The contrasting roles of BBX21 and BBX24 in photomorphogenesis arise through their different C-terminal regions and how they alter HY5 function at the post-transcriptional level.
Abstract
The Arabidopsis (Arabidopsis thaliana) BBX family comprises several positive and negative regulators of photomorphogenesis. BBX24, a member of BBX structural group IV, acts as a negative regulator of photomorphogenesis, whereas another member from the same group, BBX21, is a positive regulator. The molecular basis for the functional diversity shown by these related BBX family members is unknown. Using domain-swap lines, we show that the C-terminal regions of BBX24 and BBX21 specify their function. Because both BBX21 and BBX24 work in close association with HY5, we hypothesized that these proteins differentially regulate the levels or activity of HY5 to fulfill their opposite roles. We show that BBX21 can regulate HY5 post-transcriptionally and the two proteins can coordinate to promote photomorphogenesis. By contrast, BBX24 interferes with the binding of HY5 to the promoter of an anthocyanin biosynthetic gene, possibly by heterodimerizing with HY5 and preventing it from binding DNA. Our finding that both BBX21 and BBX24 regulate HY5 activity post-transcriptionally, in opposite ways, suggests that closely related B-box proteins execute contrasting functions through differential regulation of HY5.
Plants must integrate various external and internal signals to precisely initiate and execute proper developmental responses. Among the various environmental signals to which plants are exposed, light plays crucial regulatory roles throughout a plant’s life cycle. Light influences plant development from the very beginning, facilitating the physiological and molecular changes associated with germination. Thereon, seedling growth is modulated according to the quantity, quality, duration, and direction of available light. By carefully surveying these signals, plants are able to appropriately respond to both seasonal changes and changes in their immediate surrounding environment (Sullivan and Deng, 2003). Perhaps the most dramatic response to light is seen when a dark grown (etiolated) seedling is first exposed to light (de-etiolation). This environmental transition induces the rapid inhibition of hypocotyl elongation, unfolding of the apical hook, opening of the cotyledons, and biosynthesis of anthocyanin/chlorophyll pigments (Sullivan and Deng, 2003).
The contrasting developmental programs shown by seedlings grown in light and dark, better known as photo- and skotomorphogenesis, respectively, feature the involvement of numerous molecular players. Light of different wavelengths is perceived by a set of specialized photoreceptors including phytochromes, cryptochromes, phototropins, and UVR8. Phytochromes detect light from the red/far-red region of the spectra; cryptochromes and phototropins sense the blue region; and UVR8 specifically recognizes UV-B (Galvão and Fankhauser, 2015). The perceived light signals are then transmitted through downstream factors to induce massive transcriptional reprogramming that ultimately alters the morphological and physiological status of the plant (Ma et al., 2001).
ELONGATED HYPOCOTYL5 (HY5) is one of the central factors acting downstream to the photoreceptors to mediate light-regulated developmental responses (Oyama et al., 1997; Gangappa and Botto, 2016). Acting as a master regulator, this bZIP family transcription factor regulates the expression of numerous genes that belong to different pathways and operates in seedling development mainly by positively regulating the key features of photomorphogenesis (Oyama et al., 1997). HY5 is also known to play an important role in integrating the light signals with various other processes such as hormonal signaling, pigment accumulation, abiotic stress responses, circadian rhythm, flowering, shade avoidance response, and nutrient assimilation (Gangappa and Botto, 2016). Although HY5 was originally identified to bind directly to the promoters of its target genes on a conserved G-box motif (CACGTG), high-throughput studies have later suggested that HY5 binds to approximately one-third of all genes in Arabidopsis (Arabidopsis thaliana) (Chattopadhyay et al., 1998; Lee et al., 2007). Concordantly, HY5 has also been shown to target many ACGT-containing elements, including Z-box, C-Box, and hybrid C/G- and C/A-boxes (Song et al., 2008; Zhang et al., 2011). Having such a massive range of targets implies that it is extremely important for HY5 to regulate its target genes in a well-coordinated and systematic manner. Interestingly, in light-dark transition experiments, HY5 is constitutively bound to the promoters of several light- and circadian-regulated genes, indicating that HY5 by itself might not be sufficient to regulate transcription, and that additional cofactors may contribute to such precise regulation (Lee et al., 2007).
Post-transcriptionally, HY5 is tightly regulated by the CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1)/SUPPRESSOR OF phyA-105 (SPA) complex, which comprises a central negative regulator of photomorphogenesis (Lau and Deng, 2012; Menon et al., 2016). In the dark, HY5 interaction with the COP1/SPA complex results in ubiquitination followed by degradation of HY5 (Ang et al., 1998; Osterlund et al., 2000). However, upon light perception, activated photoreceptors directly interact with and inhibit the activity of COP1/SPA, allowing for HY5 accumulation and promotion of photomorphogenesis (Liu et al., 2011; Zuo et al., 2011; Lu et al., 2015; Sheerin et al., 2015). In addition, various other factors act in association with HY5 to fine-tune its activity and carry out light-mediated seedling development. The BBX family of proteins is well known to be among these factors.
The BBX family is composed of a group of Zn finger proteins with one or two conserved B-box motifs in tandem toward their N-terminal region (Gangappa and Botto, 2014). The 32 members of the Arabidopsis BBX family are subdivided into five structural groups based on the number and types of domains they carry (Khanna et al., 2009; Gangappa and Botto, 2014). Although several B-box proteins from different structural groups have been shown to regulate photomorphogenic development (Datta et al., 2006; Holtan et al., 2011), the six HY5 interacting BBX proteins (BBX20 to BBX25) identified thus far belong exclusively to structural group IV (Datta et al., 2007, 2008; Jiang et al., 2012; Gangappa et al., 2013a; Wei et al., 2016; Zhang et al., 2017). Similar to HY5, the accumulation of these proteins in response to light has been shown to be dependent on COP1 (Indorf et al., 2007; Datta et al., 2008; Chang et al., 2011; Fan et al., 2012; Gangappa et al., 2013a; Xu et al., 2016). Furthermore, their action largely appears to require functional HY5 to regulate inhibition of hypocotyl elongation and anthocyanin accumulation in response to light (Datta et al., 2008; Gangappa et al., 2013a; Wei et al., 2016; Xu et al., 2016).
With regard to the many similarities between these B-box proteins, it is intriguing that BBX20, BBX21, BBX22, and BBX23 have been characterized as positive regulators of photomorphogenesis, whereas BBX24 and BBX25 act as negative factors. Recently, it was proposed that BBX21 acts by directly binding the promoter of HY5 to positively regulate its transcription (Xu et al., 2016), and as overexpression of BBX20 similarly results in increased HY5 levels, it is possible that BBX20 acts in a similar manner (Wei et al., 2016). However, BBX21 also regulates HY5 at the post-transcriptional level by physically interacting and interfering with HY5 binding on the ABI5 promoter to fine-tune ABA signaling (Xu et al., 2014). HY5 on the other hand, positively regulates the transcription of BBX22 (Gangappa et al., 2013a), whereas cotransfection assays showed that both BBX24 and BBX25 act on HY5 to interfere with its transcriptional activation of BBX22 (Gangappa et al., 2013a).
Although the HY5 dependency of these group IV BBX proteins for their functioning in light signaling is clear, the nature of the opposing functions within this group is less understood. Hence, it is apt to consider whether the different modes by which these proteins modulate HY5 action might determine their opposing roles in photomorphogenesis. Here, we show that the contrasting functions of the closely related B-box proteins BBX21 and BBX24 are conferred by variations in their C-terminal regions. Furthermore, we provide evidence for post-transcriptional regulation of HY5 by BBX21 in addition to a previously reported transcriptional regulation. Finally, we uncover the mechanism by which BBX24 post-transcriptionally sequesters HY5 to modulate the expression of HY5 target genes. By comparing the post-transcriptional modulation of HY5 by BBX21 and BBX24, we demonstrate how the differential regulation of HY5 by different associated factors can ultimately result in diverse physiological consequences in plants.
RESULTS
BBX21 and BBX24/BBX25 Independently Regulate Hypocotyl Elongation and Anthocyanin Accumulation through HY5
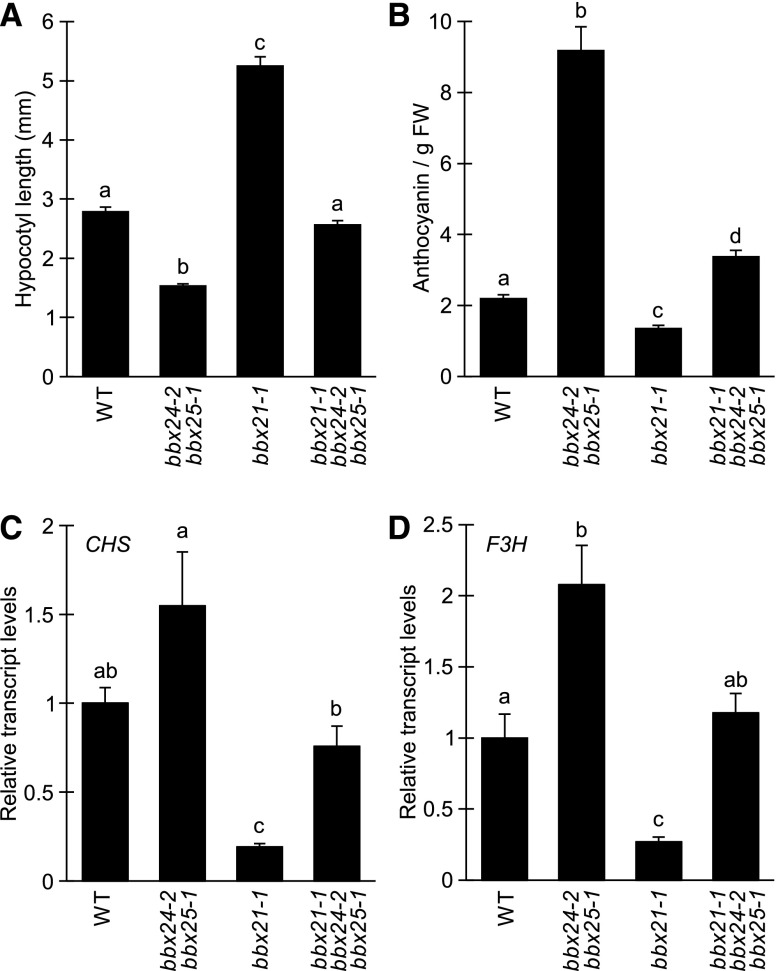
BBX21, a structural group IV BBX protein, positively regulates photomorphogenesis by inhibiting hypocotyl elongation and enhancing anthocyanin accumulation (Datta et al., 2007). In contrast, two other members of the same structural group, BBX24 and BBX25, appear to play an opposite role in photomorphogenesis (Gangappa et al., 2013a). To investigate the genetic relationship between these factors, we generated the bbx24-2bbx25-1bbx21-1 triple mutant plants and proceeded to analyze inhibition of hypocotyl elongation and anthocyanin accumulation. As previously reported, we observed a short hypocotyl and high anthocyanin accumulation in the bbx24-2bbx25-1 mutant whereas bbx21 shows opposite phenotypes (Fig. 1, A and B). Interestingly, the bbx24-2bbx25-1bbx21-1 triple mutant shows an intermediate phenotype, suggesting that these two groups of B-box proteins act largely independently of each other (Fig. 1, A and B). Furthermore, we measured the transcript levels of CHALCONE SYNTHASE (CHS) and FLAVANONE-3-HYDROXYLASE (F3H), two early genes in the anthocyanin biosynthetic pathway, and found a similar relationship where the bbx24-2bbx25-1bbx21-1 triple mutant showed transcript levels similar to the wild type (Fig. 1, C and D).
Figure 1.
BBX21 and BBX24/BBX25 independently regulate anthocyanin biosynthesis. A, Hypocotyl measurements of indicated seedlings grown for 5 d at 75 μmol m−2 s−1 of red light. Error bars represent se, n > 21. B, Anthocyanin content relative to fresh weight of 5-d-old seedlings grown in 75 μmol m−2 s−1 of red light. Error bars represent se, n = 5. C and D, CHS (C) and F3H (D) transcript levels in 4-d-old seedlings grown in 75 μmol m−2 s−1 of red light. Error bars represent se, n = 4. Statistical groups indicated by letters were determined by one-way ANOVA, P ≤ 0.05. FW, fresh weight; WT, wild type.
Although apparently acting independently of each other, BBX21 and BBX24/BBX25 are known to act upstream of HY5 to execute their regulatory roles in hypocotyl elongation and anthocyanin biosynthesis, evident by the epistatic relationship of hy5-215 to bbx24-2bbx25-1 and bbx21-1 (Supplemental Fig. S1, A and B; Datta et al., 2007; Gangappa et al., 2013a). To confirm that this relationship also applies to the regulation of CHS and F3H, we analyzed the transcript levels of these genes in dark-grown seedlings exposed to 2 h of red light. Consistent with reports of these genes being direct targets of HY5 (Shin et al., 2007), the light-induced transcription of CHS and F3H was severely impaired in the hy5-215 mutant. Furthermore, the light-induced expression of these genes was promoted and inhibited in bbx24-2bbx25-1 and bbx21-1, respectively, whereas the regulation in both bbx24-2bbx25-1hy5-215 and bbx21-1hy5-215 was similar to hy5-215 (Supplemental Fig. S1, C and D). Together, these results suggest that although BBX21 and BBX24/25 function independently, their antagonistic regulatory functions require HY5.
BBX24/BBX21 Chimera Retains Molecular Properties Shared between BBX24 and BBX21
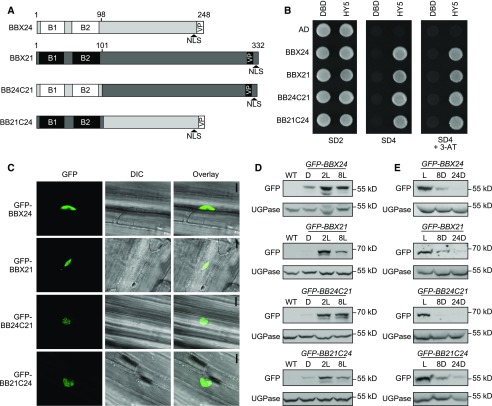
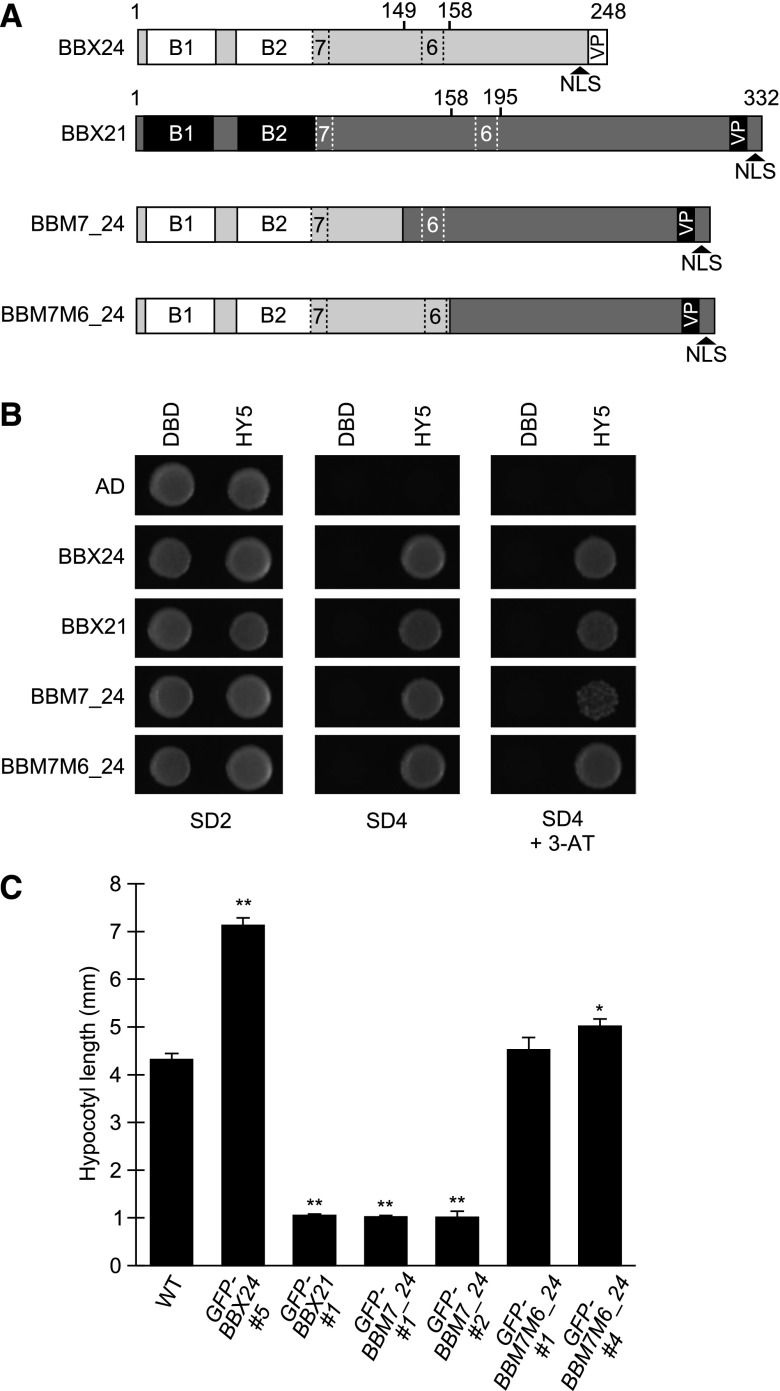
To elucidate the molecular basis of the HY5-dependency of BBX24 and how it influences its function opposite to BBX21, we compared the possible structural differences possessed by BBX24 and BBX21. To do so, the amino acid sequence of BBX24 and BBX21 was aligned using the software Clustal Omega (Sievers et al., 2011). As expected, the sequences showed high amino acid identity in the N-terminal regions (60%) comprising the two B-boxes (Fig. 2A; Supplemental Fig. S2). The residues after the second B-box (after the 98th and 101st amino acid in BBX24 and BBX21, respectively) showed only approximately 23% identity in the alignment (hereafter broadly called the “C-terminal region”), supporting the notion that the functional differences are more likely attributed to the differences in the C-terminal region (Supplemental Fig. S2). To investigate if this is the case, we generated constructs expressing chimeric BBX24 and BBX21 proteins with their C-terminal regions swapped with each other, named “BB24C21” and “BB21C24”, respectively (Fig. 2A). As the B-box regions of both BBX24 and BBX21 previously have been shown to be important for the interaction with the bZIP domain of HY5 (Datta et al., 2008; Gangappa et al., 2013a), the ability to interact with HY5 was tested to confirm the functionality of the chimeric proteins’ N-terminal domains. As previously reported using yeast two-hybrid assay (Datta et al., 2007; Jiang et al., 2012), BBX24 or BBX21 fused to the Gal4 activation domain (AD) expressed with HY5 fused to the LexA DNA binding domain (DBD) results in yeast growth on the SD4 media, indicating that these proteins interact (Fig. 2B). In addition, AD-BB24C21 and AD-BB21C24 expressed together with DBD-HY5 resulted in growth similar to that observed for BBX24 and BBX21, suggesting that their structural integrity and functional viability are intact in the yeast system (Fig. 2B). Quantitative measurements using a β-galactosidase assay further confirmed these observations (Supplemental Fig. S3).
Figure 2.
BBX24/BBX21 chimeric proteins retain expected functions. A, Schematic presentation of full-length BBX24 and BBX21 proteins and the two chimeric versions. B, Yeast two-hybrid assay using HY5 as bait and BBX24, BBX21, BB24C21, and BB21C24 as prey. C, Nuclear localization of GFP-fused domain-swapped chimeric proteins in hypocotyl cells of 4-d-old dark grown seedlings exposed to 2 h of white light (scale bar, 10 μm). D, Immunoblot analysis of GFP-fusion proteins grown for 4 d in darkness (D) and exposed to 75 μmol m−2 s−1 white light for 2 h and 8 h. E, Immunoblot analysis of seedlings grown for 4 d in 75 μmol m−2 s−1 of white light and moved to darkness for 8 h and 24 h. Anti-GFP was used to detect GFP-BBX fusion proteins and anti-UGPase was used as a loading control. Wild type served as a negative control. B1, first B-box; B2, second B-box; SD2, media lacking Trp, Leu; SD4, media lacking Trp, Leu, His, Ura; 3-AT, addition of 1 μM 3-amino-1, 2,4-triazol to the growth media; L, light; 2L, white light for 2 h; 8L, white light for 8 h; 8D, dark for 8 h; 24D, dark for 24 h; WT, wild type.
BBX21 and BBX24 have a putative nuclear localization signal (NLS) sequence at their C terminus, which has been shown to be required for nuclear localization (Yan et al., 2011; Xu et al., 2017). To verify the proper nuclear localization of the chimeric proteins in planta, we monitored the GFP fluorescence in transgenic Arabidopsis lines expressing proteins tagged with GFP at their N terminus under the control of a 35S promoter (designated GFP-BBX24, GFP-BBX21, GFP-BB24C21, and GFP-BB21C24). Visualization of hypocotyl cells of 6-d-old seedlings showed a clear nuclear localization of GFP-BBX24, GFP-BBX21, and of the chimeric GFP-BB24C21 and GFP-BB21C24 proteins (Fig. 2C).
As targets of COP1-mediated degradation, the protein levels of both BBX24 and BBX21 are stabilized by light and degraded in darkness (Datta et al., 2007; Indorf et al., 2007; Yan et al., 2011; Xu et al., 2016). Although BBX24 contains a well-defined C-terminal VP domain important for the interaction with COP1 and the degradation of BBX24 in the dark, BBX21 also harbors an uncharacterized VP pair at the C-terminal end (Holm et al., 2001; Yan et al., 2011; Fig. 2A; Supplemental Fig. S2). To validate the expected light-dependent accumulation pattern of the chimeric proteins, an immunoblot experiment was carried out with transgenic GFP-BBX24, GFP-BBX21, GFP-BB24C21, and GFP-BB21C24 seedlings. Similar to previous reports (Indorf et al., 2007; Xu et al., 2016), we observed accumulation of GFP-BBX24 and GFP-BBX21 in response to 2 h and 8 h of white light compared to darkness, consistent with light inhibition of COP1 activity (Fig. 2D). Likewise, GFP-BB24C21 and GFP-BB21C24 accumulate in response to light, showing no discernible difference to BBX24 and BBX21 (Fig. 2D). Furthermore, when seedlings grown in white light were moved to the dark, chimeric and nonchimeric protein levels gradually decreased after 8 h and 24 h, indicating that the swapped proteins retained their ability to interact with and be regulated by COP1 in the dark (Fig. 2E). From this set of control experiments, we conclude that three features shared between BBX24 and BBX21 (HY5 interaction, nuclear localization, and light regulation) are maintained in BB24C21 and BB21C24, suggesting that both the N-terminal and C-terminal parts of these two chimeras are likely to be functional.
BBX24 Functionally Mimics BBX21 when it Contains the BBX21 C-Terminal Regions
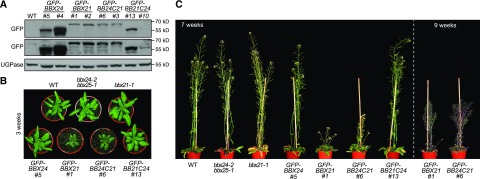
To study the role of C-terminal regions in the functional antagonism shown by BBX24 and BBX21, two independent homozygote lines expressing each of the N-terminally GFP-tagged constructs driven by a 35S promoter were identified. The expression of fusion protein of the expected size of approximately 55 kD (GFP-BBX24 and GFP-BB21C24) and approximately 68 kD (GFP-BBX21 and GFP-BB24C21) were confirmed by immunoblotting probing against the GFP-tag (Fig. 3A). The growth phenotypes of all the different lines, along with the loss-of-function mutants (bbx24-2bbx25-1 and bbx21-1), were compared in both vegetative and flowering stages. At both stages, the bbx24-2bbx25-1 and bbx21-1 mutants did not show any notable difference from the wild-type plants (Fig. 3, B and C). Although the seedling phenotypes of transgenic plants overexpressing BBX21 and its molecular basis are well characterized (Xu et al., 2016), the growth behavior in late vegetative and adult stages has not been reported. Interestingly, the GFP-BBX21 lines showed retarded growth with significantly smaller rosette size in vegetative stage (Fig. 3B) and a dwarf stature during flowering and maturation (Fig. 3C; Supplemental Fig. S4). The overall slow growth of GFP-BBX21 plants also affected its transition from vegetative to reproductive stage, causing considerably delayed flower initiation. If the differences in the C-terminal regions are responsible for the known opposing functions of BBX24 and BBX21 proteins, plants overexpressing the chimeric proteins BB24C21 and BB21C24 are expected to show phenotypes corresponding to their respective C-terminal regions. Justifying this expectation, the GFP-BB24C21 and GFP-BBX21 lines behaved similarly in terms of exhibiting slow growth, delayed flowering, and an overall dwarfed stature. In contrast, transgenic GFP-BB21C24 and GFP-BBX24 plants showed no significant growth defects (Fig. 3, B and C; Supplemental Fig. S4). These phenotypes indicate that the C-terminal region of BBX21 is important for regulating adult development, and further suggests that this region might determine the opposing functions of BBX21 and BBX24.
Figure 3.
BBX24 functionally mimics BBX21 when their C-terminal regions are interchanged. A, Immunoblot analysis of transgenic seedlings expressing GFP-BBX24, GFP-BBX21, GFP-BB24C21, and GFP-BB21C24 driven by the 35S promoter. Samples from 4-d-old dark-grown seedlings exposed to 75 μmol m−2 s−1 of white light were harvested and anti-GFP antibodies were used to visualize the fusion proteins. Upper and middle panels (labeled as GFP) represent a short and a long exposure, respectively. UGPase was used as loading control. B and C, Photograph of representative plants grown for 3 (B), 7 and 9 (C) weeks in long day conditions (16:8 light/dark) of 100 μmol m−2 s−1 white light.
C-Terminal Domains of BBX24 and BBX21 Dictate their Role in Light Signal Transduction
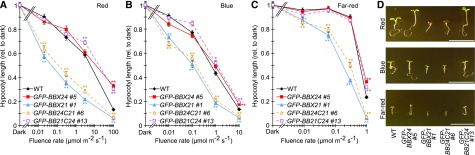
The opposite regulatory roles of BBX24 and BBX21 in photomorphogenesis are manifested in overexpressing lines via longer and shorter hypocotyl lengths, respectively, when compared to the wild type (Indorf et al., 2007; Xu et al., 2016). To investigate the contribution of C-terminal domains in determining the antagonistic roles of BBX24 and BBX21, the hypocotyl lengths of GFP-BB24C21 and GFP-BB21C24 transgenic lines were measured in red, far-red, and blue light under a wide range of intensities and compared with the phenotypes of GFP-BBX24 and GFP-BBX21 seedlings. In all light conditions, the GFP-BB24C21 lines displayed hypersensitivity to light, exhibiting shorter hypocotyls similar to GFP-BBX21. In contrast, GFP-BB21C24 showed longer hypocotyls at high intensities of light, comparable to GFP-BBX24 (Fig. 4; Supplemental Fig. S5). These results support a role for the C-terminal domains of BBX24 and BBX21 in determining their opposite functions during photomorphogenesis.
Figure 4.
C-terminal domains of BBX24 determine its role in light signal transduction. A to C, Hypocotyl measurements of 5-d-old seedlings grown at indicated fluence rate of monochromatic red (A), blue (B), and far-red (C) light. Error bars represent se, n ≥ 17. D, Photograph of representative seedlings grown in 100, 10, and 1 μmol m−2 s−1 of red, blue, and far-red light, respectively. Scale bars, 10 mm. Color-coded * and ** indicates P < 0.05 and P < 0.01, respectively, compared to wild type at each fluence rate as determined by one-way ANOVA. WT, wild type.
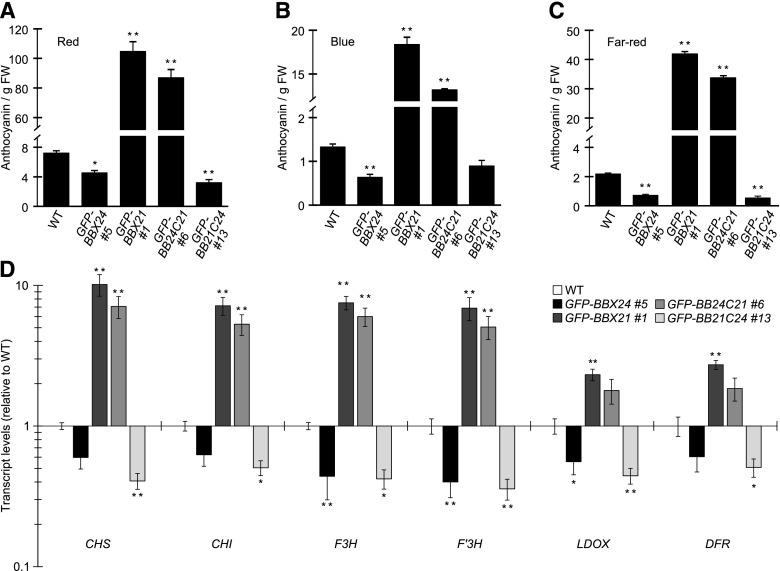
Because BBX24 and BBX21 also regulate light-mediated anthocyanin accumulation, we further asked if the C-terminal region also dictates this function. Overexpression of BBX24 has been reported to reduce anthocyanin accumulation whereas BBX21 overexpression results in increased levels (Indorf et al., 2007; Xu et al., 2016). Consistent with these observations, we noticed reduced and strongly increased anthocyanin levels in the GFP-BBX24 and GFP-BBX21 lines, respectively (Fig. 5, A to C). Furthermore, the chimeric GFP-BB24C21 lines accumulated high levels of anthocyanin similar to that of GFP-BBX21 whereas GFP-BB21C24-expressing lines had reduced anthocyanin content comparable to GFP-BBX24. These phenotypes prevailed in all monochromatic light conditions, i.e., red, far-red, and blue (Fig. 5, A and C; Supplemental Fig. S6). To further support the role of the C-terminal domain of these B-box proteins in regulation of anthocyanin accumulation, the relative expression levels of both early (CHS, CHI, and F3H) and late (F3′H, LDOX, and DFR) genes in the anthocyanin biosynthesis pathway were measured. In line with the phenotypic results, transcript levels of all six genes analyzed were reduced in the GFP-BBX24- and GFP-BB21C24-overexpressing lines whereas they were elevated in GFP-BBX21 and GFP-BB24C21 (Fig. 5D), indicating that it is the C-terminal domain that plays a crucial role in the transcriptional regulation of anthocyanin biosynthesis genes by BBX24 and BBX21.
Figure 5.
BBX24 and BBX21 regulate the transcription of anthocyanin biosynthetic genes dependent on their C-terminal domains. A to C, Anthocyanin content relative to fresh weight of 5-d-old seedlings grown in 100, 10, and 1 μmol m−2 s−1 of red (A), blue (B), and far-red (C) light, respectively. Error bars represent se, n = 4. D, Expression levels of CHS, CHI, F3H, F3′H, LDOX, and DFR in 4-d-old wild-type, GFP-BBX24, GFP-BBX21, GFP-BB24C21, and GFP-BB21C24 seedlings grown in 100 μmol m−2 s−1 of red light shown on a logarithmic scale. Error bars represent se, n = 4. * and ** indicates P < 0.05 and P < 0.01, respectively, compared to wild type as determined by one-way ANOVA, P ≤ 0.05. WT, wild type.
A Possible Role of C-Terminal Motif 6 for BBX21 Function
After mapping the regions of BBX24 and BBX21 that facilitate their opposing functions during photomorphogenic development, we further pursued to resolve the region within the C-terminal sequence. Interestingly, by searching for conserved protein motifs in a large set of plant BBX protein sequences, Motif 6 and Motif 7 (M6 and M7) of unknown function have previously been identified in proteins belonging to the structural group IV (Crocco and Botto, 2013). Although M7, located just adjacent to the second B-box, is highly similar between BBX24 and BBX21, the more C-terminally located M6 shows considerable differences (Supplemental Fig. S2). Compared to BB24C21, which contains the B-box domains and part of M7 from BBX24 followed by the C-terminal domain of BBX21 (Fig. 2A; Supplemental Fig. S2), we created two additional chimeric constructs. In addition to the two B-boxes of BBX24, these constructs further contained the full M7 and 49 additional amino acids (BBM7_24) or both M7 and M6 (BBM7M6_24) followed by the corresponding C-terminal sequence of BBX21 (Fig. 6A; Supplemental Fig. S2). To begin, we confirmed that these two chimeric proteins retained their interaction with HY5 in yeast, suggesting that the B-box domains of these proteins are fully functional (Fig. 6B). To verify the nuclear localization of BBM7_24 and BBM7M6_24 in planta, we observed the GFP fluorescence in transgenic Arabidopsis lines expressing the proteins tagged with GFP at their N terminus and under the control of a 35S promoter (designated GFP-BBM7_24 and GFP-BBM7M6_24; Supplemental Fig. S7). Together, these observations suggest that both the N-terminal B-box domains and the C-terminally located NLS of both BBM7_24 and BBM7M6_24 retain functionality, and that these proteins accumulate in the transgenic lines. To determine the possible effect of BBM7_24 and BBM7M6_24 in photomorphogenic development, we measured the hypocotyl length of two independent lines of each construct grown under red light for 5 d (Fig. 6C). Interestingly, although the two independent GFP-BBM7_24-expressing lines show a strong hypersensitivity to light similar to the GFP-BBX21 line, no hypersensitivity is observed in GFP-BBM7M6_24 #1 and a weak hyposensitivity is observed in GFP-BBM7M6_24 #4 (Fig. 6C). These results suggest that not only are the B-box domains exchangeable between BBX21 and BBX24 for BBX21 function, but also M7 and the following 49 amino acids. Interestingly, although exchanging the M6 region of BBX21 to that of BBX24 renders the protein unable to promote photomorphogenesis, we cannot rule out the possibility that M6 determines the positive functions of BBX21 in light signaling.
Figure 6.
BBX21-specific function is not dependent on Motif 7. A, Schematic presentation of full-length BBX24 and BBX21 proteins and two chimeric versions. B, Yeast two-hybrid assay using HY5 as bait and BBX24, BBX21, BBM7_24, and BBM7M6_24 as prey. C, Hypocotyl measurements of 5-d-old seedlings grown in 100 μmol m−2 s−1 monochromatic red light. Error bars represent se, n ≥ 21. * and ** indicates P < 0.05 and P < 0.01, respectively, compared to wild type as determined by one-way ANOVA, P ≤ 0.05. B1, first B-box; B2, second B-box; 7, Motif 7; 6, Motif 6; SD2, media lacking Trp, Leu; SD4, media lacking Trp, Leu, His, Ura; 3-AT, addition of 1 μM 3-amino-1, 2,4-triazol to the growth media; VP, VP domain; WT, wild type.
Transcriptional and Post-Transcriptional Regulation of HY5 by BBX21
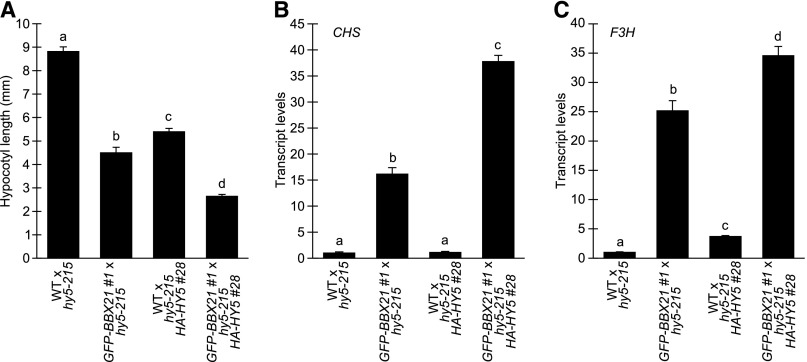
The experiments above indicated the importance of the C-terminal region in determining the functions of BBX24, antagonistic to the functions played by C-terminal region of BBX21. Although it was reported that both these proteins depend upon HY5 to regulate photomorphogenesis, how their HY5-dependency differed from each other to execute the opposite functions remained elusive. Recently, BBX21 was shown to regulate photomorphogenesis by binding to the promoter region of HY5 to directly promote its transcription (Xu et al., 2016). Although this mechanism is consistent with the genetic relationship between HY5 and BBX21 (Supplemental Fig. S1; Datta et al., 2007; Xu et al., 2016), this mechanism does not rely on the observed physical interaction between these two factors (Fig. 2B; Datta et al., 2007). We therefore hypothesized that if BBX21 operates only through transcriptional regulation of HY5, lines overexpressing HY5 should not show any additional phenotype if BBX21 is also overexpressed. On the other hand, an enhanced phenotype in the double overexpressing line might indicate that BBX21 can modulate HY5 post-transcriptionally in addition to its transcriptional regulation. To test these scenarios, we crossed hy5 35S::HA-HY5 with 35S::GFP-BBX21 and analyzed the F1 generation as propagation was not possible due to severely retarded adult growth or obvious transgene silencing. Corresponding control crosses (hy5 x Col-0, hy5 35S::HA-HY5 x Col-0, and 35S::GFP-BBX21 x hy5) were therefore made for each line to keep one functional copy of HY5. To confirm the overexpression in these F1 lines, we analyzed the transcript levels of BBX21 and HY5 by qPCR and found that the transcript levels were similar in 35S::GFP-BBX21 x hy5 35S::HA-HY5 to the corresponding controls (Supplemental Fig. S8). In agreement with previous observations (Xu et al., 2016), HY5 expression in GFP-BBX21 #1 showed a small approximately 1.5-fold increase compared to hy5-215 x Col-0 (Supplemental Fig. S8B). We then analyzed the inhibition of hypocotyl elongation in response to low red light (0.5 µM m−2 s−1), as GFP-BBX21 show a strong, but not saturated, phenotype in this condition (Fig. 4A). Interestingly, whereas both HY5 and BBX21 overexpression results in a reduced elongation in these conditions, overexpression of both factors together results in an additive phenotype (Fig. 7A). These results suggest that, although HY5 is required for BBX21 function (Supplemental Fig. S1; Xu et al., 2016), BBX21 likely acts by a mechanism independent of HY5 transcriptional regulation. To further support these observations, we analyzed the transcript levels of CHS and F3H in the different crosses. Strikingly, whereas GFP-BBX21 strongly induces CHS and F3H, HA-HY5 appears to have no or little effect, respectively (Fig. 7, B and C). However, when overexpressed together, a significant increase of both CHS and F3H is observed compared to either overexpressing line, indicating that BBX21 and HY5 act in concert to regulate these genes, consistent with a post-transcriptional mechanism by which BBX21 regulates HY5 action.
Figure 7.
BBX21 regulates HY5 activity post-transcriptionally. A, Hypocotyl measurement of indicated F1 seedlings grown for 5 d in 0.5 µmol m−2 s−1 of red light. Error bars represent se, n > 21. B and C, Relative CHS (B) and F3H (C) transcript levels in indicated F1 seedlings grown for 4 d in 0.5 μmol m−2 s−1 of red light. Error bars represent se, n = 3. Statistical groups indicated by letters were determined by one-way ANOVA, P ≤ 0.05. WT, wild type.
Nonetheless, we could also confirm that BBX21 positively regulates HY5, whereas no regulation of HY5 transcript levels was observed in the BBX24-overexpressing line (Supplemental Fig. S9). In addition, BBX24 could regulate HY5 transcription when it possessed the C-terminal regions of BBX21, whereas BBX21 lost the ability to transcriptionally regulate HY5 when it possessed the C-terminal domains from BBX24 (Supplemental Fig. S9).
BBX24 Physically Interacts with HY5 and Hinders its Binding on Target Promoters
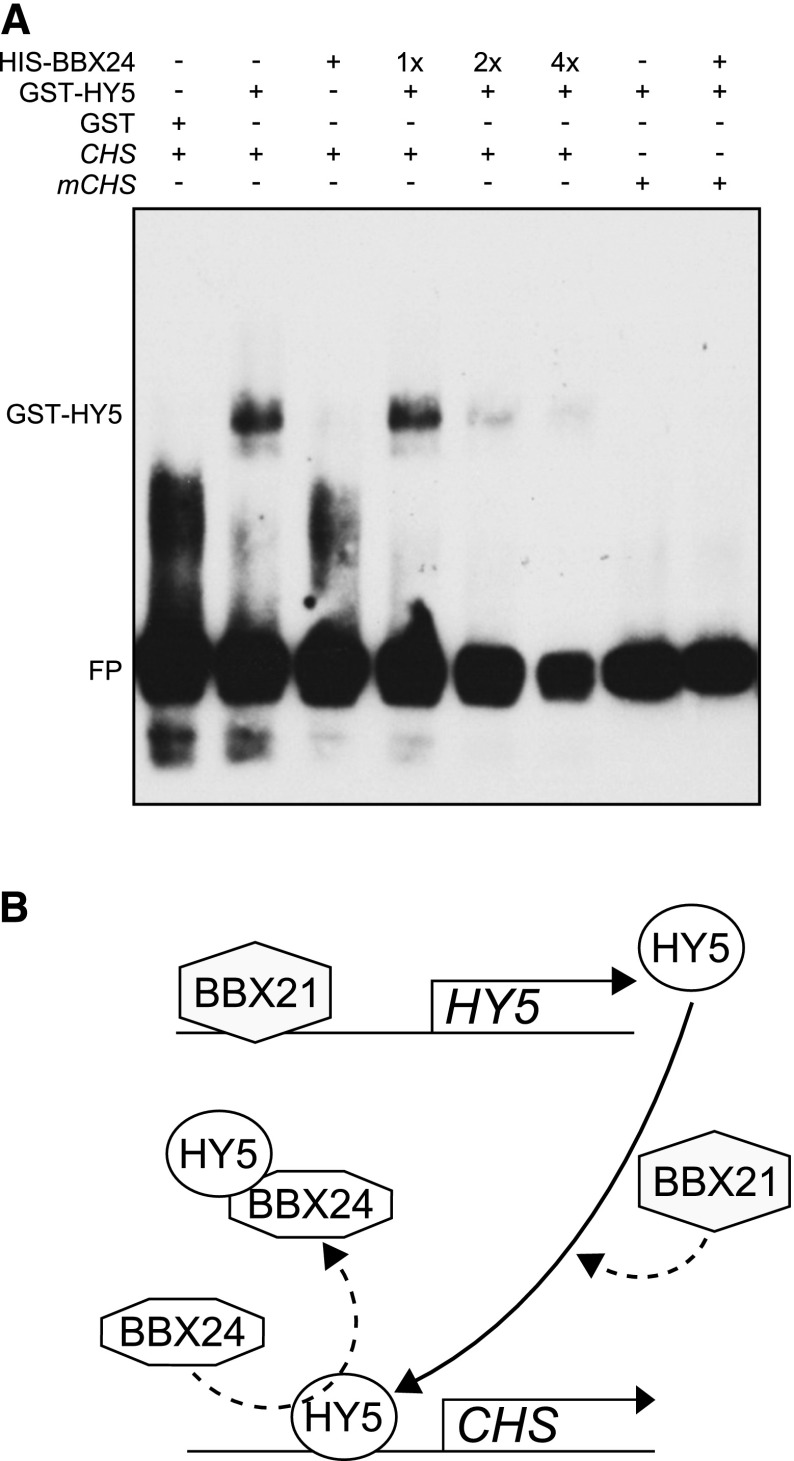
Having determined that BBX21 mainly acts to regulate HY5 at the post-transcriptional level, we also wanted to address the mechanism of BBX24 action. Several lines of evidence support a post-transcriptional mechanism by which BBX24 alters HY5 activity. First, we did not observe any effect of BBX24 overexpression on HY5 transcript levels (Supplemental Fig. S9). Second, BBX24 physically interacts with HY5 (Fig. 2B; Jiang et al., 2012), and this interaction has previously been mapped to the B-boxes of BBX24 and the bZIP domain of HY5 (Gangappa et al., 2013b). Lastly, cotransfection experiments in Arabidopsis protoplasts have revealed that BBX24 and BBX25 inhibit the ability of HY5 to promote BBX22 transcription, and this inhibition was dependent on the functionality of the B-box domains (Gangappa et al., 2013a). We therefore hypothesized that BBX24 might directly interfere with HY5 activity by inhibiting HY5 interaction with promoter regions. To test this, we performed an EMSA using a promoter region of CHS, which previously has been characterized as a direct transcriptional target of HY5 and for which we had observed regulation by BBX24 (Supplemental Fig. S1C; Fig. 5D; Shin et al., 2007; Gangappa et al., 2013a). We analyzed the promoter region of CHS and designed probes containing the HY5-binding G-box motif (CACGTG). Similar to previous reports (Shin et al., 2007), the HY5-binding to the biotin-labeled G-box containing CHS probe was lost when the G-box was mutated (Fig. 8A). Furthermore, upon the addition of an increasing amount of BBX24, we observed reduced binding of HY5 to the G-box of CHS (Fig. 8A). As no signal corresponding to the direct binding of BBX24 to the CHS promoter fragments alone, or in combination with HY5, was detected, we conclude that BBX24 does not compete with HY5 for binding to the G-Box motif. Hence, these data are consistent with the possible formation of non-DNA binding BBX24-HY5 heterodimeric complexes that prevent the binding of HY5 to its target promoters.
Figure 8.
BBX24 interferes with the binding of HY5 to its target promoters. A, EMSA shows the binding of GST-HY5 to the G-box containing promoter fragment of CHS followed by gradual decrement of binding upon increasing HIS-BBX24 (1×, 2×, and 4×). B, Proposed model based on the data from this work (dotted lines) and that of Xu et al. (2016), indicating the regulation of HY5 at transcriptional and post-transcriptional levels by BBX21 and BBX24 to modulate the expression of anthocyanin biosynthesis genes. CHS, biotin-labeled wild-type fragment; mCHS, G-box mutated CHS fragment.
DISCUSSION
The importance of the bZIP transcription factor HY5 in plant development is well documented (Gangappa and Botto, 2016). During the de-etiolation response, light-dependent inhibition of COP1 results in the accumulation of HY5, which ultimately promotes the inhibition of hypocotyl elongation, chlorophyll, and anthocyanin accumulation (Osterlund et al., 2000). However, besides the well-known regulation by COP1, HY5 action is further modulated by a number of other factors (Gangappa and Botto, 2016). In this study, we have further characterized the function of two of these factors, BBX24 and BBX21, two homologous B-box proteins that, respectively, negatively and positively regulate photomorphogenic development dependent on HY5 (Datta et al., 2007; Indorf et al., 2007; Gangappa et al., 2013a; Xu et al., 2016). Our results suggest that the C-terminal regions of BBX24 and BBX21 are responsible for their distinct functions in light-mediated seedling development. Furthermore, we demonstrate that BBX21 regulates HY5 not only at the transcriptional level but also post-transcriptionally to promote photomorphogenesis. In contrast, the physical interaction of BBX24 with HY5 interferes with the ability of HY5 to bind target promoters. These findings suggest distinct and multilayered regulation of HY5 by B-box proteins in structural group IV.
BBX24 and BBX21 Signaling Converge on HY5-Dependent Regulation of Anthocyanin Accumulation
Although belonging to the same structural group of the B-box zinc finger family, BBX24 and BBX25 have been characterized as negative regulators of photomorphogenesis whereas BBX20, BBX21, BBX22, and BBX23 positively regulate light-dependent processes (Datta et al., 2007, 2008; Gangappa et al., 2013a; Wei et al., 2016; Zhang et al., 2017). Furthermore, all these proteins are described as HY5-interacting factors that largely require HY5 for their function when grown in monochromatic light conditions (Datta et al., 2007, 2008; Jiang et al., 2012; Gangappa et al., 2013a; Wei et al., 2016; Zhang et al., 2017). Interestingly, although the functions of BBX24/BBX25 and BBX21 are clearly HY5-dependent (Supplemental Fig. S1), our analysis of the bbx24-2bbx25-1bbx21-1 mutant grown in red light suggests that BBX24/BBX25 and BBX21 act through a mechanism independent of each other (Fig. 1). Furthermore, when analyzing the regulation of anthocyanin biosynthetic genes in response to red light, we found that CHS and F3H are positively and negatively regulated by BBX21 and BBX24/25, respectively (Fig. 1, C and D). As this regulation was completely dependent on HY5 (Supplemental Figure S1, C and D), CHS and F3H represents a point of convergence where BBX24/BBX25 and BBX21 act antagonistically by modulating HY5 action to regulate anthocyanin accumulation.
Functional Diversification of BBX Proteins Is Mapped to the C-Terminal Regions
Two of the BBX proteins in structural group IV, BBX24 and BBX21, possess two tandem repeated B-box motifs in the N-terminal region. Although the C-terminal parts of these proteins are less defined, BBX24 contains a small well-characterized VP domain, located at the C terminus, which is directly responsible for its interaction with COP1 (Holm et al., 2001) and consequently required for BBX24 degradation in the dark (Yan et al., 2011). Similarly, BBX21 also interacts with COP1 (Xu et al., 2016); however, the possible role of a predicted VP domain in the C-terminal part of this protein is currently unknown (Supplemental Fig. S2; Crocco and Botto, 2013). Considering the many common features of these two proteins, it is intriguing that their function seems an opposing one in regard to photomorphogenesis. Therefore, to determine the functional relevance of the B-boxes and the C-terminal domains responsible for the negative and positive role of BBX24 and BBX21, respectively, we constructed two domain swap constructs harboring the B-box domains BBX24 and the C-terminal domain of BBX21 (BB24C21), and vice versa (BB21C24; Fig. 2A). As the structural integrity of the B-box domains of both BBX24 and BBX21 is known to be important for their interaction with HY5 (Datta et al., 2008; Gangappa et al., 2013b), the fact that BB24C21 and BB21C24 retained this ability suggests that the B-box domains of these chimeric proteins are fully functional (Fig. 2B). Likewise, the observed nuclear localization of our four constructs and the light-dependent stabilization in planta is consistent with previously published data (Datta et al., 2007; Indorf et al., 2007; Xu et al., 2016), and suggests that the known functional features of the C-terminal domains were retained (Fig. 2, C and E).
The B-box domains are known to be crucial in mediating protein-protein interactions and transcriptional regulation, and possess domain topology largely conserved across the plant kingdom (Datta et al., 2007; Qi et al., 2012; Crocco and Botto, 2013; Gangappa et al., 2013a). The highly conserved nature of B-box domains, especially within a structural group, suggests that functional diversification within each group is most likely due to changes in the C-terminal domains (Crocco and Botto, 2013). In support of this notion, our phenotypic and molecular analysis of transgenic GFP-BBX24, GFP-BBX21, GFP-BB24C21, and GFP-BB21C24 seedlings clearly suggests that the C-terminal domains strictly determine the function of BBX24 and BBX21 in regulation of hypocotyl elongation (Fig. 4) and anthocyanin accumulation (Fig. 5). Similarly, previous analysis of two B-box proteins in structural group I revealed that exchanging the B-box domains of CONSTANS LIKE1 (COL1/BBX2) with the B-box domains of CONSTANS (CO/BBX1; a central regulator of flowering time) does not confer the ability to promote flowering to COL1. In contrast, swapping the entire first exon of CO with COL1, which contains C-terminal regions after the two B-boxes, is sufficient to convey the functionality of CO to COL1 (Kim et al., 2013). Nonetheless, the B-boxes of CO are still likely to be essential for CO function as many isolated co mutants contain single amino acid substitutions within the first or second B-box domain (Robson et al., 2001). Likewise, point mutations of highly conserved residues within the first and second B-boxes of BBX24 and BBX21, likely disrupting the structural integrity of each B-box domain, have indicated that the B-boxes of both these proteins are essential for their interaction with HY5 and their ability to regulate transcription (Datta et al., 2008; Gangappa et al., 2013a, 2013b). Thus, although the B-box domains appear to be essential for the function of different groups of B-box-containing proteins, our data, and that of others, suggests that the functional diversification within each group is mainly a result of evolutionary diversification of the C-terminal domains. In this context it is interesting to note that M6 and M7 were identified in proteins belonging to the structural group IV, located C-terminally of the two N-terminal B-boxes (Supplemental Fig. S2; Crocco and Botto, 2013). By constructing two additional chimeras, BBM7_24 and BBM7M6_24, we were able to show that the highly conserved M7 was interchangeable between BBX24 and BBX21, meaning that M7 does not contribute to the divergent functions of these proteins (Fig. 6). In contrast, the BBM7M6_24 chimera, which contains the two B-boxes M7 and M6 from BBX24, followed by the corresponding C-terminal sequence of BBX21, failed to promote photomorphogenesis (Fig. 6). Although the functional relevance of these two motifs remains unknown, it is tempting to speculate that M6 plays some role in determining the function of the C-terminal domain in light signaling.
Opposing Modes of Post-Transcriptional Regulation of HY5 by BBX21 and BBX24
Functional diversification within the same family of proteins has previously been reported. In Arabidopsis, the PHYTOCHROME INTERACTING FACTORSs (PIFs) and LONG HYPOCOTYL IN FAR-RED1 (HFR1) belong to subfamily 15 of the bHLH superfamily and promote skotomorphogenesis and photomorphogenesis, respectively (Toledo-Ortiz et al., 2003; Yang et al., 2005; Leivar et al., 2008; Leivar and Quail, 2011). For these factors, the opposing roles are explained by the formation of HFR1-PIF heterodimers that inhibit the promoter binding of the PIFs to their target genes (Hornitschek et al., 2009; Lorrain et al., 2009; Shi et al., 2013, 2015). In addition, PIF4 and PIF5 directly promote the expression of HFR1, thus forming a negative feedback loop (Lorrain et al., 2009). Interestingly, as BBX24 was recently shown to associate with BBX20, which similar to BBX21 also promotes HY5 transcript levels when overexpressed (Wei et al., 2016), it is tempting to speculate that a similar regulation takes place between these two groups of B-box proteins. However, our analysis of the bbx24-2bbx25-1bbx21-1 triple mutant suggests that these factors act independently (Fig. 1). Although this genetic relationship might be masked by possible overlapping functions of BBX20, BBX21, BBX22, and BBX23, the fact that BBX24-overexpressing lines do not reduce HY5 transcript levels, is not supportive of this model (Supplemental Fig. S9A). Instead, we show that BBX21, in addition to its well-characterized transcriptional regulation of HY5 (Xu et al., 2016), is also a potent regulator of HY5 at the post-transcriptional level and acts in concert with HY5 to regulate downstream genes (Fig. 7). The fact that we only observe a 1.5-fold to 2-fold increase of HY5 transcript levels in our BBX21-overexpressing line (Supplemental Figs. S8 and S9B) and that overexpression of HY5 has been reported to not cause strong hypersensitivity to light (Ang et al., 1998), suggests that the post-transcriptional regulation of HY5 by BBX21 might play a more important role in photomorphogenesis (Fig. 8B). In contrast, BBX24 inhibits the ability of HY5 to bind to its target promoters, possibly by the formation of BBX24-HY5 heterodimers that are unable to bind HY5 target sequences (Fig. 8A). This proposed mechanism is consistent with the observed requirement of HY5 for BBX24 function (Supplemental Fig. S1). Furthermore, as one target of HY5 transcriptional activity, we have previously shown that HY5 activates the BBX22 promoter in transiently transfected protoplast assays. Coexpression of BBX24 or BBX25 with HY5 significantly reduced BBX22 promoter activity (Gangappa et al., 2013a), which is consistent with the proposed mechanism of formation of non-DNA binding BBX24-HY5 heterodimers (Fig. 8). The importance of the C-terminal domain for the antagonistic roles and the post-transcriptional regulation of HY5 in opposite ways from BBX21 also suggests a role for the C-terminal regions of BBX24 in heterodimer formation with HY5. In addition, the fact that intact B-box domains appear essential for the function of BBX24 (Gangappa et al., 2013b) implies that the B-box domains and the C-terminal regions might cooperate to mediate its regulation of HY5. Hence, it is probable that one region mediates the initial interaction with HY5 or stabilizes the interaction whereas the other region hinders the DNA binding domain of HY5, ultimately resulting in its reduced target binding.
In conclusion, our data indicate that the opposite roles played by BBX24 and BBX21 in photomorphogenesis are determined by their C-terminal regions whereas the N-terminal B-box domains are interchangeable. This also suggests that the functional diversity within B-box proteins in structural group IV is caused by divergence of the C-terminal domains. Furthermore, we have shown that both these factors act at the post-transcriptional level to regulate HY5 action in opposite ways to ultimately fine-tune the plant’s responses to changing light conditions. Our study opens up the perspective of further investigations to identify the motifs and residues in the C-terminal regions of BBX proteins responsible for the precise regulation of photomorphogenesis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The bbx21-1, bbx24-2bbx25-1, hy5-215, bbx21-1hy5-215, and bbx24-2bbx25-1hy5-215 mutant lines are all in the Col-0 background and have been described in Datta et al. (2007) and Gangappa et al. (2013a). The bbx21-1bbx24-2bbx25-1 triple mutant was obtained by crossing bbx21-1 with bbx24-2bbx25-1 followed by PCR-based genotyping in the F2 generation.
For experiments in monochromatic light, seeds were surface-sterilized and sown on 1/2 MS media containing 1% (w/v) agar without Suc. To promote synchronized germination, the seeds were stratified for 3 d at 4°C before being exposed to a 2 h white light pulse (approximately 75 μmol m−2 s−1) at 21°C. After the light pulse, the seeds were kept in darkness for 22 h before being moved to the experimental conditions. For analysis of adult development, seeds were sown directly on soil and stratified for 3 d at 4°C before being transferred to a growth cabinet at 21°C with a diurnal 16:8 long day light cycle (100 µmol m−2 s−1 white light).
Creation of Overexpressing Lines
To create the overexpression lines, the coding sequence of BBX24 and BBX21 was amplified from cDNA using the primer pairs attB1_BBX24_FP and attB2_BBX24_RP, and attB1_BBX21_FP and attB2_BBX21_RP, respectively, followed by recombination into the pDONR221 (Invitrogen) vector using the BP GATEWAY system reaction. To create the BB24C21 domain swap that constitutes amino acids 1 to 98 from BBX24 and amino acids 102 to 332 from BBX21, the primer pairs attB1_BBX24_FP and 24N_RP, and 21C_FP and attB2_BBX21_RP, were used, respectively. The two fragments were then mixed and used as template for a consecutive PCR reaction using the primer pair attB1_BBX24_FP and attB2_BBX21_RP, to create BB24C21, which was then recombined into the pDONR221 vector. Likewise, to create the BB21C24 domain swap construct that constitutes amino acids 1 to 101 from BBX21 and amino acids 99 to 248 from BBX24, the primer pairs attB1_BBX21_FP and 21N_RP, and 24C_FP and attB2_BBX24_RP, were used, respectively. The two fragments were then used as template for a consecutive PCR reaction using the primer pair attB1_BBX21_FP and attB2_BBX24_RP, to create BB21C24, which was then also recombined into the pDONR221 vector.
To create BBM7_24 containing amino acids 1 to 149 from BBX24 followed by amino acids 159 to 332 of BBX21, the primer pairs attB1_BBX24_FP and 24M21M_RP, and 24M21M_FP and attB2_BBX21_RP, were used, respectively. The two PCR fragments were purified and used together as template for a consecutive PCR reaction using the primer pair attB1_BBX24_FP and attB2_BBX21_RP, to create BBM7_24. Similarly, to create BBM7M6_24 containing amino acids 1 to 158 from BBX24 followed by amino acids 193 to 332 of BBX21, the primer pairs attB1_BBX24_FP and 24MM21_RP, and 24MM21_FP and attB2_BBX21_RP, were used, respectively, and later fused by a PCR reaction using the primer pair attB1_BBX24_FP and attB2_BBX21_RP. The BBM7_24 and BBM7M6_24 PCR fragments were then recombined into the pDONR221 vector.
The six constructs in the pDONR221 vector were then shuttled to the GATEWAY-compatible vector pB7WGF2 (Karimi et al., 2002) through an LR reaction resulting in fusion proteins containing an N-terminal eGFP driven by the 35S promoter. 35S::GFP-BBX24, 35S::GFP-BBX21, 35S::GFP-BB24C21, 35S::GFP-BB21C24, 35S::GFP-BBM7_24, and 35S::GFP-BBM7M6_24 transgenic plants were then created by transforming these vectors into Agrobacterium tumefaciens (GV3101) followed by transformation into Arabidopsis (Arabidopsis thaliana; Col-0) by the floral-dipping method (Zhang et al., 2006). Primary transformants and homozygote transgenic lines were selected by their resistance to Basta.
To create the hy5-215 35S::HA-HY5 transgenic plants, the HY5 coding sequence was amplified using the attB1_HY5_FP, attB2_HY5_RP primer pair from a cDNA template, followed by recombination into the pDONR221 vector using the GATEWAY system BP reaction. HY5 in the pDONR221 vector was then shuttled to the GATEWAY-compatible vector pGWB15 (Nakagawa et al., 2007) through an LR reaction resulting in a fusion protein containing an N-terminal 3xHA tag. This vector was then transformed into the Arabidopsis hy5-215 mutant to generate stable homozygote hy5-215 35S::HA-HY5 transgenic plants.
All primer sequences used for the cloning of these constructs are listed in Supplemental Table S1.
Hypocotyl and Anthocyanin Measurements
For measurements of hypocotyl lengths, seedlings were flattened and photographed followed by measurements of the hypocotyl length using the software ImageJ (https://imagej.nih.gov/ij/). To measure anthocyanin levels, seedlings were harvested, weighed, and frozen in liquid nitrogen. Each sample was then ground before the addition of 600 μL extraction buffer (1% (v/v) HCl in methanol). The samples were then incubated at 4°C overnight, before addition of 200 μL H2O and 650 μL chloroform. Absorbance of the aqueous phase was measured at 530 nm and 657 nm, and anthocyanin quantification was performed by the equation A530 − 0.33 A657/(tissue weight in grams).
Yeast Two-Hybrid Assay
For preparing the vectors used for the yeast assays, HY5 in the pDONR221 vector was shuttled to the GATEWAY-compatible vector pBTM116 through an LR reaction resulting in a fusion protein containing an N-terminal LexA DNA binding domain. Likewise, BBX24, BBX21, BB24C21, BB21C24, BBM7_24, and BBM7M6_24 were shuttled from the pDONR221 vector into pGAD42, resulting in an N-terminal fusion to the Gal4 activation domain. Yeast two-hybrid experiments were performed essentially as described in Dortay et al. (2008). In short, single colonies of primary transformants harboring the AD and DBD constructs were grown in liquid media lacking Trp and Leu. Cells were pelleted and OD600 was adjusted to 0.3. Cells were then dropped on SD2 (-Trp, -Leu), SD4 (-Trp, -Leu, -His, -Ura), and SD4 + 1 μM of 3-amino-1, 2,4-triazol (3-AT) and grown for 5 d at 30°C. Alternatively, β-galactosidase activity was measured in liquid cultures as described in the Yeast Protocols Handbook (Clontech).
Confocal Microscopy
Seedlings expressing GFP were grown in darkness on vertically positioned plates for 3 d or 4 d (as indicated) before being exposed to a 2 h white light pulse. Seedlings were mounted on glass slides and visualized with an LSM 780 confocal microscope (Carl Zeiss).
Immunoblotting
Extraction of total protein was carried out as described in Duek et al. (2004). In short, the harvested tissue was quickly frozen in liquid nitrogen and ground to a fine powder using a tissue lyser. After the addition of extraction buffer [50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% (w/v) sodium deoxycholate, 0.5% Triton X-100, 1 mm DTT, 50 µM each of MG132, MG115, ALLN, PS1, and 1× Complete protease inhibitor cocktail (EDTA-free, Roche)] the samples were centrifuged for 10 min at 4°C and the total protein sample was collected from the supernatant. Samples were then separated on a 10% SDS-PAGE gel and transferred to a PVDF membrane. Anti-GFP (Cat. no. ab290; Abcam) and Anti-UGPase (Cat. no. AS05 086; Agrisera) were used at a 1:2000 and 1:3000 dilution, respectively, followed by the secondary Anti-rabbit-HRP (Cat. no. A0545; Sigma-Aldrich) 1:2000.
Analysis of Transcript Levels
Samples for RNA extraction were grown for 4 d as described, before being harvested and frozen in liquid nitrogen. The samples were ground using a tissuelyser and total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions, including on-column DNAse treatment. cDNA was synthesized using Superscript III Reverse Transcriptase (Invitrogen) and a combination of oligo (dT25) and random N9 primers. Primers used for the qPCR reactions are listed in Supplemental Table S1 and the qPCR was performed with the CFX96 Real-Time System (Bio-Rad). For analysis of transcript levels, ACT2 and UBC21 were used as reference genes, and transcript levels relative to the control were calculated according to Vandesompele et al. (2002).
Statistical Analysis
Statistical analyses were performed using the software Prism7.03 (GraphPad Software). The data were log-transformed and analyzed by a one-way ANOVA, followed by Tukey’s or Dunnett’s post hoc test depending on whether one compares all data to itself (Tukey) or only to one control (Dunnett’s); P ≤ 0.05.
EMSA
Full-length coding sequences of BBX24 and HY5 were cloned into pET28a (for N-terminal 6× His tag) and pGEX-4T-1 (for N-terminal GST tag) vectors, respectively (primers are listed in Supplemental Table S1). Proteins were expressed in Escherichia coli BL21 cells by 0.1 mM of isopropyl β-d-1-thiogalactopyranoside. His-tagged proteins were purified using Ni-NTA agarose beads (Qiagen) and GST-tagged proteins were purified using Glutathione Agarose Resins (ABT). EMSA was done using biotin-labeled probes and the Light Shift Chemiluminescent EMSA Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, 50 bp of synthesized single-stranded oligonucleotides was biotinylated (Biotin 3′ End DNA Labeling Kit; Pierce), and complementary strands were annealed to obtain a double strand probe (Supplemental Table S1). A quantity of 20 fmol of biotinylated probes was incubated with 800 ng (1×) of purified GST, GST-HY5, and 6XHIS-BBX24 in the presence of 10 mm Tris-HCl pH 7.5, 1 mm DTT, 150 mm KCl, 50 ng/μL poly (dI-dC), 2.5% (v/v) glycerol, 0.05% (v/v) Nonidet P-40, 0.1 mg/mL BSA and 100 mm MgCl2 for 20 min at room temperature and separated on 6% (v/v) native polyacrylamide gels in 0.5× TBE. The gels were electroblotted onto a positively charged nylon membrane (Amersham Hybond-N+; GE Healthcare Life Sciences) in 0.5× TBE for 50 min followed by 20 min UV-B cross-linking at 302 nm using a Transilluminator (Thermo Fisher Scientific). Labeled probes were detected with the Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific), using the manufacturer’s instructions.
Accession Numbers
Sequence data from this article are found in the GenBank/EMBL data libraries under the following accession numbers: BBX21/STH2 (AT1G75540), BBX24/STO (AT1G06040), BBX25/STH (AT2G31380), HY5 (AT5G11260), CHS (AT5G13930).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The hy5 mutant is epistatic to bbx24bbx25 and bbx21.
Supplemental Figure S2. Alignment of BBX24 and BBX21.
Supplemental Figure S3. BB24C21 and BB21C24 interact with HY5 in yeast.
Supplemental Figure S4. Adult growth of second transgenic lines.
Supplemental Figure S5. Hypocotyl measurements in monochromatic light and darkness.
Supplemental Figure S6. Measurements of anthocyanin levels in second transgenic lines.
Supplemental Figure S7. Nuclear localization of GFP-BBM7_24 and GFP-BBM7M6_24.
Supplemental Figure S8. BBX21 and HY5 transcript levels in F1 crosses.
Supplemental Figure S9. BBX21 promotes HY5 transcript levels.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Neha Upadhyay for help with confocal imaging and Jan Erik Leuendorf for technical assistance.
Footnotes
Articles can be viewed without a subscription.
This project was funded by the Deutsche Forschungsgemeinschaft (DFG grant JO 1409/1-1) and DRS Postdoc Fellowship Program POINT of the Freie Universität Berlin to H.J. and grants from the Department of Biotechnology and SERB, Government of India to S.D.; N.J. and P.Y., respectively, acknowledge DST-INSPIRE and DBT, Government of India for their fellowships; and S.D. acknowledges IISER Bhopal for research support.
References
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Chang CS, Maloof JN, Wu SH (2011) COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol 156: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocco CD, Botto JF (2013) BBX proteins in green plants: insights into their evolution, structure, feature and functional diversification. Gene 531: 44–52 [DOI] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M (2007) SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GHCM, Deng X-W, Holm M (2006) Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M (2008) LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortay H, Gruhn N, Pfeifer A, Schwerdtner M, Schmülling T, Heyl A (2008) Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. J Proteome Res 7: 3649–3660 [DOI] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Fan XY, Sun Y, Cao DM, Bai MY, Luo XM, Yang HJ, Wei CQ, Zhu SW, Sun Y, Chong K, Wang ZY (2012) BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol Plant 5: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C (2015) Sensing the light environment in plants: photoreceptors and early signaling steps. Curr Opin Neurobiol 34: 46–53 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19: 460–470 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2016) The multifaceted roles of HY5 in plant growth and development. Mol Plant 9: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Crocco CD, Johansson H, Datta S, Hettiarachchi C, Holm M, Botto JF (2013a) The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25: 1243–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Holm M, Botto JF (2013b) Molecular interactions of BBX24 and BBX25 with HYH, HY5 HOMOLOG, to modulate Arabidopsis seedling development. Plant Signal Behav 8: 37–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW (2001) Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J 20: 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtan HE, Bandong S, Marion CM, Adam L, Tiwari S, Shen Y, Maloof JN, Maszle DR, Ohto MA, Preuss S, Meister R, Petracek M, et al. (2011) BBX32, an Arabidopsis B-box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol 156: 2109–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indorf M, Cordero J, Neuhaus G, Rodríguez-Franco M (2007) Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J 51: 563–574 [DOI] [PubMed] [Google Scholar]
- Jiang L, Wang Y, Li QF, Björn LO, He JX, Li SS (2012) Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res 22: 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu S-H (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-K, Park H-Y, Jang YH, Lee JH, Kim J-K (2013) The sequence variation responsible for the functional difference between the CONSTANS protein, and the CONSTANS-like (COL) 1 and COL2 proteins, resides mostly in the region encoded by their first exons. Plant Sci 199-200: 71–78 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Trevisan M, Pradervand S, Fankhauser C (2009) Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J 60: 449–461 [DOI] [PubMed] [Google Scholar]
- Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL, Yang HQ (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8: 467–478 [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon C, Sheerin DJ, Hiltbrunner A (2016) SPA proteins: SPAnning the gap between visible light and gene expression. Planta 244: 297–312 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Gibson A, Fu X, Zheng M, Kuehn R, Wang Y, Wang Y, Navarro S, Morrell JA, Jiang D, Simmons G, Bell E, et al. (2012) Involvement of the N-terminal B-box domain of Arabidopsis BBX32 protein in interaction with soybean BBX62 protein. J Biol Chem 287: 31482–31493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F, Costa MM, Hepworth SR, Vizir I, Piñeiro M, Reeves PH, Putterill J, Coupland G (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J 28: 619–631 [DOI] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Mo X, Tang C, Zhong S, Deng XW (2015) Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc Natl Acad Sci USA 112: 3817–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhong S, Mo X, Liu N, Nezames CD, Deng XW (2013) HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25: 3770–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Yoo CM, Hong AP, Kim SH, Jeong HJ, Shin SY, Kim HJ, Yun DJ, Lim CO, Bahk JD, Lee SY, Nagao RT, et al. (2008) DNA-binding study identifies C-box and hybrid C/G-box or C/A-box motifs as high-affinity binding sites for STF1 and LONG HYPOCOTYL5 proteins. Plant Physiol 146: 1862–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol 260: 289–297 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C-Q, Chien C-W, Ai L-F, Zhao J, Zhang Z, Li KH, Burlingame AL, Sun Y, Wang Z-Y (2016) The Arabidopsis B-box protein BZS1/BBX20 interacts with HY5 and mediates strigolactone regulation of photomorphogenesis. J Genet Genomics 43: 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jiang Y, Li J, Lin F, Holm M, Deng XW (2016) BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci USA 113: 7655–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li J, Gangappa SN, Hettiarachchi C, Lin F, Andersson MX, Jiang Y, Deng XW, Holm M (2014) Convergence of light and ABA signaling on the ABI5 promoter. PLoS Genet 10: e1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DB, Gao SQ, Ma YN, Wang XT, Feng L, Li LC, Xu ZS, Chen YF, Chen M, Ma YZ (2017) The G-protein β subunit AGB1 promotes hypocotyl elongation through inhibiting transcription activation function of BBX21 in Arabidopsis. Mol Plant 10: 1206–1223 [DOI] [PubMed] [Google Scholar]
- Yan H, Marquardt K, Indorf M, Jutt D, Kircher S, Neuhaus G, Rodríguez-Franco M (2011) Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol 156: 1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65: 346–358 [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin S-S, Niu Q-W, Chua N-H (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zhang X, Huai J, Shang F, Xu G, Tang W, Jing Y, Lin R (2017) A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol 174: 2487–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]