Use of integrated genomic, transcriptomic, and proteomic approaches enhanced the quality of genome assembly and annotation for sandalwood (Santalum album).
Abstract
Indian sandalwood (Santalum album) is an important tropical evergreen tree known for its fragrant heartwood-derived essential oil and its valuable carving wood. Here, we applied an integrated genomic, transcriptomic, and proteomic approach to assemble and annotate the Indian sandalwood genome. Our genome sequencing resulted in the establishment of a draft map of the smallest genome for any woody tree species to date (221 Mb). The genome annotation predicted 38,119 protein-coding genes and 27.42% repetitive DNA elements. In-depth proteome analysis revealed the identities of 72,325 unique peptides, which confirmed 10,076 of the predicted genes. The addition of transcriptomic and proteogenomic approaches resulted in the identification of 53 novel proteins and 34 gene-correction events that were missed by genomic approaches. Proteogenomic analysis also helped in reassigning 1,348 potential noncoding RNAs as bona fide protein-coding messenger RNAs. Gene expression patterns at the RNA and protein levels indicated that peptide sequencing was useful in capturing proteins encoded by nuclear and organellar genomes alike. Mass spectrometry-based proteomic evidence provided an unbiased approach toward the identification of proteins encoded by organellar genomes. Such proteins are often missed in transcriptome data sets due to the enrichment of only messenger RNAs that contain poly(A) tails. Overall, the use of integrated omic approaches enhanced the quality of the assembly and annotation of this nonmodel plant genome. The availability of genomic, transcriptomic, and proteomic data will enhance genomics-assisted breeding, germplasm characterization, and conservation of sandalwood trees.
The genus Santalum belongs to the family Santalaceae and consists of 15 extant species. It is a slow-growing, hemiparasitic tree distributed throughout tropical and temperate regions of India, Indonesia, Australia, and the Pacific Islands (Harbaugh and Baldwin, 2007). Commercially, the most valuable species is Indian sandalwood (Santalum album), which yields a unique essential oil used in perfumes, cosmetics, medicines, and incense sticks. The heartwood of this tree is treasured for its fragrance and is well known as one of the finest natural materials available for carving. Sandalwood is intertwined with Indian culture, and globally, it is the second most valuable and expensive tree after African blackwood (Dalbergia melanoxylon). Sandalwood is known to have cardiotonic, diuretic, diaphoretic, expectorant, aphrodisiac, hemostatic, anodyne, and antipyretic properties (Nambiar, 1993). Unfortunately, its population is declining from overharvesting and illegal trading, caused in part by its high commercial value. This alarming genetic erosion emphasizes the need for proper in situ conservation. Although efforts have been made for ex situ conservation, the planning and implementation of such programs have been limited due to the lack of genetic diversity existing in sandalwood populations (Rao et al., 2007; Kole, 2011).
Currently, large-scale transcriptomic data sets are available for sandalwood (Diaz-Chavez et al., 2013; Srivastava et al., 2015; Zhang et al., 2015, 2017; Celedon et al., 2016) and Santalum spicatum (Moniodis et al., 2015). Most of these genomic resources have been utilized to identify genes involved in sandalwood oil biosynthesis, cold stress response, and the hemiparasitic nature of its roots. However, a whole-genome sequence has not yet been reported for sandalwood. Our objective was to generate a draft genome assembly and annotate protein-coding genes of sandalwood based on genomic, transcriptomic, and proteomic data. We believe that this work will have a substantial impact in the near future with respect to sandalwood germplasm conservation, genetic diversity assessment, and cloning genes involved in natural essential oil production.
RESULTS
Sandalwood Genome Estimation, Sequencing, Genome Assembly, and Gene Prediction
Flow cytometry analysis of sandalwood (somatic chromosome number 2n = 20) revealed that the haploid DNA content (C value) was 0.21 pg and the deduced genome size was 203 Mb (Supplemental Fig. S1), where 1 pg is equivalent to 965 million bp (Bennett and Smith, 1976). The combination of paired-end (PE) and mate-pair (MP) libraries was prepared and sequenced on an Illumina HiSeq1000 platform that generated around 117 and 140 million reads, respectively. These data represented ∼95× of sequencing depth and sufficed for assembling into scaffolds/contigs. The overall workflow for the integrated genome assembly and annotation is depicted in Supplemental Figure S2.
The assembly of high-quality PE and MP reads into contigs was conducted with SPAdes 3.6.1, followed by scaffolding of contigs using MP data by SSPACE 3.0, which resulted in the consensus genome size of 221 Mb composed of 12,822 scaffolds/contigs (Table I). The N50 (median contig/scaffold size of genome assembly) value was 460.663 kb, with the largest scaffold being 2.9 Mb, indicating high-quality genome assembly for further downstream analyses. Based on a Core Eukaryotic Genes Mapping Approach (CEGMA), 85% (212 out of 248) were conserved core eukaryote genes (CEGs), which indicates the completeness of sandalwood genome assembly. In addition, Benchmarking Universal Single-Copy Orthologs BUSCO analysis represented 94.38% (1,359 out of 1,440) single-copy orthologs in the genome. This also provided a useful metric for describing the gene space during assembly.
Table I. Summary of sandalwood genome assembly and annotation.
| Assembly Details | Value |
|---|---|
| Total length of assembled sequence (Mb) | 220.961 |
| No. of contigs/scaffolds | 12,822 |
| Minimum length of contigs/scaffolds (bp) | 224 |
| Maximum length of contigs/scaffolds (bp) | 2,922,313 |
| Average length of contigs/scaffolds (bp) | 17,232 |
| N50 (bp) | 460,663 |
| GC content (%) | 34.38 |
| Total gene counts | 38,119 |
| Nuclear genes | 38,041 |
| Chloroplast genes | 53 |
| Mitochondria genes | 25 |
| Total non-ATGC characters | 17,338,084 |
In accordance with the National Center for Biotechnology Information (NCBI) genome submission guidelines, the assembly was finalized by removing contigs belonging to mitochondria, chloroplasts, and other vector/adapter sequences before subjecting it to gene prediction and annotation. MAKER-P-based gene prediction, supported by eudicot proteins retrieved from NCBI and RNA sequencing (RNA-seq) assembled unigenes, resulted in 38,061 genes. Of these genes, 24,479 had RNA-seq evidence and 13,582 were ab initio gene models. Based on integrated genomic, transcriptomic, and proteomic data, we identified 38,119 gene models in sandalwood.
The protein sequences of these genes were annotated by a BLASTP search against the UniProt eudicots protein database, and annotations were transferred to each gene by scripts bundled in the MAKER-P tool. The protein family (Pfam) identifier was assigned to genes using an InterProScan module. Out of 38,119 genes, 18,533 contained Pfam identifiers that were distributed across 3,159 types of Pfam domains. The Pfam domain containing proteins that were abundant in the sandalwood genome included protein kinase, PPR repeat family, gag-polypeptide of LTR copia type, protein Tyr kinase, RNA recognition motif, Myb-like DNA binding, cytochrome P450, zinc knuckle, ring finger, WD domain G-β repeat, plant mobile, and AP2.
Transcriptome and Proteome Analyses
De novo transcriptome assembly of RNA-seq reads resulted in the identification of 117,525 putative transcripts, of which 86,277 were full-length transcripts. Also, genome-guided TopHat and Cufflinks-based transcriptome assembly yielded 36,540 transcripts/isoforms comprising 23,292 genes. Out of 38,061 MAKER-P-predicted genes, 29,212 genes had RNA-seq evidence either through Cufflinks or Trinity-assembled transcripts.
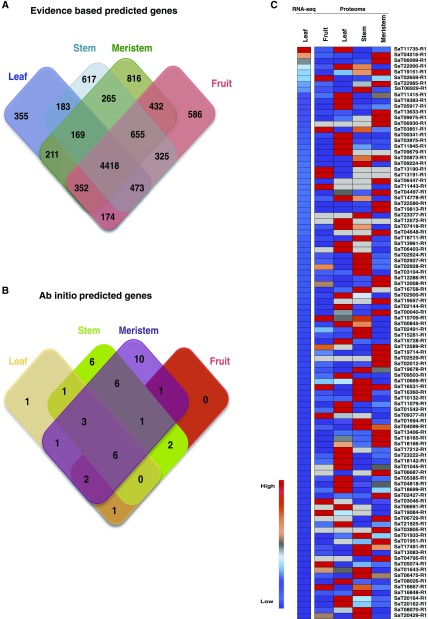
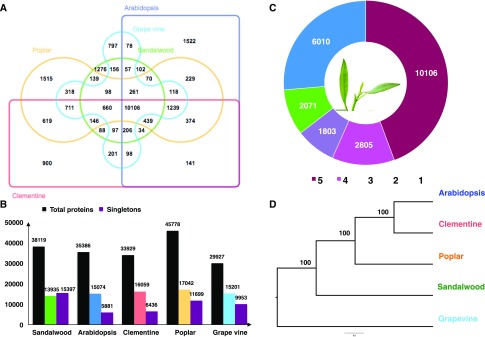
The proteome components from four different tissue samples were analyzed using mass spectrometry. In total, 1,083,446 tandem mass spectra were generated using an Orbitrap Fusion Tribrid mass spectrometer, and these were searched against the protein database created using RNA-seq and genome data. The high-confidence data set led to the identification of 72,325 peptides, mapped to 10,076 genes predicted in the sandalwood genome, thereby validating the expression of the predicted gene models (Fig. 1, A and B; Supplemental Table S1). The top 100 highly up-regulated genes based on RNA-seq Fragments Per Kilobase Million (FPKM) value and proteome (intensity-based absolute quantification [iBAQ] value) data are shown in Figure 1C.
Figure 1.
Venn diagrams illustrating the total number of proteins identified in a protein database search in four organs or tissues. A, MAKER-P-predicted genes with RNA-seq evidence. B, MAKER-P-predicted ab initio genes. C, Gene expression pattern of top 100 highly up-regulated genes.
Based on the functional annotation, ribulose bisphosphate carboxylase (small chain) was the most abundantly expressed gene, followed by chlorophyll a/b-binding protein genes in leaves. Some abundantly expressed proteins in the stem included cyclophilin-type peptidyl-prolyl cis-trans-isomerase/CLD (SaT06929-R1), uncharacterized protein containing Pfam domain ABA/WDS-induced protein (SaT09224-R1), uncharacterized protein containing Pfam domain pathogenesis-related protein Bet v I family (SaT02927-R1), and uncharacterized protein containing Pfam domain aldo/keto-reductase family (SaT01694-R1). The Bet v I protein family has been detected in the secretome fraction of chickpea (Cicer arietinum; Gupta et al., 2011). The functional roles of this molecule have been characterized under abiotic and biotic stresses (Gupta et al., 2015). Increased expression of the family of ABA/WDS proteins induced in response to water deficit also has been reported (Padmanabhan et al., 1997). Proteins with dominant expression in fruits include an uncharacterized protein containing a ubiquitin domain (SaT16667-R1), another uncharacterized protein containing a conserved thioredoxin domain (SaT05074-R1), and fatty acid desaturase (SaT02659-R1). The differential regulation of thioredoxin protein has been reported during fruit ripening in apple (Malus domestica) and tomato (Solanum lycopersicum ‘Micro-Tom’; Shi et al., 2014; Suzuki et al., 2015). Proteins abundant in leaves include proteins related to photosynthesis and glycolytic enzymes such as conserved PSII 10-kD polypeptide PsbR domain-containing protein (SaT05917-R1), chlorophyll a/b-binding protein (SaT03975-R1 and SaT07418-R1), manganese-stabilizing protein/PSII polypeptide (SaT04497-R1), and Fru-bisphosphate aldolase class I (SaT02900-R1 and SaT02144-R1).
Identification of Novel Protein-Coding Genes in the Sandalwood Genome Using Proteomics Data
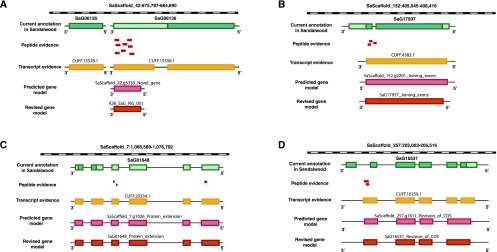
The unassigned spectra that did not map to any of the proteins represented in the computationally predicted protein database were searched against the six-frame translated genome database and three-frame translated transcript database to identify novel genes and refine the annotated gene structures. Peptides that mapped to unannotated genomic loci with no prior evidence for their protein-coding capability, termed genome search-specific peptides (GSSPs), were obtained from the searches against the six-frame translated sandalwood genome database. Details of the analysis are provided in “Materials and Methods.” The GSSPs were further filtered using stringent criteria: (1) Xcorr (cross correlation) value above 3 and (2) ion score above 30. Using this high-confidence set of GSSPs, we identified 53 novel protein-coding genes supported by evidence from at least two peptides (Supplemental Table S2.1).
Of the 53 novel protein-coding genes identified in this study, 46 were supported by RNA-seq evidence. One such example of a novel gene identified with multiple peptides as well as RNA-seq evidence is shown in Figure 2A. Additionally, orthologous evidence for these novel genes was obtained by BLAST analysis. The first line of evidence was obtained from four other plant species (Arabidopsis thaliana, Vitis vinifera, Populus trichocarpa, and Citrus clementina). In cases where no evidence could be found in these plant species, the sequences were searched against the NCBI nonredundant (nr) database. A threshold value of 60% sequence identity was applied for the analysis. To identify probable gene function(s) of the identified novel genes, we performed a functional annotation using the InterProScan tool.
Figure 2.
Reannotation of the sandalwood genome using proteomic evidence. A, Identification of a novel gene between the annotated genes SaG06135 and SaG06136. B, Evidence for the joining of exons. C, N-terminal extension of a gene, with three peptides matched to the upstream region of SaG01648. D, Revision of the coding sequence.
A significant number of uncharacterized proteins were identified in the category of novel genes with no known function. To assign functional roles to these genes, protein family domains were retrieved from the InterProScan output. The protein IOB_SaG_NG_013 contained a plant-specific domain referred to as the GRAS (GIBBERELLIC-ACID INSENSITIVE [GAI], REPRESSOR of GAI [RGA], and SCARECROW [SCR]) domain. The GRAS domain-containing proteins are transcription factors with known roles in GA3 signaling (Hofmann, 2016). Another protein, IOB_SaG_NG_014, was found to contain a protein kinase domain. The list of novel genes also included a late embryogenesis abundant (LEA) domain-containing protein. LEA proteins are known to impart a protective function under various abiotic stresses, such as drought and salinity (Gao and Lan, 2016). Proteins with cell wall-associated functions also were identified, including glycosyl hydrolase family 18 (IOB_SaG_NG_033) and plant invertase/pectin methylesterase inhibitor (IOB_SaG_NG_044; Supplemental Table S2.1).
To understand the tissue-specific/restricted distribution of the novel genes, we examined their expression. Some of the proteins that were highly enriched in the stem were a GRAS domain family protein (IOB_SaG_NG_013), a protein of the subtilase family (IOB_SaG_NG_049), and a plant protein of unknown function (IOB_SaG_NG_005). The cupin domain-containing protein (IOB_SaG_NG_031) was found to be highly enriched in fruits, whereas a Leu-rich repeat N-terminal domain-containing protein (IOB_SaG_NG_018) was found to be enriched in the shoot meristem fraction. Probing the functional roles of these proteins in sandalwood should be conducted in the future.
Revision of the Gene Structures Using Peptide Evidence
Apart from the identification of novel genes, 24,618 GSSPs were mapped to various positions in the proximity of annotated genes or extended beyond their previously defined boundaries. Based upon the genomic positions, they were categorized into the following classes: novel exons, exon extensions, protein extensions, alternate frame of translation, revision of coding sequence (coding DNA sequence), and joining of exons. Proteins reported in each of these categories were supported by evidence from at least two peptides.
We discovered novel annotations including novel exons belonging to 34 genes, 33 of which also were supported by RNA-seq evidence (Supplemental Table S2.2). We also found evidence for exon extensions in 24 genes, of which 11 were 5′ extensions and 13 were 3′ extensions (Supplemental Table S2.3). Evidence for the joining of exons also was observed in five genes (Supplemental Table S2.4), and the revised annotation for SaG17937 is depicted in Figure 2B. Junctional peptides that mapped to the exon boundaries of two distinct exons were exploited for this analysis. For example, the joining of exon 5 and exon 6 was reported in the gene d-3-phosphoglycerate dehydrogenase (SaG18379). This was supported by the gene model predicted using the AUGUSTUS tool.
We also discovered 26 instances of protein extensions (Fig. 2C), where 11 were N-terminal extensions and 15 were C-terminal extensions (Supplemental Table S2.5). Peptide evidence also was found for alternate frames of translation in four genes (Supplemental Table S2.6). In these examples, the annotated frame of translation was different from that evidenced by the identified peptides. This line of support provided by mass spectrometric data is a marked advantage over that provided by transcriptome data, which cannot accurately predict the correct frame of translation. We also report alternate translational start sites of four genes (Supplemental Table S2.7). All of these alternate start sites were supported by evidence from orthologous sequences.
Sixty-four peptides provided evidence for the revision of annotated coding sequence (Supplemental Table S2.8). This category includes examples where multiple changes in the annotated gene structures were incorporated that ultimately resulted in the revision of annotated coding sequence (Fig. 2D). For example, evidence was found for the N-terminal extension of the gene containing a gelsolin repeat (SaG16373) along with evidence for the extension of its exon boundaries. Peptide evidence in the annotated untranslated regions (UTR) also was included in this list. For example, multiple peptides were found mapping to the 3′UTR of a protein similar to DDB1A (for DNA damage-binding protein1a; SaG18842), hence extending the gene boundaries. This was further supported by the identification of orthologous sequence evidence in Citrus sinensis. All the GSSPs along with their categorization, details of peptide evidence, and revised genome coordinates are listed in Supplemental Table S2.
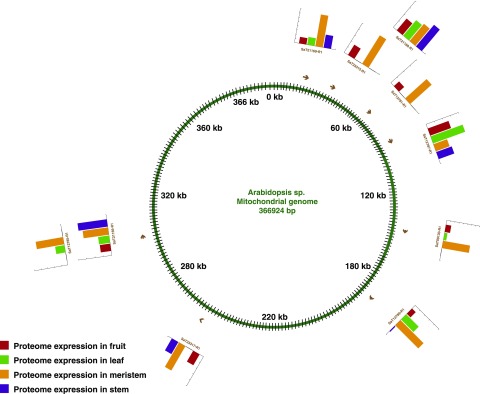
Gene Expression between Nuclear and Organellar Genomes
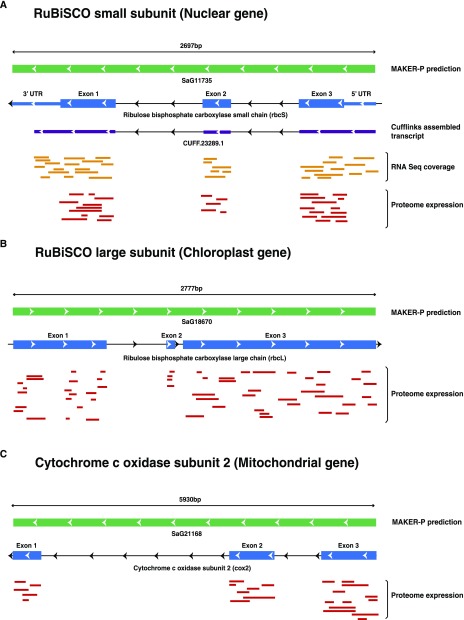
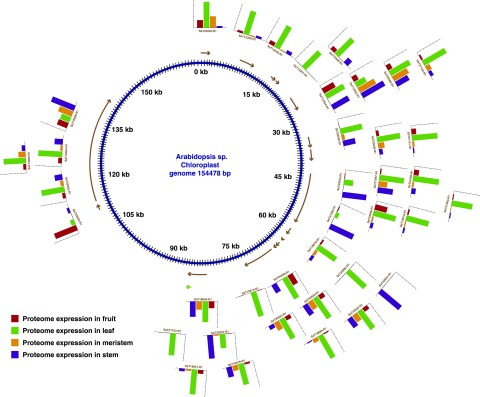
Gene prediction using an ab initio method led to the identification of 53 chloroplast and 25 mitochondrial genes. Proteomic evidence supported 38 (71.7%) chloroplast and 10 (40%) mitochondrial genes. Evidence for the 8 (32%) mitochondrial genes also was supported by RNA-seq data. As a proof of concept, we have shown RNA-seq and proteome expression data for genes encoded in the nucleus (Fig. 3A), chloroplast (Fig. 3B), and mitochondria (Fig. 3C). No RNA-seq support was available for the chloroplast genes (Supplemental Table S3). Among the mitochondrial genes, peptide evidence was found in all four tissues for proteins such as Respiratory chain NADH dehydrogenase, 30-kd subunit (SaT21169-R1), ATP synthase subunit4 (SaT12789-R1), and Cytochrome c oxidase subunit2 (SaT21168-R1). Peptide evidence in chloroplast genes was detected for proteins such as PSI P700 chlorophyll a apoprotein A2 (SaT20539-R1), ATP synthase subunit α (SaT12359-R1), and Ribosomal protein S4 (SaT21097-R1; Supplemental Table S4). The mitochondrial and chloroplast genes identified in our study, along with their protein and transcript abundance values, are represented in Arabidopsis chloroplast (Fig. 4) and mitochondrial (Fig. 5) genomes as a reference. Similarly, we looked for RNA-seq and proteome expression of ultraconserved CEGs from CEGMA mapping. Around 85.44% and 89% of CEGs had RNA-seq and peptide evidence, respectively (Supplemental Table S5). Overall, 97.37% of CEGs had either RNA-seq or peptide evidence.
Figure 3.
Transcriptome and proteome expression of genes encoded in chloroplast, mitochondrion, and nucleus. A, rbcS nuclear gene expression. B, rbcL chloroplast gene expression. C, cox2 mitochondrial gene expression.
Figure 4.
Expression pattern of chloroplast genes in sandalwood.
Figure 5.
Expression pattern of mitochondrial genes in sandalwood.
Transcription Factors of the Sandalwood Genome
The protein-protein homology analysis of the sandalwood proteome with the Plant Transcription Factor Database version 4.0 (Jin et al., 2017) revealed 58 families of transcription factors distributed across 1,414 genes. Some of the abundant transcription factors included MYB, bHLH, ERF, GRAS, NAC, C2H2, WRKY, bZIP, MYB related, and C3H (Supplemental Table S6).
Gene Family Expansion and Phylogenetic Relationships
Genome-wide analysis of orthologous genes across various species is an important component of comparative genomic studies. Knowledge of orthologous gene clusters helps in taxonomic and phylogenetic classification, thus elucidating the molecular evolution of genes and genomes across multiple species (Henikoff et al., 1997; Mushegian et al., 1998). Orthologous clustering of the proteomes of five species (sandalwood, Arabidopsis, C. clementina, P. trichocarpa, and V. vinifera) revealed 10,106 orthologous gene clusters, suggesting their conservation in the lineage after speciation. Additionally, there were 1,276, 1,522, 900, 1,515, and 797 clusters specific to sandalwood, Arabidopsis, C. clementina, P. trichocarpa, and V. vinifera, respectively.
These species-specific clusters could be multiple genes within the cluster or in-paralog clusters indicating lineage-specific gene expansion/duplication in these gene families (Fig. 6, A–C). However, 15,397, 5,881, 6,436, 11,699, and 9,953 singletons also were observed for sandalwood, Arabidopsis, C. clementina, P. trichocarpa, and V. vinifera, respectively. These are unique single-copy genes present in their respective species and do not have orthologs between and paralogs within the species. The identification of single-copy orthologs plays a necessary role in establishing the phylogenetic relationship among groups of species (Creevey et al., 2011). Based on orthologous clustering, there were 1,844 single-copy gene clusters across these five species, indicating that they have retained only one copy of these genes after a speciation event. Then, 100 single-copy gene clusters were chosen randomly to reveal the phylogenetic relationships among these species. Based on a maximum likelihood tree, sandalwood is most closely related to V. vinifera, followed by P. trichocarpa, C. clementina, and Arabidopsis (Fig. 6D).
Figure 6.
Distribution of shared gene clusters across plant species. A, Venn diagram showing the shared orthologous gene clusters among sandalwood, Arabidopsis, V. vinifera, P. trichocarpa, and C. clementina. B, Counts of total genes, clustered genes, and singletons in each genome. C, Orthologous cluster counts shared across five species. D, Phylogenetic tree showing inferred evolutionary relationships among five plant species based on 100 single-copy orthologs.
Santalol Biosynthesis Pathway in Sandalwood
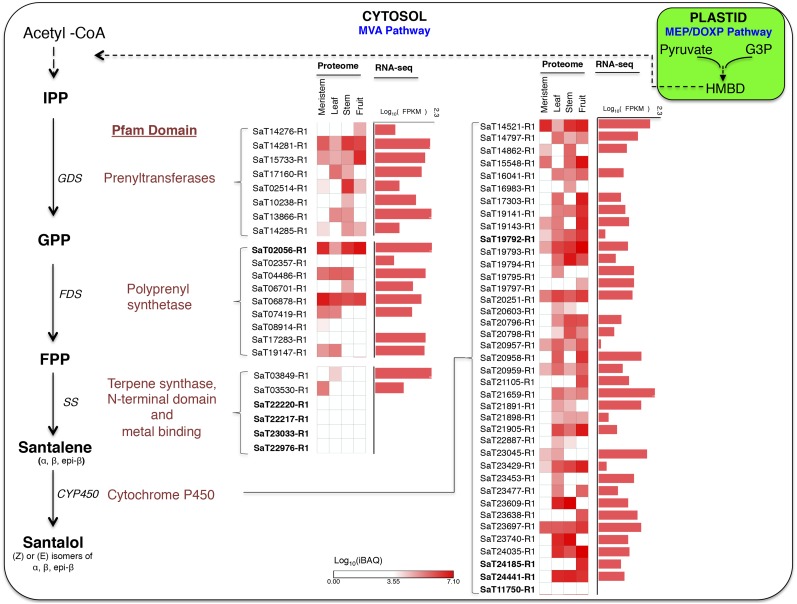
Sandalwood oil is one of the world’s most highly traded essential oils, which is composed primarily of sesquiterpene olefins and alcohols. Santalol is a bioprospecting metabolite present in sandalwood; identifying the genes responsible for santalol biosynthesis would be valuable for metabolite engineering. Although several studies have reported the genes involved in santalol biosynthesis using transcriptomic data sets, identification at a genome-wide scale was not possible due to a lack of whole-genome information. To identify the genes and encoded enzymes responsible for santalol biosynthesis at the whole-genome level, annotated genes in sandalwood were searched for Pfam domains like terpene synthase N terminal (PF01397), terpene synthase family metal-binding domain, prenyltransferases and squalene oxidase (PF00432 and PF13249), prenyltransferase like (PF13243), cytochrome P450 (PF00067), and polyprenyl synthetase (PF00348). This approach enabled us to identify four genes of PF00432, four PF13249, nine PF00348, 22 PF03936, seven PF01397, and 184 PF00067 genes.
Santalol is a sesquiterpene synthesized through the mevalonate (MVA; in the cytosol) or methylerythritol 4-phosphate (MEP; in plastids) pathway. The first step involves head-to-tail condensation of isopentenyl pyrophosphate (IPP) to geranyl pyrophosphate (GPP) with the help of geranyl diphosphate synthase. We found eight genes (PF00432 and PF13249) in sandalwood that were likely to be involved in the conversion of IPP to GPP and encoded a prenyltransferase Pfam domain.
Similarly, additional condensation of GPP produces farnesyl pyrophosphate (FPP), catalyzed by farnesyl diphosphate synthase (FDS; PF00348). We detected nine genes (SaT02056-R1, SaT02357-R1, SaT04486-R1, SaT06701-R1, SaT06878-R1, SaT07419-R1, SaT08914-R1, SaT17283-R1, and SaT19147-R1) encoding FDS with a polyprenyl synthetase Pfam domain.
Santalene synthase (SS; PF03936 and PF01397), a terpene cyclase, catalyzes the cyclization of open ring FPP into a mixture of cyclic sesquiterpenes such as α-santalene, β-santalene, epi-β-santalene, and exo-bergamotene. Six genes were predicted to encode SS (SaT03849-R1, SaT03530-R1, SaT22220-R1, SaT22217-R1, SaT23033-R1, and SaT22976-R1). We performed a BLASTP search of the predicted proteins corresponding to genes identified in our study with previously characterized genes (Jones et al., 2011; Srivastava et al., 2015). We found that SaT22220-R1, SaT22217-R1, SaT02056-R1, SaT23033-R1, and SaT22976-R1 genes were highly similar (greater than 85%) to SaSQS1 (KJ665776), SaSQS2 (KJ665777), SaFDS (KF011939), SaBS (KJ665778), SaSS (KF011938), and SaSSy (HQ343276) (Supplemental Fig. S3). A summary of cloned genes is presented in Table II.
Table II. Summary of cloned genes and their homologs in predicted genes of the sandalwood genome.
Cyclic sesquiterpenes (santalene/bergamotene) are converted into santalols by the CYP450 system (PF00067; Fig. 7). Mainly, four sesquiterpenols, α-, β-, and epi-β-santalol and α-exo-bergamotol, together constitute approximately 80% to 90% of heartwood oil obtained from mature trees (Celedon et al., 2016). Around 184 sandalwood cytochrome P450 genes were identified in the genome. Out of these, four genes (SaT19792-R1, SaT24185-R1, SaT24441-R1, and SaT11750-R1) were found to be homologous with already cloned genes reported previously (Diaz-Chavez et al., 2013; Celedon et al., 2016).
Figure 7.
Genes involved in santalol biosynthesis (terpenoid backbone biosynthesis pathway; MAP00900 from KEGG). Gene identifiers in boldface are the homologs of already cloned genes. The heat map shows iBAQ of proteome data in meristem, leaf, fruit, and stem. For RNA-seq, FPKM values are shown. GDS, Geranyl diphosphate synthase; HMBD, hydroxy methyl butenyl diphosphate.
We performed a phylogenetic analysis of these 184 genes along with the previously characterized genes (Diaz-Chavez et al., 2013; Celedon et al., 2016). Phylogenetic analysis of the CYP450 genes identified in our study and cloned genes showed that they were clustered in two major clades (Supplemental Fig. S4). Clade I had mixtures of 38 CYP450, three bergamotene oxidase, and 10 santalene/bergamotene oxidase genes clustered separately. The remaining 133 CYP450 genes clustered separately under clade II. Also, proteogenomic peptide evidence was obtained from leaves, shoot apical meristems, stems, and fruits, while RNA-seq expression (in FPKM) in leaves is shown in the pathway figure for most of the genes. The nucleotide sequences of all the genes involved in santalol biosynthesis are provided in Supplemental Text S1.
Genes identified in this study validate the findings reported earlier and will provide a foundation for the production of sandalwood oil for the essential oil industry by means of metabolic engineering (synthetic biology) under in vitro conditions. Such proof of concepts of essential oil biosynthesis have been demonstrated previously (Jones et al., 2011; Diaz-Chavez et al., 2013; Srivastava et al., 2015; Celedon et al., 2016). As discussed by Diaz Chavez et al. (2013), genes identified in their study and genes described here may be used as biomarkers to monitor the onset of oil formation in sandalwood plantations and in the development of genetic markers for tree improvement breeding strategies.
Long Noncoding RNAs in the Sandalwood Genome
Based on a Coding Potential Calculator, Cufflinks-assembled transcripts (36,540) were further categorized into protein-coding (15,615) and noncoding (20,925) transcripts. BLAST alignment of all 20,925 noncoding transcripts helped us to exclude 2,895 of them, which showed a full-length alignment to MAKER-P-predicted genes. Based on exon numbers, these noncoding transcripts were further categorized into single-exon (5,783), two-exon (1,981), and multiple-exon (10,266) noncoding transcripts. These long noncoding RNA (lncRNA) transcripts were further validated by proteome data, with 1,348 (1,205 multiple-exon, 49 two-exon, and 93 single-exon transcripts) found to have peptide evidence across the organs. Only the lncRNAs supported by two or more peptides were considered as a positive output. Furthermore, the spectra were checked manually for evidence supported by two peptides. Using these stringent parameters, we found peptide evidence for 1,348 lncRNAs. Proteome data helped us to delineate these predicted lncRNAs to be potential coding transcripts that serve as novel genes. These findings are not unprecedented, as mass spectrometry-based evidence for the coding potential of several noncoding RNAs have been reported (Kim et al., 2014; Prasad et al., 2017).
The tissue-specific distribution of these noncoding RNAs also was studied. The transcript CUFF.22264.3 contains ankyrin repeats and was found in all four organs. In Arabidopsis, the roles of ankyrin repeat-containing proteins have been implicated in disease resistance and also in antioxidation pathways (Yan et al., 2002). CUFF.2819.2 was predicted to contain a DHHC palmitoyltransferase domain and was found exclusively in the fruit tissues. This Cys-rich domain is known to play important roles in lipid modification, which, in turn, regulates the membrane localization of various target proteins (Batistič, 2012). CUFF.2769-containing Pfam domain Glc-inhibited division protein A was found exclusively in the stem. A list of all the novel coding genes identified in this study along with their peptide sequences and other associated details is provided in Supplemental Table S7.
The remaining 16,683 transcripts were considered noncoding, as we did not find any peptide evidence to support them. We also confirmed that these noncoding transcripts did not match already predicted genes. Some lncRNAs may serve as precursors for the biogenesis of small regulatory RNAs, such as microRNAs and short interfering RNAs. Based on a homology search, we identified 13 lncRNAs matching with plant stem-loop microRNA precursors (greater than 70% query coverage and identity) in miRBase. Most of these microRNAs belong to the MIR156, MIR159, MIR160, MIR162, MIR166, MIR168, MIR171, MIR172, MIR319, MIR396, and MIR399 families (Supplemental Text S2). These lncRNAs may have potential roles in many essential biological processes; therefore, understanding their function is a fascinating and new area of sandalwood research.
Repeats in the Sandalwood Genome
Repeat elements make up a substantial portion of most plant genomes, most of which are inactivated or silenced. The de novo repeat prediction revealed that 27.46% of the sandalwood genome is composed of various types of repetitive/transposable elements. Among interspersed repeats (25.65%), the unclassified repeats (17.28%) fraction was more common than retroelements (5.99%) and DNA transposons (2.38%; Supplemental Table S8). We predicted simple sequence repeats (SSRs) in the sandalwood genome using a MISA tool. In total, 153,591 SSRs were identified, composed of mono (94,089), di (44,179), tri (12,824), tetra (2,095), penta (257), and hexa (147) type repeats (Supplemental Table S9). Among mono repeats, the A/T (94.61%) type was highest, followed by C/G (5.38%). Similarly, AT/AT, AG/CT, AC/GT, and CG/CG type of di repeats were in 48.32%, 40.13%, 11.32% and 0.23% fractions, respectively. AAT/ATT and AAG/CTT were the most abundant tri repeats. (The overall distribution of all types of repeats is provided in Supplemental Table S10). Due to the unavailability of SSRs in sandalwood, cross-species SSRs were deployed for the characterization of genetic variability (Patel et al., 2016). Sandalwood is a highly valued tree, but its population is declining due to overharvesting and illegal poaching in natural habitats. Our study will supplement the necessity to develop highly reproducible genomic resources like SSRs for sandalwood characterization and conservation.
DISCUSSION
We used a workflow that integrates genomics, transcriptomics, and proteomics data to generate a well-annotated genome sequence of Indian sandalwood. This plant species is highly valued for its heartwood-derived essential oils that are often referred to as liquid gold. Despite rising global demand for sandalwood oil, supply is limited due to difficulties with sandalwood propagation. Excessive harvesting without replenishment of this invaluable tree has substantially reduced its population, leading to a global shortage and soaring market prices of its products (Kumar et al., 2012). Our efforts of generating whole-genome data and other genomic resources for sandalwood will have an immense importance. The combination of PE and MP genome-sequencing technologies helped us to assemble around 221 Mb of the sandalwood genome and predict 38,119 protein-coding genes based on mRNA and peptide evidence. The observed N50 value along with the representation of 94.38% of single-copy orthologs in the sandalwood genome assembly based on BUSCO led to a high-quality genome assembly for downstream analyses. The number of protein-coding genes is similar to those reported in other plant species.
Over the last decade, there has been a surge in available plant genome data sets. However, very few studies have put to use transcriptome- and proteome-assisted genome annotation pipelines that enable the formation of more robust, well-annotated, and reliable reference genomes. Using high-resolution mass spectrometry-based proteome analysis, two studies improved the genome annotation of Arabidopsis (Baerenfaller et al., 2008; Castellana et al., 2008). In the same plant, RNA-seq analysis allowed for the identification of several novel protein-coding genes together with novel transcribed regions, noncoding RNAs, and small RNA loci that were not annotated earlier (Cheng et al., 2017). RNA-seq-assisted genome annotation of two other members of the Brassicaceae (Arabidopsis lyrata and Brassica rapa) also has been reported (Rawat et al., 2015; Markelz et al., 2017). Among monocots, proteogenomics studies have been carried out in maize (Zea mays), wherein a total of 165 novel protein-coding genes were reported and gene models for 741 additional genes were corrected (Castellana et al., 2014). In Triticum aestivum, both RNA-seq and proteomics data have been explored for genome annotation (International Wheat Genome Sequencing Consortium, 2014; Clavijo et al., 2017). Transcriptome analysis further expanded the array of novel isoforms of known genes along with 2,253 novel genes in maize (Wang et al., 2016). In two independent studies, improved genome annotation in Oryza sativa has been reported using proteomics and RNA-seq data (Helmy et al., 2011; Watanabe et al., 2015). However, in all these cases, a better genome assembly and annotation using multiomics was achieved many years after first drafts of genome assemblies were published. For instance, the availability of a well-annotated genome sequence in humans is due to the extensive use of multiple RNA-seq- as well as mass spectrometry-derived data for genome annotation (Harrow et al., 2012; Steijger et al., 2013; Kim et al., 2014; Wilhelm et al., 2014). Similarly, using a multiomics approach and integrated data analysis, our group recently reported an improved genome assembly and annotation of Anopheles stephensi and simultaneously refined 16 other anopheline genomes (Prasad et al., 2017). A partial list of available genome sequences that made use of multiomics data for assembly and annotation is given in Supplemental Text S3.
In this study, we identified 72,325 peptides that mapped to 10,076 predicted protein-coding genes in the sandalwood genome, therefore validating the expression of the predicted gene models. Peptides that mapped to unannotated genomic loci were further used to identify novel protein-coding genes or refine the predicted gene models. We identified 53 novel protein-coding genes using peptides that mapped to intergenic regions. Apart from the identification of novel genes, several GSSPs also mapped to various positions in the proximity of annotated genes. However, these peptides extended beyond their previously defined boundaries. These peptides provided evidence for the refinement of predicted gene models to which they mapped in the sandalwood genome. Importantly, the mass spectrometry-based proteomic evidence provided an unbiased approach toward the identification of organellar proteins, which can be missed in standard transcriptome studies due to specific enrichment of mRNA using poly(A) tails. Also, the identification of chloroplast and mitochondrial genes in our data set is not unprecedented, as the integration of organellar genomes into the nuclear genome has been reported in other plant species (International Rice Genome Sequencing Project, 2005; Varshney et al., 2011; Tomato Genome Consortium, 2012). The data clearly indicate that a whole-proteome sequencing method is unbiased in capturing peptides from both nuclear and organellar genomes.
Interestingly, we also found peptide evidence for 1,348 lncRNAs that were predicted as noncoding by the Coding Potential Calculator tool. The proteome data helped us delineate these predicted lncRNAs to be potential coding transcripts, which also serve as novel genes. These findings are not unprecedented, as mass spectrometry-based evidence for the coding potential of several noncoding RNAs has been reported (Kim et al., 2014; Prasad et al., 2017). Owing to self-incompatibility and cross-pollination behavior, sandalwood exhibits a large amount of genetic diversity with respect to seed (size, shape, and vigor), bark (color, texture, and thickness), and leaf types (ovate, lanceolate, elliptic, and linear; Kulkarni and Srimathi, 1982). In spite of this, tree improvement for heartwood and oil content has not been explored. Terpenoids or isoprenoids constitute one of the most structurally diverse groups of biomolecules in plants. The building blocks of these multifaceted structures are synthesized either through the MVA or plastid-specific MEP pathway (Moniodis et al., 2015; Srivastava et al., 2015). Differential regulation of the rate-limiting enzyme in the MVA pathway, 3-hydroxy-3-methylglutaryl CoA reductase, and various terpenoid synthases has been reported under cold stress in sandalwood (Zhang et al., 2017). Sandalwood oil synthesis is regulated through the MVA and MEP pathways. This oil is composed predominantly of four sesquiterpenols, (Z)-α-santalol, (Z)-β-santalol, (Z)-epi-β-santalol, and (Z)-α-exo-bergamotol, as well as minor components such as the (E)-stereoisomer of these sesquiterpenols (Celedon et al., 2016). Together, these components constitute approximately 90% of the oil extracted from mature trees. An in-depth knowledge about the genes involved in its biosynthesis is important for metabolite engineering. This genome sequencing and annotation process paved the way for the identification of 230 genes involved in the santalol biosynthesis pathway. It must be emphasized that, in sandalwood, the CYP76F subfamily of cytochrome P450s hydroxylates santalenes and bergamotene to yield (E)-α-santalol, (E)-β-santalol, (E)-epi-β-santalol, and (E)-α-exo-bergamotol, the minor components of sandalwood oil (Diaz-Chavez et al., 2013; Celedon et al., 2016). In a recent functional characterization of SaCYP736A167, a member of another subfamily of cytochrome P450s, its role in the synthesis of (Z)-stereoisomers of the four sesquiterpenols was demonstrated (Celedon et al., 2016).
We identified several CYP450 genes (184) in our data set. However, we found a homolog SaT11750-R1 with SaCYP736A167 in our predicted gene set, which had 99.6% sequence identity at the protein level with 100% coverage. But we were unable to detect SaCYP736A167 in our transcriptome or proteome data set. Notably, SaCYP736A167 was reported to selectively accumulate in the heartwood of 15-year-old trees (Celedon et al., 2016), whereas we sampled only leaves for our transcriptome analysis. The stem segments used for our proteome analysis were young twigs (0.5 cm long) harvested from 10-year-old plants. Although other researchers have cloned and characterized a few genes related to santalol production, exploration of the remaining genes that we have identified through whole-genome-based annotation has the potential for utilization in large-scale industrial sandalwood oil production. Several complex ecological factors modulate the growth of sandalwood and are responsible, in part, for the strong genotype × environment influence on oil content. In addition to genomic data, our identification of SSRs and genes in the sandalwood genome will serve as a resource for the selection of markers to design more robust molecular breeding techniques. These aspects will hasten the process to authenticate the correlation of heartwood and oil content using phenotypic and molecular data. Plus, the data highlight the importance of using multiomics strategies to generate more robust and reliable resources of a genome sequence.
CONCLUSION
In this study, we demonstrated the feasibility of an integrative omics approach to assemble, annotate, and refine the gene models of Indian sandalwood. We utilized a proteogenomic approach to provide experimental evidence in addition to RNA-seq evidence for the predicted gene models as well as to identify novel protein-coding genes and incorporate corrections in predicted gene structures. Although RNA-seq experiments provided expression evidence for most of the predicted genes, an interface of proteomic data helped us to identify functional transcripts among many novel transcripts nominated by genomic and transcriptomic studies. Furthermore, proteogenomic analysis also enabled the identification and correction of several gene models that were missed by gene prediction algorithms. A close collaboration between genomic and proteomic groups enabled the generation of genomic resources and combined analyses using multiomics data for improved assembly and annotation of any plant genome, which will serve as a model for future genome sequencing efforts. The availability of a well-annotated genome for sandalwood will have wider implications for the conservation of tree populations by reducing pressure on the supply from native forests for sandalwood oil extraction.
MATERIALS AND METHODS
Estimation of Nuclear DNA Content
Sandalwood tree (Santalum album) growing near the Centre for Cellular and Molecular Platforms (13°04′22.7′′N, 77°34′42.1′′E) was chosen for whole-genome sequencing. The meristematic leaf tissue of sandalwood was chopped into pieces using a sterilized blade and stained using nuclear isolation buffer. The buffer was composed of hypotonic propidium iodide, 50 μg mL−1, in 3 g L−1 trisodium citrate dihydride containing 0.05% (v/v) Nonidet P-40 containing 2 mg mL−1 RNaseA stored in a dark amber bottle at 4°C. The liquid containing stained nuclei was filtered through a 30-μm nylon mesh and processed for DNA content as reported earlier (Krishan, 1975). Stained nuclei were analyzed using a BD FACS cell sorter at the Central Imaging and Flow Cytometry Facility, National Centre for Biological Sciences, Bengaluru, India. The mean sandalwood nuclei count was normalized based on nuclei counts of red blood cells from chicken (Gallus gallus). Genome size was estimated by multiplying the 1C value (pg) of sandalwood with 965 Mb (1 pg equivalent value; Bennett and Smith, 1976).
DNA and RNA Isolation
Young leaves were collected from a medium-sized sandalwood tree (10 years old). Leaves were ground in liquid nitrogen using a pestle and mortar. Homogenized powdered leaf mass was processed through the GenElute Plant Genomic DNA Miniprep kit (G2N70; Sigma-Aldrich) and the Spectrum Plant Total RNA kit (STRN50; Sigma-Aldrich) for extraction of high-quality DNA and RNA, respectively. Genomic DNA contamination in the RNA sample was removed by treatment with DNase (AM1906; Ambion). RNA integrity and quantity were verified on a Bioanalyzer using an Agilent RNA 6000 nano chip.
Illumina DNA and RNA Library Preparation and Sequencing
PE (250- to 500-bp insert size) and MP (10- to 20-kb insert size) Illumina libraries were prepared according to the manufacturer’s instructions. Libraries were sequenced using an Illumina HiSeq1000; the read length of PE was 2 × 101 bp, and that of MP was 2 × 51 bp. Similarly, a strand-specific RNA sequencing library was constructed according to the instructions from the Illumina Tru-Seq RNA sample preparation kit version 2 (RS-122-2001) and sequenced (read length of 2 × 101 bp) using the Illumina HiSeq1000 platform at the Centre for Cellular and Molecular Platforms, Bengaluru, India.
Genome Assembly and Repeat Prediction
A box plot of nucleotide base quality was generated, and low-quality bases (quality score < Q30) were trimmed using a FastX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). The raw reads of PE and MP libraries were trimmed for adapter sequence contamination and low-quality bases (PHRED score < Q30). The SPAdes assembler (Bankevich et al., 2012) was used for assembling the high-quality PE and MP reads. Contigs were further scaffolded using SSPACE (Boetzer et al., 2011), and gaps in the scaffolds were closed by the GapCloser module (Luo et al., 2012). The vector, chloroplast, and mitochondrial sequences were removed from the assembled contigs. Scaffolds were subjected to CEGMA and BUSCO to assess genome completeness (Parra et al., 2007; Simão et al., 2015). The RepeatModeller and RepeatMasker tools were used for repeat library building and repeat identification, respectively. The contigs/scaffolds were used to predict SSRs using a MIcroSAtellite (MISA) identification tool (Thiel et al., 2003).
Transcriptome Assembly
High-quality stranded RNA-seq reads were assembled into putative transcripts using Trinity version 2.1.1 (Grabherr et al., 2011). Also, RNA-seq reads were mapped to assembled sandalwood scaffolds using TopHat2 (Kim et al., 2013) and assembled by the Cufflinks suite (Trapnell et al., 2012). Transcripts assembled from Trinity and Cufflinks were used in downstream analyses for gene prediction. lncRNAs were identified with Coding Potential Calculator tools (Kong et al., 2007). Putative microRNAs were identified by performing BLAST searches between noncoding RNAs and plant stem-loop precursor RNA transcripts downloaded from miRBase (http://www.mirbase.org).
Gene Prediction and Functional Annotation
Three iterations of the SNAP (for Semi-HMM-based Nucleic Acid Parser) gene prediction tool were used to build an initial training gene set. Then, genes were predicted using MAKER-P (Campbell et al., 2014) by inputting a SNAPhmm gene model and Arabidopsis (Arabidopsis thaliana) gene model through SNAP and AUGUSTUS as gene prediction tools, respectively. Assembled sandalwood transcripts from the stranded RNA sequencing experiment, the GTF file generated from Cufflinks, and eudicot ESTs retrieved from NCBI all were used as evidence during the MAKER-P gene prediction process. The functional annotation of predicted sandalwood genes was carried out by performing a BLASTP search with the eudicot protein sequences from the UniProt database. The protein domain structures of all protein-coding genes were identified using InterProScan 5 software (Jones et al., 2014).
Extraction of Proteins from Sandalwood Tissues
Proteins were extracted from four sandalwood plant organs or tissues (shoot apical meristem, leaves, stems, and fruits) using the phenol extraction method described earlier (Wu et al., 2014). Briefly, tissues (0.25 g) were ground to fine powder using liquid nitrogen and further homogenized using 10 mL of chilled 10% TCA (v/v) prepared in acetone. Samples were transferred to precooled tubes and centrifuged (12,000g, 5 min) at 4°C. This step was repeated once, and then the pellet was air dried and resuspended in 10 mL of SDS extraction buffer (1% SDS (w/v), 1.5 m Tris-Cl [pH 8.8], 0.1 m DTT, 1 mm EDTA, and 2 mm phenylmethylsulfonyl fluoride (PMSF)), followed by incubation at room temperature for 1 h. Samples were then centrifuged to remove the cell debris (12,000g, 10 min, room temperature). To the supernatant, an equal volume of Tris-buffered phenol (pH 8) was added and mixed for 1 h at room temperature. Centrifugation (12,000g, 30 min, room temperature) was carried out to collect the phenolic phase. To invert the phases, an equal volume of wash buffer (10 mm Tris-Cl [pH 8], 1 mm EDTA, and 0.7 m Suc) was added to the phenolic phase. Proteins were precipitated overnight at −20°C using 5 volumes of 0.1 m ammonium acetate prepared in methanol. Protein pellets were retrieved through centrifugation (12,000g, 10 min) at 4°C. The pellets were washed twice in 80% acetone (v/v) containing 2% β-mercaptoethanol (v/v) and then air dried. Proteins were solubilized in 50 mm Triethylammonium bicarbonate (TEABC) with mild sonication and quantified using a bicinchoninic acid assay (Pierce).
Trypsin Digestion and Peptide Fractionation
One hundred micrograms of proteins from each tissue was subjected to reduction using 5 mm DTT at 60°C for 20 min followed by alkylation with 10 mm iodoacetamide for 10 min at room temperature in the dark. Overnight enzymatic digestion was carried out using trypsin at a ratio of 1:20 (Worthington Biochemical) at 37°C. Fractionation of the tryptic peptides was conducted using basic pH reverse-phase liquid chromatography. The peptide digests were injected using a manual injector onto an XBridge C18, 5 µm, 250- × 4.6-mm column (Waters) connected to a Hitachi (ELITE LaChrom) HPLC system. Using a gradient of 0% to 100% solvent B (10 mm TEABC in 90% acetonitrile, pH 8.5), the peptide digest was resolved in 120 min. Ninety-six fractions collected for each tissue sample were concatenated and pooled into 10 fractions, dried, and desalted using C18 StageTips.
Mass Spectrometry Analysis
The 40 basic pH reverse-phase liquid chromatography fractions (10 from each plant tissue) were subjected to liquid chromatography-tandem mass spectrometry analysis. Mass spectrometry was performed using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) coupled to the Easy-nLC1200 nano-flow UHPLC (Thermo Fisher Scientific). The peptides were reconstituted in 0.1% formic acid and loaded onto a trap column (nanoViper 2 cm, 3-μm C18 Aq; Thermo Fisher Scientific). Peptide separation was accomplished using an EASY-Spray C18 analytical column (15 cm, 75-μm PepMap C18, 2-μm C18 Aq; Thermo Fisher Scientific) set at 35°C. The solvent gradients used for the peptide separation were as follows: linear gradient of 5% to 35% solvent B (80% acetonitrile in 0.1% formic acid) over 90 min with a total run time of 120 min. The flow rate was set at 300 nL min−1. The nanoelectrospray ionization source was used to generate positively charged precursor ions. Data were acquired using a data-dependent acquisition method wherein MS1 survey scans were carried out in 400 to 1,600 mass-to-charge ratio (m/z) range (120,000 mass resolution at 200 m/z). For further peptide fragmentation using tandem mass spectrometry, the most intense precursor ions were selected at top-speed data-dependent mode with a maximum cycle time of 3 s High collision dissociation (HCD) fragmentation; collision energy, 33%; mass resolution, 30,000). Peptide charge state was set to 2 to 6, and dynamic exclusion was set to 30 s, along with an exclusion width of ±20 ppm. Internal calibration was carried out using the lock mass option (m/z 445.1200025) from ambient air.
Generation of Customized Protein Databases from Genomic and Transcriptomic Data
To identify proteins expressed in the different organs and tissues, we used genome assembly, MAKER-P-derived genes, and Trinity-assembled transcripts for the generation of a protein database. To enable the identification of novel peptides in the sandalwood genome, we searched proteomic data against the six-frame translated sandalwood genome, three-frame translated transcripts assembled from RNA-seq, and hypothetical N-terminal peptide database using a unique search workflow on Proteome Discoverer (version 2.1) software (Thermo Fisher Scientific). These peptides were analyzed manually using the Proteogenomics workflow (as described in Supplemental Fig. S2) to identify novel genes missed in the annotation pipeline. The sandalwood genome was translated into six reading frames using in-house Python scripts. The six-frame translation included stop codon-to-stop codon translation of the template sequence. Peptide sequences smaller than seven amino acids were not included in the database. A three-frame translated RNA-seq transcript sequence database was created from Trinity-assembled transcripts. A custom N-terminal tryptic peptides database was created by collecting all the peptide sequences that begin with Met and end with Lys/Arg from MAKER-P-derived proteins with evidence, genome, and RNA-seq transcripts.
Protein Identification Using the Protein Database
Data acquired on the Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) were processed to generate peak list files using Proteome Discoverer software version 2.1 (Thermo Fisher Scientific). The data were first searched against protein databases generated using MAKER-P-derived proteins (proteins with RNA-seq and EST evidence [24,479] and ab initio proteins [13,582]) using Sequest and Mascot search algorithms. Following the first-pass search, other searches were performed sequentially in the following order: (1) against a protein database with RNA-seq and EST evidence and an ab initio protein database; (2) against a six-frame translated sandalwood genome database; (3) against a three-frame translated Trinity-assembled transcripts database; and (4) against an Arabidopsis protein database. The search parameters included trypsin as the proteolytic enzyme with a maximum of two missed cleavages allowed. Oxidation of Met and acetylation of protein at the N terminus was set as a dynamic modification, while carbamidomethylation of Cys was set as a static modification. Precursor and fragment mass tolerance were set to 10 ppm and 0.05 D, respectively. A false discovery rate of 1% was set for the identification at protein, peptide, and Peptide spectral match (PSM) levels. For the quantification of proteins, a label-free quantification method (iBAQ) was followed.
Proteogenomic Analysis
We mapped GSSPs to the sandalwood genome to identify the probable coding regions. Peptides mapping to multiple locations in the genome were not considered for further analysis. The coding potential of a given genomic region was confirmed by the presence of transcript evidence and/or ortholog evidence.
Gene Family Expansion, Core Orthologous Genes, and Mining of Transcription Factors in Sandalwood
Protein sequences were retrieved for Populus trichocarpa (ftp://ftp.ensemblgenomes.org/pub/plants/release-29/fasta/populus_trichocarpa/pep/), Citrus clementina (https://www.citrusgenomedb.org/species/clementina/genome1.0), Vitis vinifera (ftp://ftp.ensemblgenomes.org/pub/plants/release-29/fasta/vitis_vinifera/pep/), and Arabidopsis (ftp://ftp.ensemblgenomes.org/pub/plants/release-29/fasta/arabidopsis_thaliana/pep/). The paralogs and orthologs among these plant species were identified using OrthoVenn (Wang et al., 2015). Groups having at least one copy of the gene from each genome were considered as core orthologous groups. Based on orthologous clustering, randomly chosen protein sequences of 100 single-copy ortholog gene groups from five species were used to infer a phylogenetic relationship. The transcription factors in the sandalwood genome were searched against the plant transcription factor database (version 4.0; http://planttfdb.cbi.pku.edu.cn/index.php).
Identification of Sandalwood Essential Oil Pathway Genes
Sandalwood essential oil typically contains 90% santalols and α-santalol. Genes involved in the biosynthesis of these constituents were identified through pathway analysis and Pfam domain annotation of sandalwood genes predicted by a gene prediction algorithm. The pathway analysis of genes was carried out by the KAAS server (Moriya et al., 2007) using most plants as reference organisms. The cloned genes of santalol biosynthesis were retrieved from the NCBI database, and BLASTP was performed to identify homologs in our study. Phylogenetic relationships were inferred among cytochrome P450 Pfam domain-containing proteins using PhyML3.0 after multiple sequence alignments using MUSCLE (Edgar, 2004; Guindon et al., 2010; Lefort et al., 2017). The phylogenetic tree was drawn using FigTree version 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Accession Numbers
The whole-genome assembly is available at NCBI/DDBJ/EMBL with the accession identifier LOCJ00000000. The version described in this article is LOCJ01000000. The raw sequence reads (PE SRR5150443 and MP SRR5150442) of whole-genome and RNA sequencing (SRR5150444) are deposited in the NCBI SRA database with the accession number SRP096167. The mass spectrometry-based proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD006283.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Analysis of cellular DNA content of sandalwood tree using flow cytometry.
Supplemental Figure S2. Flow chart showing the integrative omics approach followed in sandalwood genome assembly and annotation.
Supplemental Figure S3. Phylogenetic relationship of genes identified in sandalwood encoding for geranyl geranyl diphosphate, farnesyl diphosphate, and santalene synthase.
Supplemental Figure S4. Phylogenetic relationship of cytochrome P450 proteins identified in the sandalwood genome.
Supplemental Table S1. Summary of peptides and proteins identified from protein database search.
Supplemental Table S2. Summary of results from proteogenomics analysis.
Supplemental Table S3. List of chloroplast genes annotated in sandalwood and their corresponding homologs in Arabidopsis.
Supplemental Table S4. List of mitochondrial genes annotated in sandalwood and their corresponding homologs in Arabidopsis.
Supplemental Table S5. List of core eukaryotic genes conserved in sandalwood and their expression pattern.
Supplemental Table S6. List of transcription factors identified in the sandalwood proteome.
Supplemental Table S7. Long noncoding transcripts with proteome and RNA-seq evidence.
Supplemental Table S8. Repeat content in the sandalwood genome.
Supplemental Table S9. SSRs predicted in the sandalwood genome.
Supplemental Table S10. Overall distribution of all types of SSRs in the sandalwood genome.
Supplemental Text S1. Nucleotide sequences of genes involved in the santalol biosynthesis pathway identified in the sandalwood genome.
Supplemental Text S2. Putative sandalwood lncRNAs homologous to stem-loop microRNA in miRBase.
Supplemental Text S3. Partial list of genome sequences and annotation pipelines used for various plants and other organisms.
Acknowledgments
We thank the Next Generation Genomics Facility at the Centre for Cellular and Molecular Platforms for sequencing. We thank Yenepoya University for access to the mass spectrometry instrumentation facility.
Footnotes
We thank Karnataka Biotechnology and Information Technology Services (KBITS), Government of Karnataka, for the support to the Center for Systems Biology and Molecular Medicine at Yenepoya University under the Biotechnology Skill Enhancement Program in Multiomics Technology (BiSEP GO ITD 02 MDA 2017). P.S. is funded by the Early Career Research Award from the Science & Engineering Research Board (SERB), Government of India (award no. ECR/2016/000365). J.A. is a recipient of a Senior Research Fellowship from the Council of Scientific & Industrial Research (CSIR), Government of India. O.C. is a recipient of an INSPIRE Fellowship from the Department of Science and Technology (DST), Government of India. S.M.P. is a DST-INSPIRE Faculty (award no. LSBM-138/2015), DST, Government of India.
References
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O. (2012) Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family. Plant Physiol 160: 1597–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Smith JB (1976) Nuclear DNA amounts in angiosperms. Philos Trans R Soc Lond B Biol Sci 274: 227–274 [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W (2011) Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27: 578–579 [DOI] [PubMed] [Google Scholar]
- Campbell MS, Law M, Holt C, Stein JC, Moghe GD, Hufnagel DE, Lei J, Achawanantakun R, Jiao D, Lawrence CJ, et al. (2014) MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol 164: 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana NE, Payne SH, Shen Z, Stanke M, Bafna V, Briggs SP (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc Natl Acad Sci USA 105: 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana NE, Shen Z, He Y, Walley JW, Cassidy CJ, Briggs SP, Bafna V (2014) An automated proteogenomic method uses mass spectrometry to reveal novel genes in Zea mays. Mol Cell Proteomics 13: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celedon JM, Chiang A, Yuen MM, Diaz-Chavez ML, Madilao LL, Finnegan PM, Barbour EL, Bohlmann J (2016) Heartwood-specific transcriptome and metabolite signatures of tropical sandalwood (Santalum album) reveal the final step of (Z)-santalol fragrance biosynthesis. Plant J 86: 289–299 [DOI] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Clavijo BJ, Venturini L, Schudoma C, Accinelli GG, Kaithakottil G, Wright J, Borrill P, Kettleborough G, Heavens D, Chapman H, et al. (2017) An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res 27: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creevey CJ, Muller J, Doerks T, Thompson JD, Arendt D, Bork P (2011) Identifying single copy orthologs in metazoa. PLOS Comput Biol 7: e1002269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Chavez ML, Moniodis J, Madilao LL, Jancsik S, Keeling CI, Barbour EL, Ghisalberti EL, Plummer JA, Jones CG, Bohlmann J (2013) Biosynthesis of sandalwood oil: Santalum album CYP76F cytochromes P450 produce santalols and bergamotol. PLoS ONE 8: e75053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Lan T (2016) Functional characterization of the late embryogenesis abundant (LEA) protein gene family from Pinus tabuliformis (Pinaceae) in Escherichia coli. Sci Rep 6: 19467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Gupta S, Wardhan V, Kumar A, Rathi D, Pandey A, Chakraborty S, Chakraborty N (2015) Secretome analysis of chickpea reveals dynamic extracellular remodeling and identifies a Bet v1-like protein, CaRRP1 that participates in stress response. Sci Rep 5: 18427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Wardhan V, Verma S, Gayali S, Rajamani U, Datta A, Chakraborty S, Chakraborty N (2011) Characterization of the secretome of chickpea suspension culture reveals pathway abundance and the expected and unexpected secreted proteins. J Proteome Res 10: 5006–5015 [DOI] [PubMed] [Google Scholar]
- Harbaugh DT, Baldwin BG (2007) Phylogeny and biogeography of the sandalwoods (Santalum, Santalaceae): repeated dispersals throughout the Pacific. Am J Bot 94: 1028–1040 [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. (2012) GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res 22: 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmy M, Tomita M, Ishihama Y (2011) OryzaPG-DB: rice proteome database based on shotgun proteogenomics. BMC Plant Biol 11: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Greene EA, Pietrokovski S, Bork P, Attwood TK, Hood L (1997) Gene families: the taxonomy of protein paralogs and chimeras. Science 278: 609–614 [DOI] [PubMed] [Google Scholar]
- Hofmann NR. (2016) A structure for plant-specific transcription factors: the GRAS domain revealed. Plant Cell 28: 993–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45: D1040–D1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CG, Moniodis J, Zulak KG, Scaffidi A, Plummer JA, Ghisalberti EL, Barbour EL, Bohlmann J (2011) Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS)-a and TPS-b subfamilies, including santalene synthases. J Biol Chem 286: 17445–17454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. (2014) A draft map of the human proteome. Nature 509: 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole C. (2011) Wild Crop Relatives: Genomic and Breeding Resources: Forest Trees. Berlin/Heidelberg, Germany: Springer Science & Business Media [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: W345–W349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishan A. (1975) Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol 66: 188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni H, Srimathi R (1982) Variation in foliar characteristics in sandal. In Khosla PK, ed, Biometric Analysis in Improvement of Forest Biomass. International Book Distributors, Dehra Dun, India, pp 63–69 [Google Scholar]
- Kumar AA, Joshi G, Ram HM (2012) Sandalwood: history, uses, present status and the future. Curr Sci 103: 1408–1416 [Google Scholar]
- Lefort V, Longueville JE, Gascuel O (2017) SMS: Smart Model Selection in PhyML. Mol Biol Evol 34: 2422–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markelz RC, Covington MF, Brock MT, Devisetty UK, Kliebenstein DJ, Weinig C, Maloof JN (2017) Using RNA-seq for genomic scaffold placement, correcting assemblies, and genetic map creation in a common Brassica rapa mapping population. G3 (Bethesda) 7: 2259–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniodis J, Jones CG, Barbour EL, Plummer JA, Ghisalberti EL, Bohlmann J (2015) The transcriptome of sesquiterpenoid biosynthesis in heartwood xylem of Western Australian sandalwood (Santalum spicatum). Phytochemistry 113: 79–86 [DOI] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35: W182–W185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian AR, Garey JR, Martin J, Liu LX (1998) Large-scale taxonomic profiling of eukaryotic model organisms: a comparison of orthologous proteins encoded by the human, fly, nematode, and yeast genomes. Genome Res 8: 590–598 [DOI] [PubMed] [Google Scholar]
- Nambiar V. (1993) Indian Medicinal Plants: A Compendium of 500 Species, Vol 5 India: Orient Blackswan [Google Scholar]
- Padmanabhan V, Dias DM, Newton RJ (1997) Expression analysis of a gene family in loblolly pine (Pinus taeda L.) induced by water deficit stress. Plant Mol Biol 35: 801–807 [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I (2007) CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23: 1061–1067 [DOI] [PubMed] [Google Scholar]
- Patel DM, Fougat RS, Sakure AA, Kumar S, Kumar M, Mistry JG (2016) Detection of genetic variation in sandalwood using various DNA markers. 3 Biotech 6: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TSK, Mohanty AK, Kumar M, Sreenivasamurthy SK, Dey G, Nirujogi RS, Pinto SM, Madugundu AK, Patil AH, Advani J, et al. (2017) Integrating transcriptomic and proteomic data for accurate assembly and annotation of genomes. Genome Res 27: 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MN, Ganeshaiah K, Shaanker RU (2007) Assessing threats and mapping sandal (Santalum album L.) resources in peninsular India: identification of genetic hot-spot for in-situ conservation. Conserv Genet 8: 925–935 [Google Scholar]
- Rawat V, Abdelsamad A, Pietzenuk B, Seymour DK, Koenig D, Weigel D, Pecinka A, Schneeberger K (2015) Improving the annotation of Arabidopsis lyrata using RNA-Seq data. PLoS ONE 10: e0137391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Jiang L, Zhang L, Kang R, Yu Z (2014) Dynamic changes in proteins during apple (Malus x domestica) fruit ripening and storage. Hortic Res 1: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212 [DOI] [PubMed] [Google Scholar]
- Srivastava PL, Daramwar PP, Krithika R, Pandreka A, Shankar SS, Thulasiram HV (2015) Functional characterization of novel sesquiterpene synthases from Indian sandalwood, Santalum album. Sci Rep 5: 10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steijger T, Abril JF, Engström PG, Kokocinski F, Hubbard TJ, Guigó R, Harrow J, Bertone P (2013) Assessment of transcript reconstruction methods for RNA-seq. Nat Methods 10: 1177–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Takahashi S, Kondo T, Dohra H, Ito Y, Kiriiwa Y, Hayashi M, Kamiya S, Kato M, Fujiwara M, et al. (2015) Plastid proteomic analysis in tomato fruit development. PLoS ONE 10: e0137266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106: 411–422 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK, Schlueter JA, Donoghue MT, Azam S, Fan G, Whaley AM, et al. (2011) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol 30: 83–89 [DOI] [PubMed] [Google Scholar]
- Wang B, Tseng E, Regulski M, Clark TA, Hon T, Jiao Y, Lu Z, Olson A, Stein JC, Ware D (2016) Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun 7: 11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Coleman-Derr D, Chen G, Gu YQ (2015) OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 43: W78–W84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe KA, Homayouni A, Tufano T, Lopez J, Ringler P, Rushton P, Shen QJ (2015) Tiling Assembly: a new tool for reference annotation-independent transcript assembly and novel gene identification by RNA-sequencing. DNA Res 22: 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509: 582–587 [DOI] [PubMed] [Google Scholar]
- Wu X, Xiong E, Wang W, Scali M, Cresti M (2014) Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat Protoc 9: 362–374 [DOI] [PubMed] [Google Scholar]
- Yan J, Wang J, Zhang H (2002) An ankyrin repeat-containing protein plays a role in both disease resistance and antioxidation metabolism. Plant J 29: 193–202 [DOI] [PubMed] [Google Scholar]
- Zhang X, Berkowitz O, Teixeira da Silva JA, Zhang M, Ma G, Whelan J, Duan J (2015) RNA-Seq analysis identifies key genes associated with haustorial development in the root hemiparasite Santalum album. Front Plant Sci 6: 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Teixeira da Silva JA, Niu M, Li M, He C, Zhao J, Zeng S, Duan J, Ma G (2017) Physiological and transcriptomic analyses reveal a response mechanism to cold stress in Santalum album L. leaves. Sci Rep 7: 42165. [DOI] [PMC free article] [PubMed] [Google Scholar]