A synthetic microProtein approach unravels modes of posttranslational regulation of a wide range of protein families in Arabidopsis.
Abstract
MicroProteins are small, single-domain proteins that regulate multidomain proteins by sequestering them into novel, often nonproductive, complexes. Several microProteins have been identified in plants and animals, most of which negatively regulate transcription factors. MicroProtein candidates that potentially target a wide range of different protein classes were recently identified in a computational approach. Here, we classified all Arabidopsis (Arabidopsis thaliana) microProteins and developed a synthetic microProtein approach to target specific protein classes, such as hydrolases, receptors, and lyases, in a proof-of-concept approach. Our findings reveal that microProteins can be used to influence different physiological processes, which makes them useful tools for posttranslational regulation in plants and potentially also in animals.
The demand for crop-based products is increasing proportional to the increase in population. Major goals of modern agriculture are optimization and improvement of crops to sustain high productivity with minimal environmental impact. A variety of molecular methods have been developed for regulation of trait-specific genes at the transcriptional and translational level. RNA interference is a posttranscriptional method for targeted gene knockdown; however, off-target effects and transgenerational stability are major disadvantages of this method (Frizzi and Huang, 2010). Genome-engineering technologies, such as those involving zinc-finger nucleases, transcription activator-like effector nucleases, and CRISPR-Cas9, have been established that enable site-specific genome modifications (Gaj et al., 2013). Disadvantages of the genome-engineering technologies are also off-target effects and the complete loss of gene function, which could be detrimental or lethal to the organism. Targeted posttranslational modification for trait regulation may overcome these problems by precisely fine-tuning protein activity without altering overall plant structure and metabolism.
Recently identified small proteins termed microProteins regulate larger multidomain proteins at the posttranslational level (Staudt and Wenkel, 2011; Graeff and Wenkel, 2012; Eguen et al., 2015). MicroProteins are usually single-domain proteins that contain a protein-protein interaction domain. They have the ability to disturb the formation of protein complexes by sequestering their target proteins. In recent years, several microProteins have been identified in plants (Staudt and Wenkel, 2011; Graeff and Wenkel, 2012; Eguen et al., 2015). LITTLE ZIPPERs (ZPRs) were the first-described plant-specific microProteins. ZPRs are short proteins harboring a single Leu zipper (ZIP) domain that enables protein-protein interaction with class III homeodomain-ZIP (HD-ZIPIII) transcription factors. HD-ZIPIIIs are involved in the regulation of leaf polarity as well as shoot and vascular development. The ZPR microProteins interfere with the homodimerization of HD-ZIPIII transcription factors, and the resulting ZPR/HD-ZIPIII heterodimer cannot bind to the HD-ZIPIII cis-element to regulate gene expression (Wenkel et al., 2007; Kim et al., 2008).
So far, all characterized plant-specific microProteins are involved in the regulation of transcription factors (Eguen et al., 2015). Furthermore, it has been shown that transcription factors can be regulated by overexpression of their respective protein-protein interaction domain. SUPPRESSOR OF OVEREXPRESSOR OF CONSTANS1, AGAMOUS, and LATE ELONGATED HYPOCOTYL in Arabidopsis (Arabidopsis thaliana) and Brachypodium distachyon can be negatively regulated by overexpressing their respective protein-protein interaction domains (Seo et al., 2012). It is assumed that overexpression of these protein-protein interaction domains engages the full-length proteins into nonfunctional heterodimers. Transgenic plants ectopically expressing the protein-protein interaction domains thus resemble the loss-of-function mutants of the full-length proteins (Seo et al., 2012).
All identified plant-specific microProteins are known to regulate transcription factors, but recent findings imply that microProteins have a wider range of activities and can regulate proteins belonging to a wide range of protein families. The computational program miPFinder can identify and classify potential microProteins and their ancestors for all protein classes in any sequenced genome (Straub and Wenkel, 2017). Here, we analyzed the Arabidopsis genome with miPFinder and identified nontranscription factor microProteins that potentially regulate a wide range of different proteins. The analysis of these microProteins and the proteins they might regulate revealed that the majority of these proteins has not yet been studied. This greatly hampers in-depth studies on the respective functions. To overcome this, we targeted three well-characterized multidomain proteins belonging to three different protein classes using a synthetic microProtein approach. Phenotypes produced by ectopic overexpression of the respective protein-protein interaction domains suggest that the functions of the full-length proteins are altered. The synthetic microProteins interact with the multidomain proteins and change their biological activity. Our findings demonstrate that the microProtein concept extends beyond the regulation of transcription factors, and microProtein-mediated protein regulation might therefore be more widespread than previously anticipated.
RESULTS AND DISCUSSION
Potential MicroProteins Targeting Diverse Multidomain Proteins
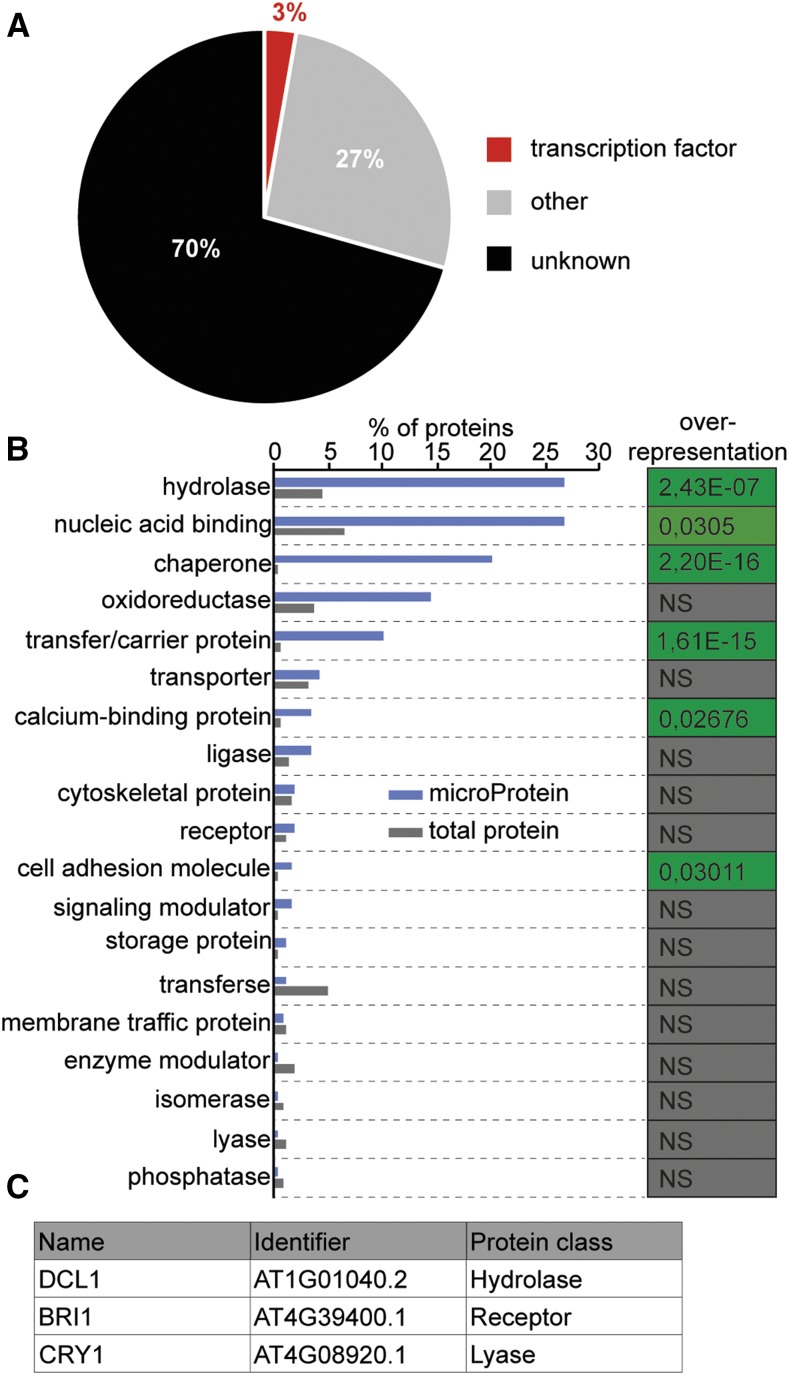
MicroProteins are characterized by their small size (<20 kD) and the presence of a single protein domain, usually a protein-protein interaction domain, that is similar or compatible to a domain of a multidomain protein. MicroProteins often act by sequestering their target proteins into multimeric protein complexes. Using the miPFinder program (Straub and Wenkel, 2017), we identified 912 microProtein candidates related to a wide range of multidomain proteins in Arabidopsis that are also conserved in at least one other plant genome. The identified microProtein candidates were clustered into 188 protein classes dependent on their putative ancestor and protein-protein interaction domain (Fig. 1A and Supplemental Table S1). However, about 70% of these microProtein candidates and their putative ancestors were proteins with unknown function. Approximately 3% of the total represents microProtein candidates, which may regulate transcription factors, including all currently known microProteins. The remaining 27% represent microProtein candidates and their putative targets with known biological activity or known protein-protein interaction domains. These 323 potential microProteins were grouped into the 19 remaining parental classes and analyzed in relation to the total amount of proteins in the respective protein class (Fig. 1B). The microProtein candidates of the hydrolase, nucleic acid binding, chaperone, transfer/carrier protein, and cell adhesion molecule protein classes were significantly overrepresented compared with the total amount of proteins. These findings indicate that microProteins might be involved in the regulation of a wide range of protein classes. Many microProteins are part of a family with two or more microProteins, such as the ZPR microProteins. Bioinformatic analyses revealed that most microProtein candidates are part of a microProtein family. In total, 831 microProtein candidates clustered into 107 microProtein families, while 81 microProtein candidates were single-copy microProteins. However, most of the microProtein candidates and their putative targets have not been investigated yet, and thus, no evidence of their function is available. Due to this, it is difficult to analyze such microProtein candidates.
Figure 1.
Classification of putative microProtein targets/ancestors. A, Overview of conserved microProtein candidates in Arabidopsis and the protein class of their putative ancestors. B, Percentage of nontranscription factor-related microProtein candidates and the protein class of their putative ancestors. Fisher’s exact test was used to test for significant overrepresentation of microProtein candidates (P < 0.05). NS, Not significant. C, Selected targets of synthetic microProteins representing different protein classes.
To determine whether microProteins can regulate a wide range of protein classes, we developed a synthetic microProtein approach. We selected DICER-LIKE1 (DCL1), BRASSINOSTEROID INSENSITIVE1 (BRI1), and CRYPTOCHROME1 (CRY1; Fig. 1C) as representative multidomain proteins from three different protein classes and examined if synthetic microProteins could regulate their activity. Proteins belonging to the hydrolase class of proteins were the largest group of proteins potentially regulated by microProteins (Fig. 1B). The identified microProtein candidates were significantly overrepresented in the hydrolase class in comparison to the total amount of proteins. The hydrolase class includes, among others, proteases, phosphatases, nucleases, and helicases. In general terms, hydrolases are enzymes that catalyze the splitting of water and thereby create novel functional groups. Hydrolases are involved in almost all major cellular processes, such as DNA replication, posttranslational modification, and photosynthesis. Because none of the potential microProteins in the hydrolase group has been studied in depth, we decided to design synthetic microProteins targeting a well-characterized multidomain protein belonging to the hydrolase class. We decided on DCL1 as a well-studied representative helicase involved in microRNA (miRNA) biogenesis. The second protein class we targeted was the class of receptor proteins. Receptors play integral roles in the perception and transduction of signals and can be divided into transmembrane, hormone, intracellular, or ionotropic receptors. As a proof of principle, we developed synthetic microProteins that interfere with the BRI1 receptor, which is involved in brassinosteroid (BR) signal transduction. We found only a single microProtein candidate, which is related to lyases, and we selected CRY1 as a representative for this family. The lyase family includes, among others, photolyases, citrate lyases, and carboxylases. These enzymes break chemical bonds of compounds to generate two products. The blue light receptor CRY1 is a well-characterized multidomain protein of the lyase protein class due to the presence of a photolyase domain.
Expression of the PAZ Domain of DCL1 Alters MiRNA Biogenesis
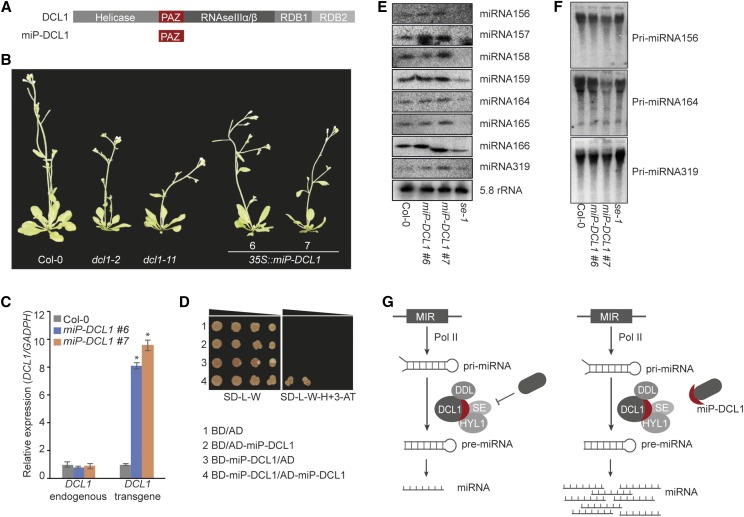
A total of 72 microProtein candidates were related to the hydrolase class, which makes it the class with the highest abundance of microProtein candidates. Members of this protein class are involved in diverse biological processes. DCL1 was selected as a representative of the hydrolase protein class. DCL1 is a type III RNase with a helicase domain that carries out primary miRNA (pri-miRNA) processing in plants and therefore plays an important role in plant development (Schauer et al., 2002; Voinnet, 2009). MiRNAs are 19- to 22-nucleotide-long RNAs involved in the regulation of gene expression in eukaryotes (Reinhart et al., 2002; Voinnet, 2009). MiRNAs are encoded by MICRORNA genes, which are transcribed into pri-miRNA by RNA polymerase II (Lee et al., 2004). Pri-miRNAs form hairpin-like secondary structures that are further processed into precursor miRNAs (premiRNAs). The processing of pri-miRNAs into premiRNAs is catalyzed by a complex composed of DCL1 and its functional partners HYPONASTIC LEAVES1 (HYL1), SERRATE (SE), and DAWDLE (DDL; Hiraguri et al., 2005; Kurihara et al., 2006; Dong et al., 2008; Yu et al., 2008; Machida et al., 2011; Liu et al., 2012). The PIWI/ARGONAUTE/ZWILLE (PAZ) domain of DCL1 mediates the interaction with SE and DDL (Yu et al., 2008; Machida et al., 2011).
We hypothesized that we could disrupt the complex formation of DCL1, HYL1, SE, and DDL by overexpressing the synthetic microProtein designed from the PAZ domain of DCL1 (Fig. 2A). As a consequence, the biogenesis of miRNAs should be affected. To verify this hypothesis, we generated transgenic plants that ectopically express the DCL1 PAZ domain (miP-DCL1). We obtained 10 individual transgenic 35S::miP-DCL1 plants (Supplemental Fig. S1), of which three showed multiple developmental defects such as abnormally shaped cotyledons, different leaf shapes, smaller rosettes, and floral defects, while the other seven more closely resembled the wild type. The dcl1 mutants have pleiotropic developmental phenotypes ranging from mild phenotypes, such as altered leaf shape and delayed floral transition, to defects in embryo development, which are often lethal (Schauer et al., 2002). For comparison, we selected two weak dcl1 mutant alleles, dcl1-2 and dcl1-11, and two independent transgenic miP-DCL1 lines for further experiments. The two independent 35S::miP-DCL1 lines showed differences in leaf shape, decreased rosette size, and defects in leaf positioning compared with wild-type Columbia (Col-0) plants. In addition, compared with Col-0 wild-type plants, they showed an overall reduction in size and resembled dcl1 mutant plants (Fig. 2B).
Figure 2.
Alteration of miRNA biogenesis by miP-DCL1. A, Domain architectures of DCL1 and miP-DCL1. RDB, RNA-binding domain. B, Images of two representative independent 35S::miP-DCL1 lines. Phenotypes were compared with Col-0 and two DCL1 mutants, dcl1-2 and dcl1-11. C, RT-qPCR results showing expression of endogenous DCL1 and transgene miP-DCL1 in 35S::miP-DCL1 lines compared with Col-0. Experimental replicates were statistically tested using a Student’s t test (*P < 0.05). D, The ability of the PAZ domain to homodimerize was tested using a yeast two-hybrid assay. Normal yeast growth was observed for the serial dilutions on nonselective SD medium lacking Leu and Trp (SD-L-W). Only positive interactors grew on selective SD medium lacking Leu, Trp, and His (SD-L-W-H) supplemented with 2.5 mm 3-aminotriazole (3-AT). E, Small RNA blot analyses display the levels of miRNAs in 35S::miP-DCL1 lines compared with Col-0 and se-1 using 5.8S rRNA as a loading control. F, Levels of pri-miRNAs in 35S::miP-DCL1 lines compared with Col-0 and se-1. G, A model depicting miP-DCL1 function. DCL1 forms a complex with HYL1, SE, and DDL to process miRNA. The complex may be regulated by an unknown suppressor protein. MiP-DCL1 enhances the DCL1 complex function by sequestering the complex inhibitor in a nonfunctional dimer.
The expression levels of endogenous DCL1 and miP-DCL1 were examined to verify that the observed phenotypes were caused by ectopic expression of miP-DCL1 (Fig. 2C). We analyzed miP-DCL1 expression using primers that amplified the region encoding the PAZ domain. Both independent transgenic lines showed significantly (P < 0.05, t test) higher expression levels of the miP-DCL1 transgene compared with wild-type Col-0 plants. Using primers that amplify fragments outside the region encoding the PAZ domain, we further showed that the endogenous expression level of DCL1 was not affected by miP-DCL1 expression, thus excluding the possibility of cosuppression. To verify the hypothesis that miP-DCL1 regulates DCL1 by sequestering it and thereby preventing it from forming a complex with its functional partners, a yeast two-hybrid test was performed. To prevent interference of the natural yeast DICER with the Arabidopsis DCL1, we decided to test only the ability of the PAZ domain to homodimerize. Therefore, the PAZ domain protein was fused to the GAL4 activation domain and to the GAL4 binding domain. To exclude autoactivation of the proteins, the interaction was tested in the presence of 2.5 mm 3-aminotriazole, a competitive inhibitor of HIS3 protein, and additionally, all constructs were tested for false-positive interaction with the empty vectors. The yeast two-hybrid assay revealed that the PAZ domain acts as a protein-protein interaction domain and is able to form homodimers (Fig. 2D). This result suggests that miP-DCL1 may sequester DCL1 into a nonfunctional complex.
Recent work shows that loss-of-function mutations in DCL1, HYL1, SE, and DDL result in pri-miRNA accumulation and reduced levels of mature miRNAs (Han et al., 2004; Kurihara and Watanabe, 2004; Kurihara et al., 2006; Lobbes et al., 2006; Yang et al., 2006; Yu et al., 2008). Therefore, we examined the abundance of three pri-miRNAs and several mature miRNAs using northern blot analysis in Col-0, se-1, and the two independent 35S::miP-DCL1 lines. Both 35S::miP-DCL1 lines surprisingly showed higher amounts of miRNAs and inversely correlated levels of pri-miRNAs compared with Col-0. The opposite effect was observed in the se-1 mutant, which showed an accumulation of pri-miRNAs and lower levels of miRNAs (Fig. 2, E and F). These results suggest that miP-DCL1 acts as an activator of DCL1 and enhances miRNA processing. MicroProteins can interact with larger proteins that have a similar or compatible protein-protein interaction domain. Our findings suggest that miP-DCL1 may sequester a yet unknown inhibitor of DCL1, which has a compatible protein-protein interaction domain, and as a consequence increases the activity of DCL1 (Fig. 2G).
BRI1 Activity Can Be Controlled through the Transmembrane-Juxtamembrane Domain of BRI1
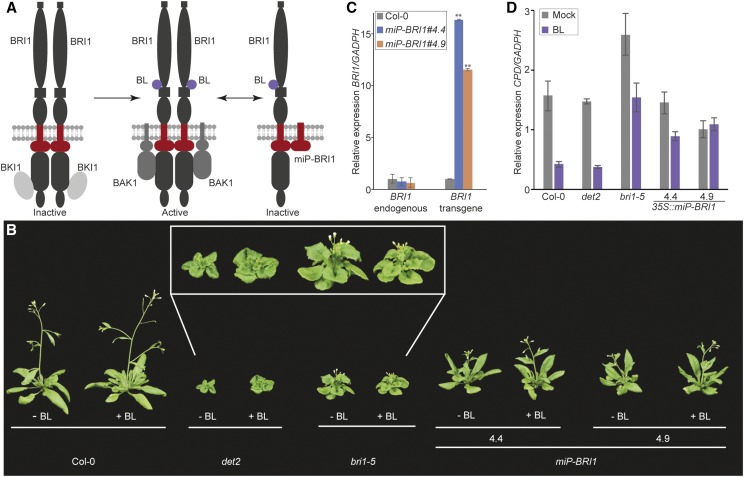
BRI1 is the major component of the BR signal transduction pathway in plants. BRI1 contains extracellular Leu-rich repeats, an island domain, and a transmembrane domain as well as intracellular parts consisting of a juxtamembrane region, a kinase domain, and a C-terminal region (Kim and Wang, 2010). BRI1 forms a ligand-independent homodimer and can interact with other proteins (Fig. 3A; Russinova et al., 2004; Wang et al., 2005). The BRI1 KINASE INHIBITOR1 (BKI1) maintains BRI1 in an inactive form through heterodimerization at the kinase domain and blocks the interaction with the coreceptor BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1). Binding of the hormone BR to the extracellular BRI1 region causes basal activation of the BRI1 kinase and dissociation of BKI1 from BRI1 (Wang and Chory, 2006). Full activation of BRI1 occurs through the interaction of BAK1 with BRI1 and their transphosphorylation (Li et al., 2002; Nam and Li, 2002). The activated BRI1 induces BR-targeted gene expression (Kim and Wang, 2010). Defects in BRI1 result in a dwarf phenotype and BR insensitivity (Clouse et al., 1996). Thus, exogenously applied BR cannot rescue the bri1 mutant phenotype. Mutants with defects in BR biosynthesis show similar dwarf phenotypes, such as DE-ETIOLATED2 (DET2), but here, the dwarf phenotype can be rescued by exogenous BR application (Fujioka et al., 1997).
Figure 3.
Inhibition of BRI1 by the synthetic microProtein miP-BRI1. A, Model of BRI1 activation and miP-BRI1 regulation. The BRI1 homodimer is inactivated by BKI1. BL binding activates BRI1 and causes dissociation of BKI1 and association of BAK1. The synthetic microProtein miP-BRI1 prevents BRI1 activation during BL binding. B, Image of representative Col-0, bri1-5, det2, and two 35S::miP-BRI1 lines treated with mock (−BL) or 100 nm BL (+BL). C, RT-qPCR showing expression of endogenous BRI1 and transgene miP-BRI1 in 35S::miP-BRI1 lines compared with Col-0. Experimental replicates were statistically tested using a Student’s t test (**P < 0.01). D, Altered CPD expression in Col-0, det2, bri1-5, and the two 35S::miP-BRI1 lines in response to 100 nm exogenous BL.
In this context, we hypothesized that a synthetic microProtein that tethers BRI1 to a nonproductive heterodimer will prevent BAK1 interaction after the basal activation of BRI1 (Fig. 3A). We designed a synthetic microProtein from the JM domain up to and including the transmembrane domain to ensure that the synthetic microProtein is able to anchor to the plasma membrane where BRI1 is localized. The synthetic microProtein was termed “miP-BRI1” and ectopically expressed in Arabidopsis from a CaMV35S promoter. Ten out of the 16 transgenic plants revealed a severe dwarf phenotype in the first generation, comparable to the dwarf phenotypes observed in bri1 mutant plants. Segregation of the 10 independent transgenic lines was examined in the next generation, but in this generation, most of the lines showed a milder dwarf phenotype that was likely caused by a reduced transgene activity as a result of transgene silencing. Two 35S::miP-BRI1 lines that showed a dwarf phenotype were selected for further experiments. These 35S::miP-BRI1 lines showed an intermediate dwarf phenotype relative to Col-0 wild type and the bri1-5 mutant plants (Fig. 3B). To verify that 35S::miP-BRI1 lines are BR insensitive, like bri1 mutant plants, the transgenic lines were grown on soil and treated once a day with 100 nm epibrassinolide (BL). Wild-type Col-0, the bri1-5 mutant, and the BR-sensitive det2 mutant were used as controls. The two 35S::miP-BRI1 lines were insensitive to exogenous BL application, like Col-0 and bri1-5, while BL-treated det2 mutant plants, as expected, showed larger rosettes and elongated petioles compared with mock-treated plants (Fig. 3B).
Expression levels of miP-BRI1 and the endogenous BRI1 gene were analyzed the same way as miP-DCL1. The 35S::miP-BRI1 lines showed significantly (P < 0.01, t test) higher expression of the transgene compared with wild-type Col-0, and the synthetic microProtein did not affect endogenous BRI expression (Fig. 2C).
Exogenous BL treatment alters expression of a large number of genes in plants. For example, CPD, which is involved in BR biosynthesis, is strongly down-regulated in Col-0 upon BL treatment. However, due to the BR insensitivity of bri1 mutants, down-regulation of CPD is less pronounced (Clouse et al., 1996). To substantiate the altered response to exogenous BL, we investigated CPD expression in BL-treated 35S::miP-BRI1 lines, the bri1-5 mutant, and Col-0. As expected, CPD expression was reduced by 75% in BL-treated Col-0 and det2 plants compared with CPD expression in mock-treated plants. In BL-treated bri1-5 mutant plants, CPD expression was reduced to a much lesser extent (approximately 40%). Both 35S::miP-BRI1 transgenic plants resembled bri1-5 mutants but showed even less BL sensitivity with a 30% reduction or less in CPD expression (Fig. 3D).
We conclude that miP-BRI1 functions as a synthetic microProtein. Plants ectopically expressing miP-BRI1 show a mild dwarf phenotype and alteration in the expression of CPD in response to BL application. Thus, miP-BRI1 influences BRI1-mediated signal transduction.
The N-Terminal Photolyase Subdomain of CRY1 Can Inhibit CRY1 Function
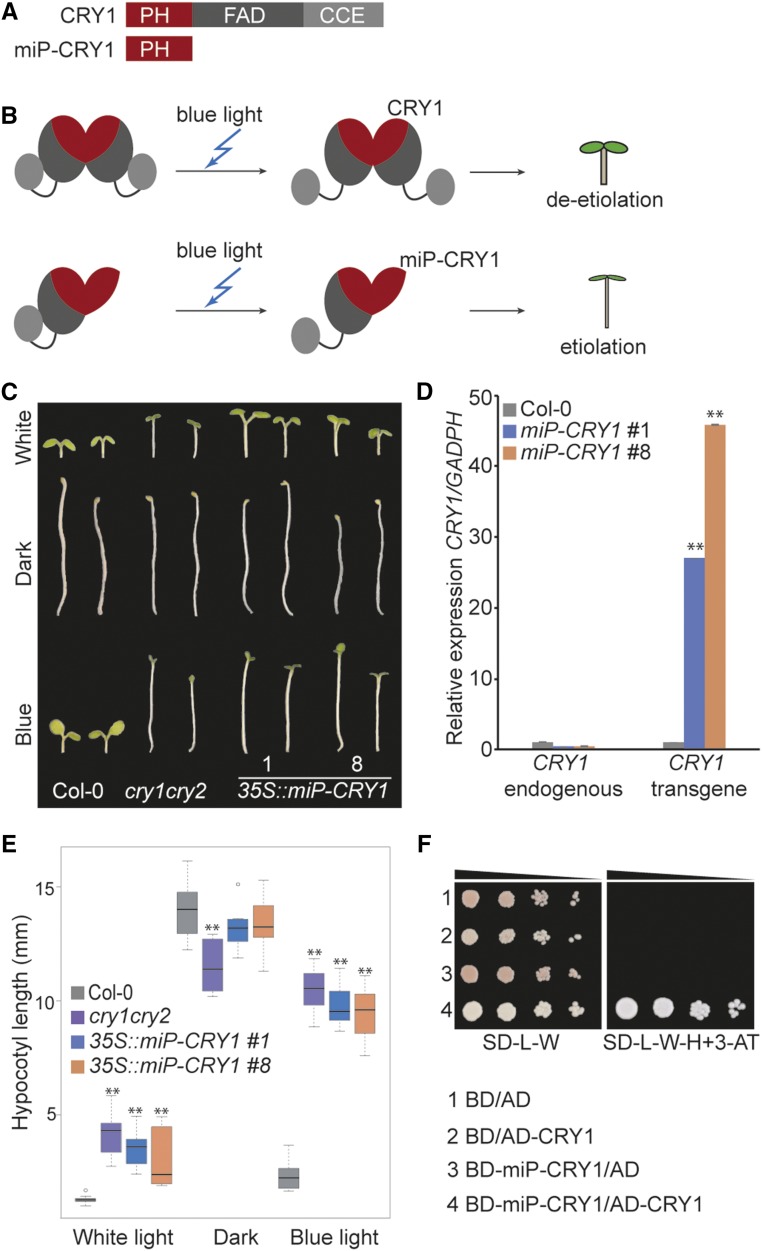
CRY1 belongs to the flavoprotein photoreceptor family found in plants and animals (Cashmore et al., 1999; Chaves et al., 2011). In plants, CRY1 and its homolog, CRY2, mediate blue light-induced responses, such as de-etiolation and photoperiodic flowering (Ahmad and Cashmore, 1993; Guo et al., 1998; Lin et al., 1998; El-Din El-Assal et al., 2001). Both CRY1 and CRY2 need to form homodimers to execute their photoreceptor activity (Sang et al., 2005; He et al., 2015). CRY1 and CRY2 monomers consist of an N-terminal photolyase homologous region domain, which is further divided in two subdomains, the N-terminal photolyase (PH) subdomain and the C-terminal FAD-binding subdomain, and a CRY C-terminal extension (CCE) domain (Fig. 4A; Liu et al., 2016). The CCE domain mediates signaling, the FAD-binding subdomain senses blue light (Yang et al., 2000), and the PH subdomain participates in homodimerization (Lin et al., 1995; Sang et al., 2005). CRY1 functions in the de-etiolation process under blue light conditions. The inactive CRY1 homodimer is activated by a blue light signal recognized by the FAD-binding subdomain. Subsequently, the CCE domain undergoes phosphorylation, which initiates a conformational change of the CRY1 oligomer (Fig. 4B; Yu et al., 2010). After that, the activated CRY1 homodimer interacts with other proteins to induce the blue light response through regulation of a large number of genes (Chaves et al., 2011; Liu et al., 2011; Liu et al., 2016). We hypothesized that a synthetic microProtein targeting the PH subdomain of CRY1 would interfere with the ability of CRY1 to homodimerize and thus cause the formation of nonfunctional heterodimeric complexes (Fig. 4, A and B). As a result, we would expect that plants overexpressing synthetic microProtein will resemble cry1 mutant plants that exhibit etiolation under blue light conditions.
Figure 4.
Inhibition of CRY1 by the synthetic microProtein miP-CRY1. A, Domain structures of CRY1 and miP-CRY1. B, Model depicting CRY1 function and miP-CRY1 inhibition. The CRY1 homodimer is activated by a blue light signal. Activated CRY1 regulates de-etiolation of the plant. MiP-CRY1 forms a nonfunctional heterodimer with CRY1 that leads to hypocotyl elongation under blue light. C, Image of representative Col-0, cry1cry2, and two independent 35S::miP-CRY1 lines grown under white light, dark, or blue light conditions. D, RT-qPCR showing expression of endogenous CRY1 and transgene miP-CRY1 in 35S::miP-CRY1 lines compared with Col-0. Experimental replicates were statistically tested using a Student’s t test (**P < 0.01). E, Boxplot of hypocotyl length under white light, dark, and blue light (n = 10; *P < 0.05, **P < 0.01, t test). F, Interaction of miP-CRY1 with CRY1 was tested using a yeast two-hybrid assay. Normal yeast growth was observed for the serial dilutions on nonselective SD medium lacking Leu and Trp (SD-L-W). Only positive interactors grew on selective SD medium lacking Leu, Trp, and His (SD-L-W-H) supplemented with 10 mm 3-aminotriazole (3-AT).
To experimentally test this hypothesis, we generated transgenic Arabidopsis plants overexpressing the coding sequence of the PH subdomain as a synthetic microProtein, termed miP-CRY1. We obtained 10 independent transgenic 35S::miP-CRY1 lines, eight of which displayed the cry1cry2 mutant phenotype, having elongated hypocotyls when grown in continuous blue light conditions. Out of those eight independent transgenic lines, we selected two lines for further characterization. To confirm the light dependence of the observed response, two 35S::miP-CRY1 lines were grown under different light conditions (white light, blue light, and darkness) and compared with Col-0 and cry1cry2 double mutant plants. We observed elongated hypocotyls (*P < 0.05, **P < 0.01, t test) for 35S::miP-CRY1 and cry1cry2 plants when grown under continuous blue light while Col-0 plants showed short hypocotyls (Fig. 4, C and E). To verify that the etiolation phenotype of the transgenic plants occurs specifically in blue light, all plants were also grown under continuous white light and in darkness. In the darkness, all analyzed plants showed growth responses comparable to wild-type plants, i.e. elongated hypocotyls. Under continuous white light, the 35S::miP-CRY1 and cry1cry2 plants showed slightly elongated hypocotyls (*P < 0.05, **P < 0.01, t test) compared with the short hypocotyls of Col-0 plants (Fig. 4, C and E).
The expression levels of the miP-CRY1 transgene and CRY1 endogenous gene were analyzed by RT-qPCR using primers amplifying the PH subdomain and endogenous gene-specific primers outside the PH domain. In both 35S::miP-CRY1 lines, the artificial microProtein was expressed at high levels (P < 0.01, t test; Fig. 4D). Furthermore, the expression of endogenous CRY1 was not affected by the miP-CRY1 transgene. Additionally, we verified our model of microProtein-mediated posttranslational regulation by observing a strong in vitro interaction between miP-CRY and CRY1 proteins in yeast two-hybrid assays (Fig. 4F).
Taken together, these results revealed that plants overexpressing miP-CRY1 behave like cry1cry2 double mutant plants, and miP-CRY1 is able to interact with the full-length CRY1 protein. We demonstrated that the PH domain of CRY1 can be used to negatively regulate CRY1 protein activity in a microProtein-type manner and thus influence blue light responses of the plant.
CONCLUSION
MicroProteins are small, single-domain proteins that regulate larger multidomain proteins by incorporating them into novel protein complexes. So far, 22 microProteins have been characterized in Arabidopsis that regulate transcription factors. Here, we show that microProtein regulation is not restricted to transcription factors and other protein families might be regulated by microProteins. We identified 912 microProtein candidates, including the 22 characterized microProteins in Arabidopsis. Only a small number (81) of those 912 microProtein candidates were single microProteins. The remaining 831 microProtein candidates were part of 107 microProtein families. For example, most of the microProtein candidates in the hydrolase class were part of two larger microProtein families. MicroProteins of a family often have the same function and regulate the same target, as seen for the ZPR microProteins (Wenkel et al., 2007). To investigate the potential microProteins, multiple loss-of-function mutants are required. The CRISPR/Cas9 method can be used to generate those mutant plants, but it can be difficult if the list of potential microProteins and potential targets is large. In addition, our bioinformatics analysis revealed that 70% of the identified microProtein candidates are of unknown function, which makes it impossible to predict their function. Of the 912 potential microProteins, 323 microProtein candidates and their putative targets have known biological activity or known protein-protein interaction domains. Because it is a challenge to investigate microProtein candidates that are part of a family, we designed a synthetic microProtein approach. Using the synthetic microProtein approach, we demonstrated that microProteins are able to regulate multidomain proteins of different classes. We showed that the multidomain proteins DCL1, BRI1, and CRY1 can be regulated by overexpressing their protein-protein interaction domains as synthetic microProteins. As a result, microProteins can inhibit any multidomain protein containing a protein-protein interaction domain that is part of a higher-order protein complex. The positive regulatory effect of miP-DCL1 on miRNA biogenesis revealed that synthetic microProteins can be used to unravel the biological activity of certain domains. Furthermore, the synthetic microProtein approach can be used as a tool to modify gene functions at the protein level.
MATERIALS AND METHODS
Classification of MicroProtein Candidates
MicroProtein candidates for Arabidopsis (Arabidopsis thaliana) TAIR10 were taken from a published list (Straub and Wenkel, 2017), and only conserved candidates were selected. The PANTHER (Protein ANalysis THrough Evolutionary Relationships; Mi et al., 2017) protein class (Mi et al., 2013) of the putative ancestor with highest sequence similarity to the microProtein was determined. Additionally, all annotated nontranscription factor-related microProtein candidates were grouped into the 19 remaining parental classes. A one-sided Fisher’s exact test for overrepresentation was performed with R Version 3.4.1.
Plant Material, Transformation, and Plant Growth
All transgenic Arabidopsis plant lines were generated in the Col-0 background. The annotation information of the protein-protein interaction domains was obtained from The Arabidopsis Information Resource (TAIR) database. To generate transgenic plant lines overexpressing the synthetic microProteins, the cDNAs, encoding the protein-protein interaction domains, were cloned into the binary vector pJAN33 containing a CaMV35S promoter, a FLAG tag, and a BASTA (glufosinate)-resistance (BAR) gene. Constructs were then transformed into Arabidopsis via floral dip. Transgenic plants were isolated from the T1 generation using BASTA selection, and the segregation ratio was observed in the T2 generation. Homozygote transgenic plant lines were used to perform the experiments. If not otherwise stated, plants were grown in the greenhouse under long-day conditions. For the BL treatment, plants were stratified for 2 d at 4°C and grown on soil under the following conditions: long day (16 h light, 8 h night), day 22°C, night 18°C, and 70% humidity. Plants were treated once a day with mock solution (ethanol) or 100 nm BL. The experiment was performed twice, and 10 plants per lines were used as replicates. To measure the hypocotyl length of 35S::miP-CRY1, cry1cry2, and Col-0, the plant lines were grown on solid culture medium (half-strength Murashige and Skoog [4.3 g/L; Duchefa], MES [0.3 g/L; Duchefa], Suc [5 g/L; Sigma], and Bacto Agar [8 g/L], pH 5.7; Duchefa), and seeds were stratified for 4 d at 4°C. After germination initiation for 1 d under continuous white light, the plant lines were grown for 6 d under continuous white light, continuous light supplemented with 10% blue light (photosynthetic active radiation of 15.16 W/m2), or in darkness. Hypocotyl length was measured after 6 d using ImageJ, and boxplots were generated using RStudio. The measurement of the hypocotyls was performed three times in independent experiments using 10 plants per plant line with identical results.
RT-qPCR
For RT-qPCR, plants were grown for 10 d on solid culture medium under continuous white light at 22°C after 2 d of stratification at 4°C. Total RNA was isolated using a Spectrum Plant Total RNA Kit (Sigma-Aldrich/Merck), following the manufacturer’s recommendations. Up to 1 µg of purified total RNA was treated with DNase (RNase-free DNase; Thermo Fisher Scientific) and then used for reverse transcription using a RevertAid RT Reverse Transcription Kit (Thermo Fisher Scientific) with oligo(dT) primers. RT-qPCR was performed using a KAPA SYBR FAST qPCR Kit (Kapa Biosystems) on a Bio-Rad CFX384 machine. Gene expression levels were calculated using the delta-Ct method and a standard curve relative to GADPH. To detect endogenous gene expression levels, primers were designed outside the protein-protein interaction domain that was used for the synthetic microProteins. To quantify the ectopic expression of the transgene, primers that bind in the transgene coding sequence were used.
To test the relative expression of BL-regulated genes, Col-0, bri1-5, det-2, and 35S::miP-BRI1 lines were grown for 10 d on solid culture medium under continuous light at 22°C. Seedlings were treated with either 100 nm BL or mock solution for 3 h. Seedlings were flash frozen immediately after 3 h of treatment, and RT-qPCR was carried out as described before. To validate the experiment, two biological replicates and three experimental replicates were used. Oligonucleotide sequences are listed in Supplemental Table S2.
Small RNA Isolation and Northern Blot Analysis
Total RNAs were isolated from 14-d-old seedlings using Trizol reagent (Invitrogen). The extracted aqueous phase was precipitated with 2-propanol twice (100% and 75%) and resolved in 50% formamide. Purified RNAs were resolved on a 5% to 15% gradient denaturing polyacrylamide gel (National Diagnostics) before transfer to a nylon membrane (Amersham). The end-labeled DNA probes were applied for hybridization of blots for 12 h (Ambion). Blots were washed twice with SSC (2×)/SDS (0.1%) for 20 min each. Hybridization signals were detected with a BAS phosphoimager (Fuji).
Yeast Two-Hybrid Assays
Protein interaction assays were performed using the Matchmaker yeast two-hybrid system (Clontech). The coding sequences of CRY1, miP-CRY1, and miP-DCL1 were recombined into pGADT7-GW or pGBKT7-GW vectors using Gateway LR Clonase II Enzyme Mix (Invitrogen). Bait constructs were transformed into the yeast strain YM4271 MATa and preys into the yeast strain PYJ654 MATα using the lithium acetate method (Schiestl and Gietz, 1989) and were selected on synthetic dropout (SD) medium lacking Leu or Trp. Mating of preys with baits was performed with three strains of each transformation for 24 h at 28°C, and successful mating was selected on SD medium lacking Leu and Trp. The yeast two-hybrid assays were performed with the generated yeast constructs on SD medium lacking His, Leu, and Trp with additional 2.5 or 10 mm 3-aminotriazole to avoid autoactivation.
Accession Numbers
Sequence data from this article can be found in the TAIR database under the following accession numbers: BRI1, AT4G39400; CRY1, AT4G08920; and DCL1, AT1G01040.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RT-qPCR of all generated transgenic lines.
Supplemental Table S1. All identified microProteins divided into the different protein classes.
Supplemental Table S2. Oligonucleotides used in this study.
Acknowledgments
We thank Poul Erik Jensen for commenting on the manuscript and Peter Brodersen for providing the dcl1-11 mutant seeds.
Footnotes
This work was supported by the European Research Council (GA336295) and the Copenhagen Plant Science Centre.
Articles can be viewed without a subscription.
References
- Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284: 760–765 [DOI] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62: 335–364 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA 105: 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguen T, Straub D, Graeff M, Wenkel S (2015) MicroProteins: small size-big impact. Trends Plant Sci 20: 477–482 [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet 29: 435–440 [DOI] [PubMed] [Google Scholar]
- Frizzi A, Huang S (2010) Tapping RNA silencing pathways for plant biotechnology. Plant Biotechnol J 8: 655–677 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, et al. (1997) The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9: 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF III (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeff M, Wenkel S (2012) Regulation of protein function by interfering protein species. Biomol Concepts 3: 71–78 [DOI] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Han MH, Goud S, Song L, Fedoroff N (2004) The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA 101: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SB, Wang WX, Zhang JY, Xu F, Lian HL, Li L, Yang HQ (2015) The CNT1 domain of Arabidopsis CRY1 alone is sufficient to mediate blue light inhibition of hypocotyl elongation. Mol Plant 8: 822–825 [DOI] [PubMed] [Google Scholar]
- Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T (2005) Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol 57: 173–188 [DOI] [PubMed] [Google Scholar]
- Kim TW, Wang ZY (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol 61: 681–704 [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Lee M, Lee I, Park HY, Seo PJ, Jung JH, Kwon EJ, Suh SW, Paek KH, et al. (2008) HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20: 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y (2006) The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR (1995) Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269: 968–970 [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95: 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Yang Z, Gomez A, Liu B, Lin C, Oka Y (2016) Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana. J Plant Res 129: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Axtell MJ, Fedoroff NV (2012) The helicase and RNaseIIIa domains of Arabidopsis Dicer-Like1 modulate catalytic parameters during microRNA biogenesis. Plant Physiol 159: 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J (2006) SERRATE: a new player on the plant microRNA scene. EMBO Rep 7: 1052–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida S, Chen HY, Adam Yuan Y (2011) Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE. Nucleic Acids Res 39: 7828–7836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD (2017) PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res (D1) 45: D183–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD (2013) Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8: 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, Yang HQ (2005) N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17: 1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16: 339–346 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Hong SY, Ryu JY, Jeong EY, Kim SG, Baldwin IT, Park CM (2012) Targeted inactivation of transcription factors by overexpression of their truncated forms in plants. Plant J 72: 162–172 [DOI] [PubMed] [Google Scholar]
- Staudt AC, Wenkel S (2011) Regulation of protein function by ‘microProteins’. EMBO Rep 12: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub D, Wenkel S (2017) Cross-species genome-wide identification of evolutionary conserved microProteins. Genome Biol Evol 9: 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang X, Chory J (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang X, Li X, Meisenhelder J, Hunter T, Yoshida S, Asami T, Chory J (2005) Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell 8: 855–865 [DOI] [PubMed] [Google Scholar]
- Wenkel S, Emery J, Hou BH, Evans MM, Barton MK (2007) A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19: 3379–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H (2006) SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J 47: 841–850 [DOI] [PubMed] [Google Scholar]
- Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC, et al. (2008) The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA 105: 10073–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, Lin C (2010) The cryptochrome blue light receptors. Arabidopsis Book 8: e0135. [DOI] [PMC free article] [PubMed] [Google Scholar]