The Arabidopsis CBP60a and two CALMODULIN-LIKE (CML) genes negatively regulate plant immunity, with the CML genes affecting amplitude and CBP60a controlling duration.
Abstract
Plant immune responses activated through the perception of microbe-associated molecular patterns, leading to pattern-triggered immunity, are tightly regulated. This results in low immune responses in the absence of pathogens and a rapid return to the resting state following an activation event. Here, we show that two CALMODULIN-LIKE genes, CML46 and CML47, negatively regulate salicylic acid accumulation and immunity in Arabidopsis (Arabidopsis thaliana). The double mutant cml46 cml47 is highly resistant to the pathogen Pseudomonas syringae pv maculicola (Pma). The effects of cml46 cml47 on Pma growth are genetically additive to that of cbp60a, a known negative regulator in the CALMODULIN-BINDING PROTEIN60 (CBP60) family. Transcriptome profiling revealed the effects of cbp60a and cml46 cml47 on both common and separate sets of genes, with the majorities of these differentially expressed genes being Pma responsive. CBP60g, a positive regulator of immunity in the CBP60 family, was found to be transcriptionally regulated by CBP60a, CML46, and CML47. Analysis of the flg22-induced mRNA levels of CBP60g in cbp60a and cml46 cml47 revealed that cml46 cml47 plants have higher induced expression while cbp60a plants retain elevated levels longer than wild-type plants. Assays for the effect of flg22 treatment on Pma growth showed that the effect is stronger in cml46 cml47 plants and lasts longer in cbp60a plants. Thus, the expression pattern of CBP60g is reflected in flg22-induced resistance to Pma.
Plants living in natural environments are exposed to various environmental cues and microorganisms both pathogenic and nonpathogenic. Although plants have evolved an effective and robust immune system that defends them against most pathogens, activating immune responses usually comes at the cost of reduced growth and seed yield. Therefore, tight control of immune responses to effectively combat pathogens while avoiding unnecessary activation is key to maximizing plant fitness.
The presence of molecules characteristic of microbes, termed microbe-associated molecular patterns (MAMPs), such as the flg22 peptide derived from bacterial flagellin, is recognized by specific pattern recognition receptors located on the cell surface, resulting in pattern-triggered immunity (PTI; Zipfel et al., 2004; Jones and Dangl, 2006; Boller and Felix, 2009). Early defense responses during PTI include rapid influxes of Ca2+, production of reactive oxygen species, activation of MAPK cascades, callose deposition, and characteristic gene expression changes (Wu et al., 2014). PTI signaling is amplified through hormone production, which, in turn, reinforces transcriptome reprogramming (Li et al., 2016).
In Arabidopsis (Arabidopsis thaliana), three members of the CALMODULIN-BINDING PROTEIN60 (CBP60) gene family have roles in immune signaling. Two of these, CBP60g and SYSTEMIC ACQUIRED RESISTANCE DEFICIENT1 (SARD1), are required for full induction of many immune-responsive genes (Wang et al., 2009, 2011; Zhang et al., 2010), including SALICYLIC ACID INDUCTION DEFICIENT2 (SID2)/ISOCHORISMATE SYNTHASE1 (ICS1), which encodes an enzyme required for the synthesis of the immunity hormone salicylic acid (SA; Wildermuth et al., 2001). CBP60g and SARD1 are partially redundant. Single mutants are somewhat more susceptible to Pseudomonas syringae pv maculicola (Pma) strain ES4326 and have reduced SA levels following infection, while cbp60g sard1 double mutants have severe defects in limiting Pma growth and greatly reduced SA levels (Wang et al., 2011). Many genes requiring CBP60g or SARD1 for full pathogen-induced expression have an enrichment of the sequence GAAATTT in their promoters (Wang et al., 2011), and electrophoretic mobility shift assay and chromatin immunoprecipitation sequencing (ChIP-seq) data suggest that CBP60g and SARD1 can bind this sequence (Zhang et al., 2010; Sun et al., 2015). SARD1 interacts physically with the TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) family transcription factor TCP8, a positive regulator of SID2/ICS1 expression shown to bind its promoter by chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assay (Wang et al., 2015). Thus, CBP60 proteins are likely transcriptional regulators.
A third CBP60 family member, CBP60a, encodes a negative regulator of immunity (Truman et al., 2013). Plants with cbp60a mutations are more resistant to Pma, and infected plants have higher levels of SA. Several immune-responsive genes, including SID2, are expressed at higher levels than the wild type in mock-infected cbp60a mutants. It seems likely that CBP60a also is a transcriptional regulator, but this hypothesis has not been tested.
CBP60g and SARD1 are strongly inducible by treatment with flg22 or Pma infection (Wang et al., 2009, 2011), while CBP60a is only slightly inducible (Truman et al., 2013). Recent work on the control of SARD1 expression found that the transcription factor WRKY70 binds the SARD1 promoter and represses SARD1 expression in the absence of infection (Zhou et al., 2018). In addition, the transcription factor TGA1 binds the SARD1 promoter in ChIP assays, and Pma-induced SARD1 expression is reduced in tga1 mutants (Sun et al., 2018).
CBP60g and CBP60a bind calmodulin (CaM) in a Ca2+-dependent manner (Wang et al., 2009; Truman et al., 2013), while SARD1 does not bind CaM (Wang et al., 2011). These observations implicate the CBP60 proteins in Ca2+ signaling, which is known to be important for immune responses (for review, see Yuan et al., 2017). For both CBP60g and CBP60a, point mutants unable to bind CaM fail to complement null mutants, indicating that CaM binding is required for their functions. CaM is a highly conserved protein in eukaryotes, and the Arabidopsis genome includes seven CaM genes, several of which encode identical proteins. In addition to CaM, many plant genomes also include many CALMODULIN-LIKE (CML) genes. There are 50 CMLs in Arabidopsis. The CMLs contain various numbers of predicted Ca2+-binding EF-hand motifs, but they vary in protein size and contain additional sequences that lack similarity to any domains of known function (Zhu et al., 2015). Thus, CMLs may affect some CaM-binding proteins or otherwise mediate cellular responses to Ca2+. We noticed that two CML genes, CML46 and CML47, are tightly coexpressed with the CBP60g/SARD1/SID2 signaling regulon (Truman and Glazebrook, 2012). This, together with the fact that CBP60a and CBP60g require CaM binding for their functions, raised the possibility that CML46 and CML47 have roles in immunity.
In this study, we found that CML46 and CML47 are both negative regulators of immunity, and the double mutation cml46 cml47 has broad impact on repression of immune signaling in the absence of pathogens and during PTI signaling. We conducted genetic analyses and found that the effect of cml46 cml47 is additive to cbp60a. A transcriptome-profiling experiment revealed that CBP60a, CML46, and CML47 regulate both overlapping and distinct sets of genes, consisting mostly of pathogen-responsive genes. Mathematical modeling of the expression kinetics of a PTI marker gene, CBP60g, revealed different impacts of mutations in cml46 cml47 and cbp60a. CBP60g expression was extended in cbp60a, while it had greater amplitude in cml46 cml47. Flg22-PTI also followed that pattern, as it was extended in cbp60a plants while it had greater amplitude than the wild type in cml46 cml47 plants. Thus, we have defined two different modes of negative regulation of immunity.
RESULTS
Arabidopsis CML46 and CML47 Are Negative Regulators of Immunity against P. syringae and Are Genetically Independent of CBP60a
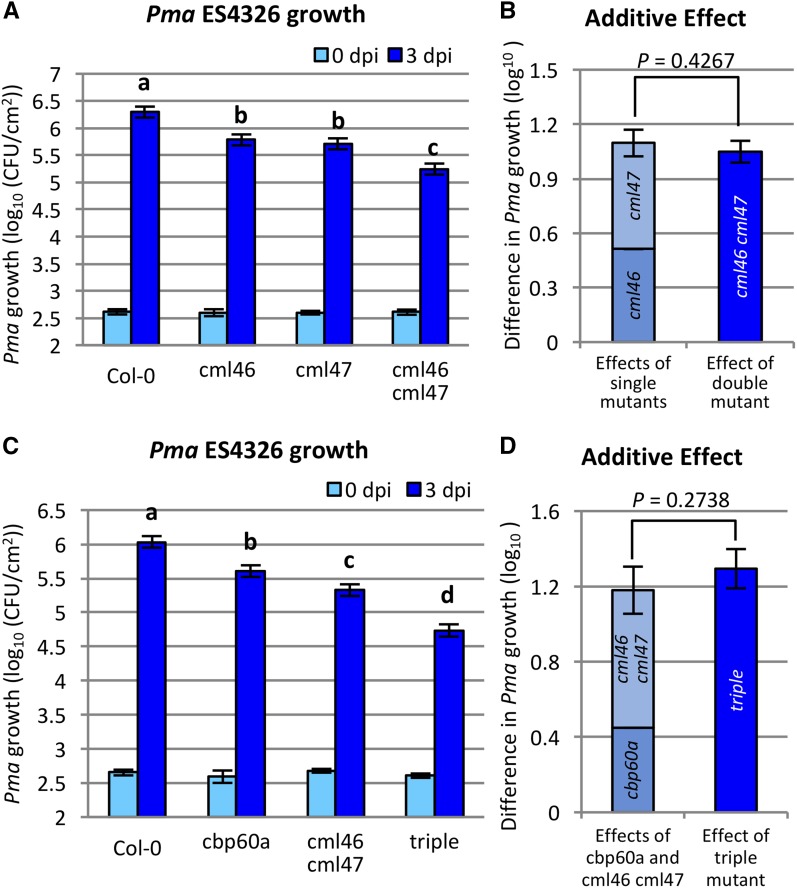
As CML46 and CML47 are closely related phylogenetically (Zhu et al., 2015) and coexpressed with CBP60g, SARD1, and SID2, we investigated their roles in plant immunity. We obtained transfer DNA (T-DNA) insertion alleles of CML46 and CML47 (Supplemental Fig. S1A). The T-DNA insertion in cml46 (SALK_127471) creates a null allele (Supplemental Fig. S1B), while cml47 (SK_31780) is a hypomorphic allele of CML47 with an approximately 10-fold reduced transcript abundance (Supplemental Fig. S1C). Both single mutants show reduced growth of Pma ES4326 at 3 d post inoculation (dpi). The enhanced disease resistance phenotype of both single mutants could be restored to the wild-type level by transgenes expressing the corresponding wild-type genes from their native promoters, although, in the case of CML46, only a subset of transgenic lines showed this complementation (Supplemental Fig. S2). The cml46 cml47 double mutant displayed a further decrease in Pma growth (Fig. 1A). The sum of enhanced resistance against Pma from cml46 and cml47 is not different from the effect of cml46 cml47 (Fig. 1B), indicating that the relationship between cml46 and cml47 is additive. Taken together, we conclude that CML46 and CML47 are both negative regulators of plant immunity and act independently. Because cml46 cml47 has a greater effect than the single mutants, we focused on the double mutant.
Figure 1.
Growth of Pma is reduced in cml46 and cml47 plants, and their effects are additive to cbp60a. A and C, Leaves of Arabidopsis wild-type Col-0, cml46, cml47, and cml46 cml47 (A) or wild-type Col-0, cbp60a, cml46 cml47, and cbp60a cml46 cml47 (triple; C) were infiltrated with Pma ES4326 suspension. Bacterial titers were determined immediately (0 dpi) or 3 dpi. Data were combined from six (A) and three (C) independent experiments, each containing four and 12 biological replicates for 0- and 3-dpi samples, respectively. Means and se for 3-dpi samples were calculated using a mixed-effect linear model. Different letters indicate significant differences between means (q < 0.05). CFU, Colony-forming units. B, Sum of effects of cml46 and cml47 compared with the effect of cml46 cml47 on Pma growth at 3 dpi, calculated from the data in A. D, Sum of effects of cbp60a and cml46 cml47 compared with the effect of cbp60a cml46 cml47 on Pma growth at 3 dpi, calculated from the data in C. Error bars represent se.
To investigate the relationship between CBP60a and the two CML genes, we crossed cbp60a-1 (SALK_124410; hereafter cbp60a) into cml46 cml47 to obtain the cbp60a cml46 cml47 triple mutant. All three mutant genotypes and Columbia-0 (Col-0) wild-type plants were tested for Pma growth. Both cbp60a and cml46 cml47 showed reduced Pma titers relative to Col-0 at 3 dpi, with the reduction in cml46 cml47 greater than that in cbp60a (Fig. 1C). The cbp60a cml46 cml47 triple mutant is even more resistant (Fig. 1C), and the effect of the triple mutant is not different from the sum of the effects of cbp60a and cml46 cml47, indicating that the effects of cbp60a and cml46 cml47 are additive (Fig. 1D). These results suggest that mutations in cbp60a and cml46 cml47 have independent effects on plant immunity that contribute to enhanced resistance against Pma.
The cbp60a and cml46 cml47 Mutants Have Elevated Salicylate Levels
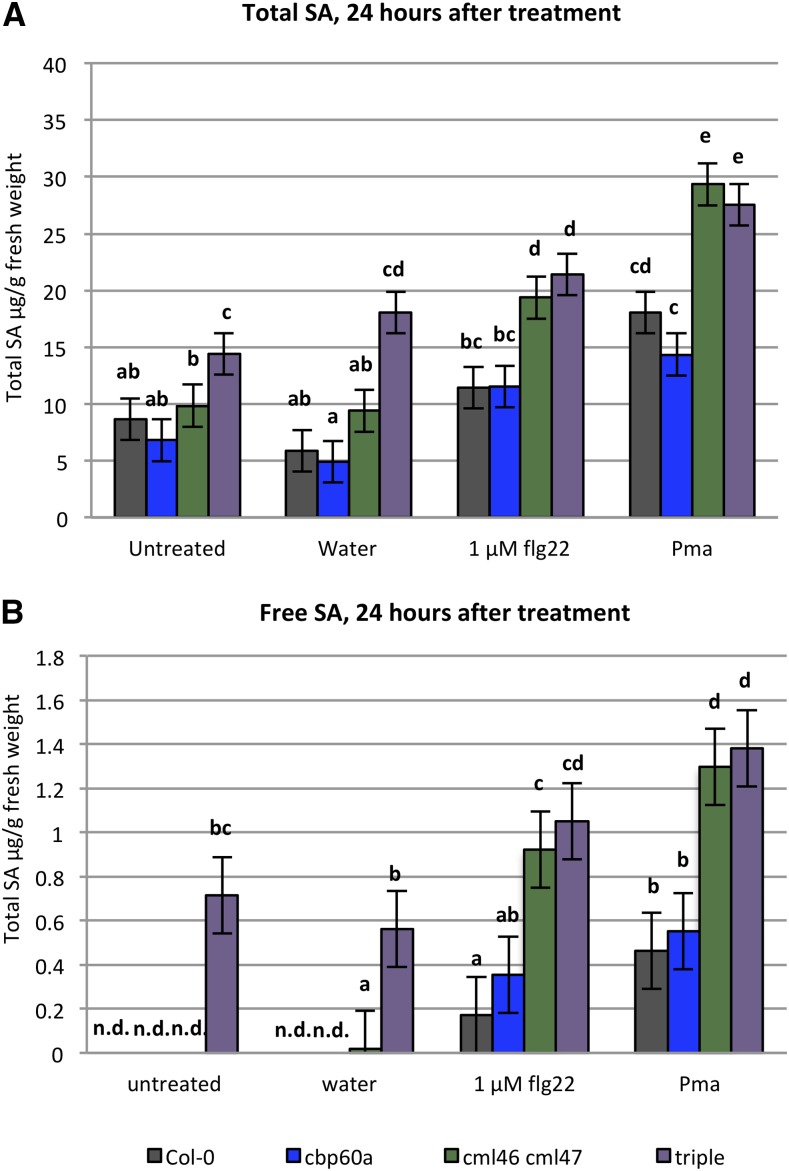
We noticed that, unlike either cbp60a or cml46 cml47, some plants of the cbp60a cml46 cml47 triple mutant exhibited spontaneous chlorosis starting from as early as 3 weeks of age and had smaller rosettes (Supplemental Fig. S3), which might indicate an autoimmune defect (van Wersch et al., 2016). Many of the known autoimmune mutants of Arabidopsis have constitutively elevated levels of SA. To test whether these mutant genotypes also accumulate higher levels of SA, we measured the free and total salicylate contents in leaves 24 h after no treatment or infiltration with water, 1 μm flg22, or Pma ES4326 using an SA biosensor assay (Defraia et al., 2008). Indeed, the triple mutant accumulated more SA than the wild type when untreated and under all three treatment conditions (Fig. 2). Interestingly, both cbp60a and cml46 cml47 have comparable levels of SA relative to the wild type in untreated or water-infiltrated leaves. This shows that CBP60a, CML46, and CML47 play redundant roles in repressing basal SA accumulation in the absence of an immune signal. However, 24 h after elicitation by either flg22 or Pma, cml46 cml47 accumulates SA to higher levels than the wild type and similar to the triple mutant, while the SA levels in cbp60a are not different from those in the wild type (Fig. 2). These results show that, in the elicited state, induced SA accumulation is higher in cml46 cml47 than in the wild type. We conclude that repression of basal SA can be achieved by either CBP60a or CML46 and CML47, while CML46 and/or CML47, but not CBP60a, limits SA accumulation in the elicited state.
Figure 2.
The cml46 cml47 and cbp60a cml46 cml47 mutants have elevated SA accumulation. Leaves of Arabidopsis wild-type Col-0, cbp60a, cml46 cml47, and cbp60a cml46 cml47 (triple) were collected for the determination of total SA (A) and free SA (B) from untreated plants or 24 h after infiltration with water, 1 μm flg22, or Pma ES4326 suspension at OD600 = 0.002. Data from four independent experiments, each containing two biological replicates, were combined using a mixed-effect linear model. Error bars represent se. Different letters indicate significant differences between means (q < 0.05). n.d., Not detected.
Transcriptome Profiling of cbp60a and cml46 cml47 Mutants
To explore the effects of the cbp60a and cml46 cml47 genotypes on global gene expression patterns, we conducted an RNA sequencing (RNA-seq) experiment. Transcriptome data were collected from mock-treated and Pma-infected plants 24 h after inoculation for a set of genotypes including the Col-0 wild type, cbp60a, cml46 cml47, cbp60g sard1, and sid2. Data were analyzed as described in “Materials and Methods” using a minimum fold change of 2 (log2 fold change > 1) and q < 0.05 as the criteria for differential expression.
Comparing mock-treated cbp60a and the wild type, 400 genes were differentially expressed, among which 364 were expressed at higher levels in cbp60a. Furthermore, 343 of these 364 genes were induced by Pma infection in wild-type plants (Supplemental Data Set S1), indicating that the primary effect of CBP60a in the absence of infection is the repression of genes that are induced during an immune response. The cml46 cml47 double mutation affected the expression of 885 genes after mock treatment. Of the 561 genes expressed at higher levels in cml46 cml47 plants compared with wild-type plants, 491 were induced by Pma in wild-type plants (Supplemental Data Set S1). Thus, CML46 and CML47 also act in the absence of infection to repress the expression of many genes induced during an immune response. However, mock-treated cml46 cml47 plants expressed 324 genes at levels lower than wild-type plants, and of these, 303 were repressed by Pma infection in wild-type plants. CML46 and CML47, therefore, act in the absence of infection to both repress the expression of genes induced during an immune response and activate the expression of genes repressed during an immune response.
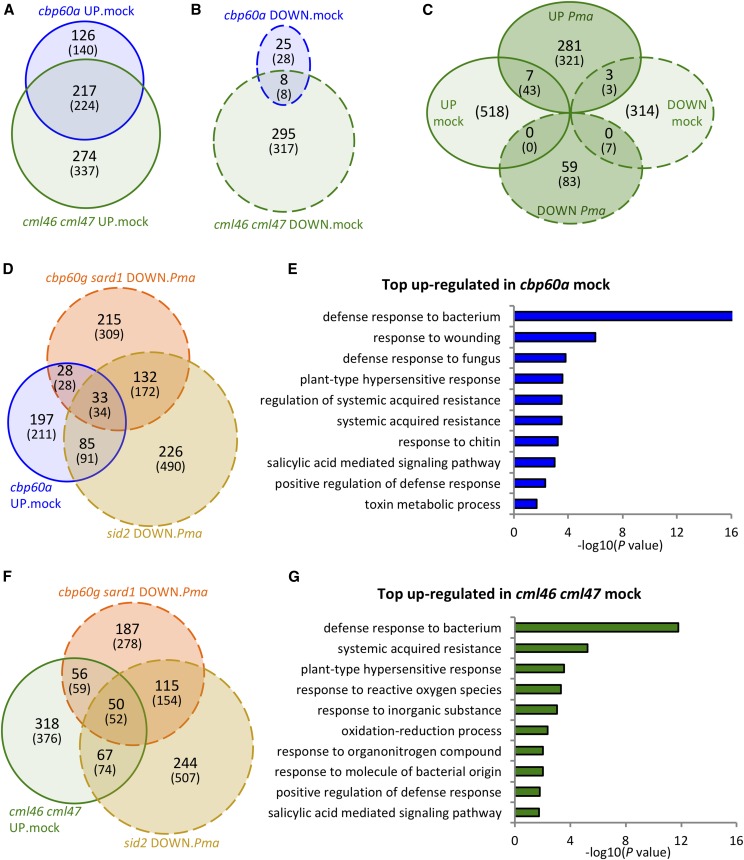
To visualize the effects of the various mutations on genes induced or repressed during an immune response, we conducted a clustering analysis. We first selected the sets of genes induced or repressed by Pma infection in wild-type plants. Within each set, we further selected the genes differentially expressed in cbp60a or cml46 cml47 plants under either mock or Pma-infected conditions, yielding 690 and 635 genes in the Pma-induced and Pma-repressed gene sets, respectively. The genes in each set were then clustered according to the similarity of expression pattern. The results are shown in Figure 3. Inspection of Figure 3A suggests that a number of Pma-induced genes are expressed at levels higher than the wild type in both mock-treated cbp60a and mock-treated cml46 cml47 plants, while many other genes are expressed at high levels in only one of these genotypes. Figure 4A shows that 217 genes are expressed at high levels in both genotypes, 126 are high only in cbp60a, and 274 are high only in cml46 cml47. Figure 3B suggests that many Pma-repressed genes are expressed at levels lower than the wild type in mock-treated cml46 cml47 plants, while very few genes are affected this way in cbp60a plants. Figure 4B shows that there are 303 such genes in cml46 cml47 plants and only 33 in cbp60a, with eight of these in common. Thus, there is extensive overlap between the inducible immune-responsive genes repressed by CBP60a or CML46 and CML47 in uninfected plants, but only CML46 and CML47 cause extensive increased expression of repressible immune-responsive genes.
Figure 3.
Hierarchical clustering analysis of Pma-responsive genes affected by cbp60a or cml46 cml47. The Pma-induced (A) or Pma-suppressed (B) genes whose expression levels were affected in cbp60a or cml46 cml47 at 24 h after mock or Pma treatment were selected for hierarchical clustering analysis. The log2 differential expression in mutants relative to the wild type following the same treatment is presented as heat maps, with each row representing one gene. Red indicates an expression level that is higher than in the wild type, and blue indicates an expression level that is lower than in the wild type. The log2 expression levels of the genes induced by Pma infiltration in the wild type are shown in orange, and those suppressed by Pma are shown in green. In total, 690 Pma-induced (A) and 635 Pma-suppressed (B) genes were included. The clustering on genes was based on Pearson correlation.
Figure 4.
Many Pma-responsive genes are differentially expressed in cbp60a and cml46 cml47. A and B, Venn diagram illustration of genes expressed higher (A) and lower (B) than wild-type levels in cbp60a and cml46 cml47 24 h after mock treatment (termed UP.mock and DOWN.mock, respectively). C, Venn diagram illustration of genes differentially expressed in cml46 cml47 24 h after mock or Pma treatment. D and F, Venn diagram illustration of genes expressed at higher than wild-type levels in cbp60a (D) and cml46 cml47 (F) 24 h after mock treatment and lower in cbp60g sard1 and sid2 24 h after Pma infection (termed DOWN.Pma). For A to D and F, numbers without parentheses indicate the counts of differentially expressed genes that are Pma induced in A, D, and F and group DOWN.Pma in C, or that are Pma repressed for B and group UP.Pma in C, in wild-type Col-0 plants. Numbers within parentheses indicate the total counts of genes differentially expressed in each intersect group between genotypes. Pma-responsive genes were selected based on absolute values of expression fold change ≥ 2 and q < 0.05 in Pma-treated than in mock-treated wild-type plants. E and G, GO enrichment analysis for GO biological processes with 364 and 561 genes up-regulated relative to wild-type levels after mock treatment in cbp60a (E) and cml46 cml47 (G), respectively. The negative log10 P values from the PANTHER overrepresentation test (x axis) for the top 10 significantly enriched GO biological processes (y axis) are reported.
As CBP60a, CBP60g, and SARD1 belong to the same gene family, it is possible that the effect of CBP60a is through the repression of CBP60g- and SARD1-dependent genes, including SID2. Among the 343 Pma-inducible genes expressed at elevated levels in cbp60a mock-treated plants, the induction by Pma infection was compromised in cbp60g sard1 and sid2 for 61 and 118 of them, respectively (Fig. 4D). For example, several genes involved in plant immunity, including PR1, FRK1, VSR6, and NUDT6, display this pattern, as their transcript levels were increased in cbp60a after mock treatment and reduced in cbp60g sard1 and sid2 after Pma infection (Supplemental Fig. S4). We conclude that some of the genes up-regulated by cbp60a in the mock treatment are CBP60g/SARD1- and/or SID2-dependent pathogen-inducible genes but that most genes with increased transcript levels in cbp60a mock-treated plants are pathogen inducible independently of CBP60g/SARD1 or SID2. To gain more insight into the functions of the genes expressed at high levels in mock-treated cbp60a plants, we carried out Gene Ontology (GO) enrichment analysis of the 364 up-regulated genes in cbp60a. These up-regulated genes were highly enriched in genes involved in defense responses to bacteria and other plant immunity-related processes (Fig. 4E). Similarly, only modest fractions of the genes expressed at high levels in cml46 cml47 mock-treated plants showed CBP60g/SARD1- and/or SID2-dependent Pma induction, and GO analysis of these genes showed that they are also enriched in genes associated with plant immunity (Fig. 4, F and G).
The effects of cbp60a and cml46 cml47 mutations in Pma-infected plants are very different from those in uninfected plants. Only 48 genes are differentially expressed between cbp60a and wild-type plants after Pma infection, including 12 and 36 genes expressed higher and lower than in the wild type, respectively (Supplemental Data Set S1). This similarity is observable in Figure 3 in the very weak signal for cbp60a versus Col-0 under Pma infection conditions. Thus, the overall gene expression profile of these plants is very similar to that of wild-type plants. In contrast to cbp60a, cml46 cml47 has substantial effects on transcript levels after Pma infection. There are 291 Pma-repressed genes with smaller repression ratios in cml46 cml47 than in the wild type after Pma infection (Figs. 3B and 4C). Only a few genes overlap between the differentially expressed genes in mock and Pma-infected samples in cml46 cml47. A similar pattern also was observed in the opposite direction: 59 Pma-induced genes (excluding CML46 and CML47) are expressed lower in cml46 cml47 than in the wild type after Pma infection (Figs. 3A and 4C). Only a few of the CML46 and CML47 regulated genes are affected in both mock and Pma-infected samples in cml46 cml47 (Fig. 4C). Therefore, cml46 cml47 affects largely separate groups of genes under mock or Pma-infected conditions.
The Kinetics of flg22-Induced CBP60g Transcript Accumulation Are Differentially Affected by cbp60a and cml46 cml47
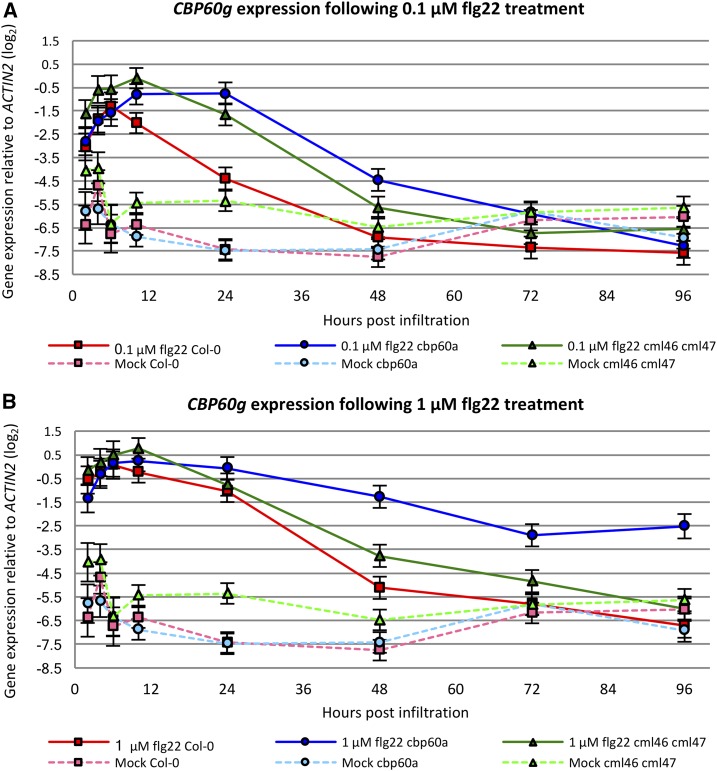
To gain further insight into the effects of cbp60a and cml46 cml47 mutations on the dynamics of immune signaling, we monitored mRNA levels of a flg22-responsive gene over time. We wanted a gene known to be important for PTI, which has a large dynamic range when induced by flg22 and a basal level that is readily detectable by quantitative reverse transcription PCR (RT-qPCR). These attributes would provide good statistical power for studying the effects of cbp60a and cml46 cml47 over time. CBP60g meets these criteria. It is important for PTI, it is induced rapidly and consistently in response to flg22 treatment, and it has both a readily detectable basal expression level and a large dynamic range when induced (Wang et al., 2009). The transcript levels of CBP60g were determined by RT-qPCR in the leaves of wild-type and mutant plants untreated and treated at 2, 4, 6, 10, 24, 48, 72, and 96 h post infiltration (hpi) with water or flg22. We first determined the basal level of CBP60g expression in the leaves of untreated wild-type, cbp60a, cml46 cml47, and cbp60a cml46 cml47 plants. Both cml46 cml47 and the triple mutant exhibit higher basal levels of CBP60g expression than the wild type and cbp60a (Supplemental Fig. S5). This increased expression of CBP60g in cml46 cml47 and cbp60a cml46 cml47 also was observed following mock infiltration (Fig. 5; Supplemental Fig. S6), consistent with the elevated expression of CBP60g in cml46 cml47 mock-treated plants in the RNA-seq experiment (Supplemental Data Set S1). When plants are infiltrated with 0.1 μm flg22, CBP60g is up-regulated transiently in the wild type, cbp60a, and cml46 cml47 (Fig. 5A), while it is maintained at high levels throughout the time course in cbp60a cml46 cml47 (Supplemental Fig. S6). In the case of the wild type, the mRNA level of CBP60g is highest at 6 and 10 hpi and has dropped markedly by 24 hpi. A similar pattern was observed in cml46 cml47, although CBP60g reaches higher mRNA levels at 10 and 24 hpi than in the wild type (Fig. 5A; Supplemental Table S1). However, CBP60g expression is sustained close to the peak level longer in cbp60a, which has higher levels at 24 and 48 hpi, than in the wild type (Fig. 5A; Supplemental Table S1). By the end point of the time course, the expression of CBP60g returns to the basal level in all three genotypes. Following treatment with 1 μm flg22, CBP60g mRNA is at high levels for longer periods of time in the wild type, cbp60a, and cml46 cml47 (Fig. 5B) and exhibits sustained high levels in cbp60a cml46 cml47 (Supplemental Fig. S6). The CBP60g mRNA level is similar in the wild type and cml46 cml47 at all time points, while it persists at higher levels in cbp60a after 48 hpi and does not return to the basal level by 96 hpi, the last time point measured (Fig. 5B; Supplemental Table S1).
Figure 5.
The kinetics of flg22-induced CBP60g expression are differentially affected by cbp60a and cml46 cml47. The transcript abundance of CBP60g in 30-d-old wild-type Col-0, cbp60a, and cml46 cml47 plants was monitored at 2, 4, 6, 10, 24, 48, 72, and 96 hpi with water (Mock; A and B), 0.1 μm flg22 (A), or 1 μm flg22 (B). Samples from five independent experiments were collected. Each experiment included all three genotypes in a balanced layout, but only a subset of the time points and treatments, with overlaps between subsets from different independent experiments. Each data point represents at least four biological replicates. Data were combined using a linear mixed-effect model for estimating means and se. Error bars represent se. The same mock data are shown in A and B.
To understand the changes of the kinetics of CBP60g transcript accumulation in wild-type, cbp60a, and cml46 cml47 plants following flg22 treatment at both low (0.1 μm) and high (1 μm) concentrations, a simple mechanistic model consisting of six first-order, ordinary differential equations (ODEs) for six combinations of the three genotypes and two flg22 concentrations was fitted to the CBP60g mRNA level data. In each ODE, the rate of the CBP60g expression level change was explained by the production and removal rates. Specifically, the ODE was formulated as
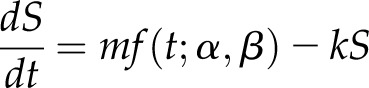 |
where S is the CBP60g mRNA level, 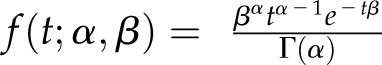 is a γ probability density function with shape parameter α and rate parameter β, representing the production rate time course, m is the amplitude parameter for the production rate, and k is the removal rate constant. The production rate term mf (t; α, β) can be interpreted as the synthesis rate of the CBP60g mRNA, and the removal rate term −kS can be interpreted as the degradation rate of the CBP60g mRNA determined by a first-order chemical reaction. Alternatively, the ODE can be interpreted in a more abstract manner: the production and removal rates represent the activation and inactivation rates of the regulatory system for CBP60g transcript accumulation, and the CBP60g mRNA level directly reports the activity of the regulatory system. We used the γ probability density function to approximate the time course of the synthesis rate, since it is a simple function with two parameters that can generate smooth unimodal time courses of various shapes and timing: we assumed that the CBP60g regulatory signals, generated by flg22 treatment, have smooth unimodal time courses. Two different sets of α and β values were employed for two different concentrations of flg22 based on the assumption that different concentrations of flg22 could generate different production rate time courses and that the production rate time courses are not affected by genotype differences. For the other parameters, different values for six different ODEs were initially assumed. The model was fitted to the data based on maximum likelihood estimation for a log-normal data error distribution. The fitted model revealed that the amplitude parameter m was greater in cml46 cml47 than in the other genotypes and the removal rate parameter k was smaller in cbp60a than in the others at each of the flg22 concentrations (Supplemental Table S2). To test the significance of the parameter value differences in the model, we performed model selection analysis on all possible model structures based on the Bayesian information criterion (BIC). Four sets of parameters m and k (two each for the two flg22 concentrations, high and low, namely, mH, mL, kH, and kL; Supplemental Table S3) with five equivalence patterns across the genotypes (Supplemental Table S4) were used to construct all the 54 possible model structures. The top 10 model structures based on BIC are shown in Supplemental Table S5. The best BIC-selected model predicts that one parameter value of m can be shared between cbp60a and the wild type, while one value of k can be shared between the wild type and cml46 cml47 at each flg22 concentration (Table I; Supplemental Fig. S7), which is consistent with the observations from the initial model (Supplemental Table S2). Thus, this mechanistic model study provides statistical support for our intuitive interpretations of the data: the CBP60g mRNA level is induced more rapidly in cml46 cml47 than in the wild type and cbp60a while there is no difference in the induction rate between the wild type and cbp60a; the CBP60g mRNA level decreases at lower rates in cbp60a than in the wild type and cml46 cml47 while there is no difference in the rates of decrease between the wild type and cml46 cml47. These notions about the induction and decrease rates are true for both low and high concentrations of flg22.
is a γ probability density function with shape parameter α and rate parameter β, representing the production rate time course, m is the amplitude parameter for the production rate, and k is the removal rate constant. The production rate term mf (t; α, β) can be interpreted as the synthesis rate of the CBP60g mRNA, and the removal rate term −kS can be interpreted as the degradation rate of the CBP60g mRNA determined by a first-order chemical reaction. Alternatively, the ODE can be interpreted in a more abstract manner: the production and removal rates represent the activation and inactivation rates of the regulatory system for CBP60g transcript accumulation, and the CBP60g mRNA level directly reports the activity of the regulatory system. We used the γ probability density function to approximate the time course of the synthesis rate, since it is a simple function with two parameters that can generate smooth unimodal time courses of various shapes and timing: we assumed that the CBP60g regulatory signals, generated by flg22 treatment, have smooth unimodal time courses. Two different sets of α and β values were employed for two different concentrations of flg22 based on the assumption that different concentrations of flg22 could generate different production rate time courses and that the production rate time courses are not affected by genotype differences. For the other parameters, different values for six different ODEs were initially assumed. The model was fitted to the data based on maximum likelihood estimation for a log-normal data error distribution. The fitted model revealed that the amplitude parameter m was greater in cml46 cml47 than in the other genotypes and the removal rate parameter k was smaller in cbp60a than in the others at each of the flg22 concentrations (Supplemental Table S2). To test the significance of the parameter value differences in the model, we performed model selection analysis on all possible model structures based on the Bayesian information criterion (BIC). Four sets of parameters m and k (two each for the two flg22 concentrations, high and low, namely, mH, mL, kH, and kL; Supplemental Table S3) with five equivalence patterns across the genotypes (Supplemental Table S4) were used to construct all the 54 possible model structures. The top 10 model structures based on BIC are shown in Supplemental Table S5. The best BIC-selected model predicts that one parameter value of m can be shared between cbp60a and the wild type, while one value of k can be shared between the wild type and cml46 cml47 at each flg22 concentration (Table I; Supplemental Fig. S7), which is consistent with the observations from the initial model (Supplemental Table S2). Thus, this mechanistic model study provides statistical support for our intuitive interpretations of the data: the CBP60g mRNA level is induced more rapidly in cml46 cml47 than in the wild type and cbp60a while there is no difference in the induction rate between the wild type and cbp60a; the CBP60g mRNA level decreases at lower rates in cbp60a than in the wild type and cml46 cml47 while there is no difference in the rates of decrease between the wild type and cml46 cml47. These notions about the induction and decrease rates are true for both low and high concentrations of flg22.
Table I. Regression results of the top model structure based on maximum likelihood estimation.
| flg22 Concentration | Genotype | Parameters |
|||||
|---|---|---|---|---|---|---|---|
|
m |
k |
α | β | ||||
| Value | 95% Confidence Interval | Value | 95% Confidence Interval | ||||
| High | Wild type | 1.263 | (1.097, 1.478) | 0.0576 | (0.0545, 0.0605) | 2.073 | 0.797 |
| cbp60a | 1.263 | (1.097, 1.478) | 0.0242 | (0.0172, 0.0293)a | |||
| cml46 cml47 | 2.053 | (1.794, 2.318)a | 0.0576 | (0.0545, 0.0605) | |||
| Low | Wild type | 0.947 | (0.701, 3.177) | 0.1168 | (0.1050, 0.4484) | 2.152 | 0.369 |
| cbp60a | 0.947 | (0.701, 3.177) | 0.0641 | (0.0394, 0.1934)a | |||
| cml46 cml47 | 2.244 | (1.761, 9.072)a | 0.1168 | (0.1050, 0.4484) | |||
Different parameter values for each of the parameters m and k are significantly different because this was the best model based on BIC among all the equivalence patterns (Supplemental Tables S3–S5).
To test whether the kinetics of CBP60g transcript accumulation in response to flg22 treatment are representative of other flg22-inducible genes, we measured the transcript levels of FRK1 (Asai et al., 2002), VSR6 (Denoux et al., 2008), and WRKY29 (Asai et al., 2002) in a subset of the samples used to monitor CBP60g expression. For both FRK1 and VSR6, slower transcript decay was observed in cbp60a plants following treatment with 1 μm flg22, and for VSR6, a higher induction rate was observed in cml46 cml47 plants following treatment with 0.1 µm flg22 (Supplemental Fig. S8, A and B; Supplemental Tables S6 and S7). WRKY29 behaved very differently. It was induced only transiently (Supplemental Fig. S8C) and was not affected by cbp60a or cml46 cml47 (Supplemental Table S8), indicating that the expression of WRKY29 is regulated quite differently from the other three genes. These observations support the idea that CBP60a and CML46 CML47 regulate the expression of a subset of pathogen- or flg22-inducible genes through different mechanisms.
PTI Induced by flg22 Is Enhanced in cbp60a and cml46 cml47 and Is Extended in cbp60a
CBP60g is a positive regulator of immunity and is induced rapidly upon flg22 treatment. It is possible that the mRNA level of CBP60g reflects the strength of flg22-PTI. If this is true, two predictions can be derived from the behavior of the CBP60g mRNA level: first, PTI might be extended in cbp60a; second, PTI might be stronger in cml46 cml47. We first demonstrated that the flg22 treatment we used triggered FLS2-dependent PTI (Supplemental Fig. S9). To test our hypotheses, we pretreated leaves with water or 0.1 μm flg22 at 72, 48, or 24 h prior to inoculation with Pma ES4326 (Fig. 6A), and the strength of PTI was quantified as the reduction of Pma growth between water- and flg22-pretreated samples 1 d after inoculation. If the growth of Pma is significantly lower in flg22-pretreated plants than in water-pretreated plants of the same genotype, then PTI is considered present. In Col-0 and cml46 cml47, PTI induced by 0.1 μm flg22 could be detected following pretreatments for 24 or 48 h (Fig. 6, F and D) but not 72 h (Fig. 6B). In contrast, flg22-PTI could still be detected in cbp60a and cbp60a cml46 cml47 even 72 h after pretreatment (Fig. 6B), indicating that flg22-PTI was extended by at least 1 d in the genetic backgrounds containing cbp60a. At each pretreatment time point, the strength of flg22-PTI in cbp60a was significantly greater than in the Col-0 wild type (Fig. 6, C, E, and G), suggesting that flg22-PTI was generally more effective in cbp60a relative to Col-0. The strength of flg22-PTI was greater in cml46 cml47 than in the wild type with 24 h of pretreatment (Fig. 6G) but not with 48 and 72 h of pretreatment (Fig. 6, E and C). It is worth noting that there was little or no Pma growth in the leaves of cbp60a cml46 cml47 plants pretreated with flg22 at 24 h prior to pathogen inoculation (Fig. 6F, compare with time-0 values in Fig. 1, A and C), which likely resulted in an underestimation of PTI in cbp60a cml46 cml47 at this time point. In fact, PTI in cbp60a cml46 cml47 was stronger than, or as strong as, PTI in cbp60a after 72 and 48 h of flg22 pretreatment, respectively (Fig. 6, C and E). This strong PTI persisting in the triple mutant is likely due to synergistic interactions between cbp60a and cml46 cml47 for compromised ability of shutting down or repressing the immune responses (Supplemental Fig. S10) and is consistent with the high expression of CBP60g induced by flg22 treatment in cbp60a cml46 cml47 (Supplemental Fig. S6). In summary, flg22-induced PTI is more effective and of longer duration in cbp60a. In cml46 cml47, it is stronger within a short window of time following flg22 pretreatment. These effects on PTI mirror the effects of these mutations on the kinetics of CBP60g transcript accumulation.
Figure 6.
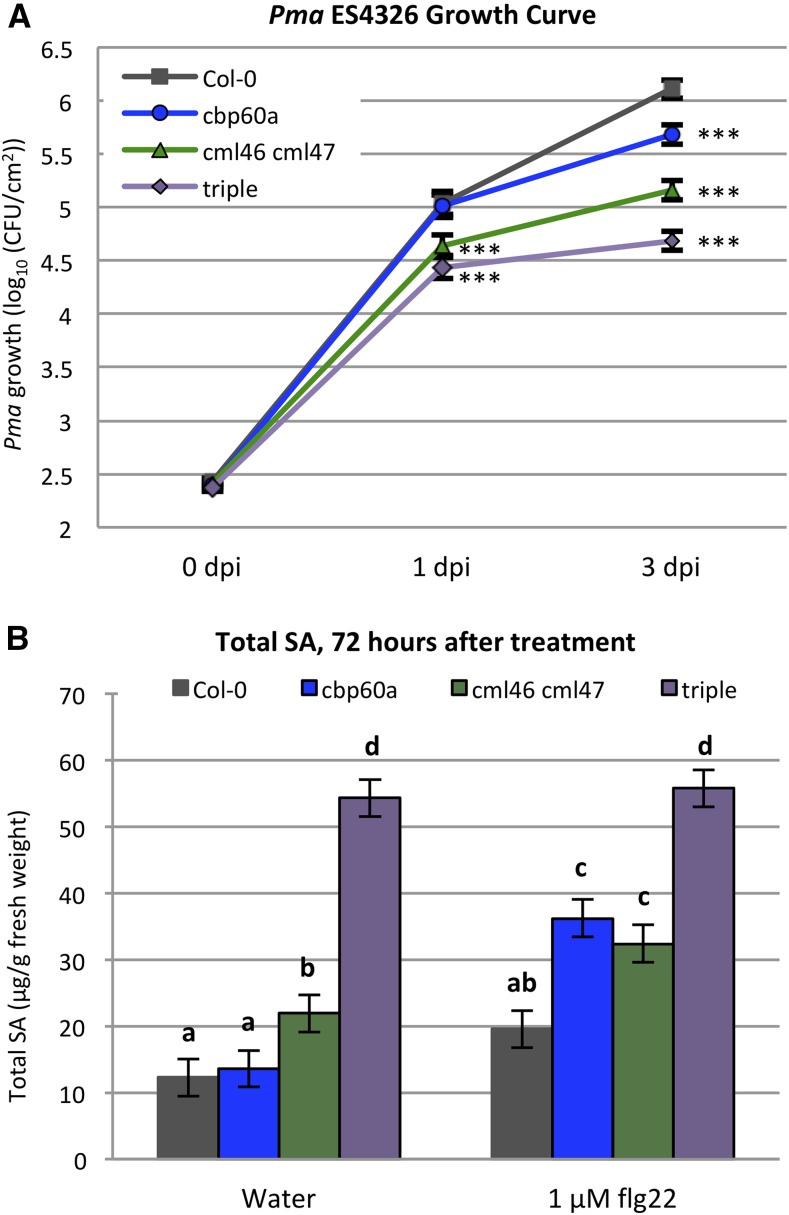
PTI induced by flg22 is enhanced in cbp60a and cml46 cml47 and extended in cbp60a. A, Description of the flg22-PTI assay on 30-d-old Arabidopsis wild-type Col-0, cbp60a, cml46 cml47, and cbp60a cml46 cml47 plants. B, D, and F, Leaves were infiltrated with 0.1 μm flg22 or water 72 (B), 48 (D), or 24 (F) h prior to inoculation with Pma ES4326 at OD600 = 0.0002. Bacterial titer was determined at 1 dpi. Data from four, five, and four independent experiments, each containing at least 12 biological replicates, for 72, 48, and 24 h pretreatment prior to inoculation, respectively, were combined separately using a linear mixed-effect model. ***, q < 0.001 for the difference of means between water- and flg22-pretreated samples. CFU, Colony-forming units. C, E, and G, Strength of PTI calculated as the difference in Pma growth between water- and flg22-pretreated samples using the data from B, D, and F. Different letters indicate significant differences between means (q < 0.05).
The Effect of cbp60a on Immunity against Pma ES4326 Is Detectable Only at a Late Stage of Infection
As the CBP60g transcript accumulation kinetics revealed that the cbp60a mutation extended the duration of the response but did not affect the signal input rate (Table I), we hypothesized that the effect of cbp60a on immunity might only be detectable at time points well after defense responses become activated. Thus, we expected that cml46 cml47 would show enhanced disease resistance throughout the course of pathogen infection while cbp60a would only provide enhanced disease resistance at a late stage of disease progression. To test this hypothesis, we monitored titers of Pma ES4326 in wild-type, cbp60a, cml46 cml47, and cbp60a cml46 cml47 plants at 1 and 3 dpi, which approximate early and late stages of Pma infection, respectively. Indeed, at 1 dpi, the Pma growth in cbp60a was not different from that in the wild type, while at 3 dpi, Pma grew to a significantly lower titer in cbp60a than in the wild type (Fig. 7A). In contrast, Pma growth was significantly lower in cml46 cml47 and cbp60a cml46 cml47 plants than in the wild type at both 1 and 3 dpi (Fig. 7A).
Figure 7.
Mutation in cbp60a does not provide enhanced resistance at an early stage of disease progression. A, Bacterial titers were determined at 0, 1, and 3 dpi with Pma ES4326 in Arabidopsis wild-type Col-0, cbp60a, cml46 cml47, and cbp60a cml46 cml47 (triple). Data from three independent experiments, each containing 8, 12, and 12 biological replicates at 0, 1, and 3 dpi, respectively, were combined using a linear mixed-effect model. Pma titers in mutant genotypes at 1 and 3 dpi were compared with the wild type at the corresponding time points using means and variances estimated from the linear model. ***, P < 0.001 from Student’s t test. CFU, Colony-forming units. B, Leaves of Arabidopsis wild-type Col-0, cbp60a, cml46 cml47, and cbp60a cml46 cml47 (triple) were collected to determine SA levels 72 h after infiltration with water or 1 μm flg22. Data from three independent experiments were combined using a linear mixed-effect model. In total, eight biological replicates were included for each genotype-treatment combination. Different letters indicate significant differences between means (q < 0.05). Error bars show se.
Enhanced resistance to Pma usually accompanies high SA accumulation. This difference between 1- and 3-dpi Pma growth in cbp60a prompted us to test whether there was a difference in SA levels at the later time point. Because Pma-infected leaves start to collapse by 3 dpi, we decided to test SA accumulation 72 h following 1 μm flg22 treatment. Although no differences were observed between the free SA levels in the wild type, cbp60a, and cml46 cml47, likely because the measured values were approaching the detection limit (Supplemental Fig. S11), both cbp60a and cml46 cml47 accumulate more total SA than the wild type and the triple mutant accumulates more SA than the other three genotypes (Fig. 7B). This differs from the relative total SA levels 24 h after flg22 treatment, when SA accumulation in cbp60a is not different from that in the wild type (Fig. 2). The SA accumulation patterns are consistent with the Pma growth phenotypes at both time points, as cml46 cml47 plants have high SA and less bacterial growth at both time points while cbp60a plants have high SA and reduced bacterial growth only at the later time point (Figs. 2 and 7). Taken together, these data support our hypothesis that cbp60a displays enhanced resistance and higher SA accumulation only at a later stage by affecting the rate at which the immune response ends, while cml46 cml47 shows greater overall expression of immunity, resulting in elevated SA accumulation and reduced Pma growth throughout the time course.
DISCUSSION
Plants have evolved highly effective inducible immune responses through coevolution with pathogens. The activation of immune responses often accompanies the production of stress-related hormones, reprogramming of the transcriptome, and down-regulation of vegetative growth and photosynthesis, representing a tradeoff between defense and growth (Vos et al., 2013). To minimize negative impacts on growth, immune responses must be repressed in the absence of pathogen attack and return to the basal level once a pathogen attack has ended. These requirements of the plant immune system are met using various regulators playing different roles. In this study, we have demonstrated that CBP60a, CML46, and CML47 negatively regulate plant immune signaling and resistance to Pma. Genetic analysis revealed additive effects between cml46 and cml47, and between cbp60a and cml46 cml47, on Pma growth at 3 dpi (Fig. 1), which suggests that these genes may have independent roles in regulating immune signaling. This hypothesis is further supported by transcriptome profiling of both mutants, showing that cbp60a and cml46 cml47 affect the expression of different, as well as overlapping, sets of genes (Figs. 3 and 4, A and B).
Transcriptome profiling of cbp60a and cml46 cml47 revealed that the majority of the genes differentially expressed in both mutants are Pma responsive, with over one-third of these genes responding to Pma infection in a CBP60g/SARD1- or SID2-dependent manner (Fig. 4, D and F). Among these are genes involved in the biosynthesis of SA, which is consistent with the observations that cbp60a and cml46 cml47 accumulate more SA than the wild type following flg22 treatment (Figs. 2 and 6B). Most of the genes differentially expressed between cbp60a and the wild type in mock-treated plants have higher transcript levels in cbp60a, suggesting that cbp60a is a negative regulator of immunity-related genes. For cml46 cml47, in addition to a number of Pma-induced genes having higher transcript levels in mock-treated plants, there are also many genes down-regulated, and the majority of them are Pma suppressed. This pattern mimics a transcriptome in an activated state of immune signaling, highlighting the roles of CML46 and CML47 as negative regulators of immunity.
Given the sizable overlap of the genes displaying altered mRNA levels in the transcriptome profiles of cbp60a and cml46 cml47, we were interested in exploring how the two mutants might differentially affect the behavior of these genes. However, the transcriptome profiling data only described effects at a single time point. Therefore, we measured the expression of one representative gene, CBP60g, over a time course following flg22 treatment in wild-type and mutant backgrounds. Using kinetic expression data of CBP60g as a readout of immune signaling during PTI, we mathematically modeled the kinetic patterns of CBP60g transcript levels and captured a slower signal decay rate in cbp60a and a greater signal amplitude in cml46 cml47. The model predicted that flg22-PTI is extended in cbp60a plants, which was supported by the observation that the flg22-PTI could be detected as long as 72 h after pretreatment prior to Pma inoculation in cbp60a plants (Fig. 6, B and C). The model also predicted that the response to flg22 might be stronger in cml46 cml47. We found that flg22-PTI was indeed enhanced in cml46 cml47 with 24 h of pretreatment (Fig. 6G), but this effect decreased to the wild-type level with longer times following pretreatment (Fig. 6, C and F).
It is probable that CBP60a and the CMLs negatively regulate immune responses through different mechanisms, such that CBP60a acts to control the duration of the activated responses while the CMLs affect the rate and amplitude of the response. This is consistent with our finding that, when naive plants were challenged with Pma, enhanced disease resistance was measurable throughout the course of disease progression in cml46 cml47 while resistance was observed only at a late time point in cbp60a (Fig. 7). Interestingly, transcriptome profiling of 24-hpi Pma-treated plants revealed that only eight Pma-inducible genes were affected by cbp60a (Fig. 4). Furthermore, SA levels in infected cbp60a plants were indistinguishable from those in the wild type at 24 h after infection (Fig. 2). These phenotypes are consistent with the observation that titers of Pma were similar in wild-type and cbp60a plants at this time point. Evidently, the derepression of some immunity-inducible genes in mock-inoculated cbp60a plants is not sufficient to cause increased resistance at early time points.
We found that the expression pattern of the immune regulatory gene, CBP60g, reflected the resistance to Pma during the PTI response. However, the expression pattern of another gene thought to be important for immunity, WRKY29, was quite different. While we do not have time-course data for the RNA-seq experiment, it seems likely that the genes with elevated expression in mock-infected cbp60a plants will not show expression patterns similar to CBP60g. The significance of these genes to resistance to Pma, the ultimate biological effect of PTI that we studied, is unclear. Little is known about which gene expression changes are causally related to the limitation of pathogen growth. The majority of genes known to be important for plant immunity have regulatory effects, not direct effects on limiting pathogen growth. There are a number of possible explanations for why the expression pattern of CBP60g correlates with Pma resistance while other genes do not. CBP60g and SARD1 play partially overlapping roles in positively regulating PTI (Wang et al., 2011). Perhaps CBP60g itself is needed for PTI resistance. It seems more likely that genes expressed with the same pattern as CBP60g are collectively critical for PTI resistance. This is plausible, as a group of genes coexpressed with CBP60g, SARD1, and ICS1 include many genes with known roles in immunity (Truman and Glazebrook, 2012). The correlation observed between CBP60g expression and PTI resistance may be due to a combination of the broad effects of CBP60g in the activation of immune responses (Sun et al., 2015) and the coordinated expression of CBP60g and other responses causal to effective PTI. If an RNA-seq data set collected over a time course following flg22 or mock treatment of wild-type and cbp60a plants was available, it would be possible to determine how many and which genes have expression patterns similar to CBP60g. It might then be worthwhile to test these for effects on PTI. Another possible explanation rests on the hypothesis that the resistance effect of PTI results from the cumulative effects of many gene expression and physiological changes. The net effect of cbp60a on PTI resistance is to extend it over time. This does not require the expression of each of the genes contributing to this net effect to be similarly extended over time.
In our previous study, we did not detect the enhanced PTI effect in cbp60a (Truman et al., 2013). This disparity is likely attributable to a combination of differences in the experimental conditions, including the concentration of flg22 used and the duration of pathogen growth. Here, we used a lower concentration of flg22 (0.1 rather than 1 μm) to avoid saturating the system. Consistent with our finding that the effect of cbp60a can only be detected after defense responses are activated (Fig. 7), the 2-dpi Pma growth used previously in the PTI assay led to lower bacterial titers in both mock- and flg22-treated cbp60a plants relative to wild-type plants, which resulted in a smaller PTI effect over 2 d of pathogen growth (Truman et al., 2013).
We have demonstrated that CBP60a is involved in ending the immune response triggered by the bacterial MAMP flg22 and affects the duration of flg22-PTI. This is not mutually exclusive with the observation that many Pma-responsive genes had higher transcript levels in cbp60a than in the wild type following mock treatment (Figs. 3 and 4A), which, in this RNA-seq experiment, was water infiltration with needleless syringes. Although not as potent as Pma or flg22 for inducing gene expression, water infiltration can trigger a smaller yet measureable pulse in the transcript levels of many genes. This transient increase in transcript levels also might be regulated by CBP60a in ways similar to those we have described in this study; hence, in mock-inoculated cbp60a, those genes had higher mRNA levels. It would be interesting to test whether this function of CBP60a is general to other MAMP treatments and whether CBP60a plays similar roles in other plant species.
It is likely that the transient nature of PTI is important for plant fitness. However, to date, we have not noticed any penalty on growth or reproduction in cbp60a, even in plants treated with flg22 or challenged with Pma. It is possible that the effect of such one-time activation of immune signaling is negligible or that an effect too small for us to notice is nevertheless sufficient to affect fitness. We did observe stunted growth and early senescence in cbp60a cml46 cml47 plants (Supplemental Fig. S3), which have high expression levels of CBP60g and high SA accumulation, suggesting that sustained high PTI responses compromise growth.
CML46 and CML47 contributed to repressing CBP60g expression in the absence of Pma or flg22 as well as following flg22 treatment. While CBP60a did not have this effect, cbp60a cml46 cml47 plants exhibited sustained high CBP60g expression under all conditions. This may result from the combined effects of the elevated expression in cml46 cml47 coupled with the reduced ability to shut down CBP60g expression caused by the cbp60a mutation. CML46 and CML47 are CaM-like proteins that are both predicted to contain a pair of EF-hand domains that bind Ca2+ (Zhu et al., 2015). Given the coexpression pattern among CML46, CML47, CBP60g, SARD1, and SID2 (Truman and Glazebrook, 2012), as well as the set of genes that are coregulated by CML46, CML47, CBP60g, and SARD1 (Fig. 4F), it is possible that either or both CML46 and CML47 might compete with the canonical CaM proteins for binding to CBP60g and, hence, modulate the activity of CBP60g as a transcription factor. Or, they might affect the activities of other CaM-binding proteins important in immune signaling. Alternatively, CML46 and CML47 could act as calcium buffers that modulate the calcium concentration available to canonical CaM proteins during immune signaling, and their coexpression pattern may reinforce the timing of the calcium-buffering effect. This calcium-buffering property of CaM has been described using hippocampal CA1 pyramidal cells (Faas et al., 2011).
CBP60a and CBP60g belong to the same family of plant-specific CaM-binding proteins. CBP60g has been shown to act as a transcription factor and directly target the promoters of various genes that it regulates (Zhang et al., 2010; Sun et al., 2015). It is possible that CBP60a exhibits similar behaviors but counteracts CBP60g by competing for common targets, which results in opposing mutant phenotypes. However, the facts that the expression of CBP60a is only induced to a fraction of the level of CBP60g after Pma challenge and that the effect of cbp60a is genetically epistatic to cbp60g (Truman et al., 2013) suggest that direct competition between CBP60a and CBP60g is unlikely to produce the observed mutant phenotypes. We hypothesize that CBP60a might directly repress the expression of CBP60g and other positive immune regulator genes by targeting their promoters and/or indirectly regulating them by activating the expression of other negative regulators. Identifying the targets of CBP60a, as well as any interaction partners of CML46 and CML47, would enhance our understanding of the negative regulatory mechanisms modulating PTI signaling.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild-type Col-0, cbp60a-1 (At5g62570, SALK_124410), cml46 (At5g39670, SALK_127471), cml47 (At3g47480, SK31780), cml46 cml47 (generated in this study by crossing cml46 and cml47), cbp60a-1 cml46 cml47 (generated in this study by crossing cbp60a-1 and cml46 cml47), cbp60g-1 sard1-2 (Wang et al., 2011), and sid2-2 (Wildermuth et al., 2001) were grown in autoclaved BM2 germinating mix (Berger) soil in a growth chamber (Conviron) at 22°C and 75% relative humidity on a 12-h-light/12-h-dark cycle at light intensities of 100 μmol m−2 s−1. Where not otherwise specified, cbp60a refers to cbp60a-1, cbp60g refers to cbp60g-1, sard1 refers to sard1-2, and sid2 refers to sid2-2.
Pathogen Growth and flg22-PTI Assays
For pathogen growth assays, Pseudomonas syringae pv maculicola ES4326 was cultured in King’s B medium supplemented with 100 mg L−1 streptomycin at 26°C until saturation. The inoculum was prepared as a suspension of Pma ES4326 at OD600 = 0.0002 (approximately 1 × 105 colony-forming units mL−1) in 5 mm MgSO4 solution. Inoculation was conducted 3 to 5 h after subjective dawn by infiltrating the inoculum into the underside of plant leaves with a needleless syringe. Inoculated plants were kept under the same growth conditions until collection for bacterial titer quantification. The collected leaf tissues were homogenized in 5 mm MgSO4, serially diluted, and spread onto King’s B agar supplemented with 100 mg L−1 streptomycin. After incubation for 2 d at 26°C, colony-forming units were counted to calculate pathogen growth.
For flg22-PTI assays, 0.1 μm flg22 or water was infiltrated into the underside of leaves at indicated time points prior to Pma inoculation. The remaining procedures were the same as in the Pma growth assay.
Mutant Complementation
To complement the cml46 and cml47 mutants, their native promoter and coding sequence, omitting the stop codon, were cloned from wild-type Col-0 with the primers listed in Supplemental Table S9 and ligated into pCR8/GW/TOPO vector (Invitrogen) as entry clones. The genomic sequences of 2,248- and 1,435-bp promoter regions upstream of the predicted translation start sites were cloned for CML46 and CML47, respectively. The correct constructs confirmed by Sanger sequencing were cloned subsequently into destination binary vectors via LR reactions. To complement cml46, the construct was cloned into pEARLEYGATE301 (Earley et al., 2006), resulting in pEG301-CML46pro::CML46. For cml47, the construct was cloned into pMDC107 (Curtis and Grossniklaus, 2003), resulting in pMDC107-CML47pro::CML47. Both destination constructs were transformed into Agrobacterium tumefaciens strain GV3101 (pMP90), which was used to transform the corresponding mutants by floral dip (Clough and Bent, 1998). Plants of the T0 and T1 generations of successfully transformed cml46 plants were selected by spraying with 300 μL of Basta in 500 mL of water (Bayer Cropscience; active ingredient, 18.19% [w/v] glufosinate ammonium). Plants of the T0 and T1 generations of successfully transformed cml47 plants were selected on Murashige and Skoog medium supplemented with 10 g L−1 Suc, 20 mg L−1 hygromycin, and 250 mg L−1 carbenicillin. Offspring plants from the T3 generation were tested for Pma growth.
Quantification of SA
Free and total SA contents were determined using an Acinetobacter sp. ADPWH_lux-based biosensor strain as described previously (Defraia et al., 2008). Plant leaf samples were collected at indicated time points following no treatment (untreated) or infiltration with water (mock), 1 μm flg22, or Pma (OD600 = 0.002). For each sample, four leaf discs from two leaves of one plant were cut with a cork borer (6 mm diameter), frozen in liquid nitrogen, and homogenized. Each sample was equivalent to approximately 17 mg of leaf tissue. Samples were mixed with 100 μL of 0.1 m sodium acetate buffer (pH 5.6) and centrifuged for 15 min at 16,000g at 4°C. The supernatant was divided into two aliquots: 80 μL of supernatant was used to determine free SA; the remaining 20 μL was diluted with 75 μL of acetate buffer, and 0.4 units of β-glucosidase (G0395; Sigma-Aldrich) in 5 μL of buffer was added. These samples were incubated at 37°C for 90 min and used to measure total SA, consisting of free SA and SA glucosides. Three technical replicates per sample were measured. For this, 20 μL of extract was combined with 50 μL of Luria-Bertani medium and 50 μL of Acinetobacter sp. ADPWH_lux culture grown in Luria-Bertani medium to OD600 = 0.4 on white 96-well microtiter plates (E150633M; Greiner Bio-One) and incubated for 1 h at 37°C. Luminescence was measured for 1 s using a luminometer (Centro LB 960; Berthold Technologies). SA content was determined in comparison with standards of known SA concentrations in extracts derived from the sid2-2 mutant, which does not accumulate pathogen-induced SA.
RT-qPCR Analysis of Gene Expression
The extraction of RNA from plant leaf tissues was performed with TRIzol reagent (Life Technologies) following the manufacturer’s protocol. Transcript abundance was determined by RT-qPCR using the SuperScript III Platinum SYBR Green One-Step RT-qPCR Kit (Invitrogen) in a LightCycler 480 instrument (Roche). The system-reported Cp (crossing point-PCR-cycle) values using the second derivative maximum method were used for data analysis. Gene expression levels were normalized to the expression levels of ACTIN2 measured from the same RT-qPCR run. Primers used for CBP60g, FRK1, VSR6, WRKY29, CML46, CML47, and ACTIN2 are listed in Supplemental Table S9.
RNA-seq Analysis
Plants of the indicated genotypes were grown to 30 d old, and the leaves were infiltrated with either a suspension of Pma ES4326 (OD600 = 0.002) in 5 mm MgSO4 or 5 mm MgSO4. Leaf tissues were collected at 24 h post treatment and frozen in liquid nitrogen until used for RNA extraction.
The preparation of RNA-seq libraries from total RNA was performed using the Smart-3SEQ method (Foley et al., 2018), which was adapted from the 3SEQ method (Beck et al., 2010). The libraries were multiplexed and sequenced as 50-bp single reads on an Illumina HiSeq 2500 instrument. For each sequenced library, reads were aligned to the Arabidopsis reference genome (TAIR10) based on gene models from Araport11_20151202 (Cheng et al., 2017) using STAR aligner (version 2.4.2a; Dobin et al., 2013), and transcript counts for each gene were determined with the unique molecular identifiers for each sample (Kivioja et al., 2011) incorporated in the Smart-3SEQ protocol.
The read-count-per-gene data were analyzed with the method described previously (Hatsugai et al., 2017) with minor modifications. Genes were not included in the downstream analysis if no transcript was detected for the gene in more than 60% of the libraries and the fifth largest transcript count value was less than 20. These criteria resulted in 18,463 genes remaining for downstream analysis using a negative binomial generalized linear model. Comparisons of gene expression between genotypes or between treatments were conducted as Z-tests with mean values and se estimated from the negative binomial generalized linear model. The two-tailed P values derived from all the comparisons were adjusted to q values through a false discovery rate-based multiple testing correction procedure (Storey and Tibshirani, 2003). Genes that have an absolute value of expression fold change ≥ 2 and q ≤ 0.05 in mutant genotypes compared with the wild type are considered differentially expressed. Genes that are expressed higher or lower in the Col-0 wild type following Pma treatment relative to mock treatment by 2-fold or greater and q ≤ 0.05 are termed Pma induced or Pma suppressed, respectively.
Hierarchical clustering analysis was performed on the Pma-induced and Pma-suppressed sets, including genes differentially expressed in cbp60a or cml46 cml47 after mock or Pma treatment. In total, 690 Pma-induced and 635 Pma-suppressed genes were selected. Hierarchical clustering was performed for genes using Pearson correlation. Visualization of the hierarchical clustering as heat maps was made using the ComplexHeatmap package (version 1.14.0; Gu et al., 2016) from the Bioconductor project in the R environment (version 3.4.1; Venables and Ripley, 2010).
GO Enrichment Analysis
The GO enrichment analysis of differentially regulated genes was performed as a PANTHER overrepresentation test (release 20170413) with the GO biological process for Arabidopsis reference in the GO database (release 20170926). The P values reported were adjusted with the Bonferroni correction for multiple testing.
Mathematical Modeling of CBP60g Expression Kinetics
The data used for modeling were a subset of the data presented in Figure 5. The data points at 72 and 96 h after 0.1 μm flg22 treatment were excluded because the expression levels of CBP60g had passed the decaying phase and remained close to the basal levels; thus, they could not be captured by our model, which does not include a basal level. Two outliers in Col-0 at 10 h after low-concentration flg22 treatment also were removed from the data set because they were abnormally low and we suspected technical errors. There were 217 observations in total used in model fitting.
To estimate σ during the maximum likelihood estimation procedure, we assumed that the CBP60g expression values followed a log-normal distribution at all nonzero time points with homogenous sd of the associated normal distributions, regardless of the genotype and treatment concentration. We applied a bootstrap approach to estimate sd values. For every nonzero time point in one case, we resampled the observations with replacement 200 times and the data in each bootstrapped set were fitted with log-normal distribution. After sorting the σ values estimated from the bootstrapped data sets, we formed a 95% point-wise confidence band from the percentiles at each time point: we found the 2.5% × 200 = fifth largest and smallest values at each time point. We inferred 48 95% confidence intervals (8 time points × 2 concentrations × 3 genotypes) and estimated the sd value that can be applied to the whole time course. We selected the value that intersects most confidence intervals and set it to 0.4 (Supplemental Fig. S12). Further analysis showed that the regression results were not sensitive to the values of σ in the sense that the equivalence pattern of the parameter sets does not change as σ changes.
We constructed the likelihood function based on the foregoing assumptions and found the parameter values that maximize the function using the fmincon function in Matlab. The likelihood of the parameter set w is defined as the probability density function of the observation y given w. Since the observations are assumed independent, we computed the likelihood function by multiplication of the probability density of all the observations given a parameter set w, which is pending to be estimated.
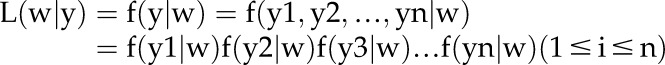 |
with 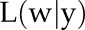 , likelihood of the parameter w given the observed data y;
, likelihood of the parameter w given the observed data y; 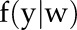 , probability density function of observation y given w;
, probability density function of observation y given w; 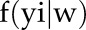 , probability density of observation yi given w. The probability density of the observation yi given parameter set w,
, probability density of observation yi given w. The probability density of the observation yi given parameter set w, 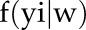 , is equal to the likelihood of observation yi for the log-normal distribution with parameters μ and σ. σ is estimated as mentioned, and the differential equation predicts the value of μ at the time point corresponding to the observation. We computed the likelihood
, is equal to the likelihood of observation yi for the log-normal distribution with parameters μ and σ. σ is estimated as mentioned, and the differential equation predicts the value of μ at the time point corresponding to the observation. We computed the likelihood 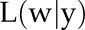 in this way and found the parameter set w that maximizes the likelihood function.
in this way and found the parameter set w that maximizes the likelihood function.
The modeling process was performed in Matlab.9.0.0.341360 (R2016a).
Statistical Analysis
Data analyses for 3-dpi Pma growth, SA measurement, and RT-qPCR experiments were performed using the lmer function in the lme4 package in the R environment (version 3.4.1; Venables and Ripley, 2010). Analyses for determining additive effects between mutants and comparing PTI effects were performed using the lme function in the nlme package in the R environment. When applicable, experiment factor and pot factor were included as random effects in the analyses. For analyses involving multiple-comparison procedures, the associated P values were adjusted to q values based on a false discovery rate approach established previously (Benjamini and Hochberg, 1995).
Accession Numbers
Accession numbers are as follows: CBP60a (At5g62570), CBP60g (At5g26920), SARD1 (At1g73805), CML46 (At5g39670), CML47 (At3g47480), and SID2/ICS1 (At1g74710). RNA-seq data have been deposited into the National Center for Biotechnology Information’s Gene Expression Omnibus under accession number GSE100187.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of T-DNA alleles of cml46 and cml47.
Supplemental Figure S2. Complementation of cml46 and cml47.
Supplemental Figure S3. Phenotypic comparisons of wild-type, cbp60a, cml46 cml47, and cbp60a cml46 cml47 plants.
Supplemental Figure S4. Expression of marker genes of plant immunity in different genotypes.
Supplemental Figure S5. Expression of CBP60g in untreated plants.
Supplemental Figure S6. Kinetics of CBP60g in wild-type and cbp60a cml46 cml47 plants following mock or flg22 treatments.
Supplemental Figure S7. Regression of the CBP60g expression kinetics data with best BIC-selected models.
Supplemental Figure S8. Mutations in cbp60a and cml46 cml47 alter kinetics of flg22-induced FRK1 and VSR6 expression but not WRKY29.
Supplemental Figure S9. PTI induced by flg22 in Col-0 and fls2 plants.
Supplemental Figure S10. Synergy between cbp60a and cml46 cml47 for shutting down flg22-PTI.
Supplemental Figure S11. Measurement of free SA at 72 h after treatments.
Supplemental Figure S12. Illustrative graph for the 95% confidence intervals of the data sd for each genotype-treatment combination at each time point.
Supplemental Table S1. q values of the CBP60g transcript level comparisons made in Figure 5.
Supplemental Table S2. Regression analysis result for CBP60g based on maximum likelihood estimation.
Supplemental Table S3. Four parameter sets for m and k.
Supplemental Table S4. Five equivalence patterns within the parameter set.
Supplemental Table S5. Top 10 BIC-selected model structures.
Supplemental Table S6. Regression analysis result for FRK1 based on maximum likelihood estimation.
Supplemental Table S7. Regression analysis result for VSR6 based on maximum likelihood estimation.
Supplemental Table S8. Regression analysis result for WRKY29 based on maximum likelihood estimation.
Supplemental Table S9. PCR primers used in this study.
Supplemental Data Set S1. RNA-seq data analysis for a set of 3,614 genes showing significant difference between the mutant and the wild type in any genotype-treatment combination.
Acknowledgments
We thank Dr. Joseph W. Foley for providing the Smart-3SEQ protocol and data analysis suggestions. We thank the University of Minnesota Genomics Center for sequencing the Smart-3SEQ libraries, the Minnesota Supercomputing Institute for data storage and computational resources, and the Arabidopsis Biological Resource Center for seeds.
Footnotes
This work was supported by grants from the National Science Foundation, IOS-1353854 to J.G., IOS-1121425 to F.K., and MCB-1518058 to F.K. and C.L.M.
Articles can be viewed without a subscription.
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Beck AH, Weng Z, Witten DM, Zhu S, Foley JW, Lacroute P, Smith CL, Tibshirani R, van de Rijn M, Sidow A, et al. (2010) 3′-end sequencing for expression quantification (3SEQ) from archival tumor samples. PLoS ONE 5: e8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defraia CT, Schmelz EA, Mou Z (2008) A rapid biosensor-based method for quantification of free and glucose-conjugated salicylic acid. Plant Methods 4: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol Plant 1: 423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Faas GC, Raghavachari S, Lisman JE, Mody I (2011) Calmodulin as a direct detector of Ca2+ signals. Nat Neurosci 14: 301–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JW, Zhu C, Jolivet P, Zhu SX, Lu P, Meaney MJ, West RB (2018) Gene-expression profiling of single cells from archival tissue with laser-capture microdissection and Smart-3SEQ. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32: 2847–2849 [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Igarashi D, Mase K, Lu Y, Tsuda Y, Chakravarthy S, Wei HL, Foley JW, Collmer A, Glazebrook J, et al. (2017) A plant effector-triggered immunity signaling sector is inhibited by pattern-triggered immunity. EMBO J 36: 2758–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kivioja T, Vähärautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J (2011) Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods 9: 72–74 [DOI] [PubMed] [Google Scholar]
- Li B, Meng X, Shan L, He P (2016) Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 19: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Busta L, Zhang Q, Ding P, Jetter R, Zhang Y (2018) TGACG-BINDING FACTOR 1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol 217: 344–354 [DOI] [PubMed] [Google Scholar]
- Sun T, Zhang Y, Li Y, Zhang Q, Ding Y, Zhang Y (2015) ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat Commun 6: 10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Glazebrook J (2012) Co-expression analysis identifies putative targets for CBP60g and SARD1 regulation. BMC Plant Biol 12: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Sreekanta S, Lu Y, Bethke G, Tsuda K, Katagiri F, Glazebrook J (2013) The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant Physiol 163: 1741–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wersch R, Li X, Zhang Y (2016) Mighty dwarfs: Arabidopsis autoimmune mutants and their usages in genetic dissection of plant immunity. Front Plant Sci 7: 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD (2010) Modern Applied Statistics with S, Ed 4 Springer, New York, NY [Google Scholar]
- Vos IA, Pieterse CMJ, van Wees SCM (2013) Costs and benefits of hormone-regulated plant defences. Plant Pathol 62: 43–55 [Google Scholar]
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog 5: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Truman W, Sato M, Nguyen V, Katagiri F, Glazebrook J (2011) CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J 67: 1029–1041 [DOI] [PubMed] [Google Scholar]
- Wang X, Gao J, Zhu Z, Dong X, Wang X, Ren G, Zhou X, Kuai B (2015) TCP transcription factors are critical for the coordinated regulation of isochorismate synthase 1 expression in Arabidopsis thaliana. Plant J 82: 151–162 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Wu S, Shan L, He P (2014) Microbial signature-triggered plant defense responses and early signaling mechanisms. Plant Sci 228: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Jauregui E, Du L, Tanaka K, Poovaiah BW (2017) Calcium signatures and signaling events orchestrate plant-microbe interactions. Curr Opin Plant Biol 38: 173–183 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu S, Ding P, Wang D, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, et al. (2010) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107: 18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Lu Y, Bethke G, Harrison BT, Hatsugai N, Katagiri F, Glazebrook J (2018) WRKY70 prevents axenic activation of plant immunity by direct repression of SARD1. New Phytol 217: 700–712 [DOI] [PubMed] [Google Scholar]
- Zhu X, Dunand C, Snedden W, Galaud JP (2015) CaM and CML emergence in the green lineage. Trends Plant Sci 20: 483–489 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]