OsWDR5a and OsTrx1/SDG723 form the core components of the COMPASS-like complex to positively regulate the flowering time and panicle branching in rice.
Abstract
Flowering time (heading date) and panicle branch number are important agronomic traits that determine yield in rice (Oryza sativa). The activation of flowering requires histone methylation, but the roles of trimethylation of Lys 4 of histone 3 (H3K4me3) in modulating heading date and panicle development are unclear. Here, we showed that the COMPASS-like complex promotes flowering and panicle branching. The rice (Oryza sativa) WD40 protein OsWDR5a interacts with the TRITHORAX-like protein OsTrx1/SET domain group protein 723 (SDG723) to form the core components of the COMPASS-like complex. Plants in which OsWDR5a or OsTrx1 expression was decreased by RNA interference produced fewer secondary branches and less grain and exhibited a delayed heading date under long-day and short-day conditions, whereas loss of OsWDR5a function resulted in embryo lethality. OsWDR5a binds to Early heading date 1 to regulate its H3K4me3 and expression levels. Together, our results show that the COMPASS-like complex promotes flowering and panicle development and suggest that modulation of H3K4me3 levels by the COMPASS-like complex is critical for rice development.
The timing of flowering is an important agricultural trait not only for successful reproduction, but also for balancing vegetative growth with reproductive growth. Heading date1 (Hd1) encodes a zinc finger protein, an ortholog of Arabidopsis CONSTANS. Hd1, which is controlled by photoperiod, promotes floral transition under short-day (SD) conditions and represses it strongly under long-day (LD) conditions. Hd1 regulates homologs of Arabidopsis (Arabidopsis thaliana) FLOWERING LOCUS T, Heading date 3a (Hd3a), and RICE FLOWERING TIME LOCUS T1 (RFT1), encoding proteins that function as SD and LD florigens in rice (Oryza sativa), respectively. In addition to the Hd1 pathway, rice has a unique, independent flowering-time pathway mediated by Early heading date 1 (Ehd1). Ehd1 encodes a B-type response regulator that promotes flowering by controlling the expression of Hd3a and RFT1 under SDs and LDs (Doi et al., 2004). Ehd1, a central integrator involved in flowering time, is activated by Ehd2 under LD and SD, and by OsMADS50 under LD conditions, whereas it is repressed by Grain number, plant height, and heading date 7 (Ghd7; Matsubara et al., 2008; Itoh et al., 2010). OsMADS56 interacts with OsMADS50 in vitro; these two MADS-box proteins form a heterodimer and function antagonistically through the OsLFL1-Ehd1 pathway under LD (Ryu et al., 2009). OsMADS51, another MADS-box gene, promotes Ehd1 expression under SD (Kim et al., 2007). In addition to being activated by MADs-box proteins, Ehd1 is suppressed by Ghd7, which encodes a CCT domain protein (Xue et al., 2008). Ghd7 is activated by EL1 (Early flowering 1)/Hd16, which encodes a casein kinase I protein (Dai and Xue, 2010), and suppressed by Ehd3 (Matsubara et al., 2011).
The activation of flowering time also requires the histone methylation modulated by SET-domain group proteins (SDG) (Sun et al., 2012; Sui et al., 2013; Choi et al., 2014; Liu et al., 2015). SDG701, related to Arabidopsis SDG2, regulates the trimethylation of Lys 4 of histone 3 (H3K4me3) at Hd3a and RFT1 (Liu et al., 2017). OsTrx1/SDG723, encoded by LOC_Os09g04890, a rice homolog of the TRITHORAX-like protein, interacts with Ehd3 to boost flowering by negatively regulating the expression of Ghd7 (Choi et al., 2014). SDG724, SDG725, and SDG708 encode proteins that belong to SET domain family II, which function in di- and trimethylation modifications of Lys 36 of H3 (H3K36me2/3). Loss of function of any of these proteins results in late flowering in both LD and SD (Sun et al., 2012; Sui et al., 2013; Choi et al., 2014; Liu et al., 2015).
The yeast (Saccharomyces cerevisiae) trithorax protein SET1 is the sole methyltransferase acting on Lys 4 of histone H3, and is responsible for the mono-, di-, and trimethylation of H3K4 (Bernstein et al., 2002; Santos-Rosa et al., 2002). Set1 forms a multiprotein complex known as COMPASS (COMplex of Proteins Associated with Set1), which functions as a H3K4 methylase in yeast (Briggs et al., 2001; Shilatifard, 2012). The COMPASS complex is highly evolutionarily conserved from yeast to humans to plants. The shared subunits between yeast COMPASS and the metazoan Set1/MLL (Mixed lineage leukemia protein)/COMPASS-like complexes include ASH2, RbBP5, and Wdr5 (Takahashi et al., 2011; Shilatifard, 2012). In mammalian cells, mammalian TRITHORAX protein MLL1 activates Hox expression and trimethylates less than 5% of human genes (Milne et al., 2002). WDR5, a WD40 protein, is conserved from yeast to mammals to plants. Earlier studies suggested that WDR5 interacts with methylated H3K4 to catalyze H3K4me3 (Wysocka et al., 2005; Dou et al., 2006), but subsequent work demonstrated that it binds an Arg-bearing motif in MLL1, which is independent of methylated H3K4 (Couture et al., 2006; Trievel and Shilatifard, 2009). WDR5 and RbBP5 stabilize the WDR5–RbBP5–MLL1 complex, whereas Ash2L has no obvious structural role in this complex. Knockdown of Ash2L, RbBP5, or WDR5 decreased the global H3K4me2/3 level (Dou et al., 2006). In Arabidopsis, the ATX1/COMPASS-like complex comprises the conserved subunits ASH2, RbBP5, and AtWDR5a (Jiang et al., 2009; Jiang et al., 2011; Ding et al., 2012b). Arabidopsis TRITHORAX-like protein ATX1 trimethylates H3K4 at specific genes involved in multiple biological processes, such as flowering time, dehydration stress responses, and flower organ development (Alvarez-Venegas et al., 2003; Pien et al., 2008; Ding et al., 2011a, 2011b; Ding et al., 2012a, 2012b). AtWDR5a associates with ATX1 to form a preinitiation complex that promotes transcription with recruitment of high levels of RNA polymerase II (Ding et al., 2011b; Ding et al., 2012b). AtWDR5a negatively regulates flowering time by inducing H3K4me3 and RNA polymerase II at FLOWERING LOCUS C (FLC; Jiang et al., 2009; Ding et al., 2012b).
In rice, over 50% of genes show H3K4 methylation (Zong et al., 2013), suggesting that H3K4 methylation has widespread functions in many biological processes. However, the roles of H3K4 methylation modulated by the COMPASS-like complex in flowering time and panicle branching are yet to be determined in rice. Here, we report that OsWDR5a interacts with OsTrx1/SDG723 and functions as a core component of the COMPASS-like complex. OsWDR5a binds Ehd1 to regulate its H3K4me3 and transcription levels. Knockdown of OsWDR5a or OsTrx1/SDG723 result in a late heading date (i.e. induction of flowering) under LD and SD conditions, and a reduction of secondary branches.
RESULTS
Characterization of an OsWDR5a Mutant in Rice
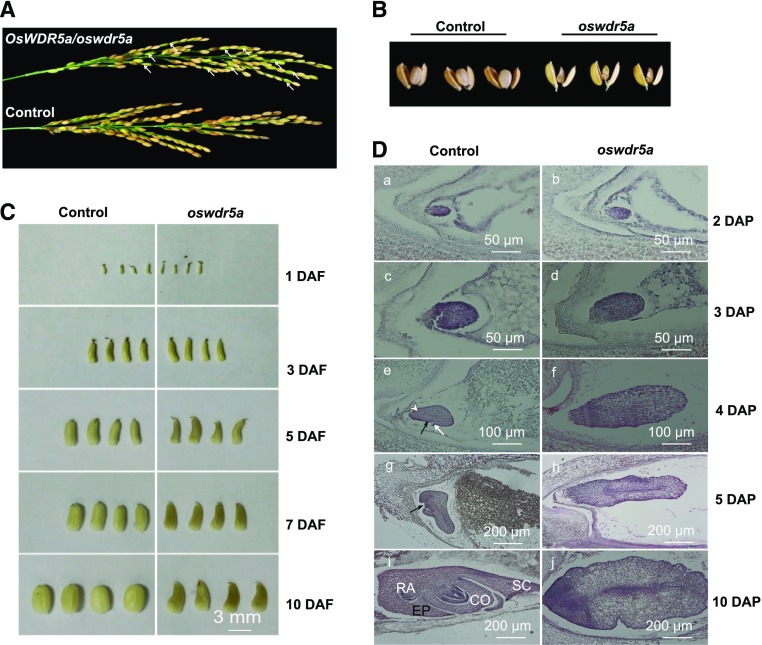
The rice genome encodes two WD40 proteins related to Arabidopsis WDR5: OsWDR5a and OsWDR5b. OsWDR5a is a 324-amino acid protein encoded by LOC_Os03g51550. Phylogenetic analysis showed that OsWDR5a and OsWDR5b are conserved in yeast, Drosophila, mammals, and plants (Supplemental Fig. S1A). To characterize the function of OsWDR5a in rice, we identified a mutant with a T-DNA insertion in this gene from the T-DNA Tagged Rice Service Center (Hsing et al., 2007). Genotypic analysis showed that the T-DNA is inserted in exon 1 of OsWDR5a (Supplemental Fig. S2A). We investigated the segregation ratio of progeny from self-pollinated F1 plants heterozygous for OsWDR5a. In the F2 population, 85 plants without T-DNA insertions and 180 heterozygous plants were obtained, but no homozygous mutants were identified. Therefore, OsWDR5a behaved like a recessive gene, with a segregation ratio of 2:1 (180:85; χ2 = 0.1377; P > 0.05) in the F2 population, indicating that the mutant phenotype is attributed to a single locus and that the loss of OsWDR5a function is lethal. Self-pollination of the F1 progeny of heterozygous OsWDR5a plants produced many green seeds, all of which aborted (Fig. 1, A and B).
Figure 1.
Loss of OsWDR5a function results in embryo lethality. A, Panicles from F1 progeny of wild-type and heterozygous OsWDR5a plants. The green seeds are indicated by arrows. B, Mature seeds from wild-type and heterozygous OsWDR5a plants. Seeds at 30 DAF are shown. C, Seed development in wild-type and heterozygous OsWDR5a plants. Seed morphology at 1 DAF to 10 DAF is shown. D, Morphogenesis of wild-type and oswdr5a mutant embryos. Embryo morphology at 2 DAP and 3 DAP (a–d). The 4 DAP wild-type embryo has a differentiating coleoptile primordium (black arrow), shoot apical meristem (white arrow), and radicle primordium (white arrowhead) (e and f). The 5 DAP wild-type embryo shows a differentiating first leaf primordium (arrow) (g and h). Morphologically complete 10 DAP embryos (i and j). SC, scutellum; CO, coleoptile; EP, epiblast; RA, radicle. Bars = 50 μm (A–D), 100 μm (E and F), and 200 μm (G–J).
F1 progenies derived from pollination of heterozygous OsWDR5a plants with wild-type pollen or from pollination of wild-type plants with heterozygous OsWDR5a pollen were indistinguishable from progenies derived from self-pollination of the wild type, suggesting that OsWDR5a might not be involved in floral organogenesis. We dissected inflorescences from wild-type and heterozygous OsWDR5a plants. Spikelets from heterozygous OsWDR5a plants contained various organs that were similar to those of the wild type (Supplemental Fig. S2B). In addition, the mature pollen grains from heterozygous OsWDR5a plants were similar to those of the wild type (Supplemental Fig. S2C). These results suggested that OsWDR5a might instead be involved in embryogenesis.
To confirm the lethal phenotype caused by the oswdr5a mutation, we generated full-length OsWDR5a cDNA fused to GFP driven by the maize (Zea mays) Ubiquitin promoter (ProUBI:GFP-OsWDR5a) and transformed this construct into the progenies of the heterozygous OsWDR5a plants. The lethal phenotype of homozygous oswdr5a plants was completely rescued by exogenous expression of OsWDR5a (Supplemental Fig. S2, D–F), indicating that GFP-OsWDR5a retains the function of OsWDR5a.
Loss of OsWDR5a Function Leads to the Termination of Embryogenesis
We investigated seed development at different days after fertilization (DAF). There was no difference between wild-type and heterozygous OsWDR5a plants during the first 3 DAF. After 5 DAF, the seeds from heterozygous OsWDR5a plants became shrunken, which persisted until 10 DAF, whereas normal, rounded seeds were observed in the wild type at this stage (Fig. 1C).
To investigate how the OsWDR5a mutation affects embryo development, we observed embryogenesis in the wild-type and heterozygous OsWDR5a plants. We detected 37 aborted embryos and 119 well-developed embryos from the progeny of heterozygous OsWDR5a plants (3:1 ratio, χ2 = 0.0769; P > 0.05), which is consistent with the high ratio of aborted seeds described above, suggesting that the loss of OsWDR5a function is lethal. At 3 d after pollination (DAP), the wild-type zygote divided quickly to form a globular embryo, in which only the epidermal cells differentiated. The globular embryos of progeny of heterozygous OsWDR5a plants were indistinguishable from those of wild type at 2 and 3 DAP (Fig. 1D, a–d). At 4 DAP, the first morphogenetic events occurred in wild type: the formation of the shoot apical meristem, coleoptile primordium, and radicle primordium. However, embryos from plants harboring a mutation in OsWDR5a failed to differentiate and resembled an undifferentiated mass of cells (Fig. 1D, e and f). We obtained more evidence for the lack of differentiation caused by the mutation of OsWDR5a at 5 to 10 DAP, when all embryonic organs had formed and embryogenesis was completed in wild type (Fig. 1D, g–j). These results indicate that the embryo lethality mediated by OsWDR5a occurs at an early stage of embryogenesis.
OsWDR5a Binds Histone H3
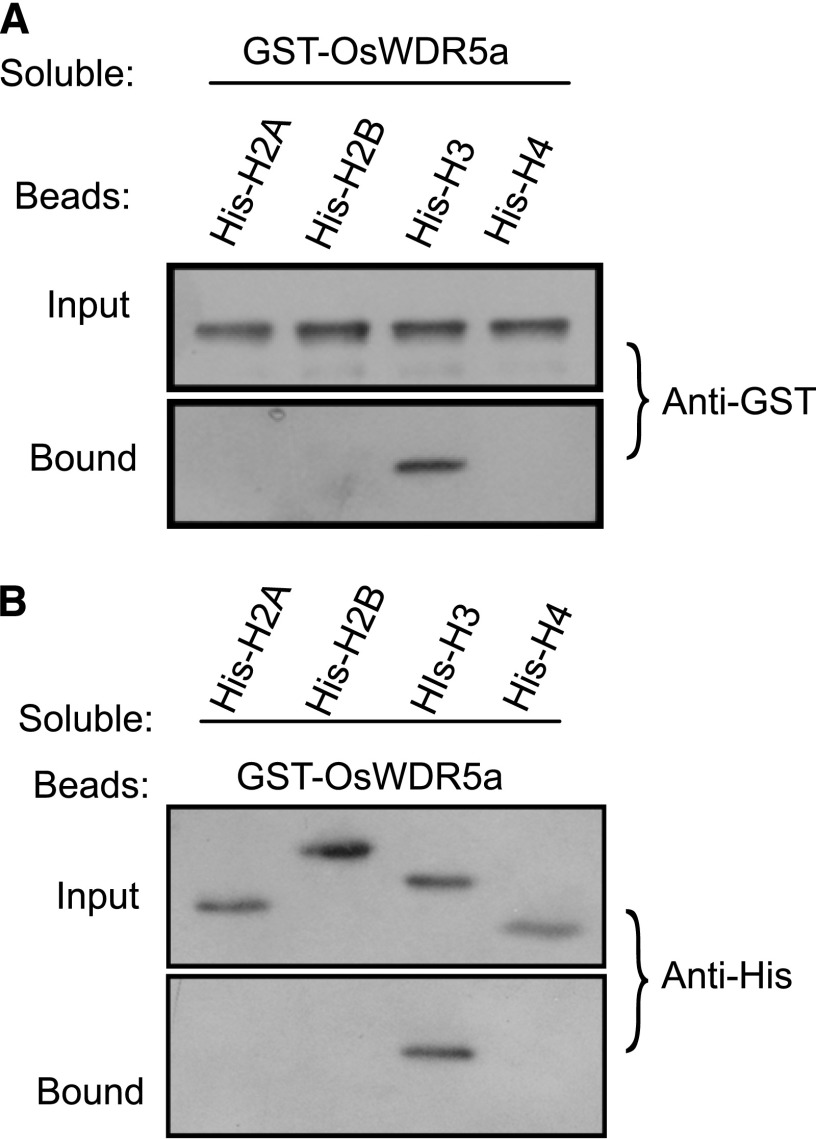
In mammalian cells, WDR5 binds the N terminus of H3 independently of H3K4 methylation (Couture et al., 2006; Song and Kingston, 2008). We investigated whether OsWDR5 can bind H3 substrate. A protein containing OsWDR5a fused to GST bound to beads containing a His fused to H3, but did not bind H2A, H2B, or H4 (Fig. 2A). In a complementary experiment, bead-attached GST-OsWDR5a bound to the soluble His-tagged H3, but not to other histones (Fig. 2B). Therefore, we concluded that OsWDR5a interacts with H3.
Figure 2.
OsWDR5a binds to histone H3. A, Beads containing His fused with H2A, H2B, H3, and H4 were assessed for their ability to bind to soluble GST-OsWDR5a and detected with the antibody to GST (Anti-GST). B, Beads containing GST fused with OsWDR5a were assessed for the ability to bind to soluble His fused with H2A, H2B, H3, and H4, and detected with the antibody to His (Anti-His).
Knockdown of OsWDR5a Results in a Late Heading Date and Small Panicles
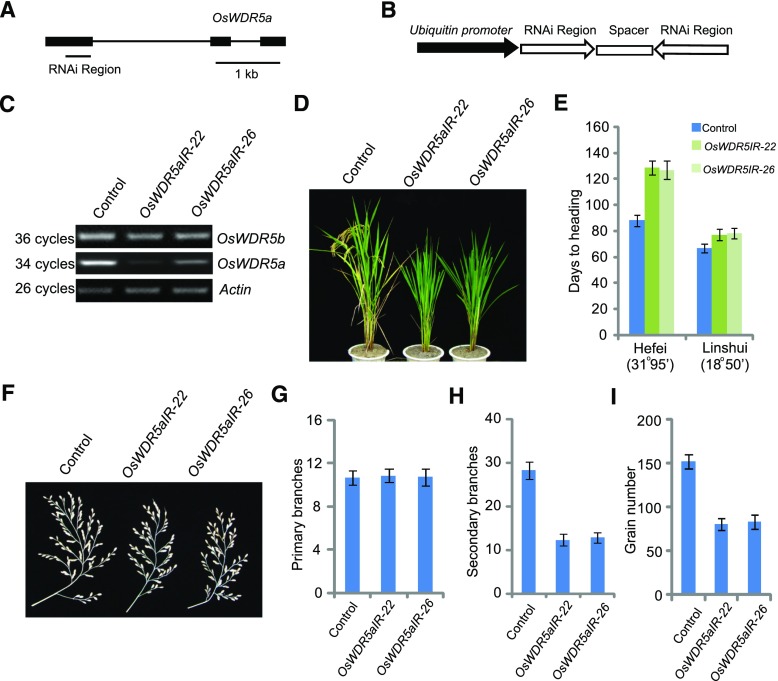
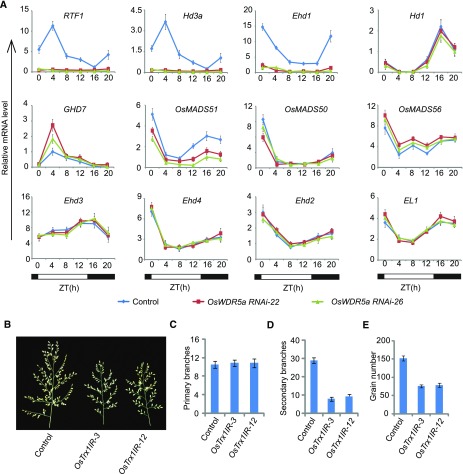
To investigate the biological roles of OsWDR5a in rice at the vegetative stage, we selected a nonconserved region at the N terminus of OsWDR5a to ensure sequence specificity and generated a knockdown mutant via RNA interference (RNAi; Fig. 3A). We introduced a hairpin construct driven by the maize Ubiquitin promoter into rice explants and successfully selected and regenerated transgenic lines (Fig. 3B). As shown in Figure 3C, the transcript levels of OsWDR5a, but not OsWDR5b, were significantly reduced in the OsWDR5aIR (RNAi plant) transformants, as revealed by RT-PCR analyses, suggesting that we successfully obtained OsWDR5-knockdown plants by RNAi. After regeneration from T1 plants, the OsWDR5aIR transformants were found to have an obvious late-heading date phenotype when grown under both LD (Hefei) and SD (Lingshui) conditions (Fig. 3, D and E). We examined the panicle branches and grain number, and found that the secondary branches and grain number, but not primary branches, were reduced in OsWDR5aIR transformants (Fig. 3, F–I).
Figure 3.
OsWDR5aIR transformants and phenotypes of OsWDR5aIR plants. A, Gene structure of OsWDR5a. Exons are indicated by boxes and introns are indicated by lines. The region used for knockdown is indicated by a line below the diagram. B, Diagram of the OsWDR5a RNAi constructs. Fragments containing the OsWDR5a fragment in the sense and antisense orientation separated by an unrelated spacer were cloned under the control of the maize Ubiquitin promoter. C, OsWDR5a and OsWDR5b transcript levels in wild-type and OsWDR5aIR plants. D, Representative image of 120-d-old wild-type and OsWDR5aIR plants grown in LD conditions (Hefei). E, Days to heading in wild-type and OsWDR5aIR plants in both LDs (Hefei) and SDs (Lingshui). Values shown are mean ± sd of heading days; 20 plants were scored per line. F, The panicles of wild-type and OsWDR5aIR plants in LDs (Hefei). G to I, The branch number (G and H), and grain number (I) of wild-type and OsWDR5aIR plants. Values shown are mean ± sd of branch number and grain number; 20 plants were scored per line.
OsWDR5b, which is encoded by LOC_Os07g38430, is closely related to OsWDR5a with 61% sequence identity and 75% similarity (Supplemental Fig. S1B). Plants with knocked-down expression of OsWDR5b displayed a similar flowering time to that of the wild type in LDs and SDs (Supplemental Fig. S3). We therefore concluded that OsWDR5a, but not OsWDR5b, is required for proper heading date and panicle development.
OsWDR5a Interacts with OsTrx1
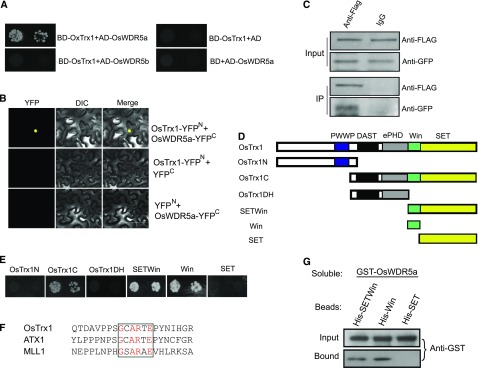
OsWDR5a is closely related to WDR5 from Arabidopsis and humans, based on the alignment of WDR5 protein sequences, suggesting that OsWDR5a might interact with rice TRITHORAX family proteins. The rice genome encodes 39 SET domain proteins, three of which belong to the TRITHORAX-like family (Ding et al., 2007; Ng et al., 2007; Avramova, 2009). We generated fusion proteins containing full-length TRITHORAX-like proteins with the binding domain (BD) for yeast two-hybrid analysis with activation domain-tagged OsWDR5a. The results revealed that OsTrx1 bound strongly to OsWDR5a, but not to OsWDR5b (Fig. 4A).
Figure 4.
OsWDR5a interacts with OsTrx1. A, Yeast two-hybrid assay revealing an interaction between OsWDR5a and OsTrx1. The growth of two dilutions (2 × 10−2 and 2 × 10−3) of yeast culture on SD medium lacking Trp, Leu, His, and adenine is shown. B, The N terminus of YFP or OsWDR5a fused to the N terminus of YFP was tested for its ability to bind to the C terminus of YFP or the C terminus of YFP fused to OsTrx1. YFP and a bright-field image (DIC) were recorded and the resulting images were merged. C, Coimmunoprecipitation of OsWDR5a and OsTrx1. GFP-OsTrx1 and FLAG-OsWDR5a were cotransformed into rice protoplasts, and the expressed proteins were immunoprecipitated using an anti-FLAG antibody and detected with anti-Flag and anti-GFP antibodies. D, Diagram of OsTrx1 showing its different domains. E, Activating domain (AD) fused to the different domains shown in C was tested for its ability to bind to BD-OsWDR5a. The growth of two dilutions (2 × 10−2 and 2 × 10−3) of yeast culture on SD medium lacking Trp, Leu, His, and adenine is shown. F, Alignment of the Win motif with other closely related TRITHORAX proteins using ClustalW. The amino acid sequence from rice OsTrx1 (amino acids 765–785), Arabidopsis ATX1 (amino acids 808–828), and human MLL1 (amino acids 3754–3774). The Win motifs are shown in-frame and the conserved amino acids are indicated in red. G, Beads containing the His-fused SETWin, Win, and SET domains of OsTrx1 were assessed for their ability to bind soluble His-fused OsWDR5a and detected with an antibody to His (anti-His).
This yeast two-hybrid interaction was confirmed by bimolecular fluorescence complementation and coimmunoprecipitation analyses. Functional YFP in the nucleus was observed when OsTrx1-YFPN (fused with the N-terminal half of yellow fluorescent protein) was coexpressed with OsWDR5a-YFPC (fused with the C-terminal half of YFP), but not in the controls (Fig. 4B). Immunoprecipitation using FLAG antibody revealed that OsTrx1 bound to OsWDR5a in rice protoplasts cotransformed with FLAG-OsWDR5a and GFP-OsTrx1 (Fig. 4C). These results indicated that OsTrx1 interacts with OsWDR5a in vitro and in vivo.
Next, we tested which domain is critical for the interaction of OsWDR5a with OsTrx1. OsTrx1 contains PWWP, DAST, ePHD, and SET domains (Fig. 4D). Both the N terminus and C terminus of OsTrx1 were fused with BD. The C terminus of OsTrx1 (OsTrx1C), but not its N terminus (OsTrx1N), bound to OsWDR5a (Fig. 4, D and E). In mammals, WDR5 preferentially recognizes the WDR5 Interaction (Win) motif, which is located in the N-terminal SET region of MLL1 and other SET1 family members (Patel et al., 2008a, 2008b; Dharmarajan et al., 2012). We detected the conserved Win motif (containing five amino acids) in OsTrx1 in an alignment with TRITHORAX proteins from mammalian cells to plants (Fig. 4F). Within OsTrx1C, the SETWin and Win motifs bound to OsWDR5a, but DAST and the ePHD domain (OsTrx1DH) or SET domain (SET) alone did not (Fig. 4E). These yeast two-hybrid interactions were further validated by a pull-down assay. Beads attached to His-SETWin and the His-Win motif bound to soluble GST-OsWDR5a, but His-SET did not (Fig. 4G), suggesting that the Win motif of OsTrx1 is sufficient for the interaction between OsTrx1 and OsWDR5a.
OsWDR5a Are Involved in the Ehd1-RFT1 Pathway
To investigate the roles of OsWDR5a in the flowering-time network, we isolated total RNA from leaves of 80-d-old plants and subjected it to quantitative RT-PCR analysis. Ehd1 and Hd1 transcript levels exhibited a strong diurnal rhythmic pattern in wild-type plants. Ehd1, but not Hd1, was markedly reduced in the OsWDR5a RNAi lines, suggesting that OsWDR5a might not be involved in the photoperiod pathway (Fig. 5A). These results are consistent to the late-flowering time phenotype of OsWDR5aIRs under LDs and SDs.
Figure 5.
Transcript levels of flowering network genes in OsWDR5aIR plants. A, Transcript levels of flowering network genes. RNA isolated from leaves of 80-d-old plants was used for RT-qPCR. The black bars indicate the dark period and the white bars indicate the light period. ZT, Zeitgeber time. The y axis shows the transcript level relative to rice Actin expression. The experiments were repeated at least three times, and each experiment included three replicates, n = 3. B, The panicles of wild-type and OsTrx1IR plants in LD conditions. C to E, The branch number (C and D), and grain number (E) of wild-type and OsTrx1IR plants. Values shown are mean ± sd of branch number and grain number; 20 plants were scored per line.
We measured the mRNA levels of RFT1 and Hd3a, and found that the expression of RFT1 and Hd3a was abolished in the OsWDR5aIR RNAi lines.
Ghd7, which acts as a negative regulator of Ehd1, was highly induced in the transformants. Next, we examined the expression of Ehd2, Ehd3, Ehd4, and OsEL1, and found that these genes were not affected in the transformants compared to the wild type. The SD flowering activator OsMADS51, but not OsMADS50 or OsMADs56, was down-regulated in OsWDR5aIRs (Fig. 5A). The up-regulated and down-regulated genes are similar to those in OsTrx1 knockdown lines (Choi et al., 2014). These results suggested that OsWDR5a and OsTrx1 function together and might be involved in regulating heading date in rice through the Ehd1-RFT1 pathway under LD conditions.
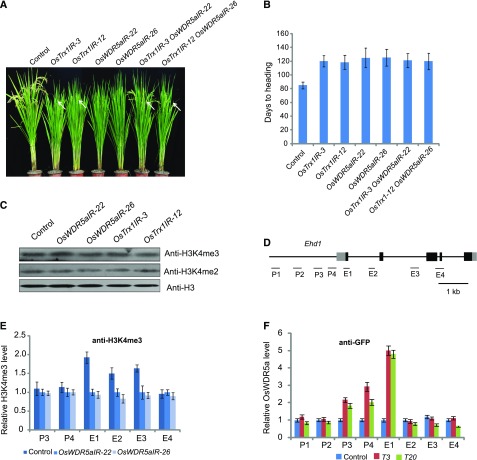
We generated an OsTrx1 knockdown mutant using the RNAi approach. RT-PCR revealed that OsTrx1 transcript levels were strongly down-regulated in two different RNAi lines (Supplemental Fig. S4, A–C). The OsTrx1IR (RNAi plant) transformants displayed a late-flowering phenotype under LD and SD conditions (Supplemental Fig. S4, D and E), suggesting that OsTrx1 might be involved in a photoperiod-independent pathway.
We then examined the panacle and grain number and found that the secondary branches and grain number, but not primary branches, were reduced in the OsTrx1IR lines (Fig. 5, B–E). These results suggested that OsTrx1 is required for proper flowering time and panicle development in rice.
OsWDR5a Is Required for H3K4me3 Deposition at Ehd1
To elucidate the possible relationship between OsTrx1 and OsWDR5a, we crossed the OsWDR5IR transformants with the OsTrx1IR lines and generated plants with knocked-down expression of both OsTrx1 and OsWDR5a. The double knockdown plants had a late-flowering phenotype similar to that of the OsTrx1IR lines under LDs (Fig. 6, A and B), suggesting that OsTrx1 functions in the same pathway as OsWDR5a. Since OsTrx1 is a homolog of ATX1, we investigated whether global H3K4me3 and H3K4me2 levels are affected by OsWDR5a and OsTrx1. Immunoblot analysis revealed no global changes in H3K4me2 or H3K4me3 in the transformants (Fig. 6C), suggesting that OsWDR5a and OsTrx1 might regulate specific genes. Next, we investigated if the reduced transcript levels of the genes were due to attenuated H3K4me3 in OsWDR5aIRs. We investigated the distribution of H3K4me3 by chromatin immunoprecipitation (ChIP), followed by quantitative PCR analysis of DNA enrichment. H3K4me3 levels were reduced at Ehd1, RFT1, and Hd3a in the OsWDR5aIR lines, indicating that OsWDR5a is required for H3K4me3 (Fig. 6, D and E; Supplemental Fig. S5, A and B). Since loss of OsTrx1 function led to up-regulation of Ghd7 (Choi et al., 2014), we investigated H3K4me3 levels at Ghd7 and found that H3K4me3 levels of Ghd7 were up-regulated in OsTrx1IR and OsWDR5aIR plants (Supplemental Fig. S5, D–F).
Figure 6.
OsWDR5a is required for H3K4me3 at Ehd1. A, Representative image of 125-d-old wild-type, OsWDR5aIR, OsTrx1IR, and OsTrx1IR OsWDR5aIR plants in LD conditions. Values shown are mean ± sd of branch number and grain number; 20 plants were scored per line. B, Days to heading of wild-type, OsWDR5aIR, OsTrx1IR, and OsTrx1IR OsWDR5aIR plants. C, Global H3K4me2 and H3K4me3 levels in wild-type, OsWDR5aIR, and OsTrx1IR plants. D, Gene structure of Ehd1. Exons are indicated by boxes and introns are indicated by lines. The locations of the gene regions analyzed by ChIP-PCR are shown below the diagram. P1 to P4 indicates regions in the promoter and E1 to E4 indicates regions in the coding region. E, Relative H3K4me3 levels at different regions of the genes in wild-type and OsWDR5aIR plants. Experiments were repeated at least three times, and the values shown indicate the mean ± se, n = 3 replicates. F, Relative OsWDR5a expression levels in plants complemented by ProUBI:GFP-OsWDR5a and the wild type. The T3 (complementary line 3 for oswdr5a) and T20 are indicated in Supplemental Figure S2. Experiments were repeated at least three times, and the values shown indicate the mean ± se, n = 3 replicates.
Next, we examined whether OsWDR5a directly targets these genes. We assessed the distribution of OsWDR5a in complemented plants harboring ProUBI:GFP-OsWDR5a using ChIP analysis with a specific GFP antibody, followed by quantitative PCR analysis of the DNA enrichment at multiple points along the Ehd1 promoter. OsWDR5a was highly enriched along the promoter and coding region of Ehd1 in two transgenic lines harboring ProUBI:GFP-OsWDR5a, but not in the control (Fig. 6F). However, OsWDR5a was not enriched along RFT1, Hd3a, or Ghd7 (Supplemental Fig. S5, C and G). These results indicated that OsWDR5a directly targets Ehd1 and activates the H3K4me3 of Ehd1.
DISCUSSION
In this study, we found that the COMPASS-like complex modulates panicle development and the induction of flowering in rice. OsWDR5a binds to OsTrx1, and these two factors form core components of the rice COMPASS-like complex. Defects in OsWDR5a and OsTrx1 result in a reduction of secondary branching and a late heading date under both LD and SD conditions, and these two factors are involved in the Ehd1-RFT1 pathway. These results were further supported by the finding that OsWDR5a induced the expression and H3K4me3 enrichment of Ehd1. Together, these results suggest that the COMPASS-like complex modulates heading date and panicle branching in rice.
WDR5, a WD40 protein, is conserved from yeast to mammals to plants. Yeast Cps30, a homolog of WDR5, is essential for COMPASS complex formation/stability and for full histone methyltransferase activity in this complex. In mammals, MLL1 and WDR5 are critical for the integrity of the MLL1 complex, and therefore, its methyltransferase activity (Dou et al., 2006, Patel et al., 2008a and Song and Kingston, 2008). WDR5 was originally identified as an interactor of methylated Lys 4 of H3; the following results indicated that this interaction is dependent on the H3 motif, but not the methylated Lys 4 of H3 (Song and Kingston, 2008; Trievel and Shilatifard, 2009). Our study also suggested that OsWDR5a bound to H3, but not the other histones. WDR5 interacts with the Win motif of MLL1, including the Arg-bearing motif, which is critical for this binding (Patel et al., 2008a, 2008b; Dharmarajan et al., 2012). Sequence alignment showed that the Win motif is conserved in mammals and plants. Although Arabidopsis WDR5a interacts with many SET domain proteins (Jiang et al., 2011), our results showed that the interaction between OsWDR5a and OsTrx1 depends on the presence of the conserved Win motif. The interaction between OsWDR5a and OsTrx1 was also supported by the similar phenotypes of OsWDR5aIR and OsTrx1IR plants and the down-regulation of flowering network genes in these lines. The OsTrx1IR plants displayed a slight late-flowering phenotype under SD (Lingshui) conditions; one possibility is that the daylight in Lingshui for rice growth was around 11 h (Supplemental File S1).
In addition to the altered flowering time and panicle branches, the loss of OsWDR5a function leads to the termination of embryo development, indicating that OsWDR5a plays two distinct roles at the vegetative phase and reproductive phase. Therefore, OsWDR5a is involved in more developmental processes than OsTrx1. Loss of OsWDR5a function resulted in termination of embryogenesis at 4 DAP (early embryo differentiation), suggesting that OsWDR5a might be involved in the initiation of embryogenesis in rice. Even though OsWDR5b is closely related to OsWDR5a, OsWDR5b was not observed to interact with SDG723. Knockdown of OsWDR5b did not result in notable changes in flowering time, suggesting that OsWDR5b evolutes the divergent functions with OsWDR5a. In mammalian cells, WDR5 residues Tyr-191, Pro-173, Phe-149, and Asp-172 line the A-pocket, and residues Tyr-191, Pro-216, and Leu-234 line the B-pocket. Both pockets are critical for MLL1 interaction (Dharmarajan et al., 2012). The Tyr-191 and Pro-216 were conserved in OsWDR5a, but not in OsWDR5b. The functions of these two amino acids in protein interaction and developmental regulation merit further study.
The COMPASS-like complex is highly evolutionarily conserved from yeast to humans to plants. In mammalian cells, knockdown of WDR5 expression led to reduced H3K4me3 levels and expression levels of Hox-a9 and Hox-c8, as was also observed in the absence of MLL1 (Wysocka et al., 2005). Defects in any components of the Arabidopsis COMPASS-like complex reduced the transcription level and H3K4me3 level at FLC, resulting in early flowering, which suggested that the Arabidopsis COMPASS-like complex suppresses flowering time (Jiang et al., 2011). By contrast, OsWDR5a and OsTrx1 promoted flowering time in rice. Arabidopsis and rice contain different flowering pathways. FLC is central regulator suppressing floral transition in Arabidopsis, which is not known in rice. Mutations in ATX1 and knockdown of WDR5a lead to early flowering due to reduced transcript levels of FLC. Rice, a model SD plant, contains two major flowering pathways: a conserved photoperiod pathway and a unique photoperiod-independent flowering-time pathway. Ehd1 is an evolutionarily unique gene in rice that does not have an ortholog in Arabidopsis (Doi et al., 2004; Tsuji et al., 2011). In this study, OsWDR5a was identified as a factor that positively regulates Ehd1 transcription and histone modification level. OsWDR5a bound to Ehd1, but not RTF1, or Hd3a. This possibility of the specificity is that the transcriptional factors interact with the subunits of the COMPASS-like complex and trigger OsWDR5a to target Ehd1. Our recent study suggested the transcriptional factors are critical for flowering time and hypocotyl elongation modulated by histone phosphrylation (Su et al., 2017; Zheng et al., 2017). A previous study suggested that OsTrx1 regulates flowering time via repressing the transcription of Ghd7 (Choi et al., 2014). Our study suggests that the H3K4me3 levels of Ghd7 were reduced in OsWDR5aIR and OsTrx1IR plants. The activated transcription levels of Ghd7 in the OsTrx1IR and OsWDR5aIR lines indicate that Ghd7 might not be the direct target of OsTrx1. Together, our findings indicate that the COMPASS-like complex promotes H3K4me3 at Ehd1 to regulate heading date and panicle branching in rice.
METHODS
Materials
The plants used in this study are in the rice (Oryza sativa ssp. japonica cv Nipponbare) background, except for the T-DNA mutant Oswdr5a, which is in the Tainung 67 background and was obtained from the T-DNA Tagged Rice Service Center (Hsing et al., 2007). The seeds of the T-DNA mutant, Oswdr5a, were a gift from Professor Keqiang Wu from National Taiwan University. All plants were regenerated from callus as described (Xu et al., 2015). T1 generation plants were generated from the seeds of self-pollinated T0 plants. The plants were grown in fields from April 20 to October at Hefei (Anhui) and from November 20 to February at Lingshui (Hainan), China. The detailed photoperiod for rice growth at Hefei and Lingshui are described in Supplemental File S1.
Plasmid Constructs
The plasmids were constructed using the DNA primers and protocols described in Supplemental File S2. All cloned DNA was confirmed by DNA sequencing.
Pollen Fertility
Ten panicles each from the wild-type (TN67) and heterozygous OsWDR5a plants were sampled at 1 or 2 d before flowering to determine the pollen fertility. Six florets per panicle were sampled from the upper, middle, and lower portions of the panicle. One anther per floret was collected, and six anthers from the same panicle were mixed and spread onto a slide. The pollen grains on the slide were stained with 1% IK-I2 and the darkness of the staining was used as an indicator of pollen fertility. Four views per slide were obtained under a microscope (Leica, M165FC), resulting in 40 views per hybrid.
Histological Analyses
Panicles from wild-type (TN67) and heterozygous OsWDR5a plants were fixed in 3.7% formaldehyde, 5% acetic acid, and 50% ethanol at 4°C overnight. The samples were auto-dehydrated in a LEICA ASP200S tissue processor, embedded in paraffin (Sigma-Aldrich), and sectioned to 8-μm thicknesses using a LEICA RM2255 rotary microtome. After dewaxing with xylene and hematoxylin-eosin staining, the sections were observed under a light microscope (Zeiss, Axioscope A1 polarized light microscope).
Protein Pull-Down Assays, Co-IP, and Immunoblot Assays
For the pull-down assay, beads were incubated with 3 µg of the fusion protein, washed, and incubated with 3 µg of soluble protein overnight at 4°C. Mock controls included extracts prepared from either the His-Tag or GST vectors. The beads were washed five times with a solution containing 20 mm Tris (pH 7.4), 150 mm NaCl, and 0.05% Tween 20, separated on an SDS-PAGE gel, and analyzed by immunoblot using an anti-GST antibody (GenScript; A00866-100, Lot: 13D000626) or an anti-His antibody (Abmart; M30111M, Lot: 273884).
For the Co-IP, OsWDR5a and OsTrx1 were fused with FLAG or GFP and cloned into the pUC19 vector. The Co-IP was performed as described previously (Su et al., 2017; Zheng et al., 2017). Briefly, 1 × 106 rice protoplasts were lysed with PEN-140 buffer (140 μm NaCl, 2.7 μm KCl, 25 μm Na2HPO4, 1.5 μm KH2PO4, 0.01 μm EDTA, and 0.05% CA-630). The supernatant was precleared with Protein G and precipitated with anti-FLAG antibody (Sigma-Aldrich; H6908, Lot: SLBQ7119V). The protein complexes were isolated by binding to protein G beads (Santa Cruz; Sc-2002) and washed five times with PEN-400 buffer (400 μm NaCl, 2.7 μm KCl, 25 μm Na2HPO4, 1.5 μm KH2PO4, 0.01 μm EDTA, and 0.05% CA-630). The samples were analyzed by protein gel blot analysis using an anti-GFP antibody (Clontech Laboratories, Inc.; JL-8, Lot: A5033481) or anti-FLAG antibody.
Transient expression in N. benthamiana Leaves and Bimolecular Fluorescence Complementation
Rice OsWDR5 and OsTrx1 were cloned into pUC-SPYCE (aa 156–239) or pUC-SPYNE (aa 1–155), respectively. Transient expression assays were performed as described previously (Lu et al., 2017). Briefly, Agrobacterium colonies were grown overnight in 10 mL of medium plus antibiotics. The cells were collected and resuspended in an equal volume of induction medium (60 mm K2HPO4, 33 mm KH2PO4, 7.5mM (NH4)SO4, 1.7 mm Na citrate · 2H2O, 10 mm MES, 1 mm MgSO4, 0.2% Glc, 0.5% glycerol, antibiotics, and 50 µg/mL acetosyringone). After shaking for 6 h, the cells were resuspended to an OD of 0.6 in infiltration medium (10 mm MgCl2, 10 mm N-morpholino-ethanesulfonic acid, 150 µm acetosyringone) and used to inject the abaxial surfaces of N. benthamiana leaves. YFP signals were visualized 24 to 48 h after inoculation by confocal laser scanning microscopy (Zeiss, LSM700).
Reverse Transcription and Quantitative PCR
Total RNA was isolated from the leaves of 80-d-old plants and reverse transcribed with oligo(dT) primers (Promega), and the amounts of individual gene transcripts were measured with gene-specific primers. Quantitative PCR analysis was performed with the CFX real-time PCR instrument (Bio-Rad) and SYBR Green mixture (Roche). The relative expression of the genes was quantitated with the 2−ΔΔCT Ct calculation, using UBIQUITIN as the reference housekeeping gene for the expression analyses. The enrichment of DNA at specific genes was quantified with the 2−ΔΔCT Ct calculation, using UBIQUITIN as the reference housekeeping gene for chromatin immunoprecipitation assays. See Supplemental File S2 for primers.
ChIP Assay
ChIP was performed as described (Lu et al., 2017; Su et al., 2017; Zheng et al., 2017). Briefly, 3 g of leaves were ground in powder, then fixed with 1% formaldehyde for 10 min and quenched in 0.125 m Gly. The pellet was homogenized in buffer I (0.4 m Suc, 10 mm Tris, pH 8.0, 5 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail) and filtered through Miracloth. After centrifugation, the pellet was extracted with buffer II (0.25 m Suc, 10 mm Tris, pH 8.0, 10 mm MgCl2, 1% Triton X-100, 5 mm β-mercaptoethanol, 0.1 mm PMSF, and protease inhibitor cocktail) and then with buffer III (1.7 m Suc, 10 mm Tris, pH 8.0, 10 mm MgCl2, 1% Triton X-100, 5 mm β-mercaptoethanol, 0.1 mm PMSF, and protease inhibitor cocktail). The nuclei were then lysed in lysis buffer (50 mm Tris, pH 8.0, 10 mm EDTA, 1% SDS, 5 mm β-mercaptoethanol, 0.1 mm PMSF, and protease inhibitor cocktail) and the extract was sonicated to fragment the DNA to a size range of 300 to 500 bp. After centrifugation, the supernatant was diluted using dilution buffer (1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris, pH 8.0, 167 mm NaCl, 0.1 mm PMSF, and protease inhibitor cocktail) and precleared with protein A or protein G magnetic beads. Specific antibodies anti-H3K4me3 (Abcam; ab8580, lot; GR273043-6) and anti-GFP (Clontech Laboratories, Inc.; JL-8, Lot: A5033481) or control IgG serum were added to the precleared supernatants for an overnight incubation at 4°C. The antibody-protein complexes were isolated by binding to protein A or protein G beads. The washed beads were heated at 65°C for 8 h with proteinase K to reverse the formaldehyde cross-linking and digest proteins. The sample was then extracted with phenol/chloroform and the DNA was precipitated in ethanol and resuspended in water. The purified DNA was analyzed by quantitative PCR with the gene-specific primers shown in Supplemental File S2.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers listed below: OsTrx1/SDG723 (Os09g0134500), OsWDR5a (Os03g0725400), and OsWDR5b (Os07g0572000).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of WDR5 from yeast, human, Arabidopsis, and rice.
Supplemental Figure S2. Characterization of OsWDR5a.
Supplemental Figure S3. Generation of OsWDR5bIRs.
Supplemental Figure S4. Generation of OsTrx1 transformants using RNA interference approach.
Supplemental Figure S5. H3K4me3 profiles at RFT1, Hd3a, and Ghd7 in wild type and OsWDR5aIRs.
Supplemental File S1. The average daylight in 10-d intervals in 2017 at Hefei and Lingshui, China.
Supplemental File S2. Plasmids and primers.
Acknowledgments
We are grateful to Dr. Peng-Cheng Wei, Hao Li, and Ya-chun Yang from Anhui Academy of Agricultural Sciences for kindly helping us with rice transformation and cultivation, Professor Keqiang Wu from National Taiwan University for kindly providing the Oswdr5a seeds, and to all members of the Ding group for their helpful discussions.
Footnotes
This work was supported by the National Natural Science Foundation of China (9143510, 31571315, and 31371306 to Y.D.) and the Strategic Priority Research Program “Molecular Mechanisms of Plant Growth and Development” of CAS (grant no. XDPB04).
Articles can be viewed without a subscription.
References
- Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z (2003) ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 13: 627–637 [DOI] [PubMed] [Google Scholar]
- Avramova Z. (2009) Evolution and pleiotropy of TRITHORAX function in Arabidopsis. Int J Dev Biol 53: 371–381 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL (2002) Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA 99: 8695–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15: 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SC, Lee S, Kim S-R, Lee Y-S, Liu C, Cao X, An G (2014) Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3. Plant Physiol 164: 1326–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture J-F, Collazo E, Trievel RC (2006) Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol 13: 698–703 [DOI] [PubMed] [Google Scholar]
- Dai C, Xue HW (2010) Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J 29: 1916–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmarajan V, Lee J-H, Patel A, Skalnik DG, Cosgrove MS (2012) Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases. J Biol Chem 287: 27275–27289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Avramova Z, Fromm M (2011a) The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J 66: 735–744 [DOI] [PubMed] [Google Scholar]
- Ding Y, Avramova Z, Fromm M (2011b) Two distinct roles of ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1) at promoters and within transcribed regions of ATX1-regulated genes. Plant Cell 23: 350–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Fromm M, Avramova Z (2012a) Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat Commun 3: 740. [DOI] [PubMed] [Google Scholar]
- Ding Y, Ndamukong I, Xu Z, Lapko H, Fromm M, Avramova Z (2012b) ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet 8: e1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang X, Su L, Zhai J, Cao S, Zhang D, Liu C, Bi Y, Qian Q, Cheng Z, et al. (2007) SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell 19: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG (2006) Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 13: 713–719 [DOI] [PubMed] [Google Scholar]
- Hsing Y-I, Chern C-G, Fan M-J, Lu P-C, Chen K-T, Lo S-F, Sun P-K, Ho S-L, Lee K-W, Wang Y-C, et al. (2007) A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol Biol 63: 351–364 [DOI] [PubMed] [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T (2010) A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet 42: 635–638 [DOI] [PubMed] [Google Scholar]
- Jiang D, Gu X, He Y (2009) Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21: 1733–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Kong NC, Gu X, Li Z, He Y (2011) Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet 7: e1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SL, Lee S, Kim HJ, Nam HG, An G (2007) OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol 145: 1484–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Wei G, Shi J, Jin J, Shen T, Ni T, Shen WH, Yu Y, Dong A (2015) SET DOMAIN GROUP 708, a histone H3 lysine 36‐specific methyltransferase, controls flowering time in rice (Oryza sativa). New Phytologist 210: 577–588 [DOI] [PubMed] [Google Scholar]
- Liu K, Yu Y, Dong A, Shen WH (2017) SET DOMAIN GROUP701 encodes a H3K4-methytransferase and regulates multiple key processes of rice plant development. New Phytol 215: 609–623 [DOI] [PubMed] [Google Scholar]
- Lu C, Tian Y, Wang S, Su Y, Mao T, Huang T, Chen Q, Xu Z, Ding Y (2017) Phosphorylation of SPT5 by CDKD; 2 is required for VIP5 recruitment and normal flowering in Arabidopsis thaliana. Plant Cell 29: 277–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang ZX, Minobe Y, Yano M (2011) Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J 66: 603–612 [DOI] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Wang Z-X, Minobe Y, Izawa T, Yano M (2008) Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol 148: 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC (2007) Plant SET domain-containing proteins: structure, function and regulation. Biochim Biophys Acta 1769: 316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Dharmarajan V, Cosgrove MS (2008a) Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J Biol Chem 283: 32158–32161 [DOI] [PubMed] [Google Scholar]
- Patel A, Vought VE, Dharmarajan V, Cosgrove MS (2008b) A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J Biol Chem 283: 32162–32175 [DOI] [PubMed] [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inzé D, Avramova Z, Dean C, Grossniklaus U (2008) ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20: 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu CH, Lee S, Cho LH, Kim SL, Lee YS, Choi SC, Jeong HJ, Yi J, Park SJ, Han CD, et al. (2009) OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ 32: 1412–1427 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Shilatifard A. (2012) The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J-J, Kingston RE (2008) WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem 283: 35258–35264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Wang S, Zhang F, Zheng H, Liu Y, Huang T, Ding Y (2017) Phosphorylation of histone H2A at serine 95: a plant-specific mark involved in flowering time regulation and H2A. Z deposition. Plant Cell 29: 2197–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui P, Shi J, Gao X, Shen W-H, Dong A (2013) H3K36 methylation is involved in promoting rice flowering. Mol Plant 6: 975–977 [DOI] [PubMed] [Google Scholar]
- Sun C, Fang J, Zhao T, Xu B, Zhang F, Liu L, Tang J, Zhang G, Deng X, Chen F, et al. (2012) The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice. Plant Cell 24: 3235–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi YH, Westfield GH, Oleskie AN, Trievel RC, Shilatifard A, Skiniotis G (2011) Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc Natl Acad Sci USA 108: 20526–20531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel RC, Shilatifard A (2009) WDR5, a complexed protein. Nat Struct Mol Biol 16: 678–680 [DOI] [PubMed] [Google Scholar]
- Tsuji H, Taoka K, Shimamoto K (2011) Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol 14: 45–52 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
- Xu R-F, Li H, Qin R-Y, Li J, Qiu C-H, Yang Y-C, Ma H, Li L, Wei P-C, Yang J-B (2015) Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep 5: 11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767 [DOI] [PubMed] [Google Scholar]
- Zheng H, Zhang F, Wang S, Su Y, Jiang P, Cheng R, Ji X, Hou S, Ding Y (2017) MLK1 and MLK2 coordinate RGA and CCA1 activity to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell 30: 67–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W, Zhong X, You J, Xiong L (2013) Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol Biol 81: 175–188 [DOI] [PubMed] [Google Scholar]