Abstract
New technologies reveal the deposition and remodeling of plant cell wall polysaccharides and their impact on plant development.
All plant cells are surrounded by complex walls that play a role in the growth and differentiation of tissues. Walls provide mechanical integrity and structure to each cell and represent an interface with neighboring cells and the environment (Somerville et al., 2004). Cell walls are composed primarily of multiple polysaccharides that can be grouped into three major classes: cellulose, pectins, and hemicelluloses. While cellulose fibrils are synthesized by the plant cells directly at the plasma membrane (PM), the matrix polysaccharides are produced in the Golgi apparatus by membrane-bound enzymes from multiple glycosyltransferase families (Oikawa et al., 2013). After secretion to the wall via exocytosis, the structures of the noncellulosic polysaccharides are modified by various apoplastic enzymes. In addition to polysaccharides, most plant cell walls contain small amounts of structural proteins such as extensins and arabinogalactan proteins.
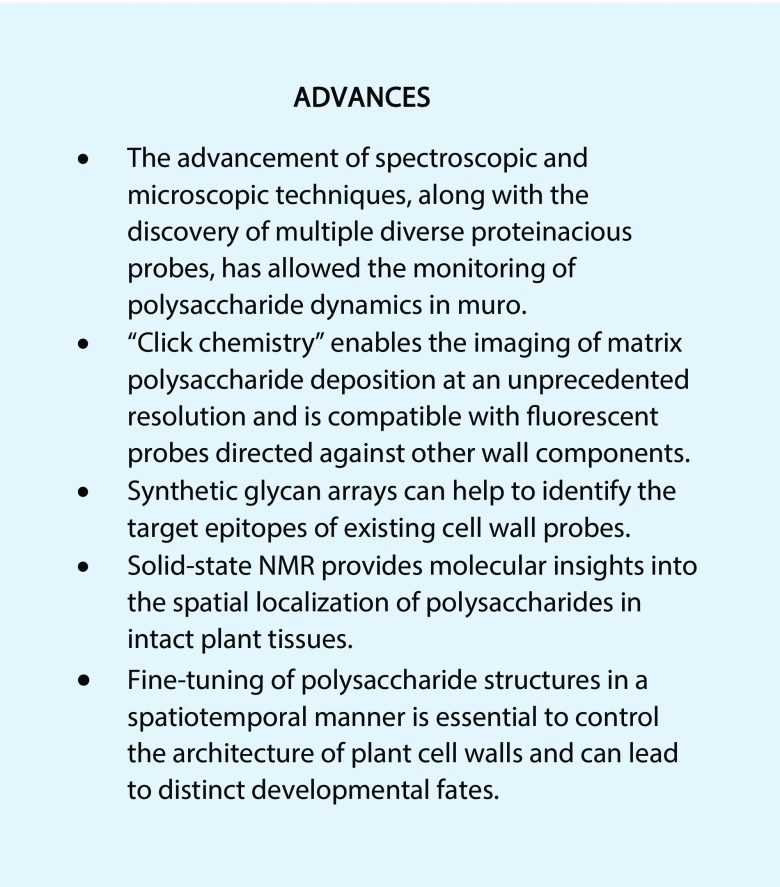
Cell walls are dynamic entities, rather than rigid and recalcitrant shells, that can be remodeled during plant development and in response to abiotic and biotic stresses. Cell expansion requires the deposition of additional material in the surrounding primary walls as well as the reorganization and loosening of existing polymers to allow for wall relaxation and controlled expansion (Cosgrove, 2005). The latest model of the primary wall structure proposes that cellulose-cellulose junctions only occur at a limited number of biomechanical hotspots, where protein catalysts must act selectively to initiate wall loosening (Cosgrove, 2018). In tissues undergoing growth, the recycling of polysaccharides via a suite of enzymes can contribute to the construction of elongating walls (Barnes and Anderson, 2018). Once elongation ceases, some cells deposit thick secondary walls that incorporate additional polysaccharides. Many secondary walls are impregnated with the polyphenol lignin and thereby become relatively fixed structures that exclude water and resist hydrolysis.
The dynamics of plant cell walls have traditionally been challenging to characterize in muro due to technical limitations and the structural complexity of their components. As an example of structural complexity, pectins can incorporate 12 different sugars in at least 25 glycosidic linkages and can be decorated further with methyl, acetyl, or phenolic groups (Atmodjo et al., 2013). While analyses of extracted carbohydrates have been instrumental for characterizing walls (Foster et al., 2010; Pettolino et al., 2012; Carpita and McCann, 2015), they do not reveal how polysaccharides are distributed across different cell layers or within a particular wall. Historically, only a few techniques were available to detect polysaccharides in living plant cells, and many of the wall-directed probes had a broad specificity and/or poorly characterized targets (Wallace and Anderson, 2012). For instance, the Calcofluor White dye has been used frequently to stain cell walls, but it fluoresces in the presence of β-glucan structures from all three major polysaccharide classes (Anderson et al., 2010). Recent technical developments, such as the identification of more specific probes, have helped elucidate the components of the plant cell wall.
In this Update, we focus on current and emerging techniques for monitoring the dynamics of polysaccharides in the cell wall (Table I). We highlight recent biological insights gained from these methods, discuss the limitations of each approach, and provide a summary of specific probes that may be used to identify different polysaccharide structures in situ (Fig. 1; Table II).
Table I. Comparison of advanced techniques for monitoring polysaccharide dynamics.
A summary is provided of technical advantages and limitations, along with key biological observations, which are discussed in the text. For the electron microscopy column, TEM or SEM indicate points specific to either transmission or scanning electron microscopy, respectively. R denotes the relative resolution of a technique and ranges from the diffraction limit of light (+) to atomic resolution (+++). S denotes the relative speed of a technique (including the typical sample preparation time) and ranges from multiple days (+) to mere seconds (+++).
| Parameter | Light Microscopy | Electron Microscopy | Atomic Force Microscopy | Solid-State NMR | X-Ray Diffraction |
|---|---|---|---|---|---|
| Advantages | Live-cell imaging | TEM: mAb-compatible | Measurement of elasticity | Atomic-scale chemical information | Highest spatial resolution |
| Many probes (see Table II) | Organelle resolution | Near-native cell walls | Intact tissues | ||
| Visualization | CESA dynamics | SEM: CESA rosette size | Pattern of polymers | Cellulose-matrix polymer location | Cellulose microfibril dimensions |
| Polymers across tissues | TEM: polymers within a cell | Stiffness of cell wall | |||
| Limitations | Epitope masking | TEM: long preparation | Uncertain polymer identity | Analysis of biomass as a whole (no cell specificity) | Works only for crystal structures |
| Probe specificity | Fixation artifacts | ||||
| R | + | ++ | ++ | +++ | +++ |
| S | +++ | + | ++ | + | + |
Figure 1.
Major classes of probes and key steps to image cell wall polysaccharides. A, Illustration of the steps required prior to microscopy (represented by the magnifying glasses) for different types of probes. Secondary antibodies (2° mAb) are commercially available and should be selected based on the final application (electron versus light microscopy) and the available equipment (e.g. fluorescence filters). Probes in colored boxes are exemplified in B to G. B and C, Sites of wall polysaccharide synthesis in Arabidopsis cotyledons expressing the yellow FP markers wave_22Y and wave_138Y, respectively (Geldner et al., 2009). FP signal and chloroplast intrinsic fluorescence are shown using Orange Hot and Cyan Hot look-up tables in Fiji (Schindelin et al., 2012). D to G, Mucilage architecture of an S4B-stained and mAb-labeled (CCRC-M36 and Alexa Fluor 488 2° mAb) Arabidopsis seed. D shows a segment of a whole seed (black) that has been brought into contact with water. The only clearly visible structures are columellae (C). S4B staining reveals cellulosic rays in E, and the antibody CCRC-M36 shows the deposition of rhamnogalacturonan in F. The comparison of the overlay G with D exemplifies the usefulness of dyes and labels. Bars = 25 µm (B–G).
Table II. Selected probes to investigate the dynamics of different plant wall polysaccharides.
Additional antibodies with known targets that are not listed here are available from PlantProbes (http://plantprobes.net) and CarboSource (https://www.ccrc.uga.edu/∼carbosource/CSS_home.html). CBMs, Carbohydrate-binding modules; Kdo, 3-deoxy-d-manno-oct-2-ulosonic acid; mAb, monoclonal antibody.
| Polysaccharide | Probe | Target | Reference |
|---|---|---|---|
| Cellulose | Calcofluor White | Various β-glucan structures | Anderson et al. (2010) |
| Pontamine Scarlet 4B (S4B) | Specific for crystalline cellulose | Anderson et al. (2010) | |
| FP-tagged CESA proteins | Proxy for cellulose synthesis | Paredez et al. (2006) | |
| CBM3a | Crystalline cellulose | Blake et al. (2006) | |
| CBM28 | Amorphous cellulose | Blake et al. (2006) | |
| Pectin | Ruthenium Red (RR) | Pectin with deesterified GalA | Hanke and Northcote (1975) |
| LM19 mAb | HG, particularly deesterified forms | Verhertbruggen et al. (2009) | |
| 2F4 mAb | Unesterified HG cross-linked by Ca2+ | Liners and Van Cutsem (1992) | |
| LM20 mAb | Methylesterified HG | Verhertbruggen et al. (2009) | |
| JIM7 mAb | Methylesterified epitopes of HG | Knox et al. (1990) | |
| CCRC-M36 mAb | RG I backbone | Ruprecht et al. (2017) | |
| Chitosan oligosaccharides (COS) | Deesterified GalA regions | Mravec et al. (2014) | |
| Oligogalacturonides | Unesterified HG cross-linked by Ca2+ | Mravec et al. (2017) | |
| Propidium iodide (PI) | Unesterified GalA units | Rounds et al. (2011) | |
| Alkyne derivative of Fuc | RG I | Anderson et al. (2012) | |
| Azido derivative of Kdo | RG II | Dumont et al. (2016) | |
| Xyloglucan | Sulforhodamine labeling | Xyloglucan oligosaccharides | Vissenberg et al. (2005) |
| CCRC-M1 mAb | FucogalatoXyG | Puhlmann et al. (1994) | |
| Mannan | LM21 mAb | Branched and unbrached mannans | Marcus et al. (2010)) |
| LM22 mAb | Unbranched mannans | Marcus et al. (2010) | |
| Xylan | CBM15-mOrange2 (OC15) | Xylan (xylohexaose) | Khatri et al. (2016) |
| LM10 mAb | Unbranched xylan | McCartney et al. (2005) | |
| LM11 mAb | Branched xylan (e.g. with arabinose) | McCartney et al. (2005) | |
| LM28 mAb | Glucuronosyl-substituted xylans | Cornuault et al. (2015) |
CELLULOSE
Visualization of Crystalline Cellulose Microfibrils
Cellulose microfibrils are aggregates of linear β-1,4-linked glucan chains, which are stabilized by intramolecular and intermolecular hydrogen bonds in the walls (Notley et al., 2004; McNamara et al., 2015). Several types of methods have been used to visualize the microfibrils in the wall (Table I). Primary wall cellulose fibrils have been estimated to be 3 nm in diameter by spectroscopic and diffraction techniques (Thomas et al., 2013), whereas larger aggregates of microfibrils in the 5- to 10-nm-diameter range have been found in conifer wood (Fernandes et al., 2011). Even fibrils with diameters up to 40 nm were observed by electron microscopy utilizing freeze-fracture techniques (McCann et al., 1990). Currently, it is not clear how much interspersed matrix polymer material contributes to these aggregate estimates. Due to this size range, microfibrils can be observed by both atomic force microscopy and scanning electron microscopy (Zhang et al., 2016), and their angles and distances can be determined (Marga et al., 2005). Crystalline cellulose is initially oriented randomly in meristematic cells (McCann et al., 1990) but is later aligned as parallel fibrils, transverse to the axis of elongation (Sugimoto et al., 2000). The orientation of microfibrils is hypothesized to mechanically restrict the direction of cell extension, leading to anisotropic growth. Parallel cellulose microfibrils are separated during cell expansion (Marga et al., 2005), presumably yielding to internal turgor pressure and being loosened via the enzymatic modification of adjoining matrix polysaccharides (Wolf and Greiner, 2012; Cosgrove, 2016). Since microfibril diameters do not appear to decrease during the elongation process, the fibrils likely are not modified directly.
Monitoring Cellulose Structure with Molecular Probes
In addition to cellulose found in a crystalline microfibril form, some walls are estimated to be composed of ∼40% amorphous cellulose (Marga et al., 2005). This substantial proportion of cellulose cannot be determined by the aforementioned microscopy tools and can only be estimated roughly by spectroscopic methods. CBMs are powerful probes for detecting the presence and dynamics of amorphous cellulose (McLean et al., 2002). CBMs are noncatalytic domains of carbohydrate-active enzymes and are thought to facilitate binding to specific substrates (Hall et al., 1995). Multiple cellulose-directed CBMs have been identified that bind either crystalline or amorphous structures (Table II; McLean et al., 2002). Indirect immunolabeling of plant sections with His-tagged versions of these CBMs (for a detailed mechanism, see Fig. 1A) has revealed the diversity of cellulose forms in various cell walls using fluorescence microscopy (Blake et al., 2006) or electron microscopy (Ruel et al., 2012). For example, collenchyma cells contain primary walls with a larger abundance of amorphous cellulose compared with microfibril-rich secondary walls. CBMs also provided hints about polymer interactions, as partial enzymatic removal of pectic polysaccharides in plant sections leads to a more intensive labeling of crystalline cellulose. This indicates a close spatial proximity of these two polymer classes (Blake et al., 2006) and has been demonstrated independently by solid-state NMR (Dick-Perez et al., 2012). However, certain probes may have broader specificities than expected. For instance, CBM3a is a common label for crystalline cellulose that also can bind xyloglucan (XyG), a Xyl-substituted glucan (Hernandez-Gomez et al., 2015). This issue could be mitigated by imaging walls before and after treatment with xyloglucanase or by using alternative probes (Table II).
Fluorescent proteins (FPs) can be tagged to CBM domains to directly analyze polysaccharide structure without the secondary or tertiary reagents shown in Figure 1A. For instance, two fluorescently tagged CBMs enabled the dual labeling of crystalline and amorphous cellulose at the surface of plant fibers following biomass deconstruction (Gourlay et al., 2015). Such chimeric protein constructs could potentially be transformed into plants to monitor polysaccharides without any incubation steps. While in vivo fluorescent CBM probes may provide greater insights into polysaccharide dynamics in growing cells, caution is necessary. For example, the direct expression of fluorescent CBM proteins in plants would have to be tested carefully and optimized to ensure that the target polymer is visualized without altering its native architecture or plant development. Another potential disadvantage is that the FP domain may interfere with the ligand-binding site of the CBM (Knox, 2012). Regardless of the presence of FP tags, binding of a CBM or any other polysaccharide-directed mAbs provides limited quantitative information; an epitope may be found in multiple polymers, or it could be masked by its neighbors or hidden by an altered conformation of the target polysaccharide (Pattathil et al., 2015). Despite these issues, proteinaceous probes such as CBMs provide an unprecedented view of the occurrence of wall polysaccharides. Alternatively, crystalline cellulose in living cells can be fluorescently stained with S4B (Fig. 1E; Table II; available as Direct Red 23) to monitor microfibril reorientation in real time (Anderson et al., 2010). In contrast to Calcofluor White, S4B is highly specific to cellulose.
Cellulose microfibrils are generated at the PM (Fig. 1C) by rosette structures harboring complexes of multiple cellulose synthase (CESA) enzymes and other proteinaceous components (Hill et al., 2014; McFarlane et al., 2014). It is believed that the synthesis of glucan chains by the CESAs propels the complex through the PM, depositing cellulose microfibrils in its wake (McFarlane et al., 2014). The estimated size of cellulose microfibrils suggests that they are produced by CESA rosettes containing a total of 18 CESA proteins, which was validated in the moss Physcomitrella patens using an improved method for electron microscopy (Nixon et al., 2016). FP-tagged CESA enzymes also have been visualized in the PM of living cells from the model plant Arabidopsis (Arabidopsis thaliana; Paredez et al., 2006). Their movement has been used as a powerful proxy to assess the speed and orientation of microfibril formation in planta. By this technique, the synthesis of cellulose in secondary walls has been estimated to be significantly faster than in primary walls (Paredez et al., 2006; Wightman et al., 2009; Watanabe et al., 2015). Since specific elements of the cytoskeleton also can be visualized with mAbs or FP probes, numerous elegant studies have shown that the direction of cellulose deposition is coupled to microtubule orientation (McFarlane et al., 2014).
Fluorescently labeled CESAs are localized not only in the PM but also in intracellular compartments such as the Golgi apparatus and other small bodies (Crowell et al., 2009). These could represent proteins that are in transit through the secretory system: from the endoplasmic reticulum, where they are synthesized, to the outer membrane of the cell. Alternatively, the intracellular CESA-containing compartments could be the result of endocytosis, presumably for protein recycling purposes, or are destined to be reinserted into the PM under specific conditions, such as salt stress (Gutierrez et al., 2009). Therefore, endomembrane trafficking is likely a key regulator of when and where cellulose can be generated at the cell surface. This also highlights the major limitation of FP probes as a proxy for assessing cellulose microfibril orientation, direction, and synthesis. Even though CESAs can be observed in real time, it is not certain when the proteins are actually producing cellulose. This may be overcome by monitoring cellulose deposition with several probes, ideally at the same time. A combined analysis of FP-CESA localization, CBM3a immunolabeling, and S4B staining in Arabidopsis roots revealed that cellulose is deposited at the cell plate earlier than was thought previously (Miart et al., 2014). Surprisingly, FP-tagged CESAs are not localized at the tip of elongating root hairs, where S4B and CBM3a signals are detected (Park et al., 2011). Hence, other proteins are likely to be responsible for the formation of these β-glucans (Park et al., 2011), and new probes are needed to further characterize their structures and to identify how they are produced.
MATRIX POLYSACCHARIDES
In contrast to the simple, linear cellulose polymers, matrix polysaccharides (such as pectins and hemicelluloses) can be branched and substituted and are thought to be synthesized in the Golgi (Fig. 1B). Once secreted to the wall, matrix polysaccharides are structurally modified by various apoplastic enzymes, including glycosyl hydrolases and carbohydrate esterases.
Pectins are structurally complex polysaccharides that are rich in α-1,4-linked GalA subunits (Atmodjo et al., 2013), such as homogalacturonan (HG), rhamnogalacturonan I (RG I) and RG II, and xylogalacturonan. Hemicellulose encompasses matrix polysaccharides that can be extracted using only alkali as a chaotropic agent (Scheller and Ulvskov, 2010) and, thus, includes XyG, xylans, mannans, and mixed-linkage glucans. In contrast to the current dogma for hemicellulose biosynthesis in the Golgi, mAbs and FP probes indicate that mixed-linkage glucans are assembled at the PM (Wilson et al., 2015). Therefore, the production of matrix polysaccharides and their subsequent remodeling in the wall are only starting to be understood. The molecular architecture of matrix polysaccharides has been revealed by the development of enhanced methods (Table I) and the identification of probes with specific target epitopes (Table II).
Monitoring Pectic Structures
HG is the most abundant pectin in primary walls and can be modified in planta via methylesterification and acetylation. Two other major pectic domains, RG I and RG II, contain Rha and are covalently linked by HG (Atmodjo et al., 2013). The Arabidopsis genome contains more than 170 HG-related enzymes (Sénéchal et al., 2014), although the precise roles of only a few have been characterized to date. HG methylesterification status plays important roles in various developmental processes and is controlled tightly by pectin methylesterases (PMEs), PME inhibitors (PMEIs), subtilisin-type Ser proteases (SBTs), and at least one E3 ubiquitin ligase (Levesque-Tremblay et al., 2015). Mutants with altered expression of these players have been characterized using mAbs directed against HG domains with different degrees of methylesterification. Some mAbs (e.g. LM19 and 2F4; Table II) preferentially bind unesterified or sparsely methylated HG, whereas other mAbs (e.g. LM20 and JIM7; Table II) recognize methylesterified pectin. Utilizing these probes has been instrumental in observing the state of HG methylesterification in cell walls. Immunolabeling coupled with transmission electron microscopy revealed that HG is synthesized in the plant Golgi apparatus in a highly methylesterified state labeled by JIM7, whereas unesterified GalA epitopes are barely detected without chemical deesterification (Zhang and Staehelin, 1992). After deposition in the wall, pectin can be deesterified by PMEs in a specific spatiotemporal manner that results in one of two contrasting roles (Levesque-Tremblay et al., 2015). Regions of unesterified GalA residues can be cross-linked via Ca2+ ions (forming egg boxes) to increase wall stiffness or cleaved by pectin-degrading enzymes (e.g. polygalacturonases) to promote wall loosening.
The impact of HG methylesterification on cell wall architecture can be examined conveniently in the mucilage capsules that coat Arabidopsis seeds (Fig. 1, D–G). This specialized cell wall contains more than 90% pectin and only minor amounts of cellulose and hemicellulose (Voiniciuc et al., 2015b). Thick, hydrophilic capsules of mucilage can be immunolabeled indirectly with the CCRC-M36 mAb (Fig. 1F; Table II) or visualized quickly with RR (Table II), a dye that selectively binds to negatively charged molecules such as unesterified GalA regions and is compatible with both light and electron microscopy (Hanke and Northcote, 1975). The RR dye was a convenient probe for identifying chemically mutagenized Arabidopsis mucilage-modified mutants (Western et al., 2001) and a large number of natural Arabidopsis variants with altered mucilage architecture (Voiniciuc et al., 2016). The presence of Ca2+ ions, which can be manipulated by distinct chemical treatments, negatively regulates the size of RR-stained mucilage capsules. For instance, flying saucer1 (fly1) mutant seeds release smaller RR-stained mucilage capsules when hydrated in water, due to a lower degree of pectin methylesterification and an increased abundance of HG egg boxes detected with the 2F4 mAb (Voiniciuc et al., 2013). The exogenous addition of Ca2+ ions further blocks the ability of fly1 mucilage to expand, consistent with the model that unesterified HG regions can form stiff gels. In stark contrast, the impaired wall architecture of the fly1 mutant was largely rescued by treating seeds with cation chelators (Voiniciuc et al., 2013) that disrupt the Ca2+ cross-links between unesterified HG chains to facilitate the loosening of matrix polysaccharides. Similar calcium-dependent phenotypes, consistent with the HG egg-box model, have been observed for two additional mucilage mutants involved in HG methylation status: sbt1.7 (Rautengarten et al., 2008) and pmei6 (Saez-Aguayo et al., 2013). While FLY1 regulates the HG methylesterification status via protein ubiquitination in the endomembrane system, SBT1.7 and PMEI6 inhibit PME activity directly in the extracellular matrix where HG is present.
Nevertheless, a new model for the role of HG in cell expansion postulates that Ca2+-bridged pectins are not as prevalent in planta as was thought previously (Hocq et al., 2017). The frequency of HG egg boxes in cell walls may have been overestimated by certain immunolabeling procedures, highlighting the need to still apply caution when using these powerful tools. For instance, 2F4 immunolabeling of egg-box cross-links requires the addition of Ca2+ ions (Liners and Van Cutsem, 1992) at concentrations that are severalfold greater than physiological levels (Hocq et al., 2017). Therefore, the effects of HG demethylesterification on cell walls also should be monitored with other techniques. Atomic force microscopy (Table I) has been used to quantify the elasticity of walls in living meristems of Arabidopsis wild-type plants as well as in transgenic lines overexpressing a PME gene or an antagonistic PMEI gene (Peaucelle et al., 2011). While the egg-box model predicts that PME activity increases the stiffness of the pectic matrix, the atomic force microscopy experiments found that demethylesterification of HG promoted wall loosening and was a prerequisite for organ initiation (Peaucelle et al., 2011). Therefore, unesterified HG in growing cells is likely targeted by pectin-cleaving enzymes such as the recently identified polygalacturonanses involved in cell expansion (PGX1, PGX2, and PGX3), which are required for the development of multiple Arabidopsis organs (Xiao et al., 2014, 2017; Rui et al., 2017).
While the HG structure is typically visualized using mAbs, these relatively large probes may have limited permeability and require multiple incubation steps that render them unsuitable for real-time imaging of pectin dynamics (Fig. 1A). Probes with considerably smaller Mr values have emerged recently for monitoring HG properties. PI is a membrane-impermeable stain that competes with Ca2+ ions to bind to demethylesterified pectins. When used at low concentrations, PI enables imaging in Arabidopsis root hairs and pollen tubes without altering cell growth (Rounds et al., 2011). Mutant pollen tubes with impaired exocytosis show repetitive bursts of growing tips at cell wall regions that accumulate PI-stained demethylesterified HG (Synek et al., 2017). Alternatively, sparsely methylated HG can be labeled in real time with fluorescently tagged COS (Table II), and the specificity of this labeling was tested using carbohydrate microarrays (Mravec et al., 2014). Sequential use of COS probes coupled to two distinct fluorophores revealed how cells in Arabidopsis root caps accumulate deesterified HG over time. Recently, another oligosaccharide probe consisting only of GalA units was used to continuously monitor the distribution of calcium-cross-linked HG in elongating pollen tubes (Mravec et al., 2017). Throughout pollen tube growth, tightly linked HG was only detected outside of the tip-growing region, consistent with the polar localization of PMEI proteins (Röckel et al., 2008). Both PI and the oligosaccharide probes can label HG in less than 20 min to provide an unprecedented resolution of HG dynamics in living cells. Recently, HG dynamics were visualized using COS conjugated with Alexa Fluor 488 (COS488) and PI probes in guard cells lacking or overexpressing the PGX3 polygalacturonase (Rui et al., 2017). Although COS488 and PI labeled only partially overlapping regions of the wall, they both supported an inverse relationship between the presence of PGX3 and unesterified HG. Additionally, COS488 and PI were more sensitive for quantifying GalA epitopes in three dimensions, compared with indirect labeling of fixed cells with mAbs (LM19, LM20, and 2F4; Rui et al., 2017). Therefore, these new probes are compelling alternatives to mAbs and should be exploited further.
Following the use of click chemistry to image polymers in animals, fungi, and bacteria, modified sugars can be metabolically incorporated into plant cell walls to visualize pectin dynamics (Anderson and Wallace, 2012). Although Fuc is part of several wall components, Arabidopsis roots were shown to primarily incorporate an alkynylated Fuc analog into a high-molecular-weight pectic polymer that is most likely RG I (Anderson et al., 2012). Since the alkynylated Fuc analog sugar can be fluorescently labeled with a membrane-impermeable compound (Fig. 1A), it can be used to identify proteins required to maintain the delivery of pectin to the cell wall. This probe showed that elongating root epidermal cells lacking the FRAGILE FIBER1 kinesin incorporate less RG I and in an uneven pattern compared with the wild type (Zhu et al., 2015). In addition, the synthesis and redistribution of another pectic polymer can now be visualized using a clickable analog of Kdo, a monosaccharide unique to RG II (Dumont et al., 2016). The Kdo-derived probe is compatible with other probes (e.g. the alkynylated Fuc analog) to simultaneously image multiple matrix polysaccharides during root growth. The RG I and RG II probes have been detected via copper-catalyzed click reactions, which are toxic to Arabidopsis seedlings and may damage the wall. However, two alternative methods to detect click-compatible analogs not requiring copper were developed recently and could be used to study the long-term dynamics of extracellular glycans (Hoogenboom et al., 2016).
Hemicellulose Dynamics in the Primary Cell Wall
XyG is a ubiquitous matrix polysaccharide in land plants and represents the most abundant hemicellulose in the primary wall (Pauly and Keegstra, 2016). Since XyG cross-links cellulose microfibrils, it was thought that this network forms the major load-bearing structure in growing cells and that its metabolism plays a major role in cell expansion. However, the Arabidopsis xxt1 xxt2 mutant lacking detectable XyG displays only minor morphological changes and vegetative growth defects (Cavalier et al., 2008). These observations question the role of XyG in extension growth. Nevertheless, xxt1 xxt2 mutant plants exhibit burst root hairs (Cavalier et al., 2008), indicating that XyG plays an important role in tip growth.
The side chain substitution pattern of XyG can vary depending on the plant species (Schultink et al., 2014). However, recent analyses revealed that XyG structures can be tissue specific (Lampugnani et al., 2013; Dardelle et al., 2015; Liu et al., 2015). Based on chemical analyses of ground plant tissue samples, fucogalactoXyG is generally found in many eudicots, but not in plant vegetative tissues from phylogenetically younger species of the Solanaceae and the grasses (Poaceae). An alternative approach to identify fucogalactoXyG at the cellular level is the use of the mAb CCRC-M1 (Table II), which was the first wall-directed mAb with a thoroughly characterized epitope specificity (Puhlmann et al., 1994). FucogalactoXyG labeled by CCRC-M1 has been visualized in specialized tissues in the grasses (Brennan and Harris, 2011), in the pollen tubes of tomato (Solanum lycopersicum; Dardelle et al., 2015) and Nicotiana alata (Lampugnani et al., 2013), and in root hairs of rice (Oryza sativa; Liu et al., 2015). The presence of this particular form of XyG in tip-growing tissues suggests that its structure and/or metabolism are important for tip growth or at the interface between the plant and the environment.
Recently, the role of XyG in stomatal guard cell function was examined through live-cell spinning-disk confocal microscopy of xxt1 xxt2 mutant cell walls (Rui and Anderson, 2016). XyG-deficient guard cell walls are less capable of longitudinal expansion during stomatal movement and show impaired organization of S4B-stained cellulose. Mutants containing point mutations in CESA genes show similar defects as well as aberrant distribution of GFP-tagged CESA proteins during stomatal movements (Rui and Anderson, 2016). While the cellulose structure was visualized with two distinct tools (S4B dye and FP-CESA), the dynamics of XyG polymers as stomata open and close have yet to be investigated.
The development of specific probes compatible with live-cell microscopy is essential for monitoring the structure of XyG in dynamic contexts, such as stomatal movements. Sulforhodamine-labeled XyG oligosaccharides have been used to visualize XyG transglucosylase/hydrolase enzymes that act specifically in elongating walls of root epidermal cells in Arabidopsis and tobacco (Nicotiana tabacum) root epidermal cells (Vissenberg et al., 2005). Since fluorescently labeled XyG was incorporated into the cell wall, this approach could be applied in other biological contexts. In addition, multiple azido or alkynyl sugar analogs of Glc and Xyl have been synthesized as potential reporters for XyG (Zhu et al., 2016), but none of them were metabolically incorporated into Arabidopsis roots. Although another study found a Glc derivative (6dAG) that is incorporated specifically at root hair tips, this probe colocalizes with callose, inhibits root growth, and leads to stunted root hairs (McClosky et al., 2016). Therefore, no suitable clickable analogs are available currently to label hemicelluloses. Even though most of the wall Fuc is present in fucogalactoXyG (Zablackis et al., 1996), an alkynylated Fuc analog only labeled RG I (Anderson et al., 2012). Apparently, the corresponding XyG:fucosyltransferase is not able to accept the Fuc analog as a donor substrate (Perrin et al., 1999; Rocha et al., 2016), while the RG I:fucosyltransferase does. These experiments highlight one of the current limitations of using click chemistry or other sugar analogs to monitor wall dynamics. Hopefully, future research with different sugar analogs will overcome the exclusion from wall polymer metabolic enzymes.
Xylan Dynamics in Secondary Cell Walls
Xylans and mannans are the two major classes of hemicelluloses that accumulate in plant secondary walls (Scheller and Ulvskov, 2010) and have backbones composed primarily of Xyl and Man, respectively. The dynamics of mannan have yet to be explored in great detail, although new insights into their structural roles were gained in recent years (Yu et al., 2014; Voiniciuc et al., 2015a). Some mAb probes are available (e.g. LM21 and LM22; Table II), but they cannot bind acetylated forms of mannan and are not sensitive to the presence of Glc in the polymer backbone (Marcus et al., 2010), which varies in nature. A major limitation of these proteinaceous probes is that their access to target epitopes can be heavily masked by pectic HG (Marcus et al., 2010). Although the masking HG could be enzymatically removed prior to immunolabeling, it would be advantageous to develop probes that surmount these challenges.
In the last 2 years, important tools for studying xylan dynamics have been developed. The de novo synthesis of various oligosaccharides has enabled the characterization of xylan-directed mAbs (Schmidt et al., 2015). While microarrays with plant polysaccharides (Moller et al., 2008) or oligosaccharides (Pedersen et al., 2012) have been probed previously with mAbs (Pattathil et al., 2010), arrays of synthetic glycans have an exceptional purity and facilitate the precise mapping of the epitopes recognized by molecular probes. A comprehensive screen of 209 wall-directed mAbs against 88 synthetic glycans, including 22 xylan oligosaccharides, identified specific epitopes for 78 probes that were uncharacterized previously (Ruprecht et al., 2017). This study also confirmed the epitopes of a few mAbs that were already characterized in detail (Table II), such as the specificity of LM28 for xylan substituted with GlcA (Cornuault et al., 2015). Since neither synthetic glycans nor specific mAbs are available currently for acetylated xylan (Ruprecht et al., 2017), the current toolbox must be expanded to detect the full diversity of hemicellulose structures found in nature. For instance, xylans also can be imaged directly using CBMs. FP-tagged CBM15 (named OC15; Table II) was developed as a sensitive probe for monitoring xylan polymers during lignocellulosic biomass conversion (Khatri et al., 2016) but also could be used in planta.
To overcome the limitation of probes and to gain greater insight into polymer interactions, solid-state NMR has emerged as a powerful technique to monitor the molecular architecture of the plant cell wall. Xylans were found to have a flattened conformation and were bound closely to cellulose microfibrils in wild-type Arabidopsis stems but not in a cellulose-deficient mutant (Simmons et al., 2016). Moreover, solid-state NMR revealed that an even pattern of substitution is essential for xylan to bind to cellulose in the 2-fold screw conformation (Grantham et al., 2017). The role of these regular motifs also is supported by mass spectrometric sequencing and molecular dynamics simulations of xylan oligomers (Martínez-Abad et al., 2017). The substitution of xylans also can be observed using matrix-assisted laser-desorption ionization (MALDI) mass spectrometry imaging, an emerging technique that employs specific glycosyl hydrolases to map variations in polysaccharide localization and structure. So far, this promising method was used only to track the composition and distribution of arabinoxylan and mixed-linkage glucans during endosperm maturation in wheat (Triticum aestivum; Veličković et al., 2014). However, MALDI mass spectrometry can detect all major classes of cell wall polysaccharides (Westphal et al., 2010) and was used successfully to screen for Arabidopsis axy mutants with altered XyG structures (Gille et al., 2011; Günl et al., 2011a, 2011b; Günl and Pauly, 2011; Schultink et al., 2015). Hence, MALDI when coupled with microscopy offers the possibility to (1) enhance the chemical wall structural resolution down to the cellular level and (2) image the dynamics of multiple polysaccharides in situ. This will be particularly advantageous for detecting wall polymers (e.g. acetylated hemicelluloses) that cannot be detected currently with existing probes (Table II).
CONCLUDING REMARKS
Advanced techniques to monitor polysaccharides (Table I), along with the genomics era of the last decade, have delivered an avalanche of new data pertaining to the mechanisms of plant cell wall synthesis and metabolism. Many genes involved in these processes have now been characterized. However, the spatiotemporal organization and regulation of wall polymer synthesis and degradation, the assembly of polymer networks, and the dynamics of wall assemblies during cell growth and differentiation remain a challenge (see Outstanding Questions box). The identification and characterization of novel polysaccharide-specific probes, coupled with sensitive techniques, will shed light on these unresolved issues. Even classic probes (e.g. S4B and FP-tagged proteins; Table II) can be visualized with increased precision by clearing plant tissues using new techniques that forgo costly and time-consuming embedding and sectioning steps (Ursache et al., 2018). Although the resolution of confocal microscopes was limited historically by diffraction to greater than 200 nm (Hell, 2007), this barrier has been surpassed in the last decade to enable the imaging of living plant cells at the nanoscale level (Komis et al., 2018). With the advent of superresolution fluorescence microscopy, polysaccharide-binding probes such as S4B can potentially be visualized in the walls of living plant cells at a resolution approaching more invasive methods such as atomic force microscopy and electron microscopy (Table I; Liesche et al., 2013). Indeed, novel membrane probes can now reveal the dynamics of organelles at a spatial resolution of 50 nm, and over tens of minutes instead of tens of seconds for FP-tagged probes (Takakura et al., 2017). Label-free in situ imaging of plant cell wall polysaccharides would be ideal and could be facilitated by further developments in techniques such as MALDI mass spectrometry imaging. Other components of the wall not covered in this Update, such as lignin and structural proteins, could or have already been monitored using similar techniques. Therefore, there has never been a better time to explore the dynamics of a diverse range of plant cell wall polymers.
Footnotes
The authors want to acknowledge support by the DFG project CASOM US98/13-1 and the Ministry of Innovation, Science, and Research within the framework of the NRW Strategieprojekt BioSC (No. 613 313/323‐400‐002 13), the Excellence Cluster CEPLAS (EXC 1028), and the BMBF project “Cornwall” (031B0193A/B).
Articles can be viewed without a subscription.
References
- Anderson CT, Carroll A, Akhmetova L, Somerville C (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CT, Wallace IS (2012) Illuminating the wall: using click chemistry to image pectins in Arabidopsis cell walls. Plant Signal Behav 7: 661–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CT, Wallace IS, Somerville CR (2012) Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proc Natl Acad Sci USA 109: 1329–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Hao Z, Mohnen D (2013) Evolving views of pectin biosynthesis. Annu Rev Plant Biol 64: 747–779 [DOI] [PubMed] [Google Scholar]
- Barnes WJ, Anderson CT (2018) Release, recycle, rebuild: cell-wall remodeling, autodegradation, and sugar salvage for new wall biosynthesis during plant development. Mol Plant 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP (2006) Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem 281: 29321–29329 [DOI] [PubMed] [Google Scholar]
- Brennan M, Harris PJ (2011) Distribution of fucosylated xyloglucans among the walls of different cell types in monocotyledons determined by immunofluorescence microscopy. Mol Plant 4: 144–156 [DOI] [PubMed] [Google Scholar]
- Carpita NC, McCann MC (2015) Characterizing visible and invisible cell wall mutant phenotypes. J Exp Bot 66: 4145–4163 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuault V, Buffetto F, Rydahl MG, Marcus SE, Torode TA, Xue J, Crépeau MJJ, Faria-Blanc N, Willats WGT, Dupree P, et al. (2015) Monoclonal antibodies indicate low-abundance links between heteroxylan and other glycans of plant cell walls. Planta 242: 1321–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2016) Catalysts of plant cell wall loosening. F1000 Res 5: F1000 Faculty Rev-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2018) Diffuse growth of plant cell walls. Plant Physiol 176: 16–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardelle F, Le Mauff F, Lehner A, Loutelier-Bourhis C, Bardor M, Rihouey C, Causse M, Lerouge P, Driouich A, Mollet JC (2015) Pollen tube cell walls of wild and domesticated tomatoes contain arabinosylated and fucosylated xyloglucan. Ann Bot 115: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick-Perez M, Wang T, Salazar A, Zabotina OA, Hong M (2012) Multidimensional solid-state NMR studies of the structure and dynamics of pectic polysaccharides in uniformly 13C-labeled Arabidopsis primary cell walls. Magn Reson Chem 50: 539–550 [DOI] [PubMed] [Google Scholar]
- Dumont M, Lehner A, Vauzeilles B, Malassis J, Marchant A, Smyth K, Linclau B, Baron A, Mas Pons J, Anderson CT, et al. (2016) Plant cell wall imaging by metabolic click-mediated labelling of rhamnogalacturonan II using azido 3-deoxy-D-manno-oct-2-ulosonic acid. Plant J 85: 437–447 [DOI] [PubMed] [Google Scholar]
- Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC (2011) Nanostructure of cellulose microfibrils in spruce wood. Proc Natl Acad Sci USA 108: E1195–E1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: carbohydrates. J Vis Exp 37: e1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M (2011) O-Acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23: 4041–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay K, Hu J, Arantes V, Penttilä M, Saddler JN (2015) The use of carbohydrate binding modules (CBMs) to monitor changes in fragmentation and cellulose fiber surface morphology during cellulase- and Swollenin-induced deconstruction of lignocellulosic substrates. J Biol Chem 290: 2938–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham NJ, Wurman-Rodrich J, Terrett OM, Lyczakowski JJ, Stott K, Iuga D, Simmons TJ, Durand-Tardif M, Brown SP, Dupree R, et al. (2017) An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat Plants 3: 859–865 [DOI] [PubMed] [Google Scholar]
- Günl M, Kraemer F, Pauly M (2011a) Oligosaccharide mass profiling (OLIMP) of cell wall polysaccharides by MALDI-TOF/MS. Methods Mol Biol 715: 43–54 [DOI] [PubMed] [Google Scholar]
- Günl M, Neumetzler L, Kraemer F, de Souza A, Schultink A, Pena M, York WS, Pauly M (2011b) AXY8 encodes an α-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 23: 4025–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl M, Pauly M (2011) AXY3 encodes a α-xylosidase that impacts the structure and accessibility of the hemicellulose xyloglucan in Arabidopsis plant cell walls. Planta 233: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Hall J, Black GW, Ferreira LM, Millward-Sadler SJ, Ali BR, Hazlewood GP, Gilbert HJ (1995) The non-catalytic cellulose-binding domain of a novel cellulase from Pseudomonas fluorescens subsp. cellulosa is important for the efficient hydrolysis of Avicel. Biochem J 309: 749–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke DE, Northcote DH (1975) Molecular visualization of pectin and DNA by ruthenium red. Biopolymers 14: 1–17 [DOI] [PubMed] [Google Scholar]
- Hell SW. (2007) Far-field optical nanoscopy. Science 316: 1153–1158 [DOI] [PubMed] [Google Scholar]
- Hernandez-Gomez MC, Rydahl MG, Rogowski A, Morland C, Cartmell A, Crouch L, Labourel A, Fontes CMGA, Willats WGT, Gilbert HJ, et al. (2015) Recognition of xyloglucan by the crystalline cellulose-binding site of a family 3a carbohydrate-binding module. FEBS Lett 589: 2297–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JL Jr, Hammudi MB, Tien M (2014) The Arabidopsis cellulose synthase complex: a proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 26: 4834–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocq L, Pelloux J, Lefebvre V (2017) Connecting homogalacturonan-type pectin remodeling to acid growth. Trends Plant Sci 22: 20–29 [DOI] [PubMed] [Google Scholar]
- Hoogenboom J, Berghuis N, Cramer D, Geurts R, Zuilhof H, Wennekes T (2016) Direct imaging of glycans in Arabidopsis roots via click labeling of metabolically incorporated azido-monosaccharides. BMC Plant Biol 16: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri V, Hébert-Ouellet Y, Meddeb-Mouelhi F, Beauregard M (2016) Specific tracking of xylan using fluorescent-tagged carbohydrate-binding module 15 as molecular probe. Biotechnol Biofuels 9: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP. (2012) In situ detection of cellulose with carbohydrate-binding modules. Methods Enzymol 510: 233–245 [DOI] [PubMed] [Google Scholar]
- Knox JP, Linstead PJ, King J, Cooper C, Roberts K (1990) Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181: 512–521 [DOI] [PubMed] [Google Scholar]
- Komis G, Novák D, Ovečka M, Šamajová O, Šamaj J (2018) Advances in imaging plant cell dynamics. Plant Physiol 176: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani ER, Moller IE, Cassin A, Jones DF, Koh PL, Ratnayake S, Beahan CT, Wilson SM, Bacic A, Newbigin E (2013) In vitro grown pollen tubes of Nicotiana alata actively synthesise a fucosylated xyloglucan. PLoS ONE 8: e77140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque-Tremblay G, Pelloux J, Braybrook SA, Müller K (2015) Tuning of pectin methylesterification: consequences for cell wall biomechanics and development. Planta 242: 791–811 [DOI] [PubMed] [Google Scholar]
- Liesche J, Ziomkiewicz I, Schulz A (2013) Super-resolution imaging with Pontamine Fast Scarlet 4BS enables direct visualization of cellulose orientation and cell connection architecture in onion epidermis cells. BMC Plant Biol 13: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liners F, Van Cutsem P (1992) Distribution of pectic polysaccharides throughout walls of suspension-cultured carrot cells: an immunocytochemical study. Protoplasma 170: 10–21 [Google Scholar]
- Liu L, Paulitz J, Pauly M (2015) The presence of fucogalactoxyloglucan and its synthesis in rice indicates conserved functional importance in plants. Plant Physiol 168: 549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus SE, Blake AW, Benians TAS, Lee KJD, Poyser C, Donaldson L, Leroux O, Rogowski A, Petersen HL, Boraston A, et al. (2010) Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J 64: 191–203 [DOI] [PubMed] [Google Scholar]
- Marga F, Grandbois M, Cosgrove DJ, Baskin TI (2005) Cell wall extension results in the coordinate separation of parallel microfibrils: evidence from scanning electron microscopy and atomic force microscopy. Plant J 43: 181–190 [DOI] [PubMed] [Google Scholar]
- Martínez-Abad A, Berglund J, Toriz G, Gatenholm P, Henriksson G, Lindström M, Wohlert J, Vilaplana F (2017) Regular motifs in xylan modulate molecular flexibility and interactions with cellulose surfaces. Plant Physiol 175: 1579–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Wells B, Roberts K (1990) Direct visualization of cross-links in the primary plant cell wall. J Cell Sci 96: 323–334 [Google Scholar]
- McCartney L, Marcus SE, Knox JP (2005) Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J Histochem Cytochem 53: 543–546 [DOI] [PubMed] [Google Scholar]
- McClosky DD, Wang B, Chen G, Anderson CT (2016) The click-compatible sugar 6-deoxy-alkynyl glucose metabolically incorporates into Arabidopsis root hair tips and arrests their growth. Phytochemistry 123: 16–24 [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Döring A, Persson S (2014) The cell biology of cellulose synthesis. Annu Rev Plant Biol 65: 69–94 [DOI] [PubMed] [Google Scholar]
- McLean BW, Boraston AB, Brouwer D, Sanaie N, Fyfe CA, Warren RAJ, Kilburn DG, Haynes CA (2002) Carbohydrate-binding modules recognize fine substructures of cellulose. J Biol Chem 277: 50245–50254 [DOI] [PubMed] [Google Scholar]
- McNamara JT, Morgan JLW, Zimmer J (2015) A molecular description of cellulose biosynthesis. Annu Rev Biochem 84: 895–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miart F, Desprez T, Biot E, Morin H, Belcram K, Höfte H, Gonneau M, Vernhettes S (2014) Spatio-temporal analysis of cellulose synthesis during cell plate formation in Arabidopsis. Plant J 77: 71–84 [DOI] [PubMed] [Google Scholar]
- Moller I, Marcus SE, Haeger A, Verhertbruggen Y, Verhoef R, Schols H, Ulvskov P, Mikkelsen JD, Knox JP, Willats W (2008) High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj J 25: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Kračun SK, Rydahl MG, Westereng B, Miart F, Clausen MH, Fangel JU, Daugaard M, Van Cutsem P, De Fine Licht HH, et al. (2014) Tracking developmentally regulated post-synthetic processing of homogalacturonan and chitin using reciprocal oligosaccharide probes. Development 141: 4841–4850 [DOI] [PubMed] [Google Scholar]
- Mravec J, Kračun SK, Rydahl MG, Westereng B, Pontiggia D, De Lorenzo G, Domozych DS, Willats WGT (2017) An oligogalacturonide-derived molecular probe demonstrates the dynamics of calcium-mediated pectin complexation in cell walls of tip-growing structures. Plant J 91: 534–546 [DOI] [PubMed] [Google Scholar]
- Nixon BT, Mansouri K, Singh A, Du J, Davis JK, Lee JG, Slabaugh E, Vandavasi VG, O’Neill H, Roberts EM, et al. (2016) Comparative structural and computational analysis supports eighteen cellulose synthases in the plant cellulose synthesis complex. Sci Rep 6: 28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notley SM, Pettersson B, Wågberg L (2004) Direct measurement of attractive van der Waals’ forces between regenerated cellulose surfaces in an aqueous environment. J Am Chem Soc 126: 13930–13931 [DOI] [PubMed] [Google Scholar]
- Oikawa A, Lund CH, Sakuragi Y, Scheller HV (2013) Golgi-localized enzyme complexes for plant cell wall biosynthesis. Trends Plant Sci 18: 49–58 [DOI] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Park S, Szumlanski AL, Gu F, Guo F, Nielsen E (2011) A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat Cell Biol 13: 973–980 [DOI] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. (2010) A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Zhang T, Cardenas CL, Hahn MG (2015) Immunological approaches to biomass characterization and utilization. Front Bioeng Biotechnol 3: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Keegstra K (2016) Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu Rev Plant Biol 67: 235–259 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Höfte H (2011) Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol 21: 1720–1726 [DOI] [PubMed] [Google Scholar]
- Pedersen HL, Fangel JU, McCleary B, Ruzanski C, Rydahl MG, Ralet MC, Farkas V, von Schantz L, Marcus SE, Andersen MCF, et al. (2012) Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J Biol Chem 287: 39429–39438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979 [DOI] [PubMed] [Google Scholar]
- Pettolino FA, Walsh C, Fincher GB, Bacic A (2012) Determining the polysaccharide composition of plant cell walls. Nat Protoc 7: 1590–1607 [DOI] [PubMed] [Google Scholar]
- Puhlmann J, Bucheli E, Swain MJ, Dunning N, Albersheim P, Darvill AG, Hahn MG (1994) Generation of monoclonal antibodies against plant cell-wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal α-(1→2)-linked fucosyl-containing epitope. Plant Physiol 104: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T (2008) A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J 54: 466–480 [DOI] [PubMed] [Google Scholar]
- Rocha J, Cicéron F, de Sanctis D, Lelimousin M, Chazalet V, Lerouxel O, Breton C (2016) Structure of Arabidopsis thaliana FUT1 reveals a variant of the GT-B class fold and provides insight into xyloglucan fucosylation. Plant Cell 28: 2352–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röckel N, Wolf S, Kost B, Rausch T, Greiner S (2008) Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J 53: 133–143 [DOI] [PubMed] [Google Scholar]
- Rounds CM, Lubeck E, Hepler PK, Winship LJ (2011) Propidium iodide competes with Ca2+ to label pectin in pollen tubes and Arabidopsis root hairs. Plant Physiol 157: 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel K, Nishiyama Y, Joseleau JP (2012) Crystalline and amorphous cellulose in the secondary walls of Arabidopsis. Plant Sci 193-194: 48–61 [DOI] [PubMed] [Google Scholar]
- Rui Y, Anderson CT (2016) Functional analysis of cellulose and xyloglucan in the walls of stomatal guard cells of Arabidopsis. Plant Physiol 170: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y, Xiao C, Yi H, Kandemir B, Wang JZ, Puri VM, Anderson CT (2017) POLYGALACTURONASE INVOLVED IN EXPANSION3 functions in seedling development, rosette growth, and stomatal dynamics in Arabidopsis thaliana. Plant Cell 29: 2413–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht C, Bartetzko MP, Senf D, Dallabernadina P, Boos I, Andersen MCF, Kotake T, Knox JP, Hahn MG, Clausen MH, et al. (2017) A synthetic glycan microarray enables epitope mapping of plant cell wall glycan-directed antibodies. Plant Physiol 175: 1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Aguayo S, Ralet MC, Berger A, Botran L, Ropartz D, Marion-Poll A, North HM (2013) PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25: 308–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schuhmacher F, Geissner A, Seeberger PH, Pfrengle F (2015) Automated synthesis of arabinoxylan-oligosaccharides enables characterization of antibodies that recognize plant cell wall glycans. Chemistry 21: 5709–5713 [DOI] [PubMed] [Google Scholar]
- Schultink A, Liu L, Zhu L, Pauly M (2014) Structural diversity and function of xyloglucan sidechain substituents. Plants (Basel) 3: 526–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultink A, Naylor D, Dama M, Pauly M (2015) The role of the plant-specific ALTERED XYLOGLUCAN9 protein in Arabidopsis cell wall polysaccharide O-acetylation. Plant Physiol 167: 1271–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sénéchal F, Wattier C, Rustérucci C, Pelloux J (2014) Homogalacturonan-modifying enzymes: structure, expression, and roles in plants. J Exp Bot 65: 5125–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons TJ, Mortimer JC, Bernardinelli OD, Pöppler AC, Brown SP, deAzevedo ER, Dupree R, Dupree P (2016) Folding of xylan onto cellulose fibrils in plant cell walls revealed by solid-state NMR. Nat Commun 7: 13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO (2000) New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol 124: 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synek L, Vukašinović N, Kulich I, Hála M, Aldorfová K, Fendrych M, Žárský V (2017) EXO70C2 is a key regulatory factor for optimal tip growth of pollen. Plant Physiol 174: 223–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakura H, Zhang Y, Erdmann RS, Thompson AD, Lin Y, McNellis B, Rivera-Molina F, Uno SN, Kamiya M, Urano Y, et al. (2017) Long time-lapse nanoscopy with spontaneously blinking membrane probes. Nat Biotechnol 35: 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LH, Forsyth VT, Sturcová A, Kennedy CJ, May RP, Altaner CM, Apperley DC, Wess TJ, Jarvis MC (2013) Structure of cellulose microfibrils in primary cell walls from collenchyma. Plant Physiol 161: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursache R, Andersen TG, Marhavý P, Geldner N (2018) A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J 93: 399–412 [DOI] [PubMed] [Google Scholar]
- Veličković D, Ropartz D, Guillon F, Saulnier L, Rogniaux H (2014) New insights into the structural and spatial variability of cell-wall polysaccharides during wheat grain development, as revealed through MALDI mass spectrometry imaging. J Exp Bot 65: 2079–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y, Marcus SE, Haeger A, Ordaz-Ortiz JJ, Knox JP (2009) An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr Res 344: 1858–1862 [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Fry SC, Pauly M, Höfte H, Verbelen JP (2005) XTH acts at the microfibril-matrix interface during cell elongation. J Exp Bot 56: 673–683 [DOI] [PubMed] [Google Scholar]
- Voiniciuc C, Dean GH, Griffiths JS, Kirchsteiger K, Hwang YT, Gillett A, Dow G, Western TL, Estelle M, Haughn GW (2013) Flying saucer1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. Plant Cell 25: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Schmidt MHW, Berger A, Yang B, Ebert B, Scheller HV, North HM, Usadel B, Günl M (2015a) MUCILAGE-RELATED10 produces galactoglucomannan that maintains pectin and cellulose architecture in Arabidopsis seed mucilage. Plant Physiol 169: 403–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Yang B, Schmidt MH, Günl M, Usadel B (2015b) Starting to gel: how Arabidopsis seed coat epidermal cells produce specialized secondary cell walls. Int J Mol Sci 16: 3452–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Zimmermann E, Schmidt MHW, Günl M, Fu L, North HM, Usadel B (2016) Extensive natural variation in Arabidopsis seed mucilage structure. Front Plant Sci 7: 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace IS, Anderson CT (2012) Small molecule probes for plant cell wall polysaccharide imaging. Front Plant Sci 3: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Meents MJ, McDonnell LM, Barkwill S, Sampathkumar A, Cartwright HN, Demura T, Ehrhardt DW, Samuels AL, Mansfield SD (2015) Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 350: 198–203 [DOI] [PubMed] [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW (2001) Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Westphal Y, Schols HA, Voragen AGJ, Gruppen H (2010) MALDI-TOF MS and CE-LIF fingerprinting of plant cell wall polysaccharide digests as a screening tool for Arabidopsis cell wall mutants. J Agric Food Chem 58: 4644–4652 [DOI] [PubMed] [Google Scholar]
- Wightman R, Marshall R, Turner SR (2009) A cellulose synthase-containing compartment moves rapidly beneath sites of secondary wall synthesis. Plant Cell Physiol 50: 584–594 [DOI] [PubMed] [Google Scholar]
- Wilson SM, Ho YY, Lampugnani ER, Van de Meene AML, Bain MP, Bacic A, Doblin MS (2015) Determining the subcellular location of synthesis and assembly of the cell wall polysaccharide (1,3; 1,4)-β-d-glucan in grasses. Plant Cell 27: 754–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Greiner S (2012) Growth control by cell wall pectins. Protoplasma (Suppl 2) 249: S169–S175 [DOI] [PubMed] [Google Scholar]
- Xiao C, Barnes WJ, Zamil MS, Yi H, Puri VM, Anderson CT (2017) Activation tagging of Arabidopsis POLYGALACTURONASE INVOLVED IN EXPANSION2 promotes hypocotyl elongation, leaf expansion, stem lignification, mechanical stiffening, and lodging. Plant J 89: 1159–1173 [DOI] [PubMed] [Google Scholar]
- Xiao C, Somerville C, Anderson CT (2014) POLYGALACTURONASE INVOLVED IN EXPANSION1 functions in cell elongation and flower development in Arabidopsis. Plant Cell 26: 1018–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Shi D, Li J, Kong Y, Yu Y, Chai G, Hu R, Wang J, Hahn MG, Zhou G (2014) CELLULOSE SYNTHASE-LIKE A2, a glucomannan synthase, is involved in maintaining adherent mucilage structure in Arabidopsis seed. Plant Physiol 164: 1842–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablackis E, York WS, Pauly M, Hantus S, Reiter WD, Chapple CC, Albersheim P, Darvill A (1996) Substitution of L-fucose by L-galactose in cell walls of Arabidopsis mur1. Science 272: 1808–1810 [DOI] [PubMed] [Google Scholar]
- Zhang GF, Staehelin LAS (1992) Functional compartmentation of the Golgi apparatus of plant cells: immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells. Plant Physiol 99: 1070–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zheng Y, Cosgrove DJ (2016) Spatial organization of cellulose microfibrils and matrix polysaccharides in primary plant cell walls as imaged by multichannel atomic force microscopy. Plant J 85: 179–192 [DOI] [PubMed] [Google Scholar]
- Zhu C, Ganguly A, Baskin TI, McClosky DD, Anderson CT, Foster C, Meunier KA, Okamoto R, Berg H, Dixit R (2015) The fragile fiber1 kinesin contributes to cortical microtubule-mediated trafficking of cell wall components. Plant Physiol 167: 780–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wu J, Chen X (2016) Metabolic labeling and imaging of N-linked glycans in Arabidopsis thaliana. Angew Chem Int Ed Engl 55: 9301–9305 [DOI] [PubMed] [Google Scholar]