaf sheath-blade boundary is marked by two structures, the ligule and the auricle (Fig. 2). The auricle can be divided into two parts, a band of small, light-colored cells separating the blade from the sheath and a flap of tissue growing out from the leaf margin that often wraps around the stem (Fig. 2). The boundary runs perpendicular to the long axis of the leaf, and the paired auricle flaps are usually directly opposite each other.
Ligules and the bands of auricle cells were generally not visible in eli-a.18 plants, but small auricle flaps were present (Fig. 2). Figure 2 presents an adaxial view of the ligular region from a nonmutant plant with the ligule cut away to show the underlying auricle cells. A small auricle develops in eli-a.18 plants at the leaf margin and extends a short distance inward (Fig. 2; Supplemental Fig. S3). Ligules were not obvious in most plants, although small rudimentary ligules have been seen. When present, rudimentary ligules were short and did not span the width of the leaf (Supplemental Fig. S3).
A range of ligule and auricle development was seen in the five eli-a alleles. Ligule and auricle development was visibly disrupted in eli-a.3, eli-a.9, eli-a.14, and eli-a.18 leaves (Supplemental Fig. S3). Ligules and auricles appeared normal in homozygous eli-a.17 plants (Fig. 2; Supplemental Fig. S3). However, heterozygous eli-a.17/eli-a.18 plants have small ligules while heterozygous eli-a.18/ELI-A plants produce normal ligules, indicating that the eli-a.17 allele is not equivalent to the nonmutant allele for ligule development (Fig. 2; Supplemental Fig. S3).
Another characteristic of leaf development in eli-a mutants was the displacement of the blade-sheath boundary, as indicated by the placement of auricle flaps at the leaf margin (Supplemental Fig. S3). In nonmutant plants, these structures are opposite one another on the leaf, and the blade-sheath boundary runs approximately perpendicular to the longitudinal axis of the leaf (Fig. 2; Supplemental Fig. S3). Displacement of the blade-sheath boundary was commonly observed in eli-a.3, eli-a.9, eli-a.14, and eli-a.18 (Supplemental Fig. S3). This aberrant boundary positioning was infrequent with nonmutant and eli-a.17 leaves.
Inflorescence Development
eli-a mutant spikes have a compact appearance with spikelets packed tightly together, particularly toward the tip (Supplemental Fig. S7; Lundqvist and Franckowiak, 2002). This characteristic is less obvious in weaker alleles like eli-a.3 and eli-a.17 (Supplemental Fig. S7). The cul2 mutation produces spikes with spikelets irregularly placed along the spike, particularly near the tip (Babb and Muehlbauer, 2003). The expression of these traits in double mutant eli-a; cul2.b plants ranges from compact spikes with an irregular arrangement of spikelets to severe disruption of spikelet formation (Supplemental Fig. S7). Thus, although the eli-a mutation partially suppresses the axillary meristem defect in cul2 mutants, the cul2 spike phenotype is not suppressed.
Secondary Cell Wall Defects in eli-a Mutants
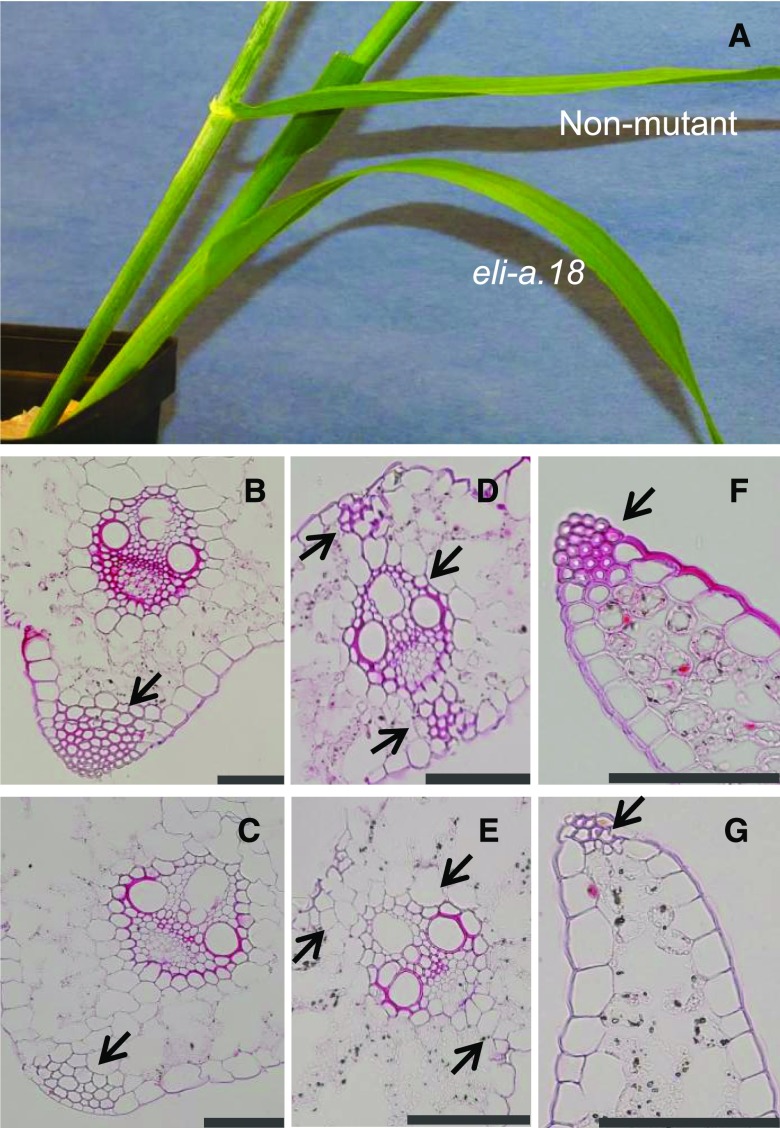
Nonmutant leaves from cv Bowman have midrib, leaf margin, and bundle sheath extension cells with thick secondary cell walls providing strength to the leaves. Stained with Safranin O, these cells appeared small with thick red cell walls (Fig. 4). Corresponding cells in Bowman-eli-a.18 leaves were larger with thin cell walls (Fig. 4). Safranin O stains lignin, and the weaker staining seen in eli-a.18 suggests reduced lignin content in eli-a.18 (Ruzin, 1999). These changes may explain the lack of structural strength and the tendency to droop downward in mutant leaves (Fig. 4). ELI-A apparently has a similar function in other tissues. Epidermal cells in the culm and cells immediately under the epidermis have thick cell walls in nonmutant plants (Supplemental Fig. S8). The corresponding cells from eli-a.18 and eli-a.3 mutant culms have thin cell walls (Supplemental Fig. S8). This may explain the weakness reported in eli-a culms (Lundqvist and Franckowiak, 2002). However, secondary cell walls did develop in the xylem and other cells within vascular bundles in eli-a.18 mutant plants, demonstrating that ELI-A is not an absolute requirement for secondary wall development (Fig. 4).
Figure 4.
Secondary cell wall development. A, Comparison of leaves from mature nonmutant cv Bowman and Bowman-eli-a.18 plants. B to G are Safranin O stained. B, Midrib from nonmutant plant. C, Midrib from Bowman-eli-a.18 plant. D, Leaf vein from nonmutant plant. E, Leaf vein from Bowman-eli-a.18 plant. F, Leaf margin from nonmutant plant. G, Leaf margin from Bowman-eli-a.18 plant. Arrows point to cell wall differences in the leaf midrib (B and C), the bundle sheath extension and mestome sheath (D and E), and the leaf margin (F and G). Bars = 100 μm.
Disrupting cell wall development may explain other characteristics of eli-a mutants. In eli-a.18 mutant plants, secondary cell wall formation in the mestome sheath and bundle sheath extensions was greatly reduced. Structural strength is but one function of secondary cell walls (for review, see Leegood, 2008). Fricke (2002) proposed that the bundle sheath regulates the flow of water and photosynthate between the leaf mesophyll and the vascular system. Other work suggests that bundle sheath extensions are an adaptation for desiccation stress (Kenzo et al., 2007). Physiological limitations imposed by mutant cell walls could explain the semidwarf stature and reduced rate of tillering in eli-a plants but would not account for the suppression of the cul2 axillary meristem trait. However, cell wall stiffness in the shoot apex influences auxin transport, CUC3 expression, and leaf primordia emergence in other systems and provides a plausible mechanism for controlling axillary meristem development (Kierzkowski et al., 2012; Nakayama et al., 2012; Fal et al., 2016).
Isolation and Characterization of the ELI-A Gene
The ELI-A gene was identified by comparing the transcriptomes of the sodium azide-generated eli-a.17 and eli-a.18 mutant alleles against nonmutant plants. RNA was isolated and sequenced from 2-week-old seedling crown tissue from cv Bowman, Bowman-cul2.b, Bowman-cul2.b-rob1, Bowman-eli.a-17, Bowman-cul2.b-rob1; eli.a-17, Bowman-eli-a.18, and Bowman-cul2.b-rob1; eli-a.18. De novo assembly of sequence reads from the cv Bowman line produced 31,976 transcripts (Supplemental Data S1). SNPs were then identified between nonmutant cv Bowman and the mutant lines. These SNPs would include any existing variation in the cv Bowman lines and mutations induced by the sodium azide treatment, including the causative mutations for eli-a.17 and eli-a.18 (Supplemental Tables S2–S7). Transcript11292 (Supplemental Data S1) contained an SNP at position 1,103 from the Bowman-cul2.b-rob1; eli-a.17 and Bowman-eli.a-17 lines and a different SNP at position 796 in the Bowman-cul2.b-rob1; eli-a.18 and Bowman-eli-a.18 lines (Supplemental Tables S2–S5). These two SNPs in Transcript11292 were not present in the cv Bowman, Bowman-cul2.b-rob1 progenitor line, or the related Bowman-cul2.b line, providing evidence that the sequence differences were not preexisting polymorphisms (Supplemental Tables S6 and S7). Both SNPs were confirmed by Sanger sequencing PCR products from the cv Bowman, Bowman-cul2.b- rob1; eli-a.17, and Bowman-cul2.b-rob1; eli-a.18 genomic DNAs.
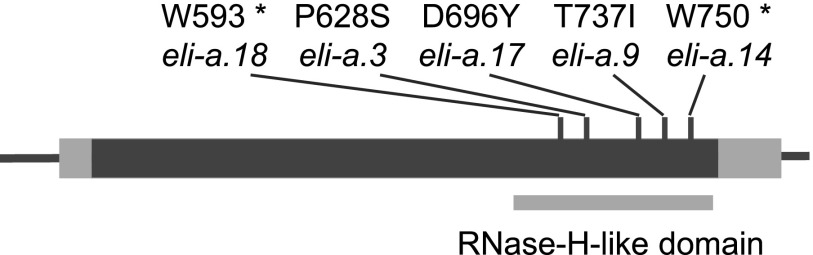
A full-length cDNA sequence, AK375036, matching Transcript11292 was identified in a BLASTn search of the GenBank nonredundant sequence database. The entire predicted coding region of AK375036 was sequenced from the eli-a.17, eli-a.18, eli-a.3, eli-a.9, and eli-a.14 alleles (Fig. 5). Cv Foma, the progenitor allele of eli-a.3 and eli-a.9, cv Kristina, the progenitor allele of eli-a.14, the Bowman-cul2.b-rob1 line, progenitor of eli-a.17 and eli-a.18, and the backcross parent cv Bowman also were sequenced. eli-a.3, eli-a.9, and eli-a.17 contained the nonconservative amino acid substitutions Pro to Ser, Thr to Ile, and Asp to Tyr, respectively. The eli-a.14 and eli-a.18 alleles contained nonsense mutations. This cDNA corresponds to gene model MLOC_58453 from the barley genome (International Barley Genome Sequencing Consortium, 2012).
Figure 5.
The ELI-A gene and locations of mutations. The dark gray box indicates the single exon in the gene, and lighter gray boxes mark the 5′ and 3′ untranslated regions. Mutations in eli-a.14 and eli-a.18 created stop codons. Prediction programs Phyre2, LOMETS, and InterProScan 5 identified an RNaseH-like domain at the C-terminal end of the peptide.
MLOC_58453 cosegregated with the liguleless phenotype in the eli-a.18 mutant and the eli-a.17 suppressor phenotype in the mapping populations described above. MLOC_58453 was mapped in the Bowman-cul2.b-rob1; eli-a.18 cv Harrington F2 population using a CAPS marker targeting the mutated base pair (Supplemental Table S1). As expected, all liguleless plants were homozygous for the mutant MLOC_58453 CAPS allele (Supplemental Fig. S4). Similarly, an SNP located within the MLOC_58453 coding region cosegregated with the suppressor phenotype in the eli-a.17 mapping population (Supplemental Fig. S4; Supplemental Table S1). MLOC_58453 has been mapped to chromosome 2HS on the barley genome assembly (International Barley Genome Sequencing Consortium, 2012).
ELI-A Is a Conserved Plant Gene Containing an RNaseH-Like Domain
Homologous ELI-A sequences were found in land plants ranging from Arabidopsis and rice to the nonvascular and primitive vascular plants Physcomitrella patens and Selaginella moellendorffii (Supplemental Table S8). A distantly related sequence was present in the green alga Chlamydomonas reinhardtii. Mutant phenotypes in Arabidopsis have not been reported in TAIR, and the two homologs, AT1G12380 and AT1G62870, are described as hypothetical proteins (https://www.arabidopsis.org/, May 2017), nor are the maize gene models homologous with ELI-A associated with a maize phenotype or classical gene (http://maizegdb.org/, May 2017). A phylogenetic tree developed from these sequences is presented in Supplemental Figure S9 (Dreeper et al., 2008). Despite the sequence conservation, there is a lack of evidence for ELI-A function outside of barley.
We examined peptide sequences of the barley ELI-A protein and in homologous rice and Arabidopsis proteins. The peptides from barley, rice, and Arabidopsis were predicted by Phyre2, LOMETS, and InterProScan 5 to contain an RNaseH-like domain (Quevillon et al., 2005; Wu and Zhang, 2007; Kelley and Sternberg, 2009). InterProScan 5 did provide additional details, but Phyre2 and LOMETS identified the putative RNaseH-like domain as a member of the Hermes transposase class. The Hermes class of RNaseH-like domains is found in hAT family transposons; hAT family transposons also contain an N-terminal BED-type zinc finger and the hAT domain (Hickman et al., 2005).
Further examination of the relationship of the ELI-A protein to members of the RNaseH-like superfamily found that the Hermes domain is a class I RNaseH that is within clade B under the classification scheme of Majorek et al. (2014). This family is composed mainly of transposases with endonuclease activity, although one member of clade B encodes the human P52rIPK protein that regulates a human RNA-dependent Ser/Thr protein kinase (Gale et al., 2002). Phyre2 detected BED-type zinc fingers in the rice and Arabidopsis peptides with moderate confidence. However, the hAT domain was not detected by Phyre2, LOMETS, or InterProScan 5 in barley, rice, or Arabidopsis. At present, the origin of ELI-A from a transposon is not known.
ELI-A Expression Pattern
The expression levels of ELI-A from eight tissues were calculated from previously published RNA sequencing (RNAseq) data (International Barley Genome Sequencing Consortium, 2012). At this level of resolution, expression was highest in 5- and 15-mm-long immature inflorescences (Supplemental Fig. S10). ELI-A expression levels were low in most other tissues. RNA was extracted from axillary buds, 5-mm-long inflorescences, and leaf blades for quantitative reverse transcription PCR (RT-qPCR) to validate the RNAseq data. Transcript levels were highest in the inflorescence, while transcript levels were below the threshold of detection in leaf tissue, consistent with results from the RNAseq data (Supplemental Fig. S10).
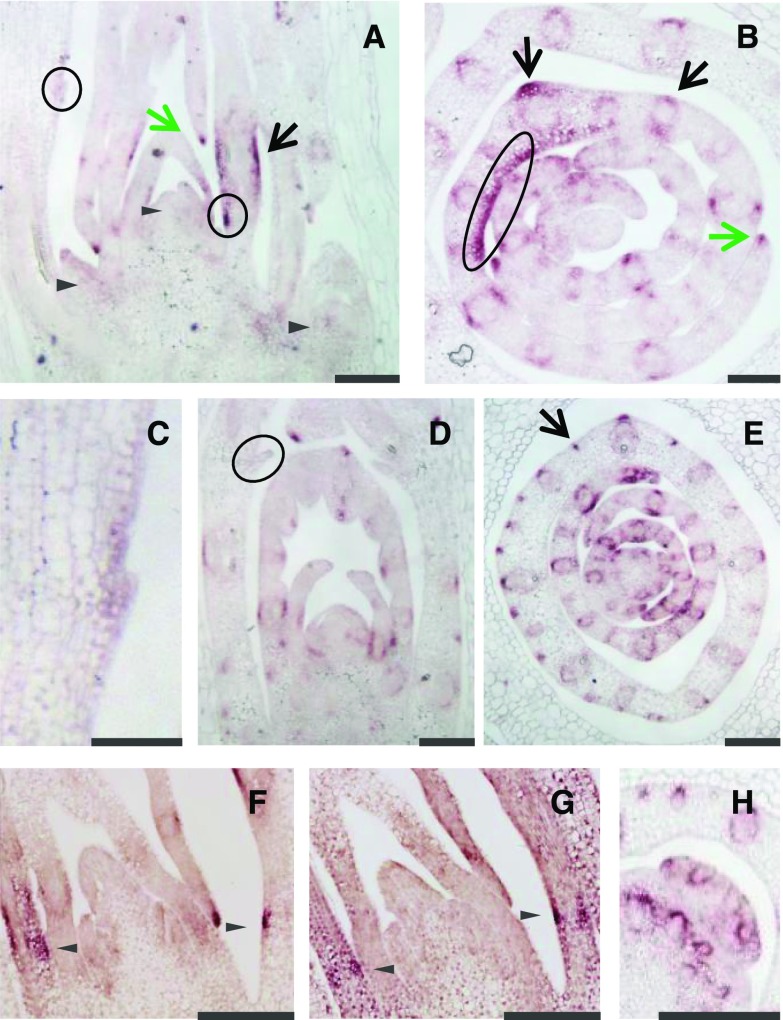
RNA in situ hybridizations were performed to further refine the distribution of ELI-A transcripts. In nonmutant, 4-d-old shoot apices, expression was strong in leaf midribs, along the leaf margin, in the bundle sheath surrounding vascular bundles, and in bundle sheath extension cells (Fig. 6). A sense control is shown in Supplemental Figure S11. ELI-A transcripts were detected in similar locations in cul2.b mutant seedlings (Fig. 6). In transverse cul2.b sections, expression was detected in small clusters of cells along the abaxial leaf surface (Fig. 6). Expression at this location was variable and also was seen in nonmutant plants (Supplemental Fig. S11). There were no consistent differences in expression between nonmutant and cul2.b plants.
Figure 6.
ELI-A expression in nonmutant and cul2.b seedlings. A to C, Four-day-old nonmutant shoot apex probed with an antisense ELI-A probe. A, Longitudinal section showing little staining in or adjacent to the shoot apical meristem and axillary buds, indicated by triangles. Short patches of staining at the adaxial and abaxial sides of leaves were seen occasionally (black arrow); this pattern is likely from vascular bundles, as seen in B and H. Staining was observed in newly forming ligules and leaf primordia that may represent developing ligules (circled). Leaf margins also were stained (green arrow). B, Transverse section showing staining in leaf margins, midribs, and around vascular bundles. Staining along a portion of the adaxial side of a developing leaf (circled) can be followed across the leaf in serial sections and may be associated with the developing ligules. C, Closeup view of the ligule circled in A. D and E, Four-day-old cul2.b shoot apex probed with an antisense ELI-A probe. D, Longitudinal section of a cul2.b shoot. E, Transverse section of a cul2.b shoot. The ELI-A staining pattern was similar to that of nonmutant shoot apices. The variable staining in small clusters of cells along the abaxial leaf surface (arrow) was seen in nonmutant plants (Supplemental Fig. S11); transcripts were not detected in the older ligules (circled). F and G, Serial sections of 4-d-old shoots probed for ELI-A or HvLg1. F, ELI-A staining was observed in the same location as the adaxial HvLg1 staining. G, HvLg1 staining was detected on the adaxial and abaxial surfaces. H, ELI-A expression in leaf axils and axillary buds is associated with vascular bundles. Bars = 100 μm in C and 200 μm in other images.
ELI-A transcripts are present in developing ligules. A low level of ELI-A transcripts was found in emerging ligules (Fig. 6; Supplemental Fig. S11) but not in older ligules (Fig. 6). In younger leaf primordia, a prominent signal was present slightly above the base of leaf primordia on the adaxial side in longitudinal sections. This signal appeared to correspond to the band of expression found on the adaxial surface of leaf primordia in transverse sections (Fig. 6). A serial section from higher up along this shoot apex showed expression continuing along the adaxial surface (Supplemental Fig. S11). This is the expected location of the preligule band, which marks the boundary between the blade and sheath. To verify this, we looked at the expression of the barley homolog of the maize Liguleless1 (Lg1) gene. The maize Lg1 gene is expressed at the preligule band at the blade-sheath boundary (Moon et al., 2013). ELI-A and HvLG1 transcripts were both found on the adaxial surface of the blade-sheath boundary in serial sections from the same shoot apex (Fig. 6). An HvLG1 sense control is shown in Supplemental Figure S11. Taken together, these results indicate that ELI-A acts like a boundary gene in the development of the blade-sheath boundary.
Weak staining was sometimes seen in axillary buds and in leaf axils adjacent to developing axillary meristems as well as within axillary buds in longitudinal sections (Fig. 6; Supplemental Fig. S11). In transverse sections, this transcript appeared to associate with developing vascular bundles rather than the leaf axil or axillary meristem (Fig. 6). ELI-A expression farther down the shoot apex where the axillary bud emerged from the shoot apex was very weak compared with expression around vascular bundles higher up the shoot apex (Supplemental Fig. S11). The expression pattern of ELI-A did not indicate a direct function in boundary formation or stem cell maintenance.
Sclerenchyma cells are found in developing leaf ribs, hypodermal sclerenchyma cells, and leaf margins from barley plants (Wenzel et al., 1997; Trivett and Evert, 1998). These are locations where ELI-A transcripts were detected in leaf primordia. In eli-a.18 mutants, cells comprising midribs have thin cell walls and lack the thick secondary cell walls of normal rib cells. Elsewhere in the leaf and in the culm, ELI-A transcripts coincided with cells having thickened secondary cell walls (Figs. 5 and 6). ELI-A transcripts were not detected in the xylem or phloem (Fig. 6; Supplemental Fig. S11). This absence of secondary cell walls can explain the weak leaves and culms in eli-a mutant plants. However, the absence of ELI-A transcripts in leaf axils from both CUL2 and cul2.b plants argues against a direct role for secondary cell walls in the suppression of the cul2 tillering phenotype by eli-a.
ELI-A Acts Like a Boundary Gene in the Leaf But Not in the Leaf Axil
A conserved set of genes are believed to control leaf and axillary meristem development. In tomato and other eudicots, the development of leaf serrations, leaflets, and axillary meristems in leaf axils is regulated by CUC, RAX, and LATERAL SUPPRESSOR (Busch et al., 2011). Grass leaves lack the serrated margins and leaflets common in eudicots, but there is a boundary between the blade and sheath consisting of the ligule and auricles (Langdale, 2005; Lewis and Hake, 2016). Laser microdissection transcriptome analysis showed maize CUC2, BOP (homologous to barley CUL4), and ELI-A homologs up-regulated at the blade-sheath boundary (Johnston et al., 2014). In situ hybridizations confirmed the maize CUC2 and BOP expression in newly forming ligules at the leaf blade-sheath and lateral organ initiation boundaries including axillary meristems (Johnston et al., 2014). This postulated genetic system may derive from a common evolutionary origin for leaves and axillary meristems, as suggested by Busch and colleagues (2011), and is consistent with expectations from the barley phytomer model proposed by Forster and coworkers (2007). Alternatively, the conserved genes may be part of a conserved genetic module that acts in leaf and axillary meristem development (Carroll, 2008).
This system of genes is expected to function in barley development. The barley BOP homolog, CUL4, is expressed in newly formed ligules at the blade-sheath junction and at axillary meristem boundaries in leaf axils (Tavakol et al., 2015). However, CUL4 is expressed in developing ligules and does not appear to specify the location of the blade-sheath boundary (Tavakol et al., 2015). Like CUL4, ELI-A is expressed in newly forming ligules but also is expressed earlier in development than CUL4, where its expression pattern overlaps the barley homolog of the maize Lg1 gene in the preligular region separating the blade from the sheath (Moon et al., 2013). In addition, the ELI-A mutants (eli-a.9, eli-a.3, eli-a.14, and eli-a.18) exhibiting a liguleless phenotype also exhibit a disrupted blade-sheath boundary (Supplemental Fig. S3). Taken together, our results show that ELI-A and CUL4 are both necessary to produce a ligule, with ELI-A acting at a similar time and place with HvLg1 to establish the leaf blade-sheath boundary.
CUL4 and ELI-A both have roles in axillary meristem development. However, their roles in axillary development appear different because the cul4 and eli-a tillering phenotypes share few characteristics. The cul4 mutation restricts axillary meristem development to a short developmental window; new axillary buds cease appearing after 3 to 4 weeks in cul4.5 plants (Tavakol et al., 2015). The eli-a mutation slows the rate of axillary meristem development. Furthermore, eli-a mutants suppress the low-tillering cul2 phenotype; cul4 does not (Babb and Muehlbauer, 2003).
RNA in situ hybridization provided further evidence for differing roles for ELI-A during leaf and axillary branch development. The ELI-A transcripts were present at the leaf blade-sheath boundary, where it participates in ligule development. Although ELI-A transcripts were occasionally detected in or adjacent to developing axillary buds in longitudinal sections, ELI-A was shown in transverse sections to be closely associated with vascular bundles rather than organ boundaries or meristematic regions. This expression pattern in the leaf axil was not similar to that of other characterized axillary meristem boundary genes, including CUL4 and the rice CUC3 and RA2 homologs (Oikawa and Kyozuka, 2009; Tavakol et al., 2015). It is possible that transient ELI-A expression in axillary meristems or organ boundaries was not detected or that the ELI-A protein is transported, as shown for the rice LAX protein (Oikawa and Kyozuka, 2009). While acknowledging these possibilities, our data support a model where ELI-A has an early role and CUL4 has a later role in creating the blade-sheath boundary during leaf development. However, during axillary meristem development, CUL4 is expressed in the leaf axil and plays a role in boundary formation. While ELI-A does not appear to be expressed in the leaf axil boundary, it still has a role in axillary meristem development. Taking these findings together, we propose that ELI-A acts like a boundary gene at the leaf blade-sheath boundary and promotes secondary cell wall formation in leaves and other tissues but acts in an unknown manner during axillary meristem development.
MATERIALS AND METHODS
Plant Materials and Populations
Barley (Hordeum vulgare) mutant alleles cul2.b, eli-a.3, eli-a.9, eli-a.14, and rob1 were obtained from the collection of mutants backcrossed to cv Bowman (Druka et al., 2011). Plants were either field grown or grown under controlled conditions in a greenhouse or growth chamber with 16 h of light at 22°C and 8 h dark at 18°C. Supplemental Table S9 provides information on the mutant alleles and barley cultivars used here.
The cul2 suppressor screen was conducted by mutagenizing Bowman-cul2.b-rob1 grain. The rob1 allele is approximately 2 cM from cul2 and produces an orange lemma phenotype that was used to track the tightly linked cul2.b allele. Approximately 20,000 Bowman-cul2.b-rob1 kernels were treated with sodium azide according to the protocol described by Döring et al. (1999). From the M2 plants, over 15,000 M3 families (∼70,000 plants) were produced and screened. M3 families segregating for plants with tillers were identified. Families were retested for the suppressor phenotype in subsequent generations.
To recover homozygous eli-a.17 plants, we conducted a single backcross of eli-a.17/eli-a.17; cul2.b-rob1/cul2.b-rob1 plants to the nonmutant cv Bowman and self-pollinated an F1 plant to generate an F2 population. Families derived from phenotypically nonmutant F2 plants were screened in the F3 and F4 generations to recover homozygous eli-a.17/eli-a.17 and eli-a.17/eli-a.17; cul2.b-rob1/cul2.b-rob1 lines (Supplemental Fig. S12). eli-a.18 mutant plants were identified by their short stature and liguleless leaves. F2 populations segregating eli-a.17 and eli-a.18 were produced by crossing the Bowman-eli-a.18; cul2.b-rob1 line with the nonmutant cv Harrington and by crossing the Bowman-eli-a.17; cul2.b-rob1 line with the nonmutant cv Steptoe.
Seedling tests for the suppression of cul2.b by eli-a.3, eli-a.9, and eli-a.14 were performed by crossing the mutants and recovering eli-a/+; cul2.b/cul2.b individuals. These plants were allowed to self-pollinate. Tillering phenotypes of suppression of cul2 by eli-a.3 and eli-a.14 were scored in the growth chamber in 3- to 4-week-old F2 plants. The suppression of cul2.b by eli-a.9 was tested in field-grown F2 families.
Morphological Characterization
Shoot apices from 1-week-old seedlings were sectioned and stained to examine axillary bud development as described previously (Babb and Muehlbauer, 2003). Axillary buds and tillers were counted on growth chamber-grown plants in weeks 2 through 6. Three replicates, three plants per replication, were counted at each time point; at least eight plants were examined in all but two time points. Leaves were removed to count axillary buds and tillers. Axillary buds were further classified as primary axillary buds, those growing in leaf axils, and secondary axillary buds, those growing in tiller axils (Dabbert et al., 2010). Tiller number was determined from field-grown plants at 4 weeks, 6 weeks, and maturity; five plants per replicate with six replicates of nonmutant and Bowman-eli-a.17, and five replicates of Bowman-eli-a.18, were randomized in the field.
Ligular regions were examined on 4- to 6-week-old plants grown in the growth chamber or greenhouse. The development of ligules, auricles, and other features was characterized from the second or third leaf. The leaf blade-sheath junction region was photographed under low-power light microscopy and with cryo-scanning electron microscopy. Scanning electron microscopy was performed on a Hitachi S3500N scanning electron microscope at 5 or 10 kV (Ahlstrand, 1996).
For histological work, plant tissues were fixed in paraformaldehyde and embedded in paraffin (Javelle et al., 2011). Sections were stained with Toluidine Blue or Safranin O (Humason, 1979; Ruzin, 1999). RNA in situ hybridizations were performed as described by Javelle et al, (2011). Probes for RNA in situ hybridizations were developed from PCR amplicons from genomic DNA using primers ELI-1393F and ELI-1877R or HvLG1-79F and HvLG1-598R and then cloned into pGEM-T Easy (Promega). Plasmids were used as templates for PCR with M13 forward and reverse primers. RNA was synthesized from the resulting amplicons with SP6 or T7 RNA polymerase to make the sense and antisense probes using the Roche DIG RNA Labeling Kit (Sigma-Aldrich).
Molecular Biology Procedures
Procedures for DNA isolation, PCR, CAPS markers, and other routine molecular techniques were described previously (Dabbert et al., 2010). PCR primers (Supplemental Table S10) were developed using the program Primer3 (Rozen and Skaletsky, 2000). Sanger sequencing was performed by the University of Minnesota Genomics Center. CAPS markers were developed from previously mapped SNP sequences (Close et al., 2009), and the program JoinMap 4 was used to calculate map distances (Van Ooijen, 2006).
Total RNA for sequencing (RNAseq) was isolated from crown tissue containing the shoot apical meristem and axillary meristems from 14-d-old seedlings grown in growth chambers using the RNeasy Plant Mini Kit (Qiagen). There were three replicates of each genotype (cv Bowman, Bowman-cul2.b, Bowman-cul2.b-rob1, Bowman-eli.a-17, Bowman-cul2.b-rob1; eli.a-17, Bowman-eli-a.18, and Bowman-cul2.b-rob1; eli-a.18), six seedlings per replicate, and tissue from each genotype was pooled. Poly(A+) RNA isolation, library construction, and Illumina sequencing were performed by the University of Minnesota Genomics Center. Fragment sizes for sequencing averaged 200 bp, after accounting for adaptor sequences, and 76-bp paired-end reads were produced.
Relative ELI-A expression levels were compared in inflorescence, axillary bud, and leaf blade tissues by RT-qPCR using the procedure described by Tavakol et al. (2015). Total RNA was isolated from 1-cm-long axillary buds from 2-week-old seedlings, 5-mm-long inflorescences, and leaf blades from 4-week-old seedlings using the RNeasy Kit (Qiagen). Approximately 250 ng of total RNA was DNase treated (RQ1 RNase-Free DNase; Promega) prior to cDNA synthesis with the ImProm-II Reverse Transcription System (Promega). One-third of the product was used for PCR. Quantitative PCR was performed on the Applied Biosystems StepOnePlus Real Time PCR System with the QuantiFast SYBR Green mix (Qiagen). GAPDH and UBI were used for normalization (Tavakol et al., 2015). Three replicates, with three to five plants each, were randomized and grown together in a growth chamber as described above. Primer sequences are shown in Supplemental Table S10.
Sequence Assembly Pipeline and SNP Analysis
Reads for all samples were quality trimmed from both ends with custom Java code using a base quality cutoff of Phred 20. Reads shorter than 30 bp were discarded. Trimmed reads from the cv Bowman sample were assembled de novo using the Trinity transcriptome assembler on default settings (release r2011-05-13; Grabherr et al., 2011). This resulted in a total of 31,976 transcript sequences (Supplemental Data S1).
Trimmed reads from each mutant sample were mapped separately to the Bowman Trinity transcripts using the Bowtie read mapper version 0.12.7 (Langmead et al., 2009). To keep mismapping and the resulting false-positive SNPs to a minimum, a strict mismatch rate of one mismatch per read was applied. Reads were mapped in all mode, which allows multimappable reads to map to all of their possible mapping locations. The –best –strata parameter was used to ensure that only the best mapping locations were reported.
For each genotype, SNP discovery was carried out using custom-written code implemented as a prototype feature in Tablet (Milne et al., 2013). The raw variant data were then filtered using a minor allele frequency of 0.9 or greater to identify homozygous SNPs with the cv Bowman reference sequence only. Several further stages of SNP filtering followed, all of which were aimed at removing false-positive SNPs. First, SNPs that were less than one read’s length from the contig start or end, or regions with zero read coverage, were removed, as a large proportion of these can be assumed to be artifacts caused by misassembly of the reference sequence (M. Bayer, unpublished data). SNPs with fewer than six instances of the alternate allele also were removed to exclude low-coverage, low-confidence variants. We called SNPs by mapping the cv Bowman reads against the cv Bowman Trinity assembly as a control set, on the assumption that any SNPs found in this largely homozygous cultivar must be artifacts caused by read mismapping or misassembly of the reference sequence. SNPs discovered in this data set were subsequently removed from all of the mutant SNP sets. The remaining robust SNPs were used for analysis.
Accession Numbers
RNAseq data have been deposited into the National Center for Biotechnology Information Short Read Archive with accession number SRP076379. ELI-A sequences were deposited in the National Center for Biotechnology Information database with accession numbers KU844110 to KU844117. Additional sequences mentioned in this article can be found in the GenBank, TAIR, or PlantGDB databases under the following accession numbers: GenBank/EMBL, Bradi5g04710 (XM_003581043), Bradi5g04720 (XM_003579296), CHLREDRAFT_180901 (XM_001692084), LOC_Os04g19140 (XM_015779144), LOC_Os02g25230 (XM_015767599), PP1S21_302V6 (XM_001756068), PP1S105_108V6 (XM_001768660), PP1S226_73V6 (XM_001777733), PP1S111_138V6 (XM_001769128), SELMODRAFT_231485 (XM_002969799), SELMODRAFT_10589 (XM_002985134), Si009424m.g (XM_004975201), Si016308m.g (XP_004952406), Sb04g014800 (XM_002453721), Solyc08g079550 (XM_004246091), Solyc03g007180 (XM_004234118), Zm00001d004164 (XR_562337), Zm00001d025091 (XM_008664864), Zm00001d015889 (XM_008647228), and Zm00001d053254 (XM_008681506); PlantGDB, Sb06g003790.1; and TAIR, AT1G12380 and AT1G62870. Original photographs used for the figures have been archived at the University of Minnesota Data Repository and can be accessed at https://doi.org/10.13020/D61H4D.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Suppression of cul2 by eli-a.17 and eli-a.18 promotes tillering.
Supplemental Figure S2. Genetic testing of eli-a alleles.
Supplemental Figure S3. Ligule development in eli-a alleles.
Supplemental Figure S4. Mapping eli-a.17 and eli-a.18 on chromosome 2HS.
Supplemental Figure S5. Axillary bud development in Bowman-eli-a.18.
Supplemental Figure S6. Rate of axillary bud and tiller appearance.
Supplemental Figure S7. eli-a spike phenotypes.
Supplemental Figure S8. Secondary cell wall development in culms.
Supplemental Figure S9. Phylogenetic tree of ELI-A homologs.
Supplemental Figure S10. ELI-A expression data.
Supplemental Figure S11. ELI-A in situ hybridizations.
Supplemental Figure S12. Crossing scheme to develop eli-a.17 and Bowman-cul2.b-rob1; eli-a.17 families.
Supplemental Table S1. CAPS markers for eli-a alleles and mapping.
Supplemental Table S2. eli-a.17; cul2.b-rob1 SNP list.
Supplemental Table S3. eli-a.18; cul2.b-rob1 SNP list.
Supplemental Table S4. eli-a.17 SNP list.
Supplemental Table S5. eli-a.18 SNP list.
Supplemental Table S6. cul2.b-rob1 SNP list.
Supplemental Table S7. cul2.b SNP list.
Supplemental Table S8. Homologous ELI-A sequences in other species.
Supplemental Table S9. Plant materials.
Supplemental Table S10. PCR primer sequences.
Supplemental Data S1. RNAseq transcript list.
Acknowledgments
We thank Bruna Bucciarelli for assistance with microscopy, Lin Li and Juan Gutierrez-Gonzalez for help with bioinformatics, Kevin Smith for providing field space, and Jerome Franckowiak for insights on the interpretation of mutants (Department of Agronomy and Plant Genetics, University of Minnesota); Gail Celio and Grant Barthel for help with scanning electron and light microscopy (University Imaging Center, University of Minnesota); Sue Miller for assistance with RT-qPCR (USDA-ARS); and for providing materials, the Nordic Genetic Resource Center, Harold Bockelman (U.S. Department of Agriculture National Small Grains Collection), and Andris Kleinhofs (Washington State University).
Footnotes
This research was supported by a grant from the United States Department of Agriculture-CSREES-NRI Plant Growth and Development program grant # 2004-03440 and funds received from the Triticeae Coordinated Agricultural Project, U.S. Department of Agriculture/National Institute for Food and Agriculture grant number 2011-68002-30029 to G.J.M.
Articles can be viewed without a subscription.
References
- Ahlstrand GG. (1996) Low-temperature low-voltage scanning microscopy (LTLVSEM) of uncoated frozen biological materials: a simple alternative. In Bailey G, Corbett J, Dimlich R, Michael J, Zaluzec N, eds, Proceedings of Microscopy Microanalysis. San Francisco Press, San Francisco, pp 918–919 [Google Scholar]
- Babb S, Muehlbauer GJ (2003) Genetic and morphological characterization of the barley uniculm2 (cul2) mutant. Theor Appl Genet 106: 846–857 [DOI] [PubMed] [Google Scholar]
- Bar M, Ori N (2014) Leaf development and morphogenesis. Development 141: 4219–4230 [DOI] [PubMed] [Google Scholar]
- Becraft PW, Bongard-Pierce DK, Sylvester AW, Poethig RS, Freeling M (1990) The liguleless-1 gene acts tissue specifically in maize leaf development. Dev Biol 141: 220–232 [DOI] [PubMed] [Google Scholar]
- Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, Zinder M, Samach A, Eshed Y, Ori N (2009) The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136: 823–832 [DOI] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M (2011) Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci USA 108: 3424–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch BL, Schmitz G, Rossmann S, Piron F, Ding J, Bendahmane A, Theres K (2011) Shoot branching and leaf dissection in tomato are regulated by homologous gene modules. Plant Cell 23: 3595–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L, Druka A, Stein N, Svensson JT, Wanamaker S, et al. (2009) Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10: 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbert T, Okagaki RJ, Cho S, Heinen S, Boddu J, Muehlbauer GJ (2010) The genetics of barley low-tillering mutants: low number of tillers-1 (lnt1). Theor Appl Genet 121: 705–715 [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring HP, Lin J, Urig H, Salamini F (1999) Clonal analysis of the development of the barley (Hordeum vulgare L.) leaf using periclinal chlorophyll chimeras. Planta 207: 335–342 [Google Scholar]
- Druka A, Franckowiak J, Lundqvist U, Bonar N, Alexander J, Houston K, Radovic S, Shahinnia F, Vendramin V, Morgante M, et al. (2011) Genetic dissection of barley morphology and development. Plant Physiol 155: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fal K, Landrein B, Hamant O (2016) Interplay between miRNA regulation and mechanical stress for CUC gene expression at the shoot apical meristem. Plant Signal Behav 11: e1127497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster BP, Franckowiak JD, Lundqvist U, Lyon J, Pitkethly I, Thomas WTB (2007) The barley phytomer. Ann Bot 100: 725–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franckowiak JD, Konishi T, Lundqvist U (1997) BGS 254, Orange lemma, rob. Barley Genet Newsl 26: 235–236 [Google Scholar]
- Fricke W. (2002) Biophysical limitation of cell elongation in cereal leaves. Ann Bot 90: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M Jr, Blakely CM, Darveau A, Romano PR, Korth MJ, Katze MG (2002) P52rIPK regulates the molecular cochaperone P58IPK to mediate control of the RNA-dependent protein kinase in response to cytoplasmic stress. Biochemistry 41: 11878–11887 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter MK, Doebley JF, Pè ME, Schmidt RJ (2004) The role of barren stalk1 in the architecture of maize. Nature 432: 630–635 [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CM, Jun JH, Nam HG, Fletcher JC (2007) BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19: 1809–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Pautot VA (2015) Beyond the divide: boundaries for patterning and stem cell regulation in plants. Front Plant Sci 6: 1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman AB, Perez ZN, Zhou L, Musingarimi P, Ghirlando R, Hinshaw JE, Craig NL, Dyda F (2005) Molecular architecture of a eukaryotic DNA transposase. Nat Struct Mol Biol 12: 715–721 [DOI] [PubMed] [Google Scholar]
- Humason GL. (1979) Animal Tissue Techniques, Ed 4 WH Freeman, San Francisco [Google Scholar]
- International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716 [DOI] [PubMed] [Google Scholar]
- Javelle M, Marco CF, Timmermans M (2011) In situ hybridization for the precise localization of transcripts in plants. J Vis Exp 57: e3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R, Wang M, Sun Q, Sylvester AW, Hake S, Scanlon MJ (2014) Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. Plant Cell 26: 4718–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Abbott J, Moritz T, Doerner P (2006) Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18: 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kenzo T, Ichie T, Watanabe Y, Hiromi T (2007) Ecological distribution of homobaric and heterobaric leaves in tree species of Malaysian lowland tropical rainforest. Am J Bot 94: 764–775 [DOI] [PubMed] [Google Scholar]
- Kierzkowski D, Nakayama N, Routier-Kierzkowska AL, Weber A, Bayer E, Schorderet M, Reinhardt D, Kuhlemeier C, Smith RS (2012) Elastic domains regulate growth and organogenesis in the plant shoot apical meristem. Science 335: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J (2003) LAX and SPA: major regulators of shoot branching in rice. Proc Natl Acad Sci USA 100: 11765–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA. (2005) The then and now of maize leaf development. Maydica 50: 459–467 [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Geisler M, Springer PS (2009) LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 136: 2423–2432 [DOI] [PubMed] [Google Scholar]
- Leegood RC. (2008) Roles of the bundle sheath cells in leaves of C3 plants. J Exp Bot 59: 1663–1673 [DOI] [PubMed] [Google Scholar]
- Lewis MW, Hake S (2016) Keep on growing: building and patterning leaves in the grasses. Curr Opin Plant Biol 29: 80–86 [DOI] [PubMed] [Google Scholar]
- Lundqvist U, Franckowiak JD (2002) BGS 623, Eligulum-a, eli-a. Barley Genet Newsl 32: 124 [Google Scholar]
- Majorek KA, Dunin-Horkawicz S, Steczkiewicz K, Muszewska A, Nowotny M, Ginalski K, Bujnicki JM (2014) The RNase H-like superfamily: new members, comparative structural analysis and evolutionary classification. Nucleic Acids Res 42: 4160–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan J, Bhattacharya J, Ranjan A (2016) Enhancing crop yield by optimizing plant developmental features. Development 143: 3283–3294 [DOI] [PubMed] [Google Scholar]
- Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, Shaw PD, Marshall D (2013) Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14: 193–202 [DOI] [PubMed] [Google Scholar]
- Moon J, Candela H, Hake S (2013) The Liguleless narrow mutation affects proximal-distal signaling and leaf growth. Development 140: 405–412 [DOI] [PubMed] [Google Scholar]
- Müller D, Schmitz G, Theres K (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N, Smith RS, Mandel T, Robinson S, Kimura S, Boudaoud A, Kuhlemeier C (2012) Mechanical regulation of auxin-mediated growth. Curr Biol 22: 1468–1476 [DOI] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, Kyozuka J (2009) Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33: W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Ruzin SE. (1999) Plant Microtechnique and Microscopy. Oxford University Press, New York [Google Scholar]
- Schmitz G, Theres K (2005) Shoot and inflorescence branching. Curr Opin Plant Biol 8: 506–511 [DOI] [PubMed] [Google Scholar]
- Sylvester AW, Cande WZ, Freeling M (1990) Division and differentiation during normal and liguleless-1 maize leaf development. Development 110: 985–1000 [DOI] [PubMed] [Google Scholar]
- Tavakol E, Okagaki R, Verderio G, Shariati J V, Hussien A, Bilgic H, Scanlon MJ, Todt NR, Close TJ, Druka A, et al. (2015) The barley Uniculme4 gene encodes a BLADE-ON-PETIOLE-like protein that controls tillering and leaf patterning. Plant Physiol 168: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivett CL, Evert RF (1998) Ontogeny of the vascular bundles and contiguous tissues in the barley leaf blade. Int J Plant Sci 159: 716–723 [Google Scholar]
- Van Ooijen JW. (2006) JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma, Wageningen, The Netherlands [Google Scholar]
- Wang Q, Hasson A, Rossmann S, Theres K (2016) Divide et impera: boundaries shape the plant body and initiate new meristems. New Phytol 209: 485–498 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59: 253–279 [DOI] [PubMed] [Google Scholar]
- Wenzel CL, Chandler PM, Cunningham RB, Passioura JB (1997) Characterization of the leaf epidermis of barley (Hordeum vulgare L. ‘Himalaya’). Ann Bot 79: 41–46 [Google Scholar]
- Wu S, Zhang Y (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35: 3375–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Wang Q, Schmitz G, Müller D, Theres K (2012) The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J 71: 61–70 [DOI] [PubMed] [Google Scholar]
- Žádníková P, Simon R (2014) How boundaries control plant development. Curr Opin Plant Biol 17: 116–125 [DOI] [PubMed] [Google Scholar]