The production of the CP47 subunit of Photosystem II requires the Pam68 protein that binds to ribosomes in the vicinity of the SecY translocon and most likely promotes chlorophyll loading into the CP47 polypeptide chain.
Abstract
Photosystem II (PSII) is a large enzyme complex embedded in the thylakoid membrane of oxygenic phototrophs. The biogenesis of PSII requires the assembly of more than 30 subunits, with the assistance of a number of auxiliary proteins. In plants and cyanobacteria, the photosynthesis-affected mutant 68 (Pam68) is important for PSII assembly. However, its mechanisms of action remain unknown. Using a Synechocystis PCC 6803 strain expressing Flag-tagged Pam68, we purified a large protein complex containing ribosomes, SecY translocase, and the chlorophyll-binding PSII inner antenna CP47. Using 2D gel electrophoresis, we identified a pigmented Pam68-CP47 subcomplex and found Pam68 bound to ribosomes. Our results show that Pam68 binds to ribosomes even in the absence of CP47 translation. Furthermore, Pam68 associates with CP47 at an early phase of its biogenesis and promotes the synthesis of this chlorophyll-binding polypeptide until the attachment of the small PSII subunit PsbH. Deletion of both Pam68 and PsbH nearly abolishes the synthesis of CP47, which can be restored by enhancing chlorophyll biosynthesis. These results strongly suggest that ribosome-bound Pam68 stabilizes membrane segments of CP47 and facilitates the insertion of chlorophyll molecules into the translated CP47 polypeptide chain.
Photosystem II (PSII) is a large protein‐cofactor complex embedded in the thylakoid membranes of oxygenic phototrophs. The key large structural components of PSII are the chlorophyll (Chl)-binding proteins D1, D2, CP43, and CP47, subjoined with other small and extrinsic subunits (Umena et al., 2011). According to this model, PSII is assembled in a stepwise manner from four preassembled smaller subcomplexes called modules (Komenda et al., 2012). Each module consists of one large Chl-binding subunit (D1, D2, CP43, or CP47) and several low molecular mass membrane polypeptides. PSII assembly is initiated through the association of D1 and D2 modules to form an assembly intermediate, termed the Reaction Center II (RCII) complex. The CP47 assembly module (CP47m) is then attached to RCII (Boehm et al., 2011), which results in a CP43-less core complex called “RC47” (Boehm et al., 2012). The active, oxygen-evolving PSII is completed by the addition of the CP43 module (Boehm et al., 2011) and attachment of the lumenal extrinsic proteins (Nixon et al., 2010). Biogenesis of PSII is a highly complex process requiring many auxiliary proteins that are not present in the fully assembled complex. A number of these assembly factors have been described (Komenda et al., 2012; Heinz et al., 2016). However, their precise functions remain mostly unknown, and only a few of them have been connected with a specific assembly step (Knoppová et al., 2014; Bečková et al., 2017).
The fully assembled PSII contains 35 Chl molecules, most of them bound to the inner PSII antennas CP47 (16) and CP43 (14). According to this model, Chl molecules are integrated directly into synthesized CP47 and CP43, and the insertion of Chl appears to be a prerequisite for the correct folding and stability of these polypeptides (for review, see Sobotka, 2014). However, little is known about how Chl proteins are produced. PSII Chl-binding subunits are integral membrane proteins most likely cotranslationally inserted into the thylakoid membrane with the assistance of the protein translocation apparatus. This process usually includes the SecYEG translocon, which forms a protein-conducting channel, and an associated insertase/foldase YidC (Sachelaru et al., 2013). Chl synthase is the last enzyme of Chl biosynthesis, and it was recently shown to physically interact with YidC insertase (Chidgey et al., 2014). This interaction suggests that Chl molecules are passed directly from Chl synthase to the nascent apoprotein chain in the vicinity of the translocon.
The small PSII subunit PsbH and two assembly factors, hypothetical chloroplast open reading frame 48 (Ycf48) and photosynthesis-affected mutant 68 (Pam68), were found to be important for the accumulation of CP47m (Komenda, 2005; Rengstl et al., 2013). Here, we identified the cyanobacterial Pam68 protein as a ribosomal factor that is in contact with the nascent CP47 in the vicinity of the SecY translocase. Our data suggest that Pam68 stabilizes membrane segments of CP47 during Chl insertion.
RESULTS
Pam68 Associates with the CP47 Protein at an Early Stage of PSII Biogenesis
To identify proteins interacting with Pam68, we constructed a Synechocystis sp. PCC 6803 strain (hereafter Synechocystis) expressing a Flag-tagged Pam68 derivative (Pam68.f protein). This protein was purified from solubilized membranes using an anti-Flag gel, and the obtained elution was analyzed by SDS-PAGE. The identities of prominent protein bands were determined by mass spectrometry (MS; Supplemental Fig. S1A). We identified CP47 and ribosomal subunits, which were missing in the control pull-down, as putative interactors (Supplemental Fig. S1A). Consistent with our previous reports, Photosystem I (PSI) subunits were the only substantial contaminants (Knoppová et al., 2014; Bečková et al., 2017). Furthermore, our control purification of the Flag-tagged ferrochelatase enzyme (FeCh) showed that the 3×Flag-tag does not bind ribosome subunits nonspecifically (Supplemental Fig. S1B).
Because membrane-bound ribosomes were present in the Pam68.f elution, we checked for the presence of SecY translocase and YidC insertase. Indeed, both these proteins coeluted with Pam68.f (Supplemental Fig. S1C). Additionally, our data support the interaction of the lumenal Ycf48 protein with Pam68, as previously suggested (Rengstl et al., 2013). Moreover, CP47 was the only PSII subunit detected in the Pam68.f elution. Remarkably, the PsbH subunit was hardly detectable even by specific antibodies, despite a high level of CP47 protein in the elution. PsbH is a component of CP47m (Boehm et al., 2011); hence, the absence of PsbH in the Pam68.f pull-down indicates that the association of CP47 with Pam68 is an early event that occurs before the attachment of PsbH to CP47.
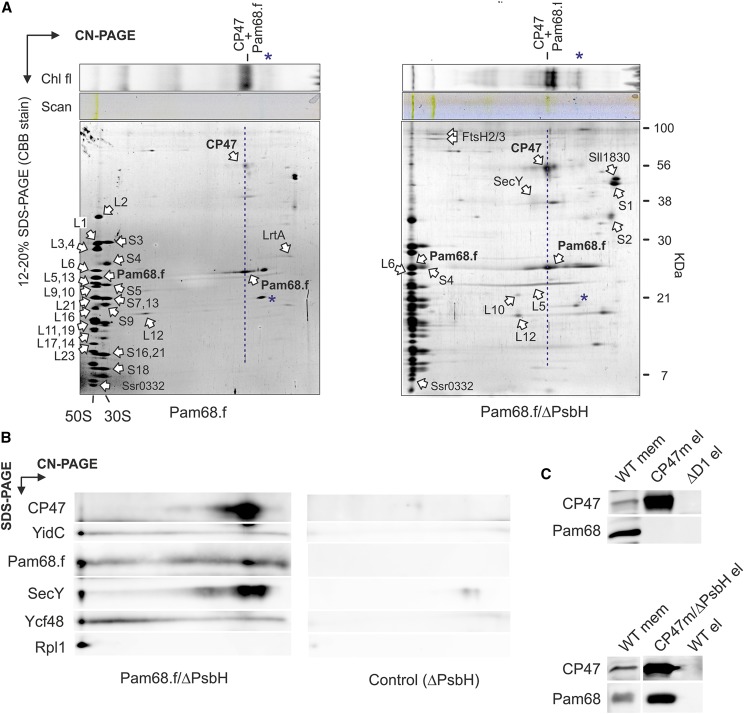
To elucidate whether Pam68.f physically interacts with unassembled CP47 in the absence of PsbH, we purified Pam68.f from the PsbH-less strain, and both elutions (Pam68.f and Pam68.f/ΔPsbH) were analyzed by 2D Clear-Native/SDS-PAGE (CN/SDS-PAGE). On the stained gels, we identified large (50S) and small (30S) ribosome subunits and two fractions of Pam68.f comigrating with 50S and with CP47, respectively. The Pam68.f-CP47 complex exhibited Chl fluorescence, and its green pigmentation was visible on the CN gel (Fig. 1A).
Figure 1.
Identification of Pam68 as a component of the CP47 assembly module. A, The Pam68.f pull-down (left-hand gel) and the Pam68.f/ΔPsbH pull-down (right-hand gel) were separated by CN-PAGE, whereas SDS-electrophoresis was used for the second dimension. Individual protein spots were cut and identified by MS (Supplemental Dataset). The dashed blue line highlights the comigration of Pam68.f with CP47; note that CP47 exhibits Chl fluorescence demonstrating the presence of Chl molecules. Asterisks mark phycobiliproteins contaminating the elutions. B, The Pam68.f/ΔPsbH pull-down prepared from a different cell culture was separated together with the control ΔPsbH elution by 2D electrophoresis and blotted. The indicated proteins were sequentially detected by specific antibodies and the separate segments of the 2D blot with the individual antibody signals are shown. C, The CP47 assembly modules containing PsbH (CP47m) and lacking PsbH (CP47m/ΔPsbH) were purified on a nickel column. The eluted proteins were separated by SDS-PAGE together with the control pull-downs of wild type and ΔD1. The separated proteins were blotted, and the blot was sequentially probed with the indicated antibodies; the separate segments of the blot with individual antibody signals are shown. Chl fl, Chl fluorescence.
In addition to the ribosome subunits, FtsH proteases, and a smeary band of SecY, the Pam68.f elutions also contained two unknown proteins (Sll1830 and Ssr0332). Whereas Sll1830 migrated as a free protein, the small Ssr0332 protein comigrated with the 50S ribosomal subunit. Another identified protein was light-repressed protein A (LrtA, Sll0947), which showed sequence similarity to the bacterial pY factor associated with stalled ribosomes (Galmozzi et al., 2016). A similar pattern of ribosomal proteins, but with higher levels of LrtA, was also obtained in the Pam68.f pull-down isolated from the ΔpsbB (ΔCP47) mutant background (Supplemental Fig. S2). This result implies that Pam68 remains associated with a pool of membrane-bound ribosomes even when no CP47 translation occurs in the cell. Notably, the electrophoretic mobility of Pam68.f proteins purified from the ΔpsbH and wild-type backgrounds were slightly different, indicating a posttranslational modification of Pam68.f upon the psbH deletion (Fig. 1A). This shift allowed us to distinguish that the spot of Pam68.f comigrating with 50S in the Pam68.f/ΔPsbH pulldown (just above the Rpl6 protein) consists of only Pam68.f, with no other (ribosomal) proteins. There is no spot in this position in the Pam68.f elution (Fig. 1A).
To better visualize the pattern of proteins on the 2D gel, the separation of Pam68.f/ΔPsbH and the control ΔPsbH pull-downs on 2D CN/SDS-PAGE was followed by immunoblotting. The immunodetection determined a fraction of YidC, Ycf48, and SecY comigrating with 50S, as expected for the isolated ribosome-translocon apparatus (Fig. 1B). However, the barely visible (SecY) or invisible (YidC, Ycf48) staining of these proteins on the gel indicates that they are substantially less abundant than Pam68. Hence, it is unlikely that they connect Pam68.f with ribosomes. CP47 was found in a spot that had the same mobility as the dissociated Pam68.f, suggesting a mutual complex.
We used an independent approach to verify the interaction between the unassembled CP47 and Pam68 proteins. We isolated CP47m and a nascent CP47m lacking PsbH (CP47m/ΔPsbH) via His-tagged CP47 from Synechocystis strains accumulating these complexes due to the absence of the D1 or D1/PsbH PSII subunits, respectively (Boehm et al., 2011; D’Haene et al., 2015). The Pam68 protein was copurified with CP47m/ΔPsbH but was not detected in the CP47m elution (Fig. 1C). Therefore, either the binding of PsbH to the CP47-Pam68 complex is considerably weaker than to CP47, or Pam68 and PsbH share a similar binding side.
N-Terminal Segment of Pam68 Is Required for the Interaction with Ribosomes
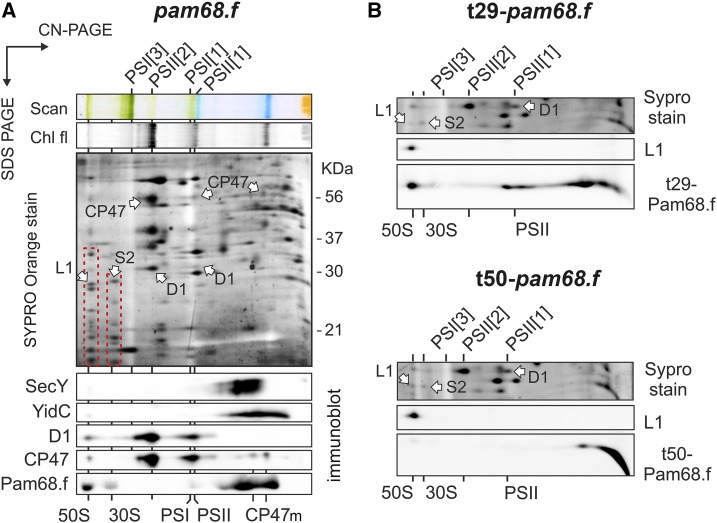
To verify that the interaction of Pam68 with ribosomes is not an artifact of the pull-down assay, solubilized membrane complexes from the pam68.f strain were separated by 2D CN/SDS-PAGE, stained by SYPRO Orange, and blotted onto a polyvinylidene fluoride (PVDF) membrane. Pam68.f comigrated with the 50S and, unexpectedly, also with the 30S subunit (Fig. 2A). On the other hand, Pam68.f spots in the region of smaller complexes did not align with either of the CP47m forms (both are known to contain PsbH; Komenda, 2005). These results suggest that complexes between Pam68.f and the CP47m forms are not detectable in the 2D gel of the pam68.f membranes (Fig. 2A). This observation is consistent with the proposed transient interaction between Pam68 and the newly synthesized CP47; the transient complex pool is apparently below the detection limit of the immunoblot analysis.
Figure 2.
2D CN/SDS-PAGE and immunodetection of membrane protein complexes from strains expressing full-length or truncated Pam68.f. A, Solubilized membrane proteins from the pam68.f strain were separated by 2D CN/SDS-PAGE. The 2D gel was stained with SYPRO Orange, blotted, and the 2D blot was sequentially probed by the indicated antibodies. Separate segments of the 2D blot with individual antibody signals are shown. The large and small ribosomal subunits are highlighted on the stained gel by red dashed boxes; protein spots belonging to Rpl1 and Rps2 were identified previously (Chidgey et al., 2014). Chl fluorescence was detected after excitation by blue light. CP47m marks two forms of the CP47 assembly module detected in the Synechocystis membrane fraction (Komenda, 2005). B, The same analysis was performed on membranes isolated from strains expressing the truncated variants t29-Pam68.f (top panel) and t50-Pam68 (bottom panel). Only a region of the SYPRO Orange stained gel around the Rpl1 protein (SYPRO stain) and separate segments of the 2D blot with signals of anti-Rpl1 and anti-Flag antibodies are shown. Complexes are designated as in (B). Chl fl, Chl fluorescence; L1, Rpl1; PSI[3], trimer of PSI; PSII[1], monomer of PSII; PSII[2], dimer of PSII; S2, Rps2.
According to this model, ribosomes can be docked to bacterial membranes via interaction with the large subunit and the SecYEG translocon, or alternatively, with YidC insertase (Prinz et al., 2000; Seitl et al., 2014). However, the interaction between the membrane-bound ribosomes and SecY or YidC in isolated thylakoids was not preserved in our 2D gel system (Fig. 2A). Therefore, it is unlikely that SecY/YidC facilitates the observed association of Pam68.f with ribosomes.
It is likely that Pam68 interacts directly with ribosomal proteins from both the 50S and 30S subunits. The 30S subunit of the membrane-docked ribosome is close to the membrane surface (approximately 10 nm; Frauenfeld et al., 2011). Theoretically, the strongly positively charged N terminus of Pam68.f is long enough (65 amino acids, approximately 20 nm; Supplemental Fig. S3) to reach the 30S subunit. To test this possibility, we constructed strains expressing variants of Pam68.f truncated either up to the V29 (t29-pam68.f strain) or the S50 amino acid residues (t50-pam68.f). The t29-Pam68.f protein still comigrated with ribosomes on the 2D gel (Fig. 2B), but the more truncated t50-Pam68.f protein was not detectable in any larger complexes, which supports the role of the Pam68 N-terminal segment in the interaction with ribosomes.
A close relationship between the cyanobacterial Pam68 and ribosomes can also be inferred from the existence of an operon of the pam68 and the rps15 genes, which is highly conserved among the cyanobacterial genomes. According to the STRING database (http://string-db.org/), there are only a few examples of sequenced cyanobacterial genomes (e.g. Gloeobacter violaceus) where these two genes are not organized in tandem. In the Synechocystis genome, the pam68 gene is transcribed from the rps15 promoter as a single mRNA with rps15 (Mitschke et al., 2011). Interestingly, the rps15-pam68 mRNA belongs to a small group of ribosomal transcripts that are significantly up-regulated under stress conditions with the strongest expression under low temperature (Kopf et al., 2014; Supplemental Fig. S4). Indeed, we found the Pam68 protein level to be high during high light or chilling stress (Supplemental Fig. S5).
Enhanced Chl Biosynthesis Rescues the Abolished CP47 Synthesis in the ΔpsbH/Δpam68 Strain
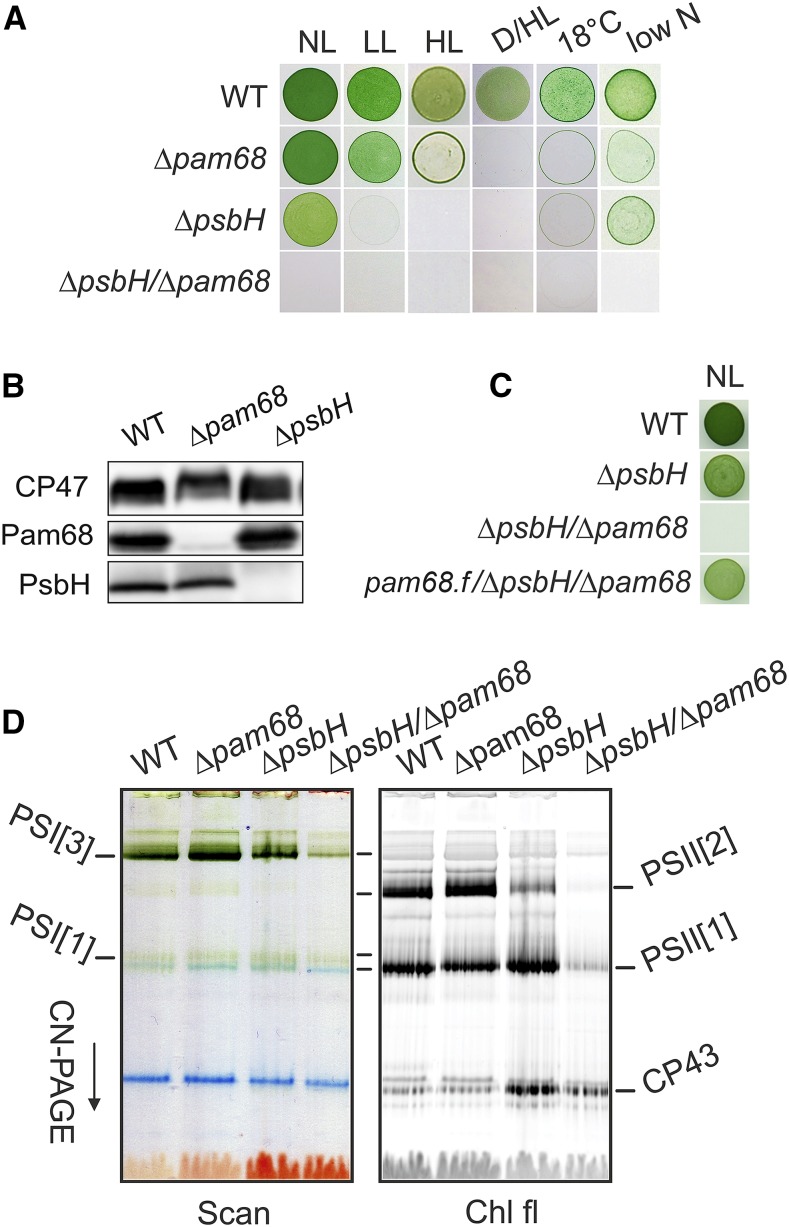
The results described above imply that Pam68 functions during the synthesis and/or folding of CP47 before it associates with PsbH, which also facilitates CP47 synthesis (Komenda, 2005). To test whether PsbH can compensate for the absence of Pam68, we characterized the Synechocystis Δpam68 and ΔpsbH mutants and the ΔpsbH/Δpam68 double mutant. Under moderate light intensities (40 µmol photons m−2 s−1), Δpam68 grew similarly as the wild-type strain and had a similar Chl content (Supplemental Fig. S6, A and B). The ΔpsbH mutation affected both the growth rate and Chl content; nevertheless, this mutant grew fairly well photoautotrophically (Supplemental Fig. S6, A and B). However, even the single Δpam68 mutant stopped proliferating on plates under more severe conditions, such as dark-/high-light fluctuation or low temperature (Fig. 3A). Moreover, the level of PsbH was merely affected in the Δpam68 strain and, vice versa, the level of Pam68 in the ΔpsbH strain remained comparable to wild type (Fig. 3B).
Figure 3.
Characterization of the Synechocystis strains lacking Pam68, PsbH, or both of these proteins. A, Autotrophic growth of the wild-type and mutant strains on agar plates under various conditions. Growth for 5 d under normal light (40 μmol photons m−2 s−1), low light (10 μmol photons m−2 s−1), high light (400 μmol photons m−2 s−1), fluctuating dark/high light conditions (5 min dark, 5 min 400 μmol photons m−2 s−1), 18°C at 40 μmol photons m−2 s−1, and low nitrogen (0.1 mm NaNO3). B, Levels of PsbH and Pam68 in the Δpam68 and ΔpsbH strains under normal light conditions. A comparable amount of Chl was loaded for each strain. C, Autotrophic growth of the pam68.f/Δpam68/ΔpsbH strain expressing the Pam68.f protein under the regulation of the psbAII promoter. D, Membranes, isolated from the wild-type and mutant strains grown as described in (A), were solubilized and separated by CN-PAGE. D/HL, dark/high light; HL, high light; LL, low light; NL, normal light; PSI[3], trimer of PSI; PSII[1], monomer of PSII; PSII[2], dimer of PSII.
Unlike the strains containing single mutations, the double mutant showed extremely slow autotrophic growth (double time approximately 20 d), accumulated only traces of Chl and died immediately after exposure to mild stress conditions (Fig. 3A; Supplemental Fig. S6). However, photoautotrophy of the ΔpsbH/Δpam68 strain can be restored by the expression of Pam68.f (Fig. 3C), which provides evidence that the poor phenotype of the double mutant is not caused by a position effect, e.g. lower levels of Rps15. To obtain enough cells of the poor-growing ΔpsbH/Δpam68 mutant, we first grew all strains with Glc supplementation. Then, we characterized the phenotype 2 d after removing Glc from the media. As revealed by the CN-PAGE separation of membrane complexes (Fig. 3D), the levels of PSI and PSII were virtually unchanged in the Δpam68 strain, but the ΔpsbH strain contained much less dimeric PSII. In the double mutant, very little PSI and only traces of the PSII complexes were detectable. Thus, both PsbH and Pam68 play distinct roles in the accumulation of PSII; the parallel elimination of both of these proteins is nearly fatal for cell viability.
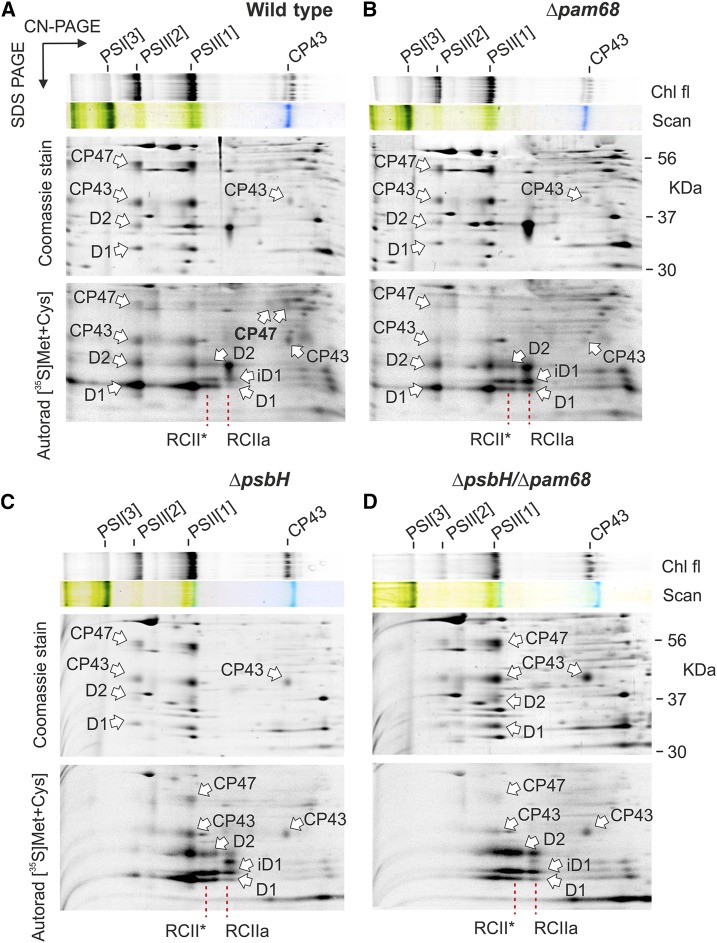
For a closer look at the role of PsbH and Pam68 in the synthesis of PSII wild type, Δpam68, ΔpsbH, and ΔpsbH/Δpam68 cells were pulse-labeled and the isolated membrane complexes analyzed by 2D CN/SDS-PAGE (Fig. 4). Consistent with the previously published analysis of Δpam68 (Rengstl et al., 2013), this strain showed less labeled CP47 and CP43 in total, and lacked the labeled unassembled CP47. In addition, we observed severe accumulation of RCIIa and RCII* assembly intermediates, which is a typical feature of cells deficient in the formation of CP47m (Knoppová et al., 2014). The obtained pattern for ΔpsbH differed from the Δpam68 strain by having only weakly labeled dimeric PSII and also less synthesized D1. A detectable pool of unassembled CP47 was also absent and both RCII complexes accumulated, which implies that the rate of CP47m formation limits the process of PSII assembly. In the ΔpsbH/Δpam68 strain, the capacity to synthesize PSII was extremely weak, almost certainly caused by the lack of CP47m because the intensively labeled RCII complexes resembled the canonical pattern of the ΔCP47 strain (Fig. 4; Komenda et al., 2004).
Figure 4.
Synthesis of PSII subunits in the Δpam68, ΔpsbH, and ΔpsbH/Δpam68 mutant strains. Wild-type (A) and the mutant Δpam68 (B), ΔpsbH (C), and ΔpsbH/Δpam68 (D) cells grown for 2 d without Glc were radiolabeled with a mixture of [35S]Met/Cys using a 30-min pulse. Isolated membrane proteins were separated by CN-PAGE on a 4% to 14% linear gradient gel, whereas 12% to 20% SDS-electrophoresis was used for the second dimension. The same amounts of Chl were loaded for each strain. Note that the ΔpsbH/Δpam68 strain contains three-times less Chl per cell than ΔpsbH, meaning that the membrane proteins from the double mutant were overloaded on a per-cell basis to obtain a detectable radioactivity signal of the assembled PSII. The 2D gels were stained with Coomassie Blue, and the labeled proteins were detected by a phosphoimager (Autorad). Chl fluorescence emitted by Chl was detected by LAS 4000 (Fuji) after excitation by blue light. Chl fl, Chl fluorescence; iD1, incompletely processed form of the D1 precursor; PSI[3], trimer of PSI; PSII[1], monomer of PSII; PSII[2], dimer of PSII; RCII*, assembly intermediate (reaction center complex) lacking CP47m (Knoppová et al., 2014); RCIIa, PSII assembly intermediate (reaction center complex) lacking CP43m (Knoppová et al., 2014).
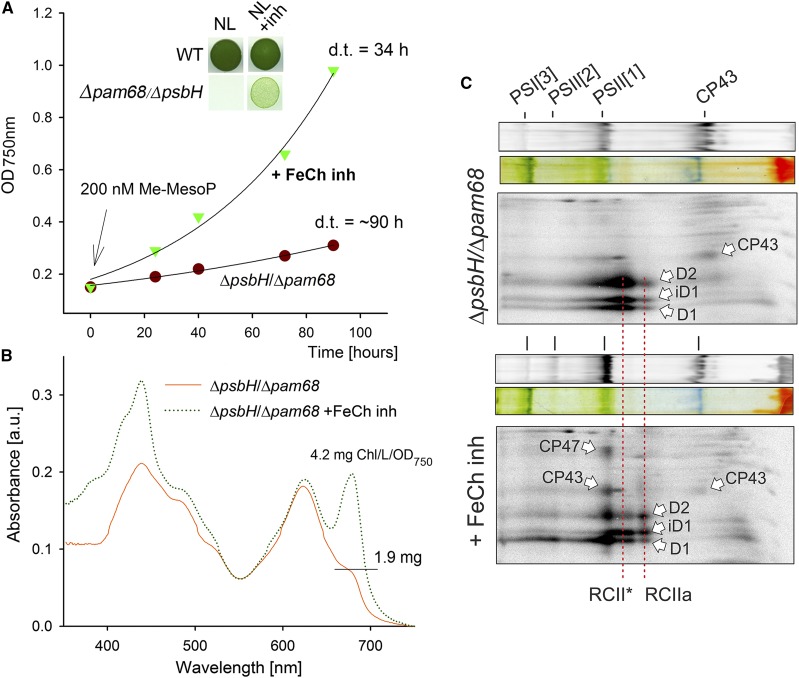
Based on the available PSII structure (Umena et al., 2011), the N-terminal segment of PsbH creates a network of hydrogen bonds with the stromal loops connecting the first four helixes of CP47 (Supplemental Fig. S7). Therefore, the PsbH protein could fix the nascent CP47 in a position that facilitates prompt insertion of Chl molecules. Moreover, the C-terminal region of Pam68 may play a similar role. To test the importance of both proteins for Chl insertion into CP47, we removed Glc from the ΔpsbH/Δpam68 liquid culture, while supplementing it with 200 nm N-methyl mesoporphyrin IX. This compound is a specific inhibitor of the FeCh enzyme and a partial inhibition of FeCh strongly enhances Chl biosynthesis (Sobotka et al., 2005). Remarkably, the ΔpsbH/Δpam68 cells treated with the FeCh inhibitor started to grow much faster than the control cells without the inhibitor (Fig. 5A). The control culture had very low Chl content on a per-cell basis, whereas the treated cells progressively built up new Chl-complexes, and in 4 d reached approximately 85% of the Chl level when compared to wild type (Fig. 5B).
Figure 5.
Abolished synthesis of CP47 in the ΔpsbH/Δpam68 mutant is rescued by enhanced Chl biosynthesis. A, The ΔpsbH/Δpam68 cells grown mixotrophically were harvested and resuspended in a growth medium without Glc. The obtained culture was divided into two flasks, with one of them supplemented with N-methyl-mesoporphyrin IX (Me-MesoP, FeCh inhibitor). The photoautotrophic growth was then monitored. The inset shows the same growth experiment but with 200 nm Me-MesoP added into the plate. B, Absorbance spectra of mutant cells growing for 4 d in the presence or absence of FeCh inhibitor. Spectra were normalized to light scattering at 750 nm. Also shown is the Chl content determined spectroscopically in methanol extract and normalized per OD750 nm. C, ΔpsbH/Δpam68 cells grown for 2 d photoautotrophically in the presence of 200 nm FeCh inhibitor were radiolabeled with a mixture of [35S]Met/Cys; incorporation of radioactivity into core PSII subunits was detected after 2D CN/SDS-PAGE. The same amounts of Chl were loaded of each sample. a.u., absorbance units; d.t., doubling time; iD1, incompletely processed form of the D1 precursor; PSI[3], trimer of PSI; PSII[1], monomer of PSII; PSII[2], dimer of PSII; RCII*, assembly intermediate (reaction center complex) lacking CP47m (Knoppová et al., 2014); RCIIa, PSII assembly intermediate (reaction center complex) lacking CP43m (Knoppová et al., 2014).
Precursors of the Chl biosynthetic pathway differed dramatically between treated and untreated cells. Whereas monovinyl-chlorophyllide was the only detectable Chl precursor in the untreated cells, the inhibitor-treated cells contained a spectrum of Chl precursors typical for wild type (Pilný et al., 2015; Supplemental Fig. S8). Because the earlier precursors upstream of chlorophyllide were below the detection level in the untreated double mutant, this chlorophyllide pool originated almost certainly from Chl recycling and not from de novo synthesis (Vavilin et al., 2005; Kopečná et al., 2015). We repeated the protein radiolabeling experiment described above using ΔpsbH/Δpam68 cells treated with the FeCh inhibitor. The assembly of PSII was restored (Fig. 5C), suggesting that boosting of the ceased Chl biosynthesis restores the formation of CP47m in the mutant lacking both Pam68 and PsbH.
DISCUSSION
The Pam68 protein was first described in the Arabidopsis (Arabidopsis thaliana) pam68-null mutant, which accumulated only approximately 10% of PSII (Armbruster et al., 2010). The function of Pam68 was originally linked to the synthesis or maturation of the D1 subunit of PSII (Armbruster et al., 2010); however, a strong relationship between Pam68 and CP47 was also suggested, based on the low level of Pam68 detected in the Synechocystis CP47-less strain (Rengstl et al., 2011). Our results agree with a recent study, which demonstrated that the lack of Pam68 in Synechocystis limits the synthesis of CP47 and CP43 (Rengstl et al., 2013). Given that the mechanism of PSII biogenesis is highly conserved, it is likely that the eukaryotic Pam68 is involved in the synthesis of CP47. Indeed, using a standard methodology (35S radiolabeling combined with 2D gel-electrophoresis), the synthesis of CP47 in the Arabidopsis pam68-null mutant was hardly detectable (Armbruster et al., 2010).
In contrast to Arabidopsis, the inactivation of pam68 in Synechocystis had no obvious effect on the PSII level under standard growth conditions, although the synthesis of CP47 and CP43 was visibly affected in both organisms (Rengstl et al., 2013; Fig. 4). However, after 3 h of high light treatment (2000 μmol photons m−2 s−1), the levels of functional PSII in the Δpam68 strain decreased by approximately 50% (Rengstl et al., 2013). In addition, we demonstrated the importance of Pam68 under fluctuating light conditions, low temperature, and nitrogen limitation (Fig. 3). These observations imply that Pam68 is essential once the synthesis of CP47 becomes difficult and limits PSII biogenesis.
The PsbH protein is required for sufficient CP47 synthesis in plants as well as in cyanobacteria (Komenda, 2005; Levey et al., 2014); the Synechocystis ΔpsbH mutant shows a noticeable growth defect even under nonstress conditions (Supplemental Fig. S6, A and B). However, the phenotype of this strain is probably quite complex, because PsbH also stabilizes electron transfer processes between QA and QB in the PSII complex (Komenda et al., 2002). It is further essential for the association of photoprotective high-light-inducible proteins to CP47 (Promnares et al., 2006; Fig. 6), and creates an environment for binding of a red Chl molecule in CP47, which is also supposed to have a protective function (D’Haene et al., 2015). However, we expect that the impaired CP47 synthesis/stability is the major reason for the slow growth of the ΔpsbH mutant (Supplemental Fig. S6A). This conclusion is supported by the fact that the growth rate of this strain can also be improved by the inhibition of FeCh (Supplemental Fig. S6C), and is consistent with the very poor phenotype of the ΔpsbH/Δpam68 double mutant. Therefore, PsbH appears to be more crucial for the biogenesis than for the functioning of fully assembled PSII complexes. Similarly, the PsbI subunit was found to be more important for attachment of CP43m to RCII, rather than for PSII activity (Dobáková et al., 2007). Other small PSII subunits may also play roles in assembly.
Figure 6.
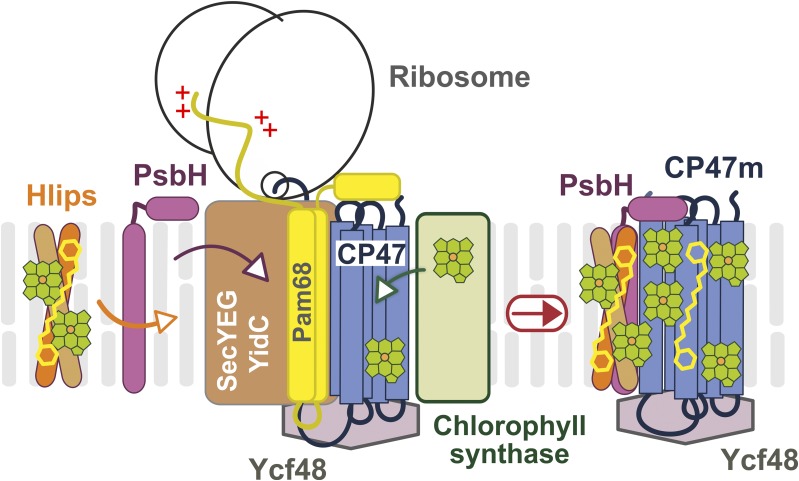
A working model of CP47m synthesis with Pam68 as a ribosome-interacting factor. The CP47 protein is translated by membrane-bound ribosomes and inserted into the membrane by the SecYEG translocon together with YidC insertase. Chl is loaded into the nascent polypeptide cotranslationally from Chl-synthase when the transmembrane segments are released from the translocase channel to YidC (Chidgey et al., 2014). The N-terminal region of the Pam68 protein is associated with the translating ribosome, whereas the C terminus segment interacts with stromal loops of the nascent CP47 chain after it emerges from the translocon. This interaction fixes the CP47 transmembrane segments in a position that facilitates the insertion of Chl molecules. A similar role can be played by the Ycf48 protein at the lumenal site of CP47 (Crawford et al., 2016). Subsequently, PsbH replaces Pam68 and recruits the photoprotective high-light-inducible proteins that associate with CP47 in the vicinity of PsbH (Promnares et al., 2006). Hlips, high-light-inducible proteins.
We present a working model of CP47m synthesis (Fig. 6). Pam68 is firmly bound to the translating ribosome via the N-terminal segment, whereas its C-terminal end interacts with the stromal loops of the nascent CP47 chain emerging from the translocon. We speculate that the coordination of Pam68 (stromal side), YidC (lateral site; Hennon et al., 2015), and Ycf48 (lumenal site; Crawford et al., 2016) fixes the CP47 helix pairs in a position that is amenable to Chl binding. The Pam68 C terminus contains highly conserved charged residues (Supplemental Fig. S3) that can form a network of hydrogen bounds resembling the interaction of the N terminus of PsbH with CP47 (see Supplemental Fig. S7). The synthesis of CP47 is impaired in the Δpam68 strain even under nonstressful conditions (Fig. 4), suggesting that Pam68 permanently assists during CP47 synthesis. Because Pam68 is particularly critical for the mutant lacking PsbH, it is probable that both proteins can work similarly as chaperones facilitating the folding of CP47 and/or the loading of Chl into the newly synthesized apo-polypeptide chain.
Based on previous data and the results of our radiolabeling experiment (Rengstl et al., 2013; Fig. 4), Pam68 appears to also facilitate the synthesis of the CP43 protein and PSI. However, the interaction of Pam68 with these proteins is either too weak to detect, or the lower levels of CP43 and PSI in the absence of Pam68 is a secondary phenotype caused by the feeble CP47 synthesis. The second possibility is more probable, as the mutant lacking CP47 has been shown to contain considerably lower cellular level of Chl in comparison with wild type, implying that the level of CP43 and PSI is lower in the absence of CP47 (Bečková et al., 2017). Although this approach is frequently used, we are aware that arresting particular PSII assembly steps to accumulate specific assembly intermediates may affect other cellular processes, including the synthesis of Chl-binding proteins. The Flag-tag technology used here has the advantage of allowing the purification of PSII assembly intermediates that only exist temporarily in the cell (such as the Pam68.f-CP47m complex) directly from the wild-type background.
The synthesis of CP47 is very sensitive to Chl availability (Hollingshead et al., 2016), which may explain why the lack of Pam68 is not tolerated under stress conditions. The Chl pathway can be temporarily switched-off after a shift to stressful conditions (Kopecná et al., 2012) and when the de novo Chl amount decreases, a fine structural stabilization of the nascent CP47 is likely to be particularly important for the smooth loading of Chls. After addition of the FeCh inhibitor, the pool of available Chl increased, and the impaired CP47 synthesis was rescued (Fig. 5). Thus, a high concentration of Chl molecules around the translated CP47 increases the chance that all Chls are inserted in time even if the orientation of CP47 is not perfect. A similar effect of FeCh inhibition was reported earlier in the Synechocystis strain harboring a mutated CP47 protein (Sobotka et al., 2005).
The observed tight binding of Pam68 to the ribosome is intriguing. A high level of LrtA protein, which associates with the 30S ribosomal subunit (Galmozzi et al., 2016), was present in the Pam68.f pull-downs prepared from strains lacking CP47 (Supplemental Fig. S2). As the potential function of LrtA is to stabilize stalled ribosomal 70S particles (Di Pietro et al., 2013), its appearance in the elution indicates that Pam68 interacts with both the SecY-bound idle ribosomes as well as with the actively translating ribosomes. In Synechocystis, Pam68 is not an abundant membrane protein (it is not detectable in the stained SDS PAGE gel with separated cellular membrane proteins), and there is only a limited pool of membrane-bound ribosomes associated with Pam68. It is possible that these ribosomes differ structurally from other ribosomes in the cell. The heterogenic nature of ribosomes is demonstrated by the variable stoichiometry among core ribosomal subunits or between the monosome/polysome arrangement of ribosomes according to environmental conditions (Xue and Barna, 2012; Slavov et al., 2015). Similarly, the plastid-encoded Rps15 is not an essential ribosomal subunit in plants, but under chilling stress, the tobacco (Tobacco nicotiana) Δrps15 knockdown showed a drastic reduction in the number of plastid ribosomes (Fleischmann et al., 2011). In Synechocystis, the rps15-pam68 operon as well as the rps18, rps20, and rps25 genes are up-regulated under cold stress, whereas many ribosomal genes are simultaneously down-regulated (Supplemental Fig. S4). This result supports the existence of a pool of modified, stress-induced type of ribosomes in cyanobacteria. It is possible that Pam68 has a higher affinity for the stress-induced type of ribosomes. Once bound to SecY, the ribosome might serve as an anchor to localize Pam68 in the vicinity of the translocon machinery. Under severe conditions with limited Chl availability and/or lowered membrane fluidity (chilling stress), Pam68 can promptly assist during the synthesis of CP47.
MATERIALS AND METHODS
Synechocystis Strains and Growth Conditions
All the Synechocystis strains used are summarized in Supplemental Table S1. The Δpam68 strain was kindly provided by Jörg Nickelsen (Ludwig-Maximilians University), and is described in Armbruster et al. (2010). The ΔpsbH/Δpam68 double mutant was prepared by transformation of the ΔpsbH strain (D’Haene et al., 2015) by genomic DNA isolated from the Δpam68 strain. The Synechocystis strain expressing the Pam68 protein fused with 3×Flag at the C terminus (the pam68.f strain) was constructed using pPD-CFLAG plasmid as described in Hollingshead et al. (2012). The pam68.f construct was further transformed into the ΔpsbH and ΔpsbB cells (Eaton-Rye and Vermaas, 1991) to express the Pam68.f protein in these genetic backgrounds. Derivatives of Pam68.f truncated at the amino acid 29 (the t29-pam68.f strain) or 50 (the t50-pam68.f strain) were constructed by PCR amplification of the Synechocystis pam68 gene lacking the 3′ part (primers are listed in Supplemental Table S2). The obtained PCR products were cloned into a pPD-CFLAG plasmid and transformed into wild type. To be able to express Pam68.f in the Δpam68/ΔpsbH mutant (already resistant to kanamycin), we replaced the kanamycin-resistance cassette in the pPD-CFLAG plasmid with an erythromycin-resistance cassette, cloned the pam68 gene into this modified construct. and fully segregated the pam68.f/Δpam68/ΔpsbH strain.
Unless stated otherwise, strains were grown photoautotrophically in liquid BG-11 medium on a rotary shaker under moderate (normal) light intensities (40 μE m−2 s−1) at 28°C. For purification of the Pam68.f protein under native conditions, 4 L of pam68.f and pam68.f/ΔpsbH cells were grown in a 10 L flask in BG-11 medium supplemented by 5 mm Glc under normal light conditions and bubbled with air. Strains lacking PSII (ΔpsbB) were supplemented with 5 mm Glc and grown under lower light intensities (10 μE m−2 s−1).
Absorption Spectra and Determination of Chl Content
Absorption spectra of the whole cells were measured at room temperature with a UV-3000 spectrophotometer (Shimadzu). Chl was extracted from cell pellets (2 mL, OD750 = approximately 0.3) with 100% (v/v) methanol, and its concentration was measured spectrophotometrically according to Porra et al. (1989).
Preparation of Cellular Membranes
Cell cultures were harvested at optical densities of 750 nm = approximately 0.5 to 0.7. Cells were pelleted, washed, and resuspended with buffer A (25 mm MES/NaOH, pH 6.5, 10 mm CaCl2, 10 mm MgCl2, 25% [v/v] glycerol) for the preparation of membranes for 2D electrophoresis and purification of Pam68.f. For nickel-affinity chromatography, the membrane fraction was prepared in 25 mm Na-P buffer, pH 7.5, 50 mm NaCl, 10% (v/v) glycerol (buffer B). Cells were broken using glass beads (0.1 mm diameter), and the membrane fraction was separated from soluble proteins by centrifugation at high speed (65,000 × g, 20 min).
Isolation of Protein Complexes by Affinity Chromatography
Cellular membranes containing approximately 1 mg/mL Chl were solubilized for 1 h with 1% (w/v) β-dodecyl-maltoside at 10°C and centrifuged for 20 min at 65,000g to remove cell debris. The Pam68.f complexes were purified using an anti-Flag-M2 agarose column (Sigma-Aldrich). To remove contaminants, the anti-Flag-resin was washed with 20 resin volumes of buffer A containing 0.04% β-dodecyl-maltoside. The Pam68.f complex was eluted with 2.5 resin volumes of buffer A containing 150 μg/mL 3×Flag peptide (Sigma-Aldrich) and 0.04% β-dodecyl-maltoside. For purification of the His-tagged proteins, solubilized membrane complexes were loaded onto a nickel-affinity chromatography column (Protino Ni–NTA-agarose; Macherey-Nagel). Proteins bound to the column were washed with buffer B containing 0.04% β-dodecyl-maltoside and increasing concentrations of imidazole (5, 10, 20, and 30 mM); His-tagged proteins were finally eluted with 150 mm imidazole.
Electrophoresis and Immunoblotting
The protein composition of the purified complexes was analyzed by electrophoresis in a denaturing 12% to 20% linear gradient polyacrylamide gel containing 7 m urea (Dobáková et al., 2009). Proteins were stained either by Coomassie Brilliant Blue or SYPRO Orange stain and subsequently transferred onto a PVDF membrane for immunodetection (see below). For native electrophoresis, solubilized membrane proteins or isolated complexes were separated on 4% to 12% CN-PAGE (Wittig et al., 2007). Individual components of protein complexes were resolved by incubating the gel strip from the first dimension in 2% (w/v) SDS and 1% (w/v) DTT for 30 min at room temperature, and proteins were separated along the second dimension by SDS-PAGE in a denaturing 12% to 20% polyacrylamide gel containing 7 m urea (Dobáková et al., 2009). Proteins were stained by Coomassie Brilliant Blue or by SYPRO Orange; in the latter case, they were subsequently transferred onto a PVDF membrane. Membranes were incubated with specific primary antibodies and then with a secondary antibody conjugated with horseradish peroxidase (Sigma-Aldrich). The following primary antibodies were used in the study: anti-SecY and anti-YidC (Linhartová et al., 2014), anti-CP47, anti-D1 and anti-PsbH (Komenda, 2005), anti-Pam68 (Armbruster et al., 2010), anti-Rpl1 (Agrisera), anti-Flag (Sigma-Aldrich), and anti-Ycf48 (which was raised in rabbit against recombinant Synechocystis Ycf48 and provided by Peter Nixon, Imperial College, London).
Protein Radiolabeling
For protein labeling, the cells were incubated with using a mixture of [35S]Met and [35S]Cys (Translabel; MP Biochemicals) as described in Dobáková et al. (2009). After separation of labeled proteins by CN-PAGE in the first dimension, the polyacrylamide gel was scanned for Chl fluorescence and then treated for second-dimension separation with 18% SDS-PAGE. The 2D gel was exposed to a Phosphorimager plate (GE Healthcare) overnight and stained by Coomassie Brilliant Blue and scanned by Storm (GE Healthcare).
Protein Identification by LC-MS/MS Analysis
Gel slices were placed in 200 μL of 40% acetonitrile, 200 mm ammonium bicarbonate and incubated at 37°C for 30 min, after which the solution was discarded. This procedure was performed twice, and the gel was subsequently dried in a vacuum centrifuge. Ten microliters of 40 mm ammonium bicarbonate in 9% acetonitrile containing 0.4 μg trypsin (proteomics grade; Sigma-Aldrich) were added to the gel slice and left to soak in the solution at 4°C for 45 min. To digest proteins, 20 μL of 9% (v/v) acetonitrile in 40 mm ammonium bicarbonate was added to the gel and incubated at 37°C overnight. Peptides were purified using ZipTip C18 pipette tips (Millipore). MS analysis was performed on a NanoAcquity UPLC (Waters) on-line coupled to the ESI Q-ToF Premier mass spectrometer (Waters). One microliter of the sample was diluted in 3% (v/v) acetonitrile/0.1% (v/v) formic acid, and tryptic peptides were desalted on a Symmetry C18 Trapping column (180 µm i.d., 20 mm length, particle size 5 μm, reverse phase; Waters) with a flow rate of 15 μL/min for 1 min. Trapping was followed by a reverse-phase UHPLC using the BEH300 C18 analytical column (75 µm i.d. 150 mm length, particle size 1.7 µm, reverse phase; Waters). The linear gradient elution ranged from 97% solvent A (0.1% formic acid) to 40% solvent B (0.1% formic acid in acetonitrile) at a flow rate of 0.4 μL/min. Eluted peptides flowed directly into the ESI source. Raw data were acquired in the data-independent MSE identity mode (Waters). Precursor ion spectra were acquired with a collision energy of 5 V and fragment ion spectra with a collision energy of 20 V to 35 V ramp in alternating 1 s scans. Data-dependent analysis mode was used for the second analysis; peptide spectra were acquired with a collision energy of 5 V and peptides with charge states of + 2, +3, and + 4 were selected for MS/MS analysis. Fragment spectra were collected with a collision energy of 20 V to 40 V ramp. In both modes, the acquired spectra were submitted for database search using the PLGS2.3 software (Waters) against Synechocystis protein databases from the Cyanobase Web site (http://genome.microbedb.jp/cyanobase/). Acetyl N-terminal, deamidation N and Q, carbamidomethyl C, and oxidation M were set as variable modifications. Identification of three consecutive y-ions or b-ions was required for a positive peptide match.
Accession Numbers
Pam68, BAA16881.1; CP47, BAA10458.1; PsbH, BAA17629.1, SecY; BAA17331.1, YidC - BAA18244.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. A list of Synechocystis strains used in this study.
Supplemental Table S2. A list of primers used to clone the pam68.f gene and its two truncated variants (t29-pam68.f and t50-pam68.f) into the pPD-CFLAG plasmid (adding of 3×Flag tag at the C terminus).
Supplemental Figure S1. Identification of proteins copurified with Pam68.f.
Supplemental Figure S2. 2D CN/SDS-PAGE of the Pam68.f complex purified from the pam68.f/ΔpsbB strain (A) and the control ΔCP47 (B).
Supplemental Figure S3. Conservation profile and the prediction of secondary structure of the Synechocystis Pam68 protein.
Supplemental Figure S4. Coexpression of the cyanobacterial pam68-rps15 operon with a subset of ribosomal genes.
Supplemental Figure S5. Accumulation of the Pam68 protein under stress conditions.
Supplemental Figure S6. Growth rate and whole cell spectra of the wild-type and mutant strains.
Supplemental Figure S7. Stromal view of the CP47–PsbH complex with indicated hydrogen bonds between the CP47 and the PsbH N terminus.
Supplemental Figure S8. Changes in the levels of Chl precursors in the ΔpsbH/Δpam68 strain after treatment with FeCh inhibitor.
Supplemental Dataset. MS data—numbers of identified trypsin peptides for 2D gel protein spots.
Footnotes
This work was supported by project 17-08755S of the Grant Agency of the Czech Republic and by the Czech Ministry of Education (projects CZ 1.05/2.1.00/19.0392 and LO1416).
Articles can be viewed without a subscription.
References
- Armbruster U, Zühlke J, Rengstl B, Kreller R, Makarenko E, Rühle T, Schünemann D, Jahns P, Weisshaar B, Nickelsen J, Leister D (2010) The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bečková M, Gardian Z, Yu J, Koník P, Nixon PJ, Komenda J (2017) Association of Psb28 and Psb27 proteins with PSII-PSI supercomplexes upon exposure of Synechocystis sp. PCC 6803 to high light. Mol Plant 10: 62–72 [DOI] [PubMed] [Google Scholar]
- Boehm M, Romero E, Reisinger V, Yu J, Komenda J, Eichacker LA, Dekker JP, Nixon PJ (2011) Investigating the early stages of photosystem II assembly in Synechocystis sp. PCC 6803: isolation of CP47 and CP43 complexes. J Biol Chem 286: 14812–14819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Yu J, Reisinger V, Bečková M, Eichacker LA, Schlodder E, Komenda J, Nixon PJ (2012) Subunit composition of CP43-less photosystem II complexes of Synechocystis sp. PCC 6803: implications for the assembly and repair of photosystem II. Philos Trans R Soc Lond B Biol Sci 367: 3444–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgey JW, Linhartová M, Komenda J, Jackson PJ, Dickman MJ, Canniffe DP, Koník P, Pilný J, Hunter CN, Sobotka R (2014) A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26: 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford TS, Eaton-Rye JJ, Summerfield TC (2016) Mutation of Gly195 of the ChlH subunit of Mg-chelatase reduces chlorophyll and further disrupts PS II assembly in a Ycf48-deficient strain of Synechocystis sp. PCC 6803. Front Plant Sci 7: 1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Haene SE, Sobotka R, Bučinská L, Dekker JP, Komenda J (2015) Interaction of the PsbH subunit with a chlorophyll bound to histidine 114 of CP47 is responsible for the red 77K fluorescence of Photosystem II. Biochim Biophys Acta 1847: 1327–1334 [DOI] [PubMed] [Google Scholar]
- Di Pietro F, Brandi A, Dzeladini N, Fabbretti A, Carzaniga T, Piersimoni L, Pon CL, Giuliodori AM (2013) Role of the ribosome-associated protein PY in the cold-shock response of Escherichia coli. MicrobiologyOpen 2: 293–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobáková M, Sobotka R, Tichý M, Komenda J (2009) Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 149: 1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobáková M, Tichý M, Komenda J (2007) Role of the PsbI protein in photosystem II assembly and repair in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 145: 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton-Rye JJ, Vermaas WFJ (1991) Oligonucleotide-directed mutagenesis of psbB, the gene encoding CP47, employing a deletion mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 17: 1165–1177 [DOI] [PubMed] [Google Scholar]
- Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schöttler MA, Bock R (2011) Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23: 3137–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R (2011) Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol 18: 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmozzi CV, Florencio FJ, Muro-Pastor MI (2016) The cyanobacterial ribosomal-associated protein LrtA Is involved in post-stress survival in Synechocystis sp. PCC 6803. PLoS One 11: e0159346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Liauw P, Nickelsen J, Nowaczyk M (2016) Analysis of photosystem II biogenesis in cyanobacteria. Biochim Biophys Acta 1857: 274–287 [DOI] [PubMed] [Google Scholar]
- Hennon SW, Soman R, Zhu L, Dalbey RE (2015) YidC/Alb3/Oxa1 family of insertases. J Biol Chem 290: 14866–14874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead S, Kopečná J, Armstrong DR, Bučinská L, Jackson PJ, Chen GE, Dickman MJ, Williamson MP, Sobotka R, Hunter CN (2016) Synthesis of chlorophyll-binding proteins in a fully segregated Δycf54 strain of the cyanobacterium Synechocystis PCC 6803. Front Plant Sci 7: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead S, Kopecná J, Jackson PJ, Canniffe DP, Davison PA, Dickman MJ, Sobotka R, Hunter CN (2012) Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J Biol Chem 287: 27823–27833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppová J, Sobotka R, Tichý M, Yu J, Koník P, Halada P, Nixon PJ, Komenda J (2014) Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell 26: 1200–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J. (2005) Autotrophic cells of the Synechocystis psbH deletion mutant are deficient in synthesis of CP47 and accumulate inactive PS II core complexes. Photosynth Res 85: 161–167 [DOI] [PubMed] [Google Scholar]
- Komenda J, Lupínková L, Kopecký J (2002) Absence of the psbH gene product destabilizes photosystem II complex and bicarbonate binding on its acceptor side in Synechocystis PCC 6803. Eur J Biochem 269: 610–619 [DOI] [PubMed] [Google Scholar]
- Komenda J, Reisinger V, Müller BC, Dobáková M, Granvogl B, Eichacker LA (2004) Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J Biol Chem 279: 48620–48629 [DOI] [PubMed] [Google Scholar]
- Komenda J, Sobotka R, Nixon PJ (2012) Assembling and maintaining the Photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol 15: 245–251 [DOI] [PubMed] [Google Scholar]
- Kopecná J, Komenda J, Bucinská L, Sobotka R (2012) Long-term acclimation of the cyanobacterium Synechocystis sp. PCC 6803 to high light is accompanied by an enhanced production of chlorophyll that is preferentially channeled to trimeric photosystem I. Plant Physiol 160: 2239–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopečná J, Pilný J, Krynická V, Tomčala A, Kis M, Gombos Z, Komenda J, Sobotka R (2015) Lack of phosphatidylglycerol inhibits chlorophyll biosynthesis at multiple sites and limits chlorophyllide reutilization in the cyanobacterium Synechocystis 6803. Plant Physiol 169: 1307–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M, Klähn S, Scholz I, Matthiessen JK, Hess WR, Voß B (2014) Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res 21: 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey T, Westhoff P, Meierhoff K (2014) Expression of a nuclear-encoded psbH gene complements the plastidic RNA processing defect in the PSII mutant hcf107 in Arabidopsis thaliana. Plant J 80: 292–304 [DOI] [PubMed] [Google Scholar]
- Linhartová M, Bučinská L, Halada P, Ječmen T, Setlík J, Komenda J, Sobotka R (2014) Accumulation of the Type IV prepilin triggers degradation of SecY and YidC and inhibits synthesis of Photosystem II proteins in the cyanobacterium Synechocystis PCC 6803. Mol Microbiol 93: 1207–1223 [DOI] [PubMed] [Google Scholar]
- Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, Hess WR (2011) An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA 108: 2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilný J, Kopečná J, Noda J, Sobotka R (2015) Detection and quantification of heme and chlorophyll precursors using a High Performance Liquid Chromatography (HPLC) system equipped with two fluorescence detectors. Bio Protoc 5: e1390 [Google Scholar]
- Porra RJ, Thompson WA, Kriedmann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies KU (2000) Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J 19: 1900–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promnares K, Komenda J, Bumba L, Nebesářová J, Vácha F, Tichý M (2006) Cyanobacterial small chlorophyll-binding protein ScpD (HliB) is located on the periphery of photosystem II in the vicinity of PsbH and CP47 subunits. J Biol Chem 281: 32705–32713 [DOI] [PubMed] [Google Scholar]
- Rengstl B, Knoppová J, Komenda J, Nickelsen J (2013) Characterization of a Synechocystis double mutant lacking the photosystem II assembly factors YCF48 and Sll0933. Planta 237: 471–480 [DOI] [PubMed] [Google Scholar]
- Rengstl B, Oster U, Stengel A, Nickelsen J (2011) An intermediate membrane subfraction in cyanobacteria is involved in an assembly network for Photosystem II biogenesis. J Biol Chem 286: 21944–21951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachelaru I, Petriman NA, Kudva R, Kuhn P, Welte T, Knapp B, Drepper F, Warscheid B, Koch HG (2013) YidC occupies the lateral gate of the SecYEG translocon and is sequentially displaced by a nascent membrane protein. J Biol Chem 288: 16295–16307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitl I, Wickles S, Beckmann R, Kuhn A, Kiefer D (2014) The C-terminal regions of YidC from Rhodopirellula baltica and Oceanicaulis alexandrii bind to ribosomes and partially substitute for SRP receptor function in Escherichia coli. Mol Microbiol 91: 408–421 [DOI] [PubMed] [Google Scholar]
- Slavov N, Semrau S, Airoldi E, Budnik B, van Oudenaarden A (2015) Differential stoichiometry among core ribosomal proteins. Cell Reports 13: 865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotka R. (2014) Making proteins green; biosynthesis of chlorophyll-binding proteins in cyanobacteria. Photosynth Res 119: 223–232 [DOI] [PubMed] [Google Scholar]
- Sobotka R, Komenda J, Bumba L, Tichý M (2005) Photosystem II assembly in CP47 mutant of Synechocystis sp. PCC 6803 is dependent on the level of chlorophyll precursors regulated by ferrochelatase. J Biol Chem 280: 31595–31602 [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Vavilin D, Brune DC, Vermaas W (2005) 15N-labeling to determine chlorophyll synthesis and degradation in Synechocystis sp. PCC 6803 strains lacking one or both photosystems. Biochim Biophys Acta 1708: 91–101 [DOI] [PubMed] [Google Scholar]
- Wittig I, Karas M, Schägger H (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics 6: 1215–1225 [DOI] [PubMed] [Google Scholar]
- Xue S, Barna M (2012) Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 13: 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]